Abstract
We have previously identified and characterized two novel nuclear RNA binding proteins, p34 and p37, which have been shown to bind 5S rRNA in Trypanosoma brucei. These two proteins are nearly identical, with one major difference, an 18-amino-acid insert in the N-terminal region of p37, as well as three minor single-amino-acid differences. Homologues to p34 and p37 have been found only in other trypanosomatids, suggesting that these proteins are unique to this ancient family. We have employed RNA interference (RNAi) studies in order to gain further insight into the interaction between p34 and p37 with 5S rRNA in T. brucei. In our p34/p37 RNAi cells, decreased expression of the p34 and p37 proteins led to morphological alterations, including loss of cell shape and vacuolation, as well as to growth arrest and ultimately to cell death. Disruption of a higher-molecular-weight complex containing 5S rRNA occurs as well as a dramatic decrease in 5S rRNA levels, suggesting that p34 and p37 serve to stabilize 5S rRNA. In addition, an accumulation of 60S ribosomal subunits was observed, accompanied by a significant decrease in overall protein synthesis within p34/p37 RNAi cells. Thus, the loss of the trypanosomatid-specific proteins p34 and p37 correlates with a diminution in 5S rRNA levels as well as a decrease in ribosome activity and an alteration in ribosome biogenesis.
Ribosomes are essential in all organisms, and their assembly is highly conserved and coordinated. Over 100 accessory proteins are necessary in order for proper processing of ribosomal RNAs and ribosome assembly to occur (8). Ribosomal proteins must be imported from the cytoplasm. The 45S precursor rRNA must be processed to yield 5.8S, small-subunit (SSU) (18S), and large-subunit (LSU) (28S) rRNAs. 5S rRNA, which is independently transcribed within the nucleoplasm, must be imported into the nucleolus by the L5 ribosomal protein for ribosome assembly to occur (16). Ribosomal subunits are subsequently exported to the cytoplasm, where the pre-40S ribosomal subunit undergoes its final processing step (29). In eukaryotes, RNA binding proteins mediate a variety of cellular activities, including mRNA maturation, trafficking, stability, and translational control of mRNA as well as having roles in ribosomal biogenesis (14).
The parasite Trypanosoma brucei and its subspecies cause human sleeping sickness (T. brucei gambiense and T. brucei rhodesiense) and nagana in livestock (T. brucei brucei) (31). These organisms continue to pose a serious threat to human health and to cause devastating economic losses (1). Little is currently known about RNA binding proteins and small nucleolar RNAs that are involved in rRNA processing and posttranscriptional modifications in T. brucei. Two proteins with homology to 5S rRNA binding proteins in higher eukaryotes, the La autoantigen and the ribosomal L5 protein, have been identified in T. brucei (19, 34) A family of nucleolar phosphoproteins termed NOPP44/46 proteins have also been identified in this organism and implicated in large ribosomal subunit formation (12, 20).
We have previously shown that two nearly identical proteins, p34 and p37, specifically interact with 5S rRNA in both life cycle stages of this parasite, the insect vector form as well as the mammalian bloodstream form (23). The interaction of p34 and p37 with 5S rRNA may suggest a role in the regulation of trypanosome ribosome biogenesis and may represent only one of the functions of these highly abundant proteins. We have used double-stranded RNA interference (dsRNAi) (24) derived from p34 and p37 to examine molecular and cellular aspects of the function(s) of these proteins.
MATERIALS AND METHODS
Cell growth, transfection, and cloning of p34/p37 dsRNAi cells.
Procyclic T. brucei strain 29-13 (kindly provided by George Cross, Rockefeller University), which contains integrated genes for the expression of T7 polymerase and the tetracycline repressor, was grown in Cunningham's medium supplemented with 15% fetal bovine serum (4). Parasites were cultured in the presence of G418 (15 μg/ml) and hygromycin (50 μg/ml) to maintain the T7 polymerase and the tetracycline repressor constructs, respectively. In the p34/p37 dsRNA construct, pZJMp34/p37 (kindly provided by Paul Englund, Johns Hopkins University), a 501-bp fragment from the central part of the p34 and p37 coding sequence, was inserted between two head-to-head T7 promoters under the control of tetracycline operators (32). Transfections were performed by electroporation as described previously (32). After 24 h of transfection, cells were sedimented and resuspended into 20 ml fresh medium, and selection was achieved by the addition of phleomycin (2.5 μg/ml). After 24 h, cells were diluted 1 to 3 and grown for about 2 weeks to create stable cell lines. Clonal cell lines were generated by extreme dilution (11).
For the induction of dsRNA expression, cells were cultured in the above-described media supplemented with 1.0 μg/ml tetracycline. Growth curves were determined as the products of cell density and total dilution. Control and experimental cells were examined microscopically at the end of all experiments to determine viability, mobility, and morphology. The cellular morphology of wild-type and p34/p37 RNAi cells at 48 h postinduction was assessed by phase-contrast microscopy or by microscopic analysis following Giemsa-Wright staining. Micrographs were obtained using a Nikon microphot-FXA microscope equipped with a Nikon FX-35DX camera at a magnification of ×1,200.
RT-PCR analysis of RNAi cells.
