Abstract
Hyphal morphogenesis in Candida albicans is regulated by multiple pathways which act by either inducing or repressing filamentation. Most notably, Tup1, Nrg1, and Rfg1 are transcriptional repressors, while Efg1, Flo8, Cph1, and Czf1 can induce filamentation. Here, we present the functional analysis of CaSFL1, which encodes the C. albicans homolog of the Saccharomyces cerevisiae SFL1 (suppressor of flocculation) gene. Deletion of CaSFL1 results in flocculation (i.e., the formation of clumps) of yeast cells, which is most pronounced in minimal medium. The flocs contained hyphae already under noninducing conditions, and filamentation could be enhanced with hypha-inducing cues at 37°C. Expression of SFL1 in a heterozygous mutant under the control of the CaMET3 promoter was shown to complement these defects and allowed switching between wild-type and mutant phenotypes. Interestingly, increased expression of SFL1 using a MET3prom-SFL1 construct prior to the induction of filamentation completely blocked germ tube formation. To localize Sfl1 in vivo, we generated a SFL1-GFP fusion. Sfl1-green fluorescent protein was found in the nucleus in both yeast cells and, to a lesser extent, hyphal cells. Using reverse transcription-PCR, we find an increased expression of ALS1, ALS3, HWP1, ECE1, and also FLO8. Our results suggest that Sfl1 functions in the repression of flocculation and filamentation and thus represents a novel negative regulator of C. albicans morphogenesis.
Among opportunistic human fungal pathogens, Candida albicans is one of the most frequently isolated species. In addition to superficial infections of the skin, vagina, or oral epithelia, C. albicans can cause systemic infections. Particularly in immunocompromised hosts, bloodstream infections pose a serious threat (3). C. albicans is a dimorphic fungus that can switch between yeast and filamentous vegetative growth modes. The ability to switch growth modes, in conjunction with the ability to induce stage-specific gene expression, is viewed to be important for its pathogenicity (34). Niche-specific responses may allow C. albicans to make the best use of its repertoire to survive as a commensal in the human host (2, 13).
Dimorphism in C. albicans is an attractive model to study morphogenetic events on the molecular level (3). In vitro-specific conditions can be used to maintain C. albicans cells either in the yeast form or by using external cues, such as serum or amino acids, to induce filament formation and mycelium development. On the molecular level, several regulators of hypha-specific gene expression have been identified. Two basic classes can be distinguished—proteins that act as inducers of hypha-specific gene expression, e.g., Efg1, Cph1, Flo8, Tec1, and Czf1, and proteins that act as negative regulators, particularly Tup1 and Nrg1 (3, 10). The cyclic AMP (cAMP) pathway appears to play a major role in regulating hyphal morphogenesis, since both Ras1 and the adenylate cyclase Cdc35 (in association with Cap1) are essential for hyphal growth and virulence (1, 14, 29). Efg1 is a downstream target of the Ras1-cAMP signaling pathway, and efg1/efg1 mutants are defective in filament formation in liquid media (32). Flo8 was shown to regulate the hypha-specific gene subset controlled by Efg1 and does so via direct interaction with Efg1. This finding is consistent with the evidence that deletion of FLO8 blocks hyphal development in C. albicans (10).
CaNrg1 is a zinc finger DNA-binding protein that represses hypha-specific genes in a CaTup1-dependent manner. Expression of NRG1 is downregulated upon hyphal induction, and deletion of either TUP1 or NRG1 enhances filamentous growth under noninducing conditions (8, 26). This indicates that relief of transcriptional repression is a major event in the dimorphic switching of C. albicans (22).
Hyphal induction media may include various additives that trigger filamentation at 37°C, e.g., serum, amino acids, or N-acetylglucosamine (GlcNAc). It has been observed that when serum is used as an inducer, cells readily aggregate and form clumps, while this is true to a much lesser extent when GlcNAc is used (4). The ability of C. albicans cells to adhere to other cells or to surfaces poses an important medical problem, particularly in biofilm formation on medical devices that allow these cells to gain access to the bloodstream of patients (25). On an industrial scale, the ability to form cell aggregates is used during beer fermentation by Saccharomyces cerevisiae to separate yeast cells from the beer. This process is termed flocculation and leads to a rapid sedimentation of lager yeast cells. Flocculation is dependent on the expression of FLO genes. There are two classes of FLO genes, which confer either cell-cell adhesion (e.g., Flo1, Flo5, Flo9, and Flo10) or cell-substrate adhesion (Flo11) (20). Different pathways are known to regulate the expression of, e.g., the FLO11 gene in S. cerevisiae, among them the Ras1-cAMP signaling pathway and the mitogen-activated protein kinase-dependent filamentous growth pathway, as well as the glucose repression pathway and the TOR pathway (33). Downstream of the cAMP-signaling pathway, Tpk2 activates Flo8 and represses Sfl1, which encodes a suppressor of flocculation (11, 15).
We were interested to determine whether flocculation in C. albicans is regulated by a similar mechanism as that for S. cerevisiae and whether flocculation and filamentation can be separated genetically. We identified a SFL1 homolog in the C. albicans genome. The deletion of SFL1 led to a flocculent phenotype best seen in minimal media. This flocculation was accompanied by filamentation under noninducing conditions. SFL1-GFP localizes to the nucleus in both yeast and hyphal cells. Overexpression of SFL1, on the other hand, led to a block in filamentation. This result suggests that Sfl1 is a negative regulator of both flocculation and filamentation in C. albicans. Consistently, hypha-specific genes, particularly ALS1 and ALS3, were found to be expressed in a sfl1 mutant under noninducing conditions.
MATERIALS AND METHODS
Strains and media.
