Abstract
The subcellular location and traffic of two selected chitin synthases (CHS) from Neurospora crassa, CHS-3 and CHS-6, labeled with green fluorescent protein (GFP), were studied by high-resolution confocal laser scanning microscopy. While we found some differences in the overall distribution patterns and appearances of CHS-3-GFP and CHS-6-GFP, most features were similar and were observed consistently. At the hyphal apex, fluorescence congregated into a conspicuous single body corresponding to the location of the Spitzenkörper (Spk). In distal regions (beyond 40 μm from the apex), CHS-GFP revealed a network of large endomembranous compartments that was predominantly comprised of irregular tubular shapes, while some compartments were distinctly spherical. In the distal subapex (20 to 40 μm from the apex), fluorescence was observed in globular bodies that appeared to disintegrate into vesicles as they advanced forward until reaching the proximal subapex (5 to 20 μm from the apex). CHS-GFP was also conspicuously found delineating developing septa. Analysis of fluorescence recovery after photobleaching suggested that the fluorescence of the Spk originated from the advancing population of microvesicles (chitosomes) in the subapex. The inability of brefeldin A to interfere with the traffic of CHS-containing microvesicles and the lack of colocalization of CHS-GFP with the endoplasmic reticulum (ER)-Golgi body fluorescent dyes lend support to the idea that CHS proteins are delivered to the cell surface via an alternative route distinct from the classical ER-Golgi body secretory pathway.
Fungal hyphae elongate and branch by a complex process based on polarized secretion. Many studies have investigated the cellular and molecular components involved in shaping fungal cells, but no detailed understanding of the mechanisms that govern and regulate polarized fungal growth has been achieved (4, 25). In the yeast Saccharomyces cerevisiae, many of the main components of the secretory pathway, including some of the enzymes involved in cell wall formation, have been extensively characterized (32). Filamentous fungi encode homologues of some key components known from the yeast secretory pathway, but despite their apparent orthology, relatively little is known about how this pathway is organized to accomplish the highly polarized growth typical of hyphae. There are some differences in cell wall synthesis between filamentous fungi and S. cerevisiae. In hyphae of septate fungi, vesicles and other components accumulate at the apex, as part of the Spitzenkörper (Spk) (14, 22-24, 28). The composition and mode of action of this pleomorphic and dynamic structure have intrigued fungal biologists for many decades.
Fungal cells have at least two types of well-defined secretory vesicles (5). It has been suggested that macrovesicles, or conventional secretory vesicles, carry the components of the amorphous phase of the cell wall, in addition to the load of extracellular enzymes (5, 27). There is a large body of evidence characterizing the chitin synthase (CHS)-carrying microvesicles as chitosomes (3, 8, 13, 30). CHS are β-glycosyltransferases that catalyze the polymerization of N-acetylglucosamine from UDP N-acetylglucosamine into chitin (47), a major structural polymer of the fungal cell wall (2). Chitin synthesis occurs in highly localized fashion both at the hyphal apices (7) and at nascent septa (29). Chitosomes are the smallest vesicles with the ability to form chitin microfibrils in vitro and have been suggested to carry and transport CHS to the cell surface at the apex of hyphae for cell wall synthesis (13, 37, 48, 55, 56). In recent years, studies on fungal CHS have concentrated mainly on gene identification. Given this wealth of information, we chose CHS as candidate markers to investigate vesicle traffic in fungal hyphae.
Fungi have multiple chs genes grouped into two divisions, with seven classes, primarily on the basis of similarities in the primary sequence of the predicted proteins (12, 16, 37, 50). Division I includes classes I, II, and III, which share a catalytic domain surrounded by a hydrophilic N-terminal region and a hydrophobic C-terminal region (12). Division II includes classes IV, V, and VII, all with a catalytic domain preceded by a cytochrome b5-like domain. In addition, classes V and VII contain an N-terminal myosin motor-like domain, suggesting a direct interaction with the actin cytoskeleton (15, 20, 58). Class VI has not been assigned to either division and includes recently identified CHS of unknown function (16). Earlier studies suggest that the various CHS have specific roles in chitin cell wall synthesis that are time or space dependent (60). In contrast to most filamentous fungi, S. cerevisiae (46) and Candida albicans (40) have only three or four CHS isozymes, respectively. S. cerevisiae Chs1p, C. albicans Chs2p, and C. albicans Chs8p belong to class I; S. cerevisiae Chs2p and C. albicans Chs1p belong to class II; and S. cerevisiae Chs3p and C. albicans Chs3p belong to class IV (46). While potential roles in hyphal growth have been suggested for some of the seven CHS classes described in filamentous fungi (9, 64, 65), we lack specific information on the cellular localization and trafficking to their sites of action in regions of active cell wall growth for most of these proteins.
The goal of this study was to elucidate the traffic of CHS-containing vesicles en route from their site of genesis to their site of exocytosis in living hyphae of Neurospora crassa. The availability of an almost-complete genome sequence for this fungus allowed the identification of seven open reading frames with high homology to previously described chs genes (10). We chose to trace the intracellular location and secretory paths of CHS-3 and CHS-6. Neurospora CHS-3 belongs to the previously reported class I CHS with known homologues in all fungi tested, including S. cerevisiae Chs1p. In contrast, CHS-6 is a newly identified CHS assigned to class VI, homologous to Aspergillus fumigatus ChsD (39) and Coccidioides posadasii CHS-6 (34) but with no apparent homologues in S. cerevisiae or C. albicans. To trace both proteins, we fused green fluorescent protein (GFP) to the carboxyl terminus of the CHS coding regions and analyzed the fate of the resulting CHS-3-GFP and CHS-6-GFP fusion proteins by high-resolution confocal laser scanning microscopy (CLSM) in living hyphae of N. crassa.
MATERIALS AND METHODS
Strains and culture conditions.
For transformations, N. crassa strain N623 (mat A his-3; FGSC 6103) was grown at 32°C for 5 to 10 days on Vogel's minimal medium (VMM) agar (59) containing 1.5% sucrose and supplemented with histidine (0.5 mg/ml). Transformed conidia were spread on VMM-FGS (0.5% fructose, 0.5% glucose, 20% sorbose) medium. Five to 10 His+ prototrophic transformants per plate were transferred to VMM tubes and incubated at 32°C for 3 days. For epifluorescence and confocal microscopy, transformants were routinely grown on VMM and observed using the “inverted agar block method” (26).
Recombinant DNA techniques and plasmid constructions.
Standard PCR and cloning procedures were used to fuse the sgfp gene to the carboxyl terminus of chs-3 and chs-6 (53). The 2.7-kb chs-3 gene (class I CHS; GenBank accession no. XM_323590) and the 2.5-kb chs-6 gene (class VI CHS; GenBank accession no. XM_324624) were amplified by PCR from N. crassa N1 (mat a; FGSC 988) genomic DNA with custom-designed primers that included XbaI and PacI restriction endonuclease sites at their respective 5′ termini (Table 1). PCR was performed in a Bio-Rad Thermal Cycler with platinum Pfu polymerase (Invitrogen) according to the manufacturer's instructions, under the following conditions: denaturation at 94°C for 1 min, followed by 30 cycles of 94°C (30 s), 61°C (30 s), and 72°C (3 min), and a final extension step at 72°C for 5 min. The amplified and gel-purified PCR products were digested with XbaI and PacI and cloned into XbaI- and PacI-digested plasmid pMF272 (19; GenBank accession no. AY598428). This yielded pJMGT5 (chs-3) and pJMG58 (chs-6). Inserts were sequenced at the Core Instrumentation Facility of the Institute for Integrative Genome Biology at the University of California, Riverside, with primers pMF272F and pMF272R2 (Table 1) designed for the upstream and downstream flanks of the multiple cloning site of pMF272.
