Abstract
Histatin 5 (Hst 5) is a salivary cationic peptide that has toxicity for Candida albicans by inducing rapid cellular ion imbalance and cell volume loss. Microarray analyses of peptide-treated cells were used to evaluate global gene responses elicited by Hst 5. The major transcriptional response of C. albicans to Hst 5 was expression of genes involved in adaptation to osmotic stress, including production of glycerol (RHR2, SKO1, and PDC11) and the general stress response (CTA1 and HSP70). The oxidative-stress genes AHP1, TRX1, and GPX1 were mildly induced by Hst 5. Cell defense against Hst 5 was dependent on the Hog1 mitogen-activated protein kinase (MAPK) pathway, since C. albicans hog1/hog1 mutants were significantly hypersensitive to Hst 5 but not to Mkc1 MAPK or Cek1 MAPK mutants. Activation of the high-osmolarity glycerol (HOG) pathway was demonstrated by phosphorylation of Hog1 MAPK as well as by glycerol production following Hst 5 treatment in a dose-dependent manner. C. albicans cells prestressed with sorbitol were less sensitive to subsequent Hst 5 treatment; however, cells treated concurrently with osmotic stress and Hst 5 were hypersensitive to Hst 5. In contrast, cells subjected to oxidative stress had no difference in sensitivity to Hst 5. These results suggest a common underlying cellular response to osmotic stress and Hst 5. The HOG stress response pathway likely represents a significant and effective challenge to physiological levels of Hst 5 and other toxic peptides in fungal cells.
Salivary histatins (Hst's) are a family of histidine-rich cationic proteins produced by human salivary gland acinar cells that are key components of the innate defense system in the oral cavity. Hst 5 has high potency against Candida albicans at physiological concentrations (10 to 30 μM) (30, 38), including azole- or amphotericin-resistant strains of Candida (39). Peptide-based therapeutics for Candida infections are promising, since peptides can be designed for species-specific targeting of C. albicans while preserving nonpathogenic microbial communities. However, better understanding of the cellular targets and mechanism of action of Hst 5 is needed before these peptides are brought into clinical use as antifungal agents.
Hst 5 is not a classical pore- or channel-forming peptide, since it has little disruptive activity in liposome model membranes (9) and cells treated with Hst 5 do not release dyes such as calcein (11). However, Hst 5 induces efflux of cellular ATP (20) and potassium and magnesium ions (42) and elevates cell permeability to small cationic dyes such as propidium iodide (40). Thus, cellular ion imbalance appears to be an early and significant consequence of Hst 5 interaction with C. albicans cells. We found that Hst 5 causes rapid and irreversible loss of C. albicans cell volume and cell cycle arrest and that these effects are attenuated or prevented by pretreatment of cells with anion channel inhibitors such as niflumic acid, 5-nitro-2-(3-phenylpropylamino) benzoic acid, and DIDS (diisothiocyanatostilbene-2,2′-disulfonic acid) (4). Further support for ionic imbalance as a primary effect of Hst 5 is the fact that single-allele deletion of the TRK1 gene, encoding the major plasma membrane K+ uptake system in C. albicans, resulted in nearly complete protection from Hst 5 killing (5). This model system, whereby the primary effect of Hst 5 is to induce ionic imbalance, suggests that the primary adaptive responses would be rapid compensatory changes in cellular ion and water transport and activation of osmotic stress response pathways (17), which potentially could minimize the ion loss and cell volume decrease caused by Hst 5.
An alternative view to the primary ionic effects of Hst 5 is based on observations that C. albicans cells grown under anaerobic conditions, respiration-deficient mutants, and cells pretreated with general metabolic inhibitors such as azide (20) are partially protected from Hst 5 toxicity. Thus, an energized cell with unimpaired mitochondrial function supports Hst 5 activity. In this alternative model, Hst 5 interferes with the mitochondrial respiratory chain and causes formation of oxygen radicals (reactive oxygen species [ROS]) as the main cause of cell death (15, 16). It has been demonstrated that ROS do not initiate apoptosis or programmed cell death pathways in Hst 5-treated cells (41); rather, direct mitochondrial and cytoplasmic membrane damage by ROS is hypothesized to be the essential step for cell killing (15). In this model, activation of oxidative stress response pathways would be the major adaptive response for protection of C. albicans from Hst 5 killing. However, an increase in ROS levels in Candida also has been observed following morphogenesis (36) or cell growth arrest (33) and in cells subjected to conditions that cause low intracellular potassium levels (45). Therefore, ROS could be generated in C. albicans cells as a result of Hst 5-induced ion loss and cell cycle arrest as well as from mitochondrial damage.
C. albicans, like Saccharomyces cerevisiae, must cope with different environmental stress conditions, including heat shock, oxidative stress, high and low osmolarity, nutrient availability, and toxins. Cells have adapted to growth under these conditions by developing a variety of protective mechanisms ranging from general stress responses to highly specific pathways (reviewed for S. cerevisiae in reference 17 and for C. albicans in references 22 and 31). The mitogen-activated protein kinase (MAPK) pathways are key elements in sensing and transmitting the responses of cells to environmental conditions by the sequential action of phosphorylation events (22). Three MAPK pathways have so far been identified in C. albicans that act in response to environmental signals. The cell integrity pathway, mediated by the Mkc1 MAPK, is involved in responses to oxidative and hypo-osmotic stress and antifungal drugs and in invasive hyphal growth and biofilm development (23, 29). The Cek1 pathway, mediated by the Cek1 and Cek2 MAPKs, homologues of S. cerevisiae Kss1 and Fus3, respectively (7), is involved in cell wall biogenesis, hyphal development, and virulence (8, 12, 28). The Hog1, or high-osmolarity glycerol, pathway is involved in adaptation to osmotic, oxidative (1, 2, 34), and heavy metal stresses (13). Recent studies have suggested that Hog1 also has a regulatory role for the Mkc1 and Cek1 MAPKs (12, 32). Although elements of the HOG pathway in S. cerevisiae are well studied, less is known about pathway components involved in stress response in C. albicans. Placing C. albicans cells in a high-osmolarity environment (ionic or nonionic) leads to rapid water loss, a decrease in cell volume, and cell cycle arrest (34). To survive such stress, cells respond with activation of the HOG pathway: Hog1 becomes phosphorylated in a Pbs2-dependent manner, translocates to the nucleus, and mediates a transcriptional response culminating in intracellular accumulation of osmolytes such as glycerol (19, 34), reorganization of the cytoskeleton, and cell wall biogenesis (1, 2, 3). Thus, the HOG pathway in Candida plays an important role in protection of this organism against osmotic stress.
Global profiling of gene expression in cells following treatment with antifungal agents is a powerful tool to shed light on the central effects of these agents. C. albicans genomic-scale DNA microarrays have been used to study global responses to antifungal agents (25) and environmental stresses (13). In the present study, we used microarray analyses of Hst 5-treated cells to gain insight into this peptide's mechanism of action by examining gene responses elicited by Hst 5. Here we provide evidence that the transcriptional response of C. albicans to Hst 5 involves the expression of genes involved in adaptation to osmotic stress. Hst 5 also induced expression of stress response genes in a Hog1-dependent manner, while expression of Hog1-independent oxidative stress genes such as CAP1 and SOD2 was only mildly elevated. Phosphorylation of the Hog1 MAPK was observed following Hst 5 treatment, and C. albicans Hog1 knockout mutants demonstrated hypersensitivity to Hst 5. Thus, our results show that the activation of Hog1-dependent genes in C. albicans cells challenged with Hst 5 involves osmotic stress response mechanisms, and they shed light on the mechanism whereby C. albicans cells mount a defensive response to toxic peptides.
MATERIALS AND METHODS
Strains and media.
The genotypes of strains used in this study are described in Table 1. The wild-type C. albicans strain CAI-4 was used for Hst 5 treatment in microarrays, quantitative reverse transcriptase PCR (RT-PCR), and Western blotting. C. albicans TK1, in which one allele of the potassium transporter TRK1 is deleted, was derived from strain CAI-4 (5). Strains CK43A (CEK1/cek1) and CK43B-16 (cek1/cek1), used for testing sensitivity to Hst 5, were kindly provided by M. Whiteway, McGill University, Montreal, Canada. To evaluate MAPK-dependent responses to Hst 5 action, we also tested strains CKY357 (mck1/mck1) and CKY358 (MCK1/mck1), generously provided by C. Kumamoto, Tufts University School of Medicine, Boston, MA. The wild-type strain RM1000, along with the Hog1 mutant strains JC50 (hog1/hog1) and JC52 (HOG1/hog1), were obtained from J. Quinn, University of Newcastle upon Tyne, Newcastle upon Tyne, United Kingdom, and C. albicans Hog1 MAPK pathway mutants CSSK21 (ssk1/ssk1) and CSSK23 (SSK1/ssk1) were generously provided by R. Calderone, Georgetown University Medical Center, Washington, DC. The C. albicans cap1/cap1 strain (MMY301) was kindly provided by W. Scott Moye-Rowley, University of Iowa. Cells were maintained either in yeast nitrogen base (YNB; Qbiogene) plus 2% glucose with the addition of uridine when required or in yeast extract-peptone-dextrose (YPD) medium (Qbiogene). Solid media were made with 1.5% Bacto agar (Difco). Cells were routinely manipulated at 30°C in order to maintain the cells in blastospore morphology.
TABLE 1.
Strains used in this study
| Strain | Genotype | Source |
|---|---|---|
| CAF2-1 | Δura3::imm434/URA3 | 14 |
| CAI-4 | Δura3::imm434/Δura3::imm434 | 14 |
| TK1 | Δura3::imm434/Δura3::imm434 TRK1/trk1Δ | 5 |
| CK43A | Δura3::imm434/Δura3::imm434 CEK1/cek1Δ::hisG-URA3-hisG | 8 |
| CK43B-16 | Δura3::imm434/Δura3::imm434 cek1Δ::hisG-URA3-hisG/cek1Δ::hisG | 8 |
| CSSK21 | Δura3::imm434/Δura3::imm434 Δssk1::hisG/Δssk1::hisG-URA3-hisG | 6 |
| CSSK23 | Δura3::imm434/Δura3::imm434 Δssk1::hisG/SSK1::URA3-hisG | 6 |
| RM1000 | ura3::imm434/ura3::imm434 his1::hisG/his1::hisG | 37 |
| JC50 | ura3::imm434/ura3::imm434 his1::hisG/his1::hisG hog1::loxP-ura3-loxP/hog1::loxP-HIS1-loxP CIp20 (URA3 HIS1) | 37 |
| JC52 | ura3::imm434/ura3::imm434 his1::hisG/his1::hisG hog1::loxP-ura3-loxP hog1::loxP-his1-loxP Clp20-HOG1 (URA3 HIS1) | 37 |
| CKY357 | Δmck1::hisG/mck1::hisG mkc1::pCK70 (URA3) | 23 |
| CKY358 | Δmkc1::hisG/Δmkc1::hisG mkc1::pCK71 (MKC1 URA3) | 23 |
| MMY301 | Δura3::imm434/Δura3::imm434 Δcap1::hisG/Δcap1::hisG-URA3-hisG | 43 |
Sample preparation and isolation of total RNA.
