Abstract
TAC1 (for transcriptional activator of CDR genes) is critical for the upregulation of the ABC transporters CDR1 and CDR2, which mediate azole resistance in Candida albicans. While a wild-type TAC1 allele drives high expression of CDR1/2 in response to inducers, we showed previously that TAC1 can be hyperactive by a gain-of-function (GOF) point mutation responsible for constitutive high expression of CDR1/2. High azole resistance levels are achieved when C. albicans carries hyperactive alleles only as a consequence of loss of heterozygosity (LOH) at the TAC1 locus on chromosome 5 (Chr 5), which is linked to the mating-type-like (MTL) locus. Both are located on the Chr 5 left arm along with ERG11 (target of azoles). In this work, five groups of related isolates containing azole-susceptible and -resistant strains were analyzed for the TAC1 and ERG11 alleles and for Chr 5 alterations. While recovered ERG11 alleles contained known mutations, 17 new TAC1 alleles were isolated, including 7 hyperactive alleles with five separate new GOF mutations. Single-nucleotide-polymorphism analysis of Chr 5 revealed that azole-resistant strains acquired TAC1 hyperactive alleles and, in most cases, ERG11 mutant alleles by LOH events not systematically including the MTL locus. TAC1 LOH resulted from mitotic recombination of the left arm of Chr 5, gene conversion within the TAC1 locus, or the loss and reduplication of the entire Chr 5. In one case, two independent TAC1 hyperactive alleles were acquired. Comparative genome hybridization and karyotype analysis revealed the presence of isochromosome 5L [i(5L)] in two azole-resistant strains. i(5L) leads to increased copy numbers of azole resistance genes present on the left arm of Chr 5, among them TAC1 and ERG11. Our work shows that azole resistance was due not only to the presence of specific mutations in azole resistance genes (at least ERG11 and TAC1) but also to their increase in copy number by LOH and to the addition of extra Chr 5 copies. With the combination of these different modifications, sophisticated genotypes were obtained. The development of azole resistance in C. albicans is therefore a powerful instrument for generating genetic diversity.
Azoles belong to a class of antifungals that are widely used for the treatment of fungal diseases and especially those caused by Candida albicans. Since azoles are fungistatic drugs for C. albicans, cells repetitively exposed to these antifungals adapt to the drug pressure and eventually become azole resistant. In C. albicans, the occurrence of azole resistance has been observed in different patient groups, mostly in human immunodeficiency virus (HIV)-positive patients with oropharyngeal candidiasis (45). Azole resistance mechanisms have been investigated at the molecular level by several authors (1, 42, 55) and fall into different categories. First, alterations such as point mutations or upregulation of the gene encoding the target of azoles, an enzyme (Erg11p) involved in ergosterol biosynthesis, can occur. Among the several nucleotide polymorphisms observed in separate ERG11 alleles, at least 12 mutations have been associated with azole resistance when separate alleles were expressed in Saccharomyces cerevisiae (1, 29). The majority of these mutations alters the binding of azoles to Erg11p (21, 22, 24, 25, 43). The upregulation of ERG11 has also been observed in several azole-resistant clinical isolates (2, 17, 24, 34, 53). That ERG11 upregulation confers azole resistance was clearly demonstrated by increasing the ERG11 copy number using a replicating vector (14). ERG11 in C. albicans is at least under the control of UPC2, which is a functional homologue of two C2H2 zinc finger transcription factors encoded by UPC2 and ECM22 in S. cerevisiae (28, 48, 51). ERG11 is inducible by fluconazole (FLC) exposure, but UPC2 disruption abolishes this property (19, 28, 48, 50).
Increased drug efflux through enhanced expression of multidrug transporter genes is also involved in azole resistance in C. albicans. This mechanism can combine with ERG11-dependent resistance mechanisms, as demonstrated by the analysis of sequential isolates exposed repeatedly to azole over time (18, 35, 43, 53). Multidrug transporters belong to both the ABC (ATP-binding cassette) transporter family and the major facilitator superfamily. Among the different ABC transporters and major facilitator superfamily members investigated in C. albicans, only Cdr1p/Cdr2p and Mdr1p have been involved in azole resistance by upregulation of the corresponding genes (1, 5).
The understanding of the transcriptional regulation of CDR1 and CDR2 has progressed in the past few years by the identification of both cis- and trans-acting elements. CDR1 and CDR2 overproduction is the most frequent azole resistance mechanism in C. albicans (34). Upregulation of CDR1 and CDR2 can be achieved transiently in vitro by treating C. albicans azole-susceptible cells with different drugs such as estradiol, progesterone, or fluphenazine. A cis-acting element, the drug response element (DRE), has been characterized in the promoters of both CDR1 and CDR2 (13). The DRE sequence (5′-CGGAA/TATCGGATA-3′) is crucial for the upregulation of these genes in azole-resistant strains but also for the transient upregulation of both genes in the presence of drugs. In addition to this element, other cis-acting elements were dissected, such as two basal responsive elements (33, 14), a negative regulatory element (17), and two steroid response elements (33). trans-acting factors regulating CDR1 and CDR2 were reported recently. Chen et al. (8) described a potential activator of CDR1 through screening of a C. albicans genomic library in an S. cerevisiae strain containing a CDR1 promoter/lacZ fusion reporter system. CaNDT80 is a homologue of a meiosis-specific transcription factor in S. cerevisiae (8). Deletion of CaNDT80 in C. albicans conferred hypersensibility to azoles and decreased the inducible expression of CDR1. Recently, our laboratory discovered a transcription factor belonging to the family of zinc finger proteins with a Zn2Cys6 motif encoded by TAC1 (for transcriptional activator of CDR genes) (11). Tac1p is able to bind in vitro to the DRE, which contains two CGG triplets typical for DNA-binding sites of Zn2Cys6 transcription factors. Tac1p was shown to be responsible for transient upregulation of CDR genes in azole-susceptible strains in the presence of inducers (11). Interestingly, TAC1 is located only 14 kb from the mating-type-like (MTL) locus on the left arm of chromosome 5 (Chr 5), where ERG11 is also located. We previously identified TAC1 hyperactive alleles from clinical azole-resistant strains, which, in contrast to wild-type alleles, conferred constitutive high CDR1 and CDR2 expression in a tac1Δ/Δ mutant (10). Hyperactivity of TAC1 was shown to be due to a gain-of-function (GOF) mutation, which substituted Asp977 for Asn977. Moreover, we demonstrated the codominance of hyperactive and wild-type TAC1 alleles and therefore the necessity for homozygosity at the TAC1 locus to acquire high-level azole resistance. Homozygosity at the TAC1 locus is often associated with loss of heterozygosity (LOH) at the MTL locus (10).
In the present study, we have analyzed, for the first time, the genetic and genomic events that lead to azole resistance through alteration of TAC1 and ERG11 in sequential clinical isolates. We isolated additional TAC1 alleles of matched azole-susceptible and azole-resistant C. albicans clinical isolates. We identified five additional GOF mutations conferring Tac1p hyperactivity as measured by the upregulation of CDR1 and CDR2. In parallel, we recovered ERG11 alleles with mutations previously shown to contribute to azole resistance (43). The analysis of Chr 5 in the different isolates established that the development of azole resistance occurred in a stepwise manner and that Chr 5 rearrangements lead not only to TAC1 homozygosity but also to LOH in other Chr 5 regions, including the MTL and ERG11 loci. Finally, we demonstrated that a further increase in azole resistance results from segmental aneuploidy on Chr 5 that increases copies of TAC1, MTL, and ERG11 via formation of isochromosome 5L [i(5L)]. This detailed analysis allowed us to develop a model describing successive genetic and genomic events leading to azole resistance in C. albicans. Such a model may represent a powerful basis for subsequent studies of azole resistance.
MATERIALS AND METHODS
Strains and media.
The C. albicans and S. cerevisiae strains used in this study are listed in Table 1. These strains were grown either in complete YEPD medium (1% Bacto peptone [Difco Laboratories, Basel, Switzerland], 0.5% yeast extract [Difco], and 2% glucose [Fluka, Buchs, Switzerland]) or in minimal medium (yeast nitrogen base [Difco] and 2% glucose [Fluka]). For growth on solid media, 2% agar (Difco) was added to either of the media. Escherichia coli DH5α was used as a host for plasmid constructions and propagation. DH5α was grown in LB (Luria-Bertani broth) or LB plates, supplemented with ampicillin (0.1 mg/ml) when required.
TABLE 1.
