Cryptococcus species are encapsulated basidiomycetous yeasts found ubiquitously in the environment, notably in pigeon guano and eucalyptus trees. Two species of this genus, Cryptococcus neoformans and Cryptococcus gattii, are human pathogens capable of causing a life-threatening meningoencephalitis. Currently, C. neoformans is divided into the varieties Cryptococcus neoformans var. grubii (serotype A) and Cryptococcus neoformans var. neoformans (serotype D), and C. gattii comprises serotypes B and C (47). Infection occurs primarily during environmental exposure via inhalation of either desiccated yeast or basidiospores. In the alveolar spaces of lungs, the fungus is initially exposed to alveolar macrophages followed by other inflammatory cells, which can lead to a successful adaptive immune response and containment or clearance of the fungal pathogen. However, C. neoformans and C. gattii can survive and proliferate in the human host, leading to dissemination to other organs, most importantly to the central nervous system, where meningoencephalitis can develop, with 100% mortality if untreated.
Cryptococcosis is recognized as a critically important opportunistic infection for an increasing number of individuals with impaired immune systems (26, 32), and C. neoformans is the species causing the vast majority of cryptococcoses (7, 14, 46). This is a particular problem in areas where treatment for human immunodeficiency virus/AIDS is limited (3, 34).
C. neoformans has biological properties considered to be virulence factors, the best-known being the capsule, growth at 37°C, and production of melanin (11). However, in recent years, new fungal factors have been identified as additional and crucial regulators of cryptococcal pathogenicity (12, 15, 22, 57, 68, 69, 72, 102). An exciting area of investigation is the biosynthetic pathway of cryptococcal sphingolipids, because it provides an extremely rich reservoir of sphingolipid molecules and fungus-specific metabolizing enzymes that regulate many cellular functions essential for fungal viability (35). Thus, studies addressing the biological and pathophysiological roles of the sphingolipid pathway during cryptococcosis may provide new insights into the development of new diagnostic and therapeutic strategies. In fungal cells, sphingolipids play key roles in cell cycle progression, apoptosis, signal transduction, and pathogenesis (16, 58, 70). Since the completion of the sequencing of the genome of the model fungal organism Saccharomyces cerevisiae, studies have identified the majority of genes and their enzyme products responsible for sphingolipid biosynthesis (21, 25). Although there are still several hypothetical steps, the knowledge pertaining to sphingolipid metabolism in S. cerevisiae has created a working scheme of the probable biosynthetic pathway and has provided a blueprint with which to examine sphingolipid metabolism in other organisms. However, since research examining fungal sphingolipid biosynthesis has been conducted almost exclusively with S. cerevisiae, which is a nonpathogenic fungus, the biological functions of fungal sphingolipids in pathogenesis are relatively unknown.
This review summarizes the current understanding of the role of sphingolipid biosynthesis in the pathobiology of cryptococcosis, focusing on the production and functions of the antigenic glycosphingolipid (GSL) glucosylceramide (GlcCer) and its role in the host-pathogen interaction. In addition, the presence of GlcCer and structurally similar molecules present in other pathogens will be discussed and highlighted for their importance as potential therapeutic targets.
SPHINGOLIPID METABOLISM OF CRYPTOCOCCUS
With the completion of the sequencing of the C. neoformans genome (56) and the current knowledge of the sphingolipid metabolism of S. cerevisiae, researchers have all the initial tools necessary to identify potential genomic sequences encoding the genes regulating sphingolipid metabolism in C. neoformans. There are three fully sequenced strains of C. neoformans, one of which (JEC21) is annotated. tBLASTx searches of each of these databases with S. cerevisiae genes that encode enzymes involved in sphingolipid synthesis revealed that C. neoformans has genomic sequences with strong similarities after translation, suggesting the existence of a similar sphingolipid biosynthetic pathway (Table 1). Although only a few genes and enzymes of the sphingolipid biosynthesis pathway in C. neoformans have been identified and characterized experimentally, they are essential to virulence and pathogenicity (34, 36, 58, 75, 84). In light of these findings, further elucidation of the sphingolipid metabolism of C. neoformans could provide new and better pharmacological targets. To further highlight the explicit differences between mammalian and fungal sphingolipid enzymes and pathways, Table 2 features a direct comparison of the enzymes found in these organisms.
TABLE 1.
Comparative homologies of Cryptococcus neoformans genomic sequences to sphingolipid-encoding genes in other organismsa
| Enzyme | Gene | Systemic name |
C. neoformans strain:
|
|||||
|---|---|---|---|---|---|---|---|---|
| JEC21
|
H99
|
B3501
|
||||||
| Annotation | Homology | Location | Homology | Location | Homology | |||
| Serine palmitoyltransferase | S. cerevisiae LCB1 | YMR296C | CNC07020 | 7.8−30 | chr1-piece5 | 5.0−60 | chr01.b3501.040506 | 4.0−62 |
| chr3-piece23 | 7.0−23 | chr03.b3501.040506 | 9.0−24 | |||||
| S. cerevisiae LCB2 | YDR062W | CNC07020 | 4.7−137 | chr1-piece5 | 5.0−60 | chr03.b3501.040506 | 1−147 | |
| chr3-piece23 | 7.0−23 | |||||||
| S. cerevisiae TSC3 | YBR058C-A | CNJ02790 | 2.7−02 | chr3-piece24 | 1.6−02 | chr05.b3501.040506 | 1.9−02 | |
| 3-Keto-dihydrosphingosine reductase | S. cerevisiae TSC10 | YBR265W | CNF02530 | 1.5−01 | chr8-piece7 | 6.0−10 | chr07.b3501.040506 | 8.0−11 |
| Sphingoid base kinase | S. cerevisiae LCB4 | YOR171C | None | None | chr5-piece15 | 4.0−07 | chr04.b3501.040623 | 8.0−11 |
| S. cerevisiae LCB5 | YLR260W | None | None | chr5-piece15 | 9.0−09 | chr04.b3501.040623 | 4.0−13 | |
| Sphingoid base phosphate lyase | S. cerevisiae DPL1 | YDR294C | None | None | chr4B-piece17 | 3.0−63 | chr12.b3501.040506 | 2.0−68 |
| Sphingoid base phosphate phosphatase | S. cerevisiae YSR2 | YJL134W | CNF02820 | 7.2−43 | chr7-piece5 | 3.0−6 | chr06.b3501.040616 | 8.0−16 |
| S. cerevisiae YSR3 | YKR053C | CNF02820 | 2.6−40 | chr7-piece5 | 2.0−5 | chr06.b3501.040616 | 3.0−9 | |
| Ceramide synthase | S. cerevisiae LAG1 | YHL003C | CNE04490 | 1.2−41 | chr2-piece7 | 2.0−15 | chr02.b3501.040506 | 1.0−16 |
| CNB00860 | 2.8−21 | chr6-piece12 | 2.0−11 | chr05.b3501.040506 | 1.0−13 | |||
| S. cerevisiae LAC1 | YKL008C | CNE04490 | 1.9−52 | chr2-piece7 | 2.0−15 | chr02.b3501.040506 | 9.0−15 | |
| CNB00860 | 9.1−24 | chr6-piece12 | 2.0−11 | chr05.b3501.040506 | 1.0−13 | |||
| S. cerevisiaec LIP1 | YMR298W | None | None | chr6-piece12 | 2.3−02 | chr07.b3501.040506 | 4.9−01 | |
| Fatty acid α-hydroxylase | S. cerevisiae SCS7 | YMR272C | None | None | chr3-piece24 | 5.0−41 | chr11.b3501.040506 | 2.0−65 |
| Dihydroceramidase | S. cerevisiae YDC1 | YPL087W | CND01740 | 1.2−37 | chr5-piece15 | 1.0−20 | chr04.b3501.040623 | 5.0−23 |
| Phytoceramidase | S. cerevisiae YPC1 | YBR183W | CND01740 | 2.4−37 | chr5-piece15 | 5.0−25 | chr04.b3501.040623 | 4.0−26 |
| Dihydrosphingosine C-4 hydroxylase | S. cerevisiae SUR2/SYR2 | YDR297W | CNC02410 | 2.9−13 | chr2-piece9 | 6.0−53 | chr02.b3501.040506 | 5.0−61 |
| Inositol phosphoryl ceramide synthase 1 (Ipc1) | C. neoformans IPC1 | CNI01040 | AY007247 | CNBI0760 | ||||
| Inositol phosphosphingolipid phospholipase C (Isc1) | C. neoformans ISC1 | None | None | DQ487762 | DQ487763 | |||
| Mannosyltransferase | S. cerevisiae CSG1 | YPL057C | CNA01170 | 9.9−01 | chr4B-piece17 | 1.0−37 | chr12.b3501.040506 | 2.0−37 |
| S. cerevisiae CSG2 | YBR036C | None | None | chr6-piece12 | 3.0−02 | chr01.b3501.040506 | 9.3−02 | |
| S. cerevisiae CSH1 | YBR161W | None | None | chr4B-piece17 | 5.0−38 | chr12.b3501.040506 | 3.0−38 | |
| Inositol phosphoryltransferase 1 (Ipt1) | S. cerevisiae IPT1 | YDR072C | CNM01330 | 9.5−01 | chr9-piece11 | 5.0−18 | chr08.b3501.040506 | 7.0−19 |
| Δ4 Desaturase | C. albicans | (Putative) | CNA06240 | 1.5−84 | chr1-piece3 | 8.0−63 | chr01.b3501.040506 | 5.0−80 |
| Δ8 Desaturase | C. albicans | (Putative) | CNB03590 | 1.3−81 | chr2-piece9 | 2.0−30 | chr02.b3501.040506 | 4.0−51 |
| C9 methyltransferase | P. pastoris | DQ070247 | CNC07040 | chr3-piece 23* | 1−133* | chr03.b3501.040506* | 1−134* | |
| Glucosylceramide synthase (Gcs1) | C. neoformans GCS1 | CNH02280 | AAX55972 | chr12.b3501.040506 | ||||
| Glucosylceramidase (Gcdase) | Mus musculus GBA | CNI03760 | 9.7−01 | chr1-piece5 | 1.1−01 | chr03.b3501.040506 | 3.6−01 | |
| Mus musculus GBA2 | CNF01870 | 2.0−01 | chr13-piece14 | 1.1−01 | chr13.b3501.040506 | 5.5−02 | ||
Genes encoding sphingolipid-metabolizing enzymes of various organisms were used to identify homologous genomic sequences in three different strains of Cryptococcus neoformans. Reference organisms whose genes were used to identify possible C. neoformans genes were Saccharomyces cerevisiae, Candida albicans, Pichia pastoris, and mouse. C. neoformans gene names are provided for those genomic sequences determined experimentally to encode enzymes involved in the C. neoformans sphingolipid biosynthesis pathway. Translated nucleotide sequences from these reference organisms were used to search the translated nucleotide databases (tBLASTx) of C. neoformans serotype D strain JEC21 (http://tigrblast.tigr.org/er-blast/index.cgi?project=cna1), C. neoformans serotype A strain H99 (http://cneo.genetics.duke.edu/blast.html), and C. neoformans serotype D strain B3501 (http://www-sequence.stanford.edu/cgi-bin/cneoformans/cneo_blast.cgi). Asterisks indicate that an amino acid sequence was used to search for the translational nucleotide sequence (tBLASTn) within C. neoformans databases, due to the lack of an available genomic sequence to serve as a query. Because JEC21 is the only strain of C. neoformans whose genome is fully annotated, genomic sequences of strains H99 and B3501 possessing homology to the reference gene are designated by chromosome location, according to the respective database. It should be noted that the genomic sequences with the highest degree of homology are provided. Therefore, the genomic sequences identified with a relatively low degree of homology may not be involved in C. neoformans sphingolipid biosynthesis.
