SUMMARY
The childhood solid muscle tumor Alveolar Rhabdomyosarcoma (ARMS) is characterized by the t(2;13)(q35;q14) chromosomal translocation, which results in the fusion of two transcription factors important for myogenesis, Pax3 and FKHR (FOX01a). The effects of myogenic differentiation on the stability of FKHR have been well characterized. However, similar studies have yet to be performed on Pax3 or the oncogenic fusion protein Pax3-FKHR. Therefore, we demonstrate in the physiologically relevant mouse primary myoblast system that the expression of Pax3 decreases nearly 95% during the first twenty-four hours of myogenic differentiation. In contrast, there is an aberrant persistence of expression of Pax3-FKHR during this same time period. These differences in protein expression levels do not result from changes on the transcriptional nor the translational level since we observed no concomitant decrease in the levels of Pax3 or Pax3-FKHR mRNA or in the ability of both proteins to be translated. Instead, a pulse-chase analysis determined that Pax3-FKHR has a half-life significantly greater than the half-life of wild type Pax3 demonstrating for the first time that Pax3-FKHR has greater post-translational protein stability relative to wild type Pax3 during early myogenic differentiation. Finally, the persistence of expression of Pax3-FKHR prevents the terminal differentiation of primary myoblasts demonstrating a biological consequence of its aberrant expression.
Keywords: alveolar rhabdomyosarcoma, Pax3, FKHR, Pax3-FKHR, myogenesis, protein stability
1. Introduction
Alveolar Rhabdomyosarcoma (ARMS) is a malignant tumor of skeletal muscle that is present primarily in the trunk and extremities of adolescents and young adults [1]. ARMS is most frequently characterized by the t(2;13)(q35;q14) chromosomal translocation [2, 3], which results in the fusion of two transcription factors Pax3 and FKHR (FOXO1a). The Pax3-FKHR fusion protein retains the Pax3 octapeptide domain, which was demonstrated to be important for mediating protein-protein interactions [4, 5], along with the paired- and homeodomain DNA binding regions of Pax3. However, the Pax3 transcriptional activation domain is replaced by two elements from FKHR - a bisected, non-functional DNA binding domain and the FKHR transcriptional activation domain.
Pax3 is a member of the paired class homeodomain family of transcription factors [6] and is expressed beginning at day 8.5 during murine embryogenesis. Mice homozygous for mutations in Pax3 fail to develop limb musculature [7-9]. In addition, it was demonstrated that ectopic expression of Pax3 in chick embryo presomitic mesoderm explants induced the expression of the myogenic transcription factor MyoD and maintained the expression of the myogenic transcription factor myf5 in the absence of inducing tissues [10]. Taken together these observations demonstrate that Pax3 is essential for myogenesis and acts in parallel with myf5 and upstream of MyoD in myogenic differentiation.
FKHR (FOXO1a) is a ubiquitously expressed protein [11] that is a member of the forkhead winged-helix family of transcription factors [12]. FKHR and its orthologs FKHRLI (FOXO3a) and AFX (FOXO4) are multifunctional transcription factors with such diverse functions as triggering apoptosis [13], regulating cell cycle progression [14, 15], controlling metabolic homeostasis [16-18], driving adipocyte differentiation [19], and inhibiting cellular transformation and tumorigenesis [20]. In addition to this broad range of functions, FKHR has been demonstrated to be essential for myogenic differentiation where its activity is crucial for the fusion of myoblasts into myotubes [21, 22].
Many of the biological activities of FKHR, including its protein stability, are regulated on a post-translational level [21, 23, 24]. In fact, the mechanism by which its protein stability is regulated throughout the induction and progression of myogenic differentiation is well known. In proliferating myoblasts FKHR resides primarily in the cytoplasm where it is rapidly degraded. Upon the induction of myogenic differentiation, FKHR is translocated to the nucleus with a subsequent 8 - 10 fold increase in its protein stability. This nuclear translocation and increase in stability are controlled by several different post-translational modifications that include phosphorylation and ubiquitination [21, 22].
