Abstract
In this report, we have characterized two metacaspases of Leishmania donovani, L. donovani metacaspase-1 (LdMC1) and LdMC2. These two proteins show 98% homology with each other, and both contain a characteristic C-terminal proline-rich domain. Both genes are transcribed in promastigotes and axenic amastigotes of L. donovani; however, LdMC1 shows increased mRNA levels in axenic amastigotes. An anti-LdMC antibody was obtained and showed reactivity with a single ∼42-kDa protein band in both promastigote and axenic amastigote parasite whole-cell lysates by Western blotting. Pulse-chase experiments suggest that LdMCs are not synthesized as proenzymes, and immunofluorescence studies show that LdMCs are associated with the acidocalcisome compartments of L. donovani. Enzymatic assays of immunoprecipitated LdMCs show that native LdMCs efficiently cleave trypsin substrates and are unable to cleave caspase-specific substrates. Consistently, LdMC activity is insensitive to caspase inhibitors and is efficiently inhibited by trypsin inhibitors, such as leupeptin, antipain, and Nα-tosyl-l-lysine-chloromethyl ketone (TLCK). In addition, our results show that LdMC activity was induced in parasites treated with hydrogen peroxide, a known trigger of programmed cell death (PCD) in Leishmania and that parasites overexpressing metacaspases are more sensitive to hydrogen peroxide-induced PCD. These findings suggest that Leishmania metacaspases are not responsible for the caspase-like activities reported in this organism and suggest a possible role for LdMCs as effector molecules in Leishmania PCD.
Metacaspases constitute a new family of caspase-related proteins recently described by Uren et al. (39). They have been identified by bioinformatic analysis and are found in plants, fungi, and parasitic protozoa but are absent in mammals (39). Metacaspases belong to the CD clan of cysteine peptidases with six other families, including caspases and paracaspases (29). Metacaspases are structurally related to caspases and show conservation of cysteine and histidine amino acid residues involved in the Cys-His catalytic dyad of the active domains of caspase (39). Caspases have been shown to play a central role in programmed cell death of mammalian cells, also called apoptosis (reviewed in reference 14). To date, no caspase gene has been identified in plants, yeasts, or protozoan parasites. However, since these organisms possess one or more metacaspases, it is conceivable that like caspases in mammalian cells, these caspase-related proteases could be involved in the PCD pathways in these organisms. In that regard, the Saccharomyces cerevisiae metacaspase YCA1 and the Norway spruce metacaspase mcII-Pa have been shown recently to be directly implicated in PCD in these organisms (4, 28). To date, however, the possible role of metacaspases in PCD of protozoan parasites remains to be demonstrated.
Little is known about the enzymatic properties of metacaspases. In contrast, the related caspase enzymes are well-characterized cysteine-dependent aspartate-specific proteases that hydrolyze peptide bonds at the C-terminal side of an aspartate (P1 residue). A number of specific synthetic substrates and inhibitors have been developed and used to determine the enzymatic properties of most mammalian caspases. Such synthetic substrates and inhibitors were also used to identify caspase-like activities in many organisms, including yeasts (26, 27, 38), plants (3, 18, 23), and protozoa (1, 6, 19, 22, 24, 34-36). However, it was shown recently that metacaspases of Arabidopsis thaliana expressed in Escherichia coli were unable to cleave caspase-specific substrates but cleaved substrates containing an arginine or lysine residues at the P1 position (40, 41). Further, these recombinant proteins were insensitive to caspase-specific inhibitors, including the pancaspase inhibitor Z-VAD-fmk. In contrast, the plant metacaspases expressed in E. coli were highly sensitive to serine protease inhibitors, including leupeptin and antipain (40). Similar substrate specificity and sensitivity to protease inhibitors were also shown for the yeast metacaspase YCA1 and the plant metacaspase mcII-Pa expressed in E. coli (4, 41). Therefore, these reports suggest that metacaspases are probably not responsible for the caspase-like activities identified in plants and yeast.
Several species of the protozoan parasite Leishmania are responsible for human diseases (leishmaniasis), varying from a mild cutaneous form to fatal visceral leishmaniasis (17). These parasites have a two-stage life cycle and reside as flagellated extracellular promastigotes in the guts of the insect vectors and as intracellular amastigote forms in the phagolysosomal compartments of mammalian macrophages. Many features of metazoan apoptosis have been observed in both developmental stages of Leishmania undergoing PCD (reviewed in references 2, 10, and 30). It was also shown that caspase-like activities were associated with parasite PCD (6, 24, 34-36). We have reported previously that a DEVDase activity was activated in live L. donovani parasites either in late stationary cells or in parasites treated with the antileishmanial drug amphotericin B (24). However, the gene encoding the DEVDase activity in Leishmania donovani or genes encoding any of the caspase-like activities reported thus far in Leishmania have yet to be identified. Of significance, no caspase gene can be found in two completed Leishmania genomes available so far (L. major and L. infantum; www.genedb.org).
In this report, we have characterized two metacaspases of Leishmania donovani. We assessed their expression in the two developmental stages of the parasite and identified their intracellular localization. Further, we have demonstrated for the first time in any protozoan parasite that the endogenous L. donovani metacaspases possess enzymatic activities and report some of their properties with regard to substrate specificity and inhibition profile. On the basis of their enzymatic properties, we concluded that metacaspases are not responsible for the caspase-like activities previously reported in Leishmania. In addition, the increased metacaspase activity measured in cells undergoing PCD and the increased sensitivity to hydrogen peroxide-induced PCD in parasites overexpressing metacaspases suggest that metacaspases are involved in Leishmania PCD.
MATERIALS AND METHODS
Abbreviations.
PCD, programmed cell death; YCA1, yeast metacaspase-1; Z, benzyloxycarbonyl; fmk, fluoromethyl ketone; Z-VAD-fmk, Z-Val-Ala-Asp-fmk; DEVD, Asp-Glu-Val-Asp; AMC, 7-amido-4-methylcoumarin; Boc-GRR-AMC, t-butyloxycarbonyl-Gly-Arg-Arg-AMC; Boc-GKR-AMC, t-butyloxycarbonyl-Gly-Lys-Arg-AMC; H-AFK-AMC, H-Ala-Phe-Lys-AMC; AFC, 7-amido-4-(trifluoromethyl)coumarin; Ac-DEVD-AMC, acetyl-Asp-Glu-Val-Asp-AMC; Ac-VEID-AMC, acetyl-Val-Glu-Ile-Asp-AMC; Ac-LEHD-AFC, acetyl-Leu-Glu-His-Asp-AFC; Z-DEVD-fmk, Z-Asp-Glu-Val-Asp(O-methyl)-fmk; Z-FA-fmk, Z-Phe-Ala-fmk; RT-PCR, reverse transcriptase PCR; LdMC1, Leishmania donovani metacaspase-1; HA, hemagglutinin; SDS, sodium dodecyl sulfate; IFA, indirect immunofluorescence assay; SDS-PAGE, SDS-polyacrylamide gel electrophoresis; TcPPase, Trypanosoma cruzi pyrophosphatase; PBS, phosphate-buffered saline; LdMC1-mut, mutant LdMC1; NRS, normal rabbit serum; AB, assay buffer; TUNEL, terminal deoxynucleotidyltransferase-mediated dUTP-biotin nick end labeling; LmjMC, Leishmania major metacaspase protein; 5′UTR, 5′ untranslated region; LdMC1mut-HA, HA-tagged mutant LdMC1; LdMC1-HA, HA-tagged LdMC1; E64, l-trans-epoxysuccinyl-leucylamide-(4-guanido)-butane; TLCK, Nα-tosyl-l-lysine-chloromethyl ketone; TPCK, Nα-tosyl-l-phenylalanine-chloromethyl ketone; TbMC4, Trypanosoma brucei metacaspase-4.
Protease substrates and inhibitors.
The fluorogenic substrates Boc-GRR-AMC, Boc-GKR-AMC, and H-AFK-AMC were obtained from Bachem (Bubendorf, Switzerland), and Ac-DEVD-AMC, Ac-VEID-AMC, and Ac-LEHD-AFC were from Calbiochem (La Jolla, CA). The caspase inhibitor Z-VAD-fmk and Z-FA-fmk were obtained from Calbiochem, and Z-DEVD-fmk was from Alexis Biochemicals (San Diego, CA). All the other protease inhibitors were obtained from Sigma (St. Louis, MO).
Parasite culture and transfection.
L. donovani promastigotes (MHOM/S.D./62/1S-CL2D) were grown as described previously (8). Axenic amastigotes of L. donovani were derived from promastigotes and maintained in culture as described previously (9). Promastigotes were transfected with plasmid constructs by electroporation and selected for growth in medium containing Geneticin (G418) up to 200 μg/ml as described previously (7).
Molecular biology methodologies and expression plasmid constructs.
Genomic DNA and mRNA were purified from Leishmania cell pellets using the Genome DNA kit (Bio 101, Carlsberg, CA) and μMACS mRNA isolation kit (Miltenyi Biotec), respectively, and following the protocols of the manufacturer. RT-PCR using the Superscript first strand synthesis system (Invitrogen) and cloning of PCR-amplified products into PCRII cloning vector (Invitrogen) were done according to the supplied protocols. LdMC1 and LdMC2 genes were amplified by PCR using the primer pair M1F4 and M1R4 and primer pair M2F3 and M2R3 (Table 1), respectively. The two reverse primers contained a HA epitope tag sequence preceding the stop codon. The resulting PCR products were cloned into the SpeI site of the pKS NEO Leishmania expression plasmid (43) to generate the pKS NEO LdMC1 and pKS NEO LdMC2 expression plasmids. The point mutations (H147A) and (C202G) were introduced in LdMC1 by PCR using standard techniques resulting in the pKS NEO LdMC1-mut plasmid construct.