At successive time points, aliquots of cell cultures (4 × 107 cells) were removed and total RNA was isolated using TRIzol reagent (Invitrogen). Reverse transcription-PCR (RT-PCR) amplification (THERMOSCRIPT RT-PCR system; Invitrogen) was performed using the T. brucei miniexon and an internal primer (jz2R), which amplify both p34 and p37 transcripts (35). β-Actin amplification with the same pool of RNA was used as an internal control (27). Densitometric analysis was performed using a GS-700 imaging densitometer in combination with the Multi-Analyst software (Bio-Rad) with the corresponding wild type as the reference.
Western analysis of RNAi cells.
Following induction of p34/p37 dsRNA, aliquots were removed at successive time points, and total cell extracts were prepared (36). Whole-cell protein extracts from 2 × 105 cells were separated by 12% sodium dodecyl sulfate-polyacrylamide gel electrophoresis followed by Western blot analysis with antibodies directed against both p34 and p37 proteins (35). β-Tubulin (Chemicon International) was used as the loading control (35). Western analysis was performed in the linear range for each antibody.
Preparation of nuclear extracts and sucrose gradient analysis.
Nuclear extracts from wild-type or clonal RNAi cells induced by tetracycline for 48 h were prepared as previously described (23). One milligram of total nuclear extract protein was layered onto a continuous 10 to 30% sucrose gradient. Following sedimentation, 28 fractions (400 μl each) were collected (23), and alternate fractions were subdivided into equal aliquots for Northern and Western analysis.
Analysis of 5S rRNA steady-state levels.
Total RNA from wild-type and clonal RNAi cells (5 × 106) at 48 h postinduction was isolated using TRIzol reagent (Invitrogen). Serial dilutions (10-fold) were prepared and utilized in Northern blot analysis (23), and densitometric analysis was performed.
Metabolic labeling of cells.
Following growth in methionine-deficient Cunningham's medium, wild-type and p34/p37 RNAi cells (5 × 107) were labeled in the presence of 100 μCi of [35S]methionine/ml culture for 1 h. Cells were sedimented, washed extensively with phosphate-buffered saline (PBS), and resuspended in 1 ml PBS. Following trichloroacetic acid (TCA) precipitation on ice, material was collected on GF/C filters and washed with TCA and then ethanol, and radioactivity was determined by liquid scintillation counting (3).
Polysome gradients.
Wild-type or p34/p37 RNAi cells (5 × 108 to 1 × 109) were harvested, resuspended in 5 ml medium supplemented with cycloheximide (100 μl/ml), and washed with ice-cold PBS-cycloheximide. The resulting pellet was prepared for polysome analysis (13). Cells were resuspended in 0.75 ml of buffer A (10 mM Tris-HCl, 300 mM KCl, 10 mM MgCl2, pH 7.4) containing protease inhibitors (1 mM phenylmethylsulfonyl fluoride, 8.5 μg/ml aprotinin, 50 μg/ml leupeptin, 1 μM pepstatin, 50 μg/ml TLCK [Nα-p-tosyl-l-lysine chloromethyl ketone], and 10 mM E-64), RNasin (40 U/μl), and 1 mM dithiothreitol. Cells were lysed by the addition of one-sixth volumes of the same buffer containing 0.2 M sucrose and 1.2% Triton N-101 and homogenized with a Dounce homogenizer. The lysate was cleared by centrifugation, and the supernatant was transferred to a fresh tube containing 10 mg/ml heparin (as a generic RNase inhibitor) and then layered on 10 to 40% sucrose gradient (made in buffer A; total volume of 10 ml). After a 2-hour centrifugation at 230,000 × g, 500-μl fractions were collected. The absorbance at 254 nm was monitored using a Teledyne ISCO UA-6 detector during the collection of fractions. Fractions were then characterized by Western analysis using p34/p37 antiserum and an affinity-purified antibody against L5 peptide 296VAAVIERIRDRAK308 (Bethyl Laboratories) in the linear range of detection for each antibody. Fractions were also characterized by Northern analysis using 5S, 18S, and 28S rRNA probes within the linear range of detection (23).
RESULTS
Expression of p34/p37 dsRNA resulted in loss of p34/p37 mRNA and protein.
Fifteen nonclonal p34/p37 RNAi cell lines were developed, and two of these (C9 and C10) were chosen for further development into clonal cell lines. Southern blot analysis of these cell lines showed that the endogenous p34/p37 gene fragment was observed in both wild-type and RNAi cell lines. An approximately 500-bp fragment corresponding to the p34/p37 RNAi fragment was detected only in the RNAi cell lines and was absent in the parental strain 29-13 cells prior to transfection (data not shown). These data confirmed that the construct containing the p34/p37 RNAi fragment was integrated into the genome of T. brucei.