The C. albicans and S. cerevisiae strains used in this study are listed in Table 1. Cells were grown either in rich medium (YPD; 2% peptone, 2% glucose, 1% yeast extract) or in minimal medium (CSM [2% glucose, 6.7 g/liter yeast nitrogen base with ammonium sulfate and without amino acids], 0.79 g/liter CSM, or SD [2% glucose, 6.7 g/liter yeast nitrogen base with ammonium sulfate and without amino acids]) at 30°C. Minimal media were supplemented with the strain-specific requirements for amino acids, adenine, and uridine. Hyphal induction was done at 37°C by adding 10% serum (calf serum; Sigma) or 0.5g/liter GlcNAc to the media.
TABLE 1.
Strains used in this study
| Strain | Genotype | Source or reference |
|---|---|---|
| SC5314 | C. albicans wild type | 17 |
| CAI-8 | ura3::λimm434/ura3::λimm434 ade2::hisG/ade2::hisG | 30 |
| CAI-8+SalexA-lexOP | RPS1/rps1::SalexA-RPS1-URA3 ade2::hisG/ade2::OP-LacZ-ADE2 | 30 |
| CAI8-SalexA-NRG1-lexOP | RPS1/rps1::SlLexA-NRG1-RPS1-URA3 ade2::hisG/ade2::OP-LacZ-ADE2 | 30 |
| CAI8-SalexA-GCN4-lexOP | RPS1/rps1::SalexA-GCN4-RPS1-URA3 ade2::hisG/ade2::OP-LacZ-ADE2 | 30 |
| SN148 | ura3::λimm434/ura3::λimm434 arg4::hisG/arg4::hisG his1::hisG/his1::hisG leu2::hisG/leu2::hisG | 27 |
| CAJ025 | SN148; SFL1/sfl1::CmLEU | This study |
| CAJ027 | SN148; SFL1/sfl1::CdHIS1 | This study |
| CAJ029 | SN148; sfl1::CmLEU2/sfl1::CdHIS1 | This study |
| CAJ035 | SN148; sfl1::CdHIS1/sfl1::CmLEU2 | This study |
| CAJ039 | SN148; sfl1::CmLEU2/sfl1::HIS1-MET3p-SFL1 | This study |
| CAJ042 | SN148; sfl1::CmLEU2/sfl1::SFL1-GFP-CdHIS1 | This study |
| CAJ049 | CAI-8; RPS1/rps1::SalexA-SFL1-RPS1-URA3 ade2::hisG/ade2::OP-LacZ-ADE2 | This study |
| CAJ066 | CAI-8; RPS1/rps1::SalexA-SFL1-RPS1-URA3 ade2::hisG/ade2::OP-LacZ-ADE2 | This study |
| CAJ071 | SN148; sfl1::CmLEU2/sfl1::URA3 | This study |
Regulation of gene expression was done using the MET3 promoter. Activation of MET3p-SFL1 expression was achieved by growing cells in media lacking methionine and cysteine, and repression of the MET3p promoter could be done by adding 3.5 mM methionine and 3.5 mM cysteine to the medium. For plasmid construction and amplification, Escherichia coli DH5α served as a host.
Disruption of CaSFL1.
The C. albicans homolog of the S. cerevisiae SFL1 was identified in the C. albicans genomic sequence (http://www.candidagenome.org) as orf19.454. Transformation was done by electroporation (24) followed by selective incubation on minimal media lacking the appropriate amino acids or uridine for 2 to 3 days at 30°C. Consecutive complete open reading frame (ORF) deletions of both alleles of CaSFL1 were achieved by using PCR-generated disruption cassettes from pFA plasmids amplified with primers S1-SFL1 and S2-SFL1 as described previously (31). All primers (Table 2) were obtained from biomers.net GmbH (Ulm, Germany). For each new strain, at least two independent mutants were generated. Verifications of correct integration at the target locus and of the absence of the target ORF in homozygous null mutants were done by diagnostic PCR. The fusion of the MET3 promoter to the SFL1 ORF was done by PCR-based gene targeting using primers S1-SFL1 and S2-MET3p-SFL1. A 3′-end fusion of SFL1 with green fluorescent protein (GFP) was generated by homologous recombination of a GFP-CdHIS1 cassette in a heterozygous SFL1 mutant, thus tagging the sole allele of SFL1 with GFP.
TABLE 2.
Oligonucleotide primers used in this study
| Primer | Sequence (5′ to 3′)a |
|---|---|
| #600 U2 | GTGTTACGAATCAATGGCACTACAGC |
| #599 U3 | GGAGTTGGATTAGATGATAAAGGTGATGG |
| #601 H2 | CAACGAAATGGCCTCCCCTACCACAG |
| #602 H3 | GGACGAATTGAAGAAAGCTGGTGCAACCG |
| #1031 5′RHO3-cDNA | CCTTAAATATCATTAAAACATGCCTCTTTGTGG |
| #1430 L2 | CGGTACCGACGTGATCACCTGGTA |
| #1431 L3 | GTTGGTGACGCGATTGTCGAAGCTG |
| #1744 S1-SFL1 | CATAACTCCAAATAAGAATAAGTTCATTTATCGCTTAAGGGAATAAGATCAAATTATATTTGCTATTGTTTTGCCAATATCACTTTAATTTTCCACCTGCgaagcttcgtacgctgcaggtc |
| #1745 S2-SFL1 | CCTGCTAGTGAAATATTCAATTTCTTGGAAACAAACACCGCTACTCCTTATTATATTGTTTATATCAAAGAAGAATTAGAATCAAGTTATATAGAACCtctgatatcatcgatgaattcgag |
| #1746 G1-SFL1 | CCAACGTTAAGTAGCCCAAGTTGC |
| #1747 G4-SFL1 | CATTCATCGTGGTGAAGCAAGCTATGG |
| #1748 I1-SFL1 | CCACCGACTTCTCATCCAATGGAATAC |
| #1749 I2-SFL1 | GCAACGTTGAGGTTGGTTGTAGACG |
| #1852 S2-SFL1-MET3p | GTTTTGATTGTAGGGGGTGCTATGATTGGTATGTGGACTTCTTGAAGTTGGAGTAGCAGTCGTTGTCGTACCAAGTGAAGACAGTACCAAATGACTcatgttttctggggagggtatttac |
| #1901 G4-SFL1-MET3p | GATTCCAGATGGTCTCCTCTAGAAG |
| #1960 CaFLO8-I1 | GATGAATGCTGTAGGTGGTGG |
| #1961 CaFLO8-I2 | GACCGGCAAACATCATTCCATTACTC |
| #1982 S1-SFL1-GFP | GAGTTTGCCACCAATCAAATCAATCAAAGATAATGATAATAAAAACGATAACGGTAATAGTGATGACAATGGGAACGATCATAAAAAGAGAAAATTAGAAggtgctggcgcaggtgcttc |
| #3127 ALS1-I1 | CACCACATCTCAACCTACAG |
| #3128 ALS1-I2 | GTTGGTTGAAGGTGAGGATGAG |
| #3129 HWP1-I1 | CTCCAGGTACTGATTCAACTCC |
| #3130 HWP1-I2 | CAGATGGTTGCATGAGTGGAAC |
| #3131 ECE1-I1 | CAACATGAAGAGAGATGTTGCTCC |
| #3132 ECE1-I2 | GACTGTCTTTCAAAGCAGATTCAGC |
| #3191 G4-CaRHO3 | GTGGTTCATTGTCATTGGCACC |
| #3204 ALS3-I1 | CGGAAATGGTCCTTATGAATCACC |
| #3205 ALS3-I2 | CAAACCACATAACCAAGTAGAATGTTG |
Capital letters correspond to genomic sequences, and lowercase letters correspond to annealing regions of pFA plasmids.
