Abstract
Class I histone deacetylases (HDACs) regulate DNA-templated processes such as transcription. They act both at specific loci and more generally across global chromatin, contributing to acetylation patterns that may underlie large-scale chromatin dynamics. Although hypoacetylation is correlated with highly condensed chromatin, little is known about the contribution of individual HDACs to chromatin condensation mechanisms. Using the ciliated protozoan Tetrahymena thermophila, we investigated the role of a specific class I HDAC, Τhd1p, in the reversible condensation of global chromatin. In this system, the normal physiological response to cell starvation includes the widespread condensation of the macronuclear chromatin and general repression of gene transcription. We show that the chromatin in Thd1p-deficient cells failed to condense during starvation. The condensation failure correlated with aberrant hyperphosphorylation of histone H1 and the overexpression of CDC2, encoding the major histone H1 kinase. Changes in the rate of acetate turnover on core histones and in the distribution of acetylated lysines 9 and 23/27 on histone H3 isoforms that were found to correlate with normal chromatin condensation were absent from Thd1p mutant cells. These results point to a role for a class I HDAC in the formation of reversible higher-order chromatin structures and global genome compaction through mechanisms involving the regulation of H1 phosphorylation and core histone acetylation/deacetylation kinetics.
The eukaryotic genome exists in two general chromatin states: highly condensed heterochromatin and less-condensed euchromatin. The fluctuation of euchromatin between various degrees of compaction is thought to serve important regulatory roles for processes such as replication and transcription, and the study of factors affecting chromatin dynamics can provide insights into related regulatory mechanisms. The fundamental repeating unit of chromatin is the nucleosome, composed of 146 bp of DNA wrapped around an octamer of four core histone types (two H2A/H2B dimers and two H3/H4 dimers) (29). The amino termini, or “tails,” of histones that protrude from the nucleosome cores make contacts with DNA and other nucleosomes in their vicinity (32), mediating higher-order chromatin structures and underlying dynamic changes in genome packaging. The contacts mediated by histone tails may be modulated through posttranslational modifications (32, 57). It is now well established that reversible modifications, such as acetylation, phosphorylation, and methylation, are associated with chromatin structure dynamics (16). Acetylation, one of the best-characterized modifications, is strongly correlated with less-condensed, transcriptionally active chromatin. Conversely, hypoacetylated regions of the genome are often found in highly condensed heterochromatin structures (44). The acetylation states are modulated by the opposing actions of histone acetyltransferases (HATs), which catalyze the transfer of acetyl moieties to specific lysine residues, and histone deacetylases (HDACs), which remove them. The steady state of chromatin acetylation in any genomic region can be maintained by different acetylation/deacetylation (turnover) kinetics rates, and distinct classes of histones that differ in these rates correlate with genomic regions of various transcriptional competencies (8).
Most HDACs identified to date fall into three phylogenetic classes, depending on their homology to the Saccharomyces cereviseae deacetylases Rpd3p (class I), Hda1 (class II), or NAD-dependent Sir2p (class III). Enzymes in these classes differ somewhat in localization, tissue-expression patterns, and lysine-specific activities (11). Class I HDACs, which are broadly conserved among eukaryotes, are commonly found in corepressor complexes, where they mediate repression by a variety of transcription factors (44). Although other factors required for HDAC-mediated gene repression have been discovered, little is known about the mechanisms of repression. One model is that histone deacetylation increases histone tail contacts with adjacent nucleosomes or with DNA, forming local structures that are refractory to transcription. There is evidence that histone deacetylation promotes the folding of nucleosomal arrays into more complex structures (3, 56, 61). This may be due to changes in overall charge or, alternatively, to the creation of posttranslational modification patterns that are recognized by downstream effector proteins which, in turn, mediate condensation. Recently, the latter model has gained substantial experimental support, and specific roles for HDACs in this context are beginning to emerge (16, 44).
An increasing body of evidence implicates HDACs as having critical roles in regional heterochromatin formation and maintenance (22, 24). In mammalian cells, the inhibition of HDAC activities by treatment with trichostatin A disrupts pericentromeric heterochromatin and centromere function (34, 52). In Schizosaccharomyces pombe, individual HDACs of all classes (I, II, and III) facilitate the formation of heterochromatin surrounding the centromeres and the mating type loci where they cooperate with methyltransferases to produce specific histone modification patterns required to recruit heterochromatin proteins (17, 21, 45, 67). Two modifications common to a broad range of heterochromatin domains are the methylation of lysine 9 and/or lysine 27 on histone H3 (H3MeLys9/27) (12, 24). Less is known about the contribution of individual HDACs to widespread chromatin condensation. Trichostatin A treatment of premeiotic and premitotic cells showed that HDAC activity is crucial for chromosome condensation (7, 33), but the individual HDAC enzymes involved in these processes have not yet been identified. The idea that HDACs help to establish a baseline acetylation state across global chromatin has some experimental support. In addition to acting locally at specific sites, the class I enzyme Rpd3p in yeast was shown to act more globally across larger domains and all types of chromatin (30, 60). Thus, class I HDACs might participate in forming more widespread, dynamic chromatin architectures, some possibly sensitive to cell cycle or environmental signals.
The ciliated protozoan Tetrahymena thermophila provides a unique opportunity to study the role of HDACs in inducible and reversible chromatin condensation. Each cell has a transcriptionally active, highly acetylated macronucleus and a transcriptionally inert, unacetylated micronucleus (1, 5, 58). Interspersed in the macronuclear chromatin are bodies of highly condensed chromatin (called chromatin bodies) whose size and number correlate with the degree of genome compaction, which fluctuates in response to nutrient availability. During prolonged nutrient starvation, macronuclear chromatin condenses while the sizes of chromatin bodies increase (26). These chromatin changes coincide with cell cycle arrest and decreased transcription of many genes (49), characteristics that are reversed upon refeeding. At the molecular level, starvation-induced chromatin condensation and gene regulation is dependent on the dephosphorylation of histone H1 and the presence of the heterochromatin protein Hhp1, an HP1-like protein that is enriched in chromatin bodies (25, 46).
We previously described Tetrahymena Thd1p, a class I HDAC that is selectively recruited to developing new macronuclei (64) and is important for the integrity of macronuclear chromatin in logarithmically dividing cells (66). Cells deficient in Thd1p contain higher amounts of macronuclear DNA, large extrusion bodies, and enlarged nucleoli (66). In the present study, we addressed whether Thd1p plays a role in normal global chromatin condensation during cell starvation. We report that Thd1p-deficient cells are defective in chromatin condensation and present evidence that Thd1p normally promotes condensation through mechanisms involving H1 phosphorylation, acetate turnover on core histones, and redistribution of specific acetylated lysine residues on histone H3 isoforms.
MATERIALS AND METHODS
Strains, cell culture, and starvation.
Tetrahymena thermophila strain CU428 [Chx/Chx(mp-r)VII] was used as the wild-type strain. Unless otherwise noted, for all experiments CU428 cells were grown at 30°C with shaking in 1% (wt/vol) enriched proteose peptone (SPP) (20) liquid medium to mid-logarithmic phase (cell density of 2 × 105 to 5 × 105 cells/ml). The mutant ΔTHD1 cells otherwise possessed the same genetic background as CU428 (66). ΔTHD1 cells have a partial gene replacement in the polyploid amitotic macronucleus that must be maintained under selection with paromomycin. For use in all experiments, ΔTHD1 cells were pregrown to mid-logarithmic phase in SPP containing 300 μg/ml paromomycin (Sigma Chemical Co.) and then transferred to medium lacking paromomycin and grown for an additional five population doublings (to reduce the possible effects of pregrowth in paromomycin) to a cell density within the range of 2 × 105 cells/ml to 5 × 105 cells/ml. This treatment was previously shown to be effective in sustaining at least a fivefold reduction in the expression of Thd1p, something that was confirmed by immunoblotting throughout the course of this study. For prolonged cell starvation, mid-logarithmically growing CU428 and ΔTHD1 cells were resuspended in 10 mM Tris, pH 7.4, at a cell density of 2.5 × 105 cells/ml and incubated at 30°C (no shaking) for 24 h.
DAPI staining.
CU428 and ΔTHD1 cells in mid-logarithmic growth (2 × 105 to 5 × 105 cells/ml) or following prolonged starvation were fixed in 4% paraformaldehyde as described previously (51), except that cells were dropped onto slides lacking polylysine coating prior to staining with 0.1 μg/ml 4′,6′-diamidino-2-phenylindole dihydrochloride (DAPI; Sigma Chemical Co). The stained cells were visualized by fluorescence microscopy. The macronuclear diameters were measured by using an ocular micrometer, and calculations of the DAPI-stained area were made based on this measurement.
Ultrastructural analyses.
Logarithmically growing cells (5 × 106 cells/ml) were washed in 40 mM HEPES buffer (pH 7.5) and fixed for 1 h at room temperature in glutaraldehyde (2.5% in 0.1 M sodium cacodylate, pH 7.2). The samples were dehydrated by three incubations in 100% ethanol and slowly infiltrated and embedded in Spurr's resin and polymerized at 70°C for 8 h. Sections were visualized by using a JEOL 1010 transmission electron microscope at 80 kV. For chromatin body analysis, the images captured on negatives were digitized by using a Hewlett-Packard ScanJet scanner. The sizes and numbers of chromatin bodies per unit area in multiple macronuclei were determined by using NIH Image 1.61 software.
Northern hybridization and reverse transcriptase PCR.
Total RNA was isolated from 5 × 106 cells (growing and starved) by extraction with Trizol reagent (Invitrogen) according to the manufacturer's protocol. The total RNA (12 μg) from each strain was resolved by electrophoresis on a 1% agarose gel containing 2.2 M formaldehyde. The RNA was transferred to Hybond N nylon membrane (Amersham) according to a standard protocol (42). Hybridization was carried out in a hybridization oven at 42°C in buffer containing 50% formamide (42). For the CYP1 probe, plasmid pBC11 carrying CYP1 (generously provided by Kathleen Karrer) was digested with PstI. A 790-kb fragment from the CYP1 gene was gel purified by using a gel extraction kit (QIAGEN) and labeled with [32P]dATP by a random priming method (Prime It II; Stratagene).
