Lipids are essential and highly abundant components of all organisms. They serve as the main building blocks of the membranes that surround and compartmentalize cells, as a store for energy and reduction power, and in posttranslational modifications that regulate the localization and function of a large number of proteins. Lipids have also emerged as important pathogenesis factors in a variety of infectious diseases. The ability to synthesize or salvage simple components like fatty acids and isoprenoids and assemble them into more complex molecules is critical for the growth and development of the pathogen and required for its ability to colonize the host and to cause disease. Here we will focus on the fatty acid metabolism of apicomplexans, a group of unicellular eukaryotes that have adapted to an obligate intracellular life style. Infections with apicomplexan parasites are the cause of several important human diseases (malaria, toxoplasmosis, and cryptosporidiosis) affecting literally millions of people around the globe. Control efforts for these diseases face an uphill battle, as effective vaccines are lacking for any of these parasites and drug treatment is not fully effective (cryptosporidiosis), limited to the acute stage (toxoplasmosis), or constantly threatened by emerging drug resistance (malaria). The completion of full genome sequencing efforts for a considerable number of apicomplexans has reinvigorated the search for targets of novel therapeutics. Lipid and fatty acid metabolism has emerged at the heart of several intensively studied areas of apicomplexan biology, including parasite adaptation to different hosts, host-parasite interaction at the cellular level, and the identification of divergent pathogen-specific targets for antiparasitic drug therapy. Here we will provide an overview of the surprisingly diverse mechanisms of fatty acid synthesis (FAS) and uptake used by the members of this important group of pathogens.
MAKE IT I: THE APICOPLAST TYPE II FATTY ACID SYNTHESIS PATHWAY
Apicomplexans were thought to lack the ability to synthesize fatty acids de novo and to depend entirely on salvaging fatty acids and several more complex lipids from their hosts (71). Initially, radioisotope experiments designed to detect de novo synthesis did not produce detectable labeling, while evidence for fatty acid and lipid uptake from the infected host cell was abundant (see below for detail). This model came into question with the discovery of the apicoplast, a chloroplast-like organelle that is likely the descendant of an endosymbiotic red alga (32, 38, 76). Plant and algal plastids are not only the home of photosynthesis but also harbor several other key biosynthetic pathways. Genomic and experimental analyses have identified pathways for the synthesis of fatty acids (73), isoprenoids (26), and heme (55) in the apicoplast. FAS in the apicoplast depends on a prokaryotic type II (FASII) system, previously characterized in bacteria and chloroplasts. A complete set of genes encoding the components of FASII have been identified in Plasmodium falciparum (51, 73), Toxoplasma gondii (73), and Eimeria tenella (15) (see Table 1 for a comparative overview of apicomplexan genes associated with fatty acid metabolism). Phylogenetic analyses strongly support the chloroplast origin of these genes, and apicoplast targeting has been experimentally demonstrated for several of the encoded proteins (73).
TABLE 1.
Comparative genomic analysis of fatty acid metabolism in the Apicomplexaa
| Pathway and gene product (localization) | ApiDB gene IDb
|
|||
|---|---|---|---|---|
| Toxoplasma gondii | Plasmodium falciparum | Theileria annulata | Cryptosporidium parvum | |
| FASII | + | + | − | − |
| ACP (plastid) | 55.m00019 | PFB0385w | ||
| ACP PPT | 20.m03796 | PFD0980w | ||
| ACP (put. mitochondrion) | 59.m03510 | PFL0415w | ||
| FabB/F | 83.m00002 | PFF1275c | ||
| FabD | 42.m03469 | PF13_0066 | ||
| FabH | 44.m00012 | PFB0505c | ||
| FabI | 50.m00011 | PFF0730c | ||
| FabG | 37.m00770 | PFI1125c | ||
| FabZ | 645.m00003 | PF13_0128 | ||
| FASI and PKS | + | − | − | + |
| FASI | 83.m00010 | cgd3_2180 | ||
| Put. PKS | 20.m03875 and additional adjacent models | cgd4_2900 | ||
| Put. PKS | 46.m01725 | |||
| SFP PPT | 33.m01284 | cgd8_4740 | ||
| ELO | + | + | − | + |
| ELO | 49.m03288 | PFF0290w | ||
| ELO | 20.m00392 | PFA0455c | cgd8_4630 | |
| ELO | 52.m01617 | PFI0980w | ||
| Malonyl-CoA | + | + | − | + |
| ACCase1 | 41.m00004 | PF14_0664 | ||
| ACCase2 | 38.m02529 | cgd8_3680 | ||
| Acetyl-CoA | ||||
| PDH, E2 subunit | 20.m00373 | PF10_0407 | ||
| Acetyl-CoA synthetase | 57.m03124 | PFF1350c | TA07295 | cgd1_3710 |
| Acyl-CoA | + | + | + | + |
| Acyl-CoA binding protein | 46.m01612, 50.m03399 | PF08_0099, PF10_0015, PF10_0016, PF14_0749 | TA19910 | cgd1_1140 |
| Acyl-CoA synthetase | 113.m00767, 50.m00056, 583.m00605, 583.m05329, 49.m03372, 44.m00014 | PF14_0761, PF14_0751, PFC0050c, PFL2570w, MAL13P1.485, PF07_0129, PFD0085c, PFL0035c, PFB0695c, PFB0685c*, PFE1250w, PFL1880w, PFF0945c | TA18970, TA15995, TA20445, TA07295 | cgd4_3400, cgd3_640, cgd1_3710 |
The genomes of four apicomplexan species were mined for a broad range of fatty acid metabolism-associated genes using the BLAST algorithm and manual inspection. Note that many of these genes have been previously annotated and published (please see text for individual references). +, present; −, absent; put., putative; PPT, phosphopantetheinyl transferase; SFP, surfactin production type.
Gene IDs are from ApiDB at http://www.apiDB.org.
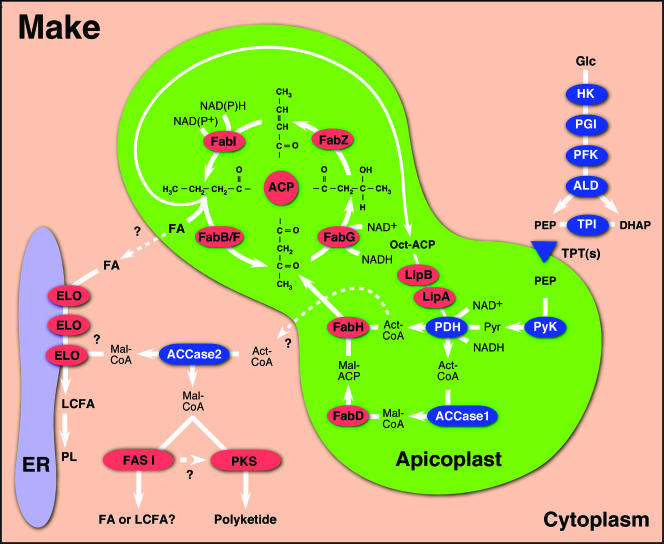
The core of the FASII system is acyl carrier protein (ACP) (Fig. 1). Acetate is transferred to the phosphopantetheine prosthetic group of ACP from acetyl coenzyme A (acetyl-CoA) by FabD. To this primer, two carbon units are added by FabH through condensation with malonyl-ACP, which are subsequently reduced, dehydrated, and again reduced by the sequential actions of FabZ, FabG, and FabI. The fully reduced acyl chain is then transferred and condensed to malonyl-ACP by FabB/F, thus initiating the next round of reduction reactions. The repetition of this process, initiated each time by malonyl condensation with the growing chain, leads to the formation of even-numbered fatty acids, typically palmitic (C16) or myristic (C14) acid.
FIG. 1.
