Abstract
Cellulolytic flagellated protists in the guts of termites produce molecular hydrogen (H2) that is emitted by the termites; however, little is known about the physiology and biochemistry of H2 production from cellulose in the gut symbiotic protists due to their formidable unculturability. In order to understand the molecular basis for H2 production, we here identified two genes encoding proteins homologous to iron-only hydrogenases (Fe hydrogenases) in Pseudotrichonympha grassii, a large cellulolytic symbiont in the phylum Parabasalia, in the gut of the termite Coptotermes formosanus. The two Fe hydrogenases were phylogenetically distinct and had different N-terminal accessory domains. The long-form protein represented a phylogenetic lineage unique among eukaryotic Fe hydrogenases, whereas the short form was monophyletic with those of other parabasalids. Active recombinant enzyme forms of these two Fe hydrogenases were successfully obtained without the specific auxiliary maturases. Although they differed in their extent of specific activity and optimal pH, both enzymes preferentially catalyzed H2 evolution rather than H2 uptake. H2 evolution, at least that associated with the short-form enzyme, was still active even under high hydrogen partial pressure. H2 evolution activity was detected in the hydrogenosomal fraction of P. grassii cells; however, the vigorous H2 uptake activity of the endosymbiotic bacteria compensated for the strong H2 evolution activity of the host protists. The results suggest that termite gut symbionts are a rich reservoir of novel Fe hydrogenases whose properties are adapted to the gut environment and that the potential of H2 production in termite guts has been largely underestimated.
H2 emission by termites, well-known insects that thrive on recalcitrant wood or cellulose, is an interesting feature, since biological H2 emission is usually extremely limited in other natural ecosystems (35). The digestion of cellulosic matter by termites depends on the presence of a dense and diverse population of microorganisms in their guts, particularly cellulolytic flagellated protists in the case of phylogenetically lower termites (3, 17, 26). Earlier studies postulated that the gut symbiotic protists phagocytose wood or cellulose particles and then depolymerize and ferment them to produce stoichiometric amounts of acetate, CO2, and H2 (16, 25, 39). The symbiotic protists, accounting for up to one-third of the total insect volume, are densely packed mainly in the voluminously dilated anterior part of the hindgut (17), where the H2 partial pressure is the highest (7). Thus, cellulolytic protists are considered to be the major producers of H2 in the gut.
Nevertheless, little is known about the detailed physiology and biochemistry of the gut symbiotic protists of termites; this is because most of these protists have formidably resisted cultivation in vitro. Culture-independent molecular studies, however, have been successfully applied to the gut dwellers of termites (for reviews, see reference 26); these studies were mainly molecular phylogenetic analyses. They revealed unexpected diversity among termite gut protists (12, 27-29). Genes involved in cellulose decomposition have also been identified previously (references 18 and 36 and references therein).
The gut symbiotic protists are unique to termites and related wood-feeding roaches and belong to the phylum Parabasalia or the order Oxymonadida (phylum Preaxostyla). The former includes most of the large gut protists that are responsible for the decomposition of highly polymerized cellulose. Parabasalids are typified by the presence of parabasal bodies and hydrogenosomes in the cell (4). Hydrogenosomes are H2-producing organelles that play a central role in anaerobic energy production (19) and are distributed in a variety of anaerobic eukaryotes; therefore, they are considered to have evolved multiple times independently from the mitochondria (8, 11).
All the known eukaryotic hydrogenases, whether they are located in the hydrogenosome or not, are classified as iron-only hydrogenases ([FeFe] hydrogenases; herein referred to simply as Fe hydrogenases), which are evolutionarily unrelated to the other major type of hydrogenases, [NiFe] hydrogenases (37). Fe hydrogenases are considered to play a role in H2 evolution by reducing protons; however, the eukaryotic enzymes are poorly understood because, thus far, only a few eukaryotic Fe hydrogenases, namely, those in Chlamydomonas and Trichomonas species, have been purified and characterized (13, 30), although a higher number of primary structures have been deduced from the genes of various anaerobic eukaryotes (1, 5, 14, 21, 38). As proposed previously (11), the gut-dwelling anaerobic eukaryotes of termites and other animals potentially harbor an enormous biodiversity of Fe hydrogenases due to their great phylogenetic diversity. Further research regarding the various anaerobic hydrogenosome-carrying eukaryotes and their hydrogenases is critical for understanding eukaryotic organelle evolution and the evolutionary origin of eukaryotic Fe hydrogenases (11).
In this study, we investigated the Fe hydrogenase genes and the characteristics of the encoded enzymes in parabasalian symbionts of the termite Coptotermes formosanus. The study aimed to understand the molecular mechanisms underlying the intensive H2 production in termites and to investigate the biodiversity of hydrogenases. The termite C. formosanus is a globally distributed and economically important pest of wood houses. It harbors only three species of gut protists, all of which belong to the phylogenetically monophyletic phlyum Parabasalia but are distantly related to the cultivated Parabasalia representatives Trichomonas vaginalis and Tritrichomonas foetus (28, 29). Among the three, the largest protist, Pseudotrichonympha grassii, is maximally responsible for the decomposition of highly polymerized cellulose (40). Recombinant enzyme forms of two distinct Fe hydrogenases of P. grassii were purified and characterized.
MATERIALS AND METHODS
Termite species and hydrogen emission rate.
The termite species used in this study was C. formosanus (Isoptera: Rhinotermitidae), and it was obtained from natural colonies in Japan. In the laboratory, the termites were maintained on infested wood blocks in polypropylene containers. Worker termites were used for all experiments.
The termites were fed an artificial diet that comprised cellulose and water in the ratio of 2:3 for 1 week. Ten termites were placed in 100-ml serum vials that contained 5 g of the artificial diet; the vials were then sealed with butyl rubber stoppers. The H2 concentration in the headspace of the vials was measured every 12 h for 72 h by gas chromatography with a Shimadzu 14C chromatograph and with a semiconductor detector and a Unibead C packed column (GL Science) operating at 60 and 80°C, respectively. The carrier was nitrogen gas flowing at 60 ml/min. Linear emission continued (R2 = 0.997) for at least 72 h.