Although the expression and stability of FKHR throughout myogenesis has been studied extensively, very little if anything is known about the expression and stability of Pax3 or the oncogenic fusion protein Pax3-FKHR during myogenic differentiation. Therefore, we decided to examine how the process of myoblast differentiation affects the expression and stability of Pax3 and Pax3-FKHR in order to gain a better understanding of the underlying molecular mechanisms that may contribute to the development of ARMS. In the present report we present evidence demonstrating that Pax3-FKHR has significantly greater post-translational protein stability than wild type Pax3 during early myogenesis. This increased stability results in the aberrant expression of Pax3-FKHR throughout myogenic differentiation, which may contribute to the development of ARMS.
2. Materials and Methods
2.1. Preparation of mouse primary myoblasts
The mouse primary myoblasts were isolated by a modification of a previously described method [25]. Briefly, the fore- and hind limbs were removed from 2 - 4 day old C57/Bl6 mice, and the skin, fat, and other non-muscle tissue was dissected away. The dissected limbs were washed once with PBS after which the cells were enzymatically dissociated by the addition of 2ml of a solution of dispase (2.4U/ml, Roche Applied Science, Indianapolis, IN) and collagenase P (10 mg/ml, Roche Applied Science). The slurry was incubated at 37°C for 45 minutes with gentle mixing every 10 - 15 minutes. After incubation, Ham’s F-10 nutrient media (Cellgro Mediatech, Inc., Herndon, VA) supplemented with 20% FBS (HyClone Laboratories, Inc., Logan, UT) and 2.5 ng/ml bFGF (Promega Corp., Madison, WI) was added to a total of 25 ml and the solution was passed through an 80 μm nylon mesh cell strainer. The resulting cells were pelleted by centrifugation at 235 × g for 5 minutes. The cell pellet was resuspended in 45 ml of the F-10 media described above supplemented with collagen (0.1 mg/ml, Type I from calf skin, Sigma, St. Louis, MO) and pre-plated two times on 3 non-collagen coated 150 mm dishes at 37°C and 5% CO2 for one hour each time. This step enriches the cell suspension for myoblasts. The supernatant was collected and the cells were pelleted by centrifugation as described above. The resulting pellet was resuspended in 25 ml of the F-10 media described above in the absence of collagen and the cells were then plated on 100 mm collagen-coated dishes (≅1 × 106 cells/100 mm dish). Myoblasts were pre-plated as described above for the first several passages of the primary cultures. Primary myoblasts isolated in this way were 99% pure as determined by staining with MyoD and desmin antibodies (data not shown).
2.2. Expression constructs
Pax3 and Pax3-FKHR were cloned into the EcoRI and XhoI sites of the retroviral expression vector MSCV-IRES-GFP (a kind gift from G. Grosveld, St. Jude Children’s Research Hospital). The MSCV-IRES-GFP retroviral vector contains the cDNA for green fluorescent protein (GFP) and an internal ribosomal entry site (IRES). The presence of the IRES allows the dual production of GFP and either Pax3 or Pax3-FKHR, all under control of the murine stem cell virus promoter (MSCV) [26].
2.3. Retroviral stocks and stable transduction of mouse primary myoblasts
Retroviral stocks were generated by the transient transfection of the ecotropic Phoenix packaging cell line [27] with 8 μg of the MSCV-IRES-GFP empty vector, or the MSCV-IRES-GFP retroviral construct containing either Pax3 or Pax3-FKHR by the Fugene™ 6 method (Roche Applied Science) according to the manufacturer’s specifications. Culture supernatants containing virus were collected between 36 and 72 hours after transfection, filtered, and subsequently used for a single transduction of mouse primary myoblasts isolated as described above. Three to seven days post-transduction primary myoblasts were harvested in F10 media supplemented with collagen (10 ng/ml) and cells expressing GFP were selected by fluorescence activated cell sort (FACS) analysis. Cells selected in this manner were cultured and expanded as described below.
2.4. Mouse primary myoblast growth and differentiation conditions
Proliferation medium for the mouse primary myoblasts consisted of Ham’s F-10 nutrient mixture supplemented with 20% FBS and 2.5 ng/ml bFGF, as described above. Differentiation medium consisted of Dulbecco’s Modification of Eagle’s Medium (DMEM, Cellgro Mediatech, Inc.) supplemented with 2% horse serum (HyClone). All media contained penicillin G (200 μg/ml) and streptomycin (200 μg/ml). DMEM was additionally supplemented with L-glutamine (2 mM, Cellgro Mediatech, Inc.). Cells were grown in a humidified incubator at 37°C in 5% CO2 (F-10 medium) or 10% CO2 (DMEM). All cells were grown on collagen-coated dishes (Becton Dickinson Labware, Bedford, MA), were passage-matched to prevent possible differences due to different passage conditions, were not used past passage 9 to prevent the cells entering crisis, and were not allowed to grow past approximately 80% confluency to maintain the cells in an undifferentiated state. To induce the differentiation of primary myoblasts, the proliferation media was removed, the cells were washed twice with PBS, the media was replaced with 10ml of differentiation media, and the cells were grown as described above until needed for further analysis.