TABLE 2.
Comparison of the genes encoding sphingolipid-metabolizing enzymes in mammals and yeast/fungia
| Enzyme nameb | Mammalian gene(s)c | Fungal gene(s)d |
|---|---|---|
| SPT | SPTLC1, SPTLC2 | LCB1, LCB2, TCS3 |
| 3-Keto-dhSph reductase | FVT-1 | TSC10 |
| Cer synthase | LASS1 to LASS6 | LAG1, LAC1, LIP1 |
| CDase | ASAH1 to ASAH3 | YDC1, YPC1 |
| FA2H | FA2H | SCS7 |
| Sphingolipid Δ4-desaturase | DEGS1, DEGS2 | Putative in C. albicans |
| Sphingolipid Δ8-desaturase | Not present | Putative in C. albicans |
| Sphingolipid C9-methyltransferase | Not present | Identified in P. pastoris |
| Gcs1 | UGCG | GCS1 |
| GCdase | GBA, GBA2 | Not identified |
| SBK | SPHK1, SPHK2 | LCB4, LCB5 |
| SBPP | SGPP1 | YSR2, YSR3 |
| SBPL | SGPL1 | DPL1 |
| dhSph C4-hydroxylase | Not identified | SUR2/SYR2 |
| dhCer hydroxylase | Not identified | Not identified |
| Mannosyltransferase | Not identified | CSG1, CSG2, CSH1 |
| Ipc1 | Not present | AUR1/IPC1 |
| Isc1 | Not present | ISC1 |
| Ipt1 | Not present | IPT1 |
| SMase | SMPD1/ASM, SMPD2, SMPD3, ENPP7 | Not present |
| SMS | SMS1, SMS2 | Not present |
| Cer kinase | CERK | Not present |
This comprehensive, but not exclusive, table lists the enzymes regulating sphingolipid biosynthesis and presents the respective genes encoding each of these products. Genes categorized as “not identified” are suspected to exist, due to the presence of a sphingolipid species or a downstream metabolite(s) requiring an intermediate. A gene is considered “not present” if, to date, there is no evidence of the sphingolipid existing in that class of organisms.
SPT, serine palmitoyltransferase; CDase, ceramidase; Gcs1, glucosylceramide synthase 1; GCdase, glucosylceramidase; SBK, sphingoid base kinase; SBPP, sphingoid base phosphate phosphatase; SBPL, sphingoid base phosphate lyase; SMase, sphingomyelinase; SMS, sphingomyelin synthase.
SPTLC1 and -2, serine palmitoyltransferase long-chain base subunits 1 and 2; FVT-1, follicular lymphoma variant translocation gene; LASS1 to LASS6, longevity assurance genes 1 to 6; ASAH1 to -3, N-acylsphingosine amidohydrolase 1 to 3 genes; DEGS1 and -2, degenerative spermatocyte homolog 1 and 2 genes; UGCG, UDP-glucose ceramide glucosyltransferase gene; GBA and GBA2, acid β-glucosidase and acid β-glucosidase-2 genes; SPHK1 and -2, sphingosine kinase 1 and 2 genes; SGPP1, sphingosine-1-phosphate phosphatase 1 gene; SGPL1, sphingosine-1-phosphate lyase 1 gene; SMPD1 to -3, sphingomyelin phosphodiesterase 1 to 3 genes; ASM, acid sphingomyelinase gene; ENPP7, ectonucleotide pyrophosphatase/phosphodiesterase 7 gene; CERK, ceramide kinase gene.
LCB1 and -2, long-chain base 1 and 2 genes; TSC3, temperature-sensitive cgs2 delta suppressor 3 gene; TSC10, temperature-sensitive cgs2 delta suppressor 10 gene; LAG1, longevity assurance gene 1; LAC1, longevity assurance gene cognate 1; LIP1, Lag1p/Lac1p-interacting protein 1 gene; YDC1, yeast dihydroceramidase 1 gene; YPC1, yeast phytoceramidase 1 gene; SCS7, suppressor of Ca2+ sensitivity 7 gene; LCB4 and -5, long-chain base 4 and 5 genes; YSR2 and -3, yeast sphingosine resistance genes 2 and 3; DPL1, dhSph phosphate lyase 1 gene; CSG1 and -2, Ca2+ sensitivity 1 and 2 genes; CSH1, CSG1/SUR1 homolog 1 gene; AUR1, aureobasidin resistance gene 1. Genes identified and characterized as encoding a sphingolipid-metabolizing enzyme(s) in Cryptococcus species are boldfaced.
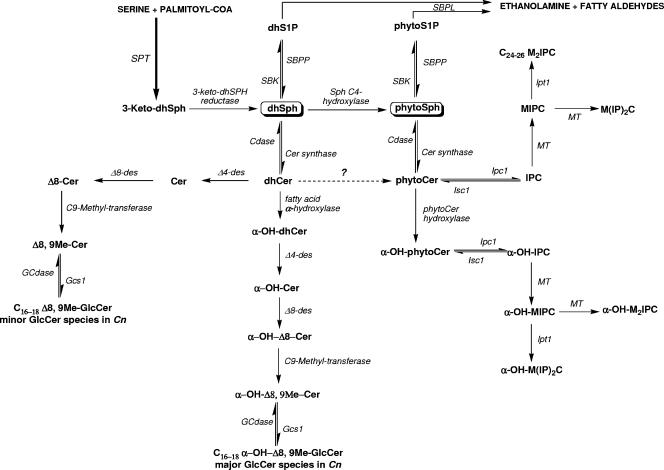
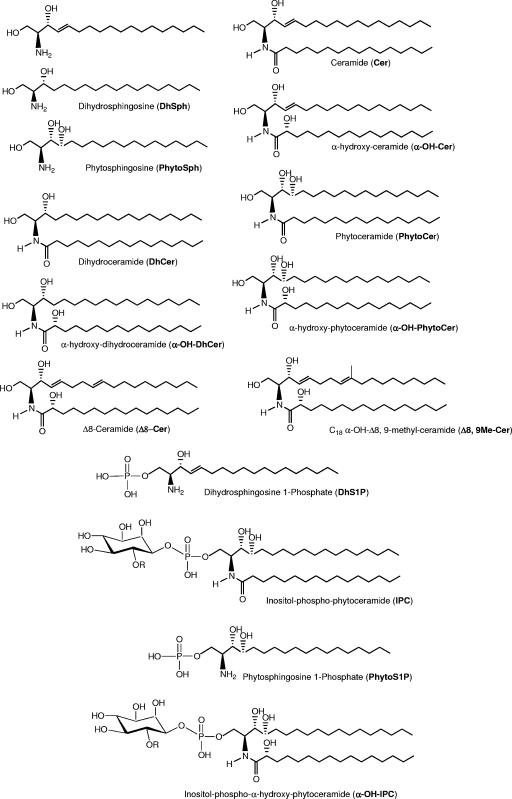
The first several steps of de novo sphingolipid biosynthesis (Fig. 1) are well conserved among all eukaryotic cells. Structures of the intermediate species referenced in Fig. 1 can be seen in detail in Fig. 2 and 3. In the initial step, serine palmitoyltransferase catalyzes the condensation of serine and palmitoyl coenzyme A to form 3-ketodihydrosphingosine. The ketone group of this molecule is then rapidly reduced in an NADPH-dependent manner, yielding dihydrosphingosine (dhSph). The molecule 3-ketodihydrosphingosine is not detected by standard methods (e.g., thin-layer chromatography or high-performance liquid chromatography) or more advanced methods (e.g., mass spectrometry) for measuring sphingolipids, and it is likely an intermediate. Therefore, dhSph is arguably the simplest biologically relevant sphingolipid (5).
FIG. 1.
Hypothetical scheme of the sphingolipid biosynthetic pathway in C. neoformans. Sphingolipid molecules are boldfaced, and enzymes are italicized. Question marks indicate enzymes or activities that have not been identified or observed. Hydroxyl groups in parentheses indicate that this group may be absent, creating the corresponding nonhydroxylated form. SPT, serine palmitoyltransferase; SBK, sphingoid base kinase; P, phosphate; SBPP, sphingoid base phosphate phosphatase; SBPL, sphingoid base phosphate lyase; Cer, ceramide; Cdase, ceramidase; des, desaturase; Gcs1, glucosylceramide synthase; GCdase, glucosylceramidase; MIPC, mannosyl-inositol phosphorylceramide; M(IP)2C, mannosyl diinositol phosphate ceramide; M2IPC, dimannosyl-inositol phosphorylceramide.
FIG. 2.
Chemical structures of C. neoformans sphingolipids.
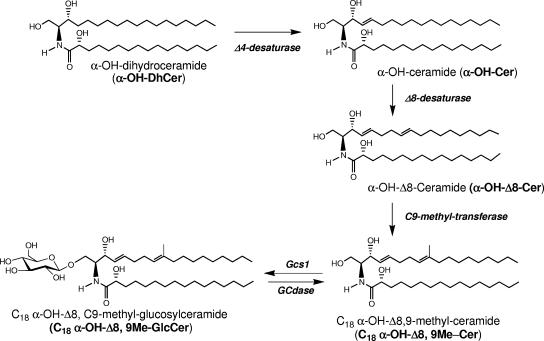
FIG. 3.
