Abstract
Fungal histidine kinase receptors (HKRs) sense and transduce many extracellular signals. We investigated the role of HKRs in morphogenetic transition, osmotolerance, oxidative stress response, and mating ability in the opportunistic yeast Candida lusitaniae. We isolated three genes, SLN1, NIK1, and CHK1, potentially encoding HKRs of classes VI, III, and X, respectively. These genes were disrupted by a transformation system based upon the “URA3 blaster” strategy. Functional analysis of disruptants was undertaken, except for the sln1 nik1 double mutant and the sln1 nik1 chk1 triple mutant, which are not viable in C. lusitaniae. The sln1 mutant revealed a high sensitivity to oxidative stress, whereas both the nik1 and chk1 mutants exhibited a more moderate sensitivity to peroxide. We also showed that the NIK1 gene was implicated in phenylpyrrole and dicarboximide compound susceptibility while HKRs seem not to be involved in resistance toward antifungals of clinical relevance. Concerning mating ability, all disruptants were still able to reproduce sexually in vitro in unilateral or bilateral crosses. The most important result of this study was that the sln1 mutant displayed a global defect of pseudohyphal differentiation, especially in high-osmolarity and oxidative-stress conditions. Thus, the SLN1 gene could be crucial for the C. lusitaniae yeast-to-pseudohypha morphogenetic transition. This implication is strengthened by a high level of SLN1 mRNAs revealed by semiquantitative reverse transcription-PCR when the yeast develops pseudohyphae. Our findings highlight a differential contribution of the three HKRs in osmotic and oxidant adaptation during the morphological transition in C. lusitaniae.
Like bacteria, fungi and plants sense and transduce many extracellular signals through histidine kinase receptors (HKRs). In eukaryotic cells, these phosphorelay proteins often carry both a histidine kinase component and a response regulator domain. In response to an external signal, the histidine kinase component autophosphorylates a conserved histidine residue. The phosphate on this histidine is then transferred to a conserved aspartic acid in the response regulator domain. Such histidine-to-aspartate phosphotransfers initiate intracellular pathways mediated in particular by mitogen-activated protein (MAP) kinases (6, 40). In fungi, it is now accepted that HKR-mediated transduction pathways are implicated in regulating diverse processes, including osmoregulation, morphogenesis, and virulence expression (62). Based on results from the fungal genome sequencing project (http//www.broad.mit.edu), a recent study provided a classification of several HKR genes identified in the Ascomycota and revealed that fungal HKRs fall into 11 classes (15). Most of these classes are encountered in each examined filamentous fungus, such as Neurospora crassa and Botryotinia fuckeliana, whereas Saccharomyces cerevisiae encodes only one HKR and the yeast species Schizosaccharomyces pombe and Candida albicans only three.
The best-documented HKR-mediated signaling pathway in fungi is the HOG cascade, modulated by an Sln1p-Ypd1p-Ssk1p phosphorelay system, which is involved in adaptation to oxidative and osmotic stresses in S. cerevisiae (61). More precisely, under high-osmolarity conditions, the autokinase activity of the S. cerevisiae HKR Sln1p (class VI) is turned off, which leads to the activation of a MAP kinase pathway via the phosphorelay elements Ypd1p and Ssk1p. Finally, phosphorylation of the MAP kinase Hog1p modulates the activation of several nuclear transcription factors of osmoresponse genes (39). In S. pombe the three HKRs (SpMak1p, class V; and SpMak2p and SpMak3p, class X) are also implicated in the response to oxidative stress and cell cycle control (5, 12). In the filamentous fungi N. crassa, B. fuckeliana, Alternaria alternata, Cochliobolus heterostrophus, Alternaria brassicicola, and Monilinia fructicola, the mutation of class III HKR (also named Nik1p-like HKR) genes is responsible for severe osmosensitivity and dicarboximide resistance (8, 21, 22, 44, 53, 70). Furthermore, the NcNIK1 gene (also referred as OS-1) was shown to be essential for hyphal development of N. crassa (2, 64). Additionally, the corresponding genes of B. fuckeliana (named BOS1) and M. fructicola (named MfOS1), have been clearly demonstrated to be virulence factors in these two plant-pathogenic species (44, 67).
Concerning human-pathogenic fungi, only a few HKR genes have been fully described. In Aspergillus fumigatus, a putative HKR gene, AfFOS1 (class IV), has been isolated, and the fos1 deletion strain showed significantly reduced virulence compared with the parental wild-type strain. Thus, the AfFos1p HKR was proposed to be a key virulence factor of A. fumigatus (19). The genome of Cryptococcus neoformans encodes seven HKRs, and it was recently demonstrated that two of these, CnTco1p (class III) and CnTco2p (unclassified HKR), are implicated in the regulation of stress responses, drug susceptibility, sexual development and virulence (9, 18). A recent work demonstrated that the HKR Drk1p (class III) senses host signals and triggers the transition from mold to yeast in the dimorphic species Histoplasma capsulatum and Blastomyces dermatitidis. This HKR was also implicated in the regulation of cell wall integrity, sporulation, and expression of virulence genes in vivo (50).
The pathogenic fungus in which the functions of HKRs have been the most extensively studied remains C. albicans, which harbors three HKRs, i.e., CaSln1p (class VI), CaNik1p (class III), and CaChk1p (class X) (17, 35). A few years ago, further studies had shown the contribution of HKRs in the morphogenesis of C. albicans (3, 13, 36, 49, 69). Moreover, deletions of CaNIK1 or CaSLN1 attenuate virulence, while deletion of CaCHK1 abolishes virulence of the strains (14, 69). More recently, it has been shown that Cachk1 null mutation could affect cell wall biosynthesis and flocculation of yeasts (13, 36) and that CaChk1p was involved in the quorum sensing of C. albicans (37). C. albicans yeast cells (round to ovoid cells) can alternatively form true hyphae (long continuous tubes with septa) or pseudohyphae (chains of distinct cells remaining attached each other) (11, 65). This morphological switching is influenced by environmental factors, such as temperature, pH, nitrogen sources, carbon sources, and physical contact with surfaces (23). It has been well demonstrated that the three HKRs CaSln1p, CaNik1p, and CaChk1p play a crucial role in true-hypha formation in C. albicans (3, 13, 49, 69), but the occurrence of HKRs in C. albicans pseudohyphal growth remains unknown. Additionally, to our knowledge, the role of HKRs in other Candida species that only form pseudohyphae has never been studied.
As described for S. cerevisiae (29, 41), C. albicans (23), and Candida glabrata (20), the budding yeast Candida lusitaniae (teleomorph Clavispora lusitaniae) can efficiently switch to pseudohyphal growth when cultured on solid medium depleted of a nitrogen source (30). It is also an emerging pathogen which is characterized by its propensity to develop resistance to antifungal agents during treatment (16, 24, 31, 32, 47, 52, 58, 60). The recent achievement of C. lusitaniae whole genome sequencing clearly reflects a growing interest of the scientific community in this opportunistic yeast. As a laboratory model, C. lusitaniae is an experimentally tractable haploid organism offering formal genetic tools based upon a complete sexual cycle reproducible in vitro (27, 51, 72) and an integrative transformation system for gene disruption using the URA3 gene as a selection marker (26, 71, 72).
In this regard, we report here on the cloning and the characterization of three genes, SLN1, NIK1, and CHK1, potentially encoding HKRs in C. lusitaniae. These genes were disrupted by homologous recombination, and functional analysis of the transformants was undertaken. The roles of HKRs in growth, osmotolerance, oxidative stress response, drug susceptibility, and mating ability were studied. The most important finding of this study is the involvement of the SLN1 gene in early steps of pseudohyphal development. The global occurrence of HKRs in the morphogenesis of C. lusitaniae is also discussed.
MATERIALS AND METHODS
Strains and standard growth conditions.
C. lusitaniae strains (Table 1) were routinely cultivated in liquid YPD medium (1% yeast extract, 2% peptone, 2% glucose) at 35°C under agitation (250 rpm). Solid media were prepared with 2% agar (Sigma).
TABLE 1.
Candida lusitaniae strains
| Strain or abbreviated genotype | Genotype | Parent | Mating type |
|---|---|---|---|
| 6936a | Wild type | MATa | |
| Cl38b | MATα | ||
| 6936 ura3[Δ360] | ura3[Δ360] | 6936 | MATa |
| PC1 (α) | ura3[Δ360] | 6936 ura3[Δ360] × Cl38 | MATα |
| sln1::GUN | ura3[Δ360]sln1Δ::REP-URA3-REP | 6936 ura3[Δ360] | MATa |
| nik1::GUN | ura3[Δ360]nik1Δ::REP-URA3-REP | 6936 ura3[Δ360] | MATa |
| chk1::GUN | ura3[Δ360]chk1Δ::REP-URA3-REP | 6936 ura3[Δ360] | MATa |
| sln1::GUNα | ura3[Δ360]sln1Δ::REP-URA3-REP | PC1 (α) | MATα |
| nik1::GUNα | ura3[Δ360]nik1Δ::REP-URA3-REP | PC1 (α) | MATα |
| chk1::GUNα | ura3[Δ360]chk1Δ::REP-URA3-REP | PC1 (α) | MATα |
| sln1::REP | ura3[Δ360]sln1Δ::REP | sln1::GUN strain | MATa |
| nik1::REP | ura3[Δ360]nik1Δ::REP | nik1::GUN strain | MATa |
| chk1::REP | ura3[Δ360]chk1Δ::REP | chk1::GUN strain | MATa |
| chk1::GUN sln1::REP | ura3[Δ360]chk1Δ::REP-URA3-REP sln1Δ::REP | sln1::REP strain | MATa |
| chk1::REP nik1::GUN | ura3[Δ360]chk1Δ::REP nik1Δ::REP-URA3-REP | chk1::REP strain | MATa |
| chk1::REP sln1::REP | ura3[Δ360]chk1Δ::REP sln1Δ::REP | chk1::GUN sln1::REP strain | MATa |
| chk1::REP nik1::REP | ura3[Δ360]chk1Δ::REP nik1Δ::REP | chk1::REP nik1::GUN strain | MATa |
| sln1+SLN1 | ura3[Δ360]sln1Δ::REP pVAX-URA3-SLN1 | sln1::REP strain | MATa |
| nik1+NIK1 | ura3[Δ360]nik1Δ::REP pVAX-URA3-NIK1 | nik1::REP strain | MATa |
| chk1+CHK1 | ura3[Δ360]chk1Δ::REP pVAX-URA3-CHK1 | chk1::REP strain | MATa |
| chk1 sln1+SLN1 | ura3[Δ360]chk1Δ::REP sln1Δ::REP, pVAX-URA3-SLN1 | chk1::REP sln1::REP strain | MATa |
| chk1 nik1+NIK1 | ura3[Δ360]chk1Δ::REP nik1Δ::REP, pVAX-URA3-NIK1 | chk1::REP nik1::REP strain | MATa |
Reference strain from Centraalbureau voor Schimmelcultures (Utrecht, The Netherlands).