A schematic outline of fatty acid synthesis in apicomplexans. Enzymes that are engaged in fatty acid synthesis are shown in red, those providing precursors in blue. Glycolysis in the cytoplasm produces phosphoenolpyruvate (PEP), which is imported into the apicoplast by triosephosphate transporters (TPTs) and converted to acetyl-CoA by the subsequent action of PK and PDH. Acetyl-CoA (Act-CoA) and malonyl-CoA (Mal-CoA) produced by ACCase are funneled into the apicoplast FASII pathway (FabD, -H, -G, -Z, -I, and -B/F), resulting in the formation of fatty acids (FA) and octanoyl-ACP (Oct-ACP). The latter is the precursor for PDH lipoylation. The activity of endoplasmic reticulum (ER)-resident ELO results in the production of long-chain fatty acids (LCFA) used for phospholipid (PL) synthesis. Substrates for ELO might be host cell-derived or apicoplast-produced fatty acids (the source of malonyl-CoA is unclear; in T. gondii and C. parvum, it might be supplied by ACCase2). FASI likely acts as an elongating enzyme, and the products of PKS are currently unknown (the presence of FASI, PKS, and ACCase2 appears linked). Note that these pathways occur in different combinations in different apicomplexans and that the compartmentalization and activity shown for some of the enzymes has not been experimentally validated yet. HK, hexokinase; PGI, phosphoglucose isomerase; PFK, phosphofructokinase; ALD, aldolase; TPI, triose phosphate isomerase; DHAP, dihydroxyacetone phosphate.
Mammalian cells lack FASII and instead rely on FASI, a gigantic protein that carries ACP, as well as the various enzymatic activities that act on the growing chain described above, on a single polypeptide. FASI and FASII differ in structure, kinetics, and susceptibility to several inhibitors, making the apicoplast FASII an attractive target for the development of parasite-specific drugs. Indeed, several lines of evidence indicate that the apicoplast FASII pathway is essential for parasite viability. Conditional knockout experiments using a tetracycline-regulated promoter in T. gondii have demonstrated that the expression of apicoplast ACP is essential for parasite survival in tissue culture (37). Furthermore, mice infected with a lethal dose of mutant parasites can be cured by tetracycline-mediated FASII suppression. Consistently, pharmacological studies using drugs that preferentially inhibit FASII over FASI enzymes (e.g., triclosan or thiolactomycin) have found strong growth inhibition in Plasmodium and Toxoplasma (39, 49, 63). The interaction of Pf- and TgFabI (P. falciparum and T. gondii FabI) with triclosan has been studied in considerable detail. Kinetic and structural data support FabI as the target enzyme in these parasites and have identified residues critical to inhibitor binding (29, 46, 49). Based on these data, several groups have embarked on structure- and screening-based programs to identify novel inhibitors of PfFabI with increased potency and specificity (8, 48). Recently FabB/F has been characterized and confirmed as the target of thiolactomycin, a FASII-specific inhibitor with demonstrated efficacy against T. gondii and P. falciparum (27, 37, 74, 75). Structural and kinetic information is now also available for recombinant PfFabG (75) and PfFabZ (56, 64). Overall, it is clear now that apicoplast FASII is essential and a validated drug target. The enzymology of the pathway is well established, and excellent reagents are available for compound screening and structure-activity relationship analysis (references 21 and 62 provide in-depth reviews of current drug development efforts focused on the Plasmodium FASII pathway).
While the apicoplast FASII pathway is clearly essential, the function of FASII and its integration into the overall parasite metabolism is less clear. The chloroplast is the main source of fatty acids in the plant cell, and bulk fatty acid production is therefore the obvious first hypothesis. Using P. falciparum-infected blood cultures, Surolia and Surolia demonstrated the incorporation of 14C-acetate into fatty acids, preferentially resulting in the production of C10 to C14 chains. Importantly, this incorporation is sensitive to triclosan inhibition (63). The observed chain length is in agreement with the substrate preference of the PfFabB/F enzyme, which most effectively elongates C6 to C10-ACP chains, with reduced activity for C12 and C14 chains and no activity for C16 or longer chains (33). In T. gondii, acetate labeling equally resulted in robust incorporation of the label into fatty acids and a variety of lipids (5, 37). However, in contrast to P. falciparum, the labeling is restricted to C16, C18, and longer-chain fatty acids. Importantly, genetic ablation of the apicoplast FASII pathway by conditional knockdown of ACP does not affect this incorporation. This suggests that FASII is not responsible for the observed incorporation (37). Consistent with this finding, incorporation is also insensitive to thiolactomycin (a FASII-specific inhibitor) but susceptible to cerulenin (a general FAS inhibitor) suggesting that other FAS systems metabolize the label (see below).
In addition to providing fatty acids, FASII is also required for the de novo synthesis of lipoic acid. Lipoic acid is an essential cofactor for a number of oxidative decarboxylases, including pyruvate dehydrogenase (PDH), the glycine cleavage system, α-ketoglutarate dehydrogenase, and branched-chain oxo-acid dehydrogenase. It is therefore possible that the critical function of apicoplast FASII might lie in lipoic acid production, as has been suggested for the FASII pathway of the plant mitochondrion (22). The FAS intermediate octanoic acid-ACP is the starting substrate for this pathway. LipB transfers octanoic acid from ACP to the E2 subunit of PDH and other enzyme complexes. LipA then introduces two sulfur atoms at the distal end of the chain to form lipoic acid (see reference 14 for a review of the initial mechanistic characterization of this pathway in Escherichia coli). Homologs of LipA and -B have been identified in Plasmodium and Toxoplasma, and both enzymes were shown to localize to the apicoplast (65, 77). Pharmacological (12) and genetic (37) disruption of FASII in T. gondii results in a loss of PDH lipoylation, supporting the essential role of FASII in this pathway. However, disruption of FASII has no effect on the lipoylation of mitochondrial enzymes. These enzymes depend on independent LplA-mediated salvage of lipoic acid from the host (2, 12, 37, 77). It therefore appears that apicoplast PDH is the only beneficiary of FASII-dependent lipoic acid synthesis. It is not clear whether apicoplast PDH is required for functions beyond providing acetyl-CoA for FASII. In most eukaryotes, PDH is an essential mitochondrial enzyme complex linking glycolysis to the tricarboxylic acid cycle. Apicomplexans appear to lack mitochondrial PDH (16, 17), and one could speculate that metabolites might be exchanged between plastid and mitochondrion, which are closely apposed through much of the intracellular development of the parasites (69, 70). However, the finding that an apicomplexan that lacks the FASII pathway, Theileria, equally lacks PDH could argue against this model and suggest that the critical role of PDH likely is to provide acetyl-CoA for fatty acid synthesis (19). It is important to bear in mind that the lack of FASII-dependent radiolabeling in T. gondii does not exclude the possibility that FASII produces bulk fatty acids. It is, e.g., possible that the Toxoplasma and Plasmodium apicoplasts differ in their ability to import acetate, thus precluding labeling. Lastly, one might consider apicoplast-specific roles for the FASII pathway. Supporting such a view is the observation that FASII inhibition results in pronounced apicoplast biogenesis defects in T. gondii (37).