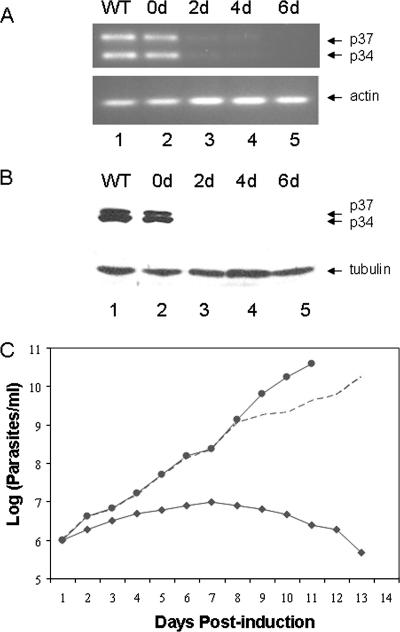
RT-PCR analysis of clonal cell line C9-3 was performed as described previously (35) (Fig. 1A) to allow us to distinguish between the p34 and p37 transcript levels. RT-PCR allowed a clear separation of p34 and p37 mRNA, compared to what was seen with Northern analysis, since only the 5′ end of the transcripts, where the major difference between the p34 and p37 resides, was amplified. Results from RT-PCR analysis showed that even without the induction of p34/p37 dsRNA expression, the steady-state levels of both p34 and p37 mRNA were decreased modestly (10 to 20%) compared to those for wild-type cells (Fig. 1A, top, compare lanes 1 and 2). Upon the initiation of p34/p37 dsRNA expression, mRNA levels of both p34 and p37 were reduced to less than 10% of the wild-type cells by 2 days (lane 3). RT-PCR of β-actin using the same pool of RNA was used as an internal control and confirmed that the decrease in mRNA level is specific to p34 and p37 transcripts (Fig. 1A, bottom).
FIG. 1.
Down-regulation of p34 and p37 in clonal p34/p37 RNAi cells results in cell death. (A) Total RNA was isolated from T. brucei and used in RT-PCR with the miniexon primer and an internal primer that will amplify both p34 and p37 mRNA and distinguish between the RT-PCR products of p34 and p37 mRNA by size. Lanes: 1, wild-type (WT) cells; 2 to 5, RNAi cells at 0 days (0d) to 6 days postinduction. The positions of the p34 and p37 RT-PCR products are shown by arrows. RT-PCR of β-actin mRNA was performed using the same pool of RNA as an internal control. (B) Each lane contains cell extract from 2.5 × 106 cells per lane from WT cells (lane 1) or RNAi cells after tetracycline induction for 0, 2, 4, and 6 days (lanes 2 through 5). Proteins were separated by 12% sodium dodecyl sulfate-polyacrylamide gel electrophoresis followed by Western blot analysis with antibody against both p34 and p37. (C) Growth curves for wild-type cells (solid circles) and clonal cell lines under uninduced (open diamonds) and induced (closed diamonds) conditions. The cell number was plotted on a logarithmic scale versus the days postinduction of p34/p37 dsRNA expression.
Western blot analysis utilizing antibody directed against both p34 and p37 is shown in Fig. 1B, top. β-Tubulin was utilized as a loading control (Fig. 1B, bottom). Again, we observed a modest decrease (10 to 20%) in the expression level of p34 protein in the RNAi cells, suggesting some leakage in the RNAi vector at time zero compared to what was seen for the wild type. After the induction of p34/p37 dsRNA, a rapid loss of both p34 and p37 proteins was observed (Fig. 1B, lanes 2 through 5). By 2 days postinduction, p34 and p37 protein levels were nearly undetectable (Fig. 1B, lane 3), and by 4 days, neither protein was detectable (Fig. 1B, lanes 4 and 5). The gene fragment that was utilized in the creation of the p34/p37 RNAi cell line overlapped a region within p34 and p37 genes which contains an RNA binding domain. To verify the specificity of RNAi to p34 and p37, we confirmed that the levels of the poly(A) binding protein, which contains several RNA binding domain motifs, were unaffected in the p34/p37 RNAi cells (data not shown).
p34 and p37 are essential in T. brucei procyclic cells.
Following the induction of p34/p37 dsRNA, growth arrest was observed within 2 to 3 days, and no viable cells were found at 2 weeks postinduction, whereas uninduced cells showed a growth rate similar to that of wild-type cells (Fig. 1C). The observed loss of cell viability correlated with Western blot analysis showing that neither p34 nor p37 protein is detectable (Fig. 1B) by 2 days postinduction.
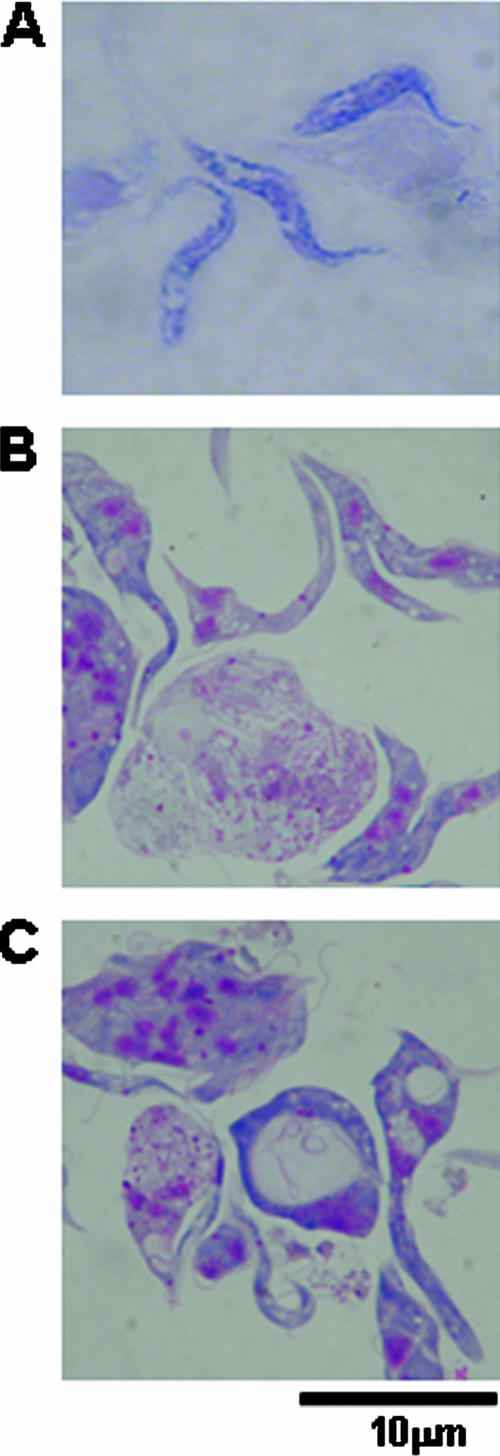
Compared to wild-type cells (Fig. 2A), RNAi cells became broader, with granules and vacuoles forming inside of the cells (Fig. 2B and C). Many cells were multinucleated and branched (Fig. 2B), suggesting problems with cellular division. Finally, observation of live cells by phase contrast showed that an increased proportion of cells formed clusters containing from 5 to more than 20 parasites (data not shown). The rate of cellular movement decreased and the pattern of movement was less fluid and clearly different from that seen for wild-type cells.