Clinical isolate; described by Francois et al. (27).
Sequence analysis.
The complete open reading frames (ORFs) of S. cerevisiae ScSln1p (GenBank accession number NP_012119) and C. glabrata CgSln1p (GenBank accession number XP_447081) were retrieved from the NCBI database. Similarity searches in the yeast databases were performed with the BLAST algorithm (4) using CaSln1p (GenBank accession number AB006362), CaNik1p (GenBank accession number AF029092), and CaChk1p (GenBank accession number AF013273) of C. albicans.
The complete ORFs of the C. lusitaniae SLN1, NIK1, and CHK1 genes and corresponding homologues from Candida guilliermondii (CguiSln1p, ORF located in supercontig 1.3: 1682656 to 1685910; CguiNik1p, supercontig 1.1, 109500 to 112860; CguiChk1p, supercontig 5, 828047 to 835330) and Candida tropicalis (CtSln1p, supercontig 1, 2247091 to 2250819; CtNik1p, supercontig 7, 629227 to 632733; CtChk1p, supercontig 2: 186020 to 193255) were retrieved from their respective genome databases, available on the Broad Institute Fungal Genome website (http://www.broad.mit.edu).
Protein alignments were generated using the ClustalW software (66), and the deduced phylogenetic tree was built with TreeView PPC software (56). Transmembrane helices and secondary structures were predicted using the TMHMM (34) and SOPMA (28) software, respectively.
DNA and RNA extraction.
Genomic DNA was extracted by following the protocol described by Scherer and Stevens (63) except that zymolyase was replaced by lyticase (60 U/ml) for production of spheroplasts. Total RNAs were extracted using the RNeasy minikit (QIAGEN) associated with the RNase-free DNase set (QIAGEN) according to the manufacturer's instructions.
PCR amplifications and plasmid constructions.
PCRs were performed with BD Advantage 2 polymerase mix (BD Biosciences). PCR conditions for amplification were those indicated by the supplier. All primers were synthesized by Invitrogen and are listed in Table 2. The PCR and endonuclease-digested products were purified using QIAquick PCR purification (QIAGEN) according to the manufacturer's instructions.
TABLE 2.
Oligonucleotides
| Primer | Sequence (5′ to 3′) | Used for PCR amplification of: |
|---|---|---|
| PHK1 | GGGTTTGAGGAAGCAGTGTGCAC | NIK1 gene (upstream primer) |
| PHK2 | GAGTGCCAGAGATGCCAAGGATC | NIK1 gene (downstream primer) |
| PHK3 | GACTAGTTACCGAGATTTCAGG | SLN1 gene (upstream primer) |
| PHK4 | CAGTTCTACCTTACCTCTACGTC | SLN1 gene (downstream primer) |
| PHK7 | CCAGTGATGGCGGACCGCAAGCCACC | CHK1 gene (upstream primer) |
| PHK8 | CCCAGAAGCACAAGGATCTCGTGCTG | CHK1 gene (downstream primer) |
| BGLUN1 | CTGACAAGATCTCCCGACGTCGCATGCTCC | GUN sequence (upstream primer, BglII site) |
| BGLUN2 | CTCAGAAGATCTCCAAGCTATTTAGGTGACAC | GUN sequence (downstream primer, BglII site) |
| MFEUN1 | CTGACACAATTGCCCGACGTCGCATGCTCC | GUN sequence (upstream primer, MfeI site) |
| MFEUN2 | CTCAGACAATTGCCAAGCTATTTAGGTGACAC | GUN sequence (downstream primer, MfeI site) |
| NEOREP1 | CTGAGACCATGGACTAGTTTGCACGCAGGTTCTCCGGCC | REP sequence (upstream primer, NcoI and SpeI site) |
| NEOREP2 | CTGAGACCGCGGATGCATGATGGATACTTTCTCGGCAGG | REP sequence (downstream, SacII and NsiI site) |
| SLRT1 | TTCATGGATGTTCAGATGCCC | 3′ SLN1 mRNA (upstream primer) |
| SLRT2 | ATTTTATGAGGCAGCATCACC | 3′ SLN1 mRNA (downstream primer) |
| NIRT1 | CAATCATTGCCTTGACGGCAC | 3′ NIK1 mRNA (upstream primer) |
| NIRT2 | TCTTCGCTAGCGAGCTGTCGG | 3′ NIK1 mRNA (downstream primer) |
| HKRT1 | TGGTTACCAGCTTTGGCCTGG | 3′ CHK1 mRNA (upstream primer) |
| HKRT2 | TCGCTCTCTATCTTCACCTGC | 3′ CHK1 mRNA (downstream primer) |
| ACT1 | GTCGGTGACGAAGCTCAGTCC | ACT1 mRNA (upstream primer) |
| ACT2 | AGCTCTGAATCTCTCGTTACC | ACT1 mRNA (downstream primer) |
The plasmid pG-ura3[Δ360] was constructed as follow. The plasmid pGEM-U (26) was digested with EcoRV (a unique site located in the central region of the URA3 gene), and the two generated extremities were cut back with 5 U of nuclease BAL31 (Fermentas) during 2 min and then ligated. We screened a plasmid, named pG-ura3[Δ360], which harbors a 360-bp deletion (nucleotide 213 to 572) located in the core of the ura3 gene.
The backbone plasmids pG-SLN1, pG-NIK1, and pG-CHK1 were built by cloning in the pGEM-T easy vector (Promega) PCR amplification fragments overlapping the SLN1 (4,523 bp), NIK1 (4,078 bp), and CHK1 (8,563 bp) genes, respectively.
For constructing the plasmid pGUN, a 327-bp cassette (nucleotides 19 to 345) of the NPTI gene (encoding prokaryotic neomycin phosphostransferase) from the pVAX1 plasmid (Invitrogen) was amplified by PCR with the primers NEOREP1 and NEOREP2 (Table 2), thus flanking the cassette with the NcoI/SpeI and SacII/NsiI restriction sites, respectively. The resulting PCR amplification fragment was then successively subcloned into the SpeI-NsiI and NcoI-SacII restriction sites, located upstream and downstream, respectively, of the URA3 gene from the pGEM-U plasmid (26), to yield the plasmid pGUN, containing the REP-URA3-REP fragment (abbreviated “GUN” sequence).
To obtain plasmids pG-ΔSLN1/GUN and pG-ΔCHK1/GUN, plasmids pG-SLN1 and pG-CHK1 were both digested with BclI (producing compatible ends with BglII) to release 2,528-bp and 1,992-bp central fragments from the SLN1 and CHK1 genes, respectively. The resulting digested plasmids were independently ligated to the GUN fragment previously amplified from pGUN with primers BGLUN1 and BGLUN2 (Table 2) and digested with BglII. In the same way, plasmid pG-ΔNIK1/GUN was constructed from plasmid pG-NIK1, replacing a 335-bp MfeI-digested central fragment from the NIK1 gene with a GUN sequence amplified with the primers MFEUN1 and MFEUN2 (Table 2). Thus, the three resulting plasmids, pG-ΔSLN1/GUN, pG-ΔNIK1/GUN, and pG-ΔCHK1/GUN, harbor 5′ end-SLN1-GUN-SLN1-3′end, 5′ end-NIK1-GUN-NIK1-3′ end, and 5′ end-CHK1-GUN-CHK1-3′ end disruption cassettes, respectively.
For construction of the complementation vectors, the plasmid pGEM-U was digested with SpeI and NotI to release a 1,536-bp fragment containing the C. lusitaniae URA3 gene. This fragment was subcloned into the SpeI/NotI restriction sites of the plasmid pVAX1, resulting in the plasmid pVAX-URA3. plasmids pG-SLN1, pG-NIK1, and pG-CHK1 were digested with NotI to release fragments overlapping the SLN1, NIK1, and CHK1 genes, respectively. Finally, these fragments were subcloned into the unique NotI restriction site from plasmid pVAX-URA3, resulting in the complementation plasmids pVAX-URA3-SLN1, pVAX-URA3-NIK1, and pVAX-URA3-CHK1, respectively.
Semiquantitative reverse transcription (RT)-PCR.
RNA (0.5 μg) was reverse transcribed by using the RevertAid H Minus First Strand cDNA synthesis kit (Fermentas) and oligo(dT)18 primers. PCR was performed with Hot Star Taq DNA polymerase (QIAGEN). One microliter of cDNA was used as a template for PCR. Oligonucleotides used in this experiment are listed in Table 2. Conditions for amplification were 15 min at 95°C, followed by 30 cycles of 30 s at 94°C, 30 s at 58°C, 30 s at 72°C, and a final extension of 5 min at 72°C. The PCR products were visualized by electrophoresis in 2% agarose gels and quantified by using a DC290 camera coupled with the Kodak 1D 3.5.3 software. Expression of the C. lusitaniae actin-encoding gene (ACT1) was used as an internal control.
Yeast transformation.
Strains were transformed by the electroporation procedure as previously described (26).
To obtain the auxotrophic strain 6936 ura3[Δ360] (MATa), the wild-type strain 6936 was transformed with the plasmid pG-ura3[Δ360] and the ura3[Δ360] genotype was selected by plating cells onto YNB solid medium supplemented with 1 mg ml−1 5-fluoroorotic acid (5-FOA) (Fermentas) and 25 μg ml−1 uracil (Fermentas).
For HKR gene disruption experiments, strains were transformed with the 5′ end-SLN1-GUN-SLN1-3′ end, 5′ end-NIK1-GUN-NIK1-3′ end, and 5′ end-CHK1-GUN-CHK1-3′ end cassettes released by digestion with NotI from plasmids pG-ΔSLN1/GUN, pG-ΔNIK1/GUN, and pG-ΔCHK1/GUN, respectively. For complementation experiments, 5-FOA-resistant strains were transformed with plasmid pVAX-URA3-SLN1, pVAX-URA3-NIK1, or pVAX-URA3-CHK1, previously linearized with endonuclease BglII, BsrGI, or ClaI, respectively, which was located in an HKR ORF, in order to obtain a greater efficiency of plasmid integration at the relevant homologous locus. Ura+ transformants were selected on YNB medium (0.67% yeast nitrogen base without amino acids [Difco Laboratoires], 2% glucose) supplemented with 1 M sorbitol. Transformants appeared after 3 days of incubation at 35°C.
Southern hybridization.