TABLE 1.
Strains, plasmids, and oligonucleotides used or generated for this study
| Plasmid, N. crassa strain,a or oligonucleotide | Description, genotype, or sequenceb | Reference or accession no. |
|---|---|---|
| Plasmids | ||
| pMF272 | Pccg-1::sgfp+ | 19 |
| pGP003 | Pccg-1::chs-3+::sgfp+ | This study |
| pGP002 | Pccg-1::chs-6+::sgfp+ | This study |
| Strains | ||
| N39 | mat A; fl | FGSC 4317 |
| N40 | mat a; fl | FGSC 4347 |
| N150 | mat A | FGSC 9013 |
| N623 | mat A his-3 | FGSC 6103 |
| N625 | mat a his-3 | FGSC 6525 |
| NMR3 | mat A his-3+::Pccg-1::chs-3+::sgfp+ | This study |
| NMR6 | mat A his-3+::Pccg-1::chs-6+::sgfp+ | This study |
| N1 | mat a | FGSC 988 |
| Oligonucleotides | ||
| MRp10 | 5′-AGAGACAAGAAAATTACCCCCTTCTT-3′ | This study |
| MRp11 | 5′-AACTACAACAGCCACAACGTCTATATC-3′ | This study |
| MRp12 | 5′-ATAATGAACGGAAGGTAGTTGTAGAAAG-3′ | This study |
| MRp13 | 5′-ATGGATATAATGTGGCTGTTGAAAG-3′ | This study |
| pMF272F | 5′-CAAATCAACACAACACTCAAACCA-3′ | 19 |
| pMF272R2 | 5′-AGATGAACTTCAGGGTCAGCTTG-3′ | This study |
| Chs3F | 5′-GAAAGTTCTAGAATGGATCCTCGAATGCATACG-3′ | This study |
| Chs3R | 5′-AGCATTTAATTAAAACACCCCTAAACATCCTCAC-3′ | This study |
| Chs6F | 5′-AAATAATCTAGAATGACTATCAATTACCTTGGG-3′ | This study |
| Chs6R | 5′-TCACCTTAATTAATCTCTGATTTGACCCCCTCGA-3′ | This study |
Transformants NMR3 and NMR6 appear homokaryotic because Southern blotting analyses revealed the presence of his-3+::chs::sgfp loci exclusively.
Restriction endonuclease sites for cloning are shown in bold type, and predicted CHS start codons are underlined.
Neurospora genetics.
Transformation of N. crassa strain N623 his-3 conidia with plasmids pJMGT5 (linearized by digestion with SspI) and pJMG58 (linearized by digestion with NdeI) was carried out by electroporation on a Bio-Rad Gene Pulser (capacitance, 25 μF; 1.5 kV; resistance, 600 Ω) as previously described (35). Prototrophic His+ transformants were screened for the expression of CHS-GFP by epifluorescence microscopy as described previously (19). None of the primary transformants showed sufficient GFP expression under a fluorescence stereomicroscope, but almost all transformants showed fluorescence at varied levels when screened under an inverted epifluorescence microscope. Transformants showing robust fluorescence were selected (strains TMR3-1 to TMR3-4 and strains TMR6-1 to TMR6-4). Potential heterokaryotic prototrophic transformants were crossed to N. crassa strain N625 (mat a his-3; FGSC 6525) on synthetic crossing medium (63). Ascospores were heat-shocked on VMM at 60°C for 1 h, germinated, picked to VMM slants, and analyzed for fluorescence. Two strains, NMR3 and NMR6, were selected for further analysis.
For genomic DNA extraction, we used the DNeasy plant extraction kit (QIAGEN Inc.). Mycelium for DNA extraction was grown at 28°C for 7 days on liquid medium with no shaking and no light, filtered, submerged in liquid nitrogen and lyophilized. Integration of chs-sgfp fusion genes was verified by PCR with two sets of primers (MRp10 and MRp11 or MRp12 and MRp13; Table 1). For confirmatory Southern blotting analyses, Neurospora genomic DNA from strains NMR3 and NMR6 was digested with NdeI, separated on a 0.8% agarose gel, blotted to charged nylon membranes, and probed with a 2.67-kb HindIII fragment from pMF272 that contains part of the his-3 gene. Probes were generated with a nonradioactive labeling kit (Roche Applied Science).
Confocal microscopy and image processing.
Growing hyphae were imaged at 20 to 22°C using an inverted Zeiss LSM-510 META CLSM fitted with an argon/2 ion laser with a GFP filter set (excitation, 488 nm; emission, 515 to 530 nm). A ×100 Ph3 Plan Neofluar oil immersion objective (N.A. 1.3) or a ×63 DIC Plan Apochromat oil immersion objective (N.A. 1.4) were used for image acquisition. A photomultiplier module allowed us to merge the confocal with the phase-contrast images, thus providing simultaneous precise imaging of fluorescently labeled proteins in the entire cell context.
Confocal images were captured using LSM-510 software (version 3.2; Carl Zeiss), evaluated with an LSM-510 image examiner (version 3.2), and further processed with Adobe Photoshop CS2 (version 9.0). Selected image series were converted into movies with the same software. For fluorescence recovery after photobleaching (FRAP) analysis, the “bleach control” command of the LSM-510 software was used. Areas to be bleached were selected with the ROI (region of interest) tool. Bleaching was applied during a time series sequence acquisition with 35 iterations and 85% laser intensity. For each strain, we analyzed at least five sequences for both subapical and apical photobleaching.
To track the movement of fluorescent particles, we used the “track object” option of Image Pro Plus v. 6.1 (Media Cybernetics). We manually traced the trajectory of the Spk and putative vesicles that remained in view for at least 3 to 4 frames. The paths and speeds of 9 and 37 vesicles were analyzed in different sequences of NMR3 and NMR6, respectively.
Fluorescent dyes and inhibitors.
Concentrated stock solutions of organelle-specific dyes and inhibitors were made in either dimethyl sulfoxide or deionized sterile water as suggested by the manufacturer, kept at −20°C, diluted in liquid VMM to the appropriate working concentration, and allowed to warm up to room temperature before being applied to growing hyphae. For each dye treatment, the agar blocks containing Neurospora that were growing mycelia were inverted onto a coverslip containing a 10-μl drop of the corresponding solution. Stained cells were imaged after a 3- to 5-min recovery period. As a marker for vacuolar content, we used a 10 μM diacetate derivative of carboxydifluorofluorescein (CDFFDA, catalog number O6151; Molecular Probes) imaged with the argon/2 ion laser (excitation maximum, 498 nm, and emission maximum, 520 nm). As a marker of endocytic organelles, we used 25 μM of the styril dye N-(3-triethylammoniumpropyl)-4-(6-[4-{diethylamino} phenyl] hexatrienyl) pyridinium dibromide (FM4-64, catalog number T3166; Molecular Probes), imaged with the argon/2 ion laser (excitation, 514 nm, and emission, 670 nm) (26). As a marker of Golgi body equivalents and the endoplasmic reticulum (ER), we used 5 μM brefeldin A (BFA) conjugated to bodipy 558/568 (catalog number B7449; Molecular Probes). Observations were made with a He/Ne-2 laser (excitation, 558 nm, and emission, 568 nm).
RESULTS
Selection of positive transformants.
After transformation of a Neurospora his-3 host strain with either pJMGT5 or pJMG58, we obtained prototrophic putative transformants that expressed CHS-3-GFP (strain TMR3) or CHS-6-GFP (strain TMR6), respectively, when screened by epifluorescence microscopy. After TMR3 and TMR6 were crossed with N. crassa N625, two homokaryotic strains, NMR3 and NMR6, showed stable fluorescence and were selected for further analysis by CSLM. Strains NMR3 and NMR6 were spot tested on plates with fluffy strains; as expected, both were mat A, reflecting the close linkage of mat and his-3.