For microarray experiments, C. albicans CAI-4 (wild-type) cells were used. For quantitative RT-PCR, total RNA from C. albicans strains CAI-4 (wild type), JC50 (hog1/hog1), and TK1 (TRK1/trk1) was isolated. Cells were grown overnight in 10 ml of YNB medium with 2% glucose and the addition of uridine when required. Preceding Hst 5 treatment, cells were harvested, washed twice with 10 mM sodium phosphate buffer (NaPB), pH 7.4, and reconstituted in the same buffer. About 3 × 108 cells were incubated with 500 μM Hst 5 (40% lethal dose) for 0 or 60 min for the microarray experiments and for 0, 30, 60, or 120 min for the quantitative RT-PCR experiments. At the indicated time points, cells were collected and stored immediately at −70°C. Hst 5 treatments were performed in duplicate from independent starter cultures. For each experiment, cells from independent cultures were treated with buffer only. Total RNA was isolated using an RNeasy mini kit from QIAGEN as previously described (4). The samples were both in-column treated with DNase I using the RNase-free DNase set from QIAGEN and off-column treated using the TURBO DNA-free set from Ambion. The absence of genomic DNA contamination was routinely confirmed by PCR using 18S rRNA primers (Table 2).
TABLE 2.
Primers used in this study
| Gene name | Primer sequence |
|---|---|
| YHB1 | Forward, 5′-TGACAGTCGAATACGAAACCA-3′ |
| Reverse, 5′-CAATGTTTCACCGGCTTCTT-3′ | |
| CSA1 | Forward, 5′-CCAGCTAACGTGCAAAGCA-3′ |
| Reverse, 5′-TCGGAAGCAGAAGCAACCT-3′ | |
| TRX1 | Forward, 5′-TCAATTGGGTTCTTTAGCACA-3′ |
| Reverse, 5′-TGACACGATTGACTTCTTCACC-3′ | |
| GAP2 | Forward, 5′-TTCCTTGCTGCATCCGATA-3′ |
| Reverse, 5′-ACCTTGAGCAGCCAATGCT-3′ | |
| HIP1 | Forward, 5′-ACAAGTGCATCTCCATTTGTCA-3′ |
| Reverse, 5′-CAACCATATACTGCCAGGTTCC-3′ | |
| AHP1 | Forward, 5′-CTGCTGTGCCTGGTGCTT-3′ |
| Reverse, 5′-TTGACGCCCTTGTCTTTGA-3′ | |
| CTA1 | Forward, 5′-TTGGTCAACACGGTCCATT-3′ |
| Reverse, 5′-CCATAAGCACCGGAACCTT-3′ | |
| HSP12 | Forward, 5′-ACCCTTGCTGAAACAGCTCA-3′ |
| Reverse, 5′-CACCGGTGACAACTCCACTC-3′ | |
| SOD2 | Forward, 5′-TTGGCTCCTGTCTCTCAAGG-3′ |
| Reverse, 5′-CCAATTTGCCATTGGTGATT-3′ | |
| GPX1 | Forward, 5′-ATGGCAAGAACCAGGCACTA-3′ |
| Reverse, 5′-CTGGATCTGCTTGTTCACCA-3′ | |
| SKO1 | Forward, 5′-TGATTCAACCGGCTACCAATAC-3′ |
| Reverse, 5′-AACCAACGCTTGAGAAGATTGA-3′ | |
| ENA21 | Forward, 5′-CTCCCACGGCAAGAGTAACTA-3′ |
| Reverse, 5′-TCGAATATCAGGGACAGT-3′ | |
| CAP1 | Forward, 5′-CTTGGTATCACCGGAATCTCA-3′ |
| Reverse, 5′-ATCAGTTCCCACACCATTGAA-3′ | |
| RHR2 | Forward, 5′-ACCACATCCACAAGGTTACCA-3′ |
| Reverse, 5′-AGCACCTTTACCTGCGGTTAT-3′ | |
| 18S rRNA | Forward, 5′-CGATGGAAGTTTGAGGCAATA-3′ |
| Reverse, 5′-CTCTCGGCCAAGGCTTATACT-3′ |
SYBR green quantitative RT-PCR assays.
To assess the levels of mRNA, 2 μg of total RNA was used per reaction for the first-strand synthesis (cDNA) using the RETROscript kit (Ambion). Quantitative real time RT-PCR experiments were carried out as described previously (24). In brief, the primers were designed according to the TaqMan criteria using the Primer 3 Input program (frodo.wi.mit.edu/cgi-bin/primer3/primer3_www.cgi) and confirmed via Amplify 1.2 Software for PCR. The primers used are listed in Table 2. Primers were custom synthesized by Integrated DNA Technologies, Inc. SYBR green PCR conditions were as follows: 2 min at 50°C plus 2 min at 95°C and then 40 cycles of 30 s at 95°C, 30 s at 55°C, and 30 s at 72°C. Amplification and detection were carried out in 96-well plates on an iCycler iQ real-time detection system (Bio-Rad). Each reaction mixture contained duplicates of the test cDNA templates, negative RNA controls, no-template controls, and positive genomic-DNA controls. All samples contained 1× SYBR green iQ Supermix (Bio-Rad) and a 150 nM concentration of both the forward and the reverse primer. Fluorescent data were collected and analyzed with iCycler iQ software. The results were analyzed using the ΔΔCT method with 18S rRNA used as a reference and cells at time zero used as a normalizer. Gene expression by Hst 5-treated cells was compared to that by buffer-treated cells using a nonpaired t test, and differences in expression levels between genes with a P value of <0.05 were considered significant.
Microarray experimental design and data analysis.
The microarray slides used in these studies were produced by a working group funded by the National Institute of Dental and Craniofacial Research (44). Methods for labeling of probes and hybridization to microarrays were adapted from protocols developed by the Institute for Genomic Research (TIGR [http://pga.tigr.org/protocols.html]). cDNA was synthesized from total RNA samples using Superscript II RT (Invitrogen) in reaction mixtures containing 0.5 mM aminoallyl-dUTP-deoxynucleoside triphosphates. After cDNA synthesis, RNA was hydrolyzed with 1 M NaOH and 0.5 M EDTA, followed by neutralization with 1 M HCl. The cDNA was purified using the QIAGEN QIAquick PCR purification kit and dried by SpeedVac. The dried cDNA was coupled with the fluorescent dye Cy3 or Cy5 (GE Healthcare) for 1 h at room temperature in the dark. Unincorporated dye was removed using a QIAGEN QIAquick PCR purification kit, and the samples were dried in a SpeedVac. The labeled cDNA samples were resuspended in a solution containing 50% formamide, 10× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate), and 0.2% sodium dodecyl sulfate. The appropriate Cy3-cDNA and Cy5-cDNA samples were mixed and heated for 2 min at 100°C before application to the microarray slide. Samples were hybridized for 20 h at 42°C, and the slides were washed, dried, and analyzed as described below. In order to balance dye bias, dye swap experiments for each of the two samples were performed.
Slides were scanned with a GenePix 4200A scanner (Axon Instruments, now part of Molecular Devices); image preprocessing data quantification was done using GenePix Pro (version 6.0); and the raw expression data (GenePix results format [GPR] files) were produced. For microarray data analysis, the TIGR microarray software suite (http://www.tm4.org/), which includes ExpressConverter, Midas, and Mev, was used. First, TIGR ExpressConverter (version 1.7) was used to convert GPR files to Mev files, which may be used in other companion software. Second, with TIGR Midas (version 2.19), the data sets were further processed using total-intensity normalization, LOWESS normalization, standard-deviation regulation, and in-slide replicate analysis so that each gene in an array received a normalized expression value. Finally, with TIGR Mev (version 3.03), one sample Student t test was performed in order to identify genes with statistically significant changes. Fold changes were derived from the averages of expression levels (log2 based) for four experiments; positive and negative values represent up- or down-regulated levels of a gene in the Hst 5-treated strain compared with the wild-type strain. Genes with P values of <0.05 are listed in Table 3; of these, five genes were found to have a >2-fold change (log2 based).
TABLE 3.