Strains used in this study
| Straina | Parental strain | Genotype | Reference |
|---|---|---|---|
| CAF2-1 | SC5314 | ura3Δ::imm434/URA3 | 15 |
| CAF4-2 | CAF2-1 | ura3Δ::imm434/ura3Δ::imm434 | 15 |
| DSY2903 | DSY2875 | tac1-1Δ::hisG/tac1-2Δ::hisG-URA-hisG | 11 |
| DSY2906 | DSY2903 | tac1-1Δ::hisG/tac1-2Δ::hisG | 11 |
| DSY2937-35 | DSY2906 | tac1-1Δ::hisG/tac1-2Δ::hisG LEU2::TAC1-1/URA3 | 10 |
| DSY347 (C33) | Related to DSY348 and DSY289 | Azole-susceptible clinical strain (MTLa/MTLα) | 44 |
| DSY348 (C82) | Azole-resistant clinical strain (MTLa/MTLα) | 44 | |
| DSY289 (C26) | Azole-resistant clinical strain (MTLa/MTLα) | 44 | |
| DSY731 | Related to DSY732 and DSY735 | Azole-susceptible clinical strain (MTLa/MTLα) | This study |
| DSY732 | Azole-resistant clinical strain (MTLα/MTLα) | This study | |
| DSY735 | Azole-resistant clinical strain (MTLα/MTLα) | This study | |
| DSY2321 | Related to DSY2322 and DSY2323 | Azole-susceptible clinical strain (MTLa/MTLα) | This study |
| DSY2322 | Azole-resistant clinical strain (MTLα/MTLα) | This study | |
| DSY2323 | Azole-resistant clinical strain (MTLα/MTLα) | This study | |
| DSY3553 (T118) | Related to DSY3565 and DSY3554 | Azole-susceptible clinical strain (MTLa/MTLα) | 12 |
| DSY3565 (D8-168) | Azole-resistant clinical strain (MTLa/MTLa) | 12 | |
| DSY3554 (D8-330) | Azole-resistant clinical strain (MTLa/MTLa) | 12 | |
| DSY3534 (isolate1) | Related to DSY3548 and DSY3549 | Azole-susceptible clinical strain (MTLa/MTLα) | 56 |
| DSY3548 (isolate16) | Azole-resistant clinical strain (MTLa/MTLα) | 56 | |
| DSY3549 (isolate17) | Azole-resistant clinical strain (MTLa/MTLα) | 56 | |
| DSY3269-451 | DSY2906 | tac1-1Δ::hisG/tac1-2Δ::hisG LEU2::TAC1-8/URA3 | This study |
| DSY3269-456 | DSY2906 | tac1-1Δ::hisG/tac1-2Δ::hisG LEU2::TAC1-9/URA3 | This study |
| DSY3016-316 | DSY2906 | tac1-1Δ::hisG/tac1-2Δ::hisG LEU2::TAC1-10/URA3 | This study |
| DSY3016-319 | DSY2906 | tac1-1Δ::hisG/tac1-2Δ::hisG LEU2::TAC1-11/URA3 | This study |
| DSY3583-4E | DSY2906 | tac1-1Δ::hisG/tac1-2Δ::hisG LEU2::TAC1-16/URA3 | This study |
| DSY3584-4B | DSY2906 | tac1-1Δ::hisG/tac1-2Δ::hisG LEU2::TAC1-17/URA3 | This study |
| DSY3585-6B | DSY2906 | tac1-1Δ::hisG/tac1-2Δ::hisG LEU2::TAC1-18/URA3 | This study |
| ACY43 | DSY2906 | tac1-1Δ::hisG/tac1-2Δ::hisG LEU2::TAC1-12/URA3 | This study |
| ACY44 | DSY2906 | tac1-1Δ::hisG/tac1-2Δ::hisG LEU2::TAC1-13/URA3 | This study |
| ACY45 | DSY2906 | tac1-1Δ::hisG/tac1-2Δ::hisG LEU2::TAC1-15/URA3 | This study |
| DSY3557-85 | DSY2906 | tac1-1Δ::hisG/tac1-2Δ::hisG LEU2::TAC1-23/URA3 | This study |
| DSY3558-87 | DSY2906 | tac1-1Δ::hisG/tac1-2Δ::hisG LEU2::TAC1-24/URA3 | This study |
| DSY3559-100 | DSY2906 | tac1-1Δ::hisG/tac1-2Δ::hisG LEU2::TAC1-25/URA3 | This study |
| DSY3156-1-12 | DSY2906 | tac1-1Δ::hisG/tac1-2Δ::hisG LEU2::TAC1-25/URA3 LEU2::TAC1-25/URA3 | This study |
| DSY3754-28 | DSY2906 | tac1-1Δ::hisG/tac1-2Δ::hisG LEU2::TAC1-19/URA3 | This study |
| DSY3754-30 | DSY2906 | tac1-1Δ::hisG/tac1-2Δ::hisG LEU2::TAC1-20/URA3 | This study |
| DSY3756-11 | DSY2906 | tac1-1Δ::hisG/tac1-2Δ::hisG LEU2::TAC1-21/URA3 | This study |
| DSY3756-13 | DSY2906 | tac1-1Δ::hisG/tac1-2Δ::hisG LEU2::TAC1-22/URA3 | This study |
| ACY13 | DSY2906 | tac1-1Δ::hisG/tac1-2Δ::hisG LEU2::TAC1-1A736V/URA3 | This study |
| ACY71 | DSY2906 | tac1-1Δ::hisG/tac1-2Δ::hisG LEU2::TAC1-1G980E/URA3 | This study |
| ACY67 | DSY2906 | tac1-1Δ::hisG/tac1-2Δ::hisG LEU2::TAC1-1T225A/URA3 | This study |
| ACY66 | DSY2906 | tac1-1Δ::hisG/tac1-2Δ::hisG LEU2::TAC1-1ΔM677/URA3 | This study |
| ACY72-1 | DSY2906 | tac1-1Δ::hisG/tac1-2Δ::hisG LEU2::TAC1-1ΔL962-N969/URA3 | This study |
Yeast transformation.
C. albicans cells were transformed as described previously (11).
Drug susceptibility testing.
Drug susceptibility testing was performed by serial dilution assays on solid agar plates and also performed using a microdilution format with microtiter plates containing twofold serial dilutions of FLC in RPMI (range, 128 to 0.06 μg/ml), as described previously (11).
Immunoblots.
C. albicans cell extracts for immunoblotting were prepared by an alkaline extraction procedure as described previously (11). Detection of Cdr1p and Cdr2p was performed as described previously (11). Signals were revealed by exposure to Kodak BioMax MR films (GE Healthcare).
Construction of TAC1 revertant strains.
The revertant strains generated in this study were obtained by transformation of the tac1Δ/Δ mutant DSY2093 with pDS178-derived plasmids containing the URA3 and LEU2 markers as described previously (13). The pDS178-derived plasmids were obtained by inserting at the BamHI and XhoI sites the PCR-amplified TAC1 open reading frame flanked by 500 bp from genomic DNAs of strains SC5314 and all the clinical strains listed in Table 1 with primers Znc2-5-BamB and Znc2-3-Xho (see Table S1 in the supplemental material for these and other cited primers). Recombinant plasmids obtained by this method that contain the distinct TAC1 alleles are listed in Table 2. For each amplified TAC1 allele, several plasmids were sequenced in order to exclude mutation artifacts due to PCR amplification.
TABLE 2.
Plasmids used in this study
| Vector | Backbone | Description | Source or reference |
|---|---|---|---|
| pBS-KS(+) | Cloning vector | Stratagene, La Jolla, CA | |
| pDS178 | pRC2312 | pRC2312-derived plasmid containing the URA3 and LEU2 markers | 13 |
| pDS1097 | pDS178 | Insertion of TAC1a from SC5314 (TAC1-1 allele) | 11 |
| pDS1227-1 | pDS178 | Insertion of TAC1 from DSY347 (TAC1-8 allele) | This study |
| pDS1227-6 | pDS178 | Insertion of TAC1 from DSY347 (TAC1-9 allele) | This study |
| pDS1151-23 | pDS178 | Insertion of TAC1 from DSY348 (TAC1-10 allele) | This study |
| pDS1151-26 | pDS178 | Insertion of TAC1 from DSY348 (TAC1-11 allele) | This study |
| pAC102 | pDS178 | Insertion of TAC1 from DSY2321 (TAC1-12 allele) | This study |
| pAC103 | pDS178 | Insertion of TAC1 from DSY2321 (TAC1-13 allele) | This study |
| pAC105 | pDS178 | Insertion of TAC1 from DSY2323 (TAC1-15 allele) | This study |
| PDS1458-4E | pDS178 | Insertion of TAC1 from DSY731 (TAC1-16 allele) | This study |
| pDS1458-4B | pDS178 | Insertion of TAC1 from DSY731 (TAC1-17 allele) | This study |
| pDS1460-6B | pDS178 | Insertion of TAC1 from DSY735 (TAC1-18 allele) | This study |
| pDS1447-1 | pDS178 | Insertion of TAC1 from DSY3534 (TAC1-19 allele) | This study |
| pDS1447-2 | pDS178 | Insertion of TAC1 from DSY3534 (TAC1-20 allele) | This study |
| pDS1446-2 | pDS178 | Insertion of TAC1 from DSY3549 (TAC1-21 allele) | This study |
| pDS1446-1 | pDS178 | Insertion of TAC1 from DSY3549 (TAC1-22 allele) | This study |
| pDS1203-1 | pDS178 | Insertion of TAC1 from DSY3553 (TAC1-23 allele) | This study |
| pDS1203-5 | pDS178 | Insertion of TAC1 from DSY3553 (TAC1-24 allele) | This study |
| pDS1204-1 | pDS178 | Insertion of TAC1 from DSY3554 (TAC1-25 allele) | This study |
| pAC141 | pBS-KS(+) | Insertion of the XhoI-BamHI fragment from pDS1097 | This study |
| pAC92 | pAC141 | Mutation of pAC141 with primers TAC1-8-C2207T-F and TAC1-8-C2207T-R, introducing the mutation A736V in TAC1-1 | This study |
| pAC97 | pDS178 | Insertion of the XhoI-BamHI fragment from pAC92 | This study |
| pAC157 | pDS178 | Insertion of the XhoI-BamHI fragment amplified with primers Zinc2-5-BAMB and gly980glu-xho-R, introducing the mutation G980E in TAC1-1 | This study |
| pAC146 | pAC141 | Mutation of pAC141 with primers Tac1-1-del18-F and Tac1-1-del18-R, introducing the deletion ΔM677 in TAC1-1 | This study |
| pAC152 | pDS178 | Insertion of the XhoI-BamHI fragment from pAC146 | This study |
| pAC147 | pAC141 | Mutation of pAC141 with primers T225A (ins-hyp)-F and T225A(ins-hyp)-R, introducing the mutation T225A in TAC1-1 | This study |
| pAC153 | pDS178 | Insertion of the XhoI-BamHI fragment from pAC147 | This study |
| pAC158 | pDS178 | Insertion of the XhoI-BamHI fragment from fusion of the two PCR fragments to delete the L962-N969 region in TAC1-1 | This study |
Insertion of an XhoI-BamHI fragment amplified with primers Zinc2-5-BAMB and Zinc2-3-Xho.
Site-directed mutagenesis of TAC1-1.
All TAC1mutations were introduced into the TAC1-1 wild-type allele. The three point mutations A736V, T225A, and ΔM677 were introduced using the QuikChange site-directed mutagenesis kit (Stratagene, Switzerland) according to the instructions of the manufacturer. For this purpose, TAC1-1 excised from pDS1097 by XhoI and BamHI was inserted into pBS-KS(+), yielding pAC141 (Table 2), which was next used as a template for PCR-mediated mutagenesis. Mutations A736V, T225A, and ΔM677 were introduced using primers pairs TAC1-8-C2207T-F and TAC1-8-C2207T-R, T225A (ins-hyp)-F and T225A (ins-hyp)-R, and Tac1-1-del18-F and Tac1-1-del18-R, respectively, thus yielding pAC92, pAC147, and pAC14 (Table 2). Mutated TAC1 alleles were next obtained by XhoI-BamHI digestion from these plasmids and ligated into pDS178 to yield pAC97, pAC153, and pAC152, respectively (Table 2). To introduce the mutation G980E, TAC1 was first amplified from pDS1097 with primers Znc2-5-BamB and gly980glu-xho-R and next introduced into pDS178 to yield pAC157. To obtain the L962-N969 deletion, the TAC1-1 5′ end was first amplified from pDS1097 with primers Znc2-5-BamB and Tac1-1-del22-R. The TAC1-1 3′ end was next amplified using primers Tac1-1-del22-F and Znc2-3-Xho. A third PCR allowed the fusion of the two previously generated PCR products, generating a mutated TAC1 allele lacking the L962-N969 region. This mutated allele was cloned into pDS178 using the XhoI-BamHI restriction sites to yield pAC158 (Table 2).
Plasmids containing either wild-type or mutated TAC1 alleles were linearized by SalI and transformed into C. albicans DSY2906 to favor integration into the genomic LEU2 locus. For each reintegration of TAC1 alleles, several independent transformants (in general four or five) were tested for different phenotypes (FLC susceptibility and Cdr1p and Cdr2p levels) and correct integration. Only a single revertant for each individual allele was selected in this study.
Counter-clamped homogeneous electric field (CHEF) analysis.