The integration of the chs-sgfp fusion genes at his-3 was confirmed by PCR and Southern blotting analyses. For chs-3-sgfp, an expected 3.2-kb PCR product was obtained with primers MRp10 and MRp11, and for chs-6-sgfp, an expected 2.1-kb PCR product was obtained with primers MRp12 and MRp13. Integration by gene replacement at his-3 was also confirmed by Southern blotting analysis for both strains. While the host strain revealed the single expected 3.9-kb band, we observed an additional 6.2-kb or 18.9-kb band for strains NMR3 and NMR6, respectively, indicative of the integration (data not shown).
The primary transformants showed variable levels of GFP, presumably reflecting the number of GFP+ nuclei in GFP+/GFP− heterokaryons. The distribution patterns of CHS-GFP were equivalent in the low- or high-expression heterokaryons and homokaryons, suggesting that the expression of either chs-3-sgfp or chs-6-sgfp from the inducible ccg-1 promoter did not significantly alter the characteristic CHS-GFP distribution described below. Preliminary results with CHS-1-GFP fusion genes that were integrated into the endogenous locus by a “knock-in” procedure to be described elsewhere (E. Sánchez-León, M. Riquelme, S. Bartnicki-Garcia, and M. Freitag, unpublished data) also suggest that expression from the ccg-1 promoter does not significantly alter CHS-GFP expression in this system. Moreover, both strain NMR3 and strain NMR6 grown at 28°C had colony extension rates (2.8 ± 0.4 [mean ± standard deviation] mm/h and 2.2 ± 0.4 mm/h, respectively) comparable to those measured for the wild-type strain (2.3 ± 0.4 mm/h) and presented colony and hyphal morphology and conidiation patterns similar to those of the wild-type strain, suggesting that the presence of the GFP-labeled CHS proteins did not interfere with normal hyphal growth.
Localization of CHS-3 and CHS-6 fusion proteins in living hyphae.
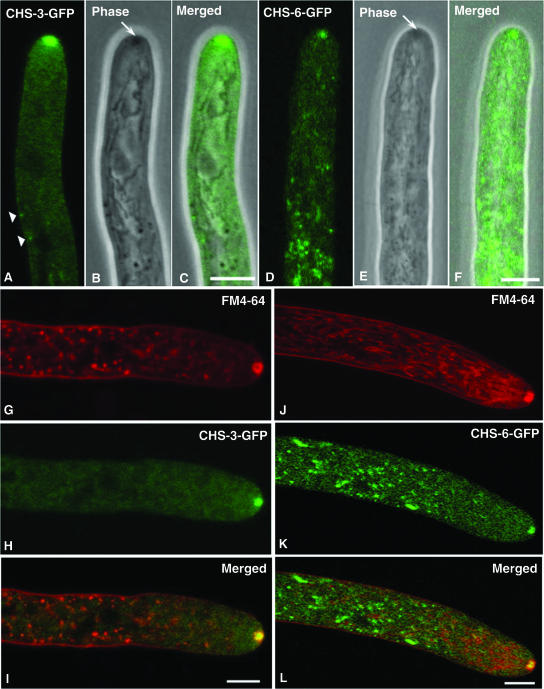
The hyphae of both strain NMR3 and strain NMR6 were extensively labeled by CHS-3-GFP and CHS-6-GFP fusion proteins, respectively. Despite the overall similarity, there were some differences in the morphology and distribution patterns of structures labeled with the two different CHS fusions (Fig. 1 and 2). The most striking common feature in strains with CHS-3-GFP and CHS-6-GFP was the presence of a conspicuous green fluorescent body at the hyphal apex. CHS-GFP fluorescence congregated into a round body (Fig. 1A, D, H, and K and Fig. 2A, C, and D), corresponding to the location of the Spk as revealed by either phase-contrast microscopy (Fig. 1B and E) or fluorescence staining with FM4-64 (Fig. 1G and J). The round apical structure labeled by CHS-GFP (Fig. 1H and K) was concentric with, but smaller than, the area stained by FM4-64 (Fig. 1G and J), suggesting that the CHS-GFP labeling was localized at the core of the Spk (Fig. 1I and L). For CHS-3-GFP, the area of fluorescence had a diameter of 0.8 ± 0.3 μm, whereas the FM4-64-stained area had a diameter of 1.3 ± 0.3 μm (n = 11 hyphae). For CHS-6-GFP, these values were 0.7 ± 0.1 μm and 1.0 ± 0.1 μm, respectively (n = 4 hyphae). Upon prolonged incubation, FM4-64 also stained the mitochondria (Fig. 1J). Except for the Spk core region, the rest of the subapical structures stained by FM4-64 did not colocalize with structures labeled by either one of the CHS-GFP fusion proteins (Fig. 1I and L).
FIG. 1.
Apical distribution of CHS-3-GFP and CHS-6-GFP in N. crassa (A to F) and comparison of CHS-GFP and FM4-64 (endocytic marker) localization (G to L). At the hyphal apex, fluorescence CSLM of GFP-labeled CHS-3 (A) and CHS-6 (D) shows a prominent apical body that coincides with the Spk, as revealed by phase-contrast microscopy (arrows in B, E) and confirmed in the overlaid images (C, F). Arrowheads in A point at fluorescent punctate structures at or near the plasma membrane; note the lack of correspondence with phase-dark structures in the merged image (C). Dual fluorescent labeling with FM4-64 (G, J) and CHS-3-GFP (H) or CHS-6-GFP (K) indicates no correspondence of labeled structures except for a concentric but only partial colocalization in the Spk. Overlaid images (I, L) show that the CHS-GFP occupies the center of a larger body stained by FM4-64. Scale bars, 5 μm.
FIG. 2.
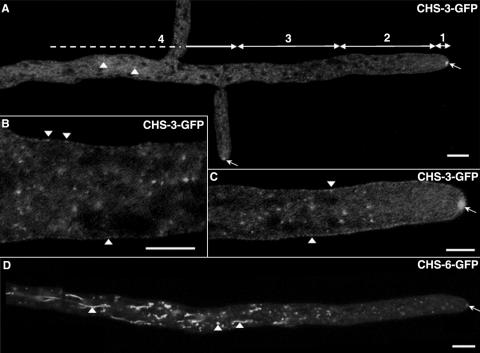
Distribution of GFP-labeled CHS-3 and CHS-6 along hyphae. Reconstruction of a Neurospora hypha, showing overall distribution of CHS-3-GFP (A). Hyphae were divided into four regions measured from the tip: 1, apex (2 to 5 μm); 2, proximal subapex (5 to 20 μm); 3, distal subapex (20 to 40 μm); and 4, distal region (>40 μm). In the distal (B) and proximal subapex (C) CHS-3-GFP shows a punctate distribution; some fluorescent dots appear to be at the plasma membrane (arrowheads in panel B, C). Reconstruction of a hypha showing overall distribution of CHS-6-GFP (D). In distal regions, both CHS-3-GFP and CHS-6-GFP are found in a highly stained network of spherical and tubular compartments (arrowheads in panel A, D). Arrows in panels A, C, and D point to the Spk. Scale bars: 10 μm (A, D); 5 μm (B, C).