Microarray analysis of Hst 5-treated C. albicans cells
| orf19 classification | Gene name | Gene producta | Fold change in expressionb |
|---|---|---|---|
| Up-regulated genes | |||
| Cell stress | |||
| 19.3707 | YHB1 | Nitric oxide dioxygenase (18); response to stress and toxin | 1.03 |
| 19.2763 | AHP1 | Putative alkyl hydroperoxide reductase, response to oxidative stress | 0.60 |
| 19.2013 | KAR2 | Hsp70; translocation of nascent polypeptides across the ER membrane | 0.56 |
| 19.2568 | IFU5 | Predicted membrane protein; response to desiccation | 0.47 |
| 19.7611 | TRX1 | Similar to thioredoxin; response to oxidative stress | 0.36 |
| 19.6515 | HSP90 | Heat shock protein; response to stress | 0.20 |
| Cellular metabolism | |||
| 19.2877 | PDC11 | Protein similar to pyruvate decarboxylase; pyruvate metabolism | 1.67 |
| 19.5811 | MET1 | Putative uroporphyrin-3 C-methyltransferase; methionine biosynthesis | 0.83 |
| 19.3934 | CAR1 | Argininase family protein; arginine catabolism to ornithine | 0.64 |
| 19.5024 | GND1 | Putative 6-phosphogluconase dehydrogenase; glucose metabolism | 0.63 |
| 19.6844 | ICL1 | Isocitrate lyase; enzyme of glyoxylate cycle (26) | 0.57 |
| 19.7408 | LEU1 | 3-Isopropylmalate dehydrase | 0.55 |
| 19.903 | GPM1 | Phosphoglycerate mutase; glycolysis; regulated by Efg1, Gnc4p | 0.53 |
| 19.3672 | GAL10 | Similar to UDP-glucose 4-epimerase; fluconazole induced | 0.35 |
| 19.4959 | None | Predicted ORF from Assembly 19 | 0.24 |
| 19.5818 | SUR2 | Similar to ceramide hydroxylase; sphingolipid biosynthesis | 0.22 |
| Transcription, signaling | |||
| 19.3838 | EFB1 | Translation elongation factor EF-1β | 0.29 |
| 19.173 | None | Putative transcription factor with zinc finger DNA-binding motif | 0.28 |
| 19.730 | RGD3 | Putative Rho GTPase-activating protein | 0.28 |
| 19.5106 | ESS1 | Prolyl isomerase; yeast hyphal switching, Cph1p pathway | 0.20 |
| Mitochondrion | |||
| 19.930 | PET9 | Mitochondrial inner membrane ATP:ADP translocator | 0.92 |
| 19.2871 | SDH12 | Succinate dehydrogenase; mitochondrial electron transport | 0.52 |
| 19.1872 | None | Predicted protein from Assembly 19; induced by nitric oxide | 0.31 |
| Cell wall protein; 19.220 | PIR1 | 1,3-β-Glucan-linked cell wall protein; Hog1 induced | 1.08 |
| Down-regulated genes | |||
| Plasma membrane transporters | |||
| 19.7114 | CSA1 | Surface antigen on elongating hyphae and buds; ion homeostasis | −1.16 |
| 19.6993 | GAP2 | General amino acid permease; amino acid transport | −1.12 |
| 19.918 | CDR11 | Putative transporter of PDR superfamily of ABC family | −0.54 |
| 19.1659 | None | Predicted protein from Assembly 19; S. cerevisiae orthologue, YDR349C | −0.29 |
| 19.3195 | None | Predicted protein from Assembly 19; S. cerevisiae orthologue, AVT5 | −0.26 |
| 19.3195 | HIP1 | Potential general amino acid permease | −0.22 |
| 19.2160 | NAG4 | Monosaccharide transporter | −0.21 |
| Cellular metabolism | |||
| 19.2909 | CTN3 | Predicted carnitine acetyltransferase; acetyl-CoA metabolism | −0.61 |
| 19.2641 | PGM2 | Similar to S. cerevisiae Pgm2p, which is phosphoglycomutase | −0.58 |
| 19.6308 | None | Predicted protein from Assembly 19; similar to endopeptidase | −0.47 |
| 19.6941 | None | Predicted protein from Assembly 19; diacylglycerol O-acyltransferase | −0.35 |
| Transcription, signaling | |||
| 19.123 | None | Predicted protein from Assembly 19; calcineurin regulatory protein | −0.56 |
| 19.3135 | None | Predicted protein from Assembly 19; S. cerevisiae orthologue, IMP2 | −0.44 |
| 19.3127 | CZF1 | Predicted transcription factor; inhibition of pheromone response | −0.27 |
| Mitochondrion | |||
| 19.4947 | None | Predicted protein from Assembly 19; S. cerevisiae orthologue, YPR098C | −0.26 |
| 19.6409 | None | ATPase activity; protein targeting to the mitochondrion | −0.24 |
ER, endoplasmic reticulum; ORF, open reading frame; CoA, coenzyme A.
Changes (n-fold) were derived from the averages of expression levels (log2 based) from four experiments as described in Materials and Methods. Positive and negative values represent up- or down-regulated levels of a gene in the Hst 5-treated strain compared with the wild-type strain. Only genes with P values of <0.05 by a one-sample Student t test were reported. Values for genes with a >2-fold change in expression are boldfaced.
Immunoblot analysis of Hog1 phosphorylation.
C. albicans strains CAI-4 (wild-type) and JC50 (hog1/hog1) were handled as for the quantitative RT-PCR experiments. Briefly, yeast strains were grown to an optical density at 600 nm of 0.8 at 30°C in 15 ml YPD cultures. Cells were collected and resuspended in prewarmed diluted (1 part medium to 3 parts 10 mM NaPB) Sabouraud dextrose medium (dSD). After a 1-h incubation, 1 ml of a CAI-4 cell suspension (2 × 107 cells) was incubated either with 100 μl of a predetermined concentration of Hst 5 for 30 min or with 32 μM Hst 5 for 0, 10, 20, 30, or 60 min. Control cells were manipulated in dSD only. As a positive control, 2 × 107 cells were treated with 400 mM NaCl or 1 M sorbitol for 10 min. JC50 cells (2 × 107) were treated with 400 mM NaCl for 10 min as a negative control. After treatment, the cells were harvested at 2,500 × g and 4°C for 2 min, washed with ice-cold NaPB, and then kept at −80°C until further analysis. For protein extraction, cells were resuspended in 250 μl of yeast breaking buffer (FastPROTEIN kit; MP Biomedicals) containing protease inhibitors (50 mM sodium fluoride, 1 mM sodium orthovanadate, 1 mM phenylmethylsulfonyl fluoride, 1 μg/ml aprotinin, 1 μg/ml pepstatin A, 1 μg/ml leupeptin, and 1 μg/ml benzamidine). Cell extracts were obtained by glass bead disruption using a FastPrep-24 instrument (MP Biomedicals) at a speed setting of 6.0, at 20 s/cycle, for three cycles and were centrifuged at 8,000 × g for 10 min to separate the cytosolic fraction from unbroken cells and organelles. Equal amounts of total protein (40 μg) were separated using 10% sodium dodecyl sulfate-polyacrylamide gel electrophoresis, transferred to polyvinylidene difluoride membranes, and probed with Hog1-specific anti-phospho-p38 (Thr180/Tyr182) monoclonal antibody 3D7 (Cell Signaling Technology, Inc.), followed by a peroxidase-conjugated goat anti-rabbit secondary antibody. The blots were developed according to the manufacturer's recommendations using the ECL kit (Amersham Pharmacia Biotech). After the anti-phospho-p38 antibody was stripped, the membrane was immunoblotted with anti-Hog1 antibody y-215 (Santa Cruz Biotechnology, Inc).
Measurement of intracellular glycerol levels.
Intracellular glycerol levels were determined spectrophotometrically with a glycerol determination kit (Sigma, St. Louis, MO; FG0100) according to the manufacturer's instructions. C. albicans cells were grown overnight in YPD medium (optical density at 600 nm, 1.2); then cells were resuspended in diluted (1 part medium to 3 parts 10 mM NaPB) dSD, because Hst 5 retains nearly complete candidacidal activity in this medium but is inactive in other media, including YPD and YNB. For each assay, 9 × 107 cells (750 μl) were treated with NaCl (0.5 M) or sorbitol (1 M) as positive controls for glycerol production or with Hst 5 (7.5 to 62 μM) for 1 h. Cells were treated with buffer alone to determine baseline glycerol production as a result of cell manipulation. Cells were then washed and resuspended in 2 ml of 0.5 M Tris-HCl (pH 7.5), and each group was divided for glycerol determination and a candidacidal assay, described below. For glycerol measurements, cells (1 ml) were incubated at 95°C for 10 min, spun down, and the supernatant used for glycerol determination. Glycerol values were normalized to C. albicans cell numbers, and basal amounts of glycerol produced in sham-treated cells (less than 10% of treatment values for all conditions) were subtracted from stimulated values. Each assay was performed in triplicate in at least three independent experiments.
Hst 5 sensitivity under stress conditions.
C. albicans (CAF2-1) was cultured under standard conditions in YNB. For Hst 5 stress conditioning, 7 μM Hst 5 was added to 106 cells at 37°C and samples were withdrawn for plating after 5 min, 15 min, 30 min, and 60 min. These samples were first diluted with buffer to stop further Hst 5 uptake and then spotted onto YPD plates, some of which were supplemented with 2 mM H2O2 or 1 M sorbitol. The growth of cells under each condition was examined following 24 h of incubation at 37°C.
For osmotic stress before Hst 5 treatment, C. albicans cells were first suspended in dSD medium (106 cells in 500 μl), then exposed to osmotic stress by addition of sorbitol (final concentration, 1 M), and finally incubated at 30°C for 1 h. Control cells were manipulated in diluted dSD alone. Cell aliquots were then removed; Hst 5 was added (7 μM to 62 μM), and cells were incubated for 1 h before being plated. To analyze the effect of Hst 5 (7 μM) at various time points, samples were withdrawn after 5 min, 15 min, 30 min, and 60 min. These samples were immediately diluted with buffer and spotted onto YPD agar plates. Growth of colonies was observed after 24 h of incubation at 37°C.
Candidacidal assay.
Killing of Candida strains by Hst 5 was performed by the microdilution plate assay, as previously described (24). Briefly, C. albicans cells were grown in YNB medium, washed twice with 10 mM NaPB (Na2HPO4-NaH2PO4 [pH 7.5]), and resuspended in NaPB at a concentration of 106 cells/ml. The cell suspensions (25 μl) were then mixed with Hst 5 (7.5 to 62 μM) for 1 h at 30°C with shaking. Cell suspensions were diluted in 10 mM NaPB, and aliquots of 500 cells were spread onto YNB agar plates and incubated for 48 h at room temperature. Assays were performed in triplicate for each strain. Cell death was calculated as [1 − (number of colonies recovered from Hst 5-treated cells/number of colonies from control cells)] × 100. For radial diffusion assays, C. albicans cells were cultured in YNB medium as described above, resuspended to 1 × 106/ml, and then trapped in thin underlay gels, which contained 9 mM sodium phosphate, 1 mM sodium citrate buffer, 1% (wt/vol) agarose, and 0.3 mg of Sabouraud dextrose broth (SDB; Difco)/ml. In some experiments, the underlay agars were supplemented with sorbitol (1 M, 0.5 M, or 0.1 M) or H2O2 (2 mM, 1 mM, or 0.5 mM). Stock Hst 5 solutions and serial twofold dilutions (7.8 to 250 μM) were prepared in distilled water. Peptide samples (5 μl) were loaded into 3-mm-diameter wells that had been punched into underlay gels. After incubation at 37°C for 3 h, a 10-ml overlay gel of 1% agarose and 6% SDB was poured onto the underlay gel. After the plates were incubated overnight at 37°C, the clear-zone diameters were measured to the nearest 0.1 mm and graphed against the peptide concentrations. Zone diameters were expressed in units (0.1 mm = 1 U).
Microarray data accession number.
The MIAME-compatible microarray data sets were submitted to Gene Expression Omnibus (GEO) under accession number GSE8473 (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE8473).
RESULTS
Genome-wide gene expression profile of Hst 5-treated cells.
To examine the global gene expression response of C. albicans cells to Hst 5, we selected a peptide concentration and treatment conditions that would provide substantial cell stress immediately following Hst 5 treatment. Hst 5 was added to 3 × 108 cells (1 ml) at a dosage (500 μM) that results in the death of approximately 30 to 40% of cells at this cell density. We anticipated that some early cell death responses would be detected by transcriptional analyses following this dose of Hst 5, but the majority of C. albicans cells survive for 1 h (20), so that adaptive responses could be assessed under high Hst 5 stress conditions. Since Hst 5 activity requires low extracellular osmolarity (10, 40), both control (sham-treated) and experimental cells were maintained in 10 mM sodium phosphate buffer throughout the assay.