MAKE IT II: ALTERNATIVE ROUTES OF FATTY ACID SYNTHESIS THROUGH FASI AND PKS-LIKE ENZYMES
As described above, acetate incorporation into fatty acids is independent of the FASII pathway in T. gondii. What other system could be responsible for this activity? An alternative pathway has been described in the related apicomplexan Cryptosporidium parvum. Cryptosporidium lacks both the apicoplast organelle and the associated FASII pathway, and instead, its genome encodes a very large (∼900-kDa) FASI-like protein (1, 83, 84). This enzyme contains four ACP domains and is organized into a loading unit, three elongation and reduction modules, and a putative terminating reductase domain (84). Phylogenetic analysis suggests that this enzyme is more closely related to bacterial polyketide synthases (PKSs) than to eukaryotic FASI (80). The massive size of this gene makes studying FASI challenging. However, in a molecular biology tour de force, Zhu and colleagues have cloned and expressed C. parvum FASI (CpFASI) in E. coli, broken down into five maltose binding fusion protein modules (82). Although the ACP units failed to be activated to holo-ACP in this system (they are apparently not recognized by the E. coli phosphopantetheinyl transferase), the enzyme activities could be measured by using CoA as an alternative acceptor. Substrate inhibition studies of the recombinant loading unit indicated fatty acids with chain lengths of C12 to C24 as potential substrates, with a pronounced preference for C16. Further studies showed that all three elongation units are active. Taken together, these data suggest that the role of CpFASI is likely not de novo synthesis but the elongation of C16 to C22 chain-length fatty acids. A second closely related gene identified in C. parvum encodes a putative PKS (81). Like CpFASI, this enzyme prefers palmitoyl-CoA acid as a starter substrate (18). The protein contains seven acyl elongation modules of varied enzyme domain composition, and the end product of the reaction catalyzed by CpPKS is currently unknown. CpPKS is even larger than FASI (∼1,300 kDa), making the identification of its product based on recombinant expression a nontrivial task. PKs are synthesized by bacteria and fungi and serve a broad range of functions, including as antibiotics, siderophores, and toxins. The only other protozoan group currently known to produce PKs is the apicomplexan sister phylum of the dinoflagellates. Dinoflagellates produce a multitude of PK toxins, and the first dinoflagellate PKS gene has recently been identified from Karenia brevis. This gene was shown to be most similar to CpPKS (53, 60), suggesting that an acquisition of bacterial PKS genes by horizontal transfer occurred before the split of apicomplexans and dinoflagellates. Alternatively, these genes might have been present in early eukaryotes but have been lost in all other groups. The T. gondii genome contains three large FASI/PKS-type genes. Preliminary analyses of the domain structure predicted one of these genes (ToxoDB gene ID 83.m00010) to encode a FASI (13, 80), while the two other genes are likely for PKSs. TgFASI could be responsible for the FASII-independent 14C-acetate incorporation observed in T. gondii, based on the putative chain length of its products and the susceptibility of the enzyme to cerulenin (based on analysis of its homolog CpFASI [82]). However, there are alternative hypotheses for the biological role of this gene. Looking at the distribution of FASI and PKS genes in the Apicomplexa, it appears that these genes are linked and restricted to species that form oocysts that are shed into the environment (Cryptosporidium, Toxoplasma, and Eimeria). One might speculate that the products of FASI and PKS serve a function in these life cycle stages, e.g., as a component of the impenetrable and highly resilient oocyst wall or as a factor required for development within the vertebrate intestinal epithelium.
MAKE IT III: ENDOPLASMIC RETICULUM-RESIDENT FATTY ACID SYNTHESIS/ELONGATION THROUGH THE ELONGATION SYSTEM
A third avenue for fatty acid synthesis is represented by the fatty acid elongation system (fatty acid elongases [ELO]). The enzymatic steps involved in the ELO process are similar to those in FASI and -II, but the growing chain is held by CoA instead of ACP. As implied by the name, the main role of this system, which is found in the endoplasmic reticulum of most eukaryotes, is to sequentially elongate fatty acids to supply the demand for long-chain fatty acids for the production of phospholipids (see reference 36 for an in-depth review). An analogous system of divergent phylogenetic origin is responsible for the synthesis of wax precursors in plants (68). Recent elegant work in the study of Trypanosoma brucei (the causative agent of sleeping sickness and nagana) has demonstrated that the ELO in this organism has been adapted to the de novo synthesis of fatty acids (34, 35). While the three yeast ELOs add two two-carbon units each to C14, C18, and C22, T. brucei ELO1 and -2 act on considerably shorter starters and elongate C4 to C10 and C10 to C14, respectively. Genetic and biochemical data support a model in which ELO1 and -2, and not the mitochondrial FASII system, are the critical source of myristic acid for the glycosylphosphatidylinositol anchor of the highly abundant variable surface glycoprotein of the bloodstream stage (35, 61). Genomic searches identify three ELO genes in both T. gondii and P. falciparum and a single gene in Cryptosporidium. The activity of these genes could be an alternative source of FASII-independent 14C-acetate incorporation in T. gondii (5, 37), as ELOs produce preferentially longer fatty acids and are sensitive to cerulenin (34). While they have not been characterized to date, it seems likely that, in contrast to T. brucei, the apicomplexan ELOs engage in conventional elongation rather than de novo synthesis.
PRECURSOR SUPPLY CHAIN FOR FATTY ACID SYNTHESIS
Fatty acid synthesis requires acetyl-CoA, malonyl-CoA, energy, and reduction power in the form of ATP and NAD(P)H. It has been proposed that cytoplasmic glycolysis supplies phosphoenolpyruvate, which is imported into the plastid and subsequently metabolized to acetyl-CoA (51) (schematic outline in Fig. 1). This genome-based model has recently received considerable experimental support. Two putative apicoplast phospho-sugar transporters have been identified in P. falciparum (47). One of them is a component of the outermost membrane, while the second one (which carries a leader sequence) likely resides in the innermost membrane. Note that the apicoplast is bounded by four membranes, a telltale of its secondary endosymbiotic origin (see reference 67 for a high-resolution electron microscopy tomographic reconstruction of the apicoplast membranes). Curiously, T. gondii harbors only a single homologue of this protein but appears to achieve targeting of this protein to multiple membranes of the apicoplast (16, 30). The homologous plant chloroplast antiporters have the ability to transport a range of C3, C5, and C6 phosphorylated carbohydrates across the inner plastid membrane (31). It has been proposed that phosphoenolpyruvate is a likely apicoplast import substrate, but this remains to be established experimentally (16, 30, 47). A strong argument for triose phosphates being imported into the apicoplast lies in the demonstration of triose phosphate-metabolizing enzymes, like pyruvate kinase, triosephosphate isomerase, and more tentatively, glyceraldehyde 3-phosphate dehydrogenase, in the apicoplast (16). Under this scenario, the apicoplast will have a supply of phosphoenolpyruvate and pyruvate, as well as dihydroxyacetone phosphate. Dihydroxyacetone phosphate likely feeds into the apicoplast isoprenoid synthesis pathway, while pyruvate is converted to acetyl-CoA by PDH to funnel into FASII. As discussed above, apicomplexans harbor a single PDH complex, and all four subunits have been localized to the apicoplast (16, 17, 40). Importantly, pyruvate kinase and PDH not only provide acetyl-CoA but also generate ATP and NAD(P)H, providing a means of shuttling reducing equivalents into the organelle. Acetyl-CoA is carboxylated to malonyl-CoA under the consumption of bicarbonate and ATP by acetyl-CoA carboxylase (ACCase). Two phylogenetically distinct ACCases have been identified in the plant chloroplast: ACCaseI, a large, multifunctional polypeptide homologous to ACCase in fungi and animals, and ACCaseII, a complex of three polypeptides homologous to the bacterial ACCase complex (52). In most plants, the chloroplast harbors an ACCaseII, while grasses have apparently rerouted a copy of the cytoplasmic ACCaseI to the plastid (a difference that has been extensively exploited for herbicide development [62]).
Large fragments of two ACCase genes have been cloned from T. gondii, both of which represent type I enzymes (23, 24). The genome predicts the two ACCase proteins to be of ∼280 (ACCase1) and 365 (ACCase2) kDa. A plastid localization of at least one of these two proteins is likely, based on the strong labeling of the apicoplast with streptavidin, which reacts to the biotin group found in ACCases (23). Western blot analysis detected only the smaller of these two proteins in extracts of T. gondii tachyzoites, and the current gene model for this gene shows a canonical apicoplast leader. Genome comparison among apicomplexans indicates that the presence of ACCaseII is linked to the presence of FASI and PKS. It is therefore tempting to speculate that ACCaseII provides malonyl-CoA for these enzymes. Further studies establishing the localization of the product and expression over the life cycle of these genes are needed to test this hypothesis. A related open question is the source of malonyl-CoA for the elongase system. In Plasmodium, the single ACCase is likely localized to the apicoplast, suggesting that this is the only compartment where malonyl-CoA production can occur. Malonyl-CoA might be exported from the apicoplast to the cytoplasm, or there may be an alternative ACCase-independent source yet to be characterized (this question remains equally unanswered for T. brucei, where the sole ACCase is likely localized to the mitochondrion [35]).