2.5. Western blot analysis
Proliferating or differentiated primary myoblasts that were isolated and transduced as described above were lysed in CelLytic™ Express (Sigma, St. Louis, MO) containing complete mini protease inhibitor cocktail (Roche Applied Science), phosphatase cocktail I specific for serine/threonine phosphatases (Sigma), and phosphatase cocktail II specific for tyrosine phosphatases (Sigma) with two rounds of sonication for 7s at power level 3 with a 550 Sonic Dismembrator (Fisher Scientific, Pittsburgh, PA). The debris was removed by centrifugation at maximum speed in a microfuge for 10 minutes at 4°C. A constant amount of total cell lysate (20μg for ectopic Pax3, 40μg for endogenous Pax3 and ectopic Pax3-FKHR) was separated by 10% SDS-PAGE, proteins were transferred to Immobilon-P membrane (Millipore, Bedford, MA), and the presence of Pax3 or Pax3-FKHR was detected using an affinity purified monospecific Pax3 antibody, as previously described [28]. To control for constant loading the blots were stripped and the presence of the housekeeping gene GAPDH was detected using a commercially available antibody (sc-32233, Santa Cruz Biotechnology, Santa Cruz, CA). Alternatively, the membrane was stained with Fast Green FCF (Sigma) to visualize the total protein transferred to the membrane (Figure 1). The presence of the Fast Green staining does not affect the efficiency of the Western blot analysis (data not shown).
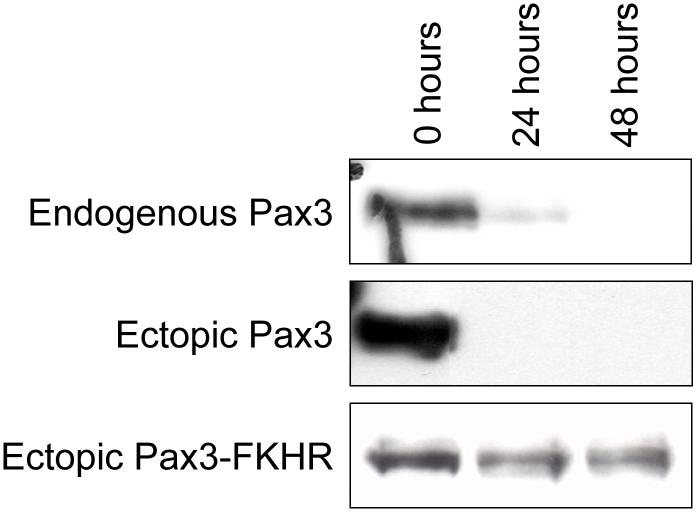
Figure 1.
Expression of Pax3 and Pax3-FKHR during myogenic differentiation. Mouse primary myoblasts (top panel) and mouse primary myoblasts stably expressing either Pax3 (middle panel) or Pax3-FKHR (bottom panel) were induced to differentiate as described in the Materials and Methods. Undifferentiated primary myoblasts (0 hours) and myoblasts from each time point of differentiation (24 and 48 hours) were lysed, equal amounts (50μg) of total cell extracts were separated by 10% SDS-PAGE, the equal loading of protein was confirmed by Fast Green staining of the membrane (data not shown), and the presence of endogenous Pax3, ectopic Pax3, or ectopic Pax3-FKHR was determined by Western blot analysis using a monospecific affinity-purified anti-Pax3 antibody, as described in the Materials and Methods.