TABLE 1.
List of oligonucleotide primers
| Oligonucleotide primer | Nucleotide sequencea |
|---|---|
| M1F1 | TGGGAATTCATGGCAGACCTTTTTGAT |
| M1R1 | CCAGAATTCTTAGCCAGGCGGGAGTGG |
| M1F2 | GCATCTCCTCATTCTCTGTAG |
| M2R1 | GAGAAGGTCAGAACGCTGGAAC |
| SLp | ACTAACGCTATATAAGTATCAGTTTC |
| M1Rv | GCAGCGAACCCAGCATCGAGC |
| M1F3 | CAAAACGAGGCACAATACAT |
| M1R3 | TATCCCGGCGCTGGGTA |
| M2F2 | CCCACACCTCCCTGTCT |
| M2R2 | CCAACCTGGACTCTGTGGT |
| TubF | GGAGCTGTTCTGCCTTGAGC |
| TubR | GGTACATGAGGCAGCACGAC |
| M1F4 | TGGACTAGTATGGCAGACCTTCTTGATATTTTG |
| M1R4 | CCAACTAGTCTACGCGTAGTCCGGCACGTCGTAGGGGTAGCCAGGCGGGAGTGGGCTAAACGT |
| M2F3 | TGGACTAGTATGGCAGACCTTTTTGATATTTGG |
| M2R3 | CCAACTAGTCTACGCGTAGTCCGGCACGTCGTAGGGGTAGCCAGGCGGGAGTGGGCTGAACG |
| M1F5 | ATGGCAGACCTTTTTGATATT |
| M1R5 | GTCAAAGACGCACGTCATGCG |
Nucleotide sequences are given in the 5′-to-3′ orientation.
Expression of LdMC1 in E. coli and antibody production.
A portion of the LdMC1 gene corresponding to the first 200 amino acid residues was amplified by PCR using the plasmid containing LdMC1 as the template and the primer pair M1F5 and M1R5 (Table 1) and subsequently cloned into the E. coli expression plasmid pCRT7CT-Topo that contains a built-in His6 sequence (Invitrogen). E. coli BL21 host cells (Invitrogen) were transformed with this plasmid, and the expressed, His-tagged LdMC1 protein was affinity purified under denaturing conditions using Ni-nitrilotriacetic acid-agarose according to the manufacturer's protocol (QIAGEN). The ∼25-kDa His-tagged LdMC1 protein was further purified by electroelution from preparative SDS-polyacrylamide gels and used to immunize a New Zealand White rabbit according to company protocol (Spring Valley Laboratories). The resulting antiserum (anti-LdMC) was analyzed by Western blotting, IFA, and immunoprecipitation as described below.
Metabolic labeling and immunoprecipitation.
Log-phase promastigotes and axenic amastigotes were metabolically labeled using [35S]methionine (100 μCi/ml; NEN Life Science Product) for 10 min and chased in complete culture medium for 60 min as previously described (11). Subsequent cell lysis, immunoprecipitation with the anti-LdMC or preimmune serum, and analysis of immunoprecipitated proteins by SDS-PAGE and fluorography were done as described previously (11).
SDS-PAGE, Western blotting, and immunofluorescence.
L. donovani promastigotes and axenic amastigotes were processed for SDS-PAGE, Western blotting, and IFA as previously described (11). For IFA, fixed and permeabilized cells were incubated with a mixture of anti-LdMC and monoclonal anti-TcPPase (kindly provided by Roberto Docampo, University of Georgia, Athens) antibodies (each diluted 1:50 in PBS containing 1% bovine serum albumin) and subsequently incubated with a mixture of rhodamine-conjugated goat anti-rabbit and fluorescein-conjugated rabbit anti-mouse antibodies (1:200 in PBS containing 1% bovine serum albumin) (Vector Laboratories). Slides were examined with an ECLIPSE TE2000-U microscope (Nikon) equipped with a digital camera (Hamamatsu C4742-95; Hamamatsu Photonics K.K.) and processed with Open Lab software (Improvision, Inc.). The images were further processed using Adobe Photoshop 5.5 (Adobe Systems).
Enzymatic assays.
Wild-type and transfected L. donovani parasites expressing HA-tagged LdMC1, LdMC2, or LdMC1-mut were lysed (108 cells/ml) in lysis buffer (50 mM Tris-HCl, pH 7.5, 1% [vol/vol] Triton X-100, 150 mM NaCl) for 30 min on ice, and the insoluble material was eliminated by centrifugation at 15,000 × g for 20 min at 4°C. For the immunoprecipitations, 5 μl of anti-LdMC antibody or its corresponding preimmune serum (NRS) or 2 μl of anti-HA antibody (Sigma) or control goat anti-rabbit antibody (Vector) or no antibody (blank) was added to 500 μl of the cleared lysates as appropriate. After 3 h of incubation at 4°C on a rotating platform, 25 μl of PBS-washed protein A-Sepharose CL-4B was added to each sample, and these samples were further incubated for 1 h at 4°C. The samples were then centrifuged for 3 min at 2,000 × g at 4°C to remove unbound proteins, and the beads were washed three times with 0.5 ml of lysis buffer and once in 0.5 ml of a buffer without detergent (50 mM Tris-HCl, pH 7.5, 150 mM NaCl). Subsequently, the beads were resuspended in 150 μl of AB (50 mM HEPES, pH 7.5, 100 mM NaCl, 10% [wt/vol] sucrose, 0.1% (vol/vol) Triton X-100) supplemented with 10 mM dithiothreitol when caspase substrates were used, and containing 50 to 75 μM of protease substrate. Following 2 h of incubation at 37°C under gentle agitation, the samples were centrifuged at 2,000 × g for 3 min and the supernatants were transferred to a 96-well microwell plate. Fluorescence of the cleaved substrates was read using a Spectramax GeminiXS fluorometer (Molecular Devices Corporation, Sunnyvale, CA) set at appropriate excitation and emission wavelength. Protease activity was expressed in relative fluorescence units and using the no-antibody sample as blank. In inhibition studies, washed bead samples were preincubated at room temperature for 15 min in AB containing various protease inhibitors at the concentrations indicated in Results. The Boc-GKR-AMC substrate was subsequently added to a final concentration of 50 μM, and the samples were further processed for enzyme activity as described above. To measure LdMC activity in Leishmania induced for PCD with hydrogen peroxide (H2O2), axenic amastigote cultures were pelleted by centrifugation as described above, resuspended in fresh culture medium containing 2 mM H2O2 (Sigma), and incubated for 60 min at 26°C. Parasites were subsequently assayed for LdMC activity as described above. To determine the pH optimum of LdMC activity using Boc-GKR-AMC as the substrate, the AB described above was adjusted to either pH 7, 7.5, or 8. To adjust to lower pHs, HEPES of the standard AB was replaced by 100 mM morpholineethanesulfonic acid (Calbiochem) and the pH was adjusted to 6 or HEPES was replaced with 100 mM glycine (Sigma) and the pH was adjusted to either 5 or 4. To adjust to pH 9, HEPES was replaced by 100 mM Tris (Sigma). HCl (5 N) or NaOH (5 N) was used to adjust the pH values of the various assay buffers.
TUNEL labeling.
Log-phase L. donovani axenic amastigotes were pelleted by centrifugation, resuspended in culture medium containing 2 mM H2O2, and incubated at 37°C for 60 min along with the untreated controls. The cells were washed twice in cold PBS and fixed in 2% p-formaldehyde for 45 min followed by permeabilization with 0.1% (vol/vol) Triton X-100 in 0.1% (wt/vol) sodium citrate for 2 min on ice. The permeabilized cells were stained with the TUNEL reaction mixture (in situ cell death detection kit fluorescein; Roche) for 60 min at 37°C and analyzed using a FACSCalibur flow cytometer (Becton Dickinson). FL1-H fluorescence was recorded on 10,000 events and analyzed using CellQuest software. Data are expressed as a percentage of cells that were TUNEL positive.
Nucleotide sequence accession numbers.
The nucleotide sequences for the Leishmania donovani metacaspase genes LdMC1 and LdMC2 have been deposited in the GenBank database under GenBank accession numbers DQ367530 and DQ367531, respectively.
RESULTS
Identification and cloning of two L. donovani metacaspases.
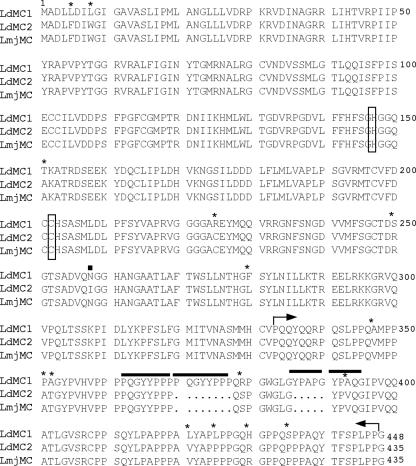
We have previously reported the presence of a caspase-like activity in L. donovani (24). The gene(s) encoding the protein(s) responsible for this activity remains to be identified. Caspase genes are not present in the Leishmania genome databases available (www.genedb.org). However, analysis of the L. major complete genome (20) (www.genedb.org) revealed the presence of a single caspase-related gene (LmjF35.1580) belonging to the metacaspase family of proteins initially described by Uren et al. (39). The LmjMC possesses the typical structural features of metacaspases, including the conservation of the histidine and cysteine amino acid residues involved in the active sites of the caspase family (39). Based on these structural characteristics and the high gene sequence homology observed between species of Leishmania, oligonucleotide primers were designed based on the L. major metacaspase to amplify by PCR its homologue in L. donovani. The forward primer started at the ATG, and the reverse primer started at the stop codon of the L. major sequence (Table 1, M1F1 and M1R1). The cloned L. donovani gene (LdMC1) showed 98% sequence homology with the LmjMC. The deduced LdMC1 protein showed 97% identity with LmjMC (Fig. 1). Both proteins possess a carboxy-terminal proline-rich domain of ∼115 amino acid residues. Within this domain, LdMC1 shows two regions of repetitive sequences (PQGYYPPP and YPAP/QG) that are not duplicated in LmjMC (Fig. 1).