For Southern blot analysis, yeast genomic DNA was digested with appropriate restriction enzymes, separated by electrophoresis into a 0.8% agarose gel, and transferred onto a Hybond N+ nylon membrane (Roche Molecular Biochemicals). Membranes were hybridized with digoxigenin-labeled probes synthesized with a PCR DIG probe synthesis kit (Roche Molecular Biochemicals), as indicated by the supplier. The SLN1, NIK1, and CHK1 DNA probes were synthesized by PCR amplification with the PHK1-PHK2, PHK3-PHK4, and PHK7-PHK8 sets of specific primers (Table 2), respectively. The REP DNA probe, which was homologous to the REP fragment contained in disruption cassettes, was synthesized by PCR amplification with the NEOREP1-NEOREP2 set of primers (Table 2). For each experiment, probe-target hybrids were visualized by a chemiluminescence assay with the DIG luminescent detection kit (Roche Molecular Biochemicals) according to the manufacturer's instructions and exposure of the blot to X-ray film for 3 h.
Sensitivity test for stress responses, methylglyoxal (MG), and antifungal compounds.
Each strain was incubated overnight at 35°C in liquid YPD medium, washed, serially diluted (102 to 105 dilutions) in distilled water, and spotted (4 μl) on solid YPD medium. This medium was supplemented with 1 or 1.5 M of NaCl, KCl, or sorbitol (Sigma-Aldrich) to test osmosensitivity of spotted cells or with 10, 15, or 20 mM MG (Sigma-Aldrich) to test sensitivity toward this compound. To test sensitivity to UV, spotted cells were exposed to UV for 4 s (960 J/m2), 8 s (1,920 J/m2), or 12 s (2880 J/m2) using a UV table (Fischer Bioblock Scientific). To test temperature sensitivity, YPD plates were incubated at 30, 35, or 40°C.
An oxidative stress tolerance study was conducted using the method outlined by Pedreño et al. (59). Exponentially growing cells (optical density at 600 nm = 0.8 to 1.3) were divided into several identical aliquots, which were treated with 2, 5, or 10 mM of H2O2 (or maintained without H2O2 as a control) and incubated at 35°C for 1 h. Viability was determined after appropriate dilution of the samples with sterile water by plating in triplicate on solid YPD. Between 100 and 1,000 colonies were counted per plate. The percentage of survival was normalized to a H2O2-treated control sample of the wild-type strain (100%).
Stock solutions of fluconazole (FLC) (ICN Biomedicals Inc.) and flucytosine (5FC) (Sigma-Aldrich) were prepared by dissolving these antifungal agents in water at concentrations of 3.2 mg ml−1 and 12.8 mg ml−1, respectively. A stock solution of amphotericin B (AmB) (Bristol-Myers Squibb) (1.6 mg ml−1) was prepared in dimethyl sulfoxide, as iprodione (100 mg ml−1) and fenpiclonil (100 mg ml−1), kindly provided by P. Leroux (INRA, Versailles, France). For FLC and 5FC antifungal susceptibility testing, we used RPMI 1640 AutoMod (modified for autoclaving) agar plates. For AmB susceptibility testing, we used antibiotic medium 3 (Difco Laboratories) supplemented with 2% agar, whereas YPD agar plates were employed to test iprodione and fenpiclonil susceptibility. A tiny yeast colony isolated on an agar plate was picked up with a toothpick and resuspended in sterile water to a final concentration of 106 cells ml−1. A 5-μl calibrated loop was then used to stake yeast cells on each drug-containing agar plate.
Mating test.
Genetic crosses were performed under the same conditions described previously (27, 51). The reference mating type tester strains were the 6936 MATa strain and the Cl38 MATα strain. To obtain an auxotrophic strain, MATα ura3[Δ360], we crossed the strain Cl38 MATα with the strain 6936 MATa ura3[Δ360]. A progeny clone named PC1(α) with the genotype ura3[Δ360] MATα was selected and transformed with plasmid pG-ΔSLN1/GUN, pG-ΔNIK1/GUN, or pG-ΔCHK1/GUN, as described in the “Yeast transformation” section, in order to generate genotypes MATα, ura3[Δ360], sln1Δ::REP-URA3-REP (abbreviated sln1::GUNα), MATα, ura3[Δ360], nik1Δ::REP-URA3-REP (abbreviated nik1::GUNα) and MATα, ura3[Δ360], chk1Δ::REP-URA3-REP (abbreviated chk1::GUNα), respectively (Table 1). The number of conjugation tubes and produced tetrads (echinulate ascospores) was evaluated on three representative replicates for each unilateral and bilateral genetic cross (27, 51).
Pseudohyphal growth study and morphological observations.
Pseudohyphal growth was triggered by spotting a 5-μl drop (106 cells) on YCB solid medium (1.17% yeast carbon base [Difco laboratories]) supplemented with sorbitol (0.5 to 1.5 M), NaCl (0.3 to 1 M), KCl (0.3 to 1 M), MG (10 to 20 mM), or H2O2 (0.5 to 5 mM).
Pseudohypha length was measured from the edge of the spotted colony after 48 h of growth with an inverted Leitz microscope fitted with a micrometer eyepiece. All pictures were taken with an Olympus BX41 microscope using DP Controller software. Pictures of spotted cells producing pseudohyphae were taken directly on agar plates at magnification ×100.
For the kinetic study, cells were observed at 3, 6, 12, and 24 h after spotting at magnification ×400.
RESULTS
Identification, cloning, and expression of genes encoding HKRs in C. lusitaniae.
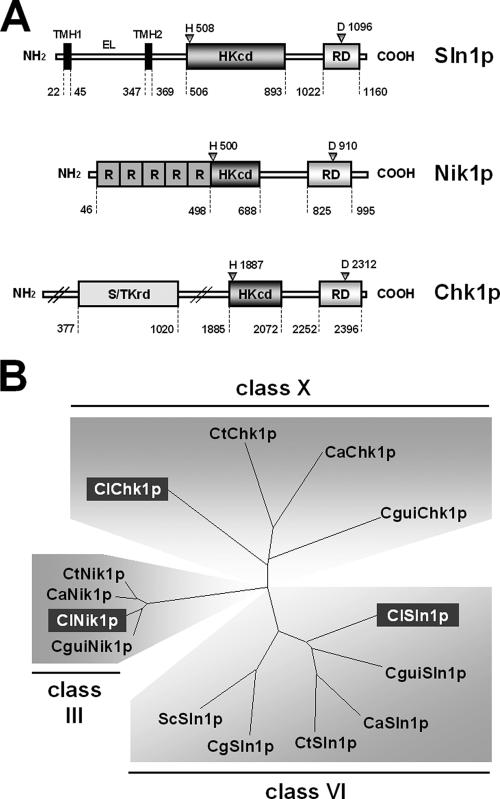
BLAST analysis of the C. lusitaniae database of the Broad Institute Fungal Genome web site revealed three genes encoding putative HKRs, i.e., SLN1, NIK1, and CHK1. The characterization of these genes and corresponding proteins is reported in Table 3 and Fig. 1A. A phylogenetic analysis is proposed in Fig. 1B by comparing C. lusitaniae Sln1p, Nik1p, and Chk1p with class III, VI, and X HKRs retrieved from C. guilliermondii (CguiSln1p, CguiNik1p, and CguiChk1p), C. tropicalis (CtSln1p, CtNik1p, and CtChk1p), C. glabrata (CgSln1p), and S. cerevisiae (ScSln1p).
TABLE 3.
Characterization of genes encoding HKRs and deduced proteins in C. lusitaniae
| Gene | Accession no.a | Supercontig: position | Protein length (aa) | MMb (kDa) | Identity (%)c
|
Classd | ||||
|---|---|---|---|---|---|---|---|---|---|---|
| Sc | Cg | Ca | Ct | Cgui | ||||||
| SLN1 | EF157661 | 3: 1560425-1556838 | 1195 | 132 | 39 | 38 | 37 | 52 | 54 | VI |
| NIK1 | EF157662 | 3: 1835363-1838611 | 1082 | 118 | 81 | 84 | 85 | III | ||
| CHK1 | EF157663 | 4: 1362474-1369664 | 2396 | 270 | 30 | 32 | 33 | X | ||
These sequence have been submitted to the GenBank database.
Molecular mass.
Abbreviations: Sc, S. cerevisiae; Cg, C. glabrata; Ca, C. albicans; Ct, C. tropicalis; Cgui, C. guilliermondii.
Described by Catlett et al. (15).
FIG. 1.
(A) Structure of the C. lusitaniae HKR proteins. The putative phosphorylable residues are indicated with triangles. EL, extracellular loop; HKcd, histidine kinase catalytic domain; R, repeated sequence; RD, receiver domain; S/TKrd, serine/threonine kinase-related domain; TMH, transmembrane helix. (B) Dendrogram generated after alignment of the predicted sequences of C. lusitaniae HKRs (ClSln1p, ClNik1p, and ClChk1p) with sequences retrieved from C. guilliermondii (CguiSln1p, CguiNik1p, and CguiChk1p), C. tropicalis (CtSln1p, CtNik1p, and CtChk1p), C. glabrata (CgSln1p), and S. cerevisiae (ScSln1p). Alignment utilizes the neighborhood-joining method from TreeView PPC software. Distances along the branches represent the divergence between two cognate sequences.
Domains commonly found in class VI hybrid HKRs are present in C. lusitaniae Sln1p. Two large hydrophobic transmembrane helices (amino acids [aa] 347 to 369) at the protein N terminus border a 302-amino-acid-long extracellular loop (Fig. 1A) predicted to be mainly fold in large helices and small sheets, as found in bacterial and plant Sln1p-related HKRs (57). The protein N terminus of C. lusitaniae Nik1p is composed of five HAMP-related repeats of about 90 aa (Fig. 1A) which are predicted to be coiled-coiled fold and have been demonstrated to be crucial for signal transduction in bacterial HKRs (7, 43). The large protein N terminus (aa 1 to 1884) of C. lusitaniae Chk1p includes a 643-aa serine/threonine kinase-related domain (Fig. 1A) which is commonly encountered in Chk1p-like (class X) fungal HKRs. The central regions of the three proteins represent histidine kinase domains with the five typical boxes (H, N, G1, F, and G2) that define the catalytic core of histidine kinases and include the presumptive autophosphorylated histidine residue. The C-terminal regions of Sln1p, Nik1p, and Chk1p are commonly composed of canonical receiver domains which exhibit acidic pockets containing phosphorylable aspartate residues (68).
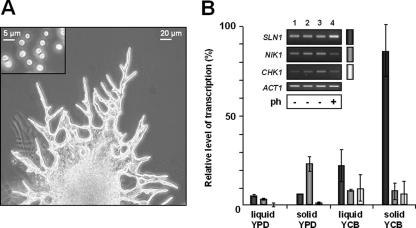
It was previously shown that pseudohyphal formation in C. lusitaniae was triggered on YCB solid medium but not in YCB liquid medium or in a complete YPD medium (30). We then compared the mRNA levels of HKR genes by semiquantitative RT-PCR using the wild-type strain 6936 in YCB or YPD, liquid or solid medium (Fig. 2A and B). The most striking result was the high level of SLN1 transcripts detected only when the 6936 strain was cultured in YCB solid medium. The accumulation of SLN1 mRNAs is concomitant with the formation of pseudohyphae. This first preliminary experiment could point out a correlation between SLN1 gene transcription and the morphological state of C. lusitaniae cells.