Genome-wide expression profiles of C. albicans cells following 60 min of incubation with Hst 5 were examined using whole-genome microarrays and compared with transcripts obtained from cells that were sham treated (treated with buffer only) for 60 min. The whole-genome profile was analyzed (as described in Materials and Methods) so that genes with significant differences (P < 0.05) in transcriptional levels between groups were identified and grouped according to functional categories (Table 3). We found a total of 40 genes whose expression was significantly altered in a reproducible manner among experiments in response to Hst 5 treatment. Gene expression levels determined from microarray experiments were confirmed by quantitative RT-PCR analysis (data not shown). Twenty-four genes were up-regulated in response to Hst 5, including 10 genes involved in cellular metabolism (mainly the glycolytic pathway), 4 genes involved in transcription and signaling, and 1 gene encoding a glucan-linked cell wall protein. Three mitochondrial gene transcripts were significantly up-regulated. Another set of genes whose transcript levels were significantly increased was the cell stress group; including two heat-shock proteins (Hsp90 and Hsp70 [KAR2]) and three genes involved in the oxidative-stress response (TRX1, AHP1, and YHB1).
According to our transcriptional profiling data, 16 genes were significantly down-regulated following Hst 5 treatment. The largest group of down-regulated genes comprised plasma membrane transporters (Table 3), including several amino acid transporters (GAP2, AVT5, HIP1, and NAG4) and a member of the ABC drug transporter family (CDR11). In addition, we observed significant reductions in the transcript levels of several genes involved in fatty acid biosynthesis and other cellular metabolic processes, as well as reductions in the transcript levels of three transcription/signaling genes. Taken together, transcript profiling of cells surviving Hst 5 treatment showed marked induction of expression of numerous cell stress genes, as well as of genes encoding cellular metabolism, mitochondrial, and cell wall proteins. We choose to focus on cell stress genes, because we hypothesized that induction of these genes contributes to recovery from Hst 5 toxic effects for surviving cells.
Hog1 MAPK is involved in the C. albicans response to Hst 5.
C. albicans responds to general environmental stress conditions by activation of multiple MAPK pathways. When cells are treated with Hst 5, osmotic stress may be the predominate effect, as a result of the cell volume decrease initiated by the efflux of ATP and potassium and magnesium ions from the cell. Thus, adaptive responses to Hst 5 by C. albicans cells will likely involve the activation of one or more MAPK pathways. In order to determine which MAPK pathways are involved in the response of Candida to Hst 5, we screened three mutant strains, each with deletions of one of the three major C. albicans MAPK genes. We expected that loss of a MAPK responsible for the protection of Candida against Hst 5 stresses would result in cells with hypersensitivity to Hst 5. C. albicans null mutants for the Cek1 (extracellular signal-regulated kinase-like MAPK), Hog1 (high-osmolarity glycerol response), or Mkc1 (cell wall integrity pathway) (Table 1) MAPK were tested for Hst 5 toxicity under our standard conditions (using 5 × 104 cells) and compared with their respective parental and gene restoration/single-allele strains.
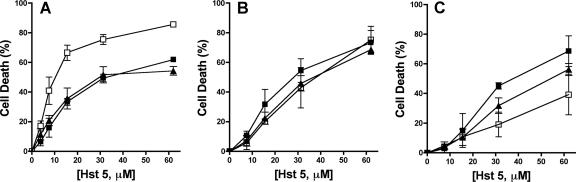
A statistically significant increase in the susceptibility of C. albicans hog1/hog1 deletion mutants over that of wild-type cells was observed at all Hst 5 dosages tested (Fig. 1A). Maximal killing by Hst 5 of wild-type cells (at 31 μM and above) was 49.5% ± 4.3%, while for hog1/hog1 cells, killing reached 75.5% ± 5.9% (Fig. 1A). Furthermore, the restoration strain (HOG1/hog1) possessed Hst 5 susceptibility equivalent to that of wild-type cells (Fig. 1A), suggesting that Hog1 and/or downstream genes activated by Hog1 reduce Hst 5 lethality. In contrast, no difference in sensitivity to Hst 5 was observed for the C. albicans Mkc1 knockout strain. Treatment of the mkc1/mkc1 strain with Hst 5 showed that these cells retained sensitivity equivalent to that of parental CAI-4 cells and the single-allele strain MKC1/mkc1 (Fig. 1B). Next, we tested the role of the Cek1 MAPK in Hst 5 killing of C. albicans. While the wild-type strain CAI-4 was highly sensitive to Hst 5, reaching 69% ± 14.6% killing, the cek1/cek1 deletion mutant cells showed reduced susceptibility to Hst 5, reaching only 39.3% ± 23.7% killing at this dosage of Hst 5 (Fig. 1C). Strain CK43A, carrying one copy of the CEK1 gene, partially regained wild-type susceptibility to Hst 5, with 56.3% ± 8% killing at 62 μM Hst 5 (Fig. 1C). Thus, CEK1 gene deletion resulted in cells with reduced sensitivity to Hst 5, in contrast to the hypersensitivity of hog1/hog1 cells. Taken together, these results suggest that Hog1 MAPK is involved in the adaptation and protection of C. albicans cells against Hst 5 toxicity, since strains lacking an intact HOG pathway exhibit increased susceptibility to Hst 5. In contrast, the Mck1 pathway appears to have no role in the protection of Candida from Hst 5. Although the basis for the reduced susceptibility of Cek1 MAPK to Hst 5 is not clear at this point, it may be a result of cross talk between the Hog1 MAPK pathway and the Cek1 MAPK pathway, since Hog1 is a negative regulator of Cek1 MAPK (12).
FIG. 1.
The HOG MAPK pathway is involved in the protection of C. albicans from Hst 5 toxicity. C. albicans stress response pathways were tested for their abilities to protect cells from Hst 5 killing. (A) The HOG pathway was tested using the wild-type strain RM1000 (filled squares), a hog1/hog1 deletion mutant (open squares), and a HOG1/hog1 restoration strain (filled triangles). (B) The Mkc1 kinase pathway was examined using the wild-type strain CAI-4 (filled squares), an mkc1/mkc1 mutant (open squares), and a single-allele MKC1/mkc1 strain (filled triangles). (C) The Cek1 MAPK pathway was tested using wild type CAI-4 (filled squares), a cek1/cek1 deletion mutant (open squares), and a CEK1/cek1 strain (filled triangles). Cells were treated with 3.8 to 62 μM Hst 5 for 1 h at 30°C, and loss of cell viability was calculated as [1 − (colonies after Hst 5 treatment/colonies after incubation with buffer only)] × 100. Each data point represents the mean ± standard deviation for at least three independent experiments. Only the C. albicans hog1/hog1 strain showed hypersensitivity to Hst 5, showing that Hog1 and/or upstream genes are involved in the reduction of Hst 5 toxicity.
Hst 5 induces transcription of Hog1-regulated osmotic-stress genes.
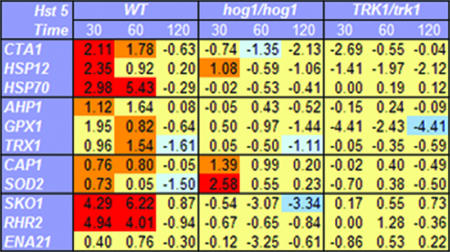
The increased susceptibility of Hog1 mutant cells to Hst 5, as well as the preponderance of cell stress genes induced upon exposure to Hst 5 as detected from microarray analyses, suggested that a major effect of this peptide is induction of osmotic and/or oxidative stress in C. albicans. Therefore, we examined transcriptional profiles by quantitative RT-PCR for selected groups of cell stress genes at earlier and later times following the exposure of cells to Hst 5 (Fig. 2). Since the time point selected for microarray analyses (60 min) may include dying cells as well as adapting cells, we examined cells at 30 min posttreatment, a time point at which Hst 5 has just accumulated in the cytosol of cells and cell death is minimal (20). We also examined late posttreatment times (120 min) at which more cells are experiencing irreversible toxic effects of Hst 5. Wild-type cells were compared with C. albicans hog1/hog1 mutant cells following Hst 5 treatment in order to identify genes specifically induced through the Hog1-dependent MAPK pathway. Transcriptional profiles of Hst 5-treated C. albicans TK1 mutants (TRK1/trk1), which have nearly complete resistance to Hst 5 (5), were used as negative controls for transcription of genes unrelated to Hst 5 response or to the Hog1-dependent MAPK pathway. Expression profiles were compared with those of cells manipulated in buffer only.
FIG. 2.
Hst 5 treatment of C. albicans causes induction of osmotic-stress genes and general stress genes. C. albicans strain CAI-4 (wild type [WT]), strain JC50 (hog1/hog1), and the Hst 5-insensitive strain TK1 (TRK1/trk1) were incubated with 500 μM Hst 5 (30% lethal dose) for 30 min, 60 min, or 120 min, followed by total-RNA isolation and cDNA synthesis (see Materials and Methods). Quantitative RT-PCR analysis was performed for each gene, and the results are shown as n-fold changes in the gene expression levels of Hst 5-treated cells compared to those for control cells treated with buffer only. Values are means from three independent experiments (standard errors are not shown). Genes whose expression was increased more than twofold upon Hst 5 treatment and was significantly different (P < 0.05) from that for untreated cells are shown in red; those whose expression was increased less than twofold but still significantly different (P < 0.05) from that for untreated cells are shown in orange. Genes whose expression was decreased more than twofold upon Hst 5 treatment and was significantly different (P < 0.05) from that for untreated cells are shown in blue; those whose expression was decreased less than twofold but still significantly are shown in light blue. Hst 5 treatment induced significant expression of stress genes (CTA1, HSP70, and HSP12) as well as of genes involved in glycerol accumulation (SKO1 and RHR2) within 30 to 60 min in wild-type C. albicans cells in a Hog1-dependent manner. Oxidative-stress genes (CAP1 and SOD1) were induced by Hst 5 in both wild-type and hog1/hog1 cells.