Two or three colonies were inoculated onto YEPD broth and incubated overnight at 30°C with agitation. A culture volume corresponding to 109 cells was pelleted at 3,000 × g for 5 min, and cells were washed in 5 ml 50 mM EDTA (pH 9). The pellet was resuspended in 330 μl 50 mM EDTA. Next, 110 μl of SCE (1 M sorbitol, 10 mM EDTA, 100 mM sodium citrate [pH 5.8], adjusted with citric acid) was added and then completed with 5% β-mercaptoethanol and Zymolyase 20T (100 units/ml) (solution I). The solution was gently mixed with 560 μl of GTG agarose and rapidly distributed into molds of 10 by 5 by 2 mm. The solidified plugs were immersed in solution II (450 mM EDTA, 10 mM Tris-HCl [pH 8], 7,5% β-mercaptoethanol) and incubated overnight at 37°C without agitation. Solution II was replaced by solution III (450 mM EDTA, 10 mM Tris-HCl [pH 8], 1% N-lauryl-Sarkosyl, and 150 mg/ml proteinase K). Plugs were incubated for 6 h at 65°C without agitation and kept for 10 min on ice. Finally, plugs were transferred in a 0.5 M EDTA (pH 9) solution and stored at 4°C for several months.
One-third of the plug was loaded into wells of 0.6% GTG agarose gel in 0.5% Tris-borate-EDTA. The gel was then placed into the electrophoresis chamber of a CHEF DR II (Bio-Rad, Zürich, Switzerland) apparatus. Migration was performed in 0.5% Tris-borate-EDTA at 14°C with the following steps: block 1, 60- to 120-min switch, 6 V/cm, and 120°C over 24 h; block 2, 120- to 300-min switch, 4.5 V/cm, and 120°C over 12 h.
Southern and Northern blotting.
Southern and Northern blotting was performed as described previously (44). Probes were labeled by random priming with [α-32P]dATP using the Mega Labeling kit (GE Healthcare) according to the instructions of the manufacturer. Radioactive signals were revealed by exposure to Kodak BioMax MR film (GE Healthcare). Signals obtained in blotted membranes were quantified by counting of radioactivity with the help of a Typhoon Trio (GE Healthcare). The TAC1 probe corresponds to the region located between the first ATG sequence and the PstI restriction site. ERG11, ACT1, MTLa, and MTLα1 probes were generated by PCR using primers CYP-CB and CYP-NS2, ACT1-RT-PCR-F and -R, MTLa-F and -R, and MTLalpha1-F and -R, respectively.
Reverse transcription-PCR analysis.
Reverse transcription was performed on 5 μg of total RNA using Superscript II (Invitrogen, Basel Switzerland) with random primers (Invitrogen) according to the instructions of the manufacturer. cDNA were used for SYBR green quantitative PCR (Quantifast SYBR green PCR kit; QIAGEN, Switzerland) of TAC1 mRNA normalized with transcripts of ACT1, using an ABI Prism 7000 detection system (Applied Biosystem, Rotkreuz, Switzerland).
SNP analysis and CGH.
Single-nucleotide polymorphism (SNP) microarray hybridization and comparative genomic hybridization (CGH) were performed as described previously (16, 46). The search for SNP markers not present on the microarray was performed by amplification of specific regions of Chr 5 from genomic DNA using different V5 and V3 primer pairs (see Table S1 in the supplemental material for details) followed by sequencing of the PCR products using an AB Prism 3130 genetic analyzer (Applied Biosystems, Rotkreuz, Switzerland). Sequences were analyzed for polymorphisms using the Contig Express software (Invitrogen, Basel, Switzerland). Results of whole-genome CGH performed on the five groups of C. albicans isolates are available online at http://www.chuv.ch/imul/imu_home/imu_recherche/imu_recherche_sanglard/imu_recherche_sanglard_suppldata.htm.
Analysis of MTL locus status.
The MTL locus status of each strain was analyzed by PCR amplifying the MTLa and MTLα1 genes, using the corresponding primers as listed in Table S1 in the supplemental material. MTL locus status was confirmed by Southern blot analysis on CHEF gels, using MTLa and MTLα1 as probes.
Nucleotide sequence accession numbers.
The TAC1 alleles recovered in this work are accessible under GenBank accession numbers EU054332 to EU054348.
RESULTS
Characteristics of sequential C. albicans isolates including azole-resistant strains.
In order to depict the mechanisms involved in the development of azole resistance in C. albicans, five groups of sequential related strains (groups A to E) (Table 1 and Fig. 1) were characterized. Groups A, B, and C, containing strains DSY731 to DSY735, DSY347 to DSY289, and DSY2321 to DSY2322, respectively, were isolated from HIV-positive patients with oropharyngeal candidiasis and treated mainly with FLC (Table 1) (44). Group D (DSY3534 to DSY3549) contains three strains chosen from 17 isolates originating from the same patient treated with FLC over 2 years for oropharyngeal candidiasis (Table 1) (56). Finally, group E (DSY3553 to DSY3554) was obtained by growing in vitro a laboratory strain, DSY3553/T118, for 330 generations on FLC-containing medium (12) (Table 1). DSY3565 and DSY3554 correspond to isolates of the 165th and the 330th generations, respectively. Each group contained closely related strains as documented in previous studies (44) or by complementary analysis performed with a Ca3 repetitive probe and multilocus sequence typing analysis (data not shown).
FIG. 1.
Expression levels of Cdr1p and Cdr2p in five groups of C. albicans isolates. Protein extracts of each strain were separated on sodium dodecyl sulfate-10% polyacrylamide gels and immunoblotted with rabbit polyclonal anti-Cdr1p and anti-Cdr2p as described previously (10). C. albicans strains were grown in liquid YEPD to mid-log phase and treated (+) or not treated (−) with fluphenazine (10 μg/ml) for 20 min. The origin of each strain is indicated with its corresponding FLC susceptibility and MTL locus status.
Each group contains one azole-susceptible strain (FLC MIC range, 0.125 to 0.25 μg/ml) and sequential strains with higher FLC MICs (range, 4 to 128 μg/ml) (Table 3). For convenience, the azole resistance threshold was set in this study at the value of 4 μg/ml FLC, which is significantly higher than the average MIC for azole-susceptible C. albicans isolates (0.125 to 0.25 μg/ml). In all groups, azole-susceptible strains exhibited a low level of Cdr1p and no Cdr2p when grown under noninducing conditions (Fig. 1) However, treating cells with fluphenazine for 20 min elevated the content of these two proteins in the azole-susceptible strains of each group (Fig. 1). In contrast, isolates with elevated FLC MICs (≥4 μg/ml) exhibited constitutively high Cdr1p and Cdr2p levels (Fig. 1).
TABLE 3.
Properties of C. albicans isolates linked to azole resistance
| Group | Origin or referencea | Strainb | FLC MIC (μg/ml) | MTL locus | TAC1 allele | TAC1 property | TAC1 mutation | Mode of acquisition of hyperactive alleles | ERG11 mutation | ERG11 upregulation | i(5L) formation |
|---|---|---|---|---|---|---|---|---|---|---|---|
| A | CHUV | DSY731 | 0.125 | a/α | TAC1-16 | Wild type | − | − | |||
| TAC1-17 | Wild type | − | − | ||||||||
| DSY732 | 16 | α/α | TAC1-18 | Hyperactive | ΔM677 | Chr 5 loss and duplication | − | − | |||
| DSY735 | 32 | α/α | TAC1-18 | Hyperactive | ΔM677 | Chr 5 loss and duplication | + | + | |||
| B | CHUV | DSY347 (C33) | 0.25 | a/α | TAC1-8 | Wild type | − | − | |||
| TAC1-9 | Wild type | − | − | ||||||||
| DSY348 (C82) | 16 | a/α | TAC1-10 | Hyperactive | A736V | Gene conversion in TAC1 | S405F | − | − | ||
| TAC1-11 | Hyperactive | − | − | ||||||||
| DSY289 (C26) | >128 | a/α | TAC1-10 | Hyperactive | A736V | Gene conversion in TAC1 | S405F Y132H | − | + | ||
| TAC1-11 | Hyperactive | ||||||||||
| C | CHUV | DSY2321 | 0.25 | a/α | TAC1-12 | Wild type | S405Fc | − | − | ||
| TAC1-13 | Wild type | − | − | ||||||||
| DSY2322 | 16 | α/α | TAC1-15 | Hyperactive | G980E | Mitotic recombination | S405F | − | − | ||
| DSY2323 | 32 | α/α | TAC1-15 | Hyperactive | G980E | Mitotic recombination | S405F | + | − | ||
| D | 56 | DSY3534 (isolate 1) | 0.125 | a/α | TAC1-19 | Wild type | − | − | |||
| TAC1-20 | Wild type | − | − | ||||||||
| DSY3548 (isolate 16) | 64 | a/α | TAC1-21 | Hyperactive | A736V | Single modification | R467K | − | − | ||
| TAC1-22 | Hyperactive | ΔL962-N969 | Single modification | − | − | ||||||
| DSY3549 (isolate 17) | 64 | a/α | TAC1-21 | Hyperactive | A736V | Single modification | R467K | − | − | ||
| TAC1-22 | Hyperactive | ΔL962-N969 | Single modification | − | − | ||||||
| E | 12 | DSY3553 (T118) | 0.125 | a/α | TAC1-23 | Wild type | − | − | |||
| TAC1-24 | Wild type | − | − | ||||||||
| DSY3565 (D8-168) | 4 | a/a | TAC1-25 | Hyperactive | T225A | Mitotic recombination | − | − | |||
| DSY3554 (D8-330) | 4 | a/a | TAC1-25 | Hyperactive | T225A | Mitotic recombination | − | − |
Since azole resistance is often associated with homozygosity at the MTL locus, analysis of the MTL locus status was performed. PCR and Southern analysis (see below) indicated that azole-susceptible strains were heterozygous at the MTL locus (Fig. 1). In contrast, azole-resistant strains of groups A and C were MTLα/MTLα, whereas those of group E were MTLa/MTLa. However, the azole-resistant strains of the two remaining groups, B and D, remained heterozygous at the MTL locus (Fig. 1).
Analysis of TAC1 alleles isolated from azole-susceptible and azole-resistant strains.
The high Cdr1p/Cdr2p levels in the azole-resistant strains described above were suspected to result from the occurrence of TAC1 hyperactive alleles in their genomes. Therefore, the TAC1 alleles of our set of 15 strains were next analyzed by measuring FLC MICs and Cdr1p/Cdr2p levels of individual transformants. Seventeen distinct TAC1 alleles, as deduced from sequencing analysis of cloned alleles (see Table 3 for details), were recovered from the 15 investigated strains. As expected, all strains heterozygous at the MTL locus contained two distinct TAC1 alleles, while all MTL homozygous strains carried two identical TAC1 alleles (Table 3). Among the cloned alleles, 10 (TAC1-8, -9, -12, -13, -16, -17, -19, -20, -23, and -24) were assigned to wild-type alleles (Tables 3 and 4), since they conferred upon DSY2906 (tac1Δ/Δ) the ability to upregulate CDR1 and CDR2 in the presence of fluphenazine (Fig. 2). The remaining seven alleles (TAC1-10, -11, -15, -18, -21, -22, and -25) conferred high constitutive CDR1 and CDR2 expression in the tac1Δ/Δ mutant even in the absence of fluphenazine and were therefore defined as hyperactive alleles (Fig. 2). The introduction of hyperactive alleles in the tac1Δ/Δ mutant resulted in an increase of the FLC MIC from 0.125 to 4 μg/ml (Fig. 2) and an ability to grow on FLC-supplemented medium (data not shown). In some cases, introduction of the same hyperactive allele in the tac1Δ/Δ mutant generated two different isolates with slight FLC MIC differences. For instance, as shown in Fig. 2, introduction of TAC1-25 (group E) not only increased the Cdr1p and Cdr2p levels in one strain compared to the other but also increased the FLC MIC from 4 to 16 μg/ml. Southern blot analysis indicated the presence of two TAC1-25 alleles in the more resistant isolate and only one copy in the other strain (data not shown). These results suggest that TAC1 copy number has an impact on CDR expression and therefore on the level of azole resistance.