For reverse transcriptase-PCR, cDNA was synthesized from total RNA by using a Protoscript first strand cDNA synthesis kit (New England Biolabs) and an oligo-dT primer according to the manufacturer's protocol. The exon-spanning primers used for amplification are presented in Table 1. The quantitation of the PCR results was carried out by using ImageQuant software to analyze the agarose gel images. The values for the band intensities were normalized to those of the control bands (RAD51) for each condition.
TABLE 1.
Primers used for PCR
| Gene | Oligonucleotide sequences |
|---|---|
| CDC2 | 5′-AATAAATAATCTGACAGTAAAAATGG-3′ |
| 5′-TTGAGGCTTCAAATCTCTATGAAG-3′ | |
| RAD51 | 5′-ACCAAGTTATGTCCTAAGTC-3′ |
| 5′-GACTTAGGACATAACTTGGTTG-3′ | |
| CYP1 | 5′-ACAGTAACCCTAATAACACC-3′ |
| 5′-GACTTAGGACATAACTTGGTTG-3′ | |
| THD1 | 5′-CCATTGGATGCTACAATTATG-3′ |
| 5′-CATGATGCAAACCACCTGA-3′ | |
| THD2 | 5′-GTTTATTTTGATATCTGCTG-3′ |
| 5′-CTAAATGCGATCCTTTAATTC-3′ | |
| THD3 | 5′-TAAGAAAGAATGGATGCACTGC-3′ |
| 5′-ATGGTGTACATCTATATCTATG-3′ | |
| THD4 | 5′-TCATCAGGTGCTGTAATTTAATC-3′ |
| 5′-ATTAATGTTAAATCCATTTCCTAC-3′ | |
| THD6 | 5′-CATGAATATGGTGATGACTTC-3′ |
| 5′-CTCAGTAATAGGAATATTTCC-3′ | |
| tSIR2 | 5′-GTAAGAATTCTCAAGTTGCG-3′ |
| 5′-TGAATAAATCAACTAAAGCTGC-3′ | |
| tGCN5 | 5′-TGCTGATAACTTTGCTATTG-3′ |
| 5′-ACTCCAGGAATATCTGAAG-3′ | |
| tNuA4 | 5′-ATGTTATTGCAACCATGAAG-3′ |
| 5′-TCTATCAAGCTCATTTTGAG-3′ |
Histone acetylation and phosphorylation analyses.
The macronuclei were isolated by using a previously described procedure (20). The purity of the isolated macronuclei was assessed by staining the nuclei with 0.2% methyl green (final concentration) and counting the numbers of macronuclei and micronuclei under the light microscope (2). For all experiments, the macronuclei preparations contained <10% micronuclei, which corresponds to <0.8% micronuclear chromatin due to the difference in genome copy number in the micronucleus and macronucleus. The total histones were acid extracted from 5 × 106 macronuclei isolated from growing and starved wild-type and ΔTHD1 cells, and histone H1 was selectively extracted from the mixture by using perchloric acid as previously described (65). Acetylated and phosphorylated histone isoforms were resolved by acid-urea polyacrylamide gel electrophoresis (PAGE) and visualized by staining with Coomassie brilliant blue R-250, as previously described (65). In some cases, the amount of protein loaded from each strain was equalized according to protein quantitation at A280 using a Beckman DU530 UV/VIS spectrophotometer. The quantitation of histone H1 phosphorylated isoforms was performed by using ImageQuant software to analyze the immunoblots. The density of each isoform band as a fraction of the total was determined.
Immunoblot analysis.
Growing or starved CU428 and ΔTHD1 cells (105) were collected by centrifugation and lysed by incubation in 30 μl of sodium dodecyl sulfate (SDS) gel loading buffer (50 mM Tris-Cl, pH 6.8, 100 mM dithiothreitol, 2% [wt/vol] SDS, 0.1% bromophenol blue, 10% [vol/vol] glycerol) and heated in a boiling water bath for 5 min. The proteins were resolved by SDS-PAGE on a 12% polyacrylamide gel, transferred to nitrocellulose membrane, and probed with a 1:5,000 dilution of anti-pan-acetyl H3 or anti-pan-acetyl H4 polyclonal antiserum (gift from C. David Allis). Histones resolved on acid-urea polyacrylamide gels (see “Histone acetylation and phosphorylation analyses”) were transferred to nitrocellulose or polyvinylidene difluoride membrane and probed with the indicated antibodies. In some cases, wide lanes were cut into multiple strips. The strips were individually probed with one of the following rabbit polyclonal antisera (Upstate Biotechnology, Inc., Millipore, or a gift from C. David Allis): anti-H3(pan), 1:5,000; anti-H3Ac(Lys9), 1:40,000 (06-942; UBI); anti-H3Ac(Lys27), 1:20,000 (07-360; UBI); anti-H4(pan), 1:30,000; anti-H4Ac(Lys5), 1:1,000 (07-327; UBI); anti-H4Ac(Lys16), 1:1,000 (06-762; UBI); and anti-H3Me3Lys27 (07-449; Millipore). Immunoreactivity was detected by chemiluminescence using horseradish peroxidase-conjugated goat anti-rabbit immunoglobulin G (1:5,000; Amersham) and an ECL plus substrate system (Amersham) following the manufacturer's protocol.
Acetate turnover assay.
The macronuclei were isolated by using a previously described protocol, except that butyrate was omitted from the isolation buffers (20). For the acetate-labeling experiments, a published protocol (50) was slightly modified: 5 × 105 macronuclei were incubated in acetyltransferase buffer (50 mM Tris, pH 8.0, 1 mM phenylmethylsulfonyl fluoride, 0.5 mM dithiothreitol) and 1.0 μM final concentration of [3H]acetyl coenzyme A ([3H]acetyl-CoA) (3.6 Ci/mmol; Amersham Biosciences) for 30 min at 30°C in a total volume of 50 μl. After 30 min, the nuclei were collected by centrifugation and washed once with acetyltransferase buffer. For analysis by acid-urea-PAGE, histones were first acid extracted following a published protocol before being resolved by acid-urea-PAGE (65).
RESULTS
Macronuclear chromatin fails to condense in starved ΔTHD1 cells.
Macronuclear chromatin normally condenses upon starvation, coincident with decreased global gene expression and cellular metabolic activity. These events require linker histone H1 for the regulation of specific genes and an HP1-like protein (Hhp1p) (25, 26, 46, 47). Previously, cells at least fivefold reduced in Thd1p protein levels (“ΔTHD1” cells) were characterized during logarithmic growth in nutrient-rich medium (66). In the present study, the effect of Thd1p was examined under starvation conditions. In order to assess the level of chromatin condensation, the area occupied by macronuclear chromatin in ΔTHD1 cells was compared to that in wild-type cells. Prior to starvation (during logarithmic growth) and following prolonged nutrient starvation (22 to 24 h), cells were fixed and the chromatin was stained with DAPI and examined by fluorescence microscopy. As shown in Fig. 1A, the DAPI-stained area in ΔTHD1 cells failed to decrease with cell starvation as it did for wild-type cells. Instead, the average area in starved ΔTHD1 cells increased (∼1.5-fold) compared to that in logarithmically growing ΔTHD1 cells (Fig. 1C). The difference in the macronuclear chromatin areas of starved wild-type and mutant cells was also observed in the same microscopic field by conjugating a wild-type strain of a different mating type [CU427; Chx/Chx(cy-s)VI] with ΔTHD1 cells (Fig. 1B). The quantitation of DAPI-stained areas of cells as represented in Fig. 1A revealed that the average area of the starved mutant macronuclei was ∼twofold larger than that of the wild-type macronuclei (Fig. 1C). As previously shown in logarithmically growing cells, there was no observed difference in the average DAPI-stained area between ΔTHD1 and wild-type cells; however, there was more size variability in the mutant cells, probably due to more variable DNA content (66). Consistent with the results of previous studies demonstrating that Thd1p is excluded from the micronucleus, no micronuclear phenotypes were observed in starved or growing ΔTHD1 cells (Fig. 1A) (66).
FIG. 1.
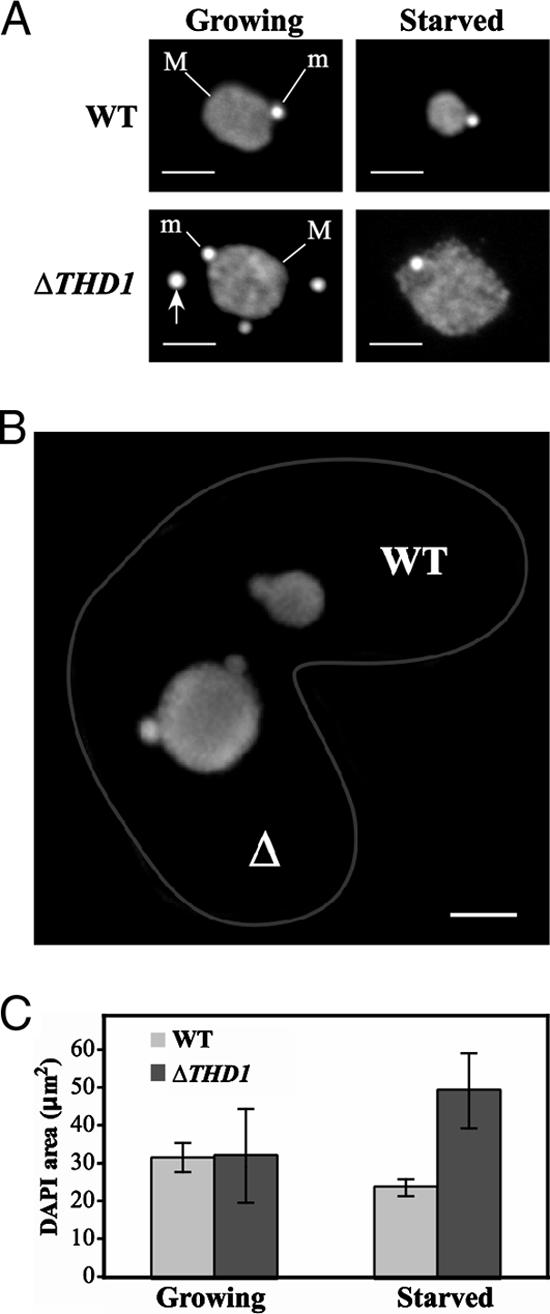
Macronuclear chromatin fails to condense in starved ΔTHD1 cells. (A) Vegetatively growing or starved wild-type (WT) or ΔTHD1 cells were fixed in paraformaldehyde, stained with the DNA-specific dye DAPI, and visualized by fluorescence microscopy. M, macronucleus; m, micronucleus. Bars, 5 μm. Arrow points to an extrusion body commonly observed in mutant cells (66). (B) A conjugating pair of wild-type (WT) and ΔTHD1 (Δ) cells. Cells were fixed, stained with DAPI, and visualized by fluorescence microscopy. The wild-type cell was distinguished by the presence of ingested fluorescent beads. Cell borders are enhanced by a gray line. Bar, 5 μm. (C) The DAPI-stained areas of 150 macronuclei in unmated cells from each strain were calculated. Bars represent the average area; standard error bars are shown. WT, wild-type.