TAKE IT: FATTY ACID SALVAGE FROM HOST CELL
There is strong evidence for the uptake of fatty acids in essentially all apicomplexans that have been analyzed. The incubation of T. gondii or P. falciparum with radio- or fluorescence-labeled fatty acids results in the incorporation of the label within the parasite (7, 20, 66). In T. gondii, such incorporation occurs in free tachyzoites, as well as in infected host cells, and has also been observed for a variety of phospholipids. Both parasites convert fatty acids taken up from the host to triacylglycerides using an endoplasmic reticulum-resident acyl-CoA:diacylglycerol acyltransferase, with these triglycerides being subsequently stored in lipid bodies (7, 50, 72).
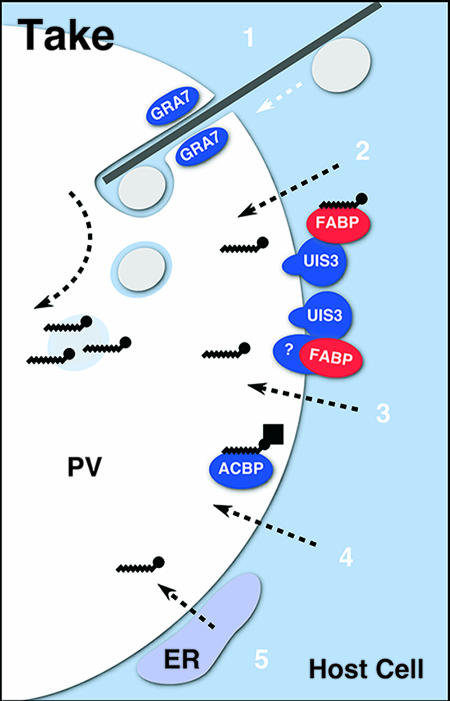
What are the cellular mechanisms of fatty acid and, more generally, lipid salvage? In considering this question, it is important to note that different apicomplexan species inhabit host cells with different architectures and metabolic capabilities. It is therefore conceivable that varied and divergent mechanisms exist (Fig. 2 shows a schematic overview of putative uptake mechanisms). A simple model could assume the unmediated transit of fatty acids across membranes, based on their hydrophobicity. Apicomplexa encode a variety of acyl-CoA synthases and acyl-CoA binding proteins that might act in salvage and transport (in P. falciparum, these genes have been amplified by gene duplication and show considerable polymorphism, hinting at strong evolutionary selection [4]). C. parvum encodes a single acyl-CoA binding protein. This protein contains an ankyrin domain in addition to the acyl-CoA binding domain and preferentially binds palmitoyl-CoA, the precursor of C. parvum FASI and PKS (78). Most interestingly, this protein, as well as a C. parvum oxysterol binding protein, has been localized to the parasitophorous vacuole (PV), an observation that led Zhu and colleagues to propose a direct role in lipid salvage from the host cell (78-80).
FIG. 2.
Apicomplexan fatty acid and lipid uptake mechanisms. Host cell cytoplasm is shown in blue, the PV in white. Parasite proteins are shown in blue, host cell proteins in red. (1) Toxoplasma has recently been shown to trap endosomal/lysosomal vesicles (gray ovals) in the PV, and host cell microtubules and the parasite protein GRA7 have been implicated in this process. These vesicles could be a source not only of sterols but also of other lipids as sources of fatty acids. (2 and 3) The Plasmodium liver schizont targets a protein to the PV membrane (UIS3) that binds host cell fatty acid binding protein (FABP). This might aid fatty acid uptake by simple diffusion (through the PV membrane or PV pores) or involve additional parasite-derived transporters (labeled “?”). (4) Cryptosporidium secretes an acyl-CoA binding protein (ACBP) into the PV. (5) The PV recruits the host cell endoplasmic reticulum (ER) and mitochondria (not shown) to its membrane, which might facilitate the salvage of fatty acids and other lipids. Note that direct biochemical evidence for a role in fatty acid uptake of these mechanisms is still largely missing; no information is available on how lipids cross the parasite's plasma membrane.
In Toxoplasma, it has been long noticed that the PV recruits and establishes intimate contact with the host cell's endoplasmic reticulum and mitochondria (58, 59), and similar findings have recently been reported for the hepatocyte stage of Plasmodium (3). Such intimate association has been shown to be crucial for lipid uptake in a variety of bacterial pathogens (54). A second such interaction with the secretory pathway of the host cell has been described for the host cell endosome/lysosome. This model is based on extensive studies of sterol uptake in T. gondii (for which the parasites lack biosynthetic pathways). Intracellular parasites readily take up fluorescently labeled sterols supplied in the medium. In experiments taking advantage of mutant host cells, Coppens and colleagues concluded that the parasite intercepts the endosome/lysosome pathway to secure low-density lipoprotein-derived cholesterol (9, 11). In more recent work, these authors identify microtubular projections from the host cell cytoplasm into the space of the PV. Host cell endocytic structures which travel along these microtubules are trapped within the vacuolar space, with the parasite protein GRA7 mediating this sequestration process (10). This pathway could be a conduit not only for cholesterol but also for other lipids. The lysosome-trapping model at least partially contradicts the previously held (and experimentally supported) view that the PV is fusion incompetent and hence isolated from the host cell secretory and endosomal pathway (25, 42, 43). Further studies are needed to unify these two models.
The most recent model for fatty acid uptake emerged from expression profiling along the Plasmodium life cycle. This work has identified numerous genes that are upregulated in the infective sporozoite (UIS) compared to the blood cell stage (28). Gene-targeting experiments have validated this finding and demonstrated that, while dispensable in the erythrocyte, they are essential for development in the hepatocyte earlier in infection (44, 45). Infection with these mutants resulted in strong protection to subsequent challenge, a fact that has rekindled the interest in the development of a live attenuated malaria vaccine. One of these genes, UIS3, encodes a type I transmembrane protein that is secreted and inserted into the PV membrane in infected hepatocytes so that its C-terminal domain is exposed to the host cell's cytoplasm (41). A yeast two-hybrid screen performed using the exposed domain as bait found strong interaction between UIS3 and liver fatty acid binding protein, a finding that was confirmed by coimmunoprecipitation from infected cells. This interaction is critical for the parasite as removing either partner by gene targeting (UIS3) or RNA interference (L-FABP) blocks development of the liver cell schizont (41, 44, 45). Based on these observations, it is likely that UIS3 promotes fatty acid uptake from the host cell. The recruitment of fatty acid-loaded L-FABP to the PV membrane could increase the availability of fatty acids to passive or active transport mechanisms. Exactly how this occurs remains to be established, but the UIS gene catalog provides an exciting list of candidate genes to dissect this important process.
WHAT IS THE REASON FOR THE SURPRISING DIVERSITY OF FATTY ACID METABOLISM ACROSS THE PHYLUM?