TABLE 4.
TAC1 alleles present in the strains used in this study
| Group | Strain | TAC1 allele | TAC1 property | Amino acida at position(s):
|
|||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 47 | 104 | 131 | 189 | 199 | 206 | 207 | 225 | 232 | 317 | 396 | 558 | 572 | 677 | 683 | 736 | 764 | 772 | 776 | 829 | 895 | 935 | 937 | 941 | 962-969 | 980 | ||||
| None | CAF2-1 | TAC1-1 | Wild type | L | F | L | F | S | R | V | T | F | V | N | I | L | M | S | A | S | N | D | E | I | S | S | S | LPFSQFNN | G |
| TAC1-2 | Wild type | K | V | L | F | N | H | A | T | F | V | S | I | L | M | S | A | S | K | N | Q | I | L | S | P | LPFSQFNN | G | ||
| B | DSY347 | TAC1-8 | Wild type | K | F | L | F | N | H | A | T | F | V | S | I | L | M | S | A | S | N | N | E | T | S | S | P | LPFSQFNN | G |
| TAC1-9 | Wild type | K | F | I | S | N | H | A | T | F | V | S | V | L | M | S | A | S | K | N | E | I | S | L | S | LPFSQFNN | G | ||
| DSY348 | TAC1-10 | Hyperactive | K | F | L | F | N | H | A | T | F | V | S | V | L | M | S | V* | S | K | N | E | T | S | S | P | LPFSQFNN | G | |
| DSY289 | TAC1-11 | Hyperactive | K | F | I | S | N | H | A | T | F | V | S | V | L | M | S | V* | S | K | N | E | I | S | L | S | LPFSQFNN | G | |
| C | DSY2321 | TAC1-12 | Wild type | K | V | L | F | S | R | A | T | F | A | S | I | L | M | S | A | S | N | D | Q | I | S | S | S | LPFSQFNN | G |
| TAC1-13 | Wild type | K | F | L | F | S | R | V | T | F | V | N | I | L | M | S | A | S | N | D | E | I | S | S | S | LPFSQFNN | G | ||
| DSY2322, DSY2323 | TAC1-15 | Hyperactive | K | V | L | F | N | H | A | T | F | V | S | I | L | M | S | A | S | K | N | Q | I | L | S | P | LPFSQFNN | E* | |
| A | DSY731 | TAC1-16 | Wild type | K | F | I | S | N | H | A | T | S | V | S | V | L | M | P | A | G | N | N | E | I | S | L | S | LPFSQFNN | G |
| TAC1-17 | Wild type | K | F | L | F | N | H | A | T | F | V | S | V | L | M | S | A | S | N | N | E | I | S | S | P | LPFSQFNN | G | ||
| DSY732, DSY735 | TAC1-18 | Hyperactive | K | F | L | F | N | H | A | T | F | V | S | V | L | — | S | A | S | N | N | E | I | S | S | P | LPFSQFNN | G | |
| D | DSY3534 | TAC1-19 | Wild type | K | F | L | F | S | R | V | T | F | V | N | I | L | M | S | A | S | N | D | E | I | S | S | S | LPFSQFNN | G |
| TAC1-20 | Wild type | K | V | L | F | N | H | A | T | F | V | S | I | F | M | S | A | S | K | N | Q | I | L | S | P | LPFSQFNN | G | ||
| DSY3548 | TAC1-21 | Hyperactive | K | F | L | F | S | R | V | T | F | V | N | I | L | M | S | V* | S | N | D | E | I | S | S | S | LPFSQFNN | G | |
| DSY3549 | TAC1-22 | Hyperactive | K | V | L | F | N | H | A | T | F | V | S | I | L | M | S | A | S | K | N | Q | I | L | S | P | — | G | |
| E | DSY3553 | TAC1-23 | Wild type | K | F | L | F | S | R | V | T | F | V | N | I | L | M | S | A | S | N | D | E | I | S | S | S | LPFSQFNN | G |
| TAC1-24 | Wild type | K | V | L | F | N | H | A | T | F | V | S | I | L | M | S | A | S | K | N | Q | I | L | S | P | LPFSQFNN | G | ||
| DSY3565, DSY3554 | TAC1-25 | Hyperactive | K | F | L | F | S | R | V | A* | F | V | N | I | L | M | S | A | S | N | D | E | I | S | S | S | LPFSQFNN | G | |
—, deletion. Asterisk indicates a change that could represent a GOF mutation.
FIG. 2.
Characteristics of TAC1 alleles from individual C. albicans isolates. Each TAC1 allele was introduced in DSY2906 (tac1Δ/Δ) by the integration of pDS178-derived plasmids (Table 2) at the LEU2 locus by SalI digestion. The FLC MIC of each strain containing the individual TAC1 allele is indicated. The different TAC1 alleles were grouped according to their origin (group A to group E). In group E, TAC1-25 was introduced in single (TAC1-25) or double (2× TAC1-25) copies. Hyperactive alleles are underlined. Experimental conditions were identical to those described in the legend to Fig. 1.
Interestingly, the results shown in Fig. 2 suggest that all hyperactive alleles were not identical in their ability to elevate Cdr1p and Cdr2p levels when in the tac1Δ/Δ mutant DSY2906. For example, the Cdr2p level conferred by TAC1-18 was constitutively high and was not changed by treatment with fluphenazine (Fig. 2, group A). In contrast, the Cdr2p levels conferred by TAC1-10 and TAC1-11 were further increased by fluphenazine addition (Fig. 2, group B).
Distinct GOF mutations are present in TAC1 hyperactive alleles.
We previously demonstrated that hyperactivity of TAC1 alleles (TAC1-5 and TAC1-7) was due to a GOF mutation located at position 977 (N977D) (10). Taken together, TAC1 alleles available for nucleotide sequence analysis (from TAC1-1 to TAC1-25) were highly polymorphic, since their open reading frames contained 29 nonsynonymous and 50 synonymous mutations. In this work, we identified five nucleotide changes that could represent GOF mutations since they occurred only in hyperactive alleles (Table 4). They included three amino acid substitutions (A225T, A736V, and G980E) and two amino acid deletions (ΔM677 and ΔL962-N969). The A225T and G980E substitutions were present in TAC1-25 and TAC1-15, respectively. TAC1-10, -11, and -21 carried the A736V substitution. The two deletions, ΔM677 and ΔL962-N969, were present in the alleles TAC1-18 and TAC1-22, respectively.
In order to demonstrate that the above-mentioned mutations were involved in TAC1 hyperactivity, they were introduced into the TAC1-1 wild-type allele. The mutated alleles were next transferred into the tac1Δ/Δ mutant to test azole resistance and Cdr1p and Cdr2p levels conferred by individual mutations in transformants. In contrast to the strain carrying TAC1-1, all strains with mutated alleles were able to grow in the presence of FLC (Fig. 3A). This feature is consistent with an increase in FLC MIC compared to that for the strain containing the wild-type TAC1-1 allele. FLC MICs increased from 1 to 8 μg/ml as a result of the three substitutions (G980E, A736V, and T225A) and from 1 to 4 μg/ml for the two deletions (ΔM677 and ΔL962-N969) (Fig. 3A). All mutations in individual TAC1-1 alleles conferred constitutively high Cdr1p and Cdr2p levels (Fig. 3B). This contrasted with the strain carrying the TAC1-1 allele, in which the increase of Cdr1p and Cdr2p still required the presence of fluphenazine (Fig. 3B).
FIG. 3.
Hyperactivity of TAC1 is mediated by distinct GOF mutations. (A) Drug susceptibility testing of C. albicans tac1Δ/Δ mutant and TAC1 revertant strains with different TAC1-1-derived alleles carrying single point mutations or deletions. Drug susceptibility testing was carried out by plating serial dilutions of overnight cultures on YEPD agar plates containing different drugs as indicated. Plates were incubated for 48 h at 35°C in the absence or presence of FLC and cyclosporine at a concentration of 1 μg/ml. MIC assays were performed as described in Materials and Methods. (B) Expression levels of Cdr1p and Cdr2p of C. albicans tac1Δ/Δ mutant and TAC1 revertant strains with different TAC1-1-derived alleles carrying single point mutations or deletions. Experimental conditions were identical to those described in the legend to Fig. 1. WT, wild type.
Importantly, all GOF mutations were not equal in their hyperactivity, since FLC MIC and Cdr1p/Cdr2p levels were not identical in all strains carrying distinct TAC1-1-derived mutated alleles. For example, the three mutations with amino acid substitutions conferred higher FLC MICs than the two mutations with amino acid deletions (8 μg/ml versus 4 μg/ml) (Fig. 3A), which is consistent with constitutive levels of Cdr1p/Cdr2p being lower in the strain carrying this 2-amino-acid deletion than in strains with other TAC1 GOF mutations (Fig. 3B).
Impact of ERG11 in the development of azole resistance.
As described above, one of the mechanisms contributing to azole resistance in clinical strains is the acquisition of GOF mutations in TAC1. In addition, ERG11, the target of azole antifungals, is an important mediator of azole resistance. Changes in ERG11 that confer azole resistance arise by mutations in the coding sequence that alter the affinity of azoles for the target and by upregulation of ERG11. We previously demonstrated by heterologous expression in S. cerevisiae that the ERG11 mutations S405F, Y132H, and R467K contribute individually or in combination to azole resistance (43). Therefore, it is likely that these mutations have the same effect in C. albicans. Table 3 summarizes the occurrence of these ERG11 mutations in the isolates of the present study (see Table S2 in the supplemental material for complete details of nucleotide polymorphisms). In the azole-resistant strains of group B (strains DSY348 and DSY289), ERG11 alleles were homozygous for the S405F substitution; however, an additional homozygous substitution (Y132H) was present in ERG11 from DSY289, consistent with observations in our previous studies (43). In strains of group C, the substitution S405F was present, although it was present in only one of the two alleles in DSY2321 but was homozygous in DSY2322 and DSY2323. In group D strains, the R467K mutation was the only substitution in ERG11 alleles recovered from the azole-resistant strains DSY3548 and DSY3549. It was homozygous, in agreement with results published by White (54). In strains of groups A and E, no ERG11 polymorphisms could be associated with azole resistance. Combined with TAC1 GOF mutations, ERG11 mutations could contribute to elevate FLC MICs compared to those for strains with a single resistance mechanism.