Biosynthetic pathway of C. neoformans GlcCer.
dhSph can be converted to phytosphingosine (PhytoSph) by hydroxylation of the 4th carbon of the backbone. PhytoSph is found abundantly in yeast, but its distribution in mammals is limited to the skin epidermis (25, 81). In yeast and fungal cells, dhSph and PhytoSph are commonly referred to as sphingoid bases, because all complex sphingolipids created de novo are derived from these molecules. These sphingoid bases can be phosphorylated, forming dhSph-1-phosphate and PhytoSph-1-phosphate, or acylated by different fatty acids (saturated, unsaturated, or hydroxylated) at their amine group to produce dihydroceramide (dhCer) and phytoceramide (PhytoCer), respectively.
Divergence between the sphingolipid synthetic pathways in mammals and analogous pathways in fungi occurs with the utilization of dhSph and following the production of dhCer. In fungi and plants, as exemplified in S. cerevisiae, N acylation of PhytoSph occurs to form PhytoCer. Additionally, C-4 hydroxylation of dhCer may also form PhytoCer. Currently, it is not known which molecule serves as the primary substrate from which PhytoCer is formed. As stated above, PhytoSph is uncommon in mammalian cells. To date, PhytoCer has been isolated only from cells comprising the skin and intestine. In mammalian cells, the C-4-C-5 bond of dhCer is reduced to the 4,5 trans-double bond of ceramide (Cer).
The synthesis of complex sphingolipids occurs via modification of the primary hydroxyl group of Cer. Mammals utilize Cer as the substrate to produce more-complex sphingolipids, such as sphingomyelin. In contrast, fungi and plants use mainly the primary hydroxyl group of PhytoCer to create complex sphingolipids, such as inositol phosphoryl PhytoCer (IPC) and its mannosylated forms. However, in some fungi, there are two different and apparently distinct populations of complex sphingolipids. As stated above, the first population includes the inositol-containing GSLs such as IPC and its mannosylated forms. These molecules are built on PhytoCer containing an N-acyl chain composed mainly of 24 to 26 carbons, considered very long chain fatty acids (VLCFA) (31, 79). The essential C. neoformans gene encoding inositol phosphorylceramide synthase 1 (Ipc1) is present in several pathogenic fungi (33). Ipc1 synthesizes IPC and diacylglycerol (DAG) by transferring inositol phosphate from phosphatidylinositol (PI) to PhytoCer. In C. neoformans, Ipc1 governs melanogenesis through the generation of DAG, a physiological activator of protein kinase C1 (Pkc1) in C. neoformans, and the Ipc1-DAG-Pkc1 pathway regulates the localization and activity of laccase, the enzyme responsible for melanin production (34, 58). Also, the production of DAG by Ipc1 has been shown to regulate the transcription of the gene encoding the antiphagocytic protein 1 (App1) (57) through the activating transcription factor 2 (Atf2) (61).
Inositol phosphosphingolipid-phospholipase C 1 (Isc1) metabolizes the reverse reaction of Ipc1, producing PhytoCer and releasing PI (80). The gene encoding Isc1 was identified in the C. neoformans var. grubii serotype A strain H99 (84). Shea et al. found that deletion of ISC1 produced a strain (Δisc1) with significantly attenuated virulence, most likely resulting from its decreased dissemination to the brain (84). Isc1 was found to protect internalized fungi from the antimicrobial activities within macrophages. These studies examining Ipc1 and Isc1 clearly established the role of PhytoCer, DAG, and this group of GSLs in the virulence of C. neoformans. Though detailed descriptions of these experiments and pathways are outside the scope of this review, interested parties can find full analyses of this branch of C. neoformans sphingolipid biology in the sources cited above (41, 43, 68, 97).
The second population of GSLs in fungi is made up of specific galactosylceramides (GalCer's) and GlcCer's, commonly called cerebrosides. The structure, function, and location of GlcCer and its role in pathogenesis and host immune system modulation are discussed in the next section. In Fig. 2 and 3 we illustrate the main chemical structures of sphingolipids present in fungi.
GlcCer SYNTHESIS AND FUNCTION IN FUNGI
GlcCer's are present in protozoans, plants, fungi, and mammalian cells. As mentioned above, the model fungal system S. cerevisiae does not synthesize GlcCer, so knowledge pertaining to the synthesis and biological function(s) of GlcCer in pathogenic fungi is limited. This section will address the structure of fungal GlcCer and the knowledge to date of GlcCer location and function in pathogenic fungi.
In C. neoformans, GlcCer has a modified sphingosine backbone, containing a Δ4 double bond in trans conformation, a Δ8 double bond, and a methyl substituent on C-9 (Fig. 3). Whereas the trans conformation of the 4,5 double bond has been determined (54) and was found to be essential for the maintenance of transmembrane asymmetry of GalCer (59), we determined by nuclear magnetic resonance the conformation of the 8,9 double bond of the sphingosine backbone found in C. neoformans GlcCer. Details of the double-bond conformation analysis can be found in Fig. S1 in the supplemental material.
Also, the fatty acid substituents in the GlcCer are long-chain fatty acids (LCFA), usually between 16 and 18 carbons long (93-95, 100) (Fig. 3). This is in contrast to the 24- to 26-carbon chain length present in the IPC-derived GSLs (Fig. 2). There has been speculation as to how these selective fatty acid lengths for different GSLs are maintained. One possible way would be if the synthesizing enzymes show specificity for a particular chain length. Another hypothesis is that compartmentalization of substrates or enzymes leads to the observed differences in substituent chain length. Studies have shown that transgenic GlcCer synthase (GCS) genes from a variety of fungi expressed in a GCS-null mutant of Pichia pastoris resulted in the production of GlcCer featuring both LCFA and VLCFA (48). This suggests that the observed difference is likely due to compartmentalization and is not a result of enzyme specificity.
Focusing on the fungal GlcCer, the structure of this molecule in fungi is fairly conserved across species, as opposed to that in plants. The general structure (Fig. 3) is that of a modified Cer (with the aforementioned backbone features) substituted at the primary carbon hydroxyl group with a β-d-glucose moiety. As mentioned, the N-acyl chain is between 16 and 18 carbons long and is hydroxylated at the α-carbon. The full name of the most abundant species of GlcCer in C. neoformans is N-2′-hydroxystearoyl-1-O-β-d-glucopyranosyl-9-methyl-4,8-sphingadiene (α-OH-C18:0 GlcCer) (75, 100). The main difference between the C. neoformans GlcCer and GlcCer species from different fungi is the presence or absence of the Δ3-double bond on the fatty acid (100). In C. neoformans, the major species of GlcCer is known as 18:0 GlcCer (51, 77, 100). Another difference between some species of fungi is the presence of α-hydroxylation on the fatty acid side chain.
The biosynthesis of GlcCer in fungi and specifically in C. neoformans is still under investigation. As mentioned above, there are C. neoformans genes that show high homology to the sphingolipid-synthesizing enzymes characterized for S. cerevisiae. Since S. cerevisiae does not synthesize GlcCer, finding these synthetic genes in the model fungal species was impossible. Thus, fungal GCS genes were identified by their similarity to mammalian GCS. Notably, Candida albicans, P. pastoris (48), C. neoformans (75, 77), Aspergillus fumigatus (9, 50), Histoplasma capsulatum (95), Paracoccidioides brasiliensis (87), and Sporothrix schenckii (92) have been found to have genes that likely encode proteins with UDP-glucose:ceramide properties. The GCS enzyme in C. neoformans (Gcs1) has been fully characterized and deleted (75) and shown to produce α-OH-C18:0 GlcCer. In addition to α-OH-C18:0 GlcCer, N-stearoyl-1-O-β- d-glucopyranosyl-9-methyl-4,8-sphingadiene (nonhydroxy-C18:0 GlcCer) and N-2′-hydroxypalmitoyl-1-O-β-d-glucopyranosyl-9-methyl-4,8-sphingadiene (α-OH-C16:0 GlcCer) are also synthesized as minor species (75). The significance of these major and minor proportions has yet to be determined, but it does indicate that there is flexibility in the enzyme's substrate specificity in vivo. The mechanisms by which these proportions are maintained are also unknown.
The particular species of Cer used by Gcs1 as substrates are quite interesting because they are not found in mammalian cells and thus are fungus specific. The genes involved in the formation of this Cer are Δ4-desaturase, Δ8-desaturase, and the C9-methyltransferase. These enzymes are not found in S. cerevisiae, but a putative Δ4-desaturase is found in C. albicans (48, 89, 100). The C. albicans Δ4-desaturase (100) was found to have significant homology to a gene on chromosome 1 of each of the three sequenced C. neoformans strains. Similarly, the C. albicans Δ8-desaturase gene showed significant homology to a gene on chromosome 2 of all three C. neoformans strains sequenced to date (Table 1). The putative homolog of the sphingolipid C9-methyltransferase has been identified in C. neoformans as a ∼1.7-kb fragment on chromosome 3, piece 23, of the H99 genome (M. Del Poeta, unpublished data) (Table 1).
Leipelt et al. (48) proposed the biosynthetic sequences of the chemical reactions leading to the formation of the Cer backbone used as a substrate for Gcs1. A double-null mutant of P. pastoris was constructed that lacked endogenous GCS and sterol glucosyltransferase activity. Using this system, Leipelt et al. expressed the human GCS gene and then separated and identified the products. The presence of GlcCer products with the backbone Δ4-desaturation alone, both Δ4- and Δ8-desaturations, and the Δ4- and Δ8-desaturations with the methyl group on C-9 suggested the order of biochemical steps outlined in Fig. 3. Furthermore, a sphingolipid C9-methyltransferase responsible for backbone methylation was found in a variety of fungi and deleted in P. pastoris (90). This enzyme was shown to use Δ4, Δ8-desaturated Cer and S-adenosylmethionine as substrates (90), as proposed by Leipelt et al. (48).
As mentioned above, Gcs1 mainly produced GlcCer with the α-hydroxylation of the N-acyl chain. In the previous reviews discussing the synthesis of GlcCer in fungi, N-acyl hydroxylation was proposed to occur in either the modified Cer (48) or the dhCer (100). Though previous studies have suggested that ceramides, not fatty acids, are the substrates for these fatty acid α-hydroxylases (41), other recent studies have suggested that some mammalian hydroxylases (fatty acid 2-hydroxylase [FA2H]) use fatty acids as substrates prior to their incorporation into GSLs (1). Homology exists between the mammalian FA2H gene and the C. neoformans genes in strains B3501 and H99 (Table 1), but it is not clear whether these represent analogous enzymes. Further study will be necessary to determine if the hydroxylation occurs on the N-acyl chain of dhCer or of modified Cer or on free fatty acid(s). On the other hand, these studies allowed the creation of a working model for the GlcCer synthesis pathway that is applicable to fungi in general and to C. neoformans specifically (Fig. 1). The only major variations seen in these structures within fungi are those mentioned above with regard to acyl chain hydroxylation and desaturation.