Promoter shutdown experiments.
To deplete cells of Sfl1, shutdown experiments of MET3 promoter-controlled SFL1 were performed. To this end, overnight cultures of the strains were grown in SD with the appropriate amino acid supplements. Starting cultures were diluted to an optical density at 600 nm of 0.1 in the same type of fresh medium with or without the addition of 3.5 mM methionine and cysteine. These yeast cultures were then incubated for 4 h at 30°C and then used for imaging. In the case of hyphal induction, 0.5 g/liter GlcNAc was added to the cultures, which were incubated for 4 to 5 h at 37°C prior to photography.
Embedded growth.
Growth conditions were as described previously (9). Cells of the strains were pregrown in YPD and then poured as dilutions thereof in YPS agar (containing 2% sucrose instead of glucose). These plates were incubated at room temperature for up to 5 days.
One-hybrid assay.
The SFL1 ORF was amplified using primers CaSFL1-OH1-MluI and CaSFL1-OH2-SphI and cloned into Clp-lexA after digestion with MluI and SphI (New England BioLabs, Germany). This generated a SalexA-SFL1 fusion construct. This plasmid was linearized by StuI and transformed into CAI-8 and CAJ043. Control strains containing NRG1 and GCN4 fusion constructs were kindly provided by Al Brown and are described in reference 30. The strains were pregrown in selective media and then inoculated in YPD and grown for 4 to 6 h at 30°C. Cultures were then adjusted to an optical density at 600 nm of 0.5. For qualitative analysis of lacZ expression, 3 μl of each cell suspension was dropped onto CSM plates and incubated for 2 days at 30°C. Colonies were then subjected to an agarose overlay (0.5 M phosphate buffer [pH 7.0], 6% N,N-dimethylformamide, 0.5% agarose, 0.1% sodium dodecyl sulfate, 20 μl 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside [X-Gal; 0.1 mg/ml stock solution]). 2-Nitrophenyl-β-d-galactopyranoside (ONPG) assays were used to quantify β-galactosidase activities for different strains. A 2-ml volume of the cell suspension was pelleted and washed with H2O, resuspended in 300 μl Z buffer (60 mM sodium phosphate monobasic dihydrate, 40 mM sodium phosphate dibasic, 10 mM potassium chloride, 1 mM magnesium sulfate), and divided into three aliquots of 100 μl. Cells were opened by using liquid nitrogen. The enzyme reaction was started by adding 160 μl ONPG-Z buffer solution (4 mg/ml ONPG in Z buffer) to each of the aliquots, followed by incubation at 37°C for 30 min. The reaction was stopped by adding 400 μl of 1 M sodium carbonate. Twofold dilutions were measured at 420 and 550 nm.
RNA isolation and RT-PCR.
Total RNA was isolated using an RNAgents total RNA isolation kit (Promega). Reverse transcription (RT)-PCR was performed with an Enhanced Avian RT first-strand synthesis kit (Sigma, Germany) using Oligo(dT)23 according to the manufacturer's protocol. Primers for RT-PCR were derived from the 3′ end of the genes tested (Table 2). As a control, RHO3 was used and was amplified from plasmids containing either a genomic or a cDNA fragment.
Microscopy.
Cells were grown to exponential phase either in minimal medium or in hypha-inducing medium for 4 to 6 h at 30°C or 37°C, respectively. Nuclear staining was done by adding 1 μl of 4′,6-diamidino-2-phenylindole dihydrochloride (DAPI; 1 mg/ml, Sigma) to 100 μl of cell suspension. Microscopy was done by using a bifunctional confocal laser scanning and fluorescence microscope on a Zeiss AxioImager platform with the appropriate filter combinations for the acquisition of GFP or DAPI fluorescence. Image acquisition and processing were done using Metamorph software (Universal Imaging Corporation).
RESULTS
Sequence analysis of Sfl1.
Based on sequence similarity, orf19.454 encodes the C. albicans homolog of the S. cerevisiae SFL1 gene. C. albicans Sfl1 encodes one of the longest ascomycetous Sfl1 proteins, with a length of 805 amino acids. A protein alignment reveals a number of conserved features (Fig. 1). Most notable is a strongly conserved domain within the N terminus that shares sequence similarity with the heat shock factor (HSF) domain. There are two regions in the C. albicans Sfl1 that are shared with Sfl1 proteins of other species: C. albicans and Aspergillus fumigatus Sfl1 proteins share an extended N-terminal sequence, and C. albicans and Kluyveromyces lactis share an internal Q-rich region. With both AfSfl1p and KlSfl1p C. albicans shares the highest degrees of sequence identity (21.5% over the entire proteins), although sequence similarities in the C-terminal halves of the proteins are rather low.