We first examined three core stress genes (the CTA1 gene, encoding catalase in Candida [orf19.6229]; HSP12, encoding a small heat shock protein [orf19.3160]; and the HSP70 gene, encoding heat shock protein Hsp70 [orf19.4980; SSA1]) that are induced in Candida by both oxidative and osmotic stresses through the HOG pathway (13, 37). Hst 5 significantly induced all three genes at 30 min, and both HSP70 and CTA1 levels remained elevated at 60 min post-Hst 5 treatment. Only HSP12 was mildly induced in hog1/hog1 cells at 30 min, perhaps reflecting the role of Hog1 repression of HSP12 under nonstress conditions (37). No other stress genes were induced in hog1/hog1 cells, and expression levels were unchanged in the Hst 5-resistant C. albicans strain TK1, demonstrating that this effect was related to Hst 5 toxicity. Furthermore, no genes showed up-regulated expression following 120 min of Hst 5 treatment in this set or other sets of genes tested, supporting the supposition that cell arrest or death is the predominate response by 120 min following exposure to Hst 5.
Oxidative stress.
Genes important for reductive detoxification of H2O2 and free radicals produced in oxidative metabolism are expressed under oxidative-stress conditions or, secondarily, in conjunction with cellular apoptosis or cell death. Therefore, we examined the transcriptional levels of a selected group of oxidative-stress genes following Hst 5 treatment to determine whether this peptide induces an oxidative-stress response (Fig. 2). Both TRX1, encoding thioredoxin (orf19.7611), and GPX1, encoding glutaredoxin (orf19.87), were mildly induced after 60 min of Hst 5 treatment, while AHP1, encoding alkyl hydroperoxide reductase (orf19.2762), was up-regulated slightly after 30 min (Fig. 2). None of these genes was induced by Hst 5 in hog1/hog1 cells, suggesting that expression is Hog1 dependent, nor was expression detected in Hst 5-resistant TRK1/trk1 cells. Likewise, the Hog1-independent gene CAP1 (orf19.1623), encoding a transcription factor induced in response to oxidative stress in C. albicans (13, 43), and the SOD2 gene (orf19.3340), encoding mitochondrial superoxide dismutase, were both induced by Hst 5 following 30 min of treatment. CAP1 levels remained elevated following 60 min of Hst 5 treatment, while SOD2 levels returned to those of sham-treated cells. However, hog1/hog1cells had nearly double the expression levels of SOD2 and CAP1 genes as wild-type cells in response to a 30-min Hst 5 treatment, showing that expression of both CAP1 and SOD2 was induced by Hst 5 independently of the HOG pathway.
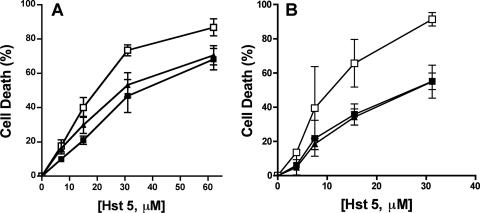
To further explore the role of the oxidative-stress response to Hst 5, we selected two C. albicans strains to examine for Hst 5 sensitivity: one strain (cap1/cap1) with inactivation of the transcriptional regulatory protein required for oxidative-stress tolerance (43) independent of Hog1 and one strain (ssk1/ssk1) with a deletion in one of the upstream phosphorelay proteins in the Sln1 branch of the HOG pathway that is important for the Hog1-mediated oxidative-stress response (6). We had previously found no difference in Hst 5 sensitivity for C. albicans sod1/sod1 cells (41), so we did not test the SOD group further. C. albicans cap1/cap1 cells had no significant difference in sensitivity to Hst 5 at lower doses (<15.7 μM), while at higher doses (>32 μM), cells were about 20% more susceptible to Hst 5 (Fig. 3A). As with the hog1/hog1 strain, deletion of SSK1 significantly increased susceptibility to Hst 5 over that of wild-type cells (Fig. 3B). However, unlike that for hog1/hog1 cells, the increase in sensitivity to Hst 5 was significant only at higher Hst 5 doses (at or above 15 μM). This biphasic response to Hst 5 exhibited by cap1/cap1 and ssk1/ssk1 cells suggests that Hst 5 elicits oxidative stresses primarily at higher doses at which cell death is the predominant phenotype. However, it appears that neither Hog1-dependent nor Hog1-independent oxidative-stress responses are involved in the response to lower doses of Hst 5.
FIG. 3.
C. albicans cells with an impaired oxidative-stress response are more sensitive to high doses of Hst 5. C. albicans oxidative-stress response pathways were tested for their sensitivities to Hst 5 killing. (A) Cells with inactivation of the transcriptional regulatory protein required for oxidative-stress tolerance independent of Hog1 were tested using a wild-type strain (filled squares), a cap1/cap1 deletion mutant (open squares), and a single-allele CAP1/cap1 strain (filled triangles). (B) The Sln1 branch of the HOG pathway was tested by comparing ssk1/ssk1 cells (open squares) with wild-type CAF2-1 (filled squares) and SSK1/ssk1 (filled triangles) cells. Cells were treated with 3.8 to 62 μM Hst 5 for 1 h at 30°C, and loss of cell viability is expressed as [1 − (colonies after Hst 5 treatment/colonies after incubation with buffer only)] × 100. Both C. albicans cap1/cap1 and ssk1/ssk1 strains showed significantly (P < 0.05) more sensitivity to Hst 5 only at higher dosages (31 μM and 62 μM).
Osmotic stress.
In order to evaluate the osmotic-stress components involved in the cellular response to Hst 5, three genes were selected based on their involvement in the Hog1-mediated osmotic-stress response in Candida. Since hyperosmotic stress demands metabolic adjustment for production of glycerol as an intracellular osmolyte, we selected the RHR2 gene (orf19.5437), which is involved in glycerol accumulation as a classical response to osmotic stress (13, 17, 34). In addition, we monitored the expression profiles of the ENA21 gene (orf19.5170), encoding a cation transporter up-regulated upon osmotic stress, and the SKO1 gene (orf19.1032), encoding a transcriptional factor that binds promoters of several genes directly involved in defense functions that relieve osmotic stress through the HOG pathway. Substantial increases in the expression levels (four- to sixfold) of both the SKO1 and RHR2 genes in Hst 5-treated cells over those for untreated cells were found at both 30 and 60 min, and expression returned to basal levels following 120 min of Hst 5 treatment, which did not occur in Hst 5-resistant TRK1/trk1 cells (Fig. 2). As expected, hog1/hog1 cells showed no significant change in the expression level of either gene following Hst 5 treatment, showing that these gene responses are Hog1 mediated. Hst 5 did not alter the expression levels of ENA21. Thus, genes encoding proteins involved in glycerol biosynthesis are significantly up-regulated by Hst 5 treatment in a Hog1-dependent manner, suggesting that production of glycerol in reaction to osmotic stress is a component of the cellular response to Hst 5. Therefore, we next examined glycerol production in Hst 5-treated cells.
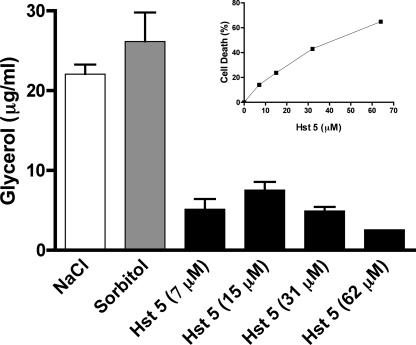
Preliminary experiments showed that cells required at least 30 min following stimulation with 0.5 M NaCl or 1 M sorbitol for glycerol production to be detected; therefore, a 60-min treatment time with Hst 5 was chosen in order to allow both Hst 5 uptake and intracellular glycerol production. Under these conditions, maximal glycerol production following 60 min of stress with 0.5 M NaCl or 1 M sorbitol was 22 to 26 μg/ml, while Hst 5 stress resulted in glycerol production of 5 to 8 μg/ml (Fig. 4). Similar levels of glycerol production (7 to 15 μg/ml) were found for cells subjected to 0.25 M NaCl or 0.5 M sorbitol (data not shown), indicating that Hst 5 induces a level of glycerol production at 60 min equivalent to one-half to one-third of that stimulated by high osmotic stress. Glycerol production was increased with increasing doses of Hst 5 (7 μM and 15 μM); however, it was diminished at higher doses (31 μM and 62 μM) as cell killing increased above 30 to 40% (Fig. 4 inset). This bell curve effect suggests that protective cell responses involving glycerol production are effective mainly at lower doses of Hst 5 and that higher Hst 5 doses inhibit cell metabolism and render the cells incapable of further glycerol production. Additionally, these results point to osmotic stress with accompanying glycerol production as an initial effect of Hst 5.
FIG. 4.
Hst 5 induces dose-dependent intracellular glycerol production in C. albicans. C. albicans CAI-4 (solid bars) cells were incubated with 7.5 to 62 μM Hst 5 for 1 h in diluted dSD. Cells were treated with buffer alone to determine baseline glycerol production as a result of cell manipulation, while cells were treated with 0.5 M NaCl (open bars) or 1 M sorbitol (shaded bars) as positive controls for glycerol production. Glycerol values were normalized to C. albicans cell numbers, and glycerol levels produced in sham-treated cells were used as basal values. Cells were divided for glycerol determination and a conventional candidacidal assay. Hst 5 treatment caused a dose-dependent increase in intracellular glycerol production up to 15 μM; then, at higher Hst 5 doses, glycerol production was reduced in proportion to cell toxicity (inset).
Hst 5 treatment induces Hog1 phosphorylation.
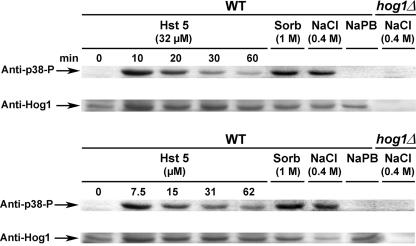
In order to assess the activation of the HOG pathway in response to Hst 5 treatment in C. albicans, phosphorylation levels of Hog1 following Hst 5 exposure were measured. An antibody that recognizes specifically the dually phosphorylated form of Hog1 was used to provide an indication of HOG pathway activation. Since we observed maximal glycerol production at lower doses of Hst 5, we examined C. albicans cells treated with Hst 5 over a range of dosages to determine whether Hog1 phosphorylation was also dose and time dependent (Fig. 5). Control cells (both the wild type and the hog1/hog1 mutant) were either treated with 0.4 M NaCl or 1 M sorbitol or manipulated in medium alone to compare positive and negative phosphorylation levels. Hog1 phosphorylation was not detected in wild-type cells manipulated in medium alone. Hog1 phosphorylation was strongly activated following a 10-min incubation with 32 μM Hst 5 (Fig. 5, upper panel). Longer (20-min) incubation with Hst 5 also resulted in phosphorylation of Hog1 (although at lower levels than those observed at 10 min), which then diminished after 30 and 60 min of incubation. Cells treated with 0.4 M NaCl or 1 M sorbitol exhibited strong activation of Hog1 phosphorylation, while no phosphorylation of Hog1 was detected in strain JC50 (hog1/hog1) following a 20-min incubation with 0.4 M NaCl (Fig. 5). Since 32 μM doses of Hst 5 cause 50% cell death after 30 min under these conditions, we questioned whether the reduction in Hog1 phosphorylation observed might be a result of the presence of dying cells unable to mount a response. Therefore, we examined cell responses to a range of Hst 5 dosages after 30 min. In agreement with our results for glycerol production, maximal levels of Hog1 phosphorylation were induced by Hst 5 at lower dosages (7 μM and 15 μM), while increasing the Hst 5 dose (31 μM and 62 μM) reduced Hog1 phosphorylation proportionally to the dosage (Fig. 5, lower panel). Thus, these data show that Hog1 phosphorylation occurs rapidly in response to treatment of cells with Hst 5 and that maximal phosphorylation is found with lower doses of Hst 5, both of which are proportional to the number of surviving cells.