Isolation of cDNA clones and 5′-end amplification.
The sequences of the cDNAs encoding hydrogenase homologs were identified by BLAST searches of the nonredundant NCBI database as part of our ongoing expressed-sequence-tag (EST) survey. The cDNA library used for the EST analysis has been established from the mixed population of gut protists in C. formosanus (18). The truncated 5′ ends of cDNAs were determined by 5′ rapid amplification of cDNA end (RACE) reactions using a Gene Racer kit (Invitrogen) with sequence-specific primers (see Table S1 in the supplemental material). The 5′ RACE products were cloned, and the DNA sequences of multiple clones were determined to confirm the identity of the overlapping cDNA region of the original clones. The entire coding sequences were amplified by reverse transcription-PCR (RT-PCR) with sequence-specific primers (see Table S1 in the supplemental material), and the PCR products were used for subsequent analyses. DNA sequencing was performed by primer walking using an automated DNA sequencer (ABI model 3700) with BigDye Terminator chemistry (Applied Biosystems).
Identification of protist origins.
Cells of each species of symbiotic protists in the gut were manually isolated with the aid of a micromanipulator (Cell-Tram; Eppendorf) as described previously (27), and the cells were washed extensively. As described previously (29), a pool of five isolated cells of each protist species was used directly for specific amplification by RT-PCR with sequence-specific primers (see Table S1 in the supplemental material) to detect the species of origin of the sequences. The amplification product from each protist species was cloned, and the DNA sequences in multiple clones were determined to confirm the identity to the corresponding cDNA sequence. Whole-cell hybridizations using oligonucleotide probes specific for 09A82 and 06B09 sequences (see Table S1 in the supplemental material) were applied for the in situ identification of the organismal origins of the Fe hydrogenase genes. Fixation and hybridization were performed as described previously (18). Due to the small amount of mRNA for Fe hydrogenase genes, hybridization signals were detected by an enzyme-linked immunoassay. The cells that hybridized with probes 5′-end labeled with digoxigenin (DIG) were detected with alkaline phosphatase-conjugated anti-DIG antibodies and colorimetric substrates by using a DIG nucleic acid detection kit (Roche) and observed by light microscopy.
Phylogenetic analysis.
The in silico-translated amino acid sequences corresponding to the genes for the Fe hydrogenase homologs were aligned with sequences of known Fe hydrogenases and those of their homologous proteins listed in the protein databases based on the Pfam seed alignment of Fe hydrogenases (http://www.sanger.ac.uk/Software/Pfam/), and the alignment was corrected manually. The program PHYML (version 2.4.4) (10) was used for inferring the maximum-likelihood (ML) tree using the Jones-Taylor-Thornton model, gamma distribution with four categories plus invariant sites, and the shape parameter and the fraction of invariable sites estimated from the data set. MrBayes (version 3.1.2) (15) was used for evaluating the posterior probabilities at the nodes of the ML tree. MrBayes was run for 1,000,000 generations with the Jones-Taylor-Thornton model, gamma-distributed rate variation, and a proportion of invariable sites; the burn-in value was set at 100,000 generations. The significance of the differences in tree topologies was examined by the approximately unbiased test implemented in CONSEL (34) with 1,000 bootstrap replicates. All the Fe hydrogenase sequences from Parabasalia were constrained to monophyly, and the topology of an ML tree inferred with MrBayes under the constraint was compared with that of an unconstrained ML tree.
Production of recombinant enzymes.
Hydrogenosome-targeting signals in the amino acid sequences of the Fe hydrogenases of the termite gut symbionts were predicted as mitochondrial targeting signals by using the programs PSORT (http://psort.ims.u-tokyo.ac.jp/), TargetP (http://www.cbs.dtu.dk/services/SignalP/), and MitoProt (http://ihg.gsf.de/ihg/mitoprot.html). The coding regions of the Fe hydrogenase cDNAs of P. grassii without the predicted hydrogenosome-targeting signals were amplified by PrimeSTAR DNA polymerase (Takara) using sequence-specific primers (see Table S1 in the supplemental material), and the amplification products were subcloned into the pET47b vector (Novagen) by using the restriction sites (SmaI and NotI) introduced into these primers. Escherichia coli BL21(DE3) cells carrying the resulting plasmids were cultured at 20°C in Luria broth for the production of His6-tagged recombinant enzymes. The cells were grown first aerobically for 12 h and then anaerobically for 40 h. The grown cells were transferred into fresh medium and grown anaerobically, and recombinant enzyme production was induced by the addition of isopropyl-β-d-thiogalactopyranoside (IPTG). IPTG was added at 48 and 60 h for the production of the 09A82 and 06B09 proteins, respectively. After the 12-h induction, the cells were harvested by centrifugation and disrupted by sonication to extract the cell lysate. The recombinant proteins were purified using a His trap nickel affinity purification column (GE Healthcare). All these manipulations were performed anaerobically in an anaerobic chamber (Bactron). Purity was assessed by measuring the absorption of the proteins at 280 and 400 nm, the latter of which was specific for Fe-S cluster proteins, as well as by performing sodium dodecyl sulfate-polyacrylamide gel electrophoresis.
Fractionation of protist cells and their cellular components.
The termites were fed an artificial diet for 1 week, and 100 termites were used for fractionating cellular components of the protist species. All of the following manipulations were performed anaerobically. Termite guts were dissected using a fine-tipped forceps and gently crushed in the solution described previously (28). After the release of the gut contents and the removal of the gut tissues, the P. grassii cells were concentrated by low-speed centrifugation at 18 × g for 5 min in a Kubota 1-13 centrifuge and the pellet was resuspended and washed three times. The concentrated P. grassii cells were mildly broken by the addition of Nonidet P-40 (Nacalai Tesque) at a final concentration of 0.8%. A fraction of the large particles (nuclei, flagella, and axostyle fragments) was obtained by centrifugation at 500 × g for 4 min. From the supernatant produced by the 500 × g centrifugation, the hydrogenosome-enriched fraction was obtained by centrifugation at 2,000 × g for 10 min. The fraction enriched with the intracellular bacteria of P. grassii was obtained by centrifugation at 10,000 × g for 10 min. The presence of the bacteria was confirmed by 4′,6-diamidino-2-phenylindole (DAPI) staining and fluorescence microscopy. Each fraction was homogenized using a sonic homogenizer (model UR-20P; Tomy Seiko) and used for the hydrogenase assay.