FIG. 1.
Protein alignment of ascomycetous Sfl1 proteins. Protein sequences of Sfl1 proteins from A. fumigatus (Af; GenBank accession number, XP_001481550), C. albicans (Ca; XP_715888), A. gossypii (Ag; NP_985683), K. lactis (Kl; XP_454089), and Candida glabrata (Cg) were aligned using the Clustal W algorithm. Matching residues are shaded. The alignment shows the N-terminal part where sequence conservation is highest.
Deletion of CaSFL1 results in flocculation and filamentation in the absence of inducers.
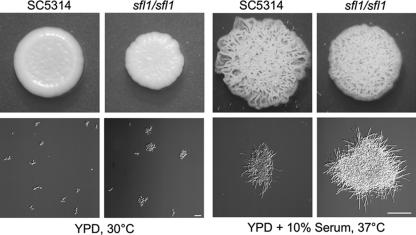
We deleted both copies of CaSFL1 in the SN148 background (27). To this end, we amplified disruption cassettes from pFA plasmids and generated independent heterozygous and homozygous sfl1 mutant strains (Table 1). Mutant strains that were of the sfl1::CdHIS1/sfl1::CmLEU2 or sfl1::CmLEU2/sfl1::URA3 genotype were phenotypically identical. Subsequently, we present the analysis of the Ura+ sfl1 mutant. To analyze cell growth and colony morphology phenotypes, we grew the mutant on solid medium and in liquid full medium at yeast and hyphal growth conditions (Fig. 2). Both the wild-type and the sfl1 mutant strains formed yeast colonies at 30°C and showed filamentous growth at 37°C in the presence of serum. We noted an increased degree of clumping for the mutant in liquid culture, both in conditions promoting yeast growth and under serum-inducing conditions. We quantified this aggregation behavior during growth in YPD (Table 3). It became evident that in YPD, a large amount of sfl1 mutant cells were found in aggregates. These aggregates appeared not to be generated due to cytokinesis defects as were observed previously, e.g., for C. albicans rho4 and cht3 mutants (12). We went on to analyze cell aggregation in SD (Fig. 3). This showed that sfl1/sfl1 cells form macroscopic flocs consisting of large amounts of cells. Microscopic observations revealed that these flocs also contained true hyphal cells.
FIG. 2.
Yeast and hyphal growth of the sfl1/sfl1 mutant strain. The wild type and the sfl1/sfl1 mutant were grown on YPD with or without 10% serum at 30°C or 37°C, respectively, in either liquid culture or on solid medium plates. Images were taken after an overnight incubation in liquid medium or after 4 days on solid medium plates. Bars, 30 μm (left) and 100 μm (right).
TABLE 3.
Cell-cell aggregates in C. albicans strains
| No. of cells in aggregates | No. of cells (%) of:
|
||
|---|---|---|---|
| SC5314 | SN148 | sfl1/sfl1 | |
| 1 to 2 | 166 (39) | 155 (30) | 60 (9) |
| 3 to 4 | 134 (32) | 204 (39) | 76 (11) |
| 5 to 10 | 96 (23) | 162 (31) | 98 (15) |
| >10 | 24 (6) | 0 (0) | 428 (65) |
| Total | 420 (100) | 521 (100) | 662 (100) |
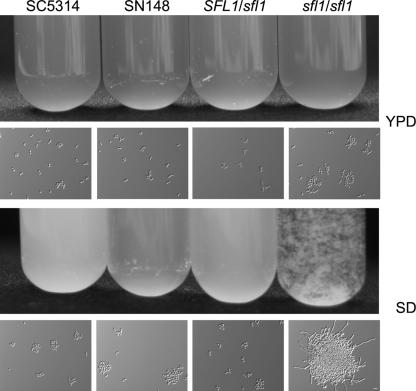
FIG. 3.
Deletion of SFL1 promotes flocculation in minimal medium. The indicated strains were grown overnight in liquid YPD (rich medium) or SD (minimal medium) at 30°C. Images of cell solutions in reagent tubes (upper panels) and microscopic images (lower panels) are shown. Bar, 10 μm.
An analysis of hyphal induction was done to determine whether the onset of filamentation occurs more rapidly in the sfl1 mutant. To this end, cells were induced to form filaments with serum. Aliquots of cells were taken in half-hourly intervals, and the average lengths of germ tubes were determined microscopically. Over a time course of 4 h, we did not find significant differences in the average lengths of the generated filaments between the wild type and the Sfl1 mutant (data not shown).
Embedded growth induces increased filamentation in a sfl1/sfl1 strain.
Several mutants in genes encoding proteins within the cAMP pathway were shown to exhibit enhanced filamentation under embedded growth conditions, e.g., the efg1/efg1 and flo8/flo8 mutants (10, 18). Interestingly, both of these mutant strains are defective in filamentation under all standard inducing conditions. Thus, we went on to examine the growth phenotype of the sfl1/sfl1 mutant under embedded conditions in YPS agar at 25°C. SFL1 strains, including the wild-type SC5314, the SN148, and the heterozygous mutant strains, started to show increased filamentation after more than 2 days under embedded conditions. In contrast, the sfl1/sfl1 strain formed consistently more abundant filaments under these conditions (Fig. 4). This result suggests that Sfl1p acts as a repressor of filamentation also under microaerophilic conditions when cells are embedded in an agarose matrix and grown at room temperature.
FIG. 4.