FIG. 1.
Amino acid sequences of L. donovani LdMC1 and LdMC2. (A) Clustal W sequence alignment of LdMC1 (GenBank accession no. DQ367530), LdMC2 (GenBank accession no. DQ367531), and Leishmania major metacaspase (LmjMC) (GenBank accession no. AAZ14381) amino acid sequences. Asterisks indicate the difference in amino acid residues between LdMC1 and LdMC2. The solid square indicates the single amino acid residue difference between LdMC2 and LmjMC. The putative catalytic His147 and Cys202 residues are boxed. Solid bars above the sequences indicate the regions of repetitive sequences (PQGYYPPP and YPAP/QG) present in LdMC1, and arrows indicate the proline-rich domains. The gaps introduced to maximize alignment are indicated by dots.
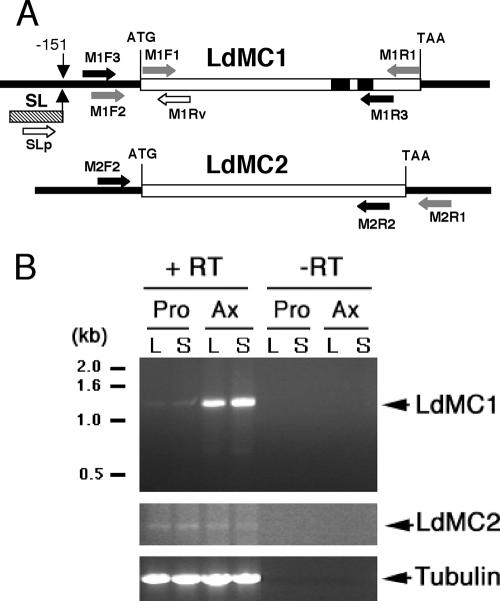
In order to demonstrate that the LdMC1 gene was transcribed in Leishmania, RT-PCR was performed using mRNA isolated from stationary-phase promastigotes and a forward primer based on the Leishmania conserved spliced leader sequence, which is present at the 5′ end of all mature mRNAs in trypanosomatids (42), and a reverse primer 253 nucleotides downstream of the LdMC1 start codon (Fig. 2A, primers SLp and M1Rv). The resulting PCR fragment was cloned and sequenced. This sequence contains the complete 5′ spliced leader sequence followed by 151 nucleotides of 5′UTR upstream of the LdMC1 start codon (not shown). The nucleotide sequence between the ATG and the downstream reverse primer in the RT-PCR fragment was identical to that of the original LdMC1 sequence. This result demonstrated that the LdMC1 gene was transcribed and spliced into a mature mRNA, since it contained the conserved 5′ spliced leader sequence. In order to confirm the nucleotide sequence of LdMC1 by an independent PCR and to analyze its 3′-end region, a forward oligonucleotide primer based on the LdMC1 5′UTR sequence obtained above (Fig. 2A, M1F2) and a reverse oligonucleotide primer (110 nucleotides after the stop codon) based on the downstream sequence of LmjMC available in the L. major genome database (www.genedb.org) were made (Fig. 2A, M2R1). The PCR performed using these two primers and L. donovani genomic DNA resulted in the amplification of a second putative L. donovani metacaspase gene, LdMC2. The sequence alignment of LdMC1, LdMC2, and the protein deduced from the reference LmjMC gene is shown in Fig. 1. LdMC2 shows 96% identity with LdMC1, possesses a single amino acid residue difference with LmjMC, and lacks the two repetitive sequences found in the C terminus of LdMC1. These results show that unlike L. major which contains a single metacaspase gene, L. donovani possesses two closely related metacaspase genes that probably also share high sequence identity in their 5′UTRs, since the M1F2 primer, based on the LdMC1 5′-flanking sequence, hybridized to the LdMC2 5′UTR.
FIG. 2.
RT-PCR of LdMC1 and LdMC2. (A) Map of the LdMC1 and LdMC2 genes and flanking sequences. The start (ATG) and stop (TAA) codons of each open reading frame (open boxes) are indicated. The two regions of repetitive sequences (PQGYYPPP and YPAP/QG) present in LdMC1 are indicated by solid boxes. Vertical arrows indicate the location of the spliced leader acceptor site at position −151. Splice leader (SL) sequence is illustrated as a hatched box. Horizontal arrows (black, gray, and white) indicate the locations of the PCR primers (Table 1). (B) Ethidium bromide-stained agarose gel of the RT-PCR products obtained with LdMC1 (top gel) LdMC2 (middle gel), and tubulin (bottom gel) gene-specific primer sets (Table 1). The reverse transcription reaction was done using mRNA isolated from either log-phase (L) or stationary-phase (S) promastigotes (Pro) or axenic amastigotes (Ax) and with reverse transcriptase (+ RT) or without reverse transcriptase (−RT). The molecular sizes (in kilobases) of protein standards are shown to the left of the gel.
Transcription of LdMC1 and LdMC2 in L. donovani.
In order to determine whether LdMC2 was also transcribed in promastigotes as was observed for LdMC1 and to assess whether the levels of LdMC1 and LdMC2 mRNAs changed in promastigotes during their growth in vitro, RT-PCR was performed using RNA isolated from parasites in the log and stationary phases of growth of the culture. In addition, since L. donovani can be adapted to grow in vitro as axenic amastigotes, a stage that closely resemble the amastigote stage of the parasite (9), RNA was also isolated from axenic amastigotes harvested in the log and stationary phases of culture. The RT-PCR was performed using total RNA from both promastigotes and axenic amastigotes as described above and with primers specific to either LdMC1 or LdMC2. The forward primers M1F3 and M2F2 were located in the 5′UTRs of LdMC1 and LdMC2 and the reverse primers M1R3 and M2R2 reflected sequences located in the C-terminal repeat regions (Fig. 2A). The specificity of the primers was verified in preliminary experiments by digestion of the PCR products obtained with each primer set with specific restriction enzymes (data not shown). RT-PCR results showed that LdMC1 and LdMC2 mRNAs were present in both stages of the parasites and in both log and stationary phases of their growth in vitro (Fig. 2B, top and middle gels). No PCR products were detected in the control reactions without RT in these experiments (Fig. 2B). However, L. donovani axenic amastigotes had significantly higher levels of LdMC1 mRNA than promastigotes did. In contrast, LdMC2 mRNA levels were similar in both parasite developmental stages (Fig. 2B, middle gel), and no differences in LdMC1 or LdMC2 mRNA levels were observed between log-phase and stationary-phase promastigotes or axenic amastigotes. For a control, a 896-bp fragment from L. major alpha-tubulin gene was amplified using TubF and TubR primers (Table 1) under identical PCR conditions. Tubulin mRNA levels were similar in promastigotes and axenic amastigotes in these experiments (Fig. 2B, bottom gel). Taken together, these results show that LdMC1 and LdMC2 genes are transcribed in both stages of the parasites and that LdMC1 has increased mRNA levels in axenic amastigotes compared to promastigotes.
Expression of LdMCs in promastigotes and axenic amastigotes.
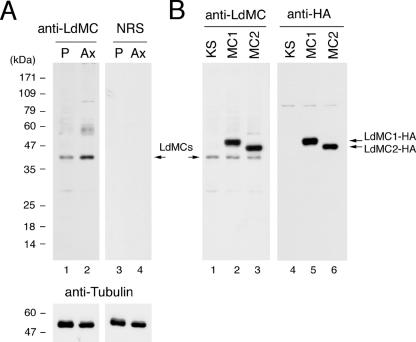
In order to assess whether LdMC proteins were expressed in the parasite and determine their localization in the cell, an anti-LdMC antibody was generated. Initial experiments showed that the expression of the full-length histidine-tagged LdMC1 protein in E. coli resulted in poor bacterial growth and limited LdMC1 expression (not shown). In contrast, a truncated LdMC1 protein, including the first 200 amino acid residues, which are highly conserved between LdMC1 and LdMC2 (98.5% identity), was successfully expressed in E. coli and used to generate an anti-LdMC antiserum in rabbits. The reactivity of the anti-LdMC antibody in Western blots with parasite cell lysates is shown in Fig. 3. The anti-LdMC antibody reacted with a ∼42-kDa protein in lysates of promastigotes and axenic amastigotes (Fig. 3A, lanes 1 and 2, respectively). The preimmune rabbit serum had no reactivity with either cell lysates, showing the specificity of the anti-LdMC antibody for LdMCs (Fig. 3A, lanes 3 and 4). The calculated masses of LdMC1 and LdMC2 are 53,760 Da and 52, 200 Da, respectively; therefore, the ∼42-kDa band in these Western blots could represent antibody reactivity with LdMC1, LdMC2, or both (LdMCs). The difference in masses between these two proteins (1,560 Da) is probably too small for these proteins to be resolved under the SDS-PAGE conditions used in these experiments. The difference between the calculated masses and their apparent molecular masses on SDS-polyacrylamide gels could correspond to their aberrant migration on SDS-polyacrylamide gels due to the presence of a proline-rich domain in these two molecules. The presence of an additional carboxy-terminal HA tag could affect the electrophoretic mobilities of LdMC1 and LdMC2 differently, resulting in a larger than expected difference in their apparent molecular mass by SDS-PAGE. These Western blot results also show that the intensity of the LdMC band in axenic amastigote lysates (Fig. 3A, lane 2) is approximately twofold more intense than that obtained with promastigote lysates (Fig. 3A, lane 1), suggesting that the steady-state level of LdMCs in log-phase axenic amastigotes is approximately twice that of log-phase promastigotes. No difference was observed in the steady-state levels of tubulin proteins between these two parasite stages (Fig. 3A, bottom gel).