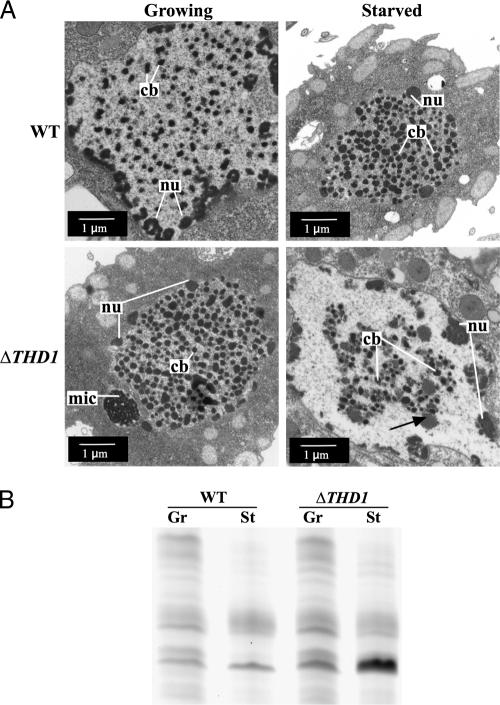
Scattered throughout the macronucleus are regions of heterochromatin called chromatin bodies that are easily identified by their high densities in transmission electron microscopy. Chromatin body size normally increases as part of the physiological response to prolonged starvation, suggesting that more chromatin is assembled into these heterochromatin structures, concomitant with the decrease in total macronuclear area (26). To test whether the enlarged macronucleus phenotype observed in starved ΔTHD1 cells correlated with smaller chromatin body size, ultrastructural analyses using transmission electron microscopy were performed on growing and starved cells (Fig. 2A). The comparison of the average sizes of chromatin bodies in growing wild-type and ΔTHD1 cells revealed no quantifiable difference (Table 2). Nucleoli, which appear as less electron-dense structures at the nuclear periphery, were carefully excluded from this analysis. In contrast to wild-type cells, chromatin bodies in starved ΔTHD1 cells failed to enlarge (Table 2). The average chromatin body area in starved ΔTHD1 cells was ∼1.5-fold smaller than that of starved wild-type cells, and there were large electron-poor regions lacking chromatin bodies altogether; similar areas were not observed in growing cells (Fig. 2A). Calculating the average area occupied by chromatin bodies as a fraction of the total chromatin area (chromatin body number × chromatin body average size/macronuclear area) revealed that, in wild-type cells, chromatin bodies occupied approximately twice the area in the starved condition as in the nutrient-rich growing condition, a significant increase in the fraction of total chromatin area comprised of chromatin bodies (P < 0.0001). However, in ΔTHD1 cells, the opposite occurred; the fraction of chromatin area occupied by chromatin bodies in the starved condition was significantly less than in the growing condition (∼threefold; P < 0.0001) (Table 2). Together, these data from cytological and ultrastructural analyses indicate that the chromatin in ΔTHD1 cells failed to condense in response to starvation. The chromatin in starved ΔTHD1 cells appeared to decondense, suggesting that Thd1p normally mediates global chromatin condensation in response to starvation.
FIG. 2.
Macronuclear chromatin bodies fail to enlarge in starved ΔTHD1 cells. (A) Growing and starved wild-type (WT) and ΔTHD1 cells were fixed and processed for ultrastructural analysis by transmission electron microscopy. A representative macronucleus is shown for each strain and condition. nu, nucleolus; cb, chromatin body; mic, micronucleus. Black arrow in bottom right panel indicates a putative nucleolus. (B) ΔTHD1 cells respond to starvation conditions in ways similar to wild-type cells. Nuclear proteins resolved by acid-urea-PAGE and stained with Coomassie brilliant blue revealed similar starvation-induced protein expression changes in wild-type and ΔTHD1 cells. WT, wild-type; Gr, growing; St, starved.
TABLE 2.
Comparison of chromatin body size and fraction of total chromatin area
| Condition and strain of cells | Avg chromatin body size (nm2) (no. of areas measured)a | % of chromatin body areab |
|---|---|---|
| Growing WT | 14 ± 0.66 (275) | 7.4 ± 0.17 |
| Starved WT | 21 ± 0.61 (385) | 13.8 ± 0.21 |
| Growing ΔTHD1 | 13 ± 0.70 (327) | 7.8 ± 0.45 |
| Starved ΔTHD1 | 14 ± 0.63 (254) | 2.4 ± 0.25 |
Chromatin bodies in randomly selected areas within 10 macronuclear transmission electron microscopy sections were measured. The averages ± standard errors of the results are reported.
For each sample, 10 macronuclear transmission electron microscopy sections were analyzed. The averages ± standard errors of the results are reported.
It was possible that chromatin condensation failure in ΔTHD1 cells resulted from insufficient starvation due to a higher rate of cell death and the accumulation of debris, which cells can use as a nutrient source. To test this possibility, cell survival during prolonged starvation was assayed by counting cell numbers at regular intervals. No significant difference was found between mutant and wild-type cell counts over a 40-h period, indicating that cell lysis in mutant cultures was not occurring at a higher rate than normal within the 24-h starvation period when the experiments were performed (data not shown). Further, the reduction of total protein synthesis is a hallmark of starved cells. The examination of proteins by SDS-PAGE or acid-urea-PAGE and Coomassie stain showed that starvation-induced changes in the expression of many proteins were occurring normally in the mutant cells (Fig. 2B), with a few proteins being misexpressed. This result indicated that ΔTHD1 cells were capable of responding to starvation signals. Taken together, these analyses indicated that starvation conditions were sufficient for ΔTHD1 cells, and that the failure of macronuclear chromatin to condense was, instead, likely due to the disruption of a Thd1p-mediated process(es).
Steady-state histone acetylation remains the same, but acetate turnover is altered in ΔTHD1 cells.
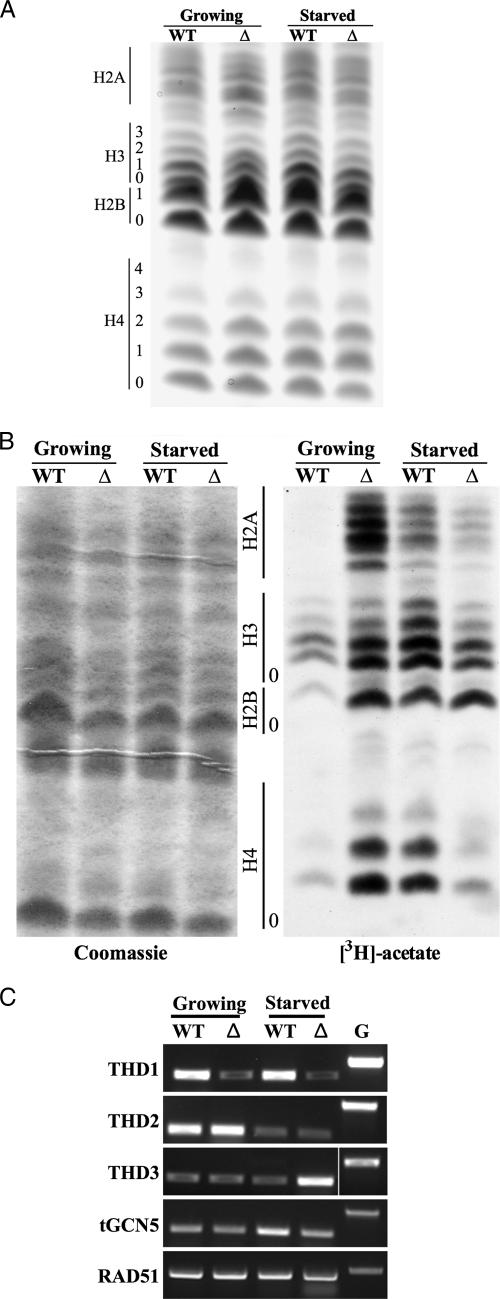
Previous work demonstrating that steady-state acetylation levels on bulk histones from growing ΔTHD1 and wild-type cells are similar prompted us to test whether the same was true in the starved condition (66). To test this possibility, the core histones were extracted from growing and starved wild-type and ΔTHD1 cells and resolved by acid-urea-PAGE and the histone isoforms were visualized by Coomassie staining. As shown in Fig. 3A, in wild-type cells there was no appreciable difference in the acetylation levels of histones H3 and H4 between growing and starved conditions. Similarly, there was no reproducible difference between mutant and wild-type cells in the overall acetylation levels of any of the core histones when compared in both growing and starved conditions; the relative proportions of different isoforms were similar in all samples (Fig. 3A). Since H2A is difficult to analyze by Coomassie stain alone, an immunoblot analysis was performed with total anti-H2A antiserum which confirmed the location of H2A molecules and revealed no difference between wild-type and mutant cells in the abundances of different isoforms (data not shown). These results were consistent with the results from immunoblot analyses of core histones resolved by SDS-PAGE using anti-acetylated histone H3 antiserum (anti-pan-acetyl H3) and anti-acetylated histone H4 (anti-pan-acetyl H4) antiserum to compare the total amounts of acetylation on histones H3 and H4 (data not shown). In sum, these analyses revealed that the steady-state level of acetylation on the core histones in mutant and wild-type cells was comparable in all conditions. There was no correlation between histone deacetylation and normal chromatin condensation in this system, and the failure of chromatin to condense in starved mutant cells did not correlate with changes in steady-state acetylation.