Hydrogenase assay.
The hydrogenases were evaluated by measuring both H2 evolution and H2 uptake activities. The H2 evolution activity was measured by gas chromatography, as described above, using reduced methyl viologen (MV) as an electron donor. The H2 uptake activity was measured by spectroscopy analysis of the H2-dependent reduction of methylene blue (MB), and the value was used to calculate the molar H2 consumed. The reactions were performed in a tightly closed quartz cuvette filled with 500 μl of the reaction mixture (50 mM buffer, 2 mM dithiothreitol, 1 mM electron carrier, and dithionite). The dithionite concentrations used for the H2 evolution and H2 uptake assays were 15 and 1 mM, respectively. The reaction mixture was bubbled for 15 min with nitrogen for the H2 evolution assay or with H2 for the H2 uptake assay. Measurement was initiated by the addition of 10 μl of the enzyme solution. All these activities were determined in triplicate. The protein concentration was determined by the DC protein assay (Bio-Rad) using bovine serum albumin as a standard.
Optimum pH was examined using 50 mM concentrations of acetate buffer, phosphate buffer, Tris-HCl buffer, and glycine-NaOH buffer at pH ranges of 5.0 to 6.5, 6.0 to 7.5, 7.0 to 9.0, and 8.5 to 9.5, respectively. In the H2 uptake assay, the specificity of the electron carrier was examined with benzyl viologen, flavin adenine dinucleotide (FAD), flavin mononucleotide (FMN), NAD, NADH, NADP, NADPH, and ferredoxin from Clostridium pasteurianum (Sigma). Only benzyl viologen, FAD, FMN, and ferredoxin were examined in the H2 evolution assay. Enzyme stability was investigated by incubating the enzyme solution at different pHs or temperatures for 5 min prior to the assay. A temperature range from 5 to 60°C and a pH range from 5 to 10 were used to investigate enzyme stability.
The effects of high H2 partial pressure on the H2 evolution activity of the recombinant 09A82 enzyme were evaluated. The headspace of a quartz cuvette containing the reaction mixture described above was first filled and compressed appropriately with nitrogen, and then the H2 partial pressure was adjusted to 10 to 50% of the final total pressure of 200 kPa. The enzyme reaction was performed at 30°C, and the increase in the headspace H2 was measured by gas chromatography.
Nucleotide sequence accession numbers.
The DNA sequences of four homologous Fe hydrogenase cDNAs have been deposited in the DDBJ database under the accession numbers AB331667 to AB331670.
RESULTS
H2 emission by the termite.
The H2 emission rate of the termite C. formosanus was measured in order to evaluate the H2 productivity of the termite. The termite continuously emitted H2 at a rate of 688 ± 360 (mean ± standard deviation) nmol h−1 g of body weight−1. Cellulose consumption by the termite was 0.16 ± 0.04 mg h−1 g−1. The H2 emission rate per gram of cellulose consumed was 4.3 mmol, which was equivalent to approximately 0.75 mol of H2 emitted per 1 M glucose. The result indicated that the termite emitted a considerable amount of H2, although the emission rate was lower than the predicted maximal stoichiometry of 4 mol of H2 per 1 M glucose for gut protists (16).
Fe hydrogenase genes and their origin.
In the EST analysis of the mixed population of gut symbiotic protists of C. formosanus, four cDNA clones (06B09, 09A82, 11B87, and 08B89) that encoded proteins homologous to known Fe hydrogenases were discovered. No sequence homologous to the [NiFe] hydrogenases was found in the ESTs examined. In 06B09, 09A82, and 11B87, the entire coding regions were identified by sequence-specific 5′ RACE and subsequent RT-PCR, while the putative translational initiation codon was found in the original cDNA clone of 08B89. The open reading frames in 06B09, 09A82, 11B87, and 08B89 encoded 550, 476, 456, and 447 amino acid residues, respectively.
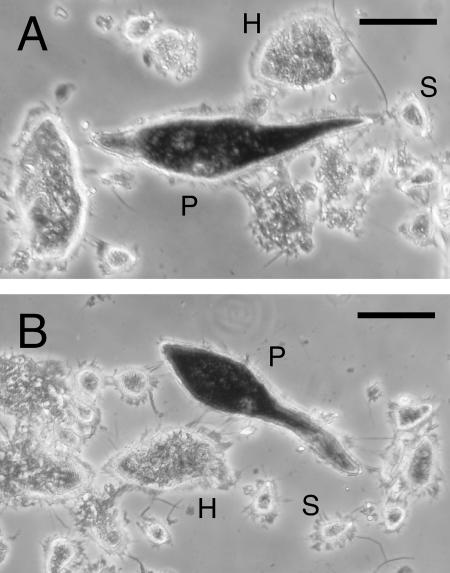
The flagellated protists in the gut of C. formosanus comprise exclusively the three parabasalian species P. grassii, Holomastigotoides mirabile, and Spirotrichonympha leidyi (28, 29). P. grassii belongs to the family Eucomonymphidae, while the latter two belong to the family Spirotrichonymphidae. The protist origins of the sequences 06B09 and 09A82 were examined by in situ hybridizations with sequence-specific probes (Fig. 1). Each probe gave positive signals only in the P. grassii cells, indicating that P. grassii was the origin of these sequences. The protist origin was also confirmed by sequence-specific RT-PCR using manually isolated cells of each protist species. The specific amplification of 06B09 and 09A82 from P. grassii and of 08B89 from H. mirabile was detected, but no amplification of 11B87 from any of the protist species was observed. The specific amplification products showed >99.6% identity to the respective cDNA sequences. Further in situ hybridization experiments are necessary to identify the protist origins of the 08B89 and 11B87 sequences.