FIG. 2.
Morphology of C. lusitaniae cells and expression analysis of genes encoding HKRs. (A) Morphology of pseudohyphae emerging from the edge of a colony when cells are plated on YCB solid medium. The morphology of budding yeast cultured in YCB liquid medium is shown in the top left of the picture. (B) Semiquantitative RT-PCR of the three genes encoding HKRs (SLN1, NIK1, and CHK1). A representative RT-PCR analysis is shown. For each target gene, the amount of transcription was compared to that of the ACT1 gene (relative level of transcription). The histogram presents the mean values of results from three independent experiments. Error bars show standard deviation. Wild-type cells were cultured during 48 h in YPD liquid medium (lane 1), in YPD solid medium (lane 2), in YCB liquid medium (lane 3), or in YCB solid medium (lane 4). ph, pseudohyphal formation.
Disruption of genes encoding HKRs in C. lusitaniae.
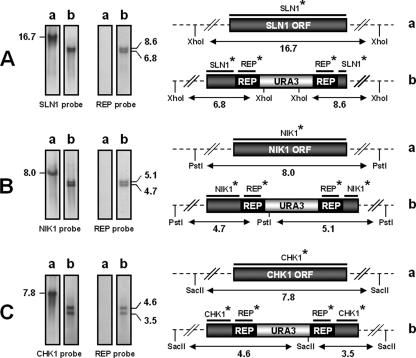
Linear DNA cassettes 5′ end-SLN1-GUN-SLN1-3′ end, 5′ end-NIK1-GUN-NIK1-3′ end, and 5′end-CHK1-GUN-CHK1-3′ end were each used to transform strain 6936 ura3[Δ360] to prototrophy. Correct insertion of the disrupting cassette was verified at each locus by Southern analysis of the genomic DNA of a subset of 20 Ura+ transformants, randomly selected from each transformation experiment, along with the DNA of the control strain, 6936 ura3[Δ360]. Genomic DNAs were digested with XhoI, PstI, or SacII and hybridized with SLN1, NIK1, and CHK1 probes according to the experiment. All fragments of the expected size are shown in Fig. 3. Southern blot analysis revealed that homologous integration of the 5′ end-SLN1-GUN-SLN1-3′ end, 5′ end-NIK1-GUN-NIK1-3′ end, and 5′ end-CHK1-GUN-CHK1-3′ end cassettes at the corresponding target locus occurred in half of transformants analyzed and was derived from gene replacement, resulting in disruption of the target gene and in the genotypes ura3[Δ360] sln1Δ::REP-URA3-REP (abbreviated sln1::GUN), ura3[Δ360] nik1Δ::REP-URA3-REP (abbreviated nik1::GUN), and ura3[Δ360] chk1Δ::REP-URA3-REP (abbreviated chk1::GUN), respectively. The molecular events were confirmed by hybridization with the REP DNA probe. For the remaining Ura+ transformants, the hybridization pattern revealed that they were derived from gene replacement at the ura3 locus (results not shown).
FIG. 3.
Southern blot hybridization and schematic representation of resident loci SLN1, NIK1, and CHK1 and of molecular events that occurred in transformants. Signals revealed by the labeled probes (each marked with an asterisk) correspond to those expected from the genomic restriction map. (A) Hybridization pattern with SLN1 and REP probes of XhoI-digested genomic DNA from 6936 ura3[Δ360] (a) or a representative sln1::GUN transformant (b). (B) Hybridization pattern with NIK1 and REP probes of PstI-digested genomic DNA from 6936 ura3[Δ360] (a) or a representative nik1::GUN transformant (b). (C) Hybridization pattern with CHK1 and REP probes of SacII-digested genomic DNA from 6936 ura3[Δ360] (a) or a representative chk1::GUN transformant (b). DNA fragment sizes are indicated in kilobases.
In order to obtain double-mutant genotypes, representative sln1::GUN, nik1::GUN, and chk1::GUN Ura+ transformants were plated onto YNB supplemented with 5-FOA and uracil. The frequency of 5-FOA-resistant colonies was about 1 × 10−5. The genetic organization of 10 5-FOA-resistant Ura− clones from each sln1::GUN, nik1::GUN, and chk1::GUN transformants was confirmed by Southern blot analysis (results not shown). The 5-FOA-resistant clones displayed the hybridization fragments expected from deletion of the URA3 gene and one of the flanking REP fragments. The genotypes ura3[Δ360] sln1Δ::REP (abbreviated sln1::REP), ura3[Δ360] nik1Δ::REP (abbreviated nik1::REP), and ura3[Δ360] chk1Δ::REP (abbreviated chk1::REP) were assigned to the 5-FOA-resistant clones.
The linear 5′ end-NIK1-GUN-NIK1-3′ end cassette was used to transform the chk1::REP strain. The double-mutant genotype ura3[Δ360] chk1Δ::REP nik1Δ::REP-URA3-REP (abbreviated chk1::REP nik1::GUN) was assigned to transformants harboring the expected hybridization profile as screened in Fig. 3. In the same way, the linear 5′ end-NIK1-GUN-NIK1-3′ end and 5′ end-CHK1-GUN-CHK1-3′ end cassettes were each used to transform the sln1::REP strain. Similarly, the double-mutant genotype ura3[Δ360] sln1Δ::REP chk1Δ::REP-URA3-REP (abbreviated chk1::GUN sln1::REP) was assigned to transformants harboring the expected hybridization profile as screened in Fig. 3.
We failed to obtain homologous integration of the 5′ end-NIK1-GUN-NIK1-3′ end cassette in the sln1::REP strain and in the chk1::REP sln1::REP strain (counterselected by cultivating the chk1::REP sln1::GUN strain on 5-FOA-containing YNB medium). In the same way, the SLN1 disruption in the nik1::REP deletant could never be achieved. Indeed, Southern blot analysis of 20 Ura+ transformants revealed that they derived only from ectopic integrations of disruption cassettes (40%) or from gene replacement at the ura3 locus (60%) (results not shown). These two last results suggested that the sln1 nik1 double mutant and the sln1 chk1 nik1 triple mutant are lethal in C. lusitaniae.
The linear DNA disruption cassettes were also used to transform the strain PC1(α) to prototrophy in order to generate sln1::GUNα, nik1::GUNα, and chk1::GUNα MATα disruptant strains.
Complementation of the sln1, nik1, and chk1 null mutant alleles.
To obtain reintegrant strains, linearized plasmids pVAX-URA3-SLN1, pVAX-URA3-NIK1, and pVAX-URA3-CHK1 were used to transform to prototrophy sln1::REP, nik1::REP, and chk1::REP mutants, respectively. We verified by Southern blotting (not shown) that homologous integration of the whole plasmids pVAX-URA3-SLN1, pVAX-URA3-NIK1, and pVAX-URA3-CHK1 occurred at the sln1, nik1, and chk1 loci, respectively, resulting in the relevant genotypes ura3[Δ360], sln1Δ::[REP pVAX-URA3-SLN1] (abbreviated sln1+SLN1), ura3[Δ360] nik1Δ::[REP pVAX-URA3-NIK1] (abbreviated nik1+NIK1), and ura3[Δ360] chk1Δ::[REP pVAX-URA3-CHK1] (abbreviated chk1+CHK1). In the same way, linearized plasmids pVAX-URA3-SLN1 and pVAX-URA3-NIK1 were used to transform to prototrophy sln1::REP chk1::REP and chk1::REP nik1::REP (counterselected by cultivating the chk1::REP nik1::GUN strain on 5-FOA-containing YNB medium) strains, respectively. By this way, we obtained chk1 sln1+SLN1 and chk1 nik1+NIK1 reintegrant strains.
Growth, osmotolerance, and oxidative stress response of mutants.
We investigated the putative involvement of the three HKRs in C. lusitaniae in perception and transduction of various environmental stresses. We started the phenotypic characterization of engineered mutants by comparing their growth kinetics. All mutants and the wild-type strain 6936 exhibited similar doubling times in liquid YPD medium (data not shown). No differences were observed in the development (colony length and aspect) of all strains cultured in solid YPD medium.
We next examined the osmotolerance of mutants. For that, drop plate assays were performed to determine the sensitivities of wild-type, sln1::GUN, nik1::GUN, chk1::GUN, chk1::GUN sln1::REP, and chk1::REP nik1::GUN strains to NaCl (1 to 1.5 M), KCl (1 to 1.5 M), and sorbitol (1 to 1.5 M). The growth of all simple and double mutants on these high-osmolarity media was similar to that of the wild-type strain (data not shown). These results imply that deletion of one or two HKR genes in C. lusitaniae has no effect on the growth and capacity of adaptation of yeast cells to hyperosmotic conditions.
Furthermore, none of the HKR mutants exhibited hypersensitivity to UV irradiation or high temperature (40°C) (data not shown), indicating that HKRs are not essential for the response to these physical stresses.
We also tested the effect of MG on HKR mutants, a metabolic by-product whose toxic action on cells can be counteracted by triggering activation of the HOG signaling pathway (1, 45). We evaluated the sensitivities of each strain by using different inoculum concentrations (102 to 105) in the presence of 10, 15, or 20 mM MG. We demonstrated that the sensitivities of disruptant strains to this compound were completely identical to that of the wild-type strain. Indeed, concentrations above and below 15 to 20 mM MG either were too toxic or had no effect on the strains (data not shown).
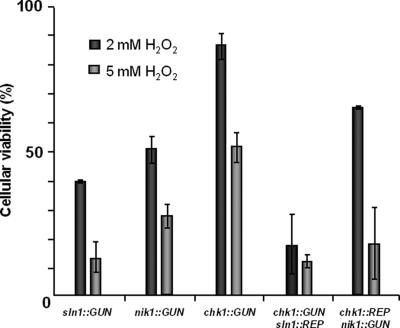
The effect of H2O2 on our engineered mutants was also studied (Fig. 4). For this purpose, YPD-grown exponential-phase cultures from all strains (wild type and simple and double disruptants) were incubated with 2, 5, or 10 mM H2O2. The percentage of survival was expressed with respect to an H2O2-treated control sample of the wild-type strain. The 10 mM H2O2 concentration was too deleterious for C. lusitaniae to estimate cell survival. Interestingly, at 2 and 5 mM H2O2, the sln1::GUN mutant exhibited the greatest sensitivity to H2O2 (39% ± 0.5% and 12% ± 5%, respectively). The nik1::GUN mutant was more resistant to H2O2 than the sln1::GUN mutant (49% ± 4% and 26% ± 5%, respectively). Moreover, the chk1::GUN strain displayed moderate sensitivity at 2 and 5 mM of peroxide (85% ± 5% and 51% ± 6%, respectively). The genetically engineered sln1+SLN1, nik1+NIK1, and chk1+CHK1 revertants showed H2O2 susceptibilities similar to that of wild-type strain 6936 (data not shown). Such a result could suggest the involvement of the three HKRs, especially Sln1p, in the regulation of oxidative stress.