FIG. 3.
Expression of LdMC1 and LdMC2 in promastigotes and axenic amastigotes. (A) Western blot showing the reactivity of the anti-LdMC antibody (anti-LdMC) or preimmune serum (NRS) with lysates of log-phase promastigotes (P) and axenic amastigotes (Ax). The Western blot membrane was reprobed with an antitubulin antibody (bottom gels). The positions of the LdMC proteins is indicated by an arrow. (B) Western blot showing the reactivity of the anti-LdMC or anti-HA antibodies with lysates of log-phase promastigotes transfected with either the control pKS NEO (KS) or pKS NEO LdMC1 (MC1) or pKS NEO LdMC2 (MC2) expression plasmid. The positions of HA-tagged proteins LdMC1-HA and LdMC2-HA and the endogenous LdMC proteins (LdMCs) are indicated by arrows. The molecular masses (in kilodaltons) of protein standards are shown to the left of the gels.
To confirm that the anti-LdMC antibody is able to react with LdMC1 and LdMC2, these two proteins were expressed as HA-tagged proteins in Leishmania. Lysates of these transfected cells were reacted with either the anti-LdMC antibody or an anti-HA antibody after SDS-PAGE and Western blotting. Results showed that the anti-LdMC antibody reacted with the endogenous ∼42-kDa LdMC proteins in lysates of either the control (parasites transfected with the pKS NEO plasmid alone) or in LdMC1- or LdMC2-transfected cells (Fig. 3B, lanes 1 to 3, respectively). This antibody also reacted with a ∼49-kDa protein in lysates of LdMC1-transfected cells (Fig. 3B, lane 2) and with a ∼47-kDa protein in lysates of LdMC2 transfectants (Fig. 3B, lane 3). The ∼49-kDa and ∼47-kDa proteins also reacted with an anti-HA antibody (Fig. 3B, lanes 5 and 6, respectively), showing that these proteins correspond to the HA-tagged LdMC1 and LdMC2 episomally expressed proteins. The anti-LdMC antibody reacted with a minor protein band of ∼32 kDa in these Western blots, probably representing a degradation fragment of the LdMC proteins. Taken together, these Western blot results show that the anti-LdMC antibody can react with both LdMC1 and LdMC2 proteins and that LdMC1, LdMC2, or both are expressed in both stages of the parasites.
Processing of LdMCs in L. donovani.
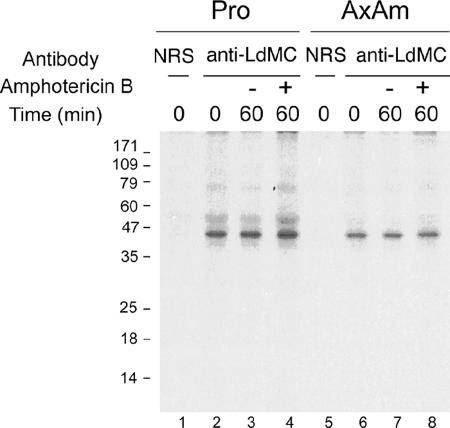
Metacaspases constitute a new family of caspase-related proteins (39). Among this family, virtually all mammalian caspases and more recently yeast and some plant metacaspases have been shown to be proteolytically processed in cells undergoing PCD (14, 26, 40, 41). To assess whether LdMCs undergo proteolytic processing, L. donovani promastigotes and axenic amastigotes were pulse-labeled with [35S]methionine for 10 min and chased for 60 min in normal culture medium supplemented either with or without amphotericin B, a drug that triggers PCD in Leishmania (24). Results showed that after 10 min of pulse-labeling, the radiolabeled ∼45-kDa LdMC proteins were immunoprecipitated from the cell lysates of both promastigotes and axenic amastigotes (Fig. 4, lanes 2 and 6). No radiolabeled proteins were immunoprecipitated from these cell lysates with the preimmune serum, showing the specificity of the anti-LdMC antibody for LdMCs (Fig. 4, lanes 1 and 5). The LdMCs were also immunoprecipitated from lysates of parasites collected after 60 min of chase without amphotericin B (Fig. 4, lanes 3 and 7). The intensity of the ∼45-kDa LdMC protein band did not decrease significantly after 60 min of chase, indicating a slow turnover of these proteins, and no labeled proteins with molecular masses of <45 kDa were immunoprecipitated at this time point, suggesting that the LdMCs do not undergo proteolytic processing in normal culture conditions. Similar results were obtained where the chase was done in the presence of 1 μg/ml of amphotericin B (Fig. 4, lanes 4 and 8), conditions that trigger cell death in >50% of the cells in L. donovani (24). Similar lack of LdMC processing has been observed when PCD was induced using hydrogen peroxide, another inducer of PCD in Leishmania (6) (not shown). Taken together, these results show that newly synthesized LdMC proteins by promastigotes or axenic amastigotes do not undergo proteolytic degradation in the presence of PCD inducers, suggesting that Leishmania metacaspases are probably not synthesized as precursor proteins.
FIG. 4.
Processing of the LdMCs in L. donovani. Autoradiogram showing the radiolabeled LdMCs immunoprecipitated with the anti-LdMC antibody (anti-LdMC) or preimmune serum (NRS) from lysates of radiolabeled promastigotes (Pro) and axenic amastigotes (AxAm). Parasites were pulse-labeled with [35S]methionine for 10 min and chased for 0 and 60 min in culture medium supplemented with amphotericin B (1 μg/ml) (+) or not supplemented with amphotericin B (−). The molecular masses (in kilodaltons) of protein standards are shown to the left of the gel.
Localization of LdMCs in Leishmania promastigotes and axenic amastigotes.
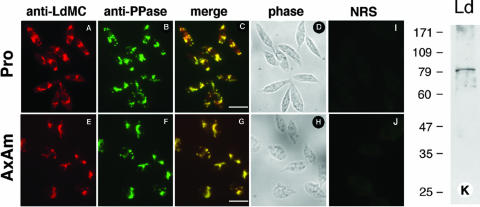
Most mammalian caspases exist as inactive precursors in the cell cytoplasm (14). In order to determine the localization of LdMCs in Leishmania, L. donovani promastigotes and axenic amastigotes were subjected to IFAs using the anti-LdMC antibody. Initial results showed that the immunofluorescence was localized in multiple intracellular vesicles detected throughout the cell body with the exclusion of the nucleus and flagellum. Such vesicular staining obtained in the anti-LdMC antibody was reminiscent of acidocalcisome compartments observed in Trypanosoma cruzi and Leishmania (12, 13). Therefore, L. donovani promastigotes and axenic amastigotes were reacted simultaneously with the anti-LdMC antibody and a monoclonal antibody specific to TcPPase, a protein associated with acidocalcisomes of T. cruzi and Leishmania (32, 33). Cells were subsequently incubated with a mixture of rhodamine-labeled anti-rabbit and fluorescein-labeled anti-mouse antibodies and observed with a UV microscope. The images of the red (anti-LdMC) and green (anti-TcPPase) channels are shown in Fig. 5A and E and Fig. 5B and F, respectively. The images of the green and red channels were merged into a single image shown in Fig. 5C and G, where yellow represents colocalization of red and green fluorescence signals. The phase-contrast images of the same fields are shown in Fig. 5D and H. No fluorescence was detected in similar preparations reacted with the preimmune serum (Fig. 5I and J). The reactivity of the anti-TcPPase antibody with L. donovani was also assessed by Western blotting. This monoclonal antibody reacted with a single ∼82-kDa protein in a promastigote cell lysate (Fig. 5K). The apparent molecular mass of the ∼82-kDa reactive protein on SDS-polyacrylamide gels is in good agreement with the calculated 83,200 Da of the TcPPase putative ortholog in L. infantum (LinJ31_V3:1240). Further, the two proteins show only one amino acid difference within the 27 residues that were used to generate the anti-TcPPase monoclonal antibody (25). An antibody isotype control showed no reactivity with L. donovani proteins in these Western blots (not shown). Taken together, these results show that LdMCs are associated with the acidocalcisome compartments in L. donovani.
FIG. 5.
Immunolocalization of LdMCs in L. donovani. L. donovani promastigotes (Pro) and axenic amastigotes (AxAm) were processed for immunofluorescence and reacted with both anti-LdMC and anti-TcPPase antibodies. The reactivity with the anti-LdMC is shown in the red channel (panels A and E), and that of the anti-TcPPase antibody is shown in the green channel (panels B and F). Merging of the red and green channels (yellow) is shown in panels C and G. The corresponding phase-contrast images are shown in panels D and H. Bars in panels C and G, 10 μm. Panel K shows the reactivity of the anti-TcPPase antibody with lysates of L. donovani promastigotes (Ld) in Western blots. The molecular masses (in kilodaltons) of protein standards are shown to the left of the blot.