FIG. 1.
Whole-cell in situ hybridizations using sequence-specific probes for 09A82 (A) and 06B09 (B). The micrographs show three parabasalid species, P. grassii (P), H. mirabile (H), and S. leidyi (S), in the gut of C. formosanus. The largest species, P. grassii, exhibited significant signal staining with both probes. No significant signal was observed in any protist species without the addition of the probes. Bars represent 50 μm.
Protein primary structure and phylogeny.
The proteins encoded by 09A82, 11B87, and 08B89 showed significant similarity to a hydrogenosomal Fe hydrogenase of T. vaginalis (TvHydA) and exhibited 54, 52, and 58% identity, respectively. These three genes encoded the so-called short-form Fe hydrogenase that comprised large and small subunits of the catalytic domain containing H-cluster binding sites and an N-terminal accessory domain containing two 4Fe-4S clusters (see Fig. S1 in the supplemental material). The protein encoded by 06B09 was somewhat similar in domain structure to the long-form putative 64-kDa Fe hydrogenase of T. vaginalis, although its level of identity was only 35%. The long-form Fe hydrogenase generally contains large and small subunits, three 4Fe-4S clusters (one of which harbors a histidine ligand for the 4Fe-4S cluster), and a plant ferredoxin-type 2Fe-2S cluster. The 06B09 protein, however, typically lacked the histidine-ligated 4Fe-4S cluster and had a shorter large subunit due to deletions of stretches of 2 to 5 amino acids in several positions in the alignment (see Fig. S1 in the supplemental material). A hydrogenosome-targeting signal in the N-terminal 12 to 24 amino acid residues that preceded arginine in each of the four proteins was predicted, similar to those in the hydrogenosomal proteins of T. vaginalis (2).
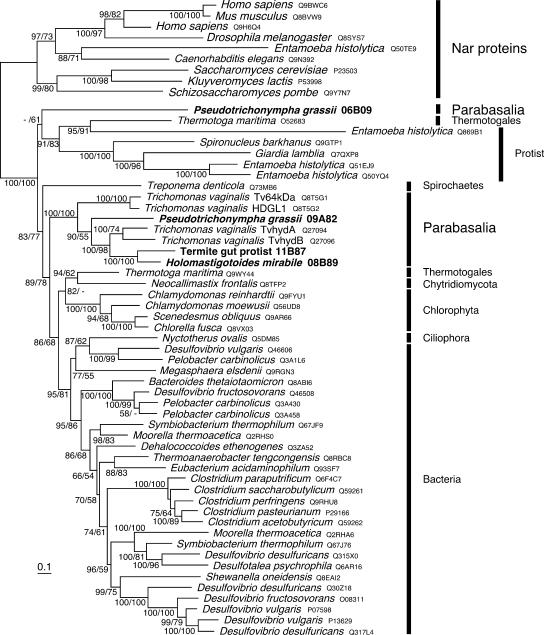
A phylogenetic analysis of the conserved catalytic large subunit domain (Fig. 2) revealed that all the four sequences obtained from the gut symbionts belonged to the eukaryotic group of Fe hydrogenases that also contains some sequences from bacteria, such as Thermotoga maritima and Treponema denticola, but does not include the sequence from the Nyctotherus species ciliate. 09A82, 11B87, and 08B89 formed a robust monophyletic cluster with the sequences from T. vaginalis, indicating that they share a common ancestor as predicted by their classification in Parabasalia. However, the 06B09 sequence was distantly related to the cluster of parabasalid sequences and formed a lineage unique among the eukaryotic Fe hydrogenases. Monophyly of 06B09 with the other sequences of parabasalids was rejected by the approximately unbiased test (P < 0.05). The phylogenetic analyses of longer regions (the large and small subunits, the large subunit plus 4Fe-4S clusters, and all of these regions) always exhibited the monophyly of the 09A82, 08B89, and 11B87 enzymes and T. vaginalis Fe hydrogenases and the distant relationship of the 06B09 protein to this monophyletic cluster (data not shown).
FIG. 2.
Phylogenetic positions of the Fe hydrogenases of termite gut symbionts. The tree was inferred by the ML method by using amino acid sequences of the large subunit. Nar-like protein sequences that are related to Fe hydrogenase but do not catalyze hydrogenase reactions were used as the out-groups. The numbers at the nodes are the Bayesian posterior probabilities and the ML bootstrap values (separated by a slash). The four sequences from the gut symbionts of C. formosanus are labeled in bold letters. Protein database accession numbers are given after the organism names. The scale bar indicates 0.10 substitutions per site.
Properties of recombinant enzymes.
The recombinant Fe hydrogenases were biochemically characterized in order to understand the mechanism underlying the intensive H2 productivity of termite gut protists. The N terminus-processed putative mature form of each of the P. grassii 09A82 and 06B09 proteins was heterologously expressed in E. coli as a His6-tagged fusion protein. The purified recombinant 09A82 and 06B09 proteins showed single bands of 51 and 60 kDa, respectively, upon sodium dodecyl sulfate-polyacrylamide gel electrophoresis, as predicted from the cDNA sequences. The short-form 09A82 protein exhibited H2 evolution activity of 2,131 ± 54 μmol of H2 min−1 mg of protein−1 and H2 uptake activity of 431 ± 12 μmol of H2 min−1 mg of protein−1 under optimum conditions (see below). The long-form 06B09 protein catalyzed only the H2 evolution activity, at a level of 121 ± 7.8 μmol of H2 min−1 mg of protein−1. The kinetic parameters of the recombinant P. grassii Fe hydrogenases are shown in Table 1. Although the 06B09 enzyme exhibited weaker activity, it demonstrated high catalytic efficiency (kcat/Km) with reduced MV. The enzymes utilized benzyl viologen as well as MV but not MB as an electron donor in the evolution reaction, and they utilized MB but not MV as an electron acceptor. FAD, FMN, NAD, NADH, NADP, NADPH, and ferredoxin of C. pasteurianum were not used as electron carriers in the reactions.
TABLE 1.