To better describe the distribution of CHS-GFP, we divided each hypha into four different regions (Fig. 2A) as previously described (45): one apical region (2 to 5 μm from the apical pole); two subapical regions, a proximal and a distal one (5 to 20 μm and 20 to 40 μm, respectively, from the apical pole); and a distal region (>40 μm from the apex). High-magnification confocal images of each region were assembled in Adobe Photoshop CS2 to obtain an overall view of CHS-GFP localization along a hypha (Fig. 2A and D). GFP-labeled structures were distributed along the hyphal tube in gradient-like fashion, from larger, more intensely fluorescent structures in the distal regions to discrete punctate structures about 0.1 μm in diameter in the distal subapex and to finely dispersed fluorescence in the proximal subapex.
At the proximal subapex, there was very little punctate vesiculoid fluorescence in the cytoplasm of either NMR3 or NMR6 (Fig. 2A and D). Some of the punctate CHS-3-GFP fluorescence was either attached or adjacent to the plasma membrane in this region (Fig. 2B and C). While punctate fluorescence at or near the plasma membrane was observed, it was more difficult to differentiate in CHS-6-GFP strains. The CHS-3-GFP fluorescent punctate structures did not correspond to phase-dark structures (Fig. 1A to F). Except for the Spk, no obvious phase-dark match was found for other CHS-3-GFP fluorescent structures.
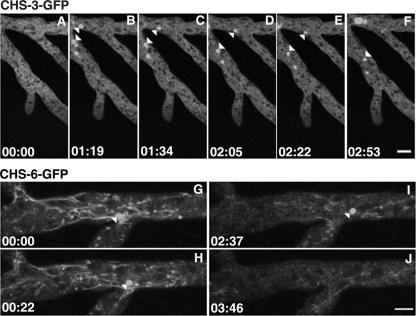
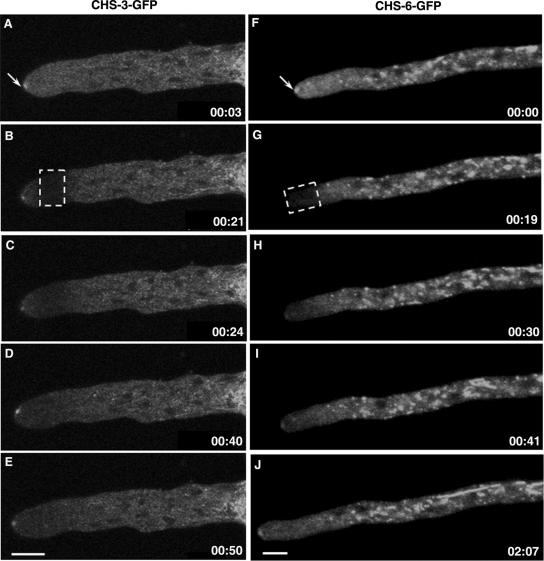
In the distal subapex, there was a higher concentration of punctate fluorescence, especially in strain NMR6 (Fig. 2D). In this region, the fluorescence of both CHS-3-GFP and CHS-6-GFP was frequently observed in globular bodies (possibly vacuoles) 1.5 to 3 μm in diameter that appeared to disintegrate into smaller bodies (putative vesicles or groups of vesicles) while advancing. These vesicles moved predominantly forward until they reached the proximal subapex, where they were no longer distinctly visible (Fig. 3A to F; see movies S3 and S4 in the supplemental material).
FIG. 3.
Progressive disintegration of GFP-labeled vacuoles and dispersal of fluorescence in strain NMR3 (A to F) and strain NMR6 (G to J). Arrowheads point at spherical bodies whose size gradually diminishes until they are no longer visible. Similarly, the CHS-6-GFP-labeled tubular compartments in NMR6 undergo progressive disintegration (G to J). Time, min:s. Scale bars, 5 μm.
In distal regions (>40 μm), fluorescence, especially of CHS-6-GFP, was mostly found in the lumen of a network of tubular and globular (>3 μm) vacuolar compartments (Fig. 3G to J).
Spk and vesicle traffic.
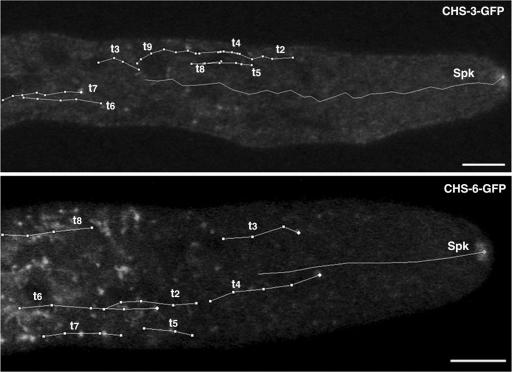
Overall, the entire array of fluorescent vesicles labeled with CHS-3-GFP and CHS-6-GFP, i.e., the discrete punctate structures about 0.1 μm in diameter, moved predominantly forward as the hyphae elongated (Fig. 4; see movies S1 and S2 in the supplemental material). The speed of vesicles varied significantly, ranging from 0.09 to 0.4 μm/s. Most of the average speed values for the traced vesicles were within the range of the Spk advancement rates obtained for N. crassa during CSLM observation (0.1 to 0.4 μm/s). Sporadically, some vesicles moved backwards at a much lower speed (0.04 μm/s).
FIG. 4.
Kinetic analysis of fluorescent structures carrying CHS-3-GFP and CHS-6-GFP. The trajectories (t) of individual particles and the Spk were followed. Fluorescent vesicles were traced moving predominantly forward until reaching the proximal subapex (15 to 20 μm from the apex) where they were no longer visible. The average speed (in μm/s) of the Spk and the different vesicles was, for strain NMR3 Spk, 0.11 ± 0.02; t2, 0.11 ± 0.06; t3, 0.13 ± 0.04; t4, 0.12 ± 0.09; t5, 0.11 ± 0.04; t6, 0.18 ± 0.05; t7, 0.12 ± 0.04; t8, 0.13 ± 0.04; and t9, 0.09 ± 0.03; and for strain NMR6 Spk, 0.24 ± 0.02; t2, 0.18 ± 0.05; t3, 0.19 ± 0.06; t4, 0.21 ± 0.02; t5, 0.18 ± 0.07; t6, 0.26 ± 0.02; t7, 0.19 ± 0.06; and t8, 0.22 ± 0.05. Time, min:s. Scale bars, 5 μm. Particle movement can be better appreciated in movies S1 and S2 in the supplemental material.
FRAP analysis.
FRAP analysis was conducted to monitor the flow of CHS-GFP towards the Spk in both strain NMR3 and strain NMR6. A small subapical area, 4 μm behind the Spk, or an apical area including the Spk was selected in hyphae of each strain and exposed to a high-intensity laser irradiation. We did FRAP analysis of eight subapical and four apical regions of CHS-3-GFP and CHS-6-GFP strains, respectively. The growth rate of hyphae (0.14 to 0.16 μm/s) was similar to that of wild-type or unbleached strains and appeared not to be affected by the photobleaching. Photobleached hyphae with CHS-3-GFP and CHS-6-GFP showed similar recovery patterns. The intensity of apical fluorescence after photobleaching was analyzed with LSM510 software. In NMR3, Spk fluorescence decreased to 56% 18 s after the pulse and recovered to 98% 20 s later, even though no change in Spk fluorescence was visually apparent (e.g., see Fig. 5A to E). Bleaching the apical area caused a drastic reduction in the apical fluorescence and the disappearance of the Spk signal (e.g., see Fig. 5G). Fluorescence intensity measurements in NMR6 showed that apical fluorescence had not disappeared completely but decreased to 16% 14 s after photobleaching and recovered to 73% of its original intensity 120 s later (e.g., see Fig. 5H and I).
FIG. 5.
FRAP analysis of the proximal subapex of a CHS-3-GFP hypha (A to E) and the apex of a CHS-6-GFP hypha (F to J). Prebleached hyphae. Arrows point at the Spk (A, F). Photobleaching was applied to the selected areas indicated by the dotted rectangles (B, G). Recovery of Spk fluorescence occurs after either apical (H to J) or subapical (C to E) bleaching. Note a temporary departure of the Spk from its centric position in the apical dome (D). Time, min:s. Scale bars, 5 μm.