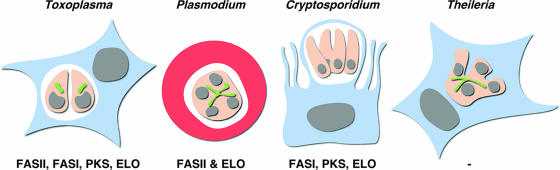
Genomic analysis leads us to a stark reversal of the initial assumption of lack of fatty acid synthesis in apicomplexans. As discussed above, members of this phylum engage three independent fatty acid synthesis machineries (FASI, FASII, and ELO). A second surprise is the diversity of fatty acid metabolism in different species of the phylum (Table 1 and Fig. 3). Theileria is the apicomplexan fatty acid minimalist. This parasite not only lacks de novo synthesis capabilities but also is unable to elongate fatty acids (19). Consistent with the lack of fatty acid synthesis, Theileria also lacks the enzymes to produce malonyl-CoA, acetyl-CoA (through PDH—there is an acetyl-CoA synthase likely used for other purposes), and ACP-holoenzyme synthase (see Table 1). It has retained a single fatty acid binding protein and several acyl-CoA synthases, pointing to their potential role in salvage. Why can Theileria thrive without pathways essential to other apicomplexans? One obvious difference is the cytoplasmic niche occupied by this parasite (57). The absence of a PV allows for a more intimate contact with the host cell and might facilitate uptake and/or manipulation of the host pathways to suit the needs of the parasite. Cryptosporidium is another apicomplexan known for its stripped-down biosynthetic metabolism: the apicoplast and its metabolic capabilities, including a FASII system, have been lost in this organism (1, 83). However, the parasite has maintained a FASI system (albeit for elongation and not de novo synthesis), a PKS, and a single ELO. Plasmodium uses an apicoplast FASII for de novo synthesis of fatty acids, and this synthesis is essential to parasite survival. Nonetheless, this parasite also engages in fatty acid uptake and elongation. The Plasmodium genome encodes a host of acyl-CoA synthases and binding proteins that appear to be under selective pressure (4). In the hepatocyte, the uptake system is further enhanced by a protein that recruits host fatty acid binding proteins to the PV (this protein appears unique to Plasmodium). Lastly, Toxoplasma boasts the broadest set of fatty acid-related genes. A full complement of pathways (not only for the synthesis of fatty acids) is likely critical to the generalist lifestyle adopted by Toxoplasma, a parasite capable of infecting a large variety of animals and cell types that likely present different nutritional challenges. A FASII pathway in T. gondii synthesizes fatty acids and lipoic acid de novo (however, it is uncertain to what extent these fatty acids contribute to the overall bulk fatty acid production in this parasite). Radiolabeling studies show the uptake and modification of host cell-derived fatty acids, likely linked to the presence of acyl-CoA synthases, binding proteins, and three ELO genes. A FASI and two PKS genes of yet-to-be-determined function add to the parasite's capabilities. These genes are likely localized outside the apicoplast, a view supported by the presence of a second ACCase (providing malonyl-CoA), and a second ACP-holoenzyme synthase (6).
FIG. 3.
Diversity of host cell niches and fatty acid synthesis pathways in apicomplexans. Schematic and highly simplified depictions of the intracellular environment of the parasite stage responsible for pathogenesis in the mammalian host is shown for each of four apicomplexan species (additional stages with different niches exist for all parasites). The host cell is shown in blue or red, parasites in pink, apicoplast in green, nuclei in gray, and the PV in white. Fatty acid synthesis pathways identified in the genome of each parasite are shown below. Note that the expression of these pathways across the life cycle has not been studied in detail and not all might be active in the stage shown. The morphology of the apicoplast in Theileria has not been studied, and the depiction is therefore hypothetical.
The diversity of the pathways throughout the phylum might be taken as an indication that the availability of (the right) fatty acids is an essential determinant to successful host adaptation for apicomplexans. Parasites inhabiting metabolically largely inactive environments like the red blood cell might depend more heavily on de novo synthesis, while those having ready access to host fatty acids lean towards uptake. It also appears that different ways to acquire fatty acids have evolved in parallel in different species, and much remains to be learned about the mechanistic detail of these processes. While the ability to mine comparatively the genomes of a variety of apicomplexans has led to the discovery of numerous pathways, the precise biological and metabolic function of each pathway remains largely unexplored. Also, our understanding of the cellular compartmentalization of the pathways and their precursor supply chains is rudimentary. The example of Plasmodium suggests that dramatic metabolic changes occur along the life cycle progression and that not all enzymes encoded in the genome might be used in the most widely studied culture models. However, strong postgenomic tools, like expression profiling, proteomics, and metabolomics, are now available to the field to tackle these questions. Combining these tools with our increased ability to genetically modify apicomplexans (currently limited to Toxoplasma, Plasmodium, and Eimeria) should permit the dissection of what is emerging as a complex yet critically important component of host-parasite interaction. Importantly, fatty acid synthesis and uptake are essential to parasite growth and pathogenesis, and ongoing work on the Plasmodium and Toxoplasma FASII pathway demonstrates that fatty acid metabolism holds promise for parasite-specific drug targets. Additional targets might emerge from a more detailed understanding of uptake pathways.
Acknowledgments
We are grateful to Giel van Dooren for many discussions and comments on the manuscript. Preliminary T. gondii genomic and/or cDNA sequence data were accessed via http://ToxoDB.org.
Genomic data were provided by The Institute for Genomic Research (supported by National Institutes of Health grant AI05093) and the Sanger Institute (Wellcome Trust). Work in our laboratory is currently funded by grants from the National Institutes of Health to B.S. (grants AI 55268 and 64671).
Footnotes
Published ahead of print on 22 August 2007.
REFERENCES
- 1.Abrahamsen, M. S., T. J. Templeton, S. Enomoto, J. E. Abrahante, G. Zhu, C. A. Lancto, M. Deng, C. Liu, G. Widmer, S. Tzipori, G. A. Buck, P. Xu, A. T. Bankier, P. H. Dear, B. A. Konfortov, H. F. Spriggs, L. Iyer, V. Anantharaman, L. Aravind, and V. Kapur. 2004. Complete genome sequence of the apicomplexan, Cryptosporidium parvum. Science 304:441-445. [DOI] [PubMed] [Google Scholar]
- 2.Allary, M., J. Z. Lu, L. Zhu, and S. T. Prigge. 2007. Scavenging of the cofactor lipoate is essential for the survival of the malaria parasite Plasmodium falciparum. Mol. Microbiol. 63:1331-1344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bano, N., J. D. Romano, B. Jayabalasingham, and I. Coppens. 2007. Cellular interactions of Plasmodium liver stage with its host mammalian cell. Int. J. Parasitol. 37:1329-1341. [DOI] [PubMed] [Google Scholar]
- 4.Bethke, L. L., M. Zilversmit, K. Nielsen, J. Daily, S. K. Volkman, D. Ndiaye, E. R. Lozovsky, D. L. Hartl, and D. F. Wirth. 2006. Duplication, gene conversion, and genetic diversity in the species-specific acyl-CoA synthetase gene family of Plasmodium falciparum. Mol. Biochem. Parasitol. 150:10-24. [DOI] [PubMed] [Google Scholar]
- 5.Bisanz, C., O. Bastien, D. Grando, J. Jouhet, E. Marechal, and M. F. Cesbron-Delauw. 2006. Toxoplasma gondii acyl-lipid metabolism: de novo synthesis from apicoplast-generated fatty acids versus scavenging of host cell precursors. Biochem. J. 394:197-205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cai, X., D. Herschap, and G. Zhu. 2005. Functional characterization of an evolutionarily distinct phosphopantetheinyl transferase in the apicomplexan Cryptosporidium parvum. Eukaryot. Cell 4:1211-1220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Charron, A. J., and L. D. Sibley. 2002. Host cells: mobilizable lipid resources for the intracellular parasite Toxoplasma gondii. J. Cell Sci. 115:3049-3059. [DOI] [PubMed] [Google Scholar]
- 8.Chhibber, M., G. Kumar, P. Parasuraman, T. N. Ramya, N. Surolia, and A. Surolia. 2006. Novel diphenyl ethers: design, docking studies, synthesis and inhibition of enoyl ACP reductase of Plasmodium falciparum and Escherichia coli. Bioorg. Med. Chem. 14:8086-8098. [DOI] [PubMed] [Google Scholar]
- 9.Coppens, I. 2006. Contribution of host lipids to Toxoplasma pathogenesis. Cell. Microbiol. 8:1-9. [DOI] [PubMed] [Google Scholar]
- 10.Coppens, I., J. D. Dunn, J. D. Romano, M. Pypaert, H. Zhang, J. C. Boothroyd, and K. A. Joiner. 2006. Toxoplasma gondii sequesters lysosomes from mammalian hosts in the vacuolar space. Cell 125:261-274. [DOI] [PubMed] [Google Scholar]
- 11.Coppens, I., A. P. Sinai, and K. A. Joiner. 2000. Toxoplasma gondii exploits host low-density lipoprotein receptor-mediated endocytosis for cholesterol acquisition. J. Cell Biol. 149:167-180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Crawford, M. J., N. Thomsen-Zieger, M. Ray, J. Schachtner, D. S. Roos, and F. Seeber. 2006. Toxoplasma gondii scavenges host-derived lipoic acid despite its de novo synthesis in the apicoplast. EMBO J. 25:3214-3222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Crawford, M. J., G. Zhu, and D. S. Roos. 2003. Both type I and type II fatty acid synthases in Toxoplasma gondii, abstr. 14C. Mol. Parasitol. Meet. XIV.