Kinetic parameters of the recombinant P. grassii Fe hydrogenases for H2 evolution and uptake activitya
| Enzyme | Reaction | Vmax (μmol of H2 min−1 mg−1) | Km (μM) | kcat (s−1) | kcat/Km (M−1 s−1) |
|---|---|---|---|---|---|
| 09A82 | Evolution | 2,033 | 229 | 1.8 × 105 | 7.9 × 108 |
| Uptake | 305 | 12.0 | 2.7 × 104 | 2.3 × 109 | |
| 06B09 | Evolution | 117 | 10.5 | 1.3 × 104 | 1.2 × 109 |
Because H2 uptake activity of the 06B09 enzyme was not detected, the parameters could not be evaluated. The parameters were determined at 30°C with MV for H2 evolution and with MB for H2 uptake and at pH 8 for the 09A82 enzyme and pH 6 for the 06B09 enzyme.
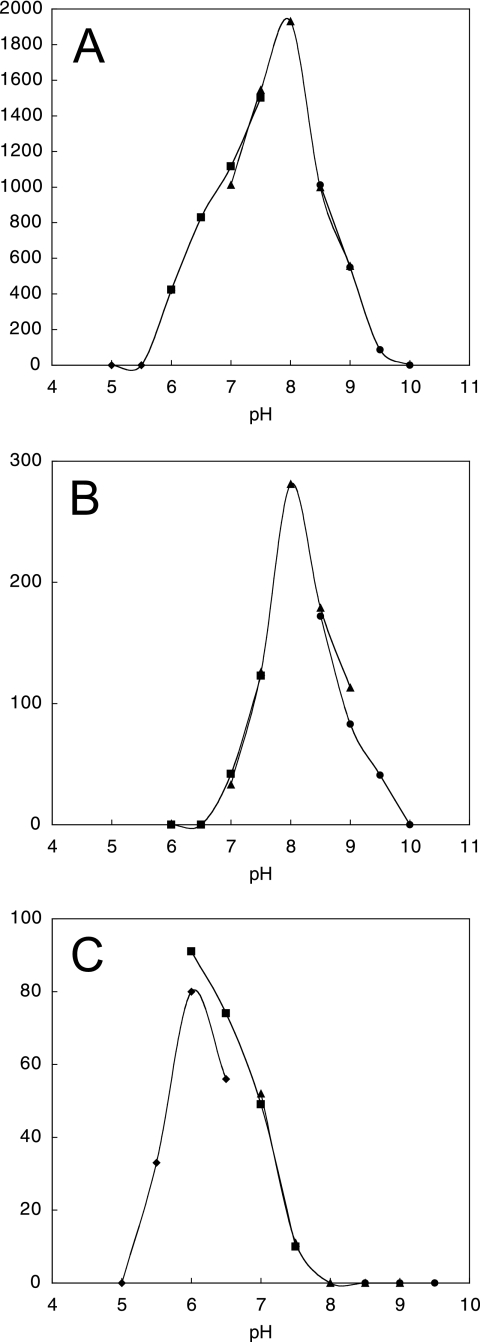
The activity of the short-form 09A82 enzyme was detected in the range of pH 6.0 to 9.5 (Fig. 3A and B) at up to 55°C, with the optimum conditions being pH 8 at 45°C for both H2 evolution and uptake activities. The activity of the long-form 06B09 enzyme was detected in the range of pH 5.5 to 7.5 (Fig. 3C) at up to 45°C, with the optimum conditions being pH 6 at 40°C. The activities of the 09A82 and 06B09 enzymes remained during incubation at pH 6.0 to 9.5 and pH 6.0 to 7.5, respectively, but the activities were lost within 5 min of incubation at 35 and 15°C, respectively. The activities were lost immediately after air was bubbled into the reaction mixture, suggesting the high sensitivity of these enzymes to oxygen.
FIG. 3.
pH profiles of hydrogenase activity. Graphs show the pH dependence of H2 evolution activity of the short-form 09A82 enzyme (A) and its H2 uptake activity (B) and the H2 evolution activity of the long-form 06B09 enzyme (C). Diamonds, squares, triangles, and circles indicate measurements using acetate, phosphate, Tris-HCl, and glycine-NaOH buffers, respectively.
Because H2 partial pressure in the gut of termites can be as high as approximately 5 kPa (7), the effect of H2 partial pressure on the H2 evolution activity of the 09A82 enzyme was examined. Approximately half the activity (53 and 46%) was retained under 10 and 20 kPa of H2, respectively, compared to the level of activity under 0 kPa (control). Even under 30 and 40 kPa of H2, 17 and 7% of the activity, respectively, was retained.
Subcellular fractionation of hydrogenase activity.
The localization and properties of native Fe hydrogenases were investigated by subcellular fractionation of the protist cells. The mixed population of hindgut microorganisms was enriched with P. grassii cells, which were used for the small-scale subcellular fractionation after the unassociated bacteria were washed out. Since P. grassii harbors a high density of intracellular bacteria (22, 23), the bacterial and organelle fractions were roughly fractionated by differential centrifugation (Table 2). The organelle-rich fraction (the 2,000 × g precipitate [ppt]) exhibited the highest H2 evolution activity at pH 8, while the highest H2 uptake activity of the bacterium-rich fraction was observed at both pH 8 and pH 6.
TABLE 2.
Fractionation of the hydrogenase activity of P. grassii cellsa
| Fraction | Value at pH 8 for:
|
Value at pH 6 for:
|
||
|---|---|---|---|---|
| Evolution | Uptake | Evolution | Uptake | |
| Cell homogenate | 33 ± 6 | 325 ± 17 | 230 ± 21 | 464 ± 32 |
| 500 × g ppt (large particles) | 67 ± 15 | 113 ± 11 | 93 ± 13 | 141 ± 23 |
| 2,000 × g ppt (hydrogenosomes) | 4,268 ± 126 | 2,778 ± 119 | 28 ± 7 | 223 ± 41 |
| 10,000 × g ppt (bacteria) | 71 ± 19 | 11,852 ± 337 | 573 ± 46 | 14,183 ± 632 |
| 10,000 × g supernatant (soluble material) | 18 ± 10 | 51 ± 13 | ND | 35 ± 15 |
The mean hydrogenase activity (± the standard deviation) measured at either pH 8 or pH 6 is expressed as nanomoles of H2 per minute per milligram. ND, not detected. The hydrogenosome-rich fraction (2,000 × g ppt) showed that only 10.9% of the observed particles stained with DAPI, whereas 79.5% of the particles in the endosymbiotic bacterial fraction (10,000 × g ppt) stained with DAPI.