FIG. 2.
Morphological changes in cells expressing p34/p37 dsRNA. (A) Micrograph of Giemsa-stained wild-type procyclic trypanosomes. (B and C) Morphological changes in p34/p37 RNAi cells at 48 h postinduction.
5S rRNA sedimentation in sucrose density gradients is altered in nuclear extracts from RNAi cells.
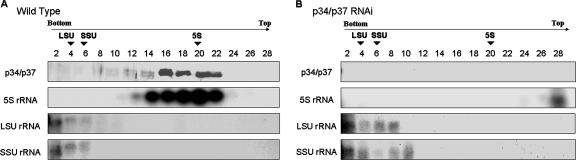
We have previously shown that p34 and p37 interact with 5S rRNA in a stable protein-RNA complex (23). Nuclear extracts of clonal RNAi-expressing cells (2 days postinduction) were separated on 10 to 30% sucrose gradients to determine the effect of decreased expression of p34 and p37 on complexes containing 5S rRNA. Results from wild-type nuclear extracts indicated that 5S rRNA and p34/p37 cosedimented near the middle of the gradient (Fig. 3A, fractions 12 to 22). Western blot analysis confirmed that p34 and p37 protein levels present in RNAi cell extracts were insufficient to be detectable within individual sucrose gradient fractions (Fig. 3B). Strikingly, the position of the 5S rRNA signal in the RNAi nuclear extracts dramatically shifted to the top of the gradient and the intensity was significantly decreased (Fig. 3B), suggesting that 5S rRNA present in the p34/p37 RNAi cells was no longer part of a higher-molecular-weight complex within the nucleus. This shift in the sedimentation profile and difference in signal intensity was specific to 5S rRNA and did not occur with either SSU rRNA (18S rRNA) or LSU rRNA (28S rRNA) (compare Fig. 3A and B). The sedimentation profile of RNA B, the T. brucei U3 homologue, was not affected in p34/p37 RNAi nuclear extracts (data not shown).
FIG. 3.
Effects of p34/p37 dsRNA expression on 5S rRNA. (A) Nuclear extracts of wild-type cells. (B) Nuclear extracts of p34/p37 RNAi cells which were sedimented through continuous 10 to 30% sucrose gradients. The positions of 5S rRNA, SSU rRNA (18S rRNA), and LSU rRNA (28S rRNA) were determined by Northern blot analysis, and the positions of p34 and p37 were detected by Western blot analysis. Fraction 2 is the bottom of the gradient. Arrows indicate the positions of bovine rRNA markers.
Loss of p34 and p37 leads to a decrease in 5S rRNA levels.
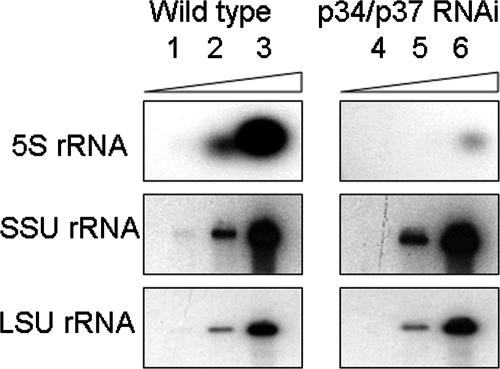
As indicated above, in addition to the change in sedimentation from nuclear extracts, we also observed a dramatic decrease in 5S rRNA levels within the p34/p37 RNAi cells. To further explore this phenomenon, we compared 5S rRNA levels from equivalent numbers of wild-type and p34/p37 RNAi cells by use of Northern blots to determine the extent of 5S rRNA loss (Fig. 4). In all comparative studies, wild-type cells were utilized as a reference, since some basal level of p34/p37 dsRNA expression occurred within uninduced RNAi cells. Increasing levels of total RNA are shown in lanes 1 to 3 for wild-type samples and in lanes 4 to 6 for p34/p37 RNAi samples. Upon quantification from four experiments, we found a (25 ± 0.42)-fold decrease in 5S rRNA levels in the RNAi cells compared to that for wild-type cells. This change was specific to 5S rRNA and was not seen for LSU rRNA (28S) or SSU rRNA (18S).
FIG. 4.
Loss of p34 and p37 leads to a decrease in 5S rRNA levels. Northern blot analysis of 10-fold serial dilutions of total RNA from equivalent numbers (5 × 106) of wild-type (lanes 1 to 3) and p34/p37 RNAi (lanes 4 to 6) cells. Results are representative of four experiments.