FIG. 3.
(A) Steady-state acetylation is unchanged in starved ΔTHD1 cells. Comparison of differentially acetylated isoforms. Core histones extracted from purified nuclei were resolved by acid-urea- PAGE and stained with Coomassie brilliant blue. The number of acetyl groups on the core histone isoforms is indicated by the numbers on the left. (B) Acetate turnover on core histones is affected in ΔTHD1 cells. Isolated macronuclei were incubated with [3H]acetyl-CoA for 30 min. Histones were then extracted, resolved by acid-urea-PAGE, stained with Coomassie brilliant blue, and subjected to autoradiography. The region of each core histone ladder is indicated. The position of the unacetylated species in each ladder is indicated by “0.” The left panel shows the Coomassie-stained and dried gel that was subjected to autoradiography. The right panel shows the autoradiograph showing the incorporation of [3H]acetate. (C) Thd1p regulates expression of HATs and HDACs. Reverse transcriptase PCR was used to assess transcript levels of several HAT and HDAC genes. The genomic lane for THD3 was relocated to the right-most position on the gel. G, genomic template; WT, wild-type; Δ, ΔTHD1.
Steady-state histone acetylation levels can be maintained by different acetate turnover rates. In general, acetylation is thought to be controlled by several kinetic classes of acetate turnover: no turnover, slow (∼30-min average half-life), and rapid turnover (∼5-min average half-life) (9, 10, 36, 58, 62, 63). There is an emerging view that differences in turnover, perhaps more than steady-state acetylation levels, may influence the transcriptional competencies of genomic loci (8, 23, 48). In Tetrahymena, histones of the various acetate turnover classes were identified in logarithmically dividing cells (36, 58, 59). To test whether differences in acetate turnover on bulk histones correlated with different chromatin states, macronuclei isolated from growing and starved ΔTHD1 and wild-type cells were isolated and incubated with [3H]acetyl-CoA for 30 min. The histones were then extracted from the labeled nuclei, resolved by acid-urea-PAGE, and fluorographed. Previously, this assay revealed that the lack of [3H]acetate incorporation into core histones, especially H2A of growing wild-type cells, was dependent on HDAC activity; the inhibition of HDAC activity by sodium butyrate permitted the rapid and stable incorporation of acetate (58). Based on this, we predicted that more acetate would incorporate into the histones of ΔTHD1 cells than into those of wild-type cells. Consistent with the results of previous reports (58), there was little to no detectable incorporation of acetate into bulk H2A species but some incorporation into H2B, H3, and H4 histones of growing wild-type cells (Fig. 3B). Consistent with our expectation, growing ΔTHD1 cells exhibited more acetate incorporation on all core histones. The experiment also revealed that, in wild-type cells alone, more acetate was incorporated into all core histones when cells were starved than in the growing condition, suggesting that, although steady-state acetylation remains the same, acetate turnover is normally altered on at least a fraction of all core histones in response to starvation (Fig. 3B). These results raise the possibility that alterations in acetate turnover on core histones are functionally linked with chromatin condensation. This possibility was supported by the results from ΔTHD1 cells. The less-condensed chromatin state in ΔTHD1 cells (starved condition) correlated with less acetate incorporation (particularly on histones H4 and H2A) than in the more-condensed chromatin state (nutrient-rich growing condition). Together, these results suggest that the acetate turnover rates on histones from bulk chromatin (especially H4 and H2A) are related to the chromatin condensation states, and that Thd1p is involved in regulating turnover.
The acetate-labeling results suggested that the balance of specific HDAC and HAT activities normally changes in response to starvation. To test the possibility that Thd1p regulates acetate turnover through affecting the expression of other HDAC or HAT genes, the transcript levels of other confirmed HDACs (E. A. Wiley, unpublished data) or proteins that have high homology to yeast HDACs or HATs were assessed by reverse transcriptase PCR. Transcripts from the confirmed HDAC gene called THD2 and the putative HDAC genes THD4 and THD6 (Wiley, unpublished) were reduced (∼two- to threefold) in starved wild-type cells in comparison to the levels in growing cells, while the transcription of a putative SIR2 homolog remained unchanged (Fig. 3C and data not shown). The respective transcript levels from these genes were not affected in ΔTHD1 cells, suggesting that their regulation was Thd1p independent. However, the class I HDAC homolog encoded by THD3 (Wiley, unpublished) was misregulated in starved ΔTHD1 cells. The transcript levels in the starved mutant cells were ∼threefold higher than in starved wild-type cells, suggesting that Thd1p regulates THD3 transcription in the starved state (Fig. 3C). As expected, THD1 expression was reduced (∼fourfold) relative to that in wild-type cells. The transcript levels from the Tetrahymena HAT GCN5 gene (tGCN5) and another putative HAT gene (encoding an NuA4 homolog) were also assessed. Both showed a similar trend in which expression was slightly increased (∼1.5-fold) in the starved wild-type cells but not in the mutant cells in comparison with expression in their growing counterparts. Overall, these results show that changes in the balance of specific HATs and HDACs are normal responses to starvation that correlate with changes in acetate turnover and chromatin condensation. For some enzymes, this regulation is dependent on Thd1p.
Distribution of acetyl moieties on histone H3 is altered in ΔTHD1 cells.
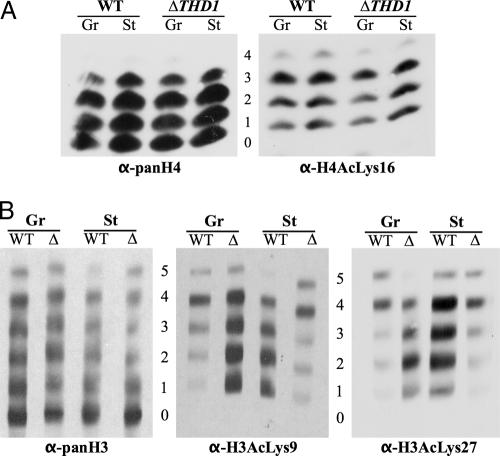
Although the steady-state histone acetylation levels were similar in all conditions, it was possible that specific acetylated lysines were differentially distributed on various histone isoforms in condensed and decondensed chromatin from mutant and wild-type cells. To explore this possibility, core histones resolved on acid-urea gels were blotted and probed with different site-specific antiacetyl antisera. In some cases, wide lanes of core histones were blotted onto nitrocellulose membrane, and multiple strips were cut from each blotted lane and probed individually with the different antisera. The antisera used were those determined to have the correct specificity for the respective sites on Tetrahymena histones by testing them on differentially acetylated histone tail peptides (data not shown). The selected antibody panel included anti-H4AcLys16, anti-H4AcLys5, anti-H3AcLys9, and anti-H3AcLys27 (note: due to some cross-reactivity of the anti-H3AcLys27 antiserum with acetylated lysine 23, we refer to this antiserum as detecting both acetylated Lys23 and -27). In the results with this panel, there was no reproducible difference between growing and starved wild-type and mutant cells with respect to acetylated H4Lys16 or H4Lys5; all isoforms in all conditions contained similar modifications (Fig. 4A and data not shown). Differences were detected in the distribution of acetylated H3Lys9 and H3Lys23/27 (Fig. 4B). In the growing wild-type cells, the acetylation of H3Lys9 was predominantly on the tetra-acetylated species. However, it was fairly equally distributed among mono-, di-, tri-, and tetra-acetylated species following starvation. This result raises the possibility that the specific distributions of H3AcLys9 are functionally related to the different states of global chromatin condensation in growing and starved cells. In support of this model, the change in H3AcLys9 distribution was reversed in ΔTHD1 cells, where starved chromatin appeared less condensed in starvation. Acetylated Lys9 in growing cells was found to be fairly equally distributed on mono-, di-, tri-, and tetra-acetylated species and then to be shifted predominantly to the tetra-acetylated species following starvation. Similar results were found with the acetylation of lysines 23/27 on histone H3 (Fig. 4B). These results suggest that a greater proportion of H3Lys9 and -23/27 acetylations on mono-, di-, and triacetylated species may be part of a molecular pathway for global chromatin condensation.
FIG. 4.
Distribution of acetyl moieties on histone H3 Lys9 and Lys27 is altered in ΔTHD1 cells. (A) Histones extracted from purified nuclei were resolved by acid-urea-PAGE, transferred to nitrocellulose membrane, and probed first with anti-H4AcLys16 antiserum. The blot was then stripped and reprobed with anti-panH4 to detect the positions of all H4 molecules. Numbers indicate the number of acetyl modifications on the H4 molecules at each position. (B) Histones extracted from purified nuclei were resolved in wide lanes by acid-urea-PAGE and transferred to nitrocellulose membrane. Multiple strips were cut from each lane and probed with the indicated antibodies. Numbers indicate the number of acetyl modifications on the H3 molecules at each position. The starved ΔTHD1 strip probed with anti-H3AcLys9 was shifted down slightly in comparison to the others. WT, wild-type; Δ, ΔTHD1; Gr, growing; St, starved; α, anti.
Histone H3 Lys27 methylation is increased in starved macronuclei.