2.6. Real-time one-step RT-PCR analysis
Proliferating or differentiated primary myoblasts that were isolated and transduced as described above were used to obtain cytoplasmic poly(A)+ mRNA using the Oligotex® mRNA Direct Isolation Kit (Qiagen, Valencia, CA) according to the manufacturer’s specifications. Final conditions for the real-time one-step RT-PCR analysis for a 25μl reaction mixture used 12.5μl of the 2X SYBR Green PCR Master Mix (Applied Biosysems, Foster City, CA), which contains the AmpliTag Gold DNA Polymerase, 100nM of the primers described below, 6U of RNase Inhibitor, 0.13μl of the MultiScribe reverse transcriptase (Applied Biosystems), and 30ng of the mRNA template. The reverse transcription reaction was carried out for 30 minutes at 48°C followed by 10 minutes at 95°C and 40 cycles of 15 seconds at 95°C and 60 seconds at 60°C. Amplification, detection, and data analysis were performed on an ABI Prism 7900HT real-time thermal cycler (Applied Biosystems). The baseline was set to where no fluorescence emissions were detectable and five cycles before the start of amplification was detected. The cycle threshold (CT) was automatically determined using the SDS software 1.3.1 (Applied Biosystems) and fell within the exponential growth region of the amplification curve. The real-time one-step RT-PCR reaction contained primers that were specific for each of the samples being amplified, as shown in Fig.3, and are as follows:
Endogenous Pax3: 5′-exon 6: 5′-CAGTCAGATGAAGGCTCC-3′; 3′-exon 7: 5′-GCTGGGGAGGCAGTAGGC-3′
Ectopic Pax3: 5′-Pax3: 5′-CTATACAGACAGCTTTGTGC-3′ 3′-GFP: 5′-GTTTACGTCGCCGTCCAG C-3′
Ectopic Pax3-FKHR: 5′-FKHR: 5′-CTGTACAAGTGCCTCTGC-3′ 3′-GFP: 5′-GTTTACGTCGCCGTCCAGC-3′
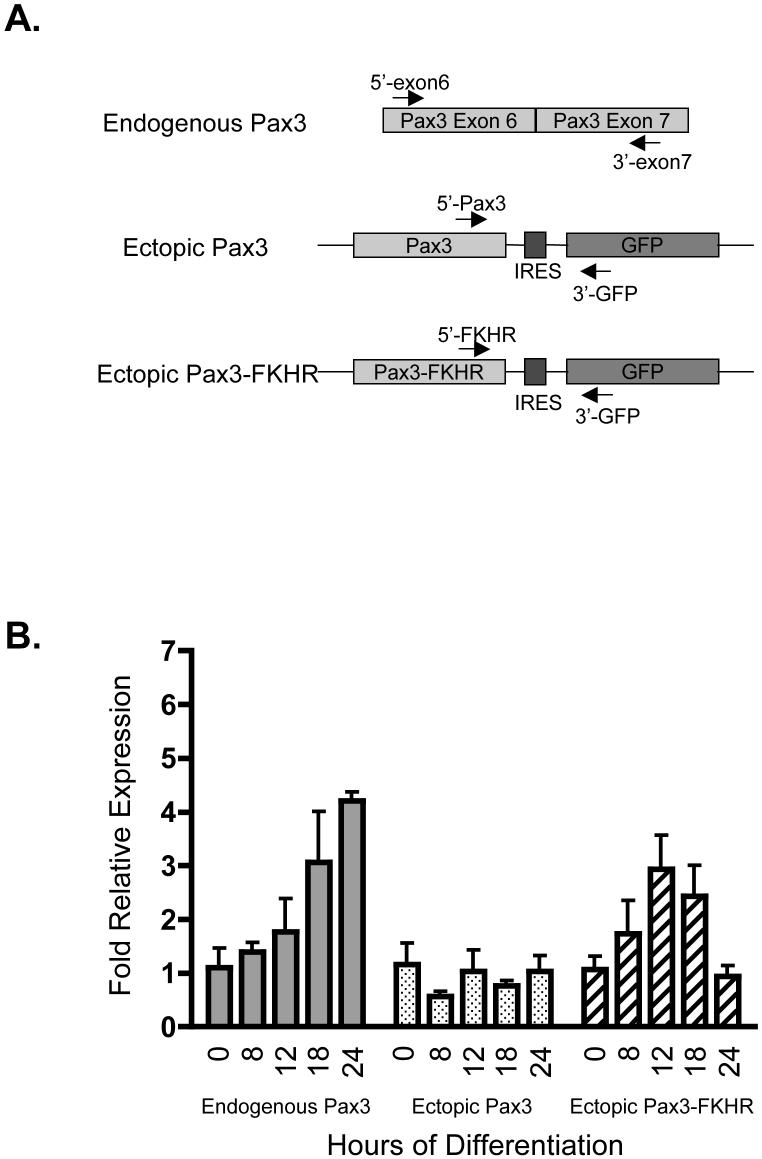
Figure 3.