FIG. 5.
Hst 5 activates the HOG pathway via Hog1 phosphorylation. C. albicans cells were collected as for glycerol assays in diluted dSD. CAI-4 cell suspensions (2 × 107 cells) were either incubated with Hst 5 (7.5, 15, 31, or 62 μM) for 30 min (lower panel) or with 32 μM Hst 5 for 0, 10, 20, 30, or 60 min (upper panel). Control cells were manipulated in dSD only; cells were treated with 400 mM NaCl or 1 M sorbitol for positive controls. Cells in which Hog1 had been deleted (hog1/hog1) were stimulated with 400 mM NaCl. Following treatment, cells were harvested and directly processed for extraction of total-cell lysates using glass bead disruption. Equal amounts of total-cell lysates (120 μg) were immunoblotted using antibodies to phospho-p38-Hog1p and Hog1p. Hst 5 (32 μM) treatment resulted in rapid phosphorylation of Hog1p after 10 min, which gradually diminished over 60 min (upper panel). Cells treated with buffered medium only (NaPB) showed no Hog1 phosphorylation at any time point. Hog1p phosphorylation was not detected in Hog1 mutants (hog1/hog1). Phosphorylation of Hog1 was strongest when cells were treated with 7.5 or 15 μM Hst 5 (intensity was equivalent in some immunoblots), while cells treated with higher doses of Hst 5 (31 or 62 μM) had reduced levels of Hog1 phosphorylation. WT, wild type.
Osmotic stress modulates Hst 5 toxicity.
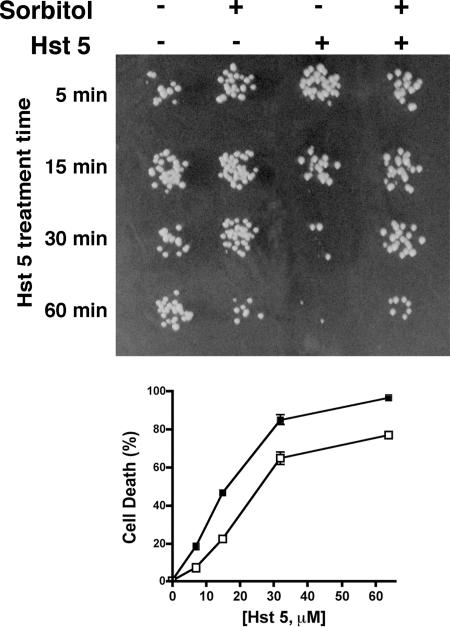
Since glycerol production is strongly stimulated by sorbitol, we questioned whether cells pretreated with sorbitol before exposure to Hst 5 would be protected from killing. We predicted that sorbitol would precondition cells for adaptation to further osmotic stress from Hst 5. Cells were incubated with 1 M sorbitol for 60 min to optimize glycerol production before addition of Hst 5; then cells were exposed to a low dose (7 μM) of Hst 5 for 5 min, 15 min, 30 min, and 60 min and were assessed for viability (Fig. 6). Pretreatment of cells with 1 M sorbitol alone for 60 min resulted in a slight loss of cell viability compared with that of cells in buffer; however, sorbitol-pretreated cells were substantially protected from Hst 5 killing following 30 min and 60 min of peptide incubation (Fig. 6, upper panel). We next examined sorbitol protection from Hst 5 killing over a range of concentrations (Fig. 6, lower panel). Cells pretreated with 1 M sorbitol were significantly (P < 0.01) protected, by 50%, from Hst 5 killing at lower doses (7 μM to 15 μM), while killing was reduced by about 25% at higher Hst 5 doses (31 μM to 62 μM). Cells pretreated with 0.5 M sorbitol were protected by 25% from Hst 5 toxicity (data not shown). In contrast, cells pretreated with 1 to 2 mM H2O2 had no change in sensitivity to Hst 5 (data not shown). Thus, cells preadapted to osmotic stress become resistant to Hst 5 toxicity.
FIG. 6.
Sorbitol-stressed cells are resistant to subsequent treatment with Hst 5. CAF4-2 cells were suspended in diluted dSD (106 cells in 500 μl), exposed to osmotic stress by addition of sorbitol (final concentration, 1 M), and incubated at 30°C for 1 h. Control cells were manipulated in dSD medium alone or with 1 M sorbitol. (Upper panel) To analyze the effects of Hst 5 at various time points, samples were withdrawn after 5, 15, 30, and 60 min of incubation with Hst 5 (7 μM), immediately diluted with buffer, and spotted onto YPD agar plates. Sorbitol protection from Hst 5 cell toxicity was evident following 30 min of incubation with 7 μM peptide (far right), while control cells treated with Hst 5 had typical loss of viability. (Lower panel) To assess concentration effects, cell aliquots were removed, Hst 5 (7 μM to 62 μM) was added, and cells were incubated for 1 h before being plated. Cell death was calculated as described in the legend to Fig. 1. Cells stressed with 1 M sorbitol before Hst 5 addition (open squares) were protected from Hst 5 compared with cells not pretreated with sorbitol (solid squares) at all Hst 5 doses, although the reduction in toxicity was more marked at lower doses.
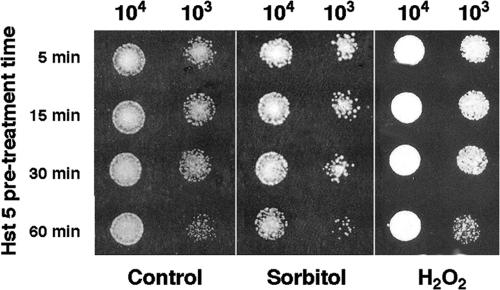
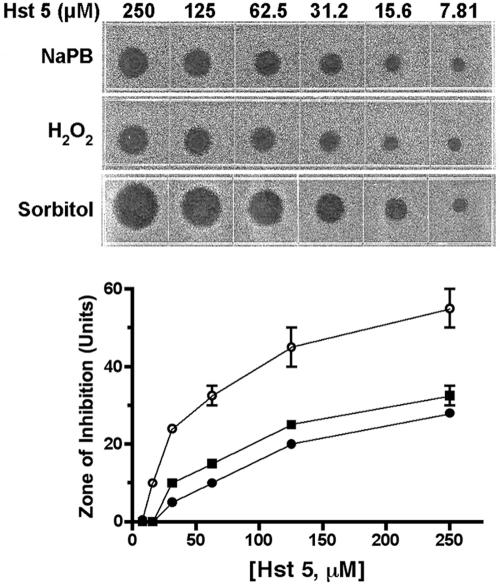
Since Hst 5 induces the production of glycerol and perhaps other stress-protective molecules, it is possible that cells treated with a low dose of Hst 5 would be protected from subsequent exposure to either osmotic or oxidative stresses. Cells were first exposed to a low dose (7 μM) of Hst 5 for 5 min, 15 min, 30 min, or 60 min and then spotted onto YPD medium containing 1 M sorbitol or 2 mM H2O2. Cells pretreated with Hst 5 and then grown under conditions of oxidative stress showed no difference in the toxicity of Hst 5 at any pretreatment time (Fig. 7, right) compared with cells grown under standard conditions (Fig. 7, left). Contrary to our prediction, cells pretreated with Hst 5 and then grown under osmotic-stress conditions (1 M sorbitol) had increased sensitivity to Hst 5 (Fig. 7, center) rather than being protected from osmotic stress. Therefore, we further examined dose-dependent Hst 5 killing under simultaneous conditions of oxidative or osmotic stress. Cells were incubated concurrently in Hst 5 with osmotic (plates containing 1 M sorbitol) or oxidative (plates containing 0.5 mM H2O2) stress (Fig. 8). No difference in sensitivity to Hst 5 at any dose was observed in C. albicans cells under concurrent oxidative-stress conditions compared with cells grown on YPD medium alone (Fig. 8). Supplementation of plates with higher concentrations of H2O2 (1 mM and 2 mM) did not alter cell sensitivity to Hst 5 (data not shown). However, cells treated concomitantly with Hst 5 and grown in plates containing 1 M sorbitol had substantially increased (at least twofold) sensitivity to Hst 5 at all peptide concentrations examined (Fig. 8). Addition of 0.5 M sorbitol slightly reduced C. albicans Hst 5 hypersensitivity compared with 1 M sorbitol, while incubation with 0.1 M sorbitol resulted in the return of typical Hst 5 sensitivity (data not shown). Thus, the level of Hst 5 killing is elevated by the presence of a concurrent osmotic stress but is unaffected by oxidative stress. These results suggest that osmotic stress facilitates Hst 5 activity, perhaps due to common interactions and/or overlapping mechanisms of action, while oxidative stress appears to play little role in Hst 5 toxicity. Taken together, our data point to osmotic stress as early and initial effects of Hst 5, while oxidative stresses are generated secondarily to cell death.
FIG. 7.
Hst 5 pretreatment increases cell sensitivity to subsequent osmotic, but not oxidative, stress. C. albicans (CAF4-2) cells (106/ml) were manipulated as for candidacidal assays. For Hst 5 stress conditioning, Hst 5 (7 μM) was added to cells at 37°C, and samples were withdrawn after 5, 15, 30, or 60 min. Hst 5-treated cells were diluted with NaPB to stop further Hst 5 uptake and then spotted either onto YPD agar plates (control) or onto YPD agar supplemented with 2 mM H2O2 or 1 M sorbitol. The growth of cells under each condition was examined following 24 h of incubation at 37°C. Cells pretreated with Hst 5 and then grown under oxidative-stress conditions (H2O2) (right) had growth profiles identical to those of cells grown in YPD alone (control) (left). Cells pretreated with Hst 5 and then grown under osmotic stress in 1 M sorbitol (center) exhibited increased sensitivity to Hst 5.
FIG. 8.