Trimethylation of H3Lys9 or H3Lys27 is an epigenetic marker for heterochromatin formation in several systems, including Tetrahymena (24, 31). An increasing body of evidence suggests that HDACs are important for the recruitment and activity of some histone methyltransferases. In Tetrahymena, H3Lys9 methylation is observed only during a stage late in the sexual conjugation pathway (54), but there is a low level of H3Lys27 trimethylation in the macronuclei of growing cells, localized primarily to chromatin bodies (31, 55). To determine whether abnormal distribution of H3Lys27 methylation correlated with the inability of ΔTHD1 cells to condense chromatin, the normal distribution of this marker was first tested in wild-type cells and then compared with the distribution in ΔTHD1 cells. Histones extracted from wild-type and ΔTHD1 cells (growing and starved) were resolved by acid-urea-PAGE and analyzed by immunoblotting with an antibody specific to trimethylated H3Lys27 (α-H3Me3Lys27). As shown in Fig. 5, the wild-type cells exhibited trimethylation of H3Lys27 primarily on the unacetylated H3 molecules under growing conditions. Following prolonged starvation, trimethylation also occurred on the mono- and diacetylated isoforms. A similar distribution pattern was observed for growing and starved ΔTHD1 cells, suggesting that Thd1p does not affect chromatin condensation through a mechanism involving the redistribution of lysine 27 trimethylation. The increase in the signal on mutant compared to wild-type histones was due to higher loading of histones in that lane (Fig. 5, α-panH3 panel).
FIG. 5.
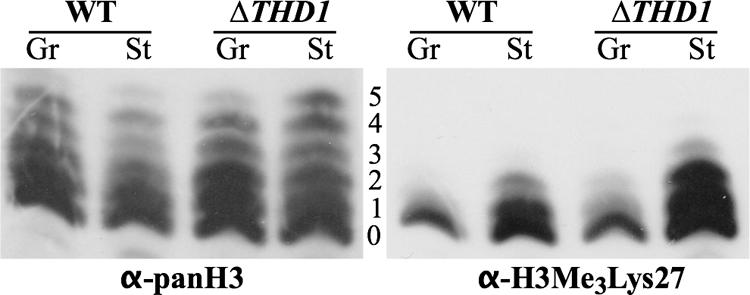
Histone H3 lysine 27 methylation increases with cell starvation. Histones extracted from purified nuclei were resolved by acid-urea-PAGE, transferred to nitrocellulose membrane, and probed first with anti-H3Me327 antiserum. The blot was then stripped and reprobed with anti-panH3 to show the positions of all H3 isoforms. The numbers indicate the number of acetyl modifications on the H3 molecules at each position. WT, wild-type; Gr, growing; St, starved; α, anti.
Linker histone H1 is hyperphosphorylated in starved ΔTHD1 cells.
Macronuclear linker histone H1 is highly phosphorylated in growing Tetrahymena cells and extensively dephosphorylated coincident with chromatin condensation during starvation (1, 19, 39). To test whether the phosphorylated state of histone H1 molecules differed in starved ΔTHD1 cells, H1 molecules were extracted and then analyzed by acid-urea-PAGE (65). In this gel system, the ladder of H1 molecules results from differences in the numbers of phosphoryl modifications. Consistent with the results of previous studies, the bulk H1 histones in the wild-type cells were dephosphorylated following starvation (Fig. 6A). However, similar dephosphorylation was not observed in ΔTHD1 cells. Whereas mono- and diphosphorylated H1 in the starved wild-type cells made up ∼85% of the total H1, they comprised only ∼45% of the total H1 from the starved mutant cells. Instead, the bulk H1 molecules from starved ΔTHD1 cells were hyperphosphorylated to at least the same degree as in the growing condition (54% versus 47% tri- and tetraphosphorylated, respectively) and more highly phosphorylated than H1 from starved wild-type cells (54% versus 14% tri- and tetraphosphorylated, respectively) (Fig. 6A). Note that, consistent with the results of previous reports (66), H1 from growing ΔTHD1 cells is less phosphorylated than H1 from growing wild-type cells (55% versus 30% mono- and diphosphorylated H1, respectively). The observed failure of ΔTHD1 H1 histones to undergo starvation-induced dephosphorylation may account at least in part for the defective chromatin changes in these cells.
FIG. 6.
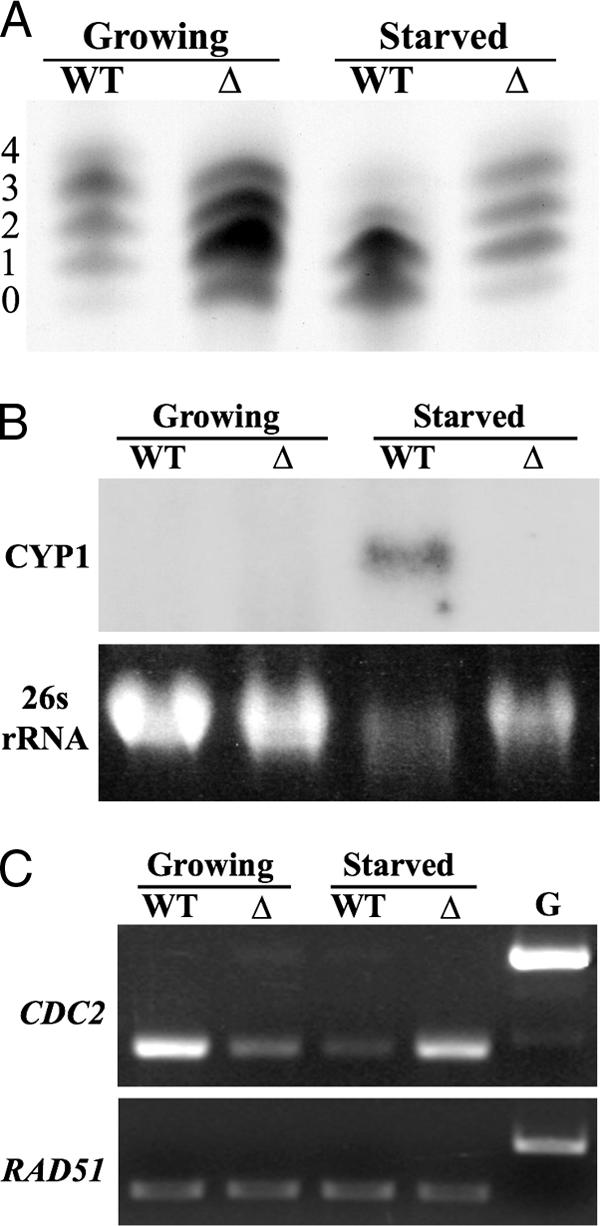
Histone H1 is hyperphosphorylated in starved ΔTHD1 cells. (A) H1 histones were isolated from core histones, resolved by acid-urea-PAGE, and stained with Coomassie brilliant blue. The numbers represent the number of phosphoryl modifications on the isoforms in each band. (B) CYP1 is not induced by starvation in ΔTHD1 cells. Total RNA from growing and starved wild-type and ΔTHD1 cells was resolved by formaldehyde-agarose gel electrophoresis and visualized by staining with ethidium bromide. RNA transferred to a nylon membrane was probed with a fragment from the CYP1 gene. The 26s rRNA stained with ethidium bromide was used as a loading control. (C) Total cDNA from each strain was used as template in PCRs with CDC2 primers or RAD51 primers as an internal control for template concentration. The amount of template used was previously determined, by dilution experiment, to yield CDC2 or RAD51 amplification products in the linear range (data not shown). G, genomic DNA used as template; WT, wild-type; Δ, THD1.
While many growth-related genes are repressed under starvation (nongrowth) conditions, a subset of genes is induced, including the cysteine protease gene CYP1 (4, 27, 49). Like macronuclear condensation, full CYP1 induction depends on the presence of chromatin proteins, such as linker histone H1 and the heterochromatin-associated protein HHP1, suggesting a role for chromatin changes (25, 47). The dephosphorylation of histone H1 is also required (13). We thus tested whether CYP1 induction was compromised in ΔTHD1 cells. The results from Northern hybridization experiments showed that, during logarithmic growth, CYP1 was repressed to the same extent in both wild-type and ΔTHD1 cells (Fig. 6B). Following prolonged starvation, the normal induction of CYP1 transcription was not observed in mutant cells. A similar result was obtained using reverse transcriptase PCR (data not shown). Immunoblot analysis revealed that the levels of histone H1 and Hhp1p in the mutant cells were comparable to the levels in the wild type, indicating that the chromatin and transcription defects in starved cells were not attributable to deficiencies in these proteins (data not shown). This result further supported the idea that normal starvation-induced chromatin changes were defective in ΔTHD1 cells.
The steady-state H1 phosphorylation levels in Tetrahymena result from the balance of opposing H1 kinase and phosphatase activities (13, 15). One known Tetrahymena H1 kinase, Cdc2p, is normally downregulated during starvation, coincident with H1 dephosphorylation by an unknown H1 phosphatase(s). We thus tested whether the high levels of H1 phosphorylation in starved mutant cells could be explained by misregulated CDC2 expression. Reverse transcriptase PCR and quantitation revealed that, indeed, the CDC2 transcript levels were ∼threefold higher in starved mutant cells than in wild-type cells (Fig. 6C). The smaller amount of CDC2 expression in growing ΔTHD1 cells compared to the amount in the wild type (∼twofold) is consistent with the previously reported H1 hypophosphorylation during logarithmic growth (66) (Fig. 6A). In general, the CDC2 transcription in mutant cells appeared to be the opposite of that in the wild type; it was upregulated instead of downregulated between growing and starved conditions. This observation suggests that Thd1p is required for the regulation of CDC2 expression in growing and starved conditions.
DISCUSSION
Hypoacetylated histones are commonly associated with more-condensed genome structures, yet little is known about the enzymes involved. Here we show that the reversible condensation of macronuclear chromatin does not correlate with histone hypoacetylation but instead may be the indirect result of HDAC action. Possible mechanistic roles for the Tetrahymena class I deacetylase enzyme Thd1p in the formation of condensed chromatin are elucidated. Our results suggest that Thd1p acts upstream of other histone modifications that correlate with chromatin condensation, some of which were also revealed in this study. These include the dephosphorylation of linker histone H1, the alteration of the acetate turnover rate on core histone tails, and the redistribution of histone H3Lys9 and -23/27 acetylation on differentially acetylated H3 molecules. Thd1p may influence these modifications in part through regulating the expression of other histone-modifying genes.
Thd1p is required for reversible global chromatin condensation.