FUNCTION AND LOCALIZATION OF GlcCer IN C. NEOFORMANS AND OTHER FUNGI
Using clinically isolated human anti-GlcCer antibodies in immunofluorescence studies, C. neoformans GlcCer has been shown to be cell wall associated (77, 96) and appears to concentrate at the site of budding daughter cells during cell division (96). This observation is supported by recent studies in which the cell cycle of C. neoformans Δgcs1 cells is arrested at the S phase under certain growing conditions (75). Since budding in yeast cells starts in S phase and concludes in G2, the localization of GlcCer at the budding site may be important to conclude the cell cycle and specifically for moving from S to G2 phase. More recently, GlcCer has been identified in extracellular vesicles that are apparently secreted through the cell wall (76), and the extracellular localization may explain its antigenic property. Interestingly, GlcCer vesicles containing the antigen polysaccharide glucuronoxylomannan (GXM), which is the principal constituent of the capsule, are transported outside the cell. The function of GlcCer in fungi is still a topic of discussion and research, since few roles of this molecule have been confirmed. One major pattern that was recognized by Saito et al. (78) was the presence of GlcCer in fungi that are characterized as more tolerant to alkaline conditions. In their study, 90 different fungal strains were divided into those that contain GlcCer and those that do not. The strains containing GlcCer grew at a higher average pH, and many of them were determined to have GCS-like genes. A GCS deletion mutant of Kluyveromyces lactis grew at a much lower pH than the wild-type strain. The GCS knockout mutant of C. neoformans (Δgcs1) created by Rittershaus et al. (75) was also found to be intolerant of a neutral/alkaline pH of 7.4 under 5% CO2. The mechanism by which this intolerance occurs is currently under investigation.
Studies addressing GlcCer function in fungi have found other recurring patterns of involvement, such as cell cycle progression and differentiation. For instance, anti-GlcCer antibodies prevented germ tube formation in Pseudallescheria boydii but did not prevent the growth of previously existing germ tubes (50, 74). In two different species of Aspergillus, inhibitor-based disruption of the GlcCer synthesis pathway resulted in defects in the cell cycle, the formation of hyphae, and spore germination (50). The Δgcs1 cells are arrested at the S and G2/M stages of the cell cycle, suggesting a role for GlcCer in exiting these stages (75). The Δgcs1 strain of C. neoformans is an interesting example of how the function of GlcCer in fungi can be related to pathogenesis. In contrast to wild-type and reconstituted strains, the C. neoformans Δgcs1 strain did not progress to lethal meningoencephalitis and appeared to be contained in granulomas in the alveolar spaces (75). Furthermore, the conditions under which the Δgcs1 strain shows impaired growth (pH 7.4, 5% CO2) are clinically relevant because they are the conditions present in the host lung environment.
As described above, the pathway by which C. neoformans and other pathogenic fungi synthesize important GSLs such as GlcCer is becoming clearer with each new experiment. The relevance and importance of studying these steps are demonstrated by the finding that the lack of GCS function in this human pathogen leads to an avirulent strain. Several of these steps exist solely in fungi, making them ideal potential targets for human therapy.
IMMUNOGENICITY OF GSLs IN OTHER ORGANISMS
The immunogenicity of GSLs has been demonstrated in a variety of infection-related clinical conditions. General indications that GSL can have significant effects on the host immune system were seen in experiments where these compounds were given exogenously. For instance, α-GalCer, originally extracted from the marine sponge Agelas mauritianus, has been found to possess antitumor activity (43) and reduces graft-versus-host disease when administered to mice (64). Also, in murine models, administration of α-GalCer shows stage-specific antimalarial activity (20). The intraperitoneal administration of β-GlcCer to mice reduced colitis and suppressed the growth of a hepatocellular carcinoma by increasing the Th2 and Th1 responses, respectively (106). An analog of GalCer containing a truncated sphingosine chain has been shown to inhibit experimental autoimmune encephalomyelitis (63) and collagen-induced arthritis (17) by a mechanism involving NK cells and the production of Th2 cytokines (17, 63).
Since GSLs have been found in several kingdoms and phyla including sponges, amphibians, plants, mammals, fungi, bacteria, and protozoa (88), the presence of these lipids has raised questions about their involvement in infectious processes. To limit our discussion in this review, we will focus on bacterial, protozoal, and fungal GSLs and their immunomodulatory role in the host.
BACTERIAL GSL
Although GSLs are found in many organisms, little is known about bacterial sphingolipids. Gram-negative bacteria have lipopolysaccharide (LPS) in their outer membranes, which serves as a potent endotoxin that elicits a variety of host immune responses. Sphingomonas species, however, lack LPS and instead contain these immunogenic GSLs (39, 40). Sphingomonas species are found in soil, on plants, and as biofilms in water (44). They have been implicated in several infections including intravascular catheter-related bacteremia, urinary tract infections, and primary biliary tract infections (38, 82).
Many scientists have addressed the significance of substitution of LPS with GSLs as a possible immune system stimulator. Despite the lack of LPS, Sphingomonas species are able to activate complement and to stimulate NK cell proliferation and production of gamma interferon, tumor necrosis factor, interleukin-6 (IL-6), and IL-1 (45, 62, 66). Moreover, mouse and human NK cells have been shown to recognize GSL from Sphingomonas, marked by the production of IL-2. NK cell-deficient mice show delayed clearance of Sphingomonas compared to that by immunocompetent mice (42, 62), emphasizing the possible role of NK cells in controlling the infection, possibly via GSL.
Structurally speaking, the sphingoid base and the fatty acid chain in all Sphingomonas species are well conserved. The sphingoid backbones of these bacterial GSLs contain dhSph's between 18 and 21 carbons long, while the length of the fatty acid chain may vary from 14 to 16 carbons and it is almost always α-hydroxylated (103). Variation occurs in the degrees of unsaturation and substitution of the sphingoid base. The sugar moiety is attached to the primary carbon of the base and may be either α-glucuronic acid or (less commonly) α-galacturonic acid (103). Methylation and reduction of the carboxyl group did not influence complement activation, suggesting that the negative charge might not be necessary for exerting its effect (66).
While glycosyltransferases are implicated in the biosynthesis of bacterial GSLs, none have been characterized to date. A putative GCS gene was found in Synechocystis species, but it did not lead to the production of GlcCer when expressed in a P. pastoris mutant lacking the GCS gene (48). Putative GCS genes have been found in Agrobacterium tumefaciens and Mesorhizobium loti, but when they were expressed in P. pastoris, the preferred sugar donor was UDP-Gal and the preferred acceptor was DAG (37). This suggests that these genes may not qualify as GCSs despite the homology, or they may not have the same substrate specificity as the fungal enzymes.
Another interesting finding linking bacterial GSLs to the host immune system involves the autoimmune diseases Guillain-Barré syndrome and miller fisher syndrome. Campylobacter jejuni has lipooligosaccharides that resemble mammalian gangliosides (complex GSLs) (30, 104, 105). Upon C. jejuni infection, the host produces anti-lipooligosaccharide antibodies that can cross-react with mammalian gangliosides, causing autoimmune neurological damage (30, 105).
PARASITIC AND PROTOZOAL GSL
Some parasites contain GSLs that have been shown to affect the host immune system as well. Antibodies to GSLs extracted from Leishmania mexicana and Leishmania braziliensis have been identified in patients with localized and diffuse cutaneous leishmaniasis, as well as in patients infected with Trypanosoma cruzi or Trypanosoma rangeli (3). Many Trypanosoma species contain neutral GSLs (97). Patients with central nervous system involvement during trypanosomiasis have been shown to have antibodies to galactocerebrosides in the cerebrospinal fluid that react with bovine galactocerebrosides (19). As in C. jejuni infection, in trypanosomiasis caused by Trypanosoma brucei, it has been suggested that anti-galactocerebroside antibodies cross-react with their mammalian counterparts (19). Furthermore, sera from patients infected with T. cruzi react with GSLs as well, although the specificity of those antibodies seems quite diverse (9).
GSLs including GlcCer from Ascaris suum have also shown significant immunomodulatory properties such as inhibition of B-cell proliferation by increasing apoptosis and altering cytokine production in vitro (23, 55). Interestingly, ganglioside presence in Entamoeba histolytica has been associated with possible antibodies found in patients with amoebiasis (85). It is also important that GSLs have been found in Plasmodium falciparum, and new therapy involves targeting GCS as antimalarial therapy, which has been shown to inhibit the growth of this intracellular protozoan in vitro (29, 73).
FUNGAL GSL
Fungal GSL synthesis has been reviewed in greater detail than the bacterial GSL pathway. Valuable reviews by Rodrigues and colleagues and by Warnecke and Heinz identify several fungi that have been found to synthesize Cer monohexosides (4, 67, 100). Although most fungi produce GlcCer, only a few produce GalCer. Interestingly, GalCer, not GlcCer, is found in S. cerevisiae, making that organism an inadequate model for the study of fungal GSL metabolism (100).
The clinical significance of fungal GlcCer was suggested when Rodrigues et al. detected immunoglobulin G1 (IgG1) antibodies to GlcCer in patients suffering from cryptococcosis, aspergillosis, histoplasmosis, paracoccidioidomycosis, and chromoblastomycosis (77). These human antibodies were able to inhibit C. neoformans growth and budding (77). A recent publication showed antibodies to acidic complex GSLs during paracoccidioidomycosis (8). Patients undergoing treatment for paracoccidioidomycosis showed first an IgM response and then IgG1, and some even had IgA and IgE production (8). Only 1 of 6 patients with invasive candidiasis and 2 of 14 patients with aspergillosis were found to have antibodies to glycosylinositolphosphorylceramide antigens (8). Although the role of these antibodies during infection is still under investigation, their presence supports the idea of an immunomodulatory role for GSLs.