DISCUSSION
This study describes the identification of novel Fe hydrogenases from symbiotic parabasalian flagellates in the termite gut. The long-form 06B09 enzyme sequence represents a phylogenetic lineage unique among the eukaryotic Fe hydrogenases and is distantly related to the sequences from the other parabasalids. The uniqueness of this sequence is also confirmed by the fact that no sequence related to the 06B09 DNA sequence was found previously in the genome of T. vaginalis (6) or in the EST database covering a taxonomically broad range of eukaryotes (24). In addition to the sequence novelty of 06B09, we observed distinct enzymatic characteristics of the short-form 09A82 Fe hydrogenase, such as the optimal pH and resistance against high H2 partial pressure, although its sequence is related to those of the Fe hydrogenases of T. vaginalis. The results suggest that symbiotic protists in termite guts are a rich reservoir of novel Fe hydrogenase biodiversity.
The levels of H2 evolution activity exhibited by the Fe hydrogenases of P. grassii, particularly that of the short-form 09A82 enzyme, were some of the highest among those demonstrated by known Fe hydrogenases. The activity of the P. grassii Fe hydrogenases, however, is probably underestimated, because the recombinant enzymes are considerably unstable. These enzymes lost their activity rapidly when exposed to ambient temperature even under strict anaerobic conditions. The loss of activity was irreversible, probably due to the decomposition of Fe-S clusters that was revealed by the decay of the absorption spectrum at around 400 nm. Electron paramagnetic resonance (EPR) spectroscopy analysis of the 09A82 enzyme indicated that only 44% of the protein formed an authentic H-cluster (the presence of an EPR signal with principal g factors of 2.032, 2.017, and 1.998 confirmed that the protein is a typical Fe hydrogenase).
It is generally believed that the production of heterologous Fe hydrogenase in E. coli is difficult since E. coli lacks auxiliary maturases for the metal clusters. In Chlamydomonas, such assembly factors have been shown to be necessary for active Fe hydrogenase production (32). In T. vaginalis, the homologous maturases are reported to localize in the hydrogenosomes (33), although their contribution to active Fe hydrogenase assembly has not been estimated. The P. grassii Fe hydrogenases probably assembled, albeit imperfectly, with the aid of unknown E. coli machinery or spontaneously without auxiliary maturases. Similarly, an Fe hydrogenase of Entamoeba histolytica is reported to be active when expressed in E. coli, although it shows only weak activity (21).
The presence of the two Fe hydrogenases in P. grassii and their distinct characteristics in terms of optimal pH and domain structure suggest that they play different physiological roles. The difference in the accessory domains in the N termini may affect the redox potentials of the Fe hydrogenases since these domains conduct electrons between the donors and the H2-activating center (20, 31). The in vivo electron donors may be ferredoxin reduced by pyruvate-ferredoxin oxidoreductase or involve NADH dehydrogenase as predicted previously for T. vaginalis (6). Further studies are necessary to address the physiological differences. The localization of both P. grassii enzymes in hydrogenosomes also needs to be ascertained.
A notable characteristic of the P. grassii Fe hydrogenases was that they preferentially catalyzed H2 evolution. Another interesting feature was that the evolution activity of the short-form 09A82 enzyme was resistant to high H2 partial pressure. In addition to their advantageous application for H2 production, these characteristics are suitable for the hydrogenosomal function of anaerobic energy production. Since the elimination of reducing equivalents is an important rate-limiting step for anaerobic fermentation, the efficient production of H2 stimulates cellulose fermentation by the protist cells. The reducing equivalents produced during the fermentation as forms of NADH and reduced ferredoxin are rapidly eliminated by reoxidizing these electron carriers through the intensive H2 production activity. Indeed, the hydrogenosome fraction of P. grassii cells exhibited strong H2 evolution activity at pH 8—the optimal pH for the short-form 09A82 enzyme. The simultaneous presence of strong H2 uptake activity in the endosymbiotic bacterial fraction (Table 2), however, appears to disturb the evolution activity. The H2 evolution activities of both P. grassii enzymes at pH 6 are lower than the evolution activity of the short-form enzyme at pH 8; similarly, H2 evolution activity in hydrogenosomes at pH 6 is probably lower than that at pH 8, and the uptake enzyme quickly utilizes the produced H2.
The potentially strong H2 evolution activity of the gut protists and the considerable H2 uptake activity of the endosymbiotic bacteria suggest that interspecies H2 transfer is involved in the symbiotic relationship between the host protist and the endosymbiont that has been shown to be a novel member of the “Bacteroidales” (22, 23). This novel aspect of the intracellular symbiotic relationship is beneficial to the host since, as shown for intracellular methanogens in anaerobic protists (9), the rapid removal of the reducing equivalent (H2) by the associated bacteria leads to enhanced fermentation by the host protists. The electron acceptor(s) for H2 utilization in the endosymbiotic bacteria remains to be identified in order to understand the true nature of this symbiotic relationship.
In addition to the H2 uptake by the endosymbiotic bacteria, the presence of hydrogenotrophic populations, such as methanogens and homoacetogens, in the gut microbial community of termites is well known (3). As apparent by the low level of activity in the unfractionated homogenate of P. grassii (Table 1), the H2 evolution ability of gut symbiotic protists is underestimated. Nevertheless, termites emit H2 at a considerable rate, as shown in this and previous studies (35), and this is a unique characteristic of natural ecosystems. Their potentially strong H2 evolution activity as well as their efficient cellulolytic ability indicates that termites and their gut symbiotic protists represent an attractive biological model of H2 production from cellulosic dead plant matter.