Efficiency of protein synthesis is significantly decreased in p34/p37 RNAi cells.
The loss of 5S rRNA combined with the rapid onset of cell death indicated that a defect in ribosome function may be occurring. Pulse-labeling with [35S]methionine was performed to determine whether protein synthesis is affected in the RNAi cells. Equivalent numbers of cells were utilized in each experiment for comparison of protein synthesis between wild-type and RNAi cells at successive times of p34/p37 dsRNA expression induction (Fig. 5). Prior to induction, protein synthesis levels were as much as 25% below those found for wild-type cells. By 2 days postinduction, concurrent with the loss of both p34 and p37 proteins (Fig. 1B), a 42% decrease in overall protein synthesis compared to that for uninduced cells was observed, which corresponds to a 57% decrease from that for the wild-type control. This difference remained static at days 3 and 4 postinduction. In contrast, the overall level of protein synthesis in the wild-type cells remained constant throughout the full 4-day course of the experiment.
FIG. 5.
Loss of p34 and p37 leads to a disruption in protein synthesis efficiency. Equivalent numbers (5 × 107) of wild-type and p34/p34 RNAi cells were pulse-labeled with [35S]methionine for 1 hour. Protein synthesis was measured as the ratio of methionine incorporated into TCA-precipitated proteins versus cell-associated counts. Shown is a graphical representation of the data, in which the lighter bars are from wild-type cells and the darker bars correspond to p34/p37 RNAi cells. The percents incorporation indicated are from four individual experiments taken at different times postinduction (T0, 0 h postinduction).
Induction of p34/p37 RNAi leads to an alteration in the sedimentation profile.
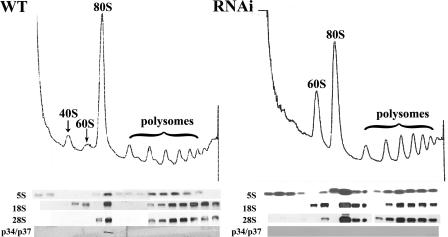
In order to analyze the effect(s) that the loss of p34 and p37 may have on the assembly of ribosomes and ribosome biogenesis, we performed polysome sedimentation analysis. Cells were treated with cycloheximide prior to lysis to block translational elongation, thus trapping ribosomes on the mRNAs. Sedimentation of the lysate on a 10 to 40% sucrose gradient resolved ribosomal subunits (40S and 60S) as well as monosomal (80S) and polysomal particles (Fig. 6). Sedimentation profiles from wild-type cell lysates exhibited both 40S and 60S peaks as well as the predominant 80S peak and up to seven well-defined polysomal peaks (Fig. 6, left). In contrast, lysates from cells depleted of p34 and p37 exhibited an increase in the 60S LSU peak and a concomitant decrease in the 80S subunit (Fig. 6, right); that is, the 60S:80S ratio is increased when the dsRNA is induced. The 40S peak was masked by an increase in low-molecular-weight soluble components; however, longer sedimentation in the presence of puromycin allowed us to verify that the 40S peak is still present (data not shown). In addition, we observed a decrease in polysomal peaks, consistent with our previous observations of a decrease in overall protein synthesis. These results suggest that the 60S subunit may be defective in its ability to be incorporated into the 80S particle.
FIG. 6.
Cells depleted of p34 and p37 by RNAi accumulate the 60S ribosomal subunit. Cell lysates of wild-type (WT) and 72-h-induced p34/p37 RNAi cells (approximately 1 × 109 cells) were treated with cycloheximide and layered onto 10 to 40% sucrose gradients. The A254 traces of wild-type (left) and p34/p37 RNAi (right) cell lysates are shown, with the top of the gradient at the left. Two hundred-microliter aliquots of each fraction from the polysomal sedimentation analysis were subjected to Northern analysis using 5S, 28S (LSU), and 18S (SSU) probes and by Western analysis using anti-p34/p37 antiserum.
As shown in Fig. 6, bottom, the identity of the peaks was confirmed by Northern analysis using probes directed against 5S rRNA and 28S rRNA (LSU, a constituent of the 60S subunit) and 18S rRNA (SSU, a constituent of the 40S subunit). Interestingly, we found that in the p34/p37 RNAi cells there was a larger fraction of the 5S rRNA associated with low-molecular-weight fractions (lanes 1 to 4) and the 80S peak (lanes 7 and 8). The p34 and p37 proteins were associated with the 80S peak in the wild-type cells (Fig. 6, bottom left) but were not detectable in other fractions. As anticipated, the p34 and p37 proteins were undetectable in the p34/p37 RNAi cells (Fig. 6, bottom right). The L5 protein, in contrast, shows similar distributions in both wild-type and RNAi cells (data not shown).
DISCUSSION
We have previously identified two nearly identical RNA binding proteins, p34 and p37 from T. brucei, which contain two consensus RNA binding domain motifs (36). Within the coding sequences of p34 and p37, the only major difference is an 18-amino-acid insert within the amino terminus of p37 which is absent in p34 and minor differences at three amino acid positions. These proteins specifically interact with 5S rRNA in both stages of the parasite life cycle (23). Furthermore, these proteins are unique to trypanosomes (9), suggesting that the interaction with 5S rRNA serves a unique function within these parasites. In the experiments presented here, we knocked down the expression of both p34 and p37 proteins by use of RNAi to further characterize their interaction with 5S rRNA.