FIG. 4.
Measurement of cell survival after oxidative stress imposed by the addition of 2 mM or 5 mM H2O2 in simple mutants (sln1::GUN, nik1::GUN, and chk1::GUN) and double mutants (chk1::GUN sln1::REP and chk1::REP nik1::GUN). The percent survival is expressed with respect to that of an H2O2-treated control sample of the wild-type strain (100%). Standard deviation bars are based on three individual replicates.
Susceptibilities of mutants to antifungal compounds.
We first monitored the effects of the clinical antifungals AmB, 5FC, and FLC. Neither hypersensitivity nor resistance toward these antifungals was observed. Indeed, the cell-mediated immunities (CMIs) of all the deletant strains were similar to that of the wild-type strain (AmB CMI, ≤1 μg ml−1; 5FC CMI, ≤4 μg ml−1; FLC CMI, ≤8 μg ml−1) (data not shown).
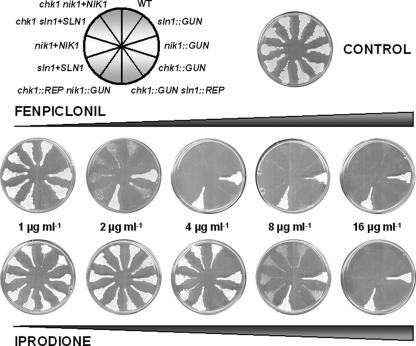
Because in various filamentous fungi, such as N. crassa and C. heterostrophus, the mutation of class III HKR genes is responsible for severe dicarboximide and phenylpyrrole resistance (53, 70), we studied the effects of these antifungals at different concentrations (1 to 16 μg ml−1) on our transformants. Figure 5 shows that wild-type strain 6936 and the chk1::GUN mutant were resistant to iprodione (dicarboximide) up to 4 μg ml−1, whereas the nik1::GUN mutant was resistant to iprodione in concentrations up to 16 μg ml−1. Interestingly, the sln1::GUN mutant was hypersusceptible to iprodione (resistance only up to 2 μg ml−1). The wild-type strain, 6936, and the chk1::GUN mutant were resistant to fenpiclonil up to 1 μg ml−1, whereas the nik1::GUN mutant was resistant to fenpiclonil to concentrations up to 16 μg ml−1. Moreover, the sln1::GUN mutant was hypersusceptible to fenpiclonil (CMI, <1 μg ml−1). The chk1::REP nik1::GUN and chk1::GUN sln1::REP double mutants presented the same antifungal susceptibility as the nik1::GUN and sln1::GUN simple mutants, respectively. The genetically engineered sln1+SLN1, nik1+NIK1, chk1 sln1+SLN1, and chk1 nik1+NIK1 revertants showed antifungal susceptibilities similar to those of wild-type strain 6936. Therefore, we concluded that in C. lusitaniae cells, NIK1 gene disruption confers fenpiclonil and iprodione resistance and that sln1 deletion increases the susceptibility to these antifungals.
FIG. 5.
Susceptibilities of C. lusitaniae mutants to iprodione and fenpiclonil. The wild-type strain and representative simple mutants (sln1::GUN, nik1::GUN, and chk1::GUN), double mutants (chk1::GUN sln1::REP and chk1::REP nik1::GUN), and reintegrant strains (sln1+SLN1, nik1+NIK1, chk1 sln1+SLN1, and chk1 nik1+NIK1) were grown for 48 h on YPD plates containing fenpiclonil or iprodione at concentrations indicated.
Mating abilities of mutants.
It was recently demonstrated that an HKR (Tco1p) promotes sexual reproduction in C. neoformans (9). We next investigated if the deletion of the SLN1, NIK1, and CHK1 genes could have an effect upon the in vitro mating ability of C. lusitaniae. We demonstrated that the simple or double disruptants, like the parental 6936 MATa wild-type strain, were still able to reproduce sexually in vitro when mated unilaterally with the appropriate opposite mating-type strain, Cl38 MATα. Moreover, all mutants exhibited normal wild-type mating in bilateral mutant crosses (sln1::GUN MATa × sln1::GUN MATα; nik1::GUN MATa × nik1::GUN MATα, and chk1::GUN MATa × chk1::GUN MATα) (data not shown). Therefore, the HKR proteins seem not to participate in the mating process of C. lusitaniae.
Pseudohyphal growth abilities of mutants.
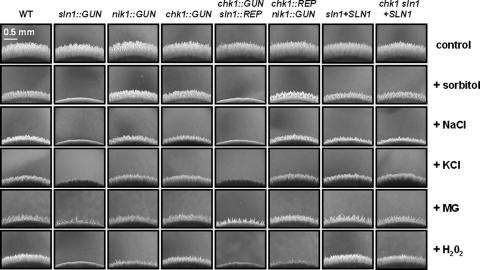
We investigated the capacity of transformants to differentiate pseudohyphae. Approximately 106 cells (contained in drops of 5 μl) of wild-type strain 6936 and of each mutant were spotted on YCB solid medium supplemented or not with sorbitol, NaCl, KCl, MG, or H2O2 (Fig. 6). The lengths of pseudohyphae emerging from the edge of the colony are reported in Table 4.
FIG. 6.
Pseudohyphal growth abilities of mutants. Cells (106; contained in drops of 5 μl) of the wild-type strain and representative simple mutants (sln1::GUN, nik1::GUN, and chk1::GUN), double mutants (chk1::GUN sln1::REP and chk1::REP nik1::GUN), and reintegrant strains (sln1+SLN1 and chk1 sln1+SLN1) were spotted on YCB solid medium supplemented or not with discriminatory concentrations of sorbitol (1 M), NaCl (0.5 M), KCl (0.5 M), MG (15 mM), and H2O2 (1 mM). Observations were done 48 h after spotting. This experiment was done in triplicate, and pictures present representative structures observed for each sample.
TABLE 4.
Pseudohyphal differentiation of C. lusitaniae strains on various supplemented YCB media
| Genotypea | Length of pseudohyphae with growth in mediumb
|
|||||
|---|---|---|---|---|---|---|
| YCB | + Sorbitol (1 M) | + NaCl (0.5 M) | + KCl (0.5 M) | + MG (15 mM) | + H202 (1 mM) | |
| WT | 560 ± 5 | 206 ± 22 | 120 ± 14 | 280 ± 14 | 240 ± 14 | 387 ± 16 |
| sln1::GUN | 440 ± 24 | 0 ± 0 | 0 ± 0 | 107 ± 8 | 0 ± 0 | 0 ± 0 |
| nik1::GUN | 580 ± 5 | 220 ± 14 | 107 ± 22 | 273 ± 21 | 240 ± 5 | 167 ± 50 |
| chk1::GUN | 567 ± 8 | 207 ± 22 | 113 ± 16 | 266 ± 8 | 247 ± 8 | 280 ± 28 |
| chk1::GUN sln1::REP | 547 ± 22 | 0 ± 0 | 0 ± 0 | 93 ± 16 | 133 ± 41 | 0 ± 0 |
| chk1::REP nik1::GUN | 580 ± 14 | 207 ± 16 | 133 ± 8 | 253 ± 16 | 233 ± 8 | 260 ± 14 |
| sln1+SLN1 | 567 ± 8 | 193 ± 22 | 107 ± 16 | 240 ± 14 | 233 ± 8 | 353 ± 22 |
| chk1 sln1+SLN1 | 566 ± 8 | 180 ± 28 | 107 ± 16 | 260 ± 14 | 213 ± 8 | 353 ± 29 |
| chk1+CHK1 | 568 ± 14 | NDc | ND | ND | ND | 373 ± 8 |
| nik1+NIK1 | 560 ± 5 | ND | ND | ND | ND | 367 ± 22 |
Abbreviated genotypes are given. WT, wild-type strain 6936.
The length of pseudohyphae was measured from the edge of the spotted colony. The values (μm) are means ± standard deviations based on three individual replicates. “+” indicates YCB medium supplemented as indicated.
ND, not determined.
Homogeneously distributed pseudohyphae were readily obtained for all strains on unsupplemented YCB medium. Engineered mutants present no significant length variation compared with the wild-type strain (560 ± 5 μm), except the sln1::GUN mutant, which displayed a reproducible reduction (440 ± 24 μm) of pseudohypha length. Interestingly, the chk1::GUN sln1::REP double mutant presented no significant reduction (547 ± 22 μm) of pseudohypha length. Figure 6 shows representative pictures of pseudohypha formation after 48 h of growth on YCB medium supplemented with the most discriminatory concentrations of sorbitol (1 M), NaCl (0.5 M), KCl (0.5 M), MG (15 mM), and H2O2 (1 mM). Indeed, concentrations above and below either were too toxic or had no effect on the pseudohyphal development of strains.
The addition of 1 M sorbitol produces a homogeneous reduction of pseudohyphal growth of the wild-type strain and of chk1::GUN, nik1::GUN, and chk1::REP nik1::GUN mutants compared to growth of the strains plated on unsupplemented YCB solid medium. The most important result of this experiment is the complete inhibition of pseudohyphal development of sln1::GUN and chk1::GUN sln1::REP mutants. Similar results were obtained with the presence of 0.5 M NaCl or KCl. The reintroduction of a functional SLN1 allele in sln1::REP (reintegrant sln1+SLN1 strain) and chk1::REP sln1::REP mutants (chk1 sln1+SLN1 reintegrant strain) was sufficient to restore pseudohyphal development similar to that of wild-type strain 6936 on YCB solid medium supplemented or not with sorbitol, NaCl, or KCl (Fig. 6). Thus, the SLN1 gene seems to play a key role in pseudohyphal development of C. lusitaniae during growth on high-osmolarity media.
We then tested the effect of MG. The addition of 15 mM MG produces a homogeneous reduction of pseudohyphal growth of the wild-type strain and of chk1::GUN, nik1::GUN, and chk1::REP nik1::GUN mutants compared to growth of strains plated on unsupplemented YCB solid medium (Fig. 6). Furthermore, sln1::GUN pseudohyphal formation was completely abolished. The chk1::GUN sln1::REP mutants formed some sporadic bunch-like pseudohyphae displaying a significant reduction of their lengths (133 ± 41 μm). Furthermore, the reintroduction of a functional SLN1 allele (sln1+SLN1 and chk1 sln1+SLN1 reintegrant strains) was sufficient to restore pseudohyphal development similar to that of wild-type strain 6936 on YCB solid medium supplemented with 15 mM MG.