CLINICAL RELEVANCE AND NEED FOR IMPROVED THERAPY
As mentioned, C. neoformans is a significant human pathogen that is often considered in the context of patients suffering from human immunodeficiency virus/AIDS. Although this is a significant problem in the United States and more so in underdeveloped countries, cryptococcosis is also a concern for many other patient populations. Individuals undergoing organ transplantation, corticosteroid treatment, or chemotherapy and patients with hematologic malignancies constitute other high-risk groups with varying degrees of susceptibility. Although C. neoformans is of little concern to immunocompetent individuals, the outbreak on Vancouver Island, where patients with functional immune systems were diagnosed with cryptococcosis (51), offers a disturbing glimpse into the possibilities of future outbreaks outside of previously defined risk groups.
CURRENT THERAPIES
Current treatments for cryptococcosis do little to alleviate these fears. The first line of treatment for pulmonary cryptococcosis is oral fluconazole. For a diagnosis of cryptococcal meningitis, amphotericin B with or without 5-flucytosine is the standard treatment (13). The problem lies in the length of treatment and the significant side effects of current antifungal treatments. Often, these treatments will continue during a treatment phase of about 10 weeks, while maintenance therapy can last 1 to 2 years or more (13). Side effects from these treatments include but are not limited to kidney failure, anemia, vomiting, shock, fever, electrolyte imbalances, hypotension/cardiac arrest (for amphotericin B, reviewed in reference 49), rash, diarrhea (for fluconazole, reviewed in reference 101), respiratory arrest, gastrointestinal hemorrhage, neuropathy, and allergic reactions (for 5-flucytosine, reviewed in reference 99). Even after these dose-limiting side effects, cryptococcosis continues to have a relatively poor prognosis and significant mortality rate (13). The need for more effective, palatable, and specific treatments is clear. Due to the multiple differences in mammalian and fungal sphingolipid pathways and their role in fungal pathogenesis, the enzymes involved may represent excellent targets for future drug therapies.
ENDOGENOUS ANTI-GlcCer ANTIBODY RESPONSE
Patients affected by cryptococcosis produce antibodies against GlcCer. Infection with C. neoformans elicits many different immune responses. Perhaps the best-characterized antibody response is that to the major capsular polysaccharide GXM (10, 28, 98). Some of these antibodies have been shown to augment current therapies, including fluconazole (65) and 5-flucytosine (28). The role of these antibodies in clearing infection varies, but their efficacy is affected by Th1 and Th2 cytokines (6, 27) and their presence is generally considered to have a more favorable prognosis (13, 24, 77).
The mechanism by which the host immune system is initially exposed to GlcCer is still unknown. The study by Rodrigues et al. (76) showed vesicles containing GXM being trafficked through the cell wall to the extracellular space. These vesicles were also found to contain GlcCer, and this remains one of the possible points during cryptococcal infection where the host immune system is exposed to this molecule. The epitope of these anti-GlcCer antibodies would also be an interesting point of study. The fact that there has been no documented autoimmune disorder from cross-reactivity of anti-cryptococcal GlcCer antibodies with human GlcCer may indicate that the portion recognized by the host immune system is one of the modifications unique to fungi. However, it is still unknown if antibodies against GlcCer do cross-react with mammalian GlcCer containing hydroxylated fatty acids. In mammalian nervous systems, α-hydroxylated GalCer's are an important component of myelin sheaths (1, 18, 60, 86). Anti-cryptococcal GlcCer antibodies have not been shown to cross-react with α-hydroxylated GalCer's; however, if this cross-reactivity does occur, it may contribute to the development of the neurological manifestations of cryptococcosis. As for future clinical applications, studies would need to be conducted to assess the potential for these anti-GlcCer antibodies to be used as diagnostic or prognostic indicators, or perhaps to chart the efficacy of other antifungal treatments. Currently, anti-GXM antibodies are being examined for their potential for passive immunization (71). The effect of anti-GlcCer antibodies on mammalian GSLs would need to be assessed before determining their effectiveness as intravenous Ig therapy or passive immunization. Additionally, there exist the same logistic concerns as for other intravenous Ig therapies, including half-life and adverse reactions to foreign antibodies. Given the hypothesis that the antibody would be helpful in preventing C. neoformans from leaving the lung, the idea of an aerosolized antibody should be explored.
POTENTIAL THERAPIES TARGETING GCS OR GlcCer
Since GlcCer is required for C. neoformans to cause infection when entering via the respiratory tract, it is hypothesized that the administration of a fungal GCS inhibitor would be effective as a preventive and/or perhaps a therapeutic agent. Although there are molecules and compounds that inhibit the mammalian GCS enzyme (83), these compounds do not inhibit the fungal enzyme (Del Poeta, unpublished), perhaps because the fungal enzyme has a substrate specificity different from that of the mammalian enzyme (75). These results suggest that the enzymatic reactions of the two enzymes are different and thus that a selective compound targeting one (fungal) but not the other (mammalian) has the potential for antifungal effects without affecting the host cells. Accordingly, specific and safe fungal GCS inhibitors could be produced to prevent or treat a fungal infection.
An alternative therapeutic strategy would be to directly target the fungal GSL. There are peptides produced by plants and insects, called defensins, that interact with fungal GlcCer's and lead to the killing of fungi in vitro (91). Additionally, anti-GlcCer antibodies have antifungal effects in vitro when added to fungal cells (77). Since GlcCer is required for C. neoformans to leave the lung, it is reasonable to hypothesize that administration of an anti-GlcCer antibody could prevent or hinder the development of cryptococcosis. This is potentially relevant for other fungal infections due to the relatively conserved structures of fungal GlcCer.
POTENTIAL FOR COMBINATION THERAPY
Alternative therapeutic strategies are critical due to general trends of pathogen resistance to drugs in modern medicine. One solution to this problem is to combine various therapies to achieve additive or synergistic effects. For instance, the administration of a Gcs1 inhibitor and/or an antibody targeting GlcCer in combination with a current antifungal agent could potentiate the effect of both. Interestingly, because of the specific function of GlcCer in promoting extracellular replication, other drug combinations may be contemplated. In addition to growing extracellularly (e.g., alveolar spaces, bloodstream), many facultatively intracellular fungi (including C. neoformans) survive and grow intracellularly within the phagolysosome of macrophages. Although an anti-GlcCer antibody treatment could be effective in reducing fungal replication in the extracellular environment, it would most likely not be able to affect the intracellular fungal component. If fungal cells, however, were forced to remain in the extracellular space, it is envisioned that an anti-GlcCer antibody could be more effective. Such a scenario could be possible by administering an antimalarial drug in combination with an anti-GlcCer antibody. Antimalarial drugs, such as chloroquine, promote extrusion of fungal cells from the intracellular to the extracellular compartment (2) because they alkalinize the phagolysosome (52, 53). Thus, treatment with an antimalarial drug would force C. neoformans cells to exist extracellularly and would make them accessible to an anti-GlcCer antibody. Importantly, the implementation of such a combination therapy (an antimalarial plus an anti-GlcCer antibody or compound) will be extremely useful in certain regions, such as Africa, where the same patients are exposed to both malaria and C. neoformans.
The need for more effective and tolerable therapies for cryptococcosis is apparent. By examining nontraditional fungal pathways such as sphingolipid synthesis, scientists and clinicians can find a rich reservoir of targets and new potential strategies for the next generation of antifungal and anticryptococcal drugs. GSLs such as GlcCer represent an exciting new direction for therapy and diagnosis due to their crucial role in pathogenesis and their well-documented interactions with the host immune system.
Supplementary Material
Acknowledgments
Special thanks to Caroline Westwater for discussions.
This work was supported in part by the Burroughs Wellcome Fund, the National Institutes of Health (grant AI56168 to M.D.P.), and the Centers of Biomedical Research Excellence Program of the National Center for Research Resources (grant RR17677, project 2 to M.D.P. and project 6 to C.L.), in part by the National Science Foundation/EPSCoR grant EPS-0132573 to C.L., and in part by C06 RR015455 from the Extramural Research Facilities Program of the National Center for Research Resources to A.B. R.R. is supported in part by a Medical Scientist Training Grant from the National Institutes of Health (GM08716). M.D.P. is a Burroughs Wellcome New Investigator in the Pathogenesis of Infectious Diseases.
Footnotes
Published ahead of print on 10 August 2007.
Supplemental material for this article may be found at http://ec.asm.org/.
REFERENCES
- 1.Alderson, N. L., M. D. Walla, and H. Hama. 2005. A novel method for the measurement of in vitro fatty acid 2-hydroxylase activity by gas chromatography-mass spectrometry. J. Lipid Res. 46:1569-1575. [DOI] [PubMed] [Google Scholar]
- 2.Alvarez, M., and A. Casadevall. 2006. Phagosome extrusion and host-cell survival after Cryptococcus neoformans phagocytosis by macrophages. Curr. Biol. 16:2161-2165. [DOI] [PubMed] [Google Scholar]
- 3.Avila, J. L., and M. Rojas. 1990. A galactosyl(α1-3)mannose epitope on phospholipids of Leishmania mexicana and L. braziliensis is recognized by trypanosomatid-infected human sera. J. Clin. Microbiol. 28:1530-1537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barreto-Bergter, E., M. R. Pinto, and M. L. Rodrigues. 2004. Structure and biological functions of fungal cerebrosides. An. Acad. Bras. Cienc. 76:67-84. [DOI] [PubMed] [Google Scholar]
- 5.Beeler, T., D. Bacikova, K. Gable, L. Hopkins, C. Johnson, H. Slife, and T. Dunn. 1998. The Saccharomyces cerevisiae TSC10/YBR265w gene encoding 3-ketosphinganine reductase is identified in a screen for temperature-sensitive suppressors of the Ca2+-sensitive csg2Δ mutant. J. Biol. Chem. 273:30688-30694. [DOI] [PubMed] [Google Scholar]
- 6.Beenhouwer, D. O., S. Shapiro, M. Feldmesser, A. Casadevall, and M. D. Scharff. 2001. Both Th1 and Th2 cytokines affect the ability of monoclonal antibodies to protect mice against Cryptococcus neoformans. Infect. Immun. 69:6445-6455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bennett, J. E., K. J. Kwon-Chung, and D. H. Howard. 1977. Epidemiologic differences among serotypes of Cryptococcus neoformans. Am. J. Epidemiol. 105:582-586. [DOI] [PubMed] [Google Scholar]
- 8.Bertini, S., A. L. Colombo, H. K. Takahashi, and A. H. Straus. 2007. Expression of antibodies directed to Paracoccidioides brasiliensis glycosphingolipids during the course of paracoccidioidomycosis treatment. Clin. Vaccine Immunol. 14:150-156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Boas, M. H., M. C. da Silva, T. G. de Oliveira, L. R. Travassos, and E. B. Bergter. 1994. Reactivity of chagasic sera with crude and highly purified glycosphingolipid fractions from Trypanosoma cruzi epimastigotes. J. Clin. Lab. Anal. 8:260-266. [DOI] [PubMed] [Google Scholar]
- 10.Casadevall, A., W. Cleare, M. Feldmesser, A. Glatman-Freedman, D. L. Goldman, T. R. Kozel, N. Lendvai, J. Mukherjee, L. A. Pirofski, J. Rivera, A. L. Rosas, M. D. Scharff, P. Valadon, K. Westin, and Z. Zhong. 1998. Characterization of a murine monoclonal antibody to Cryptococcus neoformans polysaccharide that is a candidate for human therapeutic studies. Antimicrob. Agents Chemother. 42:1437-1446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Casadevall, A., and J. R. Perfect. 1998. Cryptococcus neoformans. ASM Press, Washington, DC.