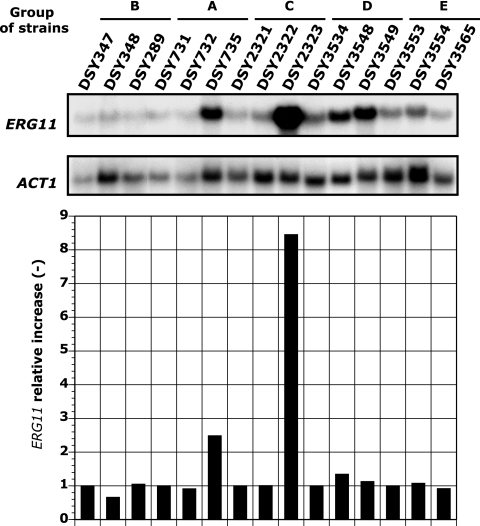
Northern blot analysis of the groups of sequential isolates revealed that ERG11 expression was elevated in two cases. As shown in Fig. 4, ERG11 expression was increased in DSY735, by approximately twofold compared to DSY732. Consistent with this expression increase, the FLC MIC was increased in DSY735 from 16 to 32 μg/ml. The results presented in Fig. 4 also revealed that ERG11 expression was increased by eightfold in DSY2323 compared to DSY2322. Such an increase might also explain the augmentation of the FLC MIC in strain DSY2323 (32 versus 16 μg/ml), in which no known resistance mechanisms other than those associated with ERG11 and TAC1 alterations were detected.
FIG. 4.
ERG11 expression levels in C. albicans isolates sequential. ERG11 expression was quantified by Northern blot analysis using a Typhoon Trio phosphorimager (GE Healthcare, Otelfingen, Switzerland). ERG11 expression was normalized using ACT1 expression (ACT1 is considered a housekeeping gene). ERG11 expression is given as the relative increase of expression compared to that in the most azole-susceptible strain of each group.
Analysis of Chr 5 status in azole-resistant strains.
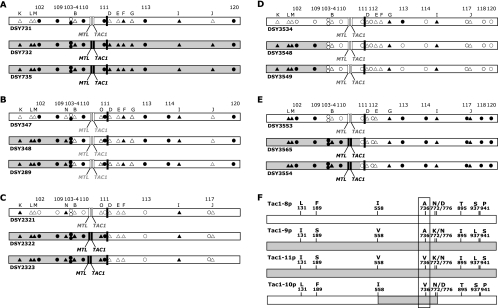
At least two azole resistance genes, ERG11 and TAC1, as well as the MTL locus are located on the same left arm of Chr 5. In this work, azole-resistant isolates of groups A, C, and E were homozygous for the MTL, TAC1, and ERG11 loci. In contrast, azole-susceptible parents were all heterozygous for the MTL and TAC1 loci. In order to determine the mechanisms that permitted LOH at the linked ERG11, TAC1, and MTL loci, we attempted to delimit the LOH event by SNP analysis on several markers present on Chr 5. SNP patterns were associated with three main Chr 5 rearrangements (see Table S2 in the supplemental material) and are illustrated in Fig. 5A to E.
FIG. 5.
Mapping of SNP on Chr 5 of C. albicans sequential isolates. (A to E) SNP on Chr 5 of isolates from groups A to E. Circles indicate SNP of Chr 5 observed by the use of an SNP microarray. The following markers were described by Forche et al. (16): 102, 1855/2172; 103, HST3; 104, SNF1; 109, 1899/2008; 110, 1445/2395; 111, 1922/2344; 112, PDE1; 113, 1969/2162; 114, DPH5; 117, 1817/2082; 118, 1341/2493; 120, 2340/2493. Triangles indicate additional markers of Chr 5 determined by multilocus sequence typing: B, orf19.1926; D, orf19.4225; E, orf19.4251; F, orf19.4288; G, orf19.429; I, orf19.6680; J, CRH12; K, orf19.971; L, ERG11; M, orf19.921; N, orf19.1971; O, orf19.3178. Empty and filled symbols indicate heterozygosity and homozygosity for the indicated markers, respectively. MTL and TAC1 loci in gray and black indicate heterozygosity and homozygosity, respectively. The gray region on Chr 5 delimitates the maximal region of LOH. (B) Schematic representation of the origin of Tac1-10p. The area in gray delimits a region spanning from codon 558 to 776 which originates from Tac1-9p and Tac1-10p.
(i) Chromosome loss and reduplication.
In group A strains, all the heterozygous SNP markers of Chr 5 in the azole-susceptible strain became homozygous in azole-resistant strains (Fig. 5A). LOH occurred across the entire Chr 5 length in DSY732 and DSY735, thus suggesting chromosomal loss and reduplication of the remaining copy. However, our results cannot exclude a mitotic nondisjunction followed by chromosome loss as an alternative LOH mechanism.
(ii) Mitotic recombination.
In the four remaining groups (B to E), azole-resistant strains differed from their azole-susceptible parents by a stretch of homozygous SNPs that extends from the left end of Chr 5 to a position prior to the centromere (Fig. 5B to E). These data indicated that in these four cases, LOH on Chr 5 had occurred through mitotic recombination or break-induced repair. The site of recombination (or chromosome break) could be mapped between SNPs 103 and B in group B strains, between SNP 111 and the centromere in group C strains, between SNPs 109 and 103 in group D strains, and between the TAC1 locus and SNP 111 in group E strains. Consequently, LOH involved ERG11 only in group B and D strains, while it involved the ERG11, MTL, and TAC1 loci in group C and E strains.
(iii) Gene conversions.
In group B strains, mitotic recombination was observed within TAC1, indicating a phenomenon of gene conversion. SNP analysis of TAC1 alleles suggests that TAC1-10 is a mosaic between TAC1-8 and TAC1-11, as illustrated in Fig. 5F. Thus, gene conversion events different from LOH events on the left part of Chr 5 as described above occurred within the TAC1 locus between TAC1-8 and TAC1-11 to yield TAC1-10. As a consequence, the azole-resistant strains DSY348 and DSY289 carry two distinct TAC1 hyperactive alleles but with the same GOF mutation (A736V).
In conclusion, our data show that LOH at the ERG11, MTL, and TAC1 loci is frequently observed in association with increased azole resistance. This is consistent with previous results obtained upon analysis of ERG11 in other resistant isolates (43, 54). Nevertheless, this phenomenon cannot account for the increase in drug resistance between some related azole-resistant strains. For example, the pairs of isolates DSY732 and DSY735, DSY348 and DSY289, and DSY2322 and DSY2323 each contain identical TAC1 hyperactive alleles and the same Chr 5 SNP profiles, but DSY735, DSY289, and DSY2323 have higher FLC MICs than DSY732, DSY348, and DSY2322, respectively.
CGH and detection of aneuploidies in azole-resistant isolates.
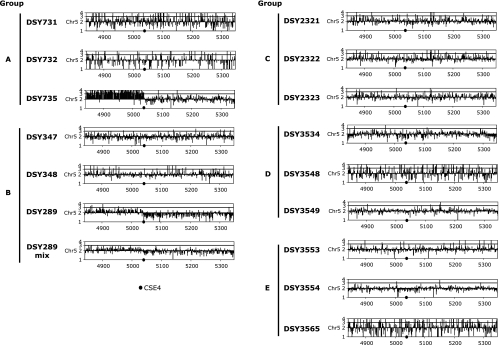
Chromosome alterations by LOH contribute to increase the copy number of mutant alleles of these genes. The copy number of mutant alleles of drug resistance genes and therefore the level of azole resistance can be increased by segmental aneuploidy and formation of i(5L), as recently demonstrated by Selmecki et al. (47). Aneuploidies are readily detected by CGH. We subjected our collection of sequential isolates to CGH analysis to address whether any of the strains had undergone whole-chromosome and/or segmental aneuploidy.
Many of the investigated isolates had CGH profiles characteristic of normal diploid strains. However, CGH revealed that most aneuploidies in the azole-resistant strains were on Chr 5. Therefore, only CGH profiles of Chr 5 are presented. We observed a single case of Chr R trisomy in isolate DSY3554 (see data at http://www.chuv.ch/imul/imu_home/imu_recherche/imu_recherche_sanglard/imu_recherche_sanglard_suppldata.htm). Strains DSY735 (group A) and DSY289 (group B) exhibited segmental aneuploidy for Chr 5 (Fig. 6). In DSY735, genes located on the left arm of Chr 5 (5L) delimited by the centromere were found in four copies, as revealed by signal intensities of CGH profiles (Fig. 6). This strongly suggests the presence of an i(5L) composed of two left-arm copies of Chr 5 in addition to the two full-length copies of Chr 5.
FIG. 6.
Mapping of gene copy number on Chr 5 by CGH. The genomes of the tested strains were hybridized against the SC5314 genome according to the protocol published by Selmecki et al. (46). Each gene on Chr 5 is represented by its relative intensity compared to signals obtained in SC5314.
In DSY289, the CGH profile of a single colony (Fig. 6, DSY289 of group B) suggested the presence of three copies for genes on the left arm of Chr 5 (5L) and apparently only one copy of genes on the right arm (5R), since the average gene copy number values plotted along the 5R axis are below the threshold value of 2 (Fig. 6, group B). These data support the hypothesis that strain DSY289 contains only one entire Chr 5 copy and one i(5L). Nevertheless, since SNP analysis of Chr 5 markers in DSY289 revealed that four markers (D, E, G, and J) on 5R remained heterozygous (Fig. 5B; see Table S2 in the supplemental material), this result does not agree with a single remaining 5R copy. We therefore concluded that Chr 5 is unstable in DSY289. To further test this hypothesis, CGH analysis was performed on a mix of several colonies of the DSY289 strain (Fig. 6, DSY289 mix of group B). In the resulting CGH profile, aneuploidies on 5R and 5L appear less dramatic, suggesting that either i(5L) or the extra Chr 5 copy is being lost quite frequently.
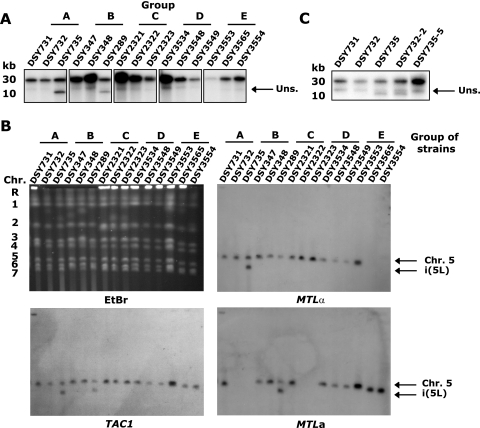
In order to test whether aneuploidy in strains DSY735 and DSY289 was indeed due to i(5L), Southern blot analysis was performed with EcoNI-digested genomic DNA (Fig. 7A) and with a CHEF karyotype (Fig. 7B). Both analyses confirmed the presence of i(5L) in DSY735 and DSY289. As shown in Fig. 7A, the CSE4 probe revealed a 10-kb band characteristic of i(5L) in the EcoNI-digested genomic DNA of both strains but not in the other strains. CHEF Southern blot analysis with a TAC1 probe revealed a signal in all strains corresponding to Chr 5, as well as an additional signal in DSY735 and DSY289 that had an electrophoretic mobility similar to that of Chr 7 (Fig. 7B), as expected for i(5L) (47). This analysis also confirmed that TAC1 is present only on Chr 5 and i(5L) and not on chromosomes of other sizes. This observation reinforces the idea that LOH events deduced from SNP analysis are the consequence of mitotic recombination between two Chr 5 copies and not the result of translocations on other chromosomes in the genome. Moreover, hybridization with MTLa and MTLα probes confirmed the MTL locus status of all the strains and determined that the i(5L) present in strain DSY735 is MTLα/MTLα and the i(5L) present in strain DSY289 is MTLa/MTLa (Fig. 7B). This latter data allowed us to deduce that the TAC1-10 allele in this strain is on i(5L), since the parental TAC1-8 allele is linked to MTLa (data not shown).