- 14.Cronan, J. E., X. Zhao, and Y. Jiang. 2005. Function, attachment and synthesis of lipoic acid in Escherichia coli. Adv. Microb. Physiol. 50:103-146. [DOI] [PubMed] [Google Scholar]
- 15.Ferguson, D. J., S. A. Campbell, F. L. Henriquez, L. Phan, E. Mui, T. A. Richards, S. P. Muench, M. Allary, J. Z. Lu, S. T. Prigge, F. Tomley, M. W. Shirley, D. W. Rice, R. McLeod, and C. W. Roberts. 2007. Enzymes of type II fatty acid synthesis and apicoplast differentiation and division in Eimeria tenella. Int. J. Parasitol. 37:33-51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fleige, T., K. Fischer, D. J. Ferguson, U. Gross, and W. Bohne. 2007. Carbohydrate metabolism in the Toxoplasma gondii apicoplast: localization of three glycolytic isoenzymes, the single pyruvate dehydrogenase complex, and a plastid phosphate translocator. Eukaryot. Cell 6:984-996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Foth, B. J., L. M. Stimmler, E. Handman, B. S. Crabb, A. N. Hodder, and G. I. McFadden. 2005. The malaria parasite Plasmodium falciparum has only one pyruvate dehydrogenase complex, which is located in the apicoplast. Mol. Microbiol. 55:39-53. [DOI] [PubMed] [Google Scholar]
- 18.Fritzler, J. M., and G. Zhu. 2007. Functional characterization of the acyl-[acyl carrier protein] ligase in the Cryptosporidium parvum giant polyketide synthase. Int. J. Parasitol. 37:307-316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gardner, M. J., R. Bishop, T. Shah, E. P. de Villiers, J. M. Carlton, N. Hall, Q. Ren, I. T. Paulsen, A. Pain, M. Berriman, R. J. Wilson, S. Sato, S. A. Ralph, D. J. Mann, Z. Xiong, S. J. Shallom, J. Weidman, L. Jiang, J. Lynn, B. Weaver, A. Shoaibi, A. R. Domingo, D. Wasawo, J. Crabtree, J. R. Wortman, B. Haas, S. V. Angiuoli, T. H. Creasy, C. Lu, B. Suh, J. C. Silva, T. R. Utterback, T. V. Feldblyum, M. Pertea, J. Allen, W. C. Nierman, E. L. Taracha, S. L. Salzberg, O. R. White, H. A. Fitzhugh, S. Morzaria, J. C. Venter, C. M. Fraser, and V. Nene. 2005. Genome sequence of Theileria parva, a bovine pathogen that transforms lymphocytes. Science 309:134-137. [DOI] [PubMed] [Google Scholar]
- 20.Gerold, P., A. Dieckmann-Schuppert, and R. T. Schwarz. 1994. Glycosylphosphatidylinositols synthesized by asexual erythrocytic stages of the malarial parasite, Plasmodium falciparum. Candidates for plasmodial glycosylphosphatidylinositol membrane anchor precursors and pathogenicity factors. J. Biol. Chem. 269:2597-2606. [PubMed] [Google Scholar]
- 21.Goodman, C. D., and G. I. McFadden. 2007. Fatty acid biosynthesis as a drug target in apicomplexan parasites. Curr. Drug Targets 8:15-30. [DOI] [PubMed] [Google Scholar]
- 22.Gueguen, V., D. Macherel, M. Jaquinod, R. Douce, and J. Bourguignon. 2000. Fatty acid and lipoic acid biosynthesis in higher plant mitochondria. J. Biol. Chem. 275:5016-5025. [DOI] [PubMed] [Google Scholar]
- 23.Jelenska, J., M. J. Crawford, O. S. Harb, E. Zuther, R. Haselkorn, D. S. Roos, and P. Gornicki. 2001. Subcellular localization of acetyl-CoA carboxylase in the apicomplexan parasite Toxoplasma gondii. Proc. Natl. Acad. Sci. USA 98:2723-2728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jelenska, J., A. Sirikhachornkit, R. Haselkorn, and P. Gornicki. 2002. The carboxyltransferase activity of the apicoplast acetyl-CoA carboxylase of Toxoplasma gondii is the target of aryloxyphenoxypropionate inhibitors. J. Biol. Chem. 277:23208-23215. [DOI] [PubMed] [Google Scholar]
- 25.Joiner, K. A., S. A. Fuhrman, H. M. Miettinen, L. H. Kasper, and I. Mellman. 1990. Toxoplasma gondii: fusion competence of parasitophorous vacuoles in Fc receptor-transfected fibroblasts. Science 249:641-646. [DOI] [PubMed] [Google Scholar]
- 26.Jomaa, H., J. Wiesner, S. Sanderbrand, B. Altincicek, C. Weidemeyer, M. Hintz, I. Turbachova, M. Eberl, J. Zeidler, H. K. Lichtenthaler, D. Soldati, and E. Beck. 1999. Inhibitors of the nonmevalonate pathway of isoprenoid biosynthesis as antimalarial drugs. Science 285:1573-1576. [DOI] [PubMed] [Google Scholar]
- 27.Jones, S. M., J. E. Urch, M. Kaiser, R. Brun, J. L. Harwood, C. Berry, and I. H. Gilbert. 2005. Analogues of thiolactomycin as potential antimalarial agents. J. Med. Chem. 48:5932-5941. [DOI] [PubMed] [Google Scholar]
- 28.Kaiser, K., K. Matuschewski, N. Camargo, J. Ross, and S. H. Kappe. 2004. Differential transcriptome profiling identifies Plasmodium genes encoding pre-erythrocytic stage-specific proteins. Mol. Microbiol. 51:1221-1232. [DOI] [PubMed] [Google Scholar]
- 29.Kapoor, M., J. Gopalakrishnapai, N. Surolia, and A. Surolia. 2004. Mutational analysis of the triclosan-binding region of enoyl-ACP (acyl-carrier protein) reductase from Plasmodium falciparum. Biochem. J. 381:735-741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Karnataki, A., A. Derocher, I. Coppens, C. Nash, J. E. Feagin, and M. Parsons. 2007. Cell cycle-regulated vesicular trafficking of Toxoplasma APT1, a protein localized to multiple apicoplast membranes. Mol. Microbiol. 63:1653-1668. [DOI] [PubMed] [Google Scholar]
- 31.Knappe, S., U. I. Flugge, and K. Fischer. 2003. Analysis of the plastidic phosphate translocator gene family in Arabidopsis and identification of new phosphate translocator-homologous transporters, classified by their putative substrate-binding site. Plant Physiol. 131:1178-1190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kohler, S., C. F. Delwiche, P. W. Denny, L. G. Tilney, P. Webster, R. J. Wilson, J. D. Palmer, and D. S. Roos. 1997. A plastid of probable green algal origin in Apicomplexan parasites. Science 275:1485-1489. [DOI] [PubMed] [Google Scholar]
- 33.Lack, G., E. Homberger-Zizzari, G. Folkers, L. Scapozza, and R. Perozzo. 2006. Recombinant expression and biochemical characterization of the unique elongating beta-ketoacyl-acyl carrier protein synthase involved in fatty acid biosynthesis of Plasmodium falciparum using natural and artificial substrates. J. Biol. Chem. 281:9538-9546. [DOI] [PubMed] [Google Scholar]
- 34.Lee, S. H., J. L. Stephens, and P. T. Englund. 2007. A fatty-acid synthesis mechanism specialized for parasitism. Nat. Rev. Microbiol. 5:287-297. [DOI] [PubMed] [Google Scholar]