Enzymatic activity of L. donovani metacaspases.
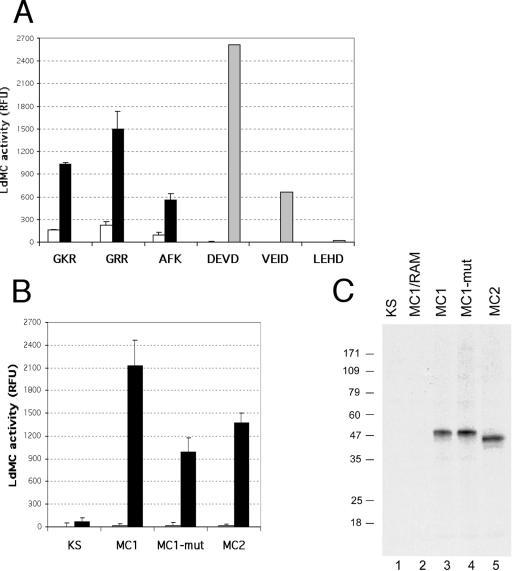
As mentioned above, the full-length LdMC1 was very poorly expressed in E. coli, and therefore, the amount of protein obtained in these conditions was insufficient to perform any enzymatic assay. However, results described above also showed that the anti-LdMC antibody could specifically immunoprecipitate LdMC proteins from Leishmania cell lysates (cf. Fig. 4). Therefore, enzymatic assays were performed by incubating anti-LdMC-immunoprecipitated material with various synthetic fluorogenic substrates. The enzyme activity was determined by measuring the amount of fluorescence generated in these assays. The detailed assay is described in Materials and Methods. Results showed that Boc-GKR-AMC, Boc-GRR-AMC, and H-AFK-AMC were cleaved by LdMC proteins immunoprecipitated with the anti-LdMC antibody from axenic amastigote lysates and that only background activity was measured in these assays when immunoprecipitation was done with the preimmune serum from the same lysates (Fig. 6A). Such background activity probably corresponds to nonspecific adsorption of LdMC proteins to antibody-bound protein A-Sepharose beads, since similar levels of ]background fluorescence were obtained with other control rabbit sera (not shown). These results demonstrate that the immunoprecipitated LdMC proteins efficiently cleave substrates containing a basic amino acid residue, arginine or lysine, at the P1 position. To determine whether LdMCs were able to cleave substrates containing an aspartic acid residue in the P1 position, three caspase-specific substrates, Ac-DEVD-AMC, Ac-VEID-AMC, and Ac-LEHD-AFC, were tested in these assays. None of these caspase substrates were cleaved by LdMCs in these assays (Fig. 6A). In order to verify that the assay conditions were correct to detect caspase activities, anti-LdMC-immunoprecipitated samples assayed with the three caspase substrates were spiked with ∼1 unit of recombinant human caspase-3 (Calbiochem). Results showed that the three caspase substrates were cleaved by recombinant caspase-3 under our experimental conditions (Fig. 6A). As expected, Ac-DEVD-AMC, which is a preferred substrate for caspase-3, was cleaved more efficiently in these assays than Ac-VEID-AMC and Ac-LEHD-AFC, preferred substrates for capsase-6 and caspase-4, respectively. These results demonstrate that L. donovani metacaspases efficiently cleave trypsin-specific substrates and are unable to cleave caspase-specific substrates.
FIG. 6.
Substrate specificity of LdMC enzyme activity. (A) LdMCs were immunoprecipitated from axenic amastigote lysates using the anti-LdMC antibody and incubated with either trypsin substrates (GKR, GRR, and AFK) or caspase substrates (DEVD, VEID, and LEHD) under appropriate reaction conditions (see Materials and Methods for details). The LdMC enzymatic activity is expressed in relative fluorescence units (RFU) (black bars). Control enzymatic assays were done using the preimmune serum during the immunoprecipitation step (white bars). Human recombinant caspase-3 was spiked into parallel assays containing the caspase substrates (gray bars). (B) Metacaspase activities of HA-tagged proteins (MC1, MC1-mut, and MC2) immunoprecipitated using the anti-HA antibody from axenic amastigote lysates of transfected parasites, and using Boc-GRR-AMC as the substrate (black bars). Parasites transfected with the pKS NEO expression plasmid was used as control (KS). Control enzymatic assays were done using the rabbit anti-mouse antibodies during the immunoprecipitation step (white bars). Results in panels A and B represent the averages of three independent assays. Standard deviations (error bars) are shown. (C) Parasites used in panel B were radiolabeled with [35S]methionine, radiolabeled HA-tagged proteins were immunoprecipitated with an anti-HA antibody or control rabbit anti-mouse antibodies (RAM), and material was analyzed by SDS-PAGE and fluorography. The molecular masses (in kilodaltons) of protein standards are shown to the left of the gel.
In order to determine whether LdMC1, LdMC2, or both had “trypsin-like” activity, enzymatic assays were done from lysates of transfected parasites expressing either LdMC1 or LdMC2 as HA-tagged proteins and using an anti-HA antibody. Results showed that Boc-GRR-AMC was efficiently cleaved by both LdMC1-HA and LdMC2-HA proteins immunoprecipitated with the anti-HA antibody (Fig. 6B). Only background protease activity was detected in assays done from lysates of control parasites transfected with the pKS NEO plasmid (Fig. 6B, KS). Similar protease activity was detected in these assays when Boc-GKR-AMC and H-AFK-AMC were used as substrates, and no detectable activity was measured when Ac-DEVD-AMC, Ac-VEID-AMC, or Ac-LEHD-AFC was used as a substrate (not shown). To assess whether the His147 and Cys202 of LdMC1, shown to be conserved with the Cys-His catalytic dyad of the active domains of caspase (39), were required for its protease activity, parasites were transfected with an expression plasmid encoding LdMC1-HA in which the His and Cys residues were substituted by Ala and Gly, respectively. The expression levels of the LdMC1mut-HA and LdMC1-HA proteins by transfected parasites were similar when analyzed by Western blotting (not shown). Results of enzymatic assays showed that Boc-GRR-AMC was efficiently cleaved by the immunoprecipitated LdMC1mut-HA (Fig. 6B, MC1-mut), although the Boc-GRR-AMC cleavage activity of LdMC1mut-HA was approximately twofold less than that of LdMC1-HA in these experiments. No detectable activity was measured in these assays when immunoprecipitations were done with the control rabbit anti-mouse antibody (Fig. 6B). To verify the specificity of the anti-HA antibody and control that similar amounts of HA-tagged proteins were immunoprecipitated in these assays, radiolabeled lysates were also used in parallel immunoprecipitation experiments. The resulting fluorograms showed that similar amounts of HA-tagged protein were immunoprecipitated in these assays (Fig. 6C, lanes 3 to 5) and that the episomally expressed HA-tagged proteins were the only proteins immunoprecipitated from the radiolabeled Leishmania lysates.
Taken together, the enzyme assays above show that L. donovani metacaspases LdMC1 and LdMC2 cleave efficiently trypsin substrates (i.e., cleavage after Lys or Arg residues) and are unable to cleave caspase substrates (i.e., cleavage after an Asp residue) and that His147 and Cys202 of LdMC1 are not essential for its “trypsin-like” activity.
Effects of protease inhibitors on LdMC activity.
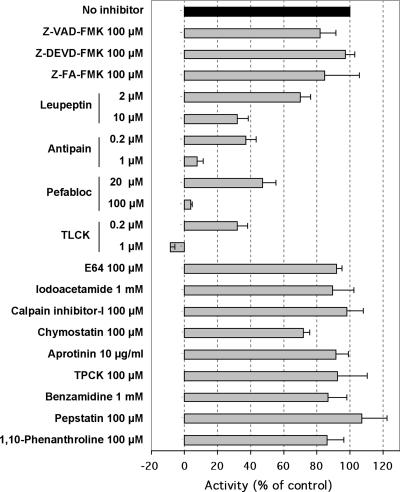
We tested the effects of various protease inhibitors on the LdMC activity (using Boc-GKR-AMC as the substrate) immunoprecipitated by the anti-LdMC antibody from axenic amastigote cell lysates (Fig. 7). Results showed that the caspase inhibitors Z-VAD-fmk and Z-DEVD-fmk did not block LdMC activity at concentrations up to 100 μM. The cathepsin B inhibitor Z-FA-fmk, often used as a negative-control inhibitor in caspase assays, similarly had no effect on LdMC activity. Inhibitors of metalloproteases, 10-phenanthroline (100 μM), aspartic proteases, and pepstatin (100 μM) and inhibitors of cysteine proteases E64 (100 μM), iodoacetamide (1 mM), and calpain inhibitor-I (100 μM) showed no inhibition of LdMC activity. In contrast, two inhibitors of trypsin and trypsin-like serine proteases, i.e., antipain and TLCK showed nearly complete (antipain) to complete (TLCK) inhibition of the LdMC activity at 1 μM concentration. Other inhibitors of serine protease had either moderate (leupeptin, Pefabloc, and chymostatin) or no inhibition (benzamidine, aprotinin, and TPCK) on LdMC activity in these assays. A similar inhibition profile was obtained for LdMC1-HA activity immunoprecipitated with the anti-HA antibody (not shown). Taken together, these results show that Leishmania metacaspases are insensitive to commonly used inhibitors of caspases and cysteine proteases and are highly sensitive to several serine protease inhibitors. Further, these results are in agreement with the substrate specificity results above and strongly suggest that the Leishmania metacaspases are not “caspase-like” proteases.