FIG. 7.
Detection of i(5L) in C. albicans isolates. (A and C) Southern blot of EcoNI-digested DNA probed with CSE4. A 10-kb fragment reveals an i(5L) structure. Uns., unspecific signal. (B) Karyotype analysis. Whole-chromosome CHEF analysis was carried out as described in Material and Methods, with ethidium bromide staining or hybridization with TAC1, MTLa, or MTLα1 probes as indicated under each panel. Arrows indicate the position of i(5L) on each blot.
To address the involvement of i(5L) in the increase of azole resistance and to confirm its instability, DSY735 [an i(5L)-containing strain] was serially passed for 8 days without FLC to obtain colonies with a reduced FLC MIC. In parallel, DSY732 (not containing i(5L)] was plated onto rich medium supplemented with 40 μg/ml of FLC for 5 days in order to obtain resistant strains. In both cases, independent colonies were analyzed for the presence of i(5L). A DSY735-derived strain (DSY735-5) lost i(5L) (Fig. 7C) and showed a decrease of the FLC MIC from 32 to 16 μg/ml. In contrast, out of four DSY732-derived strains, a single isolate, DSY732-2, gained an i(5L) (Fig. 7C) with a parallel increase in FLC MIC from 16 to 32 μg/ml. Therefore, these additional experiments confirmed the contribution of the i(5L) to the increase of azole resistance.
DISCUSSION
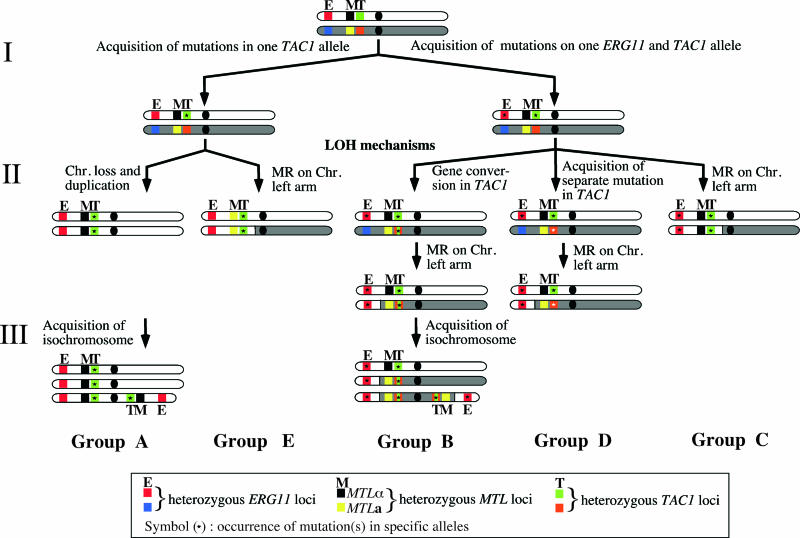
In this study, we described different series of events leading to the development of azole resistance. Our results illustrate how flexibility of the C. albicans genome allows adaptation to and/or circumvention of antifungal pressure. Several events at the level of the gene and the genome occurred during the development of azole resistance. As summarized in Fig. 8, the acquisition of mutations in drug resistance genes (step I) is a prerequisite to the development of azole resistance and is followed by different LOH events (step II), including chromosome loss and reduplication (observed in group A strains) and mitotic recombinations (observed in the other groups of strains in this study). These chromosome alterations can be accompanied by several other variations (acquisition of separate mutations and recombination within resistance genes) and acquisition of extra chromosomal elements (step III, isochromosome formation), all contributing to elevate drug resistance in C. albicans. These steps are further discussed below.
FIG. 8.
Schematic representation of Chr 5 and azole resistance gene modifications in the groups of investigated strains. After acquisition of GOF mutations on one TAC1 allele and also possible acquisition of mutations in ERG11 (step I), different LOH events occurred at these loci, including loss of Chr 5 and duplication, mitotic recombination of the left arm of Chr 5, and gene conversion in TAC1 (step II). The formation of i(5L) is an additional event (step III) leading to the increase of the copy numbers of azole resistance genes and possibly yet unidentified resistance genes on the 5L arm. For simplification, the acquisition of the TAC1 mutation (step I) is shown linked with MTLα in group E, although it is linked to MTLa. It is possible that steps I to III can be sequentially arranged in a different manner. MR, mitotic recombination. In group A, sequence analysis allow the deduction that TAC1-18 originated from TAC1-17 by acquisition of only one nonsynonymous substitution (ΔM677). In group B, TAC1-11 arose from TAC1-9 by acquisition of the A736V substitution. TAC1-10 is the result of gene conversions between TAC1-8 and TAC1-11. In group C, it is not possible to determine whether or not TAC1-15 arose from TAC1-12 or TAC1-13. In group D, TAC1-21 and TAC1-22 originated from TAC1-19 and TAC1-20, respectively, by acquisition of two distinct GOF mutations, A736V and ΔL962-ΔN969. Finally, in group E, TAC1-25 originated from TAC1-23 by acquisition of the A225T substitution.
Step I: acquisition of TAC1 and ERG11 mutations.
The first step in azole resistance seems to be the acquisition of GOF mutations in drug resistance genes. Four of the five TAC1 GOF mutations identified in this work (V736A, G980E, and deletions ΔM677 and Δ962-969) were located in the C-terminal part of the protein. These mutations, in addition to the N977D substitution reported recently (10), map in a segment containing a putative transcriptional activation domain, as deduced from protein alignments of Tac1p with other Zn2-Cys6 transcription factors. Previous studies on the S. cerevisiae Pdr1p and Pdr3p transcription factors also found that point mutations in the C-terminal region lead to enhanced expression of target genes (6, 31, 32, 49). Characterization of Pdr3p and Pdr1p has revealed the transcriptional inhibitory domain protein motifs I and II described by Poch (36) (from positions 257 to 320 in Pdr1p and from 228 to 286 in Pdr3p) (6, 23, 31, 32, 36). Even though motifs I and II cannot be clearly identified in Tac1p by MEME (multiple EM for motif elicitation) analysis (data not shown), the remaining T225 GOF mutation in Tac1p is located about 150 amino acids downstream of the DNA-binding domain, as are motifs I and II with respect to the DNA-binding domains of Pdr1p and Pdr3p. The hyperactivity of the TAC1 allele with the T225A substitution is consistent with the occurrence of a transcriptional inhibitory domain in Tac1p similar to those found in Pdr1p and Pdr3p. Deletion of this region in Pdr1p results in intrinsic hyperactivity of the transcription factor (23). Similar experiments could be carried out with TAC1 in order to verify the possible assignment of this region as a transcriptional inhibitory domain. Kolaczkowska et al. (23) have suggested that all GOF mutations in Pdr1p prevent the interaction between an inhibitory domain and the activation domain, leading to constitutive transcriptional activation of the protein. Nevertheless, the role of these mutations in the enhancement of target gene expression remains unclear. It is likely that other GOF mutations in other hyperactive TAC1 alleles from additional clinical isolates will be reported. Their identification either by cloning from clinical isolates or by random mutagenesis in TAC1 will be helpful to delimitate regions critical for the transcriptional activation of this protein. Furthermore, a one-hybrid lexA-based system might also be useful to delineate more precisely the transcriptional activation domain of this protein, as described by two other groups (30, 39).
TAC1 mutations are not sufficient to explain all increases in azole resistance in the sequential isolates in this study. The analysis of ERG11 alleles revealed several substitutions known to be responsible for azole resistance when expressed in S. cerevisiae (43). A combination of TAC1 and ERG11 point mutations in C. albicans likely contributed to FLC MIC increases among azole-resistant isolates in this study (Fig. 8, strains of groups B, C, and D). It is not yet possible to determine precisely the individual role of each of these mutations in azole resistance. Experiments are being carried out to answer this open question by sequentially restoring wild-type alleles in the resistant isolates.
Step II: LOH events associated with increased azole resistance.
In a previous study (10), we showed the codominance of the TAC1-5 and TAC1-7 alleles carrying an N977D GOF mutation with wild-type alleles. We also demonstrated the need for the strain to elevate the copy number of hyperactive alleles at the expense of remaining wild-type alleles in order to increase FLC resistance. This work extends this observation to other TAC1 hyperactive alleles. Regarding ERG11, White et al. (56) associated LOH of ERG11 mutated alleles with an increase in azole resistance. In all cases investigated here, azole-resistant strains contained only hyperactive TAC1 alleles and were homozygote for ERG11. Distinct mechanisms leading to two TAC1 hyperactive and two identical ERG11 alleles were distinguished, as summarized in Fig. 8. ERG11 LOH was obtained by mitotic recombination events either associated with LOH at TAC1 and MTL loci (in group C strains), with a chromosomal break located near the centromere, or independent of the TAC1 and MTL loci (groups B and D). For the TAC1 locus, they include Chr 5 loss and duplication (group A) and mitotic recombinations in the left part of Chr 5 (groups C and E). Gene conversion can also occur within TAC1, leading to homozygosity of the GOF mutation (group B) and resulting in the formation of two distinct TAC1 hyperactive alleles with the same GOF mutation (A736V). Together with strains of group D (two distinct TAC1 alleles with different GOF mutations), these cases illustrate that LOH of drug resistance genes in Chr 5 is not always associated with MTL homozygosity. This observation is consistent with previous studies (37, 40) showing that antifungal resistance is not necessarily linked with MTL homozygosity.
ERG11 LOH was obtained by mitotic recombination events either associated with LOH at TAC1 and MTL loci (in group C strains) with a Chr 5 break located near the centromere or independent of the TAC1 and MTL loci (groups B and D) with a chromosomal break occurring near markers 103 and 104 on 5L. Lephart and Magee (27) did not identify specific “hot spot” regions of mitotic recombination on Chr 5 segments near the major repeat sequence region. Rather, they demonstrated that mitotic recombination events could occur at a similar frequency along a 325-kbp region flanking the Chr 5 major repeat sequence. Nevertheless, we cannot exclude the possibility that under specific environmental conditions, genome rearrangements which are at specific loci can be selected.
Step III: increase of drug resistance by isochromosome formation.
TAC1 and ERG11 mutations linked to LOH were not sufficient to explain all of the azole resistance increase observed in some strains. In strains DSY735 and DSY2323, the FLC MIC was twofold higher than that in the related earlier isolates. An increase of the copy number of drug resistance genes can elevate azole resistance, as recently demonstrated by the existence of i(5L). This specific chromosome rearrangement results in an increase in the copy number of all genes on 5L, including TAC1 and ERG11 (47). Our study revealed the presence of i(5L) in strains DSY735 and DSY289. Interestingly, Northern blot analysis revealed an increase in ERG11 expression in DSY735 (approximately twofold) compared to DSY732 or DSY731, consistent with an increase of ERG11 copy number due to the i(5L). A separate measurement of TAC1 expression by reverse transcription-PCR in group A strains revealed that its transcription was increased by 2.53 (±0.29)-fold in DSY735 compared to DSY732. This result supports the idea that i(5L) formation favors the expression of drug resistance genes on this chromosome, which is coupled with decreased susceptibility to FLC. Transcript profiling experiments are currently being performed with strains of groups A and B to extend this analysis to other genes on this chromosome.