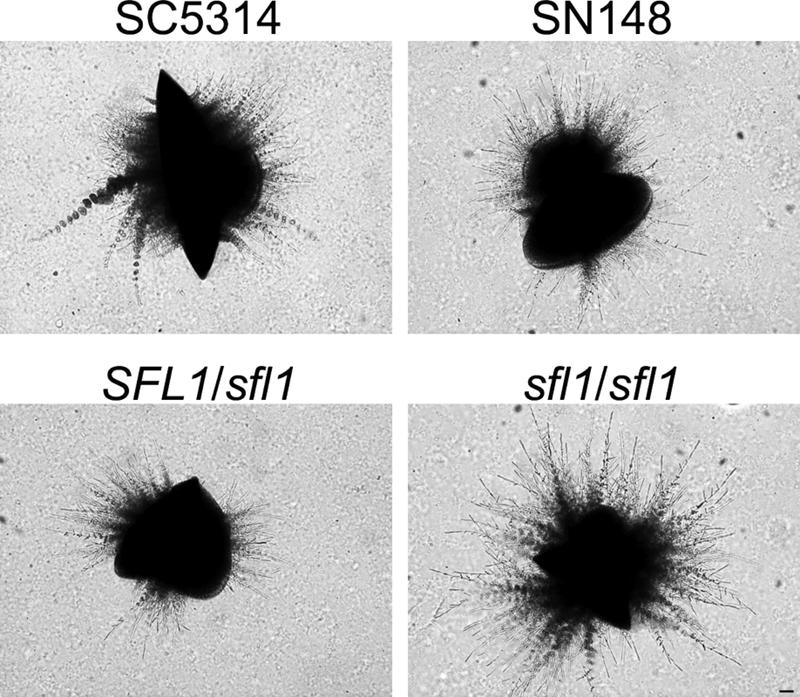
A sfl1/sfl1 mutant shows increased filamentation under embedded growth conditions. Cells of the SC5314 wild-type, SN148, heterozygous SFL1/sfl1, and homozygous sfl1/sfl1 strains were plated submersed in molten YPS agar (2%) and grown for 56 h at 25°C. Bar, 50 μm.
Upregulated expression of MET3p-SFL1 inhibits filamentation.
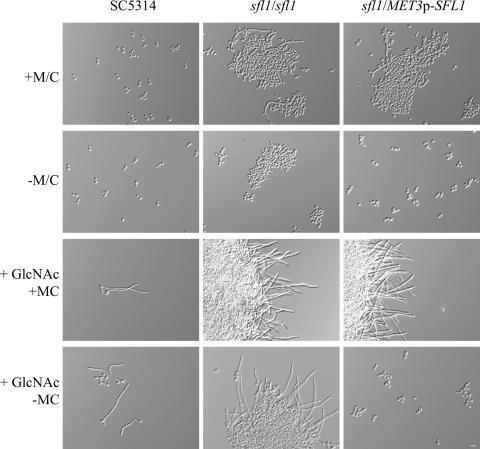
To study the effect of depleting cells of Sfl1 or of the overexpression of SFL1, we placed the single copy of SFL1 in a heterozygous SFL1/sfl1 mutant under the control of the regulatable CaMET3 promoter. The use of minimal medium supplemented with methionine and cysteine led to transcriptional repression of SFL1 and thus to the depletion of Sfl1 protein from the cells. In these cells, promoter shutdown resulted in the restoration of the sfl1 mutant phenotypes of flocculation and increased filamentation (Fig. 5). The growth of cells in SD lacking methionine and cysteine resulted in increased expression of MET3p-SFL1, which reduced the clumping phenotype, and all cells remained in the yeast phase. We analyzed the strength of different promoters by using GFP as a reporter. Use of the Ashbya gossypii TEF promoter resulted in high-level expression. Induced expression via the MET3 promoter reached half of that level while SFL1 promoter-driven expression was only half of that of the MET3 promoter. Therefore, induced expression of a MET3p-SFL1 construct results in a twofold overexpression on the protein level compared to endogenous SFL1 expression (data not shown).
FIG. 5.
MET3 promoter-controlled expression of SFL1. The wild-type, sfl1/sfl1, and MET3p-SFL1/sfl1 strains were pregrown in minimal media at 30°C with or without methionine/cysteine (+M/C and −M/C, respectively), diluted, and inoculated to the same cell density in fresh medium. Filamentation was induced with the addition of GlcNAc to the medium. Note that filamentation is blocked in cells overexpressing SFL1 (bottom right panel). Bar, 10 μm.
The promoter shutdown and inducing experiments indicate that the mutant phenotypes observed with our sfl1/sfl1 strains were due solely to the deletion of SFL1. Adding GlcNAc to minimal medium and increasing the temperature for incubation to 37°C induces C. albicans cells to form hyphal filaments. This became evident in the wild type, the sfl1/sfl1 mutant strain, and the MET3p-controlled SFL1 strain under repressing conditions. Surprisingly, however, if cells were pregrown in SD (without methionine and cysteine), which allows for full expression of SFL1 via the MET3 promoter, and were then transferred into SD plus GlcNAc medium, germ tube formation was entirely abolished. This block in filamentation required a continued high expression level of SFL1 since cells started to filament when methionine or cysteine was present in the inducing medium. Our results, therefore, indicate that overexpression of SFL1 can lead to a block in hyphal formation in C. albicans (Fig. 5).
Sfl1-GFP localizes to the nucleus.
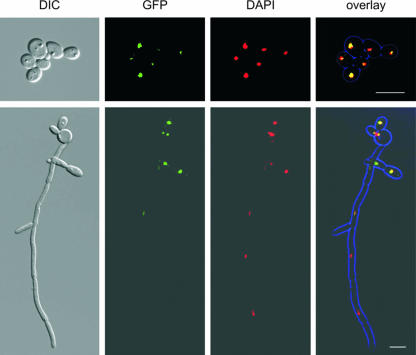
Consistent with the idea of a transcriptional regulator and DNA-binding protein should be a nuclear localization of the Sfl1 protein. Therefore, we wanted to determine the subcellular localization of Sfl1p during yeast and hyphal growth. To this end, we generated an SFL1-GFP/sfl1 strain. This strain was phenotypically like the wild-type strain, suggesting that SFL1-GFP is functional. We observed a Sfl1-GFP signal in the nuclei of yeast cells, which would be consistent with the idea of Sfl1 suppressing flocculation and filamentation under conditions that allow yeast growth. Upon filament induction, Sfl1-GFP could still be observed in the nuclei of hyphal cells but was found to be stronger in yeast or pseudohyphal cells (Fig. 6). This would be consistent with either a diminished expression of SFL1 or a reduced amount of Sfl1p in the nuclei of hyphal cells, which is in line with the notion of a derepression of Sfl1-controlled genes under hypha-inducing conditions.