Thd1p localizes to the transcriptionally active, hyperacetylated macronucleus and not to the transcriptionally silent, heterochromatic micronucleus in Tetrahymena (64). Thus, Thd1p functions within the context of an active euchromatin environment where, the current study shows, it facilitates the reversible condensation of macronuclear euchromatin. As revealed by the results using ΔTHD1 cells, a deficiency of Thd1p prevented macronuclear chromatin condensation, the normal physiological response to cell starvation (Fig. 1A, B, C). Regions of highly condensed chromatin (chromatin bodies) failed to increase in size and comprise a greater fraction of total chromatin as they do in starved wild-type cells (Table 2). Instead, transmission electron microscopy revealed large electron-poor regions within ΔTHD1 macronuclei (Fig. 2A), similar in appearance to the regions of decondensed euchromatin previously reported (37). Interestingly, the fraction of total chromatin comprised of chromatin bodies in these Thd1p-deficient cells was significantly less in the starved state than in the growing state, indicating that chromatin decondensed in response to starvation conditions, a response opposite to that in wild-type cells. These results indicate that Thd1p normally functions to condense DNA into heterochromatin-like structures in response to starvation signals.
The idea that starved chromatin is structurally different in Thd1p-deficient cells is supported by the observed lack of starvation-induced CYP1 activation (Fig. 6B), which also requires other chromatin proteins, such as linker histone H1 and the HP1-like heterochromatin-associated protein Hhp1 (25, 46). It is possible that Thd1p affects condensation indirectly through regulating the expression of such chromatin proteins. However, in Thd1p-deficient cells, the levels of Hhp1p and H1 were similar to the levels in wild-type cells, and thus, a deficiency of these proteins does not explain the lack of nuclear condensation and CYP1 activation in mutant cells (data not shown). It is also possible that the chromatin condensation proteins were mislocalized in ΔTHD1 cells. Previous reports show that HP1 interaction with HDACs in mammalian cells is important for its localization and maintenance of centromeric heterochromatin (68). However, using an immuofluorescence approach, we found no evidence supporting the mislocalization of Hhp1 in ΔTHD1 cells (data not shown). The fact that chromatin decondenses in HDAC-deficient starved cells prompts further investigation into other factors governing condensation that may cooperate with acetylation changes. Proteins that are aberrantly expressed in starved mutant cells may be good candidates (for example, the overexpressed peptide shown in Fig. 2B).
Thd1p may affect condensation through regulating acetate turnover on core histones.
The prevailing model correlating histone hypoacetylation with condensed chromatin does not appear to apply to the reversible condensation of macronuclear chromatin in Tetrahymena. Our studies on wild-type cells revealed no difference in steady-state acetylation levels on bulk histones from cells in growing and starved conditions (Fig. 3A). However, there were differences in acetate turnover (on/off) rates on all four core histones (Fig. 3B). This finding suggests a different model, invoking a role for histone acetylation/deacetylation kinetics in chromatin pathways.
In theory, steady-state acetylation levels can be maintained by a range of acetate turnover rates, and many studies have indicated that histone acetylation is highly dynamic (8, 62). It was previously shown in vegetatively growing Tetrahymena cells that, while most H2A molecules are subject to rapid acetate turnover, a fraction of H2B, H3, and H4 histones fall into slower kinetics classes (58). The current study correlates starvation (condensation) with greater acetate incorporation onto all core histones from bulk chromatin. Although the change in turnover on H2A was the most pronounced, the other core histones were affected as well. Our data could also result from changes in the rate of histone turnover. However, we think this is unlikely because the histone H3 and H4 transcript levels were similar in the wild-type and mutant cells (data not shown). The fact that more acetate incorporation into starved than into growing wild-type cells was not reflected by Coomassie staining of bulk histones from these samples (Fig. 3A) can be explained in a couple of ways. One is that the steady-state acetylation (reflected by Coomassie stain) is actually increased in starvation but only on a fraction of histones that is too small to detect by staining. Alternatively, the balance of HAT and HDAC activities is altered such that acetate turnover rates are affected but steady-state levels remain the same. In this study, we present evidence for the latter by showing that Thd1p regulates the expression of at least one other HDAC and HAT enzyme (Fig. 3C). Regardless of which is true, our results raise the possibility that increased acetate incorporation is functionally linked to starvation-induced physiological changes, such as global chromatin condensation and gene repression. In support of this, the level of acetate incorporation onto H4, H2A, and H3 was higher in the relatively more-condensed chromatin of growing ΔTHD1 cells than in the chromatin of starved ΔTHD1 cells (Fig. 3B). Together, these data suggest an inverse relationship between acetate incorporation and chromatin condensation. These findings relate to the results of recent studies proposing acetylation dynamics models in which the acetate turnover rate is more important than the steady-state acetylation levels for the transcriptional competency of genes. In these models, rapid turnover of acetylation at nontranscribed “poised” genes is an important determinant of the transcriptional efficiency upon gene induction; blocking turnover inhibited gene expression (23, 41). Although our study does not directly address turnover kinetics, a previous study using a similar assay deduced that more acetate incorporation indicated slower turnover kinetics (58). By these criteria, normal chromatin condensation in our study would correlate with slower acetate turnover. In sum, these results complement current models by demonstrating a potential link between turnover on global histones and global chromatin structures, which in turn, likely impact transcriptional competency.
We present evidence that Thd1p is involved in regulating the acetate turnover on at least a subset of core histones. In Thd1p-deficient cells, acetate incorporation increased in the growing condition compared with acetate incorporation in the wild type, suggesting a difference in turnover equilibriums. Whether this occurs regionally on specific subsets of histones or at specific histone lysine sites remains to be tested. Thd1p likely influences this equilibrium indirectly, in part by regulating the expression of other HDAC and HAT enzymes. Tetrahymena has 18 putative HDAC genes and 9 putative HAT genes. Testing the expression of only a subset of these genes revealed two that were misregulated in ΔTHD1 cells (Fig. 3C). Thd1p appears to affect the turnover equilibrium primarily on histones H2A and H4. Recent studies have supported the idea that globally acting class I HDACs, such as yeast Rpd3p, generate a highly dynamic equilibrium of acetylation/deacetylation reactions across bulk chromatin (28). As our results suggest a relationship between different acetylation/deacetylation equilibriums and chromatin structure dynamics, Thd1p, an Rpd3p homolog, may normally promote chromatin condensation through affecting the acetate turnover equilibrium rather than affecting steady-state acetylation. We cannot, however, rule out the possibility that Thd1p influences chromatin through other means and that the observed change in the turnover rate is instead a consequence of altered structures.
Thd1p affects distribution of H3Lys9 and H3Lys23/27 acetylation.
HDACs have been implicated in modifying chromatin by establishing combinations of histone markers that are bound by effector proteins. Although general hypoacetylation and the methylation of H3Lys9 and H3Lys27 are associated with heterochromatin (22), little is known about patterns related to reversible chromatin condensation. This study revealed a correlation between the distribution of acetyl marks on bulk histones and global genome compaction. Our data suggest that histones H3Lys9 and H3Lys23/27 are preferentially acetylated on a different set of the H3 isoforms from the condensed macronuclear chromatin of starved cells than of growing cells. The results shown in Fig. 4B showed that, in growing wild-type cells, H3Lys9 and -23/27 acetylation is found predominantly in combination with three other acetyl marks. This agrees with the results of previous studies showing that H3AcLys9 occurs in combination with other acetyl marks (on lysines 14, 18, and 23) in growing cells (6, 55), although the results of Taverna et al. show that monoacetylated lysine 9 forms may be more common than our results would suggest (55). After prolonged starvation, AcLys9 and AcLys23/27 on histones from the more-condensed chromatin were distributed fairly equally between mono-, di-, tri-, and tetra-acetylated isoforms. From this we speculate that the different acetylation distributions are mechanistically linked to the respective chromatin condensation states in growing and starved cells, i.e., that there may be distinct combinations and distributions of modifications that mark the genome for reversible condensation. This possibility is strengthened by the fact that the acetylation distribution patterns were opposite in growing and starved ΔTHD1 cells, in which the more condensed and decondensed chromatin states were also switched (Fig. 4B). Here, the distribution of AcLys9 and -23/27 predominantly on tetra-acetylated species still correlated with the less-condensed macronuclear chromatin. The differences in modification patterns observed in growing and starved wild-type cells could result from differential expression of the multiple HDACs and HATs; we have shown evidence for the regulation of some during starvation (Fig. 3C). The normal regulation of a subset of these genes appears dependent on Thd1p, and their misregulation may account for the aberrant acetylation patterns observed in ΔTHD1 cells.
Combinatorial modifications on histone H3 of growing cells are well characterized (55). Now, future work directed toward determining other modifications that correlate with macronuclear condensation during starvation, especially those in combination with H3AcLys9 and H3AcLys23/27, should yield further insight into potential molecular “signals” that direct chromatin dynamics. Trimethylation of histone H3Lys27, which is associated with heterochromatin in several systems, was recently detected in chromatin bodies within macronuclear chromatin (31, 55). Our study revealed that Lys27 trimethylation occurred predominantly on unacetylated H3 isoforms in growing cells but then appeared on mono- and diacetylated isoforms following starvation (Fig. 5). Although it is tempting to speculate that this shift in Lys27 methylation distribution is mechanistically linked to condensation, the shift was still observed in starved ΔTHD1 cells that failed to condense. Our results instead argue that the mechanism of starvation-induced Lys27 trimethylation is independent of Thd1p-mediated modifications.
Thd1p could facilitate chromatin condensation by promoting H1 dephosphorylation.