- 12.Chang, Y. C., C. M. Bien, H. Lee, P. J. Espenshade, and K. J. Kwon-Chung. 2007. Sre1p, a regulator of oxygen sensing and sterol homeostasis, is required for virulence in Cryptococcus neoformans. Mol. Microbiol. 64:614-629. [DOI] [PubMed] [Google Scholar]
- 13.Chayakulkeeree, M., and J. R. Perfect. 2006. Cryptococcosis. Infect. Dis. Clin. N. Am. 20:507-544. [DOI] [PubMed] [Google Scholar]
- 14.Chen, S., T. Sorrell, G. Nimmo, B. Speed, B. Currie, D. Ellis, D. Marriott, T. Pfeiffer, D. Parr, K. Byth, et al. 2000. Epidemiology and host- and variety-dependent characteristics of infection due to Cryptococcus neoformans in Australia and New Zealand. Clin. Infect. Dis. 31:499-508. [DOI] [PubMed] [Google Scholar]
- 15.Chen, S. C., L. C. Wright, R. T. Santangelo, M. Muller, V. R. Moran, P. W. Kuchel, and T. C. Sorrell. 1997. Identification of extracellular phospholipase B, lysophospholipase, and acyltransferase produced by Cryptococcus neoformans. Infect. Immun. 65:405-411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cheng, J., T. S. Park, A. S. Fischl, and X. S. Ye. 2001. Cell cycle progression and cell polarity require sphingolipid biosynthesis in Aspergillus nidulans. Mol. Cell. Biol. 21:6198-6209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chiba, A., S. Oki, K. Miyamoto, H. Hashimoto, T. Yamamura, and S. Miyake. 2004. Suppression of collagen-induced arthritis by natural killer T cell activation with OCH, a sphingosine-truncated analog of alpha-galactosylceramide. Arthritis Rheum. 50:305-313. [DOI] [PubMed] [Google Scholar]
- 18.Coetzee, T., K. Suzuki, and B. Popko. 1998. New perspectives on the function of myelin galactolipids. Trends Neurosci. 21:126-130. [DOI] [PubMed] [Google Scholar]
- 19.Courtioux, B., S. Bisser, P. M'Belesso, E. Ngoungou, M. Girard, A. Nangouma, T. Josenando, M. O. Jauberteau-Marchan, and B. Bouteille. 2005. Dot enzyme-linked immunosorbent assay for more reliable staging of patients with human African trypanosomiasis. J. Clin. Microbiol. 43:4789-4795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Couto, A. S., C. Caffaro, M. L. Uhrig, E. Kimura, V. J. Peres, E. F. Merino, A. M. Katzin, M. Nishioka, H. Nonami, and R. Erra-Balsells. 2004. Glycosphingolipids in Plasmodium falciparum. Presence of an active glucosylceramide synthase. Eur. J. Biochem. 271:2204-2214. [DOI] [PubMed] [Google Scholar]
- 21.Cowart, L. A., and L. M. Obeid. 2007. Yeast sphingolipids: recent developments in understanding biosynthesis, regulation, and function. Biochim. Biophys. Acta 1771:421-431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cox, G. M., J. Mukherjee, G. T. Cole, A. Casadevall, and J. R. Perfect. 2000. Urease as a virulence factor in experimental cryptococcosis. Infect. Immun. 68:443-448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Deehan, M. R., H. S. Goodridge, D. Blair, G. Lochnit, R. D. Dennis, R. Geyer, M. M. Harnett, and W. Harnett. 2002. Immunomodulatory properties of Ascaris suum glycosphingolipids—phosphorylcholine and non-phosphorylcholine-dependent effects. Parasite Immunol. 24:463-469. [DOI] [PubMed] [Google Scholar]
- 24.Diamond, R. D., and J. E. Bennett. 1974. Prognostic factors in cryptococcal meningitis. A study in 111 cases. Ann. Intern. Med. 80:176-181. [DOI] [PubMed] [Google Scholar]
- 25.Dickson, R. C. 1998. Sphingolipid functions in Saccharomyces cerevisiae: comparison to mammals. Annu. Rev. Biochem. 67:27-48. [DOI] [PubMed] [Google Scholar]
- 26.Dromer, F., S. Mathoulin-Pelissier, A. Fontanet, O. Ronin, B. Dupont, and O. Lortholary. 2004. Epidemiology of HIV-associated cryptococcosis in France (1985-2001): comparison of the pre- and post-HAART eras. AIDS 18:555-562. [DOI] [PubMed] [Google Scholar]
- 27.Feldmesser, M., A. Mednick, and A. Casadevall. 2002. Antibody-mediated protection in murine Cryptococcus neoformans infection is associated with pleotrophic effects on cytokine and leukocyte responses. Infect. Immun. 70:1571-1580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Feldmesser, M., J. Mukherjee, and A. Casadevall. 1996. Combination of 5-flucytosine and capsule-binding monoclonal antibody in the treatment of murine Cryptococcus neoformans infections and in vitro. J. Antimicrob. Chemother. 37:617-622. [DOI] [PubMed] [Google Scholar]
- 29.Gonzalez-Aseguinolaza, G., C. de Oliveira, M. Tomaska, S. Hong, O. Bruna-Romero, T. Nakayama, M. Taniguchi, A. Bendelac, L. Van Kaer, Y. Koezuka, and M. Tsuji. 2000. α-Galactosylceramide-activated Vα14 natural killer T cells mediate protection against murine malaria. Proc. Natl. Acad. Sci. USA 97:8461-8466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gregson, N. A., J. H. Rees, and R. A. Hughes. 1997. Reactivity of serum IgG anti-GM1 ganglioside antibodies with the lipopolysaccharide fractions of Campylobacter jejuni isolates from patients with Guillain-Barre syndrome (GBS). J. Neuroimmunol. 73:28-36. [DOI] [PubMed] [Google Scholar]
- 31.Gutierrez, A. L., L. Farage, M. N. Melo, R. S. Mohana-Borges, Y. Guerardel, B. Coddeville, J. M. Wieruszeski, L. Mendonca-Previato, and J. O. Previato. 2007. Characterization of glycoinositolphosphoryl ceramide structure mutant strains of Cryptococcus neoformans. Glycobiology 17:1-11C. [DOI] [PubMed] [Google Scholar]
- 32.Hajjeh, R. A., L. A. Conn, D. S. Stephens, W. Baughman, R. Hamill, E. Graviss, P. G. Pappas, C. Thomas, A. Reingold, G. Rothrock, L. C. Hutwagner, A. Schuchat, M. E. Brandt, R. W. Pinner, et al. 1999. Cryptococcosis: population-based multistate active surveillance and risk factors in human immunodeficiency virus-infected persons. J. Infect. Dis. 179:449-454. [DOI] [PubMed] [Google Scholar]
- 33.Heidler, S. A., and J. A. Radding. 2000. Inositol phosphoryl transferases from human pathogenic fungi. Biochim. Biophys. Acta 1500:147-152. [DOI] [PubMed] [Google Scholar]
- 34.Heung, L. J., A. E. Kaiser, C. Luberto, and M. Del Poeta. 2005. The role and mechanism of diacylglycerol-protein kinase C1 signaling in melanogenesis by Cryptococcus neoformans. J. Biol. Chem. 280:28547-28555. [DOI] [PubMed] [Google Scholar]
- 35.Heung, L. J., C. Luberto, and M. Del Poeta. 2006. Role of sphingolipids in microbial pathogenesis. Infect. Immun. 74:28-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Heung, L. J., C. Luberto, A. Plowden, Y. A. Hannun, and M. Del Poeta. 2004. The sphingolipid pathway regulates Pkc1 through the formation of diacylglycerol in Cryptococcus neoformans. J. Biol. Chem. 279:21144-21153. [DOI] [PubMed] [Google Scholar]
- 37.Hölzl, G., M. Leipelt, C. Ott, U. Zahringer, B. Lindner, D. Warnecke, and E. Heinz. 2005. Processive lipid galactosyl/glucosyltransferases from Agrobacterium tumefaciens and Mesorhizobium loti display multiple specificities. Glycobiology 15:874-886. [DOI] [PubMed] [Google Scholar]
- 38.Hsueh, P. R., L. J. Teng, P. C. Yang, Y. C. Chen, H. J. Pan, S. W. Ho, and K. T. Luh. 1998. Nosocomial infections caused by Sphingomonas paucimobilis: clinical features and microbiological characteristics. Clin. Infect. Dis. 26:676-681. [DOI] [PubMed] [Google Scholar]
- 39.Kawahara, K., N. Sato, K. Tsuge, and Y. Seto. 2006. Confirmation of the anomeric structure of galacturonic acid in the galacturonosyl-ceramide of Sphingomonas yanoikuyae. Microbiol. Immunol. 50:67-71. [DOI] [PubMed] [Google Scholar]
- 40.Kawahara, K., U. Seydel, M. Matsuura, H. Danbara, E. T. Rietschel, and U. Zahringer. 1991. Chemical structure of glycosphingolipids isolated from Sphingomonas paucimobilis. FEBS Lett. 292:107-110. [DOI] [PubMed] [Google Scholar]
- 41.Kaya, K., C. S. Ramesha, and G. A. Thompson, Jr. 1984. On the formation of alpha-hydroxy fatty acids. Evidence for a direct hydroxylation of nonhydroxy fatty acid-containing sphingolipids. J. Biol. Chem. 259:3548-3553. [PubMed] [Google Scholar]
- 42.Kinjo, Y., D. Wu, G. Kim, G. W. Xing, M. A. Poles, D. D. Ho, M. Tsuji, K. Kawahara, C. H. Wong, and M. Kronenberg. 2005. Recognition of bacterial glycosphingolipids by natural killer T cells. Nature 434:520-525. [DOI] [PubMed] [Google Scholar]
- 43.Kobayashi, E., K. Motoki, T. Uchida, H. Fukushima, and Y. Koezuka. 1995. KRN7000, a novel immunomodulator, and its antitumor activities. Oncol. Res. 7:529-534. [PubMed] [Google Scholar]
- 44.Koskinen, R., T. Ali-Vehmas, P. Kampfer, M. Laurikkala, I. Tsitko, E. Kostyal, F. Atroshi, and M. Salkinoja-Salonen. 2000. Characterization of Sphingomonas isolates from Finnish and Swedish drinking water distribution systems. J. Appl. Microbiol. 89:687-696. [DOI] [PubMed] [Google Scholar]