Osmotic stress, but not oxidative stress, increases cell susceptibility to Hst 5. For radial diffusion assays, C. albicans cells (106/ml) were trapped in thin underlay gels, some of which were supplemented with sorbitol (1 M) or H2O2 (1 mM). Twofold dilutions of Hst 5 (7.8 to 250 μM) were loaded into 3-mm-diameter wells that had been punched into underlay gels. After overnight incubation at 37°C, the clear-zone diameters indicating antifungal activity (upper panel) were measured to the nearest 0.1 mm and graphed against peptide concentrations (lower panel). Zone diameters are expressed in units (0.1 mm = 1 U). Cells grown under concurrent osmotic stress (1 M sorbitol) (open circles) were significantly more susceptible to Hst 5 at all concentrations tested than cells grown in medium alone (solid squares), while cells grown under concurrent oxidative stress (1 mM H2O2) (solid circles) showed no difference in sensitivity to Hst 5 (lower panel).
DISCUSSION
Evidence for crucial components of the toxicity of Hst 5 toward C. albicans has resulted in the development of two mechanistic models that previously appeared to be mutually exclusive. One model is based on evidence that Hst 5 induces rapid release of intracellular ATP and K+ in a Trk1p-dependent manner so that ion imbalance and irreversible cell volume loss occur in a mode resembling hyperosmotic shock. A second model is based on observations that Hst 5 associates with mitochondria, inhibits respiration, and induces the formation of ROS to produce oxidative stress (15, 16). In order to differentiate between these two models, we used genomic expression profiling of C. albicans treated with Hst 5 to examine the global responses of cells to this toxin within 1 h of treatment. Here, for the first time, we provide evidence that the transcriptional response of C. albicans to Hst 5 is that of cells undergoing adaptation to ionic or osmotic changes mediated by HOG pathways. However, osmotic stress also induces cellular changes in redox metabolism that overlap with oxidative-stress responses—both having in common the involvement of Hog1. Thus, the Hog1 MAPK pathway for stress response to Hst 5 suggests a common mechanism for unification of these two models.
The marked increase in the transcription of C. albicans cell stress genes such as YHB1, AHP1, KAR2, IFU5, TRX1, and HSP90 (Table 3) following Hst 5 treatment that we observed initially suggested that cell response occurred via activation of one or more MAPK cascades. In order to delineate the role of each MAPK pathway in Hst 5-induced stress responses, we examined the susceptibilities of three C. albicans strains, each with a mutation in the MKC1, CEK1, or HOG1 gene. Only Hog1-deleted cells exhibited hypersensitivity to Hst 5 peptide, suggesting that Hog1 is the predominant MAPK responsive to Hst 5-induced stresses. Since Hog1 MAPK is induced by both osmotic and oxidative (as well as heavy-metal) stresses in Candida (13), we selected representative groups of osmotic- and oxidative-stress response genes for further quantitative analyses following 30 min, 60 min, and 120 min of Hst 5 exposure. Hst 5 induced significant increases in the expression of two groups of genes in a Hog1-dependent manner following 30 min and 60 min of treatment: general stress genes (HSP70, HSP12, and CTA1) and genes involved in glycerol synthesis (RHR2 and SKO1).
Osmotic stress induces profound adjustments of cellular metabolism to cope with shifts in cell turgor and ion balance, including production of glycerol, which serves as a cellular osmolyte. The behavior of cells following Hst 5 treatment is very similar to this classical biochemical response to hyperosmotic stress. We observed strong induction of gene transcripts (RHR2, GND1, and GPM1) encoding enzymes of the glycolytic pathway as well as Hog 1 phosphorylation following Hst 5 treatment. Also, intracellular glycerol accumulated in a dose-dependent manner following treatment with Hst 5. At lower Hst 5 doses, cells produced larger amounts of glycerol in proportion to the Hst 5 concentration; however, at higher Hst 5 doses, glycerol accumulation was found to be inversely proportional to the Hst 5 concentration. This bell curve response may be a result of increased cellular permeability accompanying increased Hst 5 doses. Hst 5 toxicity is characterized by rapid efflux of ions and ATP from cells, which may deplete cellular energy reserves and shut down metabolic pathways needed for new glycerol synthesis. Another possibility is that Hst 5 causes unregulated ion channels through which outflow of ions and ATP occurs, and these channels may also allow the passage of glycerol, resulting in a net loss of intracellular glycerol.
Interestingly, this classical osmotic-stress response may also explain why cells grown under anaerobic conditions are more resistant to Hst 5, since glycerol accumulation is accelerated under anaerobic conditions and results in cells capable of more-rapid osmotic adaptation (21). Thus, anaerobic growth conditions or selective pressures that force cells into anaerobic growth (such as mitochondrial mutants) are preconditioned for rapid glycerol production and adaptation to the osmotic stresses produced by Hst 5. In this regard, we found that prestressing cells with sorbitol before Hst 5 treatment reduced peptide toxicity.
Osmotic stress also strongly diminishes the uptake of many amino acids, an effect resembling an amino acid starvation response. Indeed, our microarray experiments showed that a preponderance of genes down-regulated upon Hst 5 treatment were plasma membrane transporters, including GAP2 and HIP1 (general amino acid permeases), CDR11 (an ABC transporter), and NAG4 (encoding a monosaccharide transporter), suggesting energy preservation-like processes in cells treated with Hst 5. Since the cell wall plays a crucial role in maintaining cell turgor pressure, hyperosmotic stress induces cell wall remodeling and biogenesis through the HOG pathway in order to regulate cell osmolarity. In agreement with this model, we found that PIR1, encoding a Hog1-induced cell wall glycoprotein, was highly up-regulated by Hst 5 treatment.
Oxidative stresses may be generated as secondary effects of osmotic stress—for example, osmotic stress may interfere with the electron transport chain and enhance the production of ROS—or directly, due to oxidative damage and/or stimulation of redox metabolism. Hst 5 treatment caused increases in transcript levels of both Hog1-mediated (AHP1, GPX1, and TRX1) and Hog1-independent (CAP1 and SOD2) oxidative-stress genes, suggesting that both components could be involved. However, both cap1/cap1 cells and ssk1/ssk1 cells exhibited increased sensitivity to Hst 5, but only at higher doses at which osmotic-stress responses such as glycerol production are diminished. Thus, these results suggested that oxidative stress is generated secondarily to Hst 5 cell toxicity in both HOG-dependent and -independent pathways. In addition to genes encoding oxidoreductase enzymes involved in the metabolism of oxidized proteins (TRX1 and nitric oxide dioxygenase [YHB1]), increased transcript levels for MET1, involved in the biosynthesis of the oxidation-sensitive amino acid methionine, were observed, suggesting changes in cellular antioxidant capacity upon Hst 5 exposure. Interestingly, MET1 gene expression is induced in response to Hog1 activation (13), since Hog1 regulates the function of genes involved in sulfur, methionine, alcohol, and lipid metabolism, as well as glycolysis (13).
To further examine the role of oxidative and osmotic stresses in Hst 5 cytotoxicity, wild-type cells were treated with Hst 5 and grown under osmotic (sorbitol)- or oxidative (H2O2)-stress conditions. Osmotic stress substantially increased cell sensitivity to Hst 5, while oxidative stress had no effect on Hst 5 toxicity. Such facilitating interactions between the effects of sorbitol and Hst 5 further suggest a common underlying cellular response to osmotic stress and to Hst 5 toxin. Furthermore, our finding that oxidative stress did not enhance Hst 5 killing supports the view that oxidative stress is generated secondarily and is not an early or primary effect of Hst 5.
Here we report for the first time that the C. albicans response to Hst 5 involves activation of the HOG pathway. Recently it was shown that treatment of Saccharomyces cerevisiae with Pichia membranifaciens killer toxin resulted in a coordinated transcriptional response, primarily through the HOG pathway, that was related to changes in ionic homeostasis (35). Similarly, exposure of S. cerevisiae cells to bacterial endotoxin induced phosphorylation of Hog1 (27). Thus, the HOG pathway may be of significance in its ability to respond to changes in cellular osmolarity induced by a variety of toxins (as well as environmental conditions) in unicellular eukaryotes. In C. albicans, the Hog1-mediated response to Hst 5 is both time and concentration dependent. HOG pathway responses are most protective against lower Hst 5 doses, since longer incubation times and higher Hst 5 doses result in the inability of cells to mount a full protective response. However, in the native environment of the oral cavity, where Hst 5 levels are often transient with respect to colonized Candida cells and Hst concentrations are below 30 μM, the Hog1 response is likely to be a significant and effective challenge to physiological levels of Hst 5. Furthermore, adaptive activation of this pathway by cells subjected to prolonged or cyclical Hst 5 stresses may be a mechanism for the development of resistance to toxic cationic peptides. Thus, consideration of means to inactivate the HOG pathway in conjunction with doses of therapeutic peptides would enhance the efficacy of killing by Hst 5 and related natural antifungal agents. Upstream effectors of the HOG pathway, such as the osmosensor Sho1 or Sln1, are appealing candidates for this approach.
Acknowledgments
This work was supported by U.S. PHS R01 grant DE010641 from the National Institute of Dental and Craniofacial Research.
Footnotes
Published ahead of print on 22 August 2007.