Supplementary Material
Acknowledgments
We thank A. Kikuchi and Y. Shiro for assistance in EPR spectrometry and S. Noda, Y. Hongoh, S. Hattori, and T. Inoue for their useful suggestions and technical support.
J.-I.I. was supported by the Junior Research Associate Program from RIKEN. This work was partially supported by grants-in-aid for scientific research from JSPS (no. 16380065 and 19380055) and a grant from the Noda Institute for Scientific Research to M.O. and by grants for the Bioarchitect Research and the EcoMolecular Science Research programs from RIKEN.
Footnotes
Published ahead of print on 31 August 2007.
Supplemental material for this article may be found at http://ec.asm.org/.
REFERENCES
- 1.Akhmanova, A. S., F. G. Voncken, T. van Alen, A. H. van Hoek, B. Boxma, G. D. Vogels, M. Veenhuis, and J. H. Hackstein. 1998. A hydrogenosome with a genome. Nature 396:527-528. [DOI] [PubMed] [Google Scholar]
- 2.Bradley, P. J., C. J. Lahti, E. Plumper, and P. J. Johnson. 1997. Targeting and translocation of proteins into the hydrogenosome of the protist Trichomonas: similarities with mitochondrial protein import. EMBO J. 16:3484-3493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Breznak, J. A. 2000. Ecology of prokaryotic microbes in the guts of wood- and litter-feeding termites, p. 209-231. In T. Abe, D. E. Bignell, and M. Higashi (ed.), Termites: evolution, sociality, symbioses, ecology. Kluwer Academic Publishers, Dordrecht, The Netherlands.
- 4.Brugerolle, G., and J. J. Lee. 2001. Phylum Parabasalia, p. 1196-1249. In J. J. Lee, G. F. Leedale, D. J. Patterson, and P. C. Bradbury (ed.), Illustrated guide to the Protozoa, 2nd ed. Allen Press, Lawrence, KS.
- 5.Bui, E. T., and P. J. Johnson. 1996. Identification and characterization of [Fe]-hydrogenase in the hydrogenosome of Trichomonas vaginalis. Mol. Biochem. Parasitol. 76:305-310. [DOI] [PubMed] [Google Scholar]
- 6.Carlton, J. M., R. P. Hirt, J. C. Silva, et al. 2007. Draft genome sequence of the sexually transmitted pathogen Trichomonas vaginalis. Science 315:207-212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ebert, A., and A. Brune. 1997. Hydrogen concentration profiles at the oxic-anoxic interface: a microsensor study of the hindgut of the wood-feeding lower termite Reticulitermes flavipes (Kollar). Appl. Environ. Microbiol. 63:4039-4046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Embley, T. M. 2006. Multiple secondary origins of the anaerobic lifestyle in eukaryotes. Phil. Trans. R. Soc. B 361:1055-1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fenchel, T., and B. J. Finlay. 1995. Ecology and evolution in anoxic worlds. Oxford University Press, Oxford, United Kingdom.
- 10.Guindon, S., and O. Gascuel. 2003. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst. Biol. 52:696-704. [DOI] [PubMed] [Google Scholar]
- 11.Hackstein, J. H. P. 2004. Eukaryotic Fe-hydrogenases: old eukaryotic heritage or adaptive acquisitions? Biochem. Soc. Trans. 33:47-50. [DOI] [PubMed] [Google Scholar]
- 12.Hampl, V., I. Cepicka, J. Flegr, J. Tachezy, and J. Kulda. 2004. Critical analysis of the topology and rooting of the parabasalian 16S rRNA tree. Mol. Phylogenet. Evol. 32:711-723. [DOI] [PubMed] [Google Scholar]
- 13.Happe, T., and J. D. Naber. 1993. Isolation, characterization and N-terminal amino acid sequence of hydrogenase from the green alga Chlamydomonas reinhardtii. Eur. J. Biochem. 214:475-481. [DOI] [PubMed] [Google Scholar]
- 14.Horner, D. S., P. G. Foster, and T. M. Embley. 2000. Iron hydrogenases and the evolution of anaerobic eukaryotes. Mol. Biol. Evol. 17:1695-1709. [DOI] [PubMed] [Google Scholar]
- 15.Huelsenbeck, J. P., and F. Ronquist. 2001. MrBAYES: Bayesian inference of phylogenetic trees. Bioinformatics 17:754-755. [DOI] [PubMed] [Google Scholar]
- 16.Hungate, R. E. 1943. Quantitative analyses of the cellulose fermentation by termite protozoa. Ann. Entomol. Soc. Am. 36:730-739. [Google Scholar]
- 17.Inoue, T., O. Kitade, T. Yoshimura, and I. Yamaoka. 2000. Symbiotic associations with protists, p. 275-288. In T. Abe, D. E. Bignell, and M. Higashi (ed.), Termites: evolution, sociality, symbioses, ecology. Kluwer Academic Publishers, Dordrecht, The Netherlands.