Our results show that p34 and p37 are essential for T. brucei survival. Loss of p34 and p37 led to cessation of cell growth and ultimately to cell death. Phenotypically, we observed granules and vacuoles as well as multinucleate cells within the clonal cell population. Most of the cells lost normal shape (became more rounded), exhibited slower movement, and tended to cluster together in groups of 5 to 20 cells. Some of these phenotypic characteristics have also been reported upon disruption of tubulin, histone 2B, the RNA binding protein RRM1, and topoisomerase II with RNAi in T. brucei (15, 18, 25, 32), making it impossible to infer the exact functions of p34 and p37 proteins based solely on phenotype.
Significantly, we observed major changes in a complex that normally contains p34 and p37 as well as 5S rRNA. In cells expressing p34/p37 RNAi, there was a dramatic decrease in the sedimentation of 5S rRNA, suggesting that 5S rRNA was no longer part of a higher-molecular-weight complex in the nucleus. This shift in sedimentation coincided with a 25-fold decrease in 5S rRNA levels. These results are similar to those observed for yeast (Saccharomyces cerevisiae) cells depleted of the homologue to the higher eukaryotic L5 (the yeast L1 protein) in a conditional lethal mutant where a decrease in the stability as well as the level of newly synthesized 5S rRNA was observed (6). Furthermore, in yeast cells expressing 5S rRNA mutants which no longer bind the yeast L5 homologue, 5S rRNA is not stable and is rapidly degraded (17). It is believed that the yeast L5 homologue serves to stabilize and protect 5S rRNA from nuclease degradation. Our results suggest that p34 and p37 may serve a similar role in T. brucei. Given the results reported here, it may be that p34 and p37 function cooperatively with T. brucei L5 or may serve as an alternative pathway for 5S rRNA transport to the nucleolus. Conversely, the complex between p34 and p37 with 5S rRNA may serve as a storage particle for 5S rRNA, thus allowing cells to maintain an intranuclear pool of 5S rRNA. This would be similar to the situation with Xenopus oocytes, where transcription factor TFIIIA interacts with 5S rRNA in a cytoplasmic complex serving as a storage RNP for 5S rRNA in preparation for ribosome assembly toward the end of oogenesis (21, 28). Experiments are under way to distinguish between these possible mechanisms for p34 and p37 interaction with 5S rRNA in T. brucei.
We have shown here that the loss of the p34 and p37 proteins correlates with an average 60% decrease in protein synthesis and with an accumulation of 60S and a loss of 80S ribosomal subunits compared to what is seen for wild-type cells within 72 h postinduction of dsRNA production. Uninduced p34/p37 RNAi cells showed as much as a 25% drop in protein synthesis efficiency compared to what is seen for wild-type cells, even though they grew at a rate similar to the wild-type rate. In addition, after 72 h of dsRNA production, no major changes were observed in the polysomal particles that were able to form. This suggests that a modest drop in protein synthesis does not have a strong effect on cell growth, while a further loss of translation efficiency within induced p34/p37 RNAi cells later in the time course appears to be fatal. Therefore, the overall decrease in protein synthesis in p34/p37 RNAi cells may render these cells unable to keep up with cellular demands.
The incorporation of 5S rRNA into the 60S ribosomal subunit occurs late in ribosome assembly (5). Even though the exact function(s) of 5S rRNA has not been clearly defined, many potential functions have been assigned to 5S rRNA (26). In Escherichia coli, a stable and partially functional ribosome which lacked 5S rRNA was reconstituted in vitro, although these ribosomes were able to synthesize protein only at a very low rate, which was, however, dramatically increased upon the addition of 5S rRNA (7). In this case, it has been established that that 5S rRNA is critical for tRNA binding to the A-site (aminoacyl-tRNA binding site) of the ribosome (7). For yeast, it has been proposed that since 5S rRNA provides a physical connection between the different functional regions of the ribosome, it serves as a signal transducer to facilitate communication between these regions, thus helping to direct their coordination of translation events (26). Furthermore, the site of peptidyltransferase activity and the EF-G binding site may communicate through 5S rRNA via its interaction with LSU rRNA (2).
The loss of p34 and p37 resulted in a significant loss of 5S rRNA in p34/p37 RNAi cells, and in our polysomal gradients, a greater fraction of the remaining 5S rRNA was found in the lower-molecular-weight region in the p34/p37 RNAi extracts than in the wild-type extracts (Fig. 6). Taken together with the accumulation of possibly defective 60S subunits, seen here as a shift of the ratio between the 60S peak and the 80S peak, these results indicate that p34 and/or p37 may have a role in the assembly of the LSU. In the absence of p34 and p37, a step in the biogenesis of a functional 60S subunit would then be blocked, and this subunit would be rendered unable to join a 40S subunit to form the mature 80S ribosome. In one possible model, p34 and p37 could bind and stabilize 5S rRNA and direct it (perhaps via an association with other ribosomal components) to the 60S subunit. Later, binding of the mRNA or incorporation of the 40S small subunit could cause p34 and p37 to be released. In yeast, mutations of 5S that make it unstable lead to decreased assembly of 60S particles (5, 30). This observation raises the possibility that the effect of p34 and p37 on the levels of 60S particles may take place indirectly, through their effect on 5S stability. We did observe lower-molecular-weight species, suggesting the presence of partially assembled 60S particles and indicating (in accordance with the results obtained with yeast by use of mutant, unstable 5S) that a complete pool of 60S subunits is never assembled.