Previous studies have correlated histone H1 hyperphosphorylation with global mitotic chromosome condensation (38, 53). However, in Tetrahymena, H1 phosphorylation and mitosis do not occur simultaneously. Using seven potential phosphorylation sites (18, 35), Tetrahymena macronuclear H1 molecules are highly phosphorylated during interphase. During nuclear division, which occurs through an amitotic mechanism, little to no chromosome condensation occurs. Thus, the function of H1 phosphorylation in Tetrahymena is probably more related to the regulation of specific gene transcription. In support of this, H1 phosphorylation decreases during starvation and increases during heat shock (39). H1 dephosphorylation during starvation was shown to regulate (activate or repress) a number of genes, including CYP1 (15). Here we show that histone H1 in starved ΔTHD1 cells is hyperphosphorylated compared with that in wild-type cells (Fig. 6A). This result suggests that Thd1p normally regulates H1 phosphorylation during starvation, a possibility supported by the fact that CYP1 was also not activated in ΔTHD1 cells (Fig. 6B). Consistent with the results of previous studies on wild-type cells, our studies on ΔTHD1 cells show that H1 hyperphosphorylation correlates with relatively decondensed chromatin. In these cells, the failure to dephosphorylate H1 during starvation might prevent changes in gene expression that normally promote chromatin condensation. These results support models relating hyperphosphorylation of H1 to chromatin decondensation and transcription that are gaining popularity (13, 14, 40, 43).
The degree of H1 phosphorylation results from the combined action of H1 kinases and phosphatases. The normal dephosphorylation of H1 during starvation correlates with the decreased expression of CDC2, the gene encoding the major H1 kinase in Tetrahymena (15). Interestingly, H1 dephosphorylation (probably from the action of phosphatases) is necessary for the repression of CDC2, which then further enhances the dephosphorylated state of H1 (15). Here we show evidence that Thd1p is normally involved in the repression of CDC2 during starvation (Fig. 6C). Although Thd1p could act directly at the CDC2 promoter, it might also regulate CDC2 indirectly through promoting the expression or activity of an H1 phosphatase. Further studies are required to distinguish between these possibilities. Presently we favor the simplest model, in which Thd1p regulates CDC2 expression that, in turn, affects H1 phosphorylation and chromatin dynamics in growing and starved conditions. Regardless of the regulation pathway, these results demonstrate an indirect mechanism by which an HDAC influences the phosphorylation state of histone H1. Given the evidence implicating H1 phosphorylation in gene regulation, this finding provides another chromatin-based mechanism through which class I HDACs might regulate gene expression.
Taken together, our results supply evidence for a set of chromatin modifications governing structural changes that are sensitive to environmental conditions and that are regulated at least in part by a class I HDAC. Now better elucidated, we anticipate that this system can be used for further studies addressing the mechanistic role of HDACs in integrating signals to produce physiological responses involving chromatin and gene transcription dynamics.
Acknowledgments
We very gratefully acknowledge C. David Allis for providing laboratory space and intellectual input and for his generous contribution of many reagents that were critical for this study. We are grateful to Sue Ellen Gruber for technical assistance with all transmission electron microscopy preparations and imaging. We thank Yifan Liu for sharing results prior to publication and for many helpful discussions, Carey Wickham for assisting with statistical analyses, and Joshua Smith and Kersey Black for critical reading, commentary, and technical assistance in preparation of the manuscript.
This work was supported by National Science Foundation CAREER award 0545560 to E.A.W.
Footnotes
Published ahead of print on 22 August 2007.
REFERENCES
- 1.Allis, C. D., and M. A. Gorovsky. 1981. Histone phosphorylation in macro- and micronuclei of Tetrahymena thermophila. Biochemistry 20:3828-3833. [DOI] [PubMed] [Google Scholar]
- 2.Allis, C. D., L. G. Chicoine, R. Richman, and I. G. Schulman. 1985. Deposition-related histone acetylation in micronuclei of conjugating Tetrahymena. Proc. Natl. Acad. Sci. USA 82:8048-8052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Annunziato, A. T., L. L. Frado, R. L. Seale, and C. L. Woodcock. 1988. Treatment with sodium butyrate inhibits the complete condensation of interphase chromatin. Chromosoma 96:132-138. [DOI] [PubMed] [Google Scholar]
- 4.Calzone, R. J., V. A. Stathopoulos, D. Grass, M. A. Gorovsky, and R. C. Angerer. 1983. Regulation of protein synthesis in Tetrahymena: RNA sequence sets of growing and starved cells. J. Biol. Chem. 258:6899-6905. [PubMed] [Google Scholar]
- 5.Chicoine, L. G., and C. D. Allis. 1986. Regulation of histone acetylation during macronuclear differentiation in Tetrahymena: evidence for control at the level of acetylation and deacetylation. Dev. Biol. 116:477-485. [DOI] [PubMed] [Google Scholar]
- 6.Chicoine, L. G., I. G. Schulman, R. Richman, R. G. Cook, and C. D. Allis. 1986. Nonrandom utilization of acetylation sites in histones isolated from Tetrahymena. J. Biol. Chem. 261:1071-1076. [PubMed] [Google Scholar]
- 7.Cimini, D., M. Mattiuzzo, L. Torosantucci, and F. Degrassi. 2003. Histone hyperacetylation in mitosis prevents sister chromatid separation and produces chromosome segregation defects. Mol. Biol. Cell 14:3821-3833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Clayton, A. L., C. A. Hazzalin, and L. C. Mahadevan. 2006. Enhanced histone acetylation and transcription: a dynamic perspective. Mol. Cell 23:289-296. [DOI] [PubMed] [Google Scholar]
- 9.Cousens, L., D. Gallwitz, and B. M. Alberts. 1979. Different accessibilities in chromatin to histone acetylase. J. Biol. Chem. 254:1716-1723. [PubMed] [Google Scholar]
- 10.Covault, J., and R. Chalkley. 1980. The identification of distinct populations of acetylated histone. J. Biol. Chem. 255:9110-9116. [PubMed] [Google Scholar]
- 11.de Ruijter, A. J. M., A. H. van Gennip, H. N. Caron, S. Kemp, and A. B. P. van Kuilnburg. 2003. Histone deacetylases (HDACs): characterization of the classical HDAC family. Biochem. J. 370:737-749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dillon, N. 2004. Heterochromatin structure and function. Biol. Cell 96:631-637. [DOI] [PubMed] [Google Scholar]
- 13.Dou, Y., C. A. Mizzen, M. Abrams, C. D. Allis, and M. A. Gorovsky. 1999. Phosphorylation of linker histone H1 regulates gene expression in vivo by mimicking H1 removal. Mol. Cell 4:641-647. [DOI] [PubMed] [Google Scholar]
- 14.Dou, Y., J. Bowen, Y. Liu, and M. A. Gorovsky. 2002. Phosphorylation and an ATP-dependent process increase the dynamic exchange of H1 in chromatin. J. Cell Biol. 158:1161-1170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dou, Y., X. Song, Y. Liu, and M. A. Gorovsky. 2005. The HP1 phosphorylation state regulates expression of CDC2 and other genes in response to starvation in Tetrahymena thermophila. Mol. Cell. Biol. 25:3914-3922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fischle, W., Y. Wang, and C. D. Allis. 2003. Histone and chromatin cross-talk. Curr. Op. Cell Biol. 15:172-183. [DOI] [PubMed] [Google Scholar]
- 17.Freeman-Cook, L. L., E. B. Gomez, E. J. Spedale, J. Marlett, S. L. Forsburg, L. Pillus, and P. Laurenson. 2005. Conserved locus-specific silencing functions of Schizosaccharomysces pombe sir2+. Genetics 169:1243-1260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Garcia, B. A., S. Joshi, C. E. Thomas, R. K. Chitta, R. L. Diaz, S. A. Busby, P. C. Andrews, R. R. Ogorzalek Loo, J. Shabanowitz, N. L. Kelleher, C. A. Mizzen, C. D. Allis, and D. F. Hunt. 2006. Comprehensive phosphoprotein analysis of linker histone H1 from Tetrahymena thermophila. Mol. Cell Proteomics 5:1593-1609. [DOI] [PubMed] [Google Scholar]
- 19.Glover, C. V. C., K. J. Vavra, S. D. Guttman, and M. A. Gorovsky. 1981. Heat shock and deciliation induce phosphorylation of histone H1 in T. pyriformis. Cell 23:73-77. [DOI] [PubMed] [Google Scholar]
- 20.Gorovsky, M. A., M. C. Yao, J. B. Keevert, and G. L. Pleger. 1975. Isolation of micro- and macronuclei of Tetrahymena pyriformis. Methods Cell Biol. 9:311-327. [DOI] [PubMed] [Google Scholar]
- 21.Grewal, S. I., M. J. Bonaduce, and A. J. Klar. 1998. Histone deacetylase homologs regulate epigenetic inheritance of transcriptional silencing and chromosome segregation in fission yeast. Genetics 150:563-576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Grewal, S. I., and S. Jia. 2007. Heterochromatin revisited. Nat. Rev. Genet. 8:35-46. [DOI] [PubMed] [Google Scholar]
- 23.Hazzalin, C. A., and L. C. Mahadevan. 2005. Dynamic acetylation of all lysine 4-methylated histone H3 in the mouse nucleus: analysis at c-fos and c-jun. PLoS Biol. 3:e393. doi: 10.1371/journal.pbio.0030393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Horn, P. J., and C. L. Peterson. 2006. Heterochromatin assembly: a new twist on an old model. Chromosome Res. 14:83-94. [DOI] [PubMed] [Google Scholar]
- 25.Huang, H., J. F. Smothers, E. A. Wiley, and C. D. Allis. 1999. A nonessential HP1-like protein affects starvation-induced assembly of condensed chromatin and gene expression in macronuclei of Tetrahymena thermophila. Mol. Cell. Biol. 19:3624-3634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jeter, J. R., W. A. Pavlat, and I. L. Cameron. 1975. Changes in the nuclear acidic proteins and chromatin structure in starved and refed Tetrahymena. Exp. Cell Res. 93:79-88. [DOI] [PubMed] [Google Scholar]