CGH analysis performed on a mix of DSY289 individual colonies did not exhibit the same gene copy number profile as a CGH carried out from a single colony. This result raises the possibility of i(5L) and/or Chr 5 instability in DSY289, especially in the absence of antifungal drug selection. This instability was confirmed in DSY735 [an i(5L)-containing strain] grown for 8 days without FLC. In this case, loss of the i(5L) correlated with a decrease in azole resistance (Fig. 7C). Selmecki et al. (47) had already demonstrated that growing an i(5L)-containing strain for several generations in the absence of drug pressure resulted in the loss of the isochromosome. Those authors also observed a dramatic decrease of resistance, with the FLC MIC dropping from 256 to 8 μg/ml (47). In DSY735-derived strains, the loss of i(5L) resulted in a more modest decrease of the MIC FLC from 32 to 16 μg/ml. The reason for this discrepancy could be differences in FLC susceptibility measurements or subtle differences in polymorphisms and expression of drug resistance genes, among them TAC1 and ERG11. Finally, yet-unknown drug resistance genes and their linked mutations might be present on i(5L) of specific strains and thus could contribute to azole resistance independently of TAC1 and ERG11.
Chromosome alterations and azole resistance in C. albicans.
In this study, we found that the development of resistance does not result from a single event in each strain. Rather, it results from a combination of several events, including mutations in single genes and chromosomal rearrangements or nondisjunction events. The analysis of a large strain collection recently reported by Odds et al. (33) indicates that genome modifications are common in the C. albicans population, which reflect the generally clonal evolutionary pattern in an asexually reproducing species. Other studies have demonstrated a link between chromosomal rearrangements and adaptation to stress conditions such as growth on sorbose, growth in the presence of FLC, or during the course of infection (20, 26). C. albicans can therefore achieve stress adaptation and phenotypic variability by genome rearrangements. In addition, several authors demonstrated that chromosome alteration frequencies are increased under stress conditions (27, 41). This increased frequency might be a survival mechanism that circumvents the need for meiotic recombination. When returned to optimal growth conditions, such strains tend to recover their original karyotypes. This phenomenon was observed during growth of C. albicans in the presence of 5-fluoroorotic acid (52). In this condition, strains gained an extra Chr 4b copy or acquired a 260-kbp extension of Chr 5, generating a new Chr 5 copy. This additional chromosome was lost in the absence of 5-fluoroorotic acid. This phenomenon was also observed here, with the acquisition or loss of an extra i(5L) in the presence or absence of FLC selection pressure.
The development of resistance illustrated in Fig. 8 highlights that point mutations in drug resistance genes, among them TAC1 and ERG11, can be a first step towards increased resistance but is followed by chromosomal rearrangement events, which are likely affected by the genetic background of the isolated strains. In S. cerevisiae two major mechanisms are responsible for chromosomal rearrangements: nonhomologous end joining and homologous recombination (HR) (38). Potential dicentric translocations and dicentric isochromosomes were associated with cell cycle checkpoint defects, while chromosome fusions were frequent in strains with both telomerase and cell cycle defects. Finally, translocations between homologous genes were seen in strains with defects favoring HR. Moreover, a study on the LOH in a RAD family mutant (57) reported that the frequency of LOH was significantly increased in all mutants, and most events detected were chromosome loss. Other LOH events were differentially affected in each mutant. In C. albicans, similar karyotype analyses carried out with a RAD52 mutant (3, 4, 7, 9) found that HR played an essential role in the repair of DNA lesions caused by both UV light and the radiomimetic compound methyl methanesulfonate, whereas the nonhomologous end joining pathway was used only in the absence of Rad52p or after extensive DNA damage. Repair by HR is more efficient in cells in the exponential phase of growth than in the stationary phase. This raises the possibility that chromosome rearrangements favoring adaptation to stress conditions do not occur via a random process but are dependent on the genetic backgrounds and the execution of specific cell cycle growth phases. This hypothesis could also be valid for the acquisition of i(5L), and this limits such a phenomenon to specific strains. This could explain that some strains, such as DSY732, exposed to a high level of FLC can acquire i(5L), in contrast to other strains, such as FH1, in which FLC exposure in vitro results in Chr 5 trisomy, as shown recently by Coste et al. (10).
In conclusion, this study precisely dissected combinations of genome modifications that occurred during the adaptation to antifungal pressure. A distinct combination of single gene alterations or chromosome rearrangement events has occurred in each group of related strains and thus illustrates the versatility of mechanisms that facilitate the appearance of antifungal resistance in C. albicans. This study was focused on a limited number of strains originating from HIV-positive patients or from in vitro antifungal exposure but with similar resistance mechanism profiles (i.e., CDR upregulation and ERG11 alterations). Given the existence of other azole resistance mechanisms (for example, those involving MDR1 upregulation), it is likely that additional genome modifications remain to be described in the future.
Supplementary Material
Acknowledgments
This work was supported in part by an EC grant of the EURESFUN consortium (PL 518199) and by a grant from the Swiss National Foundation (3100A0-114131/1). A.C. was supported by a grant from the Faculty of Biology and Medicine of the University of Lausanne. J.B. and A.F. were supported by a grant (AI62427) from the NIH. A.S. was supported by an MPGI fellowship and by funds from the Minnesota Medical Foundation. D.D. is the recipient of a Ph.D. fellowship from the Institut National de la Recherche Agronomique.
Some strains for this study were kindly provided by T. White (University of Washington, Seattle) and J. B. Anderson (University of Toronto, Canada). Strains originating from the University Hospital Center in Lausanne were kindly provided by J. Bille and C. Durussel. We thank F. Ischer for excellent technical assistance, P. Hauser for technical help in point mutagenesis, and S. Ferrari for critical reading of the manuscript.
Footnotes
Published ahead of print on 10 August 2007.
Supplemental material for this article may be found at http://ec.asm.org/.
REFERENCES
- 1.Akins, R. A. 2005. An update on antifungal targets and mechanisms of resistance in Candida albicans. Med. Mycol. 43:285-318. [DOI] [PubMed] [Google Scholar]
- 2.Albertson, G. D., M. Niimi, R. D. Cannon, and H. F. Jenkinson. 1996. Multiple efflux mechanisms are involved in Candida albicans fluconazole resistance. Antimicrob. Agents Chemother. 40:2835-2841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Andaluz, E., T. Ciudad, J. Gomez-Raja, R. Calderone, and G. Larriba. 2006. Rad52 depletion in Candida albicans triggers both the DNA-damage checkpoint and filamentation accompanied by but independent of expression of hypha-specific genes. Mol. Microbiol. 59:1452-1472. [DOI] [PubMed] [Google Scholar]
- 4.Andaluz, E., J. Gomez-Raja, B. Hermosa, T. Ciudad, E. Rustchenko, R. Calderone, and G. Larriba. 2007. Loss and fragmentation of chromosome 5 are major events linked to the adaptation of rad52Δ/Δ strains of Candida albicans to sorbose. Fungal Genet. Biol. 44:789-798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Barker, K. S., and P. D. Rogers. 2006. Recent insights into the mechanisms of antifungal resistance. Curr. Infect. Dis. Rep. 8:449-456. [DOI] [PubMed] [Google Scholar]
- 6.Carvajal, E., H. B. van den Hazel, A. Cybularz-Kolaczkowska, E. Balzi, and A. Goffeau. 1997. Molecular and phenotypic characterization of yeast PDR1 mutants that show hyperactive transcription of various ABC multidrug transporter genes. Mol. Gen. Genet. 256:406-415. [DOI] [PubMed] [Google Scholar]
- 7.Chauhan, N., T. Ciudad, A. Rodriguez-Alejandre, G. Larriba, R. Calderone, and E. Andaluz. 2005. Virulence and karyotype analyses of rad52 mutants of Candida albicans: regeneration of a truncated chromosome of a reintegrant strain (rad52/RAD52) in the host. Infect. Immun. 73:8069-8078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen, C. G., Y. L. Yang, H. I. Shih, C. L. Su, and H. J. Lo. 2004. CaNdt80 is involved in drug resistance in Candida albicans by regulating CDR1. Antimicrob. Agents Chemother. 48:4505-4512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ciudad, T., E. Andaluz, O. Steinberg-Neifach, N. F. Lue, N. A. Gow, R. A. Calderone, and G. Larriba. 2004. Homologous recombination in Candida albicans: role of CaRad52p in DNA repair, integration of linear DNA fragments and telomere length. Mol. Microbiol. 53:1177-1194. [DOI] [PubMed] [Google Scholar]
- 10.Coste, A., V. Turner, F. Ischer, J. Morschhauser, A. Forche, A. Selmecki, J. Berman, J. Bille, and D. Sanglard. 2006. A mutation in Tac1p, a transcription factor regulating CDR1 and CDR2, is coupled with loss of heterozygosity at chromosome 5 to mediate antifungal resistance in Candida albicans. Genetics 172:2139-2156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Coste, A. T., M. Karababa, F. Ischer, J. Bille, and D. Sanglard. 2004. TAC1, transcriptional activator of CDR genes, is a new transcription factor involved in the regulation of Candida albicans ABC transporters CDR1 and CDR2. Eukaryot. Cell 3:1639-1652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cowen, L. E., D. Sanglard, D. Calabrese, C. Sirjusingh, J. B. Anderson, and L. M. Kohn. 2000. Evolution of drug resistance in experimental populations of Candida albicans. J. Bacteriol. 182:1515-1522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.de Micheli, M., J. Bille, C. Schueller, and D. Sanglard. 2002. A common drug-responsive element mediates the upregulation of the Candida albicans ABC transporters CDR1 and CDR2, two genes involved in antifungal drug resistance. Mol. Microbiol. 43:1197-1214. [DOI] [PubMed] [Google Scholar]
- 14.Du, W., M. Coaker, J. D. Sobel, and R. A. Akins. 2004. Shuttle vectors for Candida albicans: control of plasmid copy number and elevated expression of cloned genes. Curr. Genet. 45:390-398. [DOI] [PubMed] [Google Scholar]