Although ribosome assembly is highly coordinated and conserved, some organisms, such as the parasite T. brucei, contain several unique features of ribosome biogenesis. In T. brucei, the LSU rRNA, which consists of a single species in other organisms, is processed into six distinct RNAs (10, 34). Additionally, it has recently been established that a family of trypanosome-specific phosphoproteins, NOPP44/46, are involved in this processing of LSU rRNA. The loss of NOPP44/46 by RNAi led to aberrant processing of pre-LSU rRNA, disruption of large ribosomal subunit (60S) assembly, and cell death (12). Interestingly, we have previously established an interaction between the T. brucei proteins p34 and p37 with the NOPP44/46 proteins (22). At this time, we do not know whether the interaction of p34 and p37 with NOPP44/46 occurs in conjunction with that with 5S rRNA or whether the interaction of p34 and p37 with 5S rRNA is a separate and distinct phenomenon. Given the abundance of p34 and p37 in the cell, it is possible that these proteins are involved in multiple processes and have many functions throughout the parasite life cycle. Results from p34/p37 RNAi cells emphasize the vital role(s) that p34 and p37 proteins play. The interaction between these novel proteins and 5S rRNA further underscores the importance of these proteins, and they are yet another unique feature of trypanosomes.
Acknowledgments
We thank Paul Englund (Johns Hopkins University) for the p34/p37 dsRNA construct and for scientific advice. We thank George Cross (Rockefeller University) for T. brucei strain 29-13. We thank Bryan Jensen and Marilyn Parsons (Seattle Biomedical Research Institute) for assistance with initial polysome sedimentation analysis.
This work was supported by NIAID grants AI 41134 and AI57142 (to N.W. and W.R.) and NIH predoctoral training grant T32AI07614 for the support of K.M.H.
Footnotes
Published ahead of print on 22 August 2007.
REFERENCES
- 1.Barrett, M. P., R. J. S. Burchmore, A. Stitch, J. O. Lazzari, A. C. Frasch, J. J. Cazzulo, and S. Krishna. 2003. The trypanosomiases. Lancet 362:1469-1480. [DOI] [PubMed] [Google Scholar]
- 2.Bogdanov, A. A., O. A. Dontsova, S. S. Dokudovskaya, and I. N. Lavrik. 1995. Structure and function of 5S rRNA in the ribosome. Biochem. Cell Biol. 73:869-876. [DOI] [PubMed] [Google Scholar]
- 3.Bonifacino, J. S. 2004. Metabolic labeling with amino acids. p. 10.18.1-10.18.7. In F. M. Ausubel, R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl (ed.), Current protocols in molecular biology, John Wiley & Sons, Inc., New York, NY. [DOI] [PubMed]
- 4.Cunningham, I. 1977. New culture medium for maintenance of tsetse tissues and growth of trypanosomatids. J. Protozool. 24:325-329. [DOI] [PubMed] [Google Scholar]
- 5.Dechampesme, A. M., O. Koroleva, I. Leger-Silvestre, N. Gas, and S. Camier. 1999. Assembly of 5S ribosomal RNA is required at a specific step of the pre-rRNA processing pathway. J. Cell Biol. 145:1369-1380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Deshmukh, M., Y. Tsay, A. G. Paulovich, and J. L. Woolford, Jr. 1993. Yeast ribosomal protein L1 is required for stability of newly synthesized rRNA and the assembly of 60S ribosomal subunits. Mol. Cell. Biol. 13:2835-2845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dohme, F., and K. H. Nierhaus. 1976. Role of 5S RNA in assembly and function of the 50S subunit from Escherichia coli. Proc. Natl. Acad. Sci. USA 73:2221-2225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fatica, A., and D. Tollervey. 2002. Making ribosomes. Curr. Opin. Cell Biol. 14:313-318. [DOI] [PubMed] [Google Scholar]
- 9.Gomes, G. G., T. Urmenyi, E. Rondinelli, N. Williams, and R. Silva. 2004. TcRRMs and Tcp28 genes are intercalated and differentially expressed in Trypanosoma cruzi life cycle. Biochem. Biophys. Res. Commun. 322:985-992. [DOI] [PubMed] [Google Scholar]
- 10.Hasan, G., M. J. Turner, and J. S. Cordingley. 1984. Ribosomal RNA genes of Trypanosoma brucei: mapping the regions specifying the six small ribosomal RNAs. Gene 27:75-86. [DOI] [PubMed] [Google Scholar]
- 11.Huang, C. E., S. F. O'Hearn, and B. Sollner-Webb. 2002. Assembly and function of the RNA editing complex in Trypanosoma brucei requires band III protein. Mol. Cell. Biol. 22:3194-3203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jensen, B. C., D. L. Brekken, A. C. Randall, C. T. Kifer, and M. Parsons. 2005. Species specificity in ribosome biogenesis: a nonconserved phosphoprotein is required for formation of the large ribosomal subunit in Trypanosoma brucei. Eukaryot. Cell 4:30-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jensen, B. C., Q. Wang, C. T. Kifer, and M. Parsons. 2003. The NOG1 GTP-binding protein is required for biogenesis of the 60S ribosomal subunit. J. Biol. Chem. 278:32204-32211. [DOI] [PubMed] [Google Scholar]