Finally, we monitored the effect of oxidative stress. The addition of 1 mM H2O2 produces a reduction of pseudohyphal growth of the wild-type strain (387 ± 16 μm) (Fig. 6). Of all the strains, the sln1::GUN and chk1::GUN sln1::REP mutants exhibited the greatest sensitivity to peroxide, since pseudohyphal growth was completely inhibited. The nik1::GUN mutant presented a significant reduction (167 ± 50 μm) and the chk1::GUN strain a moderate reduction (280 ± 28 μm) in pseudohypha length. The reintroduction of the SLN1 allele was sufficient to restore partially (chk1 sln1+SLN1 reintegrant strain) or completely (sln1+SLN1 reintegrant strain) a pseudohyphal development similar to that of wild-type strain 6936 cultured on YCB solid medium supplemented with 1 mM H2O2. We also obtained wild-type pseudohyphal growth after the reintroduction of the NIK1 allele in the nik1::REP strain and the CHK1 allele in the chk1::REP strain (Table 4). These results demonstrate the crucial role of Sln1p and also the involvement of Nik1p and Chk1p in the regulation of oxidative stress during pseudohyphal development in C. lusitaniae.
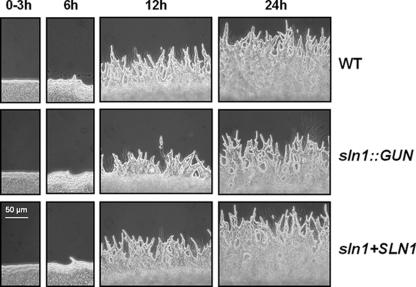
Because we observed that the sln1 deletion leads to a reduction of pseudohyphal formation in C. lusitaniae, we compared the time course of pseudohyphae differentiation in wild-type strain 6936, the sln1::GUN mutant, and the sln1+SLN1 reintegrant strain during 24 h (Fig. 7). The strains were simultaneously plated on YCB agar medium in order to induce pseudohyphal development. The first cell elongations which initiate chain formations (around the colony) of the wild-type strain and the sln1+SLN1 reintegrant strain were well detected after 6 h of incubation. However, a slight start of cell elongations was observed for the sln1::GUN mutant. After 12 h of growth, the wild-type strain and the sln1+SLN1 reintegrant strain presented readily homogeneously distributed pseudohyphae whereas the sln1::GUN mutant showed a heterogeneous distribution of shorter pseudohyphae. However, the number of branching of lateral cells along the pseudohyphae did not significantly vary between the sln1::GUN mutant and the wild-type strain. We thus concluded that the SLN1 disruption induced a delay in this morphogenetic transition and a reduction of pseudohypha growth.
FIG. 7.
Delay of pseudohyphal development of the sln1::GUN mutant. The wild-type strain, the sln1::GUN simple mutant, and the corresponding sln1+SLN1 reintegrant strain were simultaneously spotted on YCB solid medium. Observations were done at 3, 6, 12, and 24 h after spotting. This experiment was done in triplicate, and pictures present representative structures observed in each sample.
DISCUSSION
Fungal HKRs are involved in some essential cellular processes, such as osmosensing, virulence expression, oxidative stress response, and cell cycle control (62). Various investigations also report the implication of HKRs in yeast morphological switching. Indeed, in C. albicans, it has been shown that the three HKRs CaSln1p, CaNik1p, and Cahk1p play a predominant role in true-hypha formation (3, 13, 49, 69). Moreover, a recent study provides firm genetic evidence that the Nik1p-like HKRs (class III) were essential in the mold-to-yeast transition in the dimorphic pathogens H. capsulatum and B. dermatitidis (50). These findings point out a broad role of HKRs in morphological switching in nonrelated yeast species.
We undertook the characterization of HKRs in C. lusitaniae because, to our knowledge, the involvement of HKRs in yeasts which are only able to form pseudohyphae has never been fully studied. The only data available focused on the potential implication of the HOG pathway in pseudohyphal formation in S. cerevisiae (55). Moreover, C. lusitaniae represents an emerging human pathogen which is characterized by its propensity to develop resistance to antifungal agents during treatment. HKR-mediated transduction pathways, which are not encountered among the animal kingdom, could provide promising applications in antimicrobial compound research (10, 33, 46). BLAST analysis of the C. lusitaniae genome database allowed us to identify three genes, namely SLN1, NIK1, and CHK1, encoding HKRs of class VI, III, and X, respectively. An interesting fact is that each of their three homologues was found in the recently fully sequenced genomes of C. guilliermondii and C. tropicalis, as in C. albicans. In contrast, Nik1p-like and Chk1p-like HKRs were absent in C. glabrata and S. cerevisiae, each harboring a sole Sln1p-like HKR (class VI). A recent phylogeny study based on whole-genome analysis indicates that Saccharomycotina could be subdivided within two major groups: (i) species whose genomes have undergone a whole-genome duplication (referred to as the WGD clade), including C. glabrata and other yeasts from the Saccharomyces genus, and (ii) species that translate CTG as serine instead of leucine (referred to as the CTG clade), including C. guilliermondii, C. tropicalis, C. albicans, and C. lusitaniae (25). Thus, our analysis suggests that fungal species belonging to the WGD clade probably encode a unique class VI HKR, and interestingly, species belonging to the CTG clade encodes three HKRs from classes VI, III, and X, respectively.
We started this work by comparing the transcriptional regulation of the SLN1, NIK1, and CHK1 genes when C. lusitaniae grows either as budding blastospores or as pseudohyphae. We detected a high level of SLN1 transcripts when the wild-type strain was cultured in YCB solid medium, indicating that the accumulation of SLN1 mRNAs is concomitant with the formation of pseudohyphae. This experiment could point out a correlation between SLN1 gene transcription and the pseudohyphal developing process in C. lusitaniae.
We then disrupted the SLN1, NIK1, and CHK1 genes by a transformation system based upon the “URA3 blaster” strategy that we developed for C. lusitaniae in a previous study (58). In this way, we then selected sln1, nik1, and chk1 simple mutants as well as sln1 chk1 and nik1 chk1 double mutants. Nevertheless, we failed to obtain an sln1 nik1 double mutant and an sln1 chk1 nik1 triple mutant, suggesting that these deletants are not viable in C. lusitaniae, as demonstrated for C. albicans (69). Thus, since the combination of sln1 and nik1 mutations resulted in synthetic lethality, it could suggest that Sln1p and Nik1p act in two complementary pathways and that the lack of one of these proteins could be compensated by the other. Independent studies have demonstrated that (i) ScYpd1p interacts with ScSln1p HKR in S. cerevisiae (42, 48, 61) and (ii) ScYpd1p interacts with MgNik1p HKR (Hik1p, from the rice BLAST fungus Magnaporthe grisea) (48). Preliminary data in our lab showed that the C. lusitaniae YPD1 gene, encoding a histidine-containing phosphotransfer protein, is essential (unpublished result). This could suggest that Ypd1p is a cross talk key component of the Sln1p and Nik1p pathways in C. lusitaniae.
When growing as budding yeast, all mutants and the wild-type strain exhibited similar doubling times and no difference was observed between the development of strains in normal and hyperosmotic conditions. However, since the sln1 mutant was hypersensitive to oxidative stress, we hypothesized a contribution of Sln1p in oxidative stress adaptation. These results are slightly different of those described for C. albicans: (i) sln1 transformants were viable under normal and high osmolarity, but growth retardation was observed under hyperosmotic conditions (49), and (ii) only chk1 mutants were moderately hypersensitive to oxidative stress (38). C. lusitaniae also behaves differently from several filamentous fungi, such as N. crassa and M. fructicola, for which nik1 mutants are sensitive to osmotic stress (44, 53).
Important morphological changes occur during sexual reproduction of C. lusitaniae, notably during conjugation (27). Moreover, in the human pathogen C. neoformans, it has been shown that the Tco1p HKR and the cognate stress-activated Hog1p MAPK system governed several cellular events, including sexual reproduction (9). So, we investigated if the deletion of the C. lusitaniae SLN1, NIK1, and CHK1 genes modified the in vitro mating ability of the yeast. We demonstrated that all of the strains were still able to reproduce sexually when crossed with the opposite-mating-type strain.
In several filamentous fungi, the mutation of genes encoding HKRs of class III has been found to confer resistance to dicarboximides, such as iprodione, and to phenylpyrroles, such as fenpiclonil or fludioxonil (8, 21, 22, 44, 53, 70). A similar study was carried out with C. albicans sln1, chk1, and nik1 disruptants. Because the wild-type strain CAI4, used to construct the corresponding HKR mutants, was intrinsically resistant to both antifungals, it was impossible to study the specific role of CaNik1p in dicarboximide and phenylpyrrole resistance (54). We thus tested the susceptibilities of C. lusitaniae HKR disruptants and the wild-type strain 6936 to iprodione and fenpiclonil. The present study clearly demonstrated that the 6936 strain was susceptible to both antifungals whereas the nik1 genotype conferred resistance to these compounds. It has been shown that the target of these compounds was probably the osmotic stress signal transduction pathway, which involved the class III HKRs (21). Furthermore, a recent study showed that expression of MgHIK1, a NIK1 orthologue from M. grisea, could confer fungicide susceptibility on S. cerevisiae, which was intrinsically resistant (48). Moreover, two-hybrid experiments revealed an interaction between MgHik1p and the yeast phosphorelay protein Ypd1p. It was thus suggested that Nik1p-like HKRs were direct targets of the fungicides or were mediators of antifungal action which were transmitted to the Hog1p pathway via Ypd1p. To date, since disruption of the C. lusitaniae NIK1 gene confers resistance to iprodione and fenpiclonil and since sequences homologuous to those of the S. cerevisiae YPD1, HOG1, and SSK1 genes are located in the C. lusitaniae genome (not shown; personal data), similar targeting processes and signaling pathways could occur in Candida species, such as C. lusitaniae. Finally, we also found that sln1 deletion increases the susceptibilities of strains to fenpiclonil and iprodione, as was previously described for C. albicans (54). This might indicate that in Candida species, the signal transduction of Nik1p-like HKRs interacts with that of Sln1p-like HKRs. Nevertheless, Sln1p, Nik1p, and Chk1p seem not to be implicated in resistance to antifungals of clinical relevance, such as FLC, 5FC, and AmB.