Relative mRNA expression levels for Pax3 and Pax3-FKHR during early myogenic differentiation. A. Schematic of the primers used for the real time RT-PCR analysis. Primers were engineered to anneal to exon 6 and exon 7 of endogenous Pax3 (top panel), the 3′-most region of Pax3 and the 5′-most region of green fluorescent protein (GFP) for ectopic Pax3 (middle panel), and the 3′-most region of Pax3-FKHR and the 5′-most region of GFP for ectopic Pax3-FKHR (bottom panel). The arrows illustrate the direction of polymerization. B. Determination of the relative mRNA expression levels. Cytoplasmic poly(A)+ mRNA was isolated from mouse primary myoblasts or mouse primary myoblasts stably transduced with either Pax3 or Pax3-FKHR induced to differentiate for 0, 8, 12, 18, or 24 hours. Real-time one-step RT-PCR analysis was performed on 30ng of mRNA, as described in the Materials and Methods, using primers specific for endogenous Pax3, ectopic Pax3, and ectopic Pax3-FKHR. The fold relative expression levels at each time of differentiation were determined relative to the expression in undifferentiated myoblasts (time zero) and are based on the threshold cycle (CT) using the 2-ΔΔCT analysis [29], as described in the Materials and Methods. The error bars represent the standard deviation from the mean for three independent determinations.
To confirm that constant levels of mRNA were used in each reaction the real-time one-step RT-PCR was performed in parallel on each of the samples as described above using primers specific for β-actin:
actin forward: 5′-ATGGATGACGATATCGCTGCGCTGGTCG-3′
actin reverse: 5′-CTAGAAGCACTTGCGGTGCACG-3′
These primers and reaction conditions produced single DNA fragments of the expected sizes for the PCR products, as determined by agarose gel electrophoresis, confirming the specificity of the reactions (data not shown).
2.7. Determination of the fold relative gene expression
The relative change in mRNA levels of the differentiated samples relative to the undifferentiated samples were determined from the real-time RT-PCR analysis using the 2-ΔΔCT method as previously described [29]. Briefly, the CT values for each sample were normalized to the CT values for β-actin according to the following equations:
The difference between the normalized CT values relative to the undifferentiated samples (ΔΔCT) were than calculated as follows:
The fold difference in gene expression, which has been normalized to an endogenous reference (β-actin) and determined relative to the undifferentiated samples, was calculated using the following equation:
This analysis was performed in triplicate for each sample.
2.8. Pulse-chase analysis for protein stability
Primary myoblasts stably transduced with either Pax3 or Pax3-FKHR were induced to differentiate as described above. After 22.5 hours of differentiation the cells were starved by replacing the media with differentiation media lacking methionine and cysteine and incubating them as described above for 30 minutes. The cells were pulse-labeled by adding 0.5mCi of TRAN35S-LABEL (MP Biomedicals, Irvine, CA), which is a mixture of [35S]-methionine and [35S]-cysteine, and incubating them as described above for one hour. The chase was initiated by washing the cells once with PBS and replacing the media with non-radioactive differentiation media. At various time points (0, 0.5, 1.0, 2.0, 4.0, and 8.0 hours) after the initiation of the chase the cells were harvested and lysed in RIPA buffer [50mM Tris-HCl [pH 7.5], 150mM NaCl, 1% deoxycholate, 1% Triton X-100, 0.1% SDS, complete mini protease inhibitor cocktail (Roche), phosphatase cocktail I (Sigma), and phosphatase cocktail II (Sigma)], immunoprecipitated with a monospecific affinity-purified anti-Pax3 antibody, collected with Protein A magnetic beads (New England Biolabs, Beverly, MA), and the immune complex separated by 10% SDS-PAGE. The dried gel was visualized by autoradiography and the resulting bands were quantified by densitometry.