FIG. 6.
Localization of Sfl1p-GFP in yeast and hyphal cells. Strain CAJ042 expresses SFL1-GFP from its endogenous promoter in a heterozygous strain. Fluorescence images were acquired from yeast cells and hyphal cells that were induced with GlcNAc for 4 h. Cells were fixed prior to the imaging and were costained by DAPI to visualize the nuclei. DIC, differential interference contrast. Bar, 10 μm.
One-hybrid analysis with SFL1.
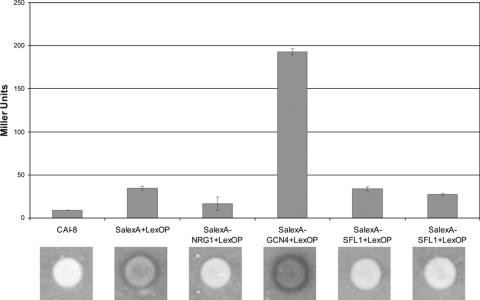
The data presented suggested that Sfl1p fulfills a role of a transcriptional repressor in C. albicans. To gain further evidence for such a role, we employed a recently developed one-hybrid system with C. albicans (30). To this end, we fused SFL1 to the Staphylococcus aureus lexA DNA-binding domain and integrated this fusion gene at the RPS1 locus in CAI8. The readout of transcriptional regulation was done using a lex operator containing Streptococcus thermophilius lacZ reporter gene construct. As controls, we used strains provided by the Brown lab that assay the Nrg1 repressor and the Gcn4 activator activity (30). We assayed the level of reporter gene expression by using an X-Gal overlay assay and quantitative analysis of β-galactosidase activity (Fig. 7). This assay confirmed that Gcn4 acts as a transcriptional activator while Nrg1 functions as a repressor. In these assays, we found no activating activity of Sfl1.
FIG. 7.
One-hybrid assay with Sfl1. A SalexA-SFL1 fusion construct was transformed into a LexOP-carrying strain in a manner similar to that done previously with GCN4 and NRG1 (30). The level of reporter gene expression was determined by the analysis of β-galactosidase activity and quantitated by the use of an X-Gal overlay assay as indicated.
A set of hypha-specific genes is derepressed in the sfl1 mutant.
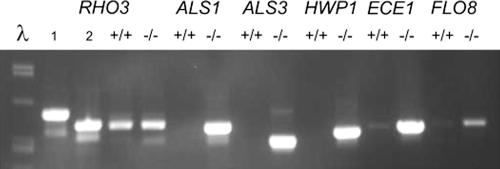
The flocculation phenotype of the sfl1/sfl1 mutant in minimal medium and the increase in filament formation provide some evidence for an altered transcriptional profile in the mutant. To gain first insight into which genes may be affected on the transcriptional level by the deletion of SFL1, we performed RT-PCR to analyze the expression of hypha-specific genes (HWP1, ECE1), agglutinin-like sequences (ALS1, ALS3), and also FLO8, which is a known positive regulator of hypha-specific gene expression in C. albicans. Also based on the regulation circuitry in S. cerevisiae, ScFLO8 is known to be regulated by ScSfl1. In this assay, we found increased expression of these genes in the sfl1/sfl1 mutant (Fig. 8).
FIG. 8.
Hypha-specific genes are upregulated in an sfl1 mutant. Cells of the wild type (SC5314; +/+) and the sfl1/sfl1 mutant (−/−) were grown in minimal medium for 6 h at RT and then collected for RNA preparation. The quality of the RNA was assessed by using the CaRHO3 gene as a control. CaRHO3 genomic DNA (lane 1) contains one intron that is missing in its corresponding cDNA (lane 2; PCR derived from a cloned cDNA fragment).
DISCUSSION
We have identified the C. albicans SFL1 gene by similarity to its S. cerevisiae homolog. Sfl1p contains an N-terminal domain with a high degree of similarity to the HSF DNA-binding domain (15). C. albicans Sfl1p shares an extended N terminus with A. fumigatus Sfl1p, the function of which, however, needs to be elucidated. Sfl1 is a conserved protein and known as a suppressor of flocculation in S. cerevisiae. In S. cerevisiae, Sfl1p acts as transcriptional repressor of flocculation (FLO) gene (15, 28). The deletion of ScSFL1 leads to floc formation and to enhanced pseudohyphal and invasive growth (28). We found that CaSFL1 regulates similar processes in C. albicans. The deletion of CaSFL1 resulted in strong flocculation in minimal medium accompanied by increased filamentation in the absence of inducers as well as enhanced filamentous growth under embedded conditions at 25°C. This result suggests that some of the transcriptional circuitry controlled by Sfl1p has been conserved between S. cerevisiae and C. albicans.
The prevailing view in the regulation of C. albicans hyphal morphogenesis is that transcriptional repression by Tup1, Nrg1, Rpg1, and Ssn6 and complexes thereof provide the cells with the means to repress specific gene sets (5-8, 21, 22, 26). The mechanistic difficulties in explaining the complex pattern of regulation of hypha-specific gene expression came from comparative transcript profiling experiments that indicated that in nrg1, tup1, and ssn6 mutant strains, overlapping but extensively different subsets of genes were regulated (16). Our studies now present one additional player in this scheme, Sfl1p. It is noteworthy that deletion of SFL1 does not induce filamentation to an extent seen, for example, in the tup1 mutant. On the other hand, the flocculation phenotype seems to be rather specific for the sfl1 mutant. Formation of cell clumps occurs in the sfl1 mutant strain under different medium conditions but is most obvious in minimal media. As flocs also contain hyphal cells in minimal media, more pronounced flocculation might be the result of hypha-specific gene expression in these cells.