- 35.Lee, S. H., J. L. Stephens, K. S. Paul, and P. T. Englund. 2006. Fatty acid synthesis by elongases in trypanosomes. Cell 126:691-699. [DOI] [PubMed] [Google Scholar]
- 36.Leonard, A. E., S. L. Pereira, H. Sprecher, and Y. S. Huang. 2004. Elongation of long-chain fatty acids. Prog. Lipid Res. 43:36-54. [DOI] [PubMed] [Google Scholar]
- 37.Mazumdar, J., E. Wilson, K. Masarek, C. Hunter, and B. Striepen. 2006. Apicoplast fatty acid synthesis is essential for organelle biogenesis and parasite survival in Toxoplasma gondii. Proc. Natl. Acad. Sci. USA 103:13192-13197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.McFadden, G. I., M. E. Reith, J. Munholland, and N. Lang-Unnasch. 1996. Plastid in human parasites. Nature 381:482. [DOI] [PubMed] [Google Scholar]
- 39.McLeod, R., S. P. Muench, J. B. Rafferty, D. E. Kyle, E. J. Mui, M. J. Kirisits, D. G. Mack, C. W. Roberts, B. U. Samuel, R. E. Lyons, M. Dorris, W. K. Milhous, and D. W. Rice. 2001. Triclosan inhibits the growth of Plasmodium falciparum and Toxoplasma gondii by inhibition of apicomplexan Fab I. Int. J. Parasitol. 31:109-113. [DOI] [PubMed] [Google Scholar]
- 40.McMillan, P. J., L. M. Stimmler, B. J. Foth, G. I. McFadden, and S. Muller. 2005. The human malaria parasite Plasmodium falciparum possesses two distinct dihydrolipoamide dehydrogenases. Mol. Microbiol. 55:27-38. [DOI] [PubMed] [Google Scholar]
- 41.Mikolajczak, S. A., V. Jacobs-Lorena, D. C. MacKellar, N. Camargo, and S. H. Kappe. 2007. L-FABP is a critical host factor for successful malaria liver stage development. Int. J. Parasitol. 37:483-489. [DOI] [PubMed] [Google Scholar]
- 42.Mordue, D. G., S. Hakansson, I. Niesman, and L. D. Sibley. 1999. Toxoplasma gondii resides in a vacuole that avoids fusion with host cell endocytic and exocytic vesicular trafficking pathways. Exp. Parasitol. 92:87-99. [DOI] [PubMed] [Google Scholar]
- 43.Mordue, D. G., and L. D. Sibley. 1997. Intracellular fate of vacuoles containing Toxoplasma gondii is determined at the time of formation and depends on the mechanism of entry. J. Immunol. 159:4452-4459. [PubMed] [Google Scholar]
- 44.Mueller, A. K., N. Camargo, K. Kaiser, C. Andorfer, U. Frevert, K. Matuschewski, and S. H. Kappe. 2005. Plasmodium liver stage developmental arrest by depletion of a protein at the parasite-host interface. Proc. Natl. Acad. Sci. USA 102:3022-3027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mueller, A. K., M. Labaied, S. H. Kappe, and K. Matuschewski. 2005. Genetically modified Plasmodium parasites as a protective experimental malaria vaccine. Nature 433:164-167. [DOI] [PubMed] [Google Scholar]
- 46.Muench, S. P., S. T. Prigge, R. McLeod, J. B. Rafferty, M. J. Kirisits, C. W. Roberts, E. J. Mui, and D. W. Rice. 2007. Studies of Toxoplasma gondii and Plasmodium falciparum enoyl acyl carrier protein reductase and implications for the development of antiparasitic agents. Acta Crystallogr. D 63:328-338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mullin, K. A., L. Lim, S. A. Ralph, T. P. Spurck, E. Handman, and G. I. McFadden. 2006. Membrane transporters in the relict plastid of malaria parasites. Proc. Natl. Acad. Sci. USA 103:9572-9577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Nicola, G., C. A. Smith, E. Lucumi, M. R. Kuo, L. Karagyozov, D. A. Fidock, J. C. Sacchettini, and R. Abagyan. 2007. Discovery of novel inhibitors targeting enoyl-acyl carrier protein reductase in Plasmodium falciparum by structure-based virtual screening. Biochem. Biophys. Res. Commun. 358:686-691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Perozzo, R., M. Kuo, A. S. Sidhu, J. T. Valiyaveettil, R. Bittman, W. R. Jacobs, Jr., D. A. Fidock, and J. C. Sacchettini. 2002. Structural elucidation of the specificity of the antibacterial agent triclosan for malarial enoyl acyl carrier protein reductase. J. Biol. Chem. 277:13106-13114. [DOI] [PubMed] [Google Scholar]
- 50.Quittnat, F., Y. Nishikawa, T. T. Stedman, D. R. Voelker, J. Y. Choi, M. M. Zahn, R. C. Murphy, R. M. Barkley, M. Pypaert, K. A. Joiner, and I. Coppens. 2004. On the biogenesis of lipid bodies in ancient eukaryotes: synthesis of triacylglycerols by a Toxoplasma DGAT1-related enzyme. Mol. Biochem. Parasitol. 138:107-122. [DOI] [PubMed] [Google Scholar]
- 51.Ralph, S. A., G. G. Van Dooren, R. F. Waller, M. J. Crawford, M. J. Fraunholz, B. J. Foth, C. J. Tonkin, D. S. Roos, and G. I. McFadden. 2004. Tropical infectious diseases: metabolic maps and functions of the Plasmodium falciparum apicoplast. Nat. Rev. Microbiol. 2:203-216. [DOI] [PubMed] [Google Scholar]
- 52.Rawsthorne, S. 2002. Carbon flux and fatty acid synthesis in plants. Prog. Lipid Res. 41:182-196. [DOI] [PubMed] [Google Scholar]
- 53.Rein, K. S., and R. V. Snyder. 2006. The biosynthesis of polyketide metabolites by dinoflagellates. Adv. Appl. Microbiol. 59:93-125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Salcedo, S. P., and D. W. Holden. 2005. Bacterial interactions with the eukaryotic secretory pathway. Curr. Opin. Microbiol. 8:92-98. [DOI] [PubMed] [Google Scholar]
- 55.Sato, S., B. Clough, L. Coates, and R. J. Wilson. 2004. Enzymes for heme biosynthesis are found in both the mitochondrion and plastid of the malaria parasite Plasmodium falciparum. Protist 155:117-125. [DOI] [PubMed] [Google Scholar]
- 56.Sharma, S. K., M. Kapoor, T. N. Ramya, S. Kumar, G. Kumar, R. Modak, S. Sharma, N. Surolia, and A. Surolia. 2003. Identification, characterization, and inhibition of Plasmodium falciparum beta-hydroxyacyl-acyl carrier protein dehydratase (FabZ). J. Biol. Chem. 278:45661-45671. [DOI] [PubMed] [Google Scholar]
- 57.Shaw, M. K. 2003. Cell invasion by Theileria sporozoites. Trends Parasitol. 19:2-6. [DOI] [PubMed] [Google Scholar]
- 58.Sinai, A. P., and K. A. Joiner. 1997. Safe haven: the cell biology of nonfusogenic pathogen vacuoles. Annu. Rev. Microbiol. 51:415-462. [DOI] [PubMed] [Google Scholar]
- 59.Sinai, A. P., P. Webster, and K. A. Joiner. 1997. Association of host cell endoplasmic reticulum and mitochondria with the Toxoplasma gondii parasitophorous vacuole membrane: a high affinity interaction. J. Cell Sci. 110:2117-2128. [DOI] [PubMed] [Google Scholar]