- 15.Fonzi, W. A., and M. Y. Irwin. 1993. Isogenic strain construction and gene mapping in Candida albicans. Genetics 134:717-728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Forche, A., G. May, and P. T. Magee. 2005. Demonstration of loss of heterozygosity by single-nucleotide polymorphism microarray analysis and alterations in strain morphology in Candida albicans strains during infection. Eukaryot. Cell. 4:156-165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Franz, R., S. L. Kelly, D. C. Lamb, D. E. Kelly, M. Ruhnke, and J. Morschhauser. 1998. Multiple molecular mechanisms contribute to a stepwise development of fluconazole resistance in clinical Candida albicans strains. Antimicrob. Agents Chemother. 42:3065-3072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Goldman, G. H., M. E. da Silva Ferreira, E. dos Reis Marques, M. Savoldi, D. Perlin, S. Park, P. C. Godoy Martinez, M. H. Goldman, and A. L. Colombo. 2004. Evaluation of fluconazole resistance mechanisms in Candida albicans clinical isolates from HIV-infected patients in Brazil. Diagn. Microbiol. Infect. Dis. 50:25-32. [DOI] [PubMed] [Google Scholar]
- 19.Henry, K. W., J. T. Nickels, and T. D. Edlind. 2000. Upregulation of ERG genes in Candida species by azoles and other sterol biosynthesis inhibitors. Antimicrob. Agents Chemother. 44:2693-2700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Janbon, G., F. Sherman, and E. Rustchenko. 1998. Monosomy of a specific chromosome determines l-sorbose utilization: a novel regulatory mechanism in Candida albicans. Proc. Natl. Acad. Sci. USA 95:5150-5155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kelly, S. L., D. C. Lamb, and D. E. Kelly. 1999. Y132H substitution in Candida albicans sterol 14alpha-demethylase confers fluconazole resistance by preventing binding to haem. FEMS Microbiol. Lett. 180:171-175. [DOI] [PubMed] [Google Scholar]
- 22.Kelly, S. L., D. C. Lamb, J. Loeffler, H. Einsele, and D. E. Kelly. 1999. The G464S amino acid substitution in Candida albicans sterol 14alpha-demethylase causes fluconazole resistance in the clinic through reduced affinity. Biochem. Biophys. Res. Commun. 262:174-179. [DOI] [PubMed] [Google Scholar]
- 23.Kolaczkowska, A., M. Kolaczkowski, A. Delahodde, and A. Goffeau. 2002. Functional dissection of Pdr1p, a regulator of multidrug resistance in Saccharomyces cerevisiae. Mol. Genet. Genomics 267:96-106. [DOI] [PubMed] [Google Scholar]
- 24.Lamb, D. C., D. E. Kelly, W. H. Schunck, A. Z. Shyadehi, M. Akhtar, D. J. Lowe, B. C. Baldwin, and S. L. Kelly. 1997. The mutation T315A in Candida albicans sterol 14alpha-demethylase causes reduced enzyme activity and fluconazole resistance through reduced affinity. J. Biol. Chem. 272:5682-5688. [DOI] [PubMed] [Google Scholar]
- 25.Lamb, D. C., D. E. Kelly, T. C. White, and S. L. Kelly. 2000. The R467K amino acid substitution in Candida albicans sterol 14α-demethylase causes drug resistance through reduced affinity. Antimicrob. Agents Chemother. 44:63-67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Legrand, M., P. Lephart, A. Forche, F. M. Mueller, T. Walsh, P. T. Magee, and B. B. Magee. 2004. Homozygosity at the MTL locus in clinical strains of Candida albicans: karyotypic rearrangements and tetraploid formation. Mol. Microbiol. 52:1451-1462. [DOI] [PubMed] [Google Scholar]
- 27.Lephart, P. R., and P. T. Magee. 2006. Effect of the major repeat sequence on mitotic recombination in Candida albicans. Genetics 174:1737-1744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.MacPherson, S., B. Akache, S. Weber, X. De Deken, M. Raymond, and B. Turcotte. 2005. Candida albicans zinc cluster protein Upc2p confers resistance to antifungal drugs and is an activator of ergosterol biosynthetic genes. Antimicrob. Agents Chemother. 49:1745-1752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Marichal, P., L. Koymans, S. Willemsens, D. Bellens, P. Verhasselt, W. Luyten, M. Borgers, F. C. Ramaekers, F. C. Odds, and H. V. Bossche. 1999. Contribution of mutations in the cytochrome P450 14α-demethylase (Erg11p, Cyp51p) to azole resistance in Candida albicans. Microbiology. 145:2701-2713. [DOI] [PubMed] [Google Scholar]
- 30.Martchenko, M., A. Levitin, and M. Whiteway. 2007. Transcriptional activation domains of the Candida albicans Gcn4p and Gal4p homologs. Eukaryot. Cell. 6:291-301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mizoguchi, H., T. Yamauchi, M. Watanabe, H. Yamanaka, A. Nishimura, and H. Hanamoto. 2002. Different missense mutations in PDR1 and PDR3 genes from clotrimazole-resistant sake yeast are responsible for pleiotropic drug resistance and improved fermentative activity. J. Biosci. Bioeng 93:221-227. [DOI] [PubMed] [Google Scholar]
- 32.Nourani, A., D. Papajova, A. Delahodde, C. Jacq, and J. Subik. 1997. Clustered amino acid substitutions in the yeast transcription regulator Pdr3p increase pleiotropic drug resistance and identify a new central regulatory domain. Mol. Gen. Genet. 256:397-405. [DOI] [PubMed] [Google Scholar]
- 33.Odds, F. C., M. E. Bougnoux, D. J. Shaw, J. M. Bain, A. D. Davidson, D. Diogo, M. D. Jacobsen, M. Lecomte, S. Y. Li, A. Tavanti, M. C. Maiden, N. A. Gow, and C. d'Enfert. 2007. Molecular phylogenetics of Candida albicans. Eukaryot. Cell 6:1041-1052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Perea, S., J. L. Lopez-Ribot, W. R. Kirkpatrick, R. K. McAtee, R. A. Santillan, M. Martinez, D. Calabrese, D. Sanglard, and T. F. Patterson. 2001. Prevalence of molecular mechanisms of resistance to azole antifungal agents in Candida albicans strains displaying high-level fluconazole resistance isolated from human immunodeficiency virus-infected patients. Antimicrob. Agents Chemother. 45:2676-2684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Perea, S., J. L. Lopez-Ribot, B. L. Wickes, W. R. Kirkpatrick, O. P. Dib, S. P. Bachmann, S. M. Keller, M. Martinez, and T. F. Patterson. 2002. Molecular mechanisms of fluconazole resistance in Candida dubliniensis isolates from human immunodeficiency virus-infected patients with oropharyngeal candidiasis. Antimicrob. Agents Chemother. 46:1695-1703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Poch, O. 1997. Conservation of a putative inhibitory domain in the GAL4 family members. Gene 184:229-235. [DOI] [PubMed] [Google Scholar]
- 37.Pujol, C., S. A. Messer, M. Pfaller, and D. R. Soll. 2003. Drug resistance is not directly affected by mating type locus zygosity in Candida albicans. Antimicrob. Agents Chemother. 47:1207-1212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Putnam, C. D., V. Pennaneach, and R. D. Kolodner. 2005. Saccharomyces cerevisiae as a model system to define the chromosomal instability phenotype. Mol. Cell. Biol. 25:7226-7238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Russell, C. L., and A. J. Brown. 2005. Expression of one-hybrid fusions with Staphylococcus aureus lexA in Candida albicans confirms that Nrg1 is a transcriptional repressor and that Gcn4 is a transcriptional activator. Fungal Genet. Biol. 42:676-683. [DOI] [PubMed] [Google Scholar]
- 40.Rustad, T. R., D. A. Stevens, M. A. Pfaller, and T. C. White. 2002. Homozygosity at the Candida albicans MTL locus associated with azole resistance. Microbiology 148:1061-1072. [DOI] [PubMed] [Google Scholar]
- 41.Rustchenko-Bulgac, E. P., F. Sherman, and J. B. Hicks. 1990. Chromosomal rearrangements associated with morphological mutants provide a means for genetic variation of Candida albicans. J. Bacteriol. 172:1276-1283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sanglard, D. 2002. Resistance of human fungal pathogens to antifungal drugs. Curr. Opin. Microbiol. 5:379-385. [DOI] [PubMed] [Google Scholar]
- 43.Sanglard, D., F. Ischer, L. Koymans, and J. Bille. 1998. Amino acid substitutions in the cytochrome P450 lanosterol 14α-demethylase (CYP51A1) from azole-resistant Candida albicans clinical isolates contributing to the resistance to azole antifungal agents. Antimicrob. Agents Chemother. 42:241-253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sanglard, D., K. Kuchler, F. Ischer, J. L. Pagani, M. Monod, and J. Bille. 1995. Mechanisms of resistance to azole antifungal agents in Candida albicans isolates from AIDS patients involve specific multidrug transporters. Antimicrob. Agents Chemother. 39:2378-2386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sanglard, D., and F. C. Odds. 2002. Resistance of Candida species to antifungal agents: molecular mechanisms and clinical consequences. Lancet Infect. Dis. 2:73-85. [DOI] [PubMed] [Google Scholar]
- 46.Selmecki, A., S. Bergmann, and J. Berman. 2005. Comparative genome hybridization reveals widespread aneuploidy in Candida albicans laboratory strains. Mol. Microbiol. 55:1553-1565. [DOI] [PubMed] [Google Scholar]
- 47.Selmecki, A., A. Forche, and J. Berman. 2006. Aneuploidy and isochromosome formation in drug-resistant Candida albicans. Science 313:367-370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Silver, P. M., B. G. Oliver, and T. C. White. 2004. Role of Candida albicans transcription factor Upc2p in drug resistance and sterol metabolism. Eukaryot. Cell 3:1391-1397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Simonics, T., Z. Kozovska, D. Michalkova-Papajova, A. Delahodde, C. Jacq, and J. Subik. 2000. Isolation and molecular characterization of the carboxy-terminal pdr3 mutants in Saccharomyces cerevisiae. Curr. Genet. 38:248-255. [DOI] [PubMed] [Google Scholar]
- 50.Song, J. L., J. B. Harry, R. T. Eastman, B. G. Oliver, and T. C. White. 2004. The Candida albicans lanosterol 14-alpha-demethylase (ERG11) gene promoter is maximally induced after prolonged growth with antifungal drugs. Antimicrob. Agents Chemother. 48:1136-1144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Vik, A., and J. Rine. 2001. Upc2p and Ecm22p, dual regulators of sterol biosynthesis in Saccharomyces cerevisiae. Mol. Cell. Biol. 21:6395-6405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wellington, M., and E. Rustchenko. 2005. 5-Fluoro-orotic acid induces chromosome alterations in Candida albicans. Yeast 22:57-70. [DOI] [PubMed] [Google Scholar]
- 53.White, T. C. 1997. Increased mRNA levels of ERG16, CDR, and MDR1 correlate with increases in azole resistance in Candida albicans isolates from a patient infected with human immunodeficiency virus. Antimicrob. Agents Chemother. 41:1482-1487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.White, T. C. 1997. The presence of an R467K amino acid substitution and loss of allelic variation correlate with an azole-resistant lanosterol 14-alpha-demethylase in Candida albicans. Antimicrob. Agents Chemother. 41:1488-1494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.White, T. C., K. A. Marr, and R. A. Bowden. 1998. Clinical, cellular, and molecular factors that contribute to antifungal drug resistance. Clin. Microbiol. Rev. 11:382-402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.White, T. C., M. A. Pfaller, M. G. Rinaldi, J. Smith, and S. W. Redding. 1997. Stable azole drug resistance associated with a substrain of Candida albicans from an HIV-infected patient. Oral Dis. 3(Suppl. 1):S102-S109. [DOI] [PubMed] [Google Scholar]
- 57.Yoshida, J., K. Umezu, and H. Maki. 2003. Positive and negative roles of homologous recombination in the maintenance of genome stability in Saccharomyces cerevisiae. Genetics 164:31-46. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.