FIG. 7.
Sensitivity of LdMC enzyme activity to protease inhibitors. Effects of different protease inhibitors on LdMC activity from lysates of axenic amastigotes using Boc-GKR-AMC as the substrate. The results are shown as percentages of the LdMC activity measured without inhibitor (control) (black bar). Results represent the averages of three independent assays. Standard deviations (error bars) are shown.
LdMC activity in L. donovani promastigotes and axenic amastigotes.
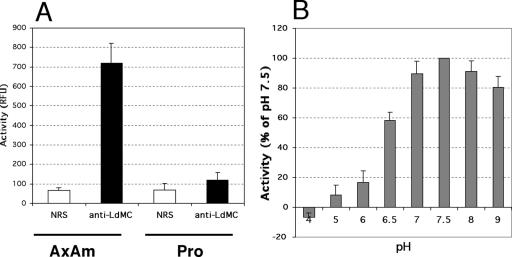
In order to compare the level of LdMC activity in the two parasite stages, enzyme assays were done with lysates of promastigotes and axenic amastigotes harvested in mid-log growth and using Boc-GKR-AMC as the substrate. Results showed that axenic amastigote cell lysates contained approximately sixfold-higher GKRase enzyme activity than lysates of promastigotes (Fig. 8A, anti-LdMC). Similar levels of background activity from both cell types were measured in these assays when immunoprecipitation was done with the preimmune serum (Fig. 8A, NRS). These LdMC activity results are consistent with higher levels of LdMC mRNAs and higher steady-state levels of LdMC proteins observed in axenic amastigotes compared to promastigotes. In these enzymatic assays, the optimal buffer pH to detect LdMC activity was 7.5 (Fig. 8B). This activity was stable over a broad range of pH values, since >50% of the activity measured at the optimum pH was still measured between pH 6.5 and 9. However, LdMCs had limited enzyme activity below pH 6 and no detectable activity at pH 4 (Fig. 8B).
FIG. 8.
Stage expression and pH optimum of LdMC enzyme activity. (A) LdMC enzyme activity in lysates of axenic amastigotes (AxAm) and promastigotes (Pro) of L. donovani. The LdMC enzyme activity is expressed in relative fluorescence units (RFU) (black bars). Control enzymatic assays were done using the preimmune serum (NRS) during the immunoprecipitation step (white bars). (B) LdMC enzyme activity in lysates of axenic amastigotes at different pHs. Relative activity is expressed as a percentage of the activity at optimal pH (7.5). The results in panels A and B were obtained using Boc-GKR-AMC as the substrate and represent the averages of three independent assays. Standard deviations (error bars) are shown.
Roles of LdMCs in H2O2 induced Leishmania PCD.
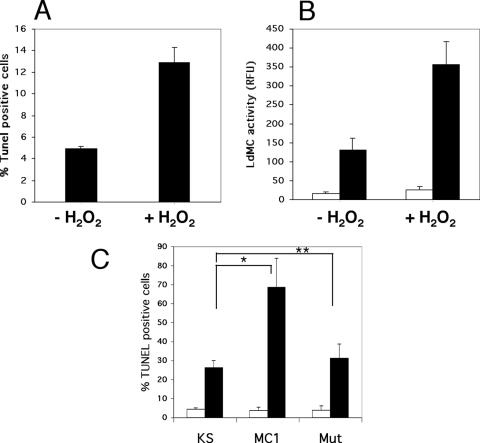
Leishmania treated with H2O2 show features of PCD, including DNA damage that can be measured by TUNEL labeling (6). In order to determine whether metacaspases were involved in H2O2-induced PCD in Leishmania, wild-type L. donovani axenic amastigotes were treated with H2O2, assayed for LdMC activity, and compared to untreated cells. The level of PCD induction in these cell cultures was assessed by TUNEL labeling of parasites and quantitated by flow cytometry. As observed in previous reports, the percent of TUNEL-positive cells increased by ∼2.6-fold in cells treated by H2O2 compared to untreated cells, rising from a background level of 4.9% in untreated cells to 13% in H2O2-treated cells (Fig. 9A). In addition, our results showed a similar ∼2.7-fold increase of LdMC activity in parasites treated with H2O2 compared to untreated controls (Fig. 9B). Similar background levels of LdMC activity were measured when the preimmune serum was used as control in these assays (Fig. 9B). The concomitant increases in metacaspase activity and in the percentage of TUNEL-positive cells in parasites treated with H2O2 suggest a possible involvement of LdMCs in H2O2-induced Leishmania PCD. To further support the potential role of LdMCs in such pathway, parasites overexpressing either LdMC1-HA, LdMC1-mut-HA, or control KS were also treated with H2O2. Results showed 69% TUNEL-positive cells in H2O2-treated Leishmania overexpressing LdMC1-HA compared to 26% in treated KS controls and 31% in treated LdMC1-mut-HA-expressing parasites (Fig. 9C). Similar background levels of TUNEL-positive cells (4 to 5%) were detected in the three untreated transfectants (Fig. 9C). Taken together, these results show that when PCD is induced in Leishmania using H2O2, these parasites show a concomitant increase of metacaspase activity, and Leishmania overexpressing metacaspases are more sensitive to H2O2-induced PCD. Therefore, these results suggest a possible role for LdMCs in Leishmania PCD.
FIG. 9.
Sensitivity of L. donovani wild type and LdMC transfectants to H2O2. (A) Percentages of TUNEL-positive cells in L. donovani axenic amastigotes either treated with 2 mM H2O2 (+ H2O2) for 60 min or untreated (− H2O2) and analyzed by flow cytometry. (B) LdMC enzyme activity in parasites treated as described above for panel A, using Boc-GRR-AMC as the substrate and expressed in relative fluorescence units (RFU) (black bars). Control enzymatic assays were done using the preimmune serum during the immunoprecipitation step (white bars). (C) Percentages of TUNEL-positive cells in L. donovani axenic amastigote transfectants (KS, MC1, and MC1-mut [Mut]) either treated with 2 mM H2O2 (black bars) for 60 min or untreated (white bars) and analyzed by flow cytometry. Significant differences (P < 0.05) (*) and nonsignificant differences (P > 0.05) (**) are indicated. Results in panels A, B, and C represent the averages of three independent assays. Standard deviations (error bars) are shown.
DISCUSSION
In this report, we cloned and characterized two metacaspase genes in L. donovani (LdMC1 and LdMC2). One characteristic feature of the Leishmania metacaspases is the presence of a carboxy-terminal proline-rich domain. Such a domain has been reported only in T. cruzi (T. cruzi metacaspase-5) (22) among the trypanosomatid family of parasites. Similar domains are also present in the yeast metacaspase YCA1 and in type I metacaspases of A. thaliana, but they are located in the amino termini of these proteins (26, 40). The role of this proline-rich domain is unknown; however, its presence in some metacaspases, including LdMCs, suggests that it may be involved in protein-protein interactions (21, 31). The major difference between the two L. donovani metacaspases is the presence of two amino acid repeats in the C terminus of LdMC1 that results in a slightly larger proline-rich domain in LdMC1 than in LdMC2. Two putative metacaspase genes can also be identified in the genome of L. infantum, another Leishmania species responsible for visceral leishmaniasis. In contrast, L. major, the species responsible for cutaneous leishmaniasis, for which the complete genome is known possesses only a single metacaspase gene (www.genedb.org). Among the trypanosomatids with completely sequenced genomes, Leishmania metacaspases show the most identity with Trypanosoma brucei metacaspase-5 (52%) and Trypanosoma cruzi metacaspase-5 (61%), organisms that possess 5 and 17 metacaspase genes, respectively. The significance of this range in metacaspase gene copy number within the trypanosomatid family is not known.
Our data showed that LdMC1 and LdMC2 genes were transcribed in promastigotes and axenic amastigotes of L. donovani; however, the level of LdMC1 mRNA was significantly higher in axenic amastigotes. RNA stability is likely the mechanism involved in the increased LdMC1 mRNA levels observed in axenic amastigotes, since gene expression is controlled exclusively at the posttranscriptional level in Leishmania (5), but this remains to be demonstrated for LdMC1. Such increased LdMC1 mRNA levels observed in axenic amastigotes could explain the significant higher levels of metacaspase activity measured in this form of the parasite. Since the anti-LdMC antibody reacts with both LdMC1 and LdMC2 and does not discriminate between LdMC1 and LdMC2 proteins, the respective contributions of LdMC1 and LdMC2 to the LdMC activity measured in our current assays are not known. Further, the increased levels of LdMC activity in axenic amastigotes also suggests that metacaspases may play a more important role in the mammalian stage of this parasite than in the insect stage. Further studies will be needed to define such a role(s).