2.9 Immunofluorescence
To determine the expression and cellular localization of Pax3, Pax3-FKHR, and Myosin Heavy Chain (MHC), primary myoblasts stably infected with either the MSCV-IRES-GFP or the MSCV-IRES-GFP construct containing Pax3 or Pax3-FKHR were plated on two-chamber collagen treated microscope slides (2 × 105 cells/chamber) in proliferation medium. The cells were induced to differentiate by changing the medium to differentiation medium and both undifferentiated and differentiated myoblasts were analyzed by immunofluorescence. Cells were fixed and permeabilized as previously described [28] and subsequently incubated with a mixture of a 1:500 dilution of a mono-specific affinity-purified rabbit anti-Pax3 antibody [28] and a 1:500 dilution of an anti-MHC antibody (clone MF20, Developmental Studies Hybridoma Bank, The University of Iowa, Iowa City, IA) for one hour. After washing with PBS, the cells were incubated with a mixture of the Alexa Fluor® 546 goat anti-rabbit antibody and the FITC-conjugated goat anti-mouse antibody (1:500 dilution for each antibody) for 30 minutes, washed with PBS, and mounted with the Vectashield mounting medium that contained DAPI nuclear stain (0.1 μg/ml, Sigma, St. Louis, MO). Slides were examined using a Leica TCS NT SP Confocal microscope.
3. Results
3.1. Expression of Pax3 and Pax3-FKHR during early myogenesis
In the present study we were interested in investigating some of the underlying molecular mechanisms that contribute to the development of ARMS from its native cell type (i.e. - myoblasts) instead of analyzing how one of the key factors, the oncogenic fusion protein Pax3-FKHR, acts in an established tumor environment (i.e. - ARMS cell lines). Therefore, we performed all of our studies in the physiologically relevant mouse primary myoblasts by creating primary myoblasts that stably express either Pax3 or Pax3-FKHR. A western blot analysis on proliferating mouse primary myoblasts using a Pax3-specific antibody demonstrated the expression of endogenous Pax3 (Figure 1, top panel), which is consistent with literature reports [5]. A similar analysis on proliferating primary myoblasts stably transduced with either Pax3 or Pax3-FKHR demonstrated the expression of Pax3, and Pax3-FKHR (Figure 1, middle and bottom panels). The levels of Pax3-FKHR protein expression that we observed are consistent with the endogenous levels of Pax3-FKHR present in the ARMS cell line Rh30 [4]. Upon the induction of differentiation we found that the expression of Pax3 from the wild-type cells or cells stably transduced with Pax3 was greatly reduced after twenty-four hours and was completely gone after forty-eight hours of differentiation (Figure 1, top and middle panels). In contrast to the observed decrease in the expression of Pax3 there were minimal changes in the levels of expression of Pax3-FKHR over the first forty-eight hours of differentiation (Figure 1, bottom panel).
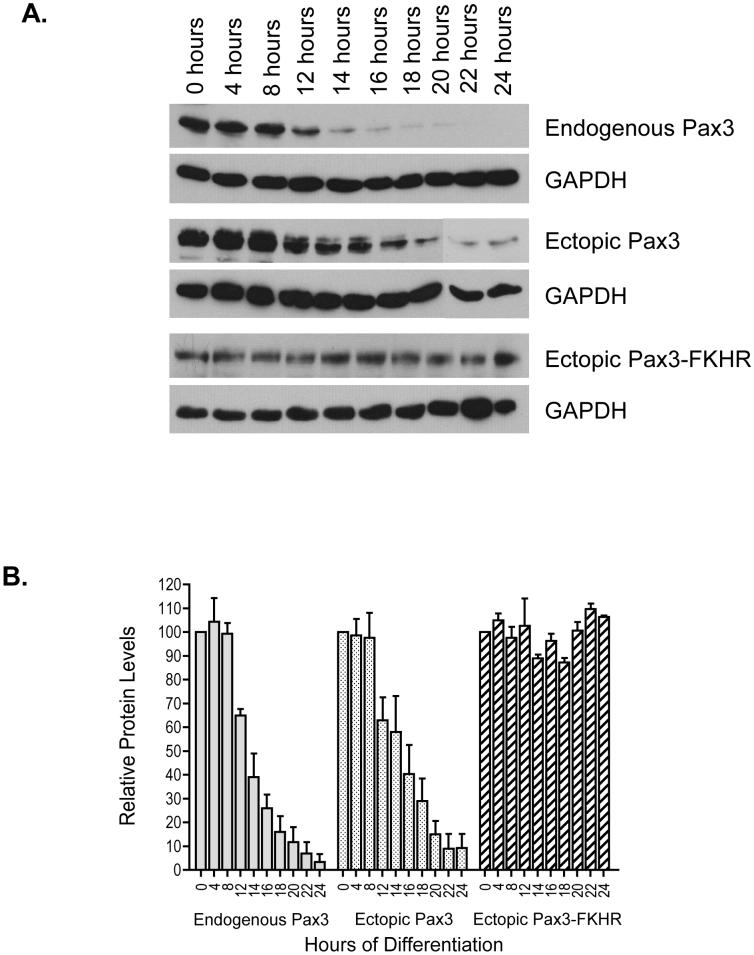
Because we observed such a drastic decrease in the expression of Pax3 after the first twenty-four hours of differentiation, we were interested in further characterizing the pattern of expression for Pax3 and Pax3-FKHR during this time period. Primary myoblasts or primary myoblasts stably transduced with Pax3 or Pax3-FKHR were induced to differentiate, total cell extracts were made at a variety of time points during the first twenty-four hours of differentiation, and a Western blot analysis was performed to determine the expression levels of Pax3 or Pax3-FKHR. We observed