- 14.Kressler, D., P. Linder, and J. De la Cruz. 1999. Protein trans-acting factors involved in ribosome biogenesis in Saccharomyces cerevisiae. Mol. Cell. Biol. 19:7897-7912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.LaCount, D. J., S. Bruse, K. L. Hill, and J. E. Donelson. 2000. Double-stranded RNA interference in Trypanosoma brucei using head-to-head promoters. Mol. Biochem. Parasitol. 111:67-76. [DOI] [PubMed] [Google Scholar]
- 16.Larson, D. E., P. Zahradka, and B. H. Sells. 1991. Control points in eucaryotic ribosome biogenesis. Biochem. Cell Biol. 69:5-22. [DOI] [PubMed] [Google Scholar]
- 17.Lee, Y., and R. N. Nazar. 1997. Ribosomal 5 S rRNA maturation in Saccharomyces cerevisiae. J. Biol. Chem. 272:15206-15212. [DOI] [PubMed] [Google Scholar]
- 18.Manger, I. D., and J. C. Boothroyd. 2001. Targeted disruption of an essential RNA-binding protein perturbs cell division in Trypanosoma brucei. Mol. Biochem. Parasitol. 116:239-245. [DOI] [PubMed] [Google Scholar]
- 19.Michaeli, S., and N. Agabian. 1990. A Trypanosoma brucei small RNP particle containing the 5S rRNA. Mol. Biochem. Parasitol. 41:7-15. [DOI] [PubMed] [Google Scholar]
- 20.Parsons, M., J. A. Ledbetter, G. L. Schieven, A. E. Nel, and S. B. Kanner. 1994. Developmental regulation of pp44/46, tyrosine-phosphorylated proteins associated with tyrosine/serine kinase activity in Trypanosoma brucei. Mol. Biochem. Parasitol. 63:69-78. [DOI] [PubMed] [Google Scholar]
- 21.Pelham, H. R. B., and D. D. Brown. 1980. A specific transcription factor that can bind either the 5S rRNA gene or 5S rRNA. Proc. Natl. Acad. Sci. USA 77:4170-4174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pitula, J., J. Park, M. Parsons, W. T. Ruyechan, and N. Williams. 2002. Two families of RNA binding proteins from Trypanosoma brucei associate in a direct protein-protein interaction. Mol. Biochem. Parasitol. 122:81-89. [DOI] [PubMed] [Google Scholar]
- 23.Pitula, J., W. T. Ruyechan, and N. Williams. 2002. Two novel RNA binding proteins from Trypanosoma brucei are associated with 5S rRNA. Biochem. Biophys. Res. Commun. 290:569-576. [DOI] [PubMed] [Google Scholar]
- 24.Sharp, P. 2001. RNA interference. Genes Dev. 15:485-490. [DOI] [PubMed] [Google Scholar]
- 25.Shi, H., A. Djikeng, T. Mark, E. Wirtz, C. Tschudi, and E. Ullu. 2000. Genetic interference in Trypanosoma brucei by heritable and inducible double-stranded RNA. RNA 6:1069-1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Smith, M. W., A. Meskauskas, P. Wang, P. V. Sergiev, and J. D. Dinman. 2001. Saturation mutagenesis of 5S rRNA in Saccharomyces cerevisiae. Mol. Cell. Biol. 21:8264-8275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Stojdl, D., and M. Clarke. 1996. Trypanosoma brucei: analysis of cytoplasmic Ca2+ during differentiation of bloodstream stages in vitro. Exp. Parasitol. 83:134-146. [DOI] [PubMed] [Google Scholar]
- 28.Szymański, M., M. Z. Baraciszewska, V. A. Erdmann, and J. Barciszewski. 2003. 5S rRNA: structure and interactions. Biochem. J. 371:641-651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tschochner, H., and E. Hurt. 2003. Pre-ribosomes on the road from the nucleolus to the cytoplasm. Trends Cell Biol. 13:255-263. [DOI] [PubMed] [Google Scholar]
- 30.Van Ryk, D. I., Y. Lee, and R. N. Nazar. 1992. Unbalanced ribosome in Saccharomyces cerevisiae expressing mutant 5S rRNAs. J. Biol. Chem. 23:16177-16181. [PubMed] [Google Scholar]
- 31.Vickerman, K. 1985. Developmental cycles and biology of pathogenic trypanosomes. Br. Med. Bull. 41:105-114. [DOI] [PubMed] [Google Scholar]
- 32.Wang, Z., J. Morris, M. Drew, and P. T. Englund. 2000. Inhibition of Trypanosoma brucei gene expression by RNA interference using an integratable vector with opposing T7 promoters. J. Biol. Chem. 275:40174-40179. [DOI] [PubMed] [Google Scholar]
- 33.Reference deleted.
- 34.White, T. C., G. Rudenko, and P. Borst. 1986. Three small RNAs within the 10 kb trypanosome rRNA transcription unit are analogous to domain VII of other eukaryotic 28S rRNAs. Nucleic Acids Res. 14:9471-9489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhang, J., W. T. Ruyechan, and N. Williams. 1998. Developmental regulation of two nuclear RNA binding proteins, p34 and p37, from Trypanosoma brucei. Mol. Biochem. Parasitol. 92:79-88. [DOI] [PubMed] [Google Scholar]
- 36.Zhang, J., and N. Williams. 1997. Purification, cloning, and expression of two closely related Trypanosoma brucei nucleic acid binding proteins. Mol. Biochem. Parasitol. 87:145-158. [DOI] [PubMed] [Google Scholar]