To date, none of the native trypanosomatid metacaspases (i.e., purified from parasite lysates) have been characterized with regard to their enzymatic activity and substrate specificity. In this report, using a specific anti-LdMC antibody, we have been able to specifically immunoprecipitate the native L. donovani metacaspases from parasite cell lysates and assess their enzymatic properties. These native L. donovani metacaspases were unable to cleave any of the caspase-specific substrates tested and were insensitive to caspase-specific inhibitors, such as Z-VAD-fmk and Z-DEVD-fmk, up to 100 μM concentration. Similar substrates and inhibitors have been used in several reports to characterize caspase-like activities in Leishmania (6, 24, 34-36). In contrast, native L. donovani metacaspases efficiently cleaved substrates with either a lysine or arginine amino acid residue at their P1 position and were efficiently inhibited by leupeptin and antipain, two arginine inhibitors, and by the P1-lysine inhibitor TLCK, confirming the lysine/arginine specificity of LdMCs. Therefore, our results strongly suggest that Leishmania metacaspases are not responsible for the caspase-like activities reported in these organisms. The substrate specificity of LdMCs agrees with previous reports showing that plants (A. thaliana type I and II), yeast (YCA-1), and more recently L. major (LmjMCA) metacaspases when expressed in bacteria or yeast also have lysine/arginine substrate specificity and do not cleave caspase substrates (15, 40, 41). In contrast to these recombinant metacaspases which lose enzymatic activity when their predicted catalytic cysteine is mutated to an alanine residue, the LdMC1(H147A, C202G) mutant retains significant enzyme activity when purified from parasite lysates. This suggests that His147 and Cys202 of LdMC1 are not absolutely required for its trypsin-like activity and that parasite-specific posttranslational modifications of LdMC1(H147A, C202G) mutant may be involved in its activity, since the LmjMCA(C202A) mutant expressed in yeast or E. coli does not have enzyme activity (15). Further studies are needed to identify the catalytic amino acid residue(s) of the L. donovani metacaspases. This will help clarify the nature of these proteases.
Most mammalian caspases are synthesized as catalytically inactive zymogens that are processed into active proteases upon activation of the cell death pathway (14). Similarly, the yeast metacaspase YCA-1 has been shown to follow such a caspase-like processing in cells undergoing PCD (26). A. thaliana and more recently L. major metacaspases were also shown to be proteolytically processed when expressed in bacteria or yeast, and mutation of the conserved catalytic cysteine residue to alanine prevented their proteolytic processing and suggests autocatalytic processing (15, 40, 41). In contrast, our results show that L. donovani metacaspases do not appear to be processed by the parasites under normal growth conditions or upon treatment with inducers of PCD. This suggests that LdMCs could be synthesized in active forms and that the parasites must have a mechanism(s) to control/isolate metacaspase activity inside the cell. Since our results show that LdMCs are associated with acidocalcisomes, one such mechanism could be sequestration of LdMCs in these acidic compartments. Further, we also showed that LdMCs are poorly active in acidic pH in vitro, which strongly suggests that they would be poorly active in acidocalcisomes, which are acidic compartments in Leishmania (12). To our knowledge, LdMCs represent the first proteases to be associated with acidocalcisomes in any protozoan parasite. Whether LdMCs are released from acidocalcisomes during PCD and/or whether they play an active role in the physiology of these cellular compartments remains to be elucidated.
As in yeast and plants, the role of metacaspases in protozoan parasites is still poorly understood. Since none of the few metacaspases characterized thus far are able to cleave caspase substrates, they are probably not directly responsible for the caspase-like activities reported in these organisms. However, there is some evidence that the yeast metacaspase YCA1 could be involved in the cell death pathway of this organism by acting upstream of a caspase-like enzyme (41). Recently, a metacaspase of Norway spruce (mcII-Pa) was shown to accumulate in the nuclei of the embryo suspensor cells during embryogenesis, resulting in nuclear disassembly and cell death, therefore showing the direct involvement of metacaspases in plant PCD (4). In protozoan parasites, the heterologous expression of TbMC4 in yeast leads to growth inhibition, mitochondrial dysfunction, and cell death, suggesting a possible role of TbMC4 in control of cell proliferation coupled to mitochondrial function (37). Whether TbMC4 exhibits a similar function in T. brucei itself remains to be demonstrated. Recently, a triple null T. brucei mutant, deficient in T. brucei metacaspase-2, -3, and -5 genes, was shown to have no difference compared to the wild type in its cell death kinetics after treatment with the PCD inducer prostaglandin D2, suggesting that these metacaspases are not required as effector molecules in prostaglandin D2-induced cell death of T. brucei (16). In the related trypanosomatid parasite T. cruzi, the overexpression of T. cruzi metacaspase-5 in the insect stage of T. cruzi renders these parasites more susceptible to fresh human serum-induced cell death, suggesting that T. cruzi metacaspase-5 has a role in T. cruzi PCD (22). Gonzalez et al. (15) reported recently that the L. major metacaspase was able to complement the function of its homologue (YCA1) in the yca1-deficient yeast mutant, suggesting conservation of function between yeast and Leishmania metacaspases. However, the role of L. major metacaspase in L. major PCD remains to be demonstrated. Our results above strongly suggest a role of the LdMCs in the L. donovani PCD pathway, since metacaspase activity is significantly increased (∼2.7-fold) in parasites undergoing PCD and parasites overexpressing LdMC1 are more sensitive to H2O2-induced PCD. Approaches such as dominant-negative expression and gene knockout as a means to alter or delete the functions of LdMCs in the parasite should help to confirm the involvement of metacaspases in Leishmania PCD. Such approaches will also be helpful to assess other potential functions of these new “trypsin-like” proteases in Leishmania.
In summary, this represents the first report of the enzymatic properties of native metacaspases purified from a protozoan parasite. Leishmania metacaspases share some similarities with yeast and plant homologues, such as the conservation of the cysteine and histidine residues shown to be involved in the Cys-His catalytic dyad of related caspases. Similar to yeast and plant homologues, Leishmania metacaspases do not have caspase-like activity but show enzymatic characteristics of trypsin-like proteases. However, Leishmania metacaspases have some unique characteristics. They possess a distinct C-terminal proline-rich domain, and L. donovani enzymes do not appear to have autocatalytic activities or to be subjected to a caspase-like processing. In contrast to plants where metacaspases have been found in the cytoplasm or associated with the nucleus (4), LdMCs are localized in unique acidocalcisome compartments, which could represent a form of sequestration of inactive enzymes in the cell. Finally, our results suggest the involvement of LdMCs in the L. donovani PCD pathway representing a significant step forward in the molecular characterization of this pathway in protozoan parasites.
Acknowledgments
We thank G. Matlashewski for providing the expression plasmid pKS NEO and R. Docampo for providing the anti-TcPPase antibody. We also thank H. Nakhasi, R. Duncan, and S. Kumar for their critical review of the manuscript.
The findings and conclusions in this article have not been formally disseminated by the Food and Drug Administration and should not be construed to represent any agency determination and policy.
Footnotes
Published ahead of print on 22 August 2007.
REFERENCES
- 1.Al-Olayan, E. M., G. T. Williams, and H. Hurd. 2002. Apoptosis in the malaria protozoan, Plasmodium berghei: a possible mechanism for limiting intensity of infection in the mosquito. Int. J. Parasitol. 32:1133-1143. [DOI] [PubMed] [Google Scholar]
- 2.Arnoult, D., P. Parone, J. C. Martinou, B. Antonsson, J. Estaquier, and J. C. Ameisen. 2002. Mitochondrial release of apoptosis-inducing factor occurs downstream of cytochrome c release in response to several proapoptotic stimuli. J. Cell Biol. 159:923-929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bozhkov, P. V., L. H. Filonova, M. F. Suarez, A. Helmersson, A. P. Smertenko, B. Zhivotovsky, and S. von Arnold. 2004. VEIDase is a principal caspase-like activity involved in plant programmed cell death and essential for embryonic pattern formation. Cell Death Differ. 11:175-182. [DOI] [PubMed] [Google Scholar]