- 60.Snyder, R. V., M. A. Guerrero, C. D. Sinigalliano, J. Winshell, R. Perez, J. V. Lopez, and K. S. Rein. 2005. Localization of polyketide synthase encoding genes to the toxic dinoflagellate Karenia brevis. Phytochemistry 66:1767-1780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Stephens, J. L., S. H. Lee, K. S. Paul, and P. T. Englund. 2007. Mitochondrial fatty acid synthesis in Trypanosoma brucei. J. Biol. Chem. 282:4427-4436. [DOI] [PubMed] [Google Scholar]
- 62.Surolia, A., T. N. Ramya, V. Ramya, and N. Surolia. 2004. ′FAS't inhibition of malaria. Biochem. J. 383:401-412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Surolia, N., and A. Surolia. 2001. Triclosan offers protection against blood stages of malaria by inhibiting enoyl-ACP reductase of Plasmodium falciparum. Nat. Med. 7:167-173. [DOI] [PubMed] [Google Scholar]
- 64.Swarnamukhi, P. L., S. K. Sharma, P. Bajaj, N. Surolia, A. Surolia, and K. Suguna. 2006. Crystal structure of dimeric FabZ of Plasmodium falciparum reveals conformational switching to active hexamers by peptide flips. FEBS Lett. 580:2653-2660. [DOI] [PubMed] [Google Scholar]
- 65.Thomsen-Zieger, N., J. Schachtner, and F. Seeber. 2003. Apicomplexan parasites contain a single lipoic acid synthase located in the plastid. FEBS Lett. 547:80-86. [DOI] [PubMed] [Google Scholar]
- 66.Tomavo, S., J. F. Dubremetz, and R. T. Schwarz. 1992. A family of glycolipids from Toxoplasma gondii. Identification of candidate glycolipid precursor(s) for Toxoplasma gondii glycosylphosphatidylinositol membrane anchors. J. Biol. Chem. 267:11721-11728. [PubMed] [Google Scholar]
- 67.Tomova, C., W. J. Geerts, T. Muller-Reichert, R. Entzeroth, and B. M. Humbel. 2006. New comprehension of the apicoplast of sarcocystis by transmission electron tomography. Biol. Cell 98:535-545. [DOI] [PubMed] [Google Scholar]
- 68.Trenkamp, S., W. Martin, and K. Tietjen. 2004. Specific and differential inhibition of very-long-chain fatty acid elongases from Arabidopsis thaliana by different herbicides. Proc. Natl. Acad. Sci. USA 101:11903-11908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.van Dooren, G. G., M. Marti, C. J. Tonkin, L. M. Stimmler, A. F. Cowman, and G. I. McFadden. 2005. Development of the endoplasmic reticulum, mitochondrion and apicoplast during the asexual life cycle of Plasmodium falciparum. Mol. Microbiol. 57:405-419. [DOI] [PubMed] [Google Scholar]
- 70.van Dooren, G. G., L. M. Stimmler, and G. I. McFadden. 2006. Metabolic maps and functions of the Plasmodium mitochondrion. FEMS Microbiol. Rev. 30:596-630. [DOI] [PubMed] [Google Scholar]
- 71.Vial, H. J., and M. L. Ancelin. 1992. Malarial lipids. An overview. Subcell. Biochem. 18:259-306. [PubMed] [Google Scholar]
- 72.Vielemeyer, O., M. T. McIntosh, K. A. Joiner, and I. Coppens. 2004. Neutral lipid synthesis and storage in the intraerythrocytic stages of Plasmodium falciparum. Mol. Biochem. Parasitol. 135:197-209. [DOI] [PubMed] [Google Scholar]
- 73.Waller, R. F., P. J. Keeling, R. G. Donald, B. Striepen, E. Handman, N. Lang-Unnasch, A. F. Cowman, G. S. Besra, D. S. Roos, and G. I. McFadden. 1998. Nuclear-encoded proteins target to the plastid in Toxoplasma gondii and Plasmodium falciparum. Proc. Natl. Acad. Sci. USA 95:12352-12357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Waller, R. F., S. A. Ralph, M. B. Reed, V. Su, J. D. Douglas, D. E. Minnikin, A. F. Cowman, G. S. Besra, and G. I. McFadden. 2003. A type II pathway for fatty acid biosynthesis presents drug targets in Plasmodium falciparum. Antimicrob. Agents Chemother. 47:297-301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Wickramasinghe, S. R., K. A. Inglis, J. E. Urch, S. Muller, D. M. van Aalten, and A. H. Fairlamb. 2006. Kinetic, inhibition and structural studies on 3-oxoacyl-ACP reductase from Plasmodium falciparum, a key enzyme in fatty acid biosynthesis. Biochem. J. 393:447-457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Wilson, R. J., P. W. Denny, P. R. Preiser, K. Rangachari, K. Roberts, A. Roy, A. Whyte, M. Strath, D. J. Moore, P. W. Moore, and D. H. Williamson. 1996. Complete gene map of the plastid-like DNA of the malaria parasite Plasmodium falciparum. J. Mol. Biol. 261:155-172. [DOI] [PubMed] [Google Scholar]
- 77.Wrenger, C., and S. Muller. 2004. The human malaria parasite Plasmodium falciparum has distinct organelle-specific lipoylation pathways. Mol. Microbiol. 53:103-113. [DOI] [PubMed] [Google Scholar]
- 78.Zeng, B., X. Cai, and G. Zhu. 2006. Functional characterization of a fatty acyl-CoA-binding protein (ACBP) from the apicomplexan Cryptosporidium parvum. Microbiology 152:2355-2363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Zeng, B., and G. Zhu. 2006. Two distinct oxysterol binding protein-related proteins in the parasitic protist Cryptosporidium parvum (Apicomplexa). Biochem. Biophys. Res. Commun. 346:591-599. [DOI] [PubMed] [Google Scholar]
- 80.Zhu, G. 2004. Current progress in the fatty acid metabolism in Cryptosporidium parvum. J. Eukaryot. Microbiol. 51:381-388. [DOI] [PubMed] [Google Scholar]
- 81.Zhu, G., M. J. LaGier, F. Stejskal, J. J. Millership, X. Cai, and J. S. Keithly. 2002. Cryptosporidium parvum: the first protist known to encode a putative polyketide synthase. Gene 298:79-89. [DOI] [PubMed] [Google Scholar]
- 82.Zhu, G., Y. Li, X. Cai, J. J. Millership, M. J. Marchewka, and J. S. Keithly. 2004. Expression and functional characterization of a giant type I fatty acid synthase (CpFAS1) gene from Cryptosporidium parvum. Mol. Biochem. Parasitol. 134:127-135. [DOI] [PubMed] [Google Scholar]
- 83.Zhu, G., M. J. Marchewka, and J. S. Keithly. 2000. Cryptosporidium parvum appears to lack a plastid genome. Microbiology 146:315-321. [DOI] [PubMed] [Google Scholar]
- 84.Zhu, G., M. J. Marchewka, K. M. Woods, S. J. Upton, and J. S. Keithly. 2000. Molecular analysis of a type I fatty acid synthase in Cryptosporidium parvum. Mol. Biochem. Parasitol. 105:253-260. [DOI] [PubMed] [Google Scholar]