- 45.Krziwon, C., U. Zahringer, K. Kawahara, B. Weidemann, S. Kusumoto, E. T. Rietschel, H. D. Flad, and A. J. Ulmer. 1995. Glycosphingolipids from Sphingomonas paucimobilis induce monokine production in human mononuclear cells. Infect. Immun. 63:2899-2905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kwon-Chung, K. J., and J. E. Bennett. 1984. Epidemiologic differences between the two varieties of Cryptococcus neoformans. Am. J. Epidemiol. 120:123-130. [DOI] [PubMed] [Google Scholar]
- 47.Kwon-Chung, K. J., and A. Varma. 2006. Do major species concepts support one, two or more species within Cryptococcus neoformans? FEMS Yeast Res. 6:574-587. [DOI] [PubMed] [Google Scholar]
- 48.Leipelt, M., D. Warnecke, U. Zahringer, C. Ott, F. Muller, B. Hube, and E. Heinz. 2001. Glucosylceramide synthases, a gene family responsible for the biosynthesis of glucosphingolipids in animals, plants, and fungi. J. Biol. Chem. 276:33621-33629. [DOI] [PubMed] [Google Scholar]
- 49.Lemke, A., A. F. Kiderlen, and O. Kayser. 2005. Amphotericin B. Appl. Microbiol. Biotechnol. 68:151-162. [DOI] [PubMed] [Google Scholar]
- 50.Levery, S. B., M. Momany, R. Lindsey, M. S. Toledo, J. A. Shayman, M. Fuller, K. Brooks, R. L. Doong, A. H. Straus, and H. K. Takahashi. 2002. Disruption of the glucosylceramide biosynthetic pathway in Aspergillus nidulans and Aspergillus fumigatus by inhibitors of UDP-Glc:ceramide glucosyltransferase strongly affects spore germination, cell cycle, and hyphal growth. FEBS Lett. 525:59-64. [DOI] [PubMed] [Google Scholar]
- 51.Levery, S. B., M. S. Toledo, R. L. Doong, A. H. Straus, and H. K. Takahashi. 2000. Comparative analysis of ceramide structural modification found in fungal cerebrosides by electrospray tandem mass spectrometry with low energy collision-induced dissociation of Li+ adduct ions. Rapid Commun. Mass Spectrom. 14:551-563. [DOI] [PubMed] [Google Scholar]
- 52.Levitz, S. M., T. S. Harrison, A. Tabuni, and X. Liu. 1997. Chloroquine induces human mononuclear phagocytes to inhibit and kill Cryptococcus neoformans by a mechanism independent of iron deprivation. J. Clin. Investig. 100:1640-1646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Levitz, S. M., S. H. Nong, K. F. Seetoo, T. S. Harrison, R. A. Speizer, and E. R. Simons. 1999. Cryptococcus neoformans resides in an acidic phagolysosome of human macrophages. Infect. Immun. 67:885-890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Li, L., X. Tang, K. G. Taylor, D. B. DuPre, and M. C. Yappert. 2002. Conformational characterization of ceramides by nuclear magnetic resonance spectroscopy. Biophys. J. 82:2067-2080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lochnit, G., R. D. Dennis, U. Zahringer, and R. Geyer. 1997. Structural analysis of neutral glycosphingolipids from Ascaris suum adults (Nematoda:Ascaridida). Glycoconj. J. 14:389-399. [DOI] [PubMed] [Google Scholar]
- 56.Loftus, B. J., E. Fung, P. Roncaglia, D. Rowley, P. Amedeo, D. Bruno, J. Vamathevan, M. Miranda, I. J. Anderson, J. A. Fraser, J. E. Allen, I. E. Bosdet, M. R. Brent, R. Chiu, T. L. Doering, M. J. Donlin, C. A. D'Souza, D. S. Fox, V. Grinberg, J. Fu, M. Fukushima, B. J. Haas, J. C. Huang, G. Janbon, S. J. Jones, H. L. Koo, M. I. Krzywinski, J. K. Kwon-Chung, K. B. Lengeler, R. Maiti, M. A. Marra, R. E. Marra, C. A. Mathewson, T. G. Mitchell, M. Pertea, F. R. Riggs, S. L. Salzberg, J. E. Schein, A. Shvartsbeyn, H. Shin, M. Shumway, C. A. Specht, B. B. Suh, A. Tenney, T. R. Utterback, B. L. Wickes, J. R. Wortman, N. H. Wye, J. W. Kronstad, J. K. Lodge, J. Heitman, R. W. Davis, C. M. Fraser, and R. W. Hyman. 2005. The genome of the basidiomycetous yeast and human pathogen Cryptococcus neoformans. Science 307:1321-1324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Luberto, C., B. Martinez-Marino, D. Taraskiewicz, B. Bolanos, P. Chitano, D. L. Toffaletti, G. M. Cox, J. R. Perfect, Y. A. Hannun, E. Balish, and M. Del Poeta. 2003. Identification of App1 as a regulator of phagocytosis and virulence of Cryptococcus neoformans. J. Clin. Investig. 112:1080-1094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Luberto, C., D. L. Toffaletti, E. A. Wills, S. C. Tucker, A. Casadevall, J. R. Perfect, Y. A. Hannun, and M. Del Poeta. 2001. Roles for inositol-phosphoryl ceramide synthase 1 (IPC1) in pathogenesis of C. neoformans. Genes Dev. 15:201-212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Malewicz, B., J. T. Valiyaveettil, K. Jacob, H. S. Byun, P. Mattjus, W. J. Baumann, R. Bittman, and R. E. Brown. 2005. The 3-hydroxy group and 4,5-trans double bond of sphingomyelin are essential for modulation of galactosylceramide transmembrane asymmetry. Biophys. J. 88:2670-2680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Marcus, J., and B. Popko. 2002. Galactolipids are molecular determinants of myelin development and axo-glial organization. Biochim. Biophys. Acta 1573:406-413. [DOI] [PubMed] [Google Scholar]
- 61.Mare, L., R. Iatta, M. T. Montagna, C. Luberto, and M. Del Poeta. 2005. APP1 transcription is regulated by inositol-phosphorylceramide synthase 1-diacylglycerol pathway and is controlled by ATF2 transcription factor in Cryptococcus neoformans. J. Biol. Chem. 280:36055-36064. [DOI] [PubMed] [Google Scholar]
- 62.Mattner, J., K. L. Debord, N. Ismail, R. D. Goff, C. Cantu III, D. Zhou, P. Saint-Mezard, V. Wang, Y. Gao, N. Yin, K. Hoebe, O. Schneewind, D. Walker, B. Beutler, L. Teyton, P. B. Savage, and A. Bendelac. 2005. Exogenous and endogenous glycolipid antigens activate NKT cells during microbial infections. Nature 434:525-529. [DOI] [PubMed] [Google Scholar]
- 63.Miyamoto, K., S. Miyake, and T. Yamamura. 2001. A synthetic glycolipid prevents autoimmune encephalomyelitis by inducing TH2 bias of natural killer T cells. Nature 413:531-534. [DOI] [PubMed] [Google Scholar]
- 64.Morecki, S., S. Panigrahi, G. Pizov, E. Yacovlev, Y. Gelfand, O. Eizik, and S. Slavin. 2004. Effect of KRN7000 on induced graft-vs-host disease. Exp. Hematol. 32:630-637. [DOI] [PubMed] [Google Scholar]
- 65.Mukherjee, J., M. Feldmesser, M. D. Scharff, and A. Casadevall. 1995. Monoclonal antibodies to Cryptococcus neoformans glucuronoxylomannan enhance fluconazole efficacy. Antimicrob. Agents Chemother. 39:1398-1405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Münstermann, M., A. Wiese, K. Brandenburg, U. Zahringer, L. Brade, K. Kawahara, and U. Seydel. 1999. Complement activation by bacterial surface glycolipids: a study with planar bilayer membranes. J. Membr. Biol. 167:223-232. [DOI] [PubMed] [Google Scholar]
- 67.Nimrichter, L., M. D. Cerqueira, E. A. Leitao, K. Miranda, E. S. Nakayasu, S. R. Almeida, I. C. Almeida, C. S. Alviano, E. Barreto-Bergter, and M. L. Rodrigues. 2005. Structure, cellular distribution, antigenicity, and biological functions of Fonsecaea pedrosoi ceramide monohexosides. Infect. Immun. 73:7860-7868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Noverr, M. C., G. M. Cox, J. R. Perfect, and G. B. Huffnagle. 2003. Role of PLB1 in pulmonary inflammation and cryptococcal eicosanoid production. Infect. Immun. 71:1538-1547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Noverr, M. C., S. M. Phare, G. B. Toews, M. J. Coffey, and G. B. Huffnagle. 2001. Pathogenic yeasts Cryptococcus neoformans and Candida albicans produce immunomodulatory prostaglandins. Infect. Immun. 69:2957-2963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Obeid, L. M., and Y. A. Hannun. 1995. Ceramide: a stress signal and mediator of growth suppression and apoptosis. J. Cell. Biochem. 58:191-198. [DOI] [PubMed] [Google Scholar]
- 71.Oscarson, S., M. Alpe, P. Svahnberg, A. Nakouzi, and A. Casadevall. 2005. Synthesis and immunological studies of glycoconjugates of Cryptococcus neoformans capsular glucuronoxylomannan oligosaccharide structures. Vaccine 23:3961-3972. [DOI] [PubMed] [Google Scholar]
- 72.Panepinto, J., L. Liu, J. Ramos, X. Zhu, T. Valyi-Nagy, S. Eksi, J. Fu, H. A. Jaffe, B. Wickes, and P. R. Williamson. 2005. The DEAD-box RNA helicase Vad1 regulates multiple virulence-associated genes in Cryptococcus neoformans. J. Clin. Investig. 115:632-641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Pankova-Kholmyansky, I., and E. Flescher. 2006. Potential new antimalarial chemotherapeutics based on sphingolipid metabolism. Chemotherapy 52:205-209. [DOI] [PubMed] [Google Scholar]