- 4.Bozhkov, P. V., M. F. Suarez, L. H. Filonova, G. Daniel, A. A. Zamyatnin, Jr., S. Rodriguez-Nieto, B. Zhivotovsky, and A. Smertenko. 2005. Cysteine protease mcII-Pa executes programmed cell death during plant embryogenesis. Proc. Natl. Acad. Sci. USA 102:14463-14468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Clayton, C. E. 2002. Life without transcriptional control? From fly to man and back again. EMBO J. 21:1881-1888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Das, M., S. B. Mukherjee, and C. Shaha. 2001. Hydrogen peroxide induces apoptosis-like death in Leishmania donovani promastigotes. J. Cell Sci. 114:2461-2469. [DOI] [PubMed] [Google Scholar]
- 7.Debrabant, A., E. Ghedin, and D. M. Dwyer. 2000. Dissection of the functional domains of the Leishmania surface membrane 3′-nucleotidase/nuclease, a unique member of the class I nuclease family. J. Biol. Chem. 275:16366-16372. [DOI] [PubMed] [Google Scholar]
- 8.Debrabant, A., M. Gottlieb, and D. M. Dwyer. 1995. Isolation and characterization of the gene encoding the surface membrane 3′-nucleotidase/nuclease of Leishmania donovani. Mol. Biochem. Parasitol. 71:51-63. [DOI] [PubMed] [Google Scholar]
- 9.Debrabant, A., M. B. Joshi, P. F. Pimenta, and D. M. Dwyer. 2004. Generation of Leishmania donovani axenic amastigotes: their growth and biological characteristics. Int. J. Parasitol. 34:205-217. [DOI] [PubMed] [Google Scholar]
- 10.Debrabant, A., N. Lee, S. Bertholet, R. Duncan, and H. L. Nakhasi. 2003. Programmed cell death in trypanosomatids and other unicellular organisms. Int. J. Parasitol. 33:257-267. [DOI] [PubMed] [Google Scholar]
- 11.Debrabant, A., N. Lee, G. Pogue, D. Dwyer, and H. Nakhasi. 2002. Expression of calreticulin P-domain results in impairment of secretory pathway in Leishmania donovani and reduced parasite survival in macrophages. Int. J. Parasitol. 32:1423-1434. [DOI] [PubMed] [Google Scholar]
- 12.Docampo, R., W. de Souza, K. Miranda, P. Rohloff, and S. N. Moreno. 2005. Acidocalcisomes—conserved from bacteria to man. Nat. Rev. Microbiol. 3:251-261. [DOI] [PubMed] [Google Scholar]
- 13.Docampo, R., and S. N. Moreno. 2001. The acidocalcisome. Mol. Biochem. Parasitol. 114:151-159. [DOI] [PubMed] [Google Scholar]
- 14.Earnshaw, W. C., L. M. Martins, and S. H. Kaufmann. 1999. Mammalian caspases: structure, activation, substrates, and functions during apoptosis. Annu. Rev. Biochem. 68:383-424. [DOI] [PubMed] [Google Scholar]
- 15.Gonzalez, I. J., C. Desponds, C. Schaff, J. C. Mottram, and N. Fasel. 2007. Leishmania major metacaspase can replace yeast metacaspase in programmed cell death and has arginine-specific cysteine peptidase activity. Int. J. Parasitol. 37:161-172. [DOI] [PubMed] [Google Scholar]
- 16.Helms, M. J., A. Ambit, P. Appleton, L. Tetley, G. H. Coombs, and J. C. Mottram. 2006. Bloodstream form Trypanosoma brucei depend upon multiple metacaspases associated with RAB11-positive endosomes. J. Cell Sci. 119:1105-1117. [DOI] [PubMed] [Google Scholar]
- 17.Herwaldt, B. L. 1999. Leishmaniasis. Lancet 354:1191-1199. [DOI] [PubMed] [Google Scholar]
- 18.Hoeberichts, F. A., and E. J. Woltering. 2003. Multiple mediators of plant programmed cell death: interplay of conserved cell death mechanisms and plant-specific regulators. Bioessays 25:47-57. [DOI] [PubMed] [Google Scholar]
- 19.Hurd, H., V. Carter, and A. Nacer. 2005. Interactions between malaria and mosquitoes: the role of apoptosis in parasite establishment and vector response to infection. Curr. Top. Microbiol. Immunol. 289:185-217. [DOI] [PubMed] [Google Scholar]
- 20.Ivens, A. C., C. S. Peacock, E. A. Worthey, L. Murphy, G. Aggarwal, et al. 2005. The genome of the kinetoplastid parasite, Leishmania major. Science 309:436-442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kay, B. K., M. P. Williamson, and M. Sudol. 2000. The importance of being proline: the interaction of proline-rich motifs in signaling proteins with their cognate domains. FASEB J. 14:231-241. [PubMed] [Google Scholar]
- 22.Kosec, G., V. E. Alvarez, F. Aguero, D. Sanchez, M. Dolinar, B. Turk, V. Turk, and J. J. Cazzulo. 2006. Metacaspases of Trypanosoma cruzi: possible candidates for programmed cell death mediators. Mol. Biochem. Parasitol. 145:18-28. [DOI] [PubMed] [Google Scholar]
- 23.Lam, E., and O. del Pozo. 2000. Caspase-like protease involvement in the control of plant cell death. Plant Mol. Biol. 44:417-428. [DOI] [PubMed] [Google Scholar]
- 24.Lee, N., S. Bertholet, A. Debrabant, J. Muller, R. Duncan, and H. L. Nakhasi. 2002. Programmed cell death in the unicellular protozoan parasite Leishmania. Cell Death Differ. 9:53-64. [DOI] [PubMed] [Google Scholar]
- 25.Luo, S., M. Vieira, J. Graves, L. Zhong, and S. N. Moreno. 2001. A plasma membrane-type Ca2+-ATPase co-localizes with a vacuolar H+-pyrophosphatase to acidocalcisomes of Toxoplasma gondii. EMBO J. 20:55-64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Madeo, F., E. Herker, C. Maldener, S. Wissing, S. Lachelt, M. Herlan, M. Fehr, K. Lauber, S. J. Sigrist, S. Wesselborg, and K. U. Frohlich. 2002. A caspase-related protease regulates apoptosis in yeast. Mol. Cell 9:911-917. [DOI] [PubMed] [Google Scholar]
- 27.Madeo, F., E. Herker, S. Wissing, H. Jungwirth, T. Eisenberg, and K. U. Frohlich. 2004. Apoptosis in yeast. Curr. Opin. Microbiol. 7:655-660. [DOI] [PubMed] [Google Scholar]
- 28.Mazzoni, C., E. Herker, V. Palermo, H. Jungwirth, T. Eisenberg, F. Madeo, and C. Falcone. 2005. Yeast caspase 1 links messenger RNA stability to apoptosis in yeast. EMBO Rep. 6:1076-1081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mottram, J. C., M. J. Helms, G. H. Coombs, and M. Sajid. 2003. Clan CD cysteine peptidases of parasitic protozoa. Trends Parasitol. 19:182-187. [DOI] [PubMed] [Google Scholar]
- 30.Nguewa, P. A., M. A. Fuertes, B. Valladares, C. Alonso, and J. M. Perez. 2004. Programmed cell death in trypanosomatids: a way to maximize their biological fitness? Trends Parasitol. 20:375-380. [DOI] [PubMed] [Google Scholar]
- 31.Renfranz, P. J., and M. C. Beckerle. 2002. Doing (F/L)PPPPs: EVH1 domains and their proline-rich partners in cell polarity and migration. Curr. Opin. Cell Biol. 14:88-103. [DOI] [PubMed] [Google Scholar]
- 32.Rodrigues, C. O., D. A. Scott, and R. Docampo. 1999. Presence of a vacuolar H+-pyrophosphatase in promastigotes of Leishmania donovani and its localization to a different compartment from the vacuolar H+-ATPase. Biochem. J. 340:759-766. [PMC free article] [PubMed] [Google Scholar]
- 33.Scott, D. A., W. de Souza, M. Benchimol, L. Zhong, H. G. Lu, S. N. Moreno, and R. Docampo. 1998. Presence of a plant-like proton-pumping pyrophosphatase in acidocalcisomes of Trypanosoma cruzi. J. Biol. Chem. 273:22151-22158. [DOI] [PubMed] [Google Scholar]
- 34.Sen, N., B. B. Das, A. Ganguly, T. Mukherjee, S. Bandyopadhyay, and H. K. Majumder. 2004. Camptothecin-induced imbalance in intracellular cation homeostasis regulates programmed cell death in unicellular hemoflagellate Leishmania donovani. J. Biol. Chem. 279:52366-52375. [DOI] [PubMed] [Google Scholar]
- 35.Sen, N., B. B. Das, A. Ganguly, T. Mukherjee, G. Tripathi, S. Bandyopadhyay, S. Rakshit, T. Sen, and H. K. Majumder. 2004. Camptothecin induced mitochondrial dysfunction leading to programmed cell death in unicellular hemoflagellate Leishmania donovani. Cell Death Differ. 11:924-936. [DOI] [PubMed] [Google Scholar]
- 36.Singh, G., K. G. Jayanarayan, and C. S. Dey. 2005. Novobiocin induces apoptosis-like cell death in topoisomerase II over-expressing arsenite resistant Leishmania donovani. Mol. Biochem. Parasitol. 141:57-69. [DOI] [PubMed] [Google Scholar]
- 37.Szallies, A., B. K. Kubata, and M. Duszenko. 2002. A metacaspase of Trypanosoma brucei causes loss of respiration competence and clonal death in the yeast Saccharomyces cerevisiae. FEBS Lett. 517:144-150. [DOI] [PubMed] [Google Scholar]
- 38.Thrane, C., U. Kaufmann, B. M. Stummann, and S. Olsson. 2004. Activation of caspase-like activity and poly (ADP-ribose) polymerase degradation during sporulation in Aspergillus nidulans. Fungal Genet. Biol. 41:361-368. [DOI] [PubMed] [Google Scholar]
- 39.Uren, A. G., K. O'Rourke, L. A. Aravind, M. T. Pisabarro, S. Seshagiri, E. V. Koonin, and V. M. Dixit. 2000. Identification of paracaspases and metacaspases: two ancient families of caspase-like proteins, one of which plays a key role in MALT lymphoma. Mol. Cell 6:961-967. [DOI] [PubMed] [Google Scholar]
- 40.Vercammen, D., B. van de Cotte, G. De Jaeger, D. Eeckhout, P. Casteels, K. Vandepoele, I. Vandenberghe, J. Van Beeumen, D. Inze, and F. Van Breusegem. 2004. Type II metacaspases Atmc4 and Atmc9 of Arabidopsis thaliana cleave substrates after arginine and lysine. J. Biol. Chem. 279:45329-45336. [DOI] [PubMed] [Google Scholar]
- 41.Watanabe, N., and E. Lam. 2005. Two Arabidopsis metacaspases AtMCP1b and AtMCP2b are arginine/lysine-specific cysteine proteases and activate apoptosis-like cell death in yeast. J. Biol. Chem. 280:14691-14699. [DOI] [PubMed] [Google Scholar]
- 42.Wilson, K., S. Hanson, S. Landfear, and B. Ullman. 1991. Nucleotide sequence of the Leishmania donovani medRNA gene. Nucleic Acids Res. 19:5787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhang, W. W., H. Charest, E. Ghedin, and G. Matlashewski. 1996. Identification and overexpression of the A2 amastigote-specific protein in Leishmania donovani. Mol. Biochem. Parasitol. 78:79-90. [DOI] [PubMed] [Google Scholar]