In order to determine if HKRs could be involved in the yeast-to-pseudohypha morphological transition in C. lusitaniae, we compared the development of the HKR disruptants with that of the wild-type strain on YCB plates, an appropriate medium efficiently triggering pseudohyphal differentiation (30). Homogeneously distributed pseudohyphae were readily obtained for all mutants, but a reproducible reduction of about 20% of pseudohypha length was observed for the sln1 genotype. More precisely, we showed that the defect of pseudohyphae forming by the sln1 mutant was due to a global delay of the development process. Interestingly, the disruption of the CHK1 gene in the sln1 genotype partially restored wild-type-like pseudohyphal development. These results underlined a couple of conclusions compared to C. albicans true hyphal development. First, although HKRs are all clearly implicated in a dramatic defect in true hypha formation when deleted in C. albicans, only one gene, namely SLN1, appears to be crucial for pseudohyphal differentiation in our model. This clearly illustrates that the information leading the yeast switching toward true hyphae or pseudohyphae is controlled by a set of specific genes. HKRs seem thus to be more closely involved in the true hyphal shaping of C. albicans than they are in the pseudohyphal shaping of C. lusitaniae. Another possibility is that the HKRs in C. albicans behave similarly to those in C. lusitaniae when C. albicans is undergoing pseudohyphal formation. Secondly, similar effects were observed when the CHK1 gene was deleted in the sln1 strain, since it partially restored wild-type pseudohyphal development in C. lusitaniae and partly restored true hyphal growth in C. albicans. This supports the hypothesis that Candida sp. Sln1p-like HKRs would be able to regulate negative effectors of Chk1p-like HKRs (69). Moreover, pseudohyphal development on the hyperosmotic surface and oxidant environment of the sln1 disruptant is dramatically affected comparing to that of the wild-type strain. Interestingly, these growth conditions have no effect (hyperosmotic condition) or have a lesser effect (oxidant stress) on other HKR-disrupted mutants. This implies a differential contribution of the three HKRs in osmotic and oxidant adaptation during the morphological transition.
In summary, this study reveals that C. lusitaniae, like other species belonging to the CTG clade of the subphylum Saccharomycotina, harbors three HKR genes. The NIK1 gene, which encodes a class III HKR, is clearly involved in dicarboximide and phenylpyrrole resistance when the corresponding gene is mutated. The SLN1 gene encoding a transmembrane class VI HKR appears to be crucial (i) for oxidative stress adaptation during C. lusitaniae strain growth as budding yeast and (ii) in the early steps of pseudohyphal development, especially in hyperosmotic and oxidant conditions. Future studies will aim to characterize downstream elements, such as histidine-containing phosphotransfer protein and response regulators, in order to clarify fungal HKR-mediated pathways and cross talk events that regulate the morphological transition of this emerging pathogen.
Acknowledgments
We acknowledge the Broad Institute Fungal Genome Initiative for making the complete genome sequence of Candida lusitaniae available. We thank Susan Fox for critical reading of the manuscript.
Footnotes
Published ahead of print on 27 July 2007.
REFERENCES
- 1.Aguilera, J., S. Rodriguez-Vargas, and J. A. Prieto. 2005. The HOG MAP kinase pathway is required for the induction of methylglyoxal-responsive genes and determines methylglyoxal resistance in Saccharomyces cerevisiae. Mol. Microbiol. 56:228-239. [DOI] [PubMed] [Google Scholar]
- 2.Alex, L. A., K. A. Borkovich, and M. I. Simon. 1996. Hyphal development in Neurospora crassa: involvement of a two-component histidine kinase. Proc. Natl. Acad. Sci. USA 93:3416-3421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Alex, L. A., C. Korch, C. P. Selitrennikoff, and M. I. Simon. 1998. COS1, a two-component histidine kinase that is involved in hyphal development in the opportunistic pathogen Candida albicans. Proc. Natl. Acad. Sci. USA 95:7069-7073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Altschul, S. F., T. L. Madden, A. A. Schaffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Aoyama, K., H. Aiba, and T. Mizuno. 2001. Genetic analysis of the His-to-Asp phosphorelay implicated in mitotic cell cycle control: involvement of histidine-kinase genes of Schizosaccharomyces pombe. Biosci. Biotechnol. Biochem. 65:2347-2352. [DOI] [PubMed] [Google Scholar]
- 6.Appleby, J. L., J. S. Parkinson, and R. B. Bourret. 1996. Signal transduction via the multi-step phosphorelay: not necessarily a road less traveled. Cell 86:845-848. [DOI] [PubMed] [Google Scholar]
- 7.Aravind, L., and C. P. Ponting. 1999. The cytoplasmic helical linker domain of receptor histidine kinase and methyl-accepting proteins is common to many prokaryotic signalling proteins. FEMS Microbiol. Lett. 176:111-116. [DOI] [PubMed] [Google Scholar]
- 8.Avenot, H., P. Simoneau, B. Iacomi-Vasilescu, and N. Bataille-Simoneau. 2005. Characterization of mutations in the two-component histidine kinase gene AbNIK1 from Alternaria brassicicola that confer high dicarboximide and phenylpyrrole resistance. Curr. Genet. 47:234-243. [DOI] [PubMed] [Google Scholar]
- 9.Bahn, Y. S., K. Kojima, G. M. Cox, and J. Heitman. 2006. A unique fungal two-component system regulates stress responses, drug sensitivity, sexual development, and virulence of Cryptococcus neoformans. Mol. Biol. Cell 17:3122-3135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Barrett, J. F., and J. A. Hoch. 1998. Two-component signal transduction as a target for microbial anti-infective therapy. Antimicrob. Agents Chemother. 42:1529-1536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Berman, J. 2006. Morphogenesis and cell cycle progression in Candida albicans. Curr. Opin. Microbiol. 9:595-601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Buck, V., J. Quinn, T. Soto Pino, H. Martin, J. Saldanha, K. Makino, B. A. Morgan, and J. B. Millar. 2001. Peroxide sensors for the fission yeast stress-activated mitogen-activated protein kinase pathway. Mol. Biol. Cell 12:407-419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Calera, J. A., and R. Calderone. 1999. Flocculation of hyphae is associated with a deletion in the putative CaHK1 two-component histidine kinase gene from Candida albicans. Microbiology 145:1431-1442. [DOI] [PubMed] [Google Scholar]
- 14.Calera, J. A., X. J. Zhao, F. De Bernardis, M. Sheridan, and R. Calderone. 1999. Avirulence of Candida albicans CaHK1 mutants in a murine model of hematogenously disseminated candidiasis. Infect. Immun. 67:4280-4284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Catlett, N. L., O. C. Yoder, and B. G. Turgeon. 2003. Whole-genome analysis of two-component signal transduction genes in fungal pathogens. Eukaryot. Cell 2:1151-1161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chapeland-Leclerc, F., J. Bouchoux, A. Goumar, C. Chastin, J. Villard, and T. Noel. 2005. Inactivation of the FCY2 gene encoding purine-cytosine permease promotes cross-resistance to flucytosine and fluconazole in Candida lusitaniae. Antimicrob. Agents Chemother. 49:3101-3108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chauhan, N., J. P. Latge, and R. Calderone. 2006. Signalling and oxidant adaptation in Candida albicans and Aspergillus fumigatus. Nat. Rev. Microbiol. 4:435-444. [DOI] [PubMed] [Google Scholar]
- 18.Chun, C. D., O. W. Liu, and H. D. Madhani. 2007. A link between virulence and homeostatic responses to hypoxia during infection by the human fungal pathogen Cryptococcus neoformans. PLoS Pathog. 3:e22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Clemons, K. V., T. K. Miller, C. P. Selitrennikoff, and D. A. Stevens. 2002. fos-1, a putative histidine kinase as a virulence factor for systemic aspergillosis. Med. Mycol. 40:259-262. [DOI] [PubMed] [Google Scholar]
- 20.Csank, C., and K. Haynes. 2000. Candida glabrata displays pseudohyphal growth. FEMS Microbiol. Lett. 189:115-120. [DOI] [PubMed] [Google Scholar]
- 21.Cui, W., R. E. Beever, S. L. Parkes, P. L. Weeds, and M. D. Templeton. 2002. An osmosensing histidine kinase mediates dicarboximide fungicide resistance in Botryotinia fuckeliana (Botrytis cinerea). Fungal Genet. Biol. 36:187-198. [DOI] [PubMed] [Google Scholar]
- 22.Dry, I. B., K. H. Yuan, and D. G. Hutton. 2004. Dicarboximide resistance in field isolates of Alternaria alternata is mediated by a mutation in a two-component histidine kinase gene. Fungal Genet. Biol. 41:102-108. [DOI] [PubMed] [Google Scholar]
- 23.Ernst, J. F. 2000. Transcription factors in Candida albicans—environmental control of morphogenesis. Microbiology 146:1763-1774. [DOI] [PubMed] [Google Scholar]
- 24.Favel, A., A. Michel-Nguyen, F. Peyron, C. Martin, L. Thomachot, A. Datry, J. P. Bouchara, S. Challier, T. Noel, C. Chastin, and P. Regli. 2003. Colony morphology switching of Candida lusitaniae and acquisition of multidrug resistance during treatment of a renal infection in a newborn: case report and review of the literature. Diagn. Microbiol. Infect. Dis. 47:331-339. [DOI] [PubMed] [Google Scholar]
- 25.Fitzpatrick, D. A., M. E. Logue, J. E. Stajich, and G. Butler. 2006. A fungal phylogeny based on 42 complete genomes derived from supertree and combined gene analysis. BMC Evol. Biol. 6:99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Francois, F., F. Chapeland-Leclerc, J. Villard, and T. Noel. 2004. Development of an integrative transformation system for the opportunistic pathogenic yeast Candida lusitaniae using URA3 as a selection marker. Yeast 21:95-106. [DOI] [PubMed] [Google Scholar]
- 27.François, F., T. Noel, R. Pepin, A. Brulfert, C. Chastin, A. Favel, and J. Villard. 2001. Alternative identification test relying upon sexual reproductive abilities of Candida lusitaniae strains isolated from hospitalized patients. J. Clin. Microbiol. 39:3906-3914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Geourjon, C., and G. Deleage. 1995. SOPMA: significant improvements in protein secondary structure prediction by consensus prediction from multiple alignments. Comput. Appl. Biosci. 11:681-684. [DOI] [PubMed] [Google Scholar]