In S. cerevisiae, Sfl1p recruits the Ssn6-Tup1 complex via an Ssn6 interaction domain (11). This domain is only rather weakly conserved on the amino acid level between different fungal Sfl1 proteins. However, it leads to a testable hypothesis in C. albicans, namely, that Sfl1p might repress hypha- and/or flocculation-specific genes via a Sfl1-Ssn6-Tup1 complex. We analyzed the potential of CaSfl1p to act as a repressor of transcription using a recently developed one-hybrid system, which proved useful to identify Gcn4 as a transcriptional activator (30). In these assays, CaSfl1 was only weakly downregulating reporter gene transcription and certainly not activating it. Consistent with the idea of a repressor could be a differential localization of CaSfl1 in the nucleus of yeast cells but not of hyphal cells. Even though we found evidence to support this idea, we cannot rule out a nucleocytoplasmic shuttling of Sfl1 that could contribute to its specific activity. Such a shuttling mechanism is quite common and has been described, e.g., for the Msn2/4 global stress response regulator in S. cerevisiae (19).
In S. cerevisiae, Sfl1 is regulated via phosphorylation by Tpk2 (28). By analogy, this could place Sfl1 downstream of the cAMP-signaling pathway in concert with Flo8. Furthermore, in S. cerevisiae, Sfl1p regulates the expression of FLO8 in a negative manner (23). A similar scenario seems likely for C. albicans, since overexpression of SFL1 using the MET3 promoter resulted in the inability of these cells to form hyphae under GlcNAC-induced filamentation. This result is consistent with the role of CaFlo8 as an inducer of hypha-specific genes, as the flo8/flo8 mutant is blocked for hypha-specific gene expression and filamentation (10). First insights into the target genes of Sfl1 came from RT-PCR, which showed expression of the adhesins ALS1 and ALS3 as well as of other hypha-specific genes. FLO8 appears to be upregulated in a sfl1 mutant. Interestingly, the deletion of SFL1 points toward a role of Sfl1 that is not exclusive for filamentation, but it reveals a novel role in flocculation. This function could be important for cell-cell adhesion or cell-surface adhesion, such as that seen in biofilm formation. Determining the Sfl1 regulon, particularly with respect to adhesins and hyphal specific genes, therefore, will be an essential task to understand the role of Sfl1.
In conclusion, our functional analysis of the C. albicans SFL1 gene provides evidence that Sfl1 is a conserved negative regulator of flocculation and filamentation in C. albicans that may regulate a specific gene set required for flocculation but may regulate hypha-specific genes in concert with other known repressors, particularly Tup1 and Nrg1.
Acknowledgments
We thank Al Brown for making available the C. albicans “one-hybrid-kit.”
This research was supported by the EU-Signalpath Marie Curie Training Network.
Footnotes
Published ahead of print on 31 August 2007.
REFERENCES
- 1.Bahn, Y.-S., and P. Sundstrom. 2001. CAP1, an adenylate cyclase-associated protein gene, regulates bud-hypha transitions, filamentous growth, and cyclic AMP levels and is required for virulence of Candida albicans. J. Bacteriol. 183:3211-3223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barelle, C. J., C. L. Priest, D. M. Maccallum, N. A. Gow, F. C. Odds, and A. J. Brown. 2006. Niche-specific regulation of central metabolic pathways in a fungal pathogen. Cell. Microbiol. 8:961-971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Berman, J., and P. E. Sudbery. 2002. Candida albicans: a molecular revolution built on lessons from budding yeast. Nat. Rev. Genet. 3:918-930. [DOI] [PubMed] [Google Scholar]
- 4.Biswas, S., M. Roy, and A. Datta. 2003. N-Acetylglucosamine-inducible CaGAP1 encodes a general amino acid permease which co-ordinates external nitrogen source response and morphogenesis in Candida albicans. Microbiology 149:2597-2608. [DOI] [PubMed] [Google Scholar]
- 5.Braun, B. R., W. S. Head, M. X. Wang, and A. D. Johnson. 2000. Identification and characterization of TUP1-regulated genes in Candida albicans. Genetics 156:31-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Braun, B. R., and A. D. Johnson. 1997. Control of filament formation in Candida albicans by the transcriptional repressor TUP1. Science 277:105-109. [DOI] [PubMed] [Google Scholar]
- 7.Braun, B. R., and A. D. Johnson. 2000. TUP1, CPH1 and EFG1 make independent contributions to filamentation in Candida albicans. Genetics 155:57-67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Braun, B. R., D. Kadosh, and A. D. Johnson. 2001. NRG1, a repressor of filamentous growth in C. albicans, is down-regulated during filament induction. EMBO J. 20:4753-4761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brown, D. H., Jr., A. D. Giusani, X. Chen, and C. A. Kumamoto. 1999. Filamentous growth of Candida albicans in response to physical environmental cues and its regulation by the unique CZF1 gene. Mol. Microbiol. 34:651-662. [DOI] [PubMed] [Google Scholar]
- 10.Cao, F., S. Lane, P. P. Raniga, Y. Lu, Z. Zhou, K. Ramon, J. Chen, and H. Liu. 2006. The Flo8 transcription factor is essential for hyphal development and virulence in Candida albicans. Mol. Biol. Cell 17:295-307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Conlan, R. S., and D. Tzamarias. 2001. Sfl1 functions via the co-repressor Ssn6-Tup1 and the cAMP-dependent protein kinase Tpk2. J. Mol. Biol. 309:1007-1015. [DOI] [PubMed] [Google Scholar]