- 27.Karrer, K. M., S. L. Peiffer, and M. E. DiTomas. 1993. Two distinct gene subfamilies within the family of cysteine protease genes. Proc. Natl. Acad. Sci. USA 90:3063-3067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Katan-Khaykovich, Y., and K. Struhl. 2002. Dynamics of global histone acetylation and deacetylation in vivo: rapid restoration of normal histone acetylation status upon removal of activators and repressors. Genes Dev. 16:743-752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kornberg, R. D., and Y. L. Lorch. 1999. Twenty-five years of the nucleosome, fundamental particle of the eukaryotic chromosome. Cell 98:285-294. [DOI] [PubMed] [Google Scholar]
- 30.Kurdistani, S. K., D. Robyr, S. Tavazole, and M. Grunstein. 2002. Genome-wide binding map of the histone deacetylase Rpd3 in yeast. Nat. Genet. 31:248-254. [DOI] [PubMed] [Google Scholar]
- 31.Liu, Y., S. D. Taverna, T. Muratore, J. Shabanowitz, D. Hunt, and C. D. Allis. 2007. RNAi-dependent H3K27 methylation is required for heterochromatin formation and DNA elimination in Tetrahymena. Genes Dev. 21:1520-1545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Luger, K., A. W. Mader, R. K. Richmond, D. F. Sargent, and T. J. Richmond. 1997. Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature 389:251-260. [DOI] [PubMed] [Google Scholar]
- 33.Magnaghi-Jaulin, L., and C. Jaulin. 2006. Histone deacetylase activity is necessary for chromosome condensation during meiotic maturation in Xenopus laevis. Chromosome Res. 14:319-332. [DOI] [PubMed] [Google Scholar]
- 34.Maison, C., D. Bailly, A. H. F. M. Peters, J.-P. Quivy, D. Roche, A. Taddei, M. Lachner, T. Jenuwein, and G. Almouzni. 2002. Higher-order structure in pericentric heterochromatin involves a distinct pattern of histone modification and an RNA component. Nat. Gen. 30:329-334. [DOI] [PubMed] [Google Scholar]
- 35.Mizzen, C. A., Y. Dou, Y. Liu, R. G. Cook, M. A. Gorovsky, and C. D. Allis. 1999. Identification and mutation of phosphorylation sites in a linker histone. Phosphorylation of macronuclear H1 is not essential for viability in tetrahymena. J. Biol. Chem. 274:14533-14536. [DOI] [PubMed] [Google Scholar]
- 36.Moore, M., V. Jackson, L. Sealy, and R. Chalkley. 1979. Comparative studies on highly metabolically active histone acetylation. Biochim. Biophys. Acta 561:248-260. [DOI] [PubMed] [Google Scholar]
- 37.Nilsson, J. R. 1970. Macronuclear change in Tetrahymena pyriformis during the cell cycle and in response to alterations in environmental conditions. C. R. Trav. Lab. Carlsberg 37:285-300. [PubMed] [Google Scholar]
- 38.Roberge, M., J. Th'ng, J. Hamaguchi, and E. M. Bradbury. 1990. The topoisomerase II inhibitor VM-26 induces marked changes in histone H1 kinase activity, histones H1 and H3 phosphorylation, and chromosome condensation in G2 phase and mitotic BHK cells. J. Cell Biol. 111:1753-1762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Roth, S. Y., I. G. Schulman, R. Richman, R. G. Cook, and C. D. Allis. 1988. Characterization of phosphorylation sites in histone H1 in the amitotic macronucleus of Tetrahymena during different physiological states. J. Cell Biol. 107:2473-2482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Roth, S. Y., and C. D. Allis. 1992. Chromatin condensation: does histone H1 dephosphorylation play a role? Trends Biochem. Sci. 17:93-98. [DOI] [PubMed] [Google Scholar]
- 41.Sakamoto, S., R. Potla, and A. C. Lamer. 2004. Histone deacetylase activity is required to recruit RNA polymerase II to the promoters of selected interferon-stimulated early response genes. J. Biol. Chem. 279:40362-40367. [DOI] [PubMed] [Google Scholar]
- 42.Sambrook, J., and D. W. Russell. 2002. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY.
- 43.Sarg, B., W. Helliger, H. Talasz, B. Forg, and H. H. Lindner. 2006. Histone H1 phosphorylation occurs site-specifically during interphase and mitosis. J. Biol. Chem. 281:6573-6580. [DOI] [PubMed] [Google Scholar]
- 44.Shahbazian, M. D., and M. Grunstein. 2007. Functions of site-specific histone acetylation and deacetylation. Annu. Rev. Biochem. 22:26.1-26.26. [DOI] [PubMed] [Google Scholar]
- 45.Shankaranarayana, G. D., M. R. Motamedi, D. Moazed, and S. I. Grewal. 2003. Sir2 regulates histone H3 lysine 9 methylation and heterochromatin assembly in fission yeast. Curr. Biol. 13:1240-1246. [DOI] [PubMed] [Google Scholar]
- 46.Shen, X., L. Yu, J. W. Weir, and M. A. Gorovsky. 1995. Linker histones are not essential and affect chromatin condensation in vivo. Cell 82:47-56. [DOI] [PubMed] [Google Scholar]
- 47.Shen, X., and M. A. Gorovsky. 1996. Linker histone H1 regulates specific gene expression but not global transcription in vivo. Cell 86:475-483. [DOI] [PubMed] [Google Scholar]
- 48.Spencer, V. A., and J. R. Davie. 2001. Dynamically acetylated histone association with transcriptionally active and competent genes in the avian adult β-globin gene domain. J. Biol Chem. 276:34810-34815. [DOI] [PubMed] [Google Scholar]
- 49.Stargell, L. A., K. M. Karrer, and M. A. Gorovsky. 1990. Transcriptional regulation of gene expression in Tetrahymena thermophila. Nucleic Acids Res. 18:6637-6639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Strahl, B. D., R. Ohba, R. G. Cook, and C. D. Allis. 1999. Methylation of histone H3 at lysine 4 is highly conserved and correlates with transcriptionally active nuclei in Tetrahymena. Proc. Natl. Acad. Sci. USA 96:14967-14972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Stuart, K. R., and E. S. Cole. 2000. Nuclear and cytoskeletal fluorescence microscopy techniques. Methods Cell Biol. 62:291-311. [DOI] [PubMed] [Google Scholar]
- 52.Taddei, A., C. Maison, D. Roche, and G. Almouzni. 2001. Reversible disruption of pericentromeric heterochromatin and centromere function by inhibiting deacetylases. Nat. Cell Biol. 3:114-120. [DOI] [PubMed] [Google Scholar]
- 53.Talasz, H., W. Helliger, B. Puschendorf, and H. Lindner. 1996. In vivo phosphorylation of histone H1 variants during the cell cycle. Biochemistry 35:1761-1767. [DOI] [PubMed] [Google Scholar]
- 54.Taverna, S. D., R. S. Coyne, and C. D. Allis. 2002. Methylation of histone H3 at lysine 9 targets programmed DNA elimination in Tetrahymena. Cell 110:701-711. [DOI] [PubMed] [Google Scholar]
- 55.Taverna, S. D., B. M. Ueberheide, Y. Liu, A. J. Tackett, R. L. Diaz, J. Shabanowitz, B. T. Chait, D. F. Hunt, and C. D. Allis. 2007. Long-distance combinatorial linkage between methylation and acetylation on histone H3 N termini. Proc. Natl. Acad. Sci. USA 104:2086-2091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tse, C., T. Sera, A. P. Wolffe, and J. C. Hansen. 1998. Disruption of higher-order folding by core histone acetylation dramatically enhances transcription of nucleosomal arrays by RNA polymerase III. Mol. Cell. Biol. 18:4629-4638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.van Holde, K. E. 1988. Chromatin. Springer, New York, NY.
- 58.Vavra, K. J., C. D. Allis, and M. A. Gorovsky. 1982. Regulation of histone acetylation in Tetrahymena macro- and micronuclei. J. Biol. Chem. 257:2591-2598. [PubMed] [Google Scholar]
- 59.Vavra, K. J., M. Colavito-Shepanski, and M. A. Gorovsky. 1982. Histone acetylation and the deoxyribonuclease I sensitivity of the Tetrahymena ribosomal gene. Biochemistry 21:1772-1781. [DOI] [PubMed] [Google Scholar]
- 60.Vogelauer, M., J. Wu, N. Suka, and M. Grunstein. 2000. Global histone acetylation and deacetylation in yeast. Nature 408:495-498. [DOI] [PubMed] [Google Scholar]
- 61.Walia, H., H. Y. Chen, J. M. Sun, L. T. Holth, and J. R. Davie. 1998. Histone acetylation is required to maintain the unfolded nucleosome structure associated with transcribing DNA. J. Biol. Chem. 273:14516-14522. [DOI] [PubMed] [Google Scholar]
- 62.Waterborg, J. H. 2002. Dynamics of histone acetylation in vivo. A function for acetylation turnover? Biochem. Cell Biol. 80:363-378. [DOI] [PubMed] [Google Scholar]
- 63.Waterborg, J. H. 2001. Dynamics of histone acetylation in Saccharomyces cerevisiae. Biochemistry 40:2599-2605. [DOI] [PubMed] [Google Scholar]
- 64.Wiley, E. A., R. Ohba, M. C. Yao, and C. D. Allis. 2000. Developmentally regulated Rpd3p homolog specific to the transcriptionally active macronucleus of vegetative Tetrahymena thermophila. Mol. Cell. Biol. 20:8319-8328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wiley, E. A., C. A. Mizzen, and C. D. Allis. 2000. Isolation and characterization of in vivo modified histones and an activity gel assay for identification of histone acetyltransferases. Methods Cell Biol. 62:379-394. [DOI] [PubMed] [Google Scholar]
- 66.Wiley, E. A., T. Myers, K. Parker, T. Braun, and M. C. Yao. 2005. The class I histone deacetylase Thd1p affects nuclear integrity in Tetrahymena thermophila. Eukaryot. Cell 4:981-990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Yamada, T., W. Fischle, T. Sugiyama, C. D. Allis, and S. I. Grewal. 2005. The nucleation and maintenance of heterochromatin by a histone deacetylase in fission yeast. Mol. Cell 20:173-185. [DOI] [PubMed] [Google Scholar]
- 68.Zhang, C. L., T. A. McKinsey, and E. N. Olson. 2002. Association of class II histone deacetylases with heterochromatin protein 1: potential role for histone methylation in control of muscle differentiation. Mol. Cell. Biol. 22:7302-7312. [DOI] [PMC free article] [PubMed] [Google Scholar]