- 29.Gimeno, C. J., P. O. Ljungdahl, C. A. Styles, and G. R. Fink. 1992. Unipolar cell divisions in the yeast S. cerevisiae lead to filamentous growth: regulation by starvation and RAS. Cell 68:1077-1090. [DOI] [PubMed] [Google Scholar]
- 30.Goumar, A., A. Brulfert, C. Martin, J. Villard, and T. Noël. 2004. Selection and genetic analysis of pseudohyphae defective mutants with attenuated virulence in Candida lusitaniae. J. Med. Mycol. 14:3-11. [Google Scholar]
- 31.Guinet, R., J. Chanas, A. Goullier, G. Bonnefoy, and P. Ambroise-Thomas. 1983. Fatal septicemia due to amphotericin B-resistant Candida lusitaniae. J. Clin. Microbiol. 18:443-444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hawkins, J. L., and L. M. Baddour. 2003. Candida lusitaniae infections in the era of fluconazole availability. Clin. Infect. Dis. 36:e14-e18. [DOI] [PubMed] [Google Scholar]
- 33.Koretke, K. K., A. N. Lupas, P. V. Warren, M. Rosenberg, and J. R. Brown. 2000. Evolution of two-component signal transduction. Mol. Biol. Evol. 17:1956-1970. [DOI] [PubMed] [Google Scholar]
- 34.Krogh, A., B. Larsson, G. von Heijne, and E. L. Sonnhammer. 2001. Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J. Mol. Biol. 305:567-580. [DOI] [PubMed] [Google Scholar]
- 35.Kruppa, M., and R. Calderone. 2006. Two-component signal transduction in human fungal pathogens. FEMS Yeast Res. 6:149-159. [DOI] [PubMed] [Google Scholar]
- 36.Kruppa, M., T. Goins, J. E. Cutler, D. Lowman, D. Williams, N. Chauhan, V. Menon, P. Singh, D. Li, and R. Calderone. 2003. The role of the Candida albicans histidine kinase CHK1 gene in the regulation of cell wall mannan and glucan biosynthesis. FEMS Yeast Res. 3:289-299. [DOI] [PubMed] [Google Scholar]
- 37.Kruppa, M., B. P. Krom, N. Chauhan, A. V. Bambach, R. L. Cihlar, and R. A. Calderone. 2004. The two-component signal transduction protein Chk1p regulates quorum sensing in Candida albicans. Eukaryot. Cell 3:1062-1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Li, D., V. Gurkovska, M. Sheridan, R. Calderone, and N. Chauhan. 2004. Studies on the regulation of the two-component histidine kinase gene CHK1 in Candida albicans using the heterologous lacZ reporter gene. Microbiology 150:3305-3313. [DOI] [PubMed] [Google Scholar]
- 39.Li, S., A. Ault, C. L. Malone, D. Raitt, S. Dean, L. H. Johnston, R. J. Deschenes, and J. S. Fassler. 1998. The yeast histidine protein kinase, Sln1p, mediates phosphotransfer to two response regulators, Ssk1p and Skn7p. EMBO J. 17:6952-6962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Loomis, W. F., G. Shaulsky, and N. Wang. 1997. Histidine kinases in signal transduction pathways of eukaryotes. J. Cell Sci. 110:1141-1145. [DOI] [PubMed] [Google Scholar]
- 41.Lorenz, M. C., and J. Heitman. 1998. Regulators of pseudohyphal differentiation in Saccharomyces cerevisiae identified through multicopy suppressor analysis in ammonium permease mutant strains. Genetics 150:1443-1457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lu, J. M., R. J. Deschenes, and J. S. Fassler. 2003. Saccharomyces cerevisiae histidine phosphotransferase Ypd1p shuttles between the nucleus and cytoplasm for SLN1-dependent phosphorylation of Ssk1p and Skn7p. Eukaryot. Cell 2:1304-1314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lupas, A., M. Van Dyke, and J. Stock. 1991. Predicting coiled coils from protein sequences. Science 252:1162-1164. [DOI] [PubMed] [Google Scholar]
- 44.Ma, Z., Y. Luo, and T. Michailides. 2006. Molecular characterization of the two-component histidine kinase gene from Monilinia fructicola. Pest Manag. Sci. 62:991-998. [DOI] [PubMed] [Google Scholar]
- 45.Maeta, K., S. Izawa, and Y. Inoue. 2005. Methylglyoxal, a metabolite derived from glycolysis, functions as a signal initiator of the high osmolarity glycerol-mitogen-activated protein kinase cascade and calcineurin/Crz1-mediated pathway in Saccharomyces cerevisiae. J. Biol. Chem. 280:253-260. [DOI] [PubMed] [Google Scholar]
- 46.Matsushita, M., and K. D. Janda. 2002. Histidine kinases as targets for new antimicrobial agents. Bioorg. Med. Chem. 10:855-867. [DOI] [PubMed] [Google Scholar]
- 47.Minari, A., R. Hachem, and I. Raad. 2001. Candida lusitaniae: a cause of breakthrough fungemia in cancer patients. Clin. Infect. Dis. 32:186-190. [DOI] [PubMed] [Google Scholar]
- 48.Motoyama, T., T. Ohira, K. Kadokura, A. Ichiishi, M. Fujimura, I. Yamaguchi, and T. Kudo. 2005. An Os-1 family histidine kinase from a filamentous fungus confers fungicide-sensitivity to yeast. Curr. Genet. 47:298-306. [DOI] [PubMed] [Google Scholar]
- 49.Nagahashi, S., T. Mio, N. Ono, T. Yamada-Okabe, M. Arisawa, H. Bussey, and H. Yamada-Okabe. 1998. Isolation of CaSLN1 and CaNIK1, the genes for osmosensing histidine kinase homologues, from the pathogenic fungus Candida albicans. Microbiology 144:425-432. [DOI] [PubMed] [Google Scholar]
- 50.Nemecek, J. C., M. Wuthrich, and B. S. Klein. 2006. Global control of dimorphism and virulence in fungi. Science 312:583-588. [DOI] [PubMed] [Google Scholar]
- 51.Noël, T., A. Favel, A. Michel-Nguyen, A. Goumar, K. Fallague, C. Chastin, F. Leclerc, and J. Villard. 2005. Differentiation between atypical isolates of Candida lusitaniae and Candida pulcherrima by determination of mating type. J. Clin. Microbiol. 43:1430-1432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Noël, T., F. Francois, P. Paumard, C. Chastin, D. Brethes, and J. Villard. 2003. Flucytosine-fluconazole cross-resistance in purine-cytosine permease-deficient Candida lusitaniae clinical isolates: indirect evidence of a fluconazole uptake transporter. Antimicrob. Agents Chemother. 47:1275-1284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ochiai, N., M. Fujimura, T. Motoyama, A. Ichiishi, R. Usami, K. Horikoshi, and I. Yamaguchi. 2001. Characterization of mutations in the two-component histidine kinase gene that confer fludioxonil resistance and osmotic sensitivity in the os-1 mutants of Neurospora crassa. Pest Manag. Sci. 57:437-442. [DOI] [PubMed] [Google Scholar]
- 54.Ochiai, N., M. Fujimura, M. Oshima, T. Motoyama, A. Ichiishi, H. Yamada-Okabe, and I. Yamaguchi. 2002. Effects of iprodione and fludioxonil on glycerol synthesis and hyphal development in Candida albicans. Biosci. Biotechnol. Biochem. 66:2209-2215. [DOI] [PubMed] [Google Scholar]
- 55.O'Rourke, S. M., and I. Herskowitz. 1998. The Hog1 MAPK prevents cross talk between the HOG and pheromone response MAPK pathways in Saccharomyces cerevisiae. Genes Dev. 12:2874-2886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Page, R. D. 1996. TreeView: an application to display phylogenetic trees on personal computers. Comput. Appl. Biosci. 12:357-358. [DOI] [PubMed] [Google Scholar]
- 57.Papon, N., J. Bremer, A. Vansiri, G. Glevarec, M. Rideau, and J. Creche. 2006. Molecular cloning and expression of a cDNA encoding a hybrid histidine kinase receptor in tropical periwinkle Catharanthus roseus. Plant Biol. (Stuttgart) 8:731-736. [DOI] [PubMed] [Google Scholar]
- 58.Papon, N., T. Noel, M. Florent, S. Gibot-Leclerc, D. Jean, C. Chastin, J. Villard, and F. Chapeland-Leclerc. 2007. Molecular mechanism of flucytosine resistance in Candida lusitaniae: contribution of the FCY2, FCY1, and FUR1 genes to 5-fluorouracil and fluconazole cross-resistance. Antimicrob. Agents Chemother. 51:369-371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Pendreno, Y., P. Gonzalez-Parraga, S. Conesa, M. Martinez-Esparza, A. Aguinaga, J. A. Hernandez, and J. C. Arguelles. 2006. The cellular resistance against oxidative stress (H2O2) is independent of neutral trehalase (Ntc1p) activity in Candida albicans. FEMS Yeast Res. 6:57-62. [DOI] [PubMed] [Google Scholar]
- 60.Pfaller, M. A., S. A. Messer, and R. J. Hollis. 1994. Strain delineation and antifungal susceptibilities of epidemiologically related and unrelated isolates of Candida lusitaniae. Diagn. Microbiol. Infect. Dis. 20:127-133. [DOI] [PubMed] [Google Scholar]
- 61.Posas, F., S. M. Wurgler-Murphy, T. Maeda, E. A. Witten, T. C. Thai, and H. Saito. 1996. Yeast HOG1 MAP kinase cascade is regulated by a multistep phosphorelay mechanism in the SLN1-YPD1-SSK1 “two-component” osmosensor. Cell 86:865-875. [DOI] [PubMed] [Google Scholar]
- 62.Santos, J. L., and K. Shiozaki. 2001. Fungal histidine kinases. Sci. STKE 2001:RE1. [DOI] [PubMed] [Google Scholar]
- 63.Scherer, S., and D. A. Stevens. 1987. Application of DNA typing methods to epidemiology and taxonomy of Candida species. J. Clin. Microbiol. 25:675-679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Schumacher, M. M., C. S. Enderlin, and C. P. Selitrennikoff. 1997. The osmotic-1 locus of Neurospora crassa encodes a putative histidine kinase similar to osmosensors of bacteria and yeast. Curr. Microbiol. 34:340-347. [DOI] [PubMed] [Google Scholar]
- 65.Sudbery, P., N. Gow, and J. Berman. 2004. The distinct morphogenic states of Candida albicans. Trends Microbiol. 12:317-324. [DOI] [PubMed] [Google Scholar]
- 66.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Viaud, M., S. Fillinger, W. Liu, J. S. Polepalli, P. Le Pecheur, A. R. Kunduru, P. Leroux, and L. Legendre. 2006. A class III histidine kinase acts as a novel virulence factor in Botrytis cinerea. Mol. Plant-Microbe Interact. 19:1042-1050. [DOI] [PubMed] [Google Scholar]
- 68.West, A. H., and A. M. Stock. 2001. Histidine kinases and response regulator proteins in two-component signaling systems. Trends Biochem. Sci. 26:369-376. [DOI] [PubMed] [Google Scholar]
- 69.Yamada-Okabe, T., T. Mio, N. Ono, Y. Kashima, M. Matsui, M. Arisawa, and H. Yamada-Okabe. 1999. Roles of three histidine kinase genes in hyphal development and virulence of the pathogenic fungus Candida albicans. J. Bacteriol. 181:7243-7247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Yoshimi, A., M. Tsuda, and C. Tanaka. 2004. Cloning and characterization of the histidine kinase gene Dic1 from Cochliobolus heterostrophus that confers dicarboximide resistance and osmotic adaptation. Mol. Genet. Genomics 271:228-236. [DOI] [PubMed] [Google Scholar]
- 71.Young, L. Y., C. M. Hull, and J. Heitman. 2003. Disruption of ergosterol biosynthesis confers resistance to amphotericin B in Candida lusitaniae. Antimicrob. Agents Chemother. 47:2717-2724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Young, L. Y., M. C. Lorenz, and J. Heitman. 2000. A STE12 homolog is required for mating but dispensable for filamentation in Candida lusitaniae. Genetics 155:17-29. [DOI] [PMC free article] [PubMed] [Google Scholar]