REFERENCES
- 1.Alonso-Monge, R., F. Navarro-García, G. Molero, R. Diez-Orejas, J. Pla, M. Sanchez, and C. Nombela. 1999. Role of the mitogen-activated protein kinase Hog1p in morphogenesis and virulence of Candida albicans. J. Bacteriol. 181:3058-3068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alonso-Monge, R., F. Navarro-García, E. Román, A. I. Negredo, B. Eisman, C. Nombela, and J. Pla. 2003. The Hog1 mitogen-activated protein kinase is essential in the oxidative stress response and chlamydospore formation in Candida albicans. Eukaryot. Cell 2:351-361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Arana, D. M., C. Nombela, R. Alonso-Monge, and J. Pla. 2005. The Pbs2 MAP kinase kinase is essential for the oxidative stress response in the fungal pathogen Candida albicans. Microbiology 151:1033-1049. [DOI] [PubMed] [Google Scholar]
- 4.Baev, D., X. S. Li, J. Dong, P. Keng, and M. Edgerton. 2002. Human salivary histatin 5 causes disordered volume regulation and cell cycle arrest in Candida albicans. Infect. Immun. 70:4777-4784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Baev, D., A. Rivetta, S. Vylkova, J. N. Sun, G. F. Zeng, C. L. Slayman, and M. Edgerton. 2004. The TRK1 potassium transporter is the critical effector for killing of Candida albicans by the cationic protein, histatin 5. J. Biol. Chem. 279:55060-55072. [DOI] [PubMed] [Google Scholar]
- 6.Calera, J. A., X. J. Zhao, and R. Calderone. 2000. Defective hyphal development and avirulence caused by a deletion of the SSK1 response regulator gene in Candida albicans. Infect. Immun. 68:518-525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen, J., J. Chen, S. Lane, and H. Liu. 2002. A conserved mitogen-activated protein kinase pathway is required for mating in Candida albicans. Mol. Microbiol. 46:1335-1344. [DOI] [PubMed] [Google Scholar]
- 8.Csank, C., K. Schroppel, E. Leberer, D. Harcus, O. Mohamed, S. Meloche, D. Y. Thomas, and M. Whiteway. 1998. Roles of the Candida albicans mitogen-activated protein kinase homolog, Cek1p, in hyphal development and systematic candidiasis. Infect. Immun. 66:2713-2721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Den Hertog, A. L., H. W. Wong Fong Sang, R. Kraayenhof, J. G. M. Bolscher, W. Van't Hof, E. S. I. Veerman, and A. V. N. Amerongen. 2004. Interactions of histatin 5 and histatin 5-derived peptides with liposome membranes: surface effects, translocation and permeabilization. Biochem. J. 379:665-672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dong, J., S. Vylkova, X. S. Li, and M. Edgerton. 2003. Calcium blocks fungicidal activity of human salivary histatin 5 through disruption of binding with Candida albicans. J. Dent. Res. 82:748-752. [DOI] [PubMed] [Google Scholar]
- 11.Edgerton, M., S. E. Koshlukova, T. E. Lo, B. G. Chrzan, R. M. Staubinger, and P. A. Raj. 1998. Candidacidal activity of salivary histatins. Identification of a histatin 5-binding protein on Candida albicans. J. Biol. Chem. 273:20438-20447. [DOI] [PubMed] [Google Scholar]
- 12.Eisman, B., R. Alonso-Monge, E. Román, D. Arana, C. Nombela, and J. Pla. 2006. The Cek1 and Hog1 mitogen-activated protein kinases play complementary roles in cell wall biogenesis and chlamydospore formation in the fungal pathogen Candida albicans. Eukaryot. Cell 5:347-358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Enjalbert, B., D. A. Smith, M. J. Cornell, I. Alam, S. Nickolls, A. J. P. Brown, and J. Quinn. 2006. Role of the Hog1 stress-activated protein kinase in the global transcriptional response to stress in the fungal pathogen Candida albicans. Mol. Biol. Cell 17:1018-1032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fonzi, W. A., and M. Y. Irwin. 1993. Isogenic strain construction and gene mapping in Candida albicans. Genetics 134:717-728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Helmerhorst, E. J., P. Breeuwer, W. Van't Hof, E. Walgreen-Weterings, A. V. Amerongen, and T. Abee. 1999. The cellular target of histatin 5 on Candida albicans is the energized mitochondrion. J. Biol. Chem. 274:7286-7291. [DOI] [PubMed] [Google Scholar]
- 16.Helmerhorst, E. J., R. F. Troxler, and F. G. Oppenheim. 2001. The human salivary peptide histatin 5 exerts its antifungal activity through the formation of reactive oxygen species. Proc. Natl. Acad. Sci. USA 98:14637-14642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hohmann, S. 2002. Osmotic stress signaling and osmoadaptation in yeasts. Microbiol. Mol. Biol. Rev. 66:300-372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hromatka, B. C., S. M. Noble, and A. D. Johnson. 2005. Transcriptional response of Candida albicans to nitric oxide and the role of the YHB1 gene in nitrosative stress and virulence. Mol. Biol. Cell 16:4814-4826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kayingo, G., and B. Wong. 2005. The MAP kinase Hog1p differentially regulates stress-induced production and accumulation of glycerol and d-arabitol in Candida albicans. Microbiology 151:2987-2999. [DOI] [PubMed] [Google Scholar]
- 20.Koshlukova, S. E., T. L. Lloyd, M. W. B. Araujo, and M. Edgerton. 1999. Salivary histatin 5 induced non-lytic release of ATP from Candida albicans leading to cell death. J. Biol. Chem. 274:18872-18879. [DOI] [PubMed] [Google Scholar]
- 21.Krantz, M., B. Norlander, H. Valadi, M. Johannson, L. Gustafsson, and S. Hohmann. 2004. Anaerobicity prepares Saccharomyces cerevisiae cells for faster adaptation to osmotic shock. Eukaryot. Cell 3:1381-1390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kruppa, M., and R. Calderone. 2006. Two-component signal transduction in human fungal pathogens. FEMS Yeast Res. 6:149-159. [DOI] [PubMed] [Google Scholar]
- 23.Kumamoto, C. A. 2005. A contact-activated kinase signals Candida albicans invasive growth and biofilm development. Proc. Natl. Acad. Sci. USA 102:5576-5581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li, X. S., J. N. Sun, K. Okamoto-Shibayama, and M. Edgerton. 2006. Candida albicans cell wall Ssa proteins bind and facilitate import of salivary histatin 5 required for toxicity. J. Biol. Chem. 281:22453-22463. [DOI] [PubMed] [Google Scholar]
- 25.Liu, T. T., R. E. Lee, K. S. Barker, R. E. Lee, L. Wei, R. Homayouni, and P. D. Rogers. 2005. Genome-wide expression profiling of the response to azole, polyene, echinocandin, and pyrimidine antifungal agents in Candida albicans. Antimicrob. Agents Chemother. 49:2226-2236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lorenz, M. C., and G. R. Fink. 2001. The glyoxylate cycle is required for fungal virulence. Nature 412:83-86. [DOI] [PubMed] [Google Scholar]
- 27.Marques, J. M., R. J. Rodrigues, A. C. de Magalhaes-Sant'ana, and T. Goncalves. 2006. Saccharomyces cerevisiae Hog1 protein phosphorylation upon exposure to bacterial endotoxin. J. Biol. Chem. 281:24687-24694. [DOI] [PubMed] [Google Scholar]
- 28.Monge, A. R., E. Román, C. Nombela, and J. Pla. 2006. The MAP kinase signal transduction network in Candida albicans. Microbiology 152:905-912. [DOI] [PubMed] [Google Scholar]
- 29.Navarro-García, F., B. Eisman, S. M. Fiuza, C. Nombela, and J. Pla. 2005. The MAP kinase Mkc1p is activated under different stress conditions in Candida albicans. Microbiology 151:2737-2749. [DOI] [PubMed] [Google Scholar]
- 30.Oppenheim, F. G. 1989. Human saliva: clinical chemistry and microbiology, vol. 1, p. 151-160. CRC Press, Inc., Boca Raton, FL. [Google Scholar]
- 31.Román, E., D. M. Arana, C. Nombela, R. Alonso-Monge, and J. Pla. 2007. MAP kinase pathways as regulators of fungal virulence. Trends Microbiol. 15:181-190. [DOI] [PubMed] [Google Scholar]
- 32.Román, E., C. Nombela, and J. Pla. 2005. The Sho1 adaptor protein links oxidative stress to morphogenesis and cell wall biosynthesis in the fungal pathogen Candida albicans. Mol. Cell. Biol. 25:10611-10627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sander, C. S., U. C. Hipler, U. Wollina, and P. Elsner. 2002. Inhibitory effect of terbinaline on reactive oxygen species generation by Candida albicans. Mycoses 45:152-155. [DOI] [PubMed] [Google Scholar]
- 34.San José, C., R. Monge, R. Perez-Diaz, J. Pla, and C. Nombela. 1996. The mitogen-activated protein kinase homolog Hog1 gene controls glycerol accumulation in the pathogenic fungus Candida albicans. J. Bacteriol. 178:5850-5852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Santos, A., M. M. Alvarez, M. San Mauro, C. Abrusci, and D. Marquina. 2005. The transcriptional response of Saccharomyces cerevisiae to Pichia membranifaciens killer toxin. J. Biol. Chem. 280:41881-41892. [DOI] [PubMed] [Google Scholar]
- 36.Schröter, R. C., U. C. Hipler, A. Wilmer, W. Kunkel, and U. Wollina. 2000. Generation of reactive oxygen species by Candida albicans in relation to morphogenesis. Arch. Dermatol. Res. 292:260-264. [DOI] [PubMed] [Google Scholar]
- 37.Smith, D. A., S. Nicholls, B. A. Morgan, A. J. P. Brown, and J. A. Quinn. 2004. A conserved stress-activated protein kinase regulates a core stress response in the human pathogen Candida albicans. Mol. Biol. Cell 15:4179-4190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Troxler, R. F., G. D. Offner, T. Xu., J. C. Vanderspek, and F. G. Oppenheim. 1990. Structural relationship between human salivary histatins. J. Dent. Res. 69:2-6. [DOI] [PubMed] [Google Scholar]
- 39.Tsai, H., and L. A. Bobek. 1997. Human salivary histatin-5 exerts potent fungicidal activity against Cryptococcus neoformans. Biochim. Biophys. Acta 1336:367-369. [DOI] [PubMed] [Google Scholar]
- 40.Vylkova, S., X. S. Li, J. C. Berner, and M. Edgerton. 2006. Distinct antifungal mechanisms: β-defensins require Candida albicans Ssa1 protein, while Trk1p mediates activity of cysteine-free cationic peptides. Antimicrob. Agents Chemother. 50:324-331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wunder, D., J. Dong, D. Baev, and M. Edgerton. 2004. Human salivary histatin 5 fungicidal action does not induce programmed cell death pathways in Candida albicans. Antimicrob. Agents Chemother. 48:110-115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Xu, Y., I. Ambudkar, H. Yamagishi, W. Swaim, T. J. Walsh, and B. C. O'Connell. 1999. Histatin 3-mediated killing of Candida albicans: effect of extracellular salt concentration on binding and internalization. Antimicrob. Agents Chemother. 43:2256-2262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhang, X., M. De Micheli, S. T. Coleman, D. Sanglard, and W. S. Moye-Rowley. 2000. Analysis of the oxidative stress regulation of the Candida albicans transcription factor, Cap1p. Mol. Microbiol. 36:618-629. [DOI] [PubMed] [Google Scholar]
- 44.Zhao, X., S. H. Oh, K. M. Yeater, and L. L. Hoyer. 2005. Analysis of the Candida albicans Als2p and Als4p adhesins suggests the potential for compensatory function within the ALS family. Microbiology 151:1619-1630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhou, X., W. Yin, S. Q. Doi, S. W. Robinson, K. Takeyasu, and X. Fan. 2003. Stimulation of Na,K-ATPase by low potassium requires reactive oxygen species. Am. J. Physiol. Cell Physiol. 285:C319-C326. [DOI] [PubMed] [Google Scholar]