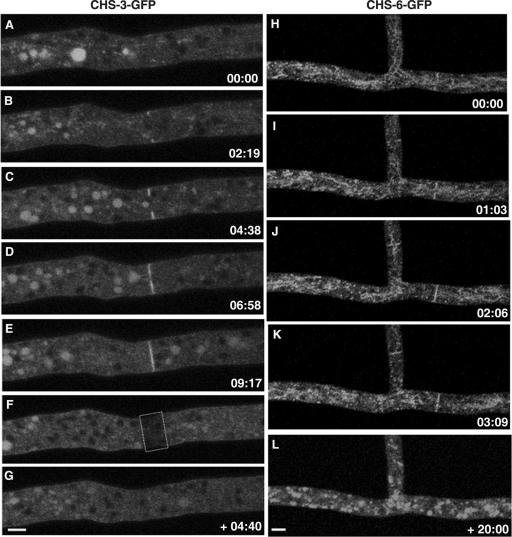
Localization of CHS-3 and CHS-6 in septa.
As in the growing apex, septa are zones of intense cell wall synthesis. Both CHS-3-GFP (Fig. 6A to E; see movie S5 in the supplemental material) and CHS-6-GFP (Fig. 6H to K) were found at the developing septa. Septum development lasted about 3 to 6 min as observed in different septa of strains NMR3 and NMR6. After the completion of septum formation, FRAP experiments showed that there was no additional accumulation of CHS-3-GFP in the photobleached septum (Fig. 6F and G; see movie S5 in the supplemental material). In addition, we observed that CHS-6-GFP fluorescence did not persist in the septa for longer than 10 to 20 min after septum formation had been completed (Fig. 6). Furthermore, existing septa identified by phase-contrast optics in far distal—and therefore older—regions of hyphae showed very low or no fluorescence (data not shown).
FIG. 6.
Localization of CHS-GFP during cross-wall synthesis. CSLM time series show the progressive and localized accumulation of both CHS-3-GFP (A to E) and CHS-6-GFP (H to K) during septum formation. In a FRAP analysis, the formed septum disappeared after photobleaching (F) and no fluorescence reappeared in the septum even after 4 min following FRAP (G). Septum completion in strain NMR6 lasts ∼3 to 6 min (H to K); afterwards, the CHS-6-GFP signal dissipates over time; 20 minutes after the septum had been completed, septum fluorescence had vanished (L). Time, min:s. Scale bars, 5 μm.
Lack of colocalization of CHS-GFP with compartments of the conventional secretory pathway.
Strains NMR3, NMR6, and N150 (wild type; mat A; FGSC 9013) were stained with specific vital fluorescent dyes to determine whether CHS-GFP colocalized with various endomembranous compartments (i.e., the ER, Golgi body equivalents, vacuoles, and endocytic compartments) involved in the conventional secretory routes. All dyes tested were incorporated into actively growing hyphae ∼5 to 10 min after application.
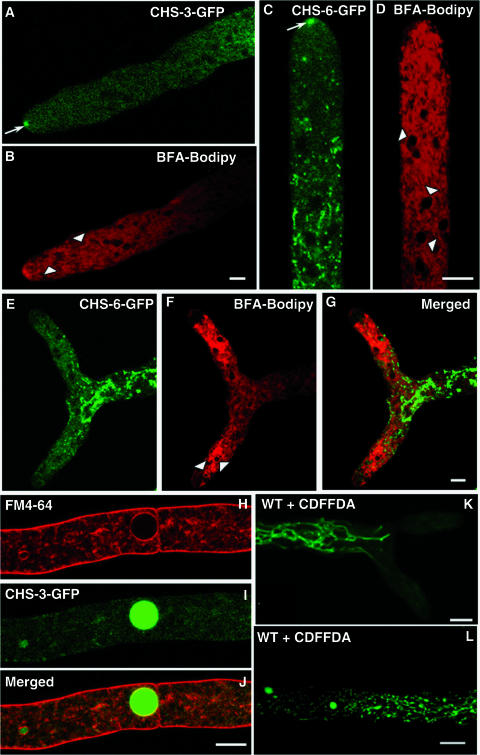
The putative Golgi body equivalents and ER stained by the BFA bodipy 558/568 conjugate did not colocalize with the GFP-labeled endomembranous system (Fig. 7A to G). BFA interferes with Golgi body-dependent secretion and also with endosomal/post-Golgi body trafficking (11, 18, 21, 52), although the effects of this inhibitor can be tissue and species specific (21, 54). In Neurospora, BFA caused a decrease in hyphal elongation and an increase in branching, but the treatment did not interfere with the traffic of CHS-containing microvesicles to the Spk. For a short, 5-min initial period, the dye stained putative Golgi equivalents present at the hyphal apex and proximal subapex (Fig. 7B). During the following 5 to 20 min, as a result of the effect of the BFA, the dye was redistributed and stained the ER surrounding the nuclei (Fig. 7D). During that period, GFP fluorescence was still very prominent at the Spk of both strain NMR3 and strain NMR6 (Fig. 7A and C). In some hyphae, growth ceased upon prolonged exposure to BFA-bodipy. This caused the disappearance of the Spk concomitantly with a movement of the dye away from the apical region, while staining the ER more strongly (Fig. 7E to G). The lack of colocalization of CHS-GFP-labeled and bodipy-stained membranes showed clearly that CHS-GFP is not transported by a conventional ER-Golgi body pathway.
FIG. 7.
Distinct localization of CHS-GFP and conventional secretory compartments. (A to G) Comparative distribution of fluorescence from CHS-3-GFP (A) and CHS-6-GFP (C, E) with Golgi body structures revealed after staining with 5 μM BFA-bodipy 558/568 conjugate. After a short period (∼5 min), the dye stained apical and subapical Golgi body equivalents (arrowheads in B). Note the preservation of CHS-GFP in the Spk (arrow in panel A). Over 5 to 20 min, the dye stained the ER (arrowheads in D). During that period, GFP fluorescence was still very prominent at the Spk of strain NMR6 (arrow in panel C). After a longer exposure (20 to 25 min), the hyphae stopped growing (E to G), the Spk disappeared, and both fluorescence and dye moved away from the apical region and strongly stained the ER (arrowheads in panel F), but the separation between fluorescent CHS-6-GFP and the bodipy-stained structures, particularly the ER around the nuclei, remained clearly evident (G). Scale bars, 5 μm. (H to L) Relationship between CHS-3-GFP and the vacuolar system. Localization of CHS-3-GFP fluorescence in the vacuoles of NMR3 in hyphae stained with FM4-64 (H to J); note that CHS-3-GFP is found in the lumen of the vacuole (I), whereas FM4-64 stains the vacuolar membrane (H, J). Scale bars, 5 μm. For comparison, the vacuolar system in wild-type hyphae of N. crassa is shown after staining with 10 μM CDFFDA (K, L). Scale bars, 10 μm.
Evidence of a possible relationship between CHS and the vacuole system is shown in Fig. 7H to L. CHS-GFP fluorescence was found in the lumen of vacuoles but, significantly, not in their membranes (Fig. 7H to J). The membrane, but not the lumen, was stained by the endocytic marker FM4-64. Also the distribution of CHS-GFP in distal regions is reminiscent of the globular and tubular vacuolar system as revealed by CDFFDA staining (Fig. 7K and L).