- 18.Inoue, T., S. Moriya, M. Ohkuma, and T. Kudo. 2005. Molecular cloning and characterization of a cellulase gene from a symbiotic protist of the lower termite, Coptotermes formosanus. Gene 349:67-75. [DOI] [PubMed] [Google Scholar]
- 19.Müller, M. 1993. The hydrogenosome. J. Gen. Microbiol. 139:2879-2889. [DOI] [PubMed] [Google Scholar]
- 20.Nicolet, Y., C. Piras, P. Legrand, C. E. Hatchikian, and J. C. Fontecilla-Camps. 1999. Desulfovibrio desulfuricans iron hydrogenase: the structure shows unusual coordination to an active site Fe binuclear center. Structure 7:13-23. [DOI] [PubMed] [Google Scholar]
- 21.Nixon, J. E., J. Field, A. G. McArthur, M. L. Sogin, N. Yarlett, B. J. Loftus, and J. Samuelson. 2003. Iron-dependent hydrogenases of Entoamoeba histolytica and Giardia lamblia: activity of the recombinant entamoebic enzyme and evidence for lateral gene transfer. Biol. Bull. 204:1-9. [DOI] [PubMed] [Google Scholar]
- 22.Noda, S., T. Iida, O. Kitade, H. Nakajima, T. Kudo, and M. Ohkuma. 2005. Endosymbiotic Bacteroidales bacteria of the flagellated protist Pseudotrichonympha grassii in the gut of the termite Coptotermes formosanus. Appl. Environ. Microbiol. 71:8811-8817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Noda, S., O. Kitade, T. Inoue, M. Kawai, M. Kanuka, K. Hiroshima, Y. Hongoh, R. Constantino, V. Uys, J.-H. Zhong, T. Kudo, and M. Ohkuma. 2007. Cospeciation in the triplex symbiosis of termite gut protists (Pseudotrichonympha spp.), their hosts, and their bacterial endosymbionts. Mol. Ecol. 16:1257-1266. [DOI] [PubMed] [Google Scholar]
- 24.O'Brien, E. A., L. B. Koski, Y. Zhang, L. Yang, E. Wang, M. W. Gray, G. Burger, and B. F. Lang. 2007. TBestDB: a taxonomically broad database of expressed sequence tags (ESTs). Nucleic Acids Res. 35:D445-D451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Odelson, D. A., and J. A. Breznak. 1985. Nutrition and growth characteristics of Trichomitopsis termopsidis, a cellulolytic protozoan from termites. Appl. Environ. Microbiol. 49:614-621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ohkuma, M. 2003. Termite symbiotic systems: efficient bio-recycling of lignocellulose. Appl. Microbiol. Biotechnol. 61:1-9. [DOI] [PubMed] [Google Scholar]
- 27.Ohkuma, M., T. Iida, O. Kuniyo, H. Yuzawa, S. Noda, E. Viscogliosi, and T. Kudo. 2005. Molecular phylogeny of parabasalids inferred from small subunit rRNA sequences, with emphasis on the Hypermastigea. Mol. Phylogenet. Evol. 35:646-655. [DOI] [PubMed] [Google Scholar]
- 28.Ohkuma, M., K. Ohtoko, T. Iida, M. Tokura, S. Moriya, R. Usami, K. Horikoshi, and T. Kudo. 2000. Phylogenetic identification of hypermastigotes, Pseudotrichonympha, Spirotrichonympha, Holomastigotoides, and parabasalian symbionts in the hindgut of termites. J. Eukaryot. Microbiol. 47:249-259. [DOI] [PubMed] [Google Scholar]
- 29.Ohkuma, M., K. Saita, T. Inoue, and T. Kudo. 2007. Comparison of four protein phylogeny of parabasalian symbionts in termite guts. Mol. Phylogenet. Evol. 42:847-853. [DOI] [PubMed] [Google Scholar]
- 30.Payne, M. J., A. Chapman, and R. Cammack. 1993. Evidence for an [Fe]-type hydrogenase in the parasitic protozoan Trichomonas vaginalis. FEBS Lett. 317:101-104. [DOI] [PubMed] [Google Scholar]
- 31.Peters, J. W., W. N. Lanzilotta, B. J. Lemon, and L. C. Seefeldt. 1998. X-ray crystal structure of the Fe-only hydrogenase (CpI) from Clostridium pasteurianum to 1.8 angstrom resolution. Science 282:1853-1858. [DOI] [PubMed] [Google Scholar]
- 32.Posewitz, M. C., P. W. King, S. L. Smolinski, L. Zhang, M. Seibert, and M. L. Ghirardi. 2004. Discovery of two novel radical S-adenosylmethionine proteins required for the assembly of an active Fe hydrogenase. J. Biol. Chem. 279:25711-25720. [DOI] [PubMed] [Google Scholar]
- 33.Pütz, S., P. Dolezal, G. Gelius-Dietrich, L. Bohacova, J. Tachezy, and K. Henze. 2006. Fe-hydrogenase maturases in hydrogenosomes of Trichomonas vaginalis. Eukaryot. Cell 5:579-586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shimodaira, H., and M. Hasegawa. 2001. CONSEL: for assessing the confidence of phylogenetic tree selection. Bioinformatics 17:1246-1247. [DOI] [PubMed] [Google Scholar]
- 35.Sugimoto, A., T. Inoue, I. Tayasu, L. Miller, S. Takeichi, and T. Abe. 1998. Methane and hydrogen production in a termite-symbiont system. Ecol. Res. 13:241-257. [Google Scholar]
- 36.Todaka, N., S. Moriya, K. Saita, T. Hondo, I. Kiuch, H. Takasu, M. Ohkuma, C. Piero, Y. Hayashizaki, and T. Kudo. 2007. Environmental cDNA analysis of the genes involved in lignocellulose digestion in the symbiotic protist community of Reticulitermes speratus. FEMS Microbiol. Ecol. 59:592-599. [DOI] [PubMed] [Google Scholar]
- 37.Vignais, P. M., B. Billoud, and J. Meyer. 2001. Classification and phylogeny of hydrogenases. FEMS Microbiol. Rev. 25:455-501. [DOI] [PubMed] [Google Scholar]
- 38.Voncken, F. G., B. Boxma, A. H. van Hoek, A. S. Akhmanova, G. D. Vogels, M. Huynen, M. Veenhuis, and J. H. Hackstein. 2002. A hydrogenosomal [Fe]-hydrogenase from the anaerobic chytrid Neocallimastix sp. L2. Gene 284:103-112. [DOI] [PubMed] [Google Scholar]
- 39.Yamin, M. A. 1981. Cellulose metabolism by the flagellate Trichonympha from a termite is independent of endosymbiotic bacteria. Science 211:58-59. [DOI] [PubMed] [Google Scholar]
- 40.Yoshimura, T., J.-I. Azuma, K. Tsunoda, and M. Takahashi. 1993. Cellulose metabolism of the symbiotic protozoa in termite, Coptotermes formosanus Shiraki (Isoptera: Rhinotermitidae). I. Effect of degree of polymerization of cellulose. Mokuzai Gakkaishi 39:221-226. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.