- 12.Dünkler, A., and J. Wendland. 2007. Candida albicans Rho-type GTPase-encoding genes required for polarized cell growth and cell separation. Eukaryot. Cell 6:844-854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Enjalbert, B., D. M. MacCallum, F. C. Odds, and A. J. Brown. 2007. Niche-specific activation of the oxidative stress response by the pathogenic fungus Candida albicans. Infect. Immun. 75:2143-2151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Feng, Q., E. Summers, B. Guo, and G. Fink. 1999. Ras signaling is required for serum-induced hyphal differentiation in Candida albicans. J. Bacteriol. 181:6339-6346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fujita, A., Y. Kikuchi, S. Kuhara, Y. Misumi, S. Matsumoto, and H. Kobayashi. 1989. Domains of the SFL1 protein of yeasts are homologous to Myc oncoproteins or yeast heat-shock transcription factor. Gene 85:321-328. [DOI] [PubMed] [Google Scholar]
- 16.Garcia-Sanchez, S., A. L. Mavor, C. L. Russell, S. Argimon, P. Dennison, B. Enjalbert, and A. J. Brown. 2005. Global roles of Ssn6 in Tup1- and Nrg1-dependent gene regulation in the fungal pathogen, Candida albicans. Mol. Biol. Cell 16:2913-2925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gillum, A. M., E. Y. Tsay, and D. R. Kirsch. 1984. Isolation of the Candida albicans gene for orotidine-5′-phosphate decarboxylase by complementation of S. cerevisiae ura3 and E. coli pyrF mutations. Mol. Gen. Genet. 198:179-182. [DOI] [PubMed] [Google Scholar]
- 18.Giusani, A. D., M. Vinces, and C. A. Kumamoto. 2002. Invasive filamentous growth of Candida albicans is promoted by Czf1p-dependent relief of Efg1p-mediated repression. Genetics 160:1749-1753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gorner, W., E. Durchschlag, M. T. Martinez-Pastor, F. Estruch, G. Ammerer, B. Hamilton, H. Ruis, and C. Schuller. 1998. Nuclear localization of the C2H2 zinc finger protein Msn2p is regulated by stress and protein kinase A activity. Genes Dev. 12:586-597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Guo, B., C. A. Styles, Q. Feng, and G. R. Fink. 2000. A Saccharomyces gene family involved in invasive growth, cell-cell adhesion, and mating. Proc. Natl. Acad. Sci. USA 97:12158-12163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hwang, C. S., J. H. Oh, W. K. Huh, H. S. Yim, and S. O. Kang. 2003. Ssn6, an important factor of morphological conversion and virulence in Candida albicans. Mol. Microbiol. 47:1029-1043. [DOI] [PubMed] [Google Scholar]
- 22.Kadosh, D., and A. D. Johnson. 2005. Induction of the Candida albicans filamentous growth program by relief of transcriptional repression: a genome-wide analysis. Mol. Biol. Cell 16:2903-2912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kim, T. S., S. B. Lee, and H. S. Kang. 2004. Glucose repression of STA1 expression is mediated by the Nrg1 and Sfl1 repressors and the Srb8-11 complex. Mol. Cell. Biol. 24:7695-7706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Köhler, G. A., T. C. White, and N. Agabian. 1997. Overexpression of a cloned IMP dehydrogenase gene of Candida albicans confers resistance to the specific inhibitor mycophenolic acid. J. Bacteriol. 179:2331-2338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kojic, E. M., and R. O. Darouiche. 2004. Candida infections of medical devices. Clin. Microbiol. Rev. 17:255-267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Murad, A. M., P. Leng, M. Straffon, J. Wishart, S. Macaskill, D. MacCallum, N. Schnell, D. Talibi, D. Marechal, F. Tekaia, C. d'Enfert, C. Gaillardin, F. C. Odds, and A. J. Brown. 2001. NRG1 represses yeast-hypha morphogenesis and hypha-specific gene expression in Candida albicans. EMBO J. 20:4742-4752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Noble, S. M., and A. D. Johnson. 2005. Strains and strategies for large-scale gene deletion studies of the diploid human fungal pathogen Candida albicans. Eukaryot. Cell 4:298-309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Robertson, L. S., and G. R. Fink. 1998. The three yeast A kinases have specific signaling functions in pseudohyphal growth. Proc. Natl. Acad. Sci. USA 95:13783-13787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rocha, C. R., K. Schroppel, D. Harcus, A. Marcil, D. Dignard, B. N. Taylor, D. Y. Thomas, M. Whiteway, and E. Leberer. 2001. Signaling through adenylyl cyclase is essential for hyphal growth and virulence in the pathogenic fungus Candida albicans. Mol. Biol. Cell 12:3631-3643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Russell, C. L., and A. J. Brown. 2005. Expression of one-hybrid fusions with Staphylococcus aureus lexA in Candida albicans confirms that Nrg1 is a transcriptional repressor and that Gcn4 is a transcriptional activator. Fungal Genet. Biol 42:676-683. [DOI] [PubMed] [Google Scholar]
- 31.Schaub, Y., A. Dunkler, A. Walther, and J. Wendland. 2006. New pFA-cassettes for PCR-based gene manipulation in Candida albicans. J. Basic Microbiol. 46:416-429. [DOI] [PubMed] [Google Scholar]
- 32.Stoldt, V. R., A. Sonneborn, C. E. Leuker, and J. F. Ernst. 1997. Efg1p, an essential regulator of morphogenesis of the human pathogen Candida albicans, is a member of a conserved class of bHLH proteins regulating morphogenetic processes in fungi. EMBO J. 16:1982-1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Verstrepen, K. J., and F. M. Klis. 2006. Flocculation, adhesion and biofilm formation in yeasts. Mol. Microbiol. 60:5-15. [DOI] [PubMed] [Google Scholar]
- 34.Whiteway, M., and U. Oberholzer. 2004. Candida morphogenesis and host-pathogen interactions. Curr. Opin. Microbiol. 7:350-357. [DOI] [PubMed] [Google Scholar]