DISCUSSION
By tagging chs genes with sgfp, we have succeeded in determining the intracellular location of two CHS in Neurospora and obtained a first glimpse of CHS trafficking inside the cell. Despite some differences in the patterns of distribution between CHS-3 and CHS-6, four major conclusions can be drawn about the intracellular location of these two CHS: (i) most of the GFP-tagged CHS in distal regions is found in rather large compartments, some tubular, some spherical; (ii) the subapical, round, GFP-labeled structures diminish dramatically in size as they approach the apex; (iii) in the proximal subapex, the GFP label is finely dispersed into putative microvesicles that are likely chitosomes; and (iv) most strikingly, at the apex, there is a dense accumulation of CHS-GFP into a spherical body that coincides with the position of the Spk.
A salient finding in the present study was the localization of CHS-3 and CHS-6 in the Spk, which has previously not been observed or reported. Thus far, A. fumigatus ChsD, a class VI CHS, has been shown to be expressed during hyphal growth (39), but no localization data were reported. In Ustilago maydis, the class I CHS-3 and CHS-4—both labeled with fluorescent proteins—have been localized at septa and at cortical regions of the cytoplasm (61) but were not found at the Spk.
Given the fragile nature of the Spk (33), it is not surprising that it may not have survived procedures employed previously for autoradiography or electron microscopy. Another crucial factor may be the fungal growth rate: the high elongation rate and large diameter of the N. crassa hyphae produce a well-defined Spk. In slow-growing fungi, the Spk is much smaller and harder to see, and this may help explain why CHS-GFP fusions, although present at tips, were not detected in the Spk in studies with Colletotrichum graminicola (1). On the other hand, our finding that both CHS-3-GFP and CHS-6-GFP accumulate at growing septa but are turned over fairly rapidly coincides with previous reports on the localization of CHS at the septa of C. graminicola (1), A. nidulans (58), and U. maydis (51, 61, 62).
Our results show that CHS-GFP accumulates at the core of the Spk, presumably the same region in which microvesicles have been detected in earlier studies by transmission electron microscopy (23, 38). To better discern whether the CHS-GFP distribution indeed coincides with the core identified by transmission electron microscopy, we plan to further characterize the CHS-GFP distribution at the ultrastructural level by immunoelectron microscopy. The dimensions of the smallest CHS-GFP fluorescent vesicles cannot be determined from our confocal images with high confidence, but they are probably smaller than 100 nm in diameter, a size in the size range of chitosomes, which measure 40 to 70 nm and are known to contain most of the CHS in fungal cells (6). In a separate, ongoing study (J. Verdín-Ramos, M. Riquelme, E. Castro-Longoria, and S. Bartnicki-García, unpublished data), cell extracts of strains NMR3 and NMR6 were separated by isopycnic sedimentation and analyzed by Western blotting with a GFP-specific monoclonal antibody. Several bands of a higher molecular weight than that of GFP alone were detected in cell fractions, with a density around 1.13 g/ml, the characteristic buoyant density of chitosomes of N. crassa (36).
FRAP analysis indicated that the fluorescence of the Spk originates from the finely dispersed fluorescence in the proximal subapex, i.e., it originated in the population of CHS-3-GFP- and CHS-6-GFP-labeled microvesicles. These findings lend credence to the long-standing prediction that the Spk is directly involved in guiding cell wall formation (4, 8).
Our observations that CHS-6-GFP and CHS-3-GFP do not accumulate at the plasma membrane in N. crassa hyphal tips support the idea that, upon reaching the cell surface, CHS turns over rapidly (56). Our dynamic images of CHS-GFP traffic, plus the identification of the GFP-labeled particles as microvesicles (likely chitosomes, because of their low specific gravity), support previous findings obtained by immunolocalization in N. crassa (56) and by autoradiography in Mucor rouxii (55).
Our findings on the localization of CHS-3 and CHS-6 in N. crassa, plus previous studies on the three CHS from S. cerevisiae, Chs1p, Chs2p (31), and Chs3p (17), show that they all can be detected in chitosomes. This raises the question as to whether these different CHS belong to one multienzyme complex transported within the same population of chitosomes or whether there exist different populations of microvesicles specialized in the transport of a specific class of CHS. The different distribution and traffic patterns of CHS-3 and CHS-6 along the hyphae of N. crassa might be construed as an indication of the existence of different populations of chitosomes.
Three observations argue against the involvement of the conventional ER-to-Golgi body secretory pathway in the intracellular traffic of CHS but in favor of a new model for an unconventional, separate, CHS secretory pathway: (i) the inability of BFA to diminish the fluorescence reaching the Spk; (ii) the lack of colocalization between Golgi body equivalents and ER membranes and CHS-GFP-stained membranes; and (iii) results from previous studies showing that the ER microvesicles and chitosomes of N. crassa (30) belong to different populations that can be separated by high-performance ultracentrifugation (31, 56). Both the gradual decrease in size of the GFP-labeled globular vacuoles from the subapical region of the hyphae to the apex and the findings on the intracellular movement point to a progressive dispersion of the CHS-containing structures as they move towards the growing tip. The fact that the CHS-GFP appear to be not in the membrane but in the lumen of these membranous structures suggests that the CHS-GFP accumulates in the interior of these compartments, to be discharged once it reaches its final destination. There are numerous recent reports of proteins exported by unconventional secretory routes (41-44, 57), including some routes involving multivesicular bodies that have received much attention lately (42). The ability of chitosomes to dissociate reversibly upon treatment with digitonin (49) suggests that chitosome microvesicles may not be generated by budding from a membranous structure, but rather by self-assembly from 16S subunits. Such self-assembly may occur either in the cytosol or in vacuoles, the latter giving rise to multivesicular bodies (13).
In conclusion, our in vivo studies on the localization and movement of Neurospora CHS-GFP have already yielded valuable insights into the pathway that delivers proteins for cell wall synthesis to the Spk in fungal hyphae. Future studies on the structure and localization of Neurospora CHS at the resolution level of the electron microscope will characterize the structures revealed here by fluorescence microscopy. This initial study sets the stage for further work that will test the unconventional transport model proposed here and that could eventually result in a more complete understanding of how the different components of the CHS family are transported and integrated into microfibril-assembling complexes.
Supplementary Material
Acknowledgments
This work was supported by CONACYT (Consejo Nacional de Ciencia y Tecnología, México) grants DG/2003-1158 and U-45818Q. The Freitag lab is supported by start-up funds from the Computational and Genome Biology Initiative at Oregon State University.
We thank Eric Selker (University of Oregon, Eugene) for support of this work.
Footnotes
Published ahead of print on 20 July 2007.
Supplemental material for this article may be found at http://ec.asm.org/.
REFERENCES
- 1.Amnuaykanjanasin, A., and L. Epstein. 2006. A class Vb chitin synthase in Colletotrichum graminicola is localized in the growing tips of multiple cell types, in nascent septa, and during septum conversion to an end wall after hyphal breakage. Protoplasma 227:155-164. [DOI] [PubMed] [Google Scholar]
- 2.Bartnicki-Garcia, S. 1968. Cell wall chemistry, morphogenesis and taxonomy of fungi. Annu. Rev. Microbiol. 22:87-107. [DOI] [PubMed] [Google Scholar]
- 3.Bartnicki-Garcia, S. 2006. Chitosomes: past, present and future. FEMS Yeast Res. 6:957-965. [DOI] [PubMed] [Google Scholar]
- 4.Bartnicki-Garcia, S. 2002. Hyphal tip growth: outstanding questions, p. 29-58. In H. D. Osiewacz (ed.), Molecular biology of fungal development. Marcel Dekker, Inc., New York, NY.
- 5.Bartnicki-Garcia, S. 1990. Role of vesicles in apical growth and a new mathematical model of hyphal morphogenesis, p. 211-232. In I. B. Heath (ed.), Tip growth in plant and fungal cells. Academic Press, San Diego, CA.