- 74.Pinto, M. R., M. L. Rodrigues, L. R. Travassos, R. M. Haido, R. Wait, and E. Barreto-Bergter. 2002. Characterization of glucosylceramides in Pseudallescheria boydii and their involvement in fungal differentiation. Glycobiology 12:251-260. [DOI] [PubMed] [Google Scholar]
- 75.Rittershaus, P. C., T. B. Kechichian, J. C. Allegood, A. H. Merrill, Jr., M. Hennig, C. Luberto, and M. Del Poeta. 2006. Glucosylceramide synthase is an essential regulator of pathogenicity of Cryptococcus neoformans. J. Clin. Investig. 116:1651-1659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Rodrigues, M. L., L. Nimrichter, D. L. Oliveira, S. Frases, K. Miranda, O. Zaragoza, M. Alvarez, A. Nakouzi, M. Feldmesser, and A. Casadevall. 2007. Vesicular polysaccharide export in Cryptococcus neoformans is a eukaryotic solution to the problem of fungal trans-cell wall transport. Eukaryot. Cell 6:48-59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Rodrigues, M. L., L. R. Travassos, K. R. Miranda, A. J. Franzen, S. Rozental, W. de Souza, C. S. Alviano, and E. Barreto-Bergter. 2000. Human antibodies against a purified glucosylceramide from Cryptococcus neoformans inhibit cell budding and fungal growth. Infect. Immun. 68:7049-7060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Saito, K., N. Takakuwa, M. Ohnishi, and Y. Oda. 2006. Presence of glucosylceramide in yeast and its relation to alkali tolerance of yeast. Appl. Microbiol. Biotechnol. 71:515-521. [DOI] [PubMed] [Google Scholar]
- 79.Sakaki, T., U. Zahringer, D. C. Warnecke, A. Fahl, W. Knogge, and E. Heinz. 2001. Sterol glycosides and cerebrosides accumulate in Pichia pastoris, Rhynchosporium secalis and other fungi under normal conditions or under heat shock and ethanol stress. Yeast 18:679-695. [DOI] [PubMed] [Google Scholar]
- 80.Sawai, H., Y. Okamoto, C. Luberto, C. Mao, A. Bielawska, N. Domae, and Y. A. Hannun. 2000. Identification of ISC1 (YER019w) as inositol phosphosphingolipid phospholipase C in Saccharomyces cerevisiae. J. Biol. Chem. 275:39793-39798. [DOI] [PubMed] [Google Scholar]
- 81.Schürer, N. Y., G. Plewig, and P. M. Elias. 1991. Stratum corneum lipid function. Dermatologica 183:77-94. [DOI] [PubMed] [Google Scholar]
- 82.Selmi, C., and M. E. Gershwin. 2004. Bacteria and human autoimmunity: the case of primary biliary cirrhosis. Curr. Opin. Rheumatol. 16:406-410. [DOI] [PubMed] [Google Scholar]
- 83.Shayman, J. A., and A. Abe. 2000. Glucosylceramide synthase: assay and properties. Methods Enzymol. 311:42-49. [DOI] [PubMed] [Google Scholar]
- 84.Shea, J. M., T. B. Kechichian, C. Luberto, and M. Del Poeta. 2006. The cryptococcal enzyme inositol phosphosphingolipid-phospholipase C confers resistance to the antifungal effects of macrophages and promotes fungal dissemination to the central nervous system. Infect. Immun. 74:5977-5988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Sorice, M., T. Griggi, G. Nicodemo, T. Garofalo, M. Marangi, S. Sanguigni, S. I. Becker, and D. Mirelman. 1996. Evidence for the existence of ganglioside molecules in the antigen of Entamoeba histolytica. Parasite Immunol. 18:133-137. [DOI] [PubMed] [Google Scholar]
- 86.Stoffel, W., and A. Bosio. 1997. Myelin glycolipids and their functions. Curr. Opin. Neurobiol. 7:654-661. [DOI] [PubMed] [Google Scholar]
- 87.Takahashi, H. K., S. B. Levery, M. S. Toledo, E. Suzuki, M. E. Salyan, S. Hakomori, and A. H. Straus. 1996. Isolation and possible composition of glucosylceramides from Paracoccidioides brasiliensis. Braz. J. Med. Biol. Res. 29:1441-1444. [PubMed] [Google Scholar]
- 88.Tan, R. X., and J. H. Chen. 2003. The cerebrosides. Nat. Prod. Rep. 20:509-534. [DOI] [PubMed] [Google Scholar]
- 89.Ternes, P., S. Franke, U. Zahringer, P. Sperling, and E. Heinz. 2002. Identification and characterization of a sphingolipid Δ4-desaturase family. J. Biol. Chem. 277:25512-25518. [DOI] [PubMed] [Google Scholar]
- 90.Ternes, P., P. Sperling, S. Albrecht, S. Franke, J. M. Cregg, D. Warnecke, and E. Heinz. 2006. Identification of fungal sphingolipid C9-methyltransferases by phylogenetic profiling. J. Biol. Chem. 281:5582-5592. [DOI] [PubMed] [Google Scholar]
- 91.Thevissen, K., D. C. Warnecke, I. E. Francois, M. Leipelt, E. Heinz, C. Ott, U. Zahringer, B. P. Thomma, K. K. Ferket, and B. P. Cammue. 2004. Defensins from insects and plants interact with fungal glucosylceramides. J. Biol. Chem. 279:3900-3905. [DOI] [PubMed] [Google Scholar]
- 92.Toledo, M. S., S. B. Levery, J. Glushka, A. H. Straus, and H. K. Takahashi. 2001. Structure elucidation of sphingolipids from the mycopathogen Sporothrix schenckii: identification of novel glycosylinositol phosphorylceramides with core Manα1→6Ins linkage. Biochem. Biophys. Res. Commun. 280:19-24. [DOI] [PubMed] [Google Scholar]
- 93.Toledo, M. S., S. B. Levery, A. H. Straus, E. Suzuki, M. Momany, J. Glushka, J. M. Moulton, and H. K. Takahashi. 1999. Characterization of sphingolipids from mycopathogens: factors correlating with expression of 2-hydroxy fatty acyl (E)-Δ3-unsaturation in cerebrosides of Paracoccidioides brasiliensis and Aspergillus fumigatus. Biochemistry 38:7294-7306. [DOI] [PubMed] [Google Scholar]
- 94.Toledo, M. S., S. B. Levery, A. H. Straus, and H. K. Takahashi. 2000. Dimorphic expression of cerebrosides in the mycopathogen Sporothrix schenckii. J. Lipid Res. 41:797-806. [PubMed] [Google Scholar]
- 95.Toledo, M. S., S. B. Levery, E. Suzuki, A. H. Straus, and H. K. Takahashi. 2001. Characterization of cerebrosides from the thermally dimorphic mycopathogen Histoplasma capsulatum: expression of 2-hydroxy fatty N-acyl (E)-Δ3-unsaturation correlates with the yeast-mycelium phase transition. Glycobiology 11:113-124. [DOI] [PubMed] [Google Scholar]
- 96.Toledo, M. S., E. Suzuki, S. B. Levery, A. H. Straus, and H. K. Takahashi. 2001. Characterization of monoclonal antibody MEST-2 specific to glucosylceramide of fungi and plants. Glycobiology 11:105-112. [DOI] [PubMed] [Google Scholar]
- 97.Uemura, A., S. Watarai, Y. Kushi, T. Kasama, Y. Ohnishi, and H. Kodama. 2006. Analysis of neutral glycosphingolipids from Trypanosoma brucei. Vet. Parasitol. 140:264-272. [DOI] [PubMed] [Google Scholar]
- 98.Vecchiarelli, A., and A. Casadevall. 1998. Antibody-mediated effects against Cryptococcus neoformans: evidence for interdependency and collaboration between humoral and cellular immunity. Res. Immunol. 149:321-333. [DOI] [PubMed] [Google Scholar]
- 99.Vermes, A., H. J. Guchelaar, and J. Dankert. 2000. Flucytosine: a review of its pharmacology, clinical indications, pharmacokinetics, toxicity and drug interactions. J. Antimicrob. Chemother. 46:171-179. [DOI] [PubMed] [Google Scholar]
- 100.Warnecke, D., and E. Heinz. 2003. Recently discovered functions of glucosylceramides in plants and fungi. Cell. Mol. Life Sci. 60:919-941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Washton, H. 1989. Review of fluconazole: a new triazole antifungal agent. Diagn. Microbiol. Infect. Dis. 12:229S-233S. [DOI] [PubMed] [Google Scholar]
- 102.Waterman, S. R., M. Hacham, G. Hu, X. Zhu, Y. D. Park, S. Shin, J. Panepinto, T. Valyi-Nagy, C. Beam, S. Husain, N. Singh, and P. R. Williamson. 2007. Role of a CUF1/CTR4 copper regulatory axis in the virulence of Cryptococcus neoformans. J. Clin. Investig. 117:794-802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Yabuuchi, E., Y. Kosako, N. Fujiwara, T. Naka, I. Matsunaga, H. Ogura, and K. Kobayashi. 2002. Emendation of the genus Sphingomonas Yabuuchi et al. 1990 and junior objective synonymy of the species of three genera, Sphingobium, Novosphingobium and Sphingopyxis, in conjunction with Blastomonas ursincola. Int. J. Syst. Evol. Microbiol. 52:1485-1496. [DOI] [PubMed] [Google Scholar]
- 104.Yuki, N., and T. Miyatake. 1998. Guillain-Barre syndrome and Fisher's syndrome following Campylobacter jejuni infection. Ann. N. Y. Acad. Sci. 845:330-340. [DOI] [PubMed] [Google Scholar]
- 105.Yuki, N., T. Taki, F. Inagaki, T. Kasama, M. Takahashi, K. Saito, S. Handa, and T. Miyatake. 1993. A bacterium lipopolysaccharide that elicits Guillain-Barre syndrome has a GM1 ganglioside-like structure. J. Exp. Med. 178:1771-1775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Zigmond, E., S. Preston, O. Pappo, G. Lalazar, M. Margalit, Z. Shalev, L. Zolotarov, D. Friedman, R. Alper, and Y. Ilan. 2007. Beta-glucosylceramide: a novel method for enhancement of natural killer T lymphoycte plasticity in murine models of immune-mediated disorders. Gut 56:82-89. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.