- 6.Bartnicki-Garcia, S., C. E. Bracker, E. Reyes, and J. Ruiz Herrera. 1978. Isolation of chitosomes from taxonomically diverse fungi and synthesis of chitin microfibrils in vitro. Exp. Mycol. 2:173-192. [Google Scholar]
- 7.Bartnicki-Garcia, S., and E. Lippman. 1969. Fungal morphogenesis: cell wall construction in Mucor rouxii. Science 165:302-304. [DOI] [PubMed] [Google Scholar]
- 8.Bartnicki-Garcia, S., J. Ruiz-Herrera, and C. E. Bracker. 1979. Chitosomes and chitin synthesis, p. 149-168. In J. H. Burnett and A. P. J. Trinci (ed.), Fungal walls and hyphal growth. Cambridge University Press, Cambridge, United Kingdom.
- 9.Borgia, P. T., N. Iartchouk, P. J. Riggle, K. R. Winter, Y. Koltin, and C. E. Bulawa. 1996. The chsB gene of Aspergillus nidulans is necessary for normal hyphal growth and development. Fungal Genet. Biol. 20:193-203. [DOI] [PubMed] [Google Scholar]
- 10.Borkovich, K. A., L. A. Alex, O. Yarden, M. Freitag, G. E. Turner, N. D. Read, S. Seiler, D. Bell-Pedersen, J. Paietta, N. Plesofsky, M. Plamann, M. Goodrich-Tanrikulu, U. Schulte, G. Mannhaupt, F. E. Nargang, A. Radford, C. Selitrennikoff, J. E. Galagan, J. C. Dunlap, J. J. Loros, D. Catcheside, H. Inoue, R. Aramayo, M. Polymenis, E. U. Selker, M. S. Sachs, G. A. Marzluf, I. Paulsen, R. Davis, D. J. Ebbole, A. Zelter, E. R. Kalkman, R. O'Rourke, F. Bowring, J. Yeadon, C. Ishii, K. Suzuki, W. Sakai, and R. Pratt. 2004. Lessons from the genome sequence of Neurospora crassa: tracing the path from genomic blueprint to multicellular organism. Microbiol. Mol. Biol. Rev. 68:1-108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bourett, T. M., and R. J. Howard. 1996. Brefeldin A-induced structural changes in the endomembrane system of a filamentous fungus, Magnaporthe grisea. Protoplasma 190:151-163. [Google Scholar]
- 12.Bowen, A. R., J. L. Chen-Wu, M. Momany, R. Young, P. J. Szaniszlo, and P. W. Robbins. 1992. Classification of fungal chitin synthases. Proc. Natl. Acad. Sci. USA 89:519-523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bracker, C. E., J. Ruiz-Herrera, and S. Bartnicki-Garcia. 1976. Structure and transformation of chitin synthetase particles (chitosomes) during microfibril synthesis in vitro. Proc. Natl. Acad. Sci. USA 73:4570-4574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Brunswik, H. 1924. Untersuchungen über die Geschlechts- und Kernverhältnisse bei der Hymenomyzetengattung Coprinus. Bot. Abh. 5:1-152. [Google Scholar]
- 15.Chigira, Y., K. Abe, K. Gomi, and T. Nakajima. 2002. chsZ, a gene for a novel class of chitin synthase from Aspergillus oryzae. Curr. Genet. 41:261-267. [DOI] [PubMed] [Google Scholar]
- 16.Choquer, M., M. Boccara, I. R. Gonçalves, M. Soulié, and A. Vidal-Cros. 2004. Survey of the Botrytis cinerea chitin synthase multigenic family through the analysis of six euascomycetes genomes. Eur. J. Biochem. 271:2153-2164. [DOI] [PubMed] [Google Scholar]
- 17.Chuang, J. S., and R. W. Schekman. 1996. Differential trafficking and timed localization of two chitin synthase proteins, Chs2p and Chs3p. J. Cell Biol. 135:597-610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cole, L., D. Davies, G. J. Hyde, and A. E. Ashford. 2000. Brefeldin A affects growth, endoplasmic reticulum, Golgi bodies, tubular vacuole system, and secretory pathway in Pisolithus tinctorius. Fungal Genet. Biol. 29:95-106. [DOI] [PubMed] [Google Scholar]
- 19.Freitag, M., P. C. Hickey, N. B. Raju, E. U. Selker, and N. D. Read. 2004. GFP as a tool to analyze the organization, dynamics and function of nuclei and microtubules in Neurospora crassa. Fungal Genet. Biol. 41:897-910. [DOI] [PubMed] [Google Scholar]
- 20.Fujiwara, M., H. Horiuchi, A. Ohta, and M. Takagi. 1997. A novel fungal gene encoding chitin synthase with a myosin motor-like domain. Biochem. Biophys. Res. Commun. 236:75-78. [DOI] [PubMed] [Google Scholar]
- 21.Geldner, N. 2004. The plant endosomal system—its structure and role in signal transduction and plant development. Planta 219:547-560. [DOI] [PubMed] [Google Scholar]
- 22.Girbardt, M. 1957. Der Spitzenkörper von Polystictus versicolor. Planta 50:47-59. [Google Scholar]
- 23.Girbardt, M. 1969. Die Ultrastruktur der Apikalregion von Pilzhyphen. Protoplasma 67:413-441. [Google Scholar]
- 24.Grove, S. N., and C. E. Bracker. 1970. Protoplasmic organization of hyphal tips among fungi: vesicles and Spitzenkörper. J. Bacteriol. 104:989-1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Harris, S. D., N. D. Read, R. W. Roberson, B. Shaw, S. Seiler, M. Plamann, and M. Momany. 2005. Polarisome meets Spitzenkörper: microscopy, genetics, and genomics converge. Eukaryot. Cell 4:225-229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hickey, P. C., S. M. Swift, M. G. Roca, and N. D. Read. 2005. Live-cell imaging of filamentous fungi using vital fluorescent dyes. Methods Microbiol. 34:63-87. [Google Scholar]
- 27.Holcomb, C. L., W. J. Hansen, T. Etcheverry, and R. Schekman. 1988. Secretory vesicles externalize the major plasma membrane ATPase in yeast. J. Cell Biol. 106:641-648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Howard, R. J. 1981. Ultrastructural analysis of hyphal tip cell growth in fungi: Spitzenkörper, cytoskeleton and endomembranes after freeze-substitution. J. Cell Sci. 48:89-103. [DOI] [PubMed] [Google Scholar]
- 29.Hunsley, D., and G. W. Gooday. 1974. The structure and development of septa in Neurospora crassa. Protoplasma 82:125-146. [DOI] [PubMed] [Google Scholar]
- 30.Leal-Morales, C., C. E. Bracker, and S. Bartnicki-Garcia. 1994. Distribution of chitin synthetase and various membrane marker enzymes in chitosomes and other organelles of the slime mutant of Neurospora crassa. Exp. Mycol. 18:168-179. [Google Scholar]
- 31.Leal-Morales, C. A., C. E. Bracker, and S. Bartnicki-Garcia. 1994. Subcellular localization, abundance and stability of chitin synthetases 1 and 2 from Saccharomyces cerevisiae. Microbiology 140:2207-2216. [DOI] [PubMed] [Google Scholar]
- 32.Lesage, G., and H. Bussey. 2006. Cell wall assembly in Saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev. 70:317-343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.López-Franco, R., and C. E. Bracker. 1996. Diversity and dynamics of the Spitzenkörper in growing hyphal tips of higher fungi. Protoplasma 195:90-111. [Google Scholar]
- 34.Mandel, M. A., J. N. Galgiani, S. Kroken, and M. J. Orbach. 2006. Coccidioides posadasii contains single chitin synthase genes corresponding to classes I to VII. Fungal Genet. Biol. 43:775-788. [DOI] [PubMed] [Google Scholar]
- 35.Margolin, B. S., M. Freitag, and E. U. Selker. 1997. Improved plasmids for gene targeting at the his-3 locus of Neurospora crassa by electroporation. Fungal Genet. Newslett. 44:34-36. [Google Scholar]
- 36.Martinez, J. P., G. Gimenez, and S. Bartnicki Garcia. 1987. Intracellular localization of UDP-N-acetylglucosamine in Neurospora crassa wild-type and slime mutant strains. Exp. Mycol. 11:278-286. [Google Scholar]
- 37.Martín-Udíroz, M., M. P. Madrid, and M. I. G. Roncero. 2004. Role of chitin synthase genes in Fusarium oxysporum. Microbiol. Mol. Biol. Rev. 150:3175-3187. [DOI] [PubMed] [Google Scholar]
- 38.McClure, W. K., D. Park, and P. M. Robinson. 1968. Apical organization in the somatic hyphae of fungi. J. Gen. Microbiol. 50:177-182. [DOI] [PubMed] [Google Scholar]
- 39.Mellado, E., C. A. Specht, P. W. Robbins, and D. W. Holden. 1996. Cloning and characterization of chsD, a chitin synthase-like gene of Aspergillus fumigatus. FEMS Microbiol. Lett. 143:69-76. [DOI] [PubMed] [Google Scholar]
- 40.Munro, C. A., D. A. Schofield, G. W. Gooday, and N. A. Gow. 1998. Regulation of chitin synthesis during dimorphic growth of Candida albicans. Microbiology 144:391-401. [DOI] [PubMed] [Google Scholar]
- 41.Nickel, W. 2003. The mystery of nonclassical protein secretion: a current view on cargo proteins and potential export routes. Eur. J. Biochem. 270:2109. [DOI] [PubMed] [Google Scholar]
- 42.Nickel, W. 2005. Unconventional secretory routes: direct protein export across the plasma membrane of mammalian cells. Traffic 6:607-614. [DOI] [PubMed] [Google Scholar]
- 43.Rayner, J. C., and H. R. Pelham. 1997. Transmembrane domain-dependent sorting of proteins to the ER and plasma membrane in yeast. EMBO J. 16:1832-1841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Reggiori, F., and H. R. Pelham. 2001. Sorting of proteins into multivesicular bodies: ubiquitin-dependent and -independent targeting. EMBO J. 20:5176-5186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Riquelme, M., R. Roberson, D. McDaniel, and S. Bartnicki-Garcia. 2002. The effect of ropy-1 mutation on cytoplasmic organization and intracellular motility in mature Neurospora crassa. Fungal Genet. Biol. 37:171-179. [DOI] [PubMed] [Google Scholar]
- 46.Roncero, C. 2002. The genetic complexity of chitin synthesis in fungi. Curr. Genet. 41:367-378. [DOI] [PubMed] [Google Scholar]
- 47.Ruiz-Herrera, J. 1992. Fungal cell wall: structure, synthesis and assembly. CRC Press, Boca Raton, FL.
- 48.Ruiz-Herrera, J., and S. Bartnicki-Garcia. 1974. Synthesis of cell wall microfibrils in vitro by a “soluble” chitin synthetase from Mucor rouxii. Science 186:357-359. [DOI] [PubMed] [Google Scholar]
- 49.Ruiz-Herrera, J., S. Bartnicki-Garcia, and C. E. Bracker. 1980. Dissociation of chitosomes by digitonin into 16 S subunits with chitin synthetase activity. Biochim. Biophys. Acta 629:201-206. [DOI] [PubMed] [Google Scholar]
- 50.Ruiz-Herrera, J., J. M. Gonzalez-Prieto, and R. Ruiz-Medrano. 2002. Evolution and phylogenetic relationships of chitin synthases from yeasts and fungi. FEMS Yeast Res. 1:247-256. [DOI] [PubMed] [Google Scholar]
- 51.Ruiz-Herrera, J., B. Xoconostle-Cazare, C. G. Reynaga-Pena, C. Leon-Ramirez, and A. Carabez-Trejo. 2006. Immunolocalization of chitin synthases in the phytopathogenic dimorphic fungus Ustilago maydis. FEMS Yeast Res. 6:999-1009. [DOI] [PubMed] [Google Scholar]
- 52.Rupes, I., W. Z. Mao, H. Åström, and M. Raudaskoski. 1995. Effects of nocodazole and brefeldin A on microtubule cytoskeleton and membrane organization in the homobasidiomycete Schizophyllum commune. Protoplasma 185:212-221. [Google Scholar]
- 53.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 54.Satiat-Jeunemaitre, B., L. Cole, T. Bourett, R. Howard, and C. Hawes. 1996. Brefeldin A effects in plant and fungal cells: something new about vesicle trafficking? J. Microsc. 181:162-177. [DOI] [PubMed] [Google Scholar]
- 55.Sentandreu, R., A. Martinez-Ramon, and J. Ruiz-Herrera. 1984. Localization of chitin synthase in Mucor rouxii by an autoradiographic method. J. Gen. Microbiol. 130:1193-1199. [DOI] [PubMed] [Google Scholar]
- 56.Sietsma, J. H., A. Beth Din, V. Ziv, K. A. Sjollema, and O. Yarden. 1996. The localization of chitin synthase in membranous vesicles (chitosomes) in Neurospora crassa. Microbiology 142:1591-1596. [DOI] [PubMed] [Google Scholar]
- 57.Stoorvogel, W., M. J. Kleijmeer, H. J. Geuze, and G. Raposo. 2002. The biogenesis and functions of exosomes. Traffic 3:321-330. [DOI] [PubMed] [Google Scholar]
- 58.Takeshita, N., A. Ohta, and H. Horiuchi. 2005. CsmA, a class V chitin synthase with a myosin motor-like domain, is localized through direct interaction with the actin cytokeleton in Aspergillus nidulans. Mol. Biol. Cell 16:1961-1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Vogel, H. J. 1956. A convenient growth medium for Neurospora (medium N). Microbiol. Gen. Bull. 13:42-43. [Google Scholar]
- 60.Wang, Q., H. Liu, and P. J. Szaniszlo. 2002. Compensatory expression of five chitin synthase genes, a response to stress stimuli, in Wangiella (Exophiala) dermatitidis, a melanized fungal pathogen of humans. Microbiology 148:2811-2817. [DOI] [PubMed] [Google Scholar]
- 61.Weber, I., D. Assmann, E. Thines, and G. Steinberg. 2006. Polar localizing class V myosin chitin synthases are essential during early plant infection in the plant pathogenic fungus Ustilago maydis. Plant Cell 18:225-242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Weber, I., C. Gruber, and G. Steinberg. 2003. A class-V myosin required for mating, hyphal growth, and pathogenicity in the dimorphic plant pathogen Ustilago maydis. Plant Cell 15:2826-2842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Westergaard, M., and H. K. Mitchell. 1947. Neurospora V. A synthetic medium favoring sexual reproduction. Am. J. Bot. 34:573-577. [Google Scholar]
- 64.Yanai, K., N. Kojima, N. Takaya, H. Horiuchi, A. Ohta, and M. Takagi. 1994. Isolation and characterization of two chitin synthase genes from Aspergillus nidulans. Biosci. Biotechnol. Biochem. 58:1828-1835. [DOI] [PubMed] [Google Scholar]
- 65.Yarden, O., and C. Yanofsky. 1991. Chitin synthase 1 plays a major role in cell wall biogenesis in Neurospora crassa. Genes Dev. 5:2420-2430. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.