no changes in Pax3 protein levels, with either endogenous Pax3 or in the cells stably transduced with Pax3, during the first eight hours of differentiation. This, however, was followed by a consistent decrease in Pax3 protein levels between twelve and twenty-four hours of differentiation (Figure 2A) resulting in a nearly 95% loss of Pax3 protein expression. The minor differences in Pax3 protein expression at twenty-four hours of differentiation represents the slight variability that we observed between independent determinations from individual experiments. We quantified the samples by densitometry and normalized the Pax3 protein levels for the levels of GAPDH to account for differences in protein loading and then expressed each value relative to the amount of Pax3 at 0 hours of differentiation (Figure 2B). This analysis demonstrated a 90 - 95% decrease in the expression of Pax3 by twenty-four hours of myogenic differentiation regardless of whether it was derived from an endogenous or ectopic source of DNA (Figure 2B). This result suggests that the decrease in protein levels is independent of its DNA source. Similar results were also observed in primary myoblasts stably expressing a FLAG-epitope tagged Pax3 that was subsequently analyzed with a Western blot using an anti-FLAG antibody (data not shown), thereby confirming that the decrease in Pax3 protein levels is the same for endogenous and ectopically expressed Pax3.
Figure 2.
Expression of Pax3 and Pax3-FKHR during early myogenic differentiation. A. A representative Western blot analysis of Pax3 and Pax3-FKHR protein expression. Mouse primary myoblasts (top panels) or mouse primary myoblasts stably expressing either Pax3 (middle panels) or Pax3-FKHR (bottom panels) were induced to differentiate as described in the Materials and Methods. Undifferentiated primary myoblasts (0 hours) and myoblasts from the indicated time points of the initial twenty-four hours of differentiation were lysed, equal amounts of total cell extracts (endogenous Pax3 and ectopic Pax3-FKHR - 40μg; ectopic Pax3 - 20μg) were separated by 10% SDS-PAGE, and the presence of the proteins was determined by Western blot analysis using a monospecific affinity-purified anti-Pax3 antibody. As a control for protein loading the presence of the housekeeping gene GAPDH was determined by Western blot analysis. B. Quantification of Pax3 and Pax3-FKHR protein levels. The protein levels of Pax3 and Pax3-FKHR were quantified by densitometric analysis and normalized for the protein levels of GAPDH. The normalized values for each time point were plotted relative to the protein levels for undifferentiated primary myoblasts (0 hours), which was given a relative value of 100%. The errors bars represent the standard deviation from the mean of three independent determinations.
Consistent with the results described above (Figure 1) and in direct contrast to the expression of wild-type Pax3, there was no significant difference in the expression levels of Pax3-FKHR throughout the first twenty-four hours of differentiation either by direct Western blot analysis (Figure 2A) or after quantifying the values as described for Pax3 (Figure 2B). The observed differences in protein levels did not result from differences in gel loading as confirmed by a Western blot analysis of the same membranes for the housekeeping gene GAPDH (Figure 2A). Therefore, these results directly demonstrate that the expression of Pax3-FKHR, but not wild-type Pax3, persists throughout the first twenty-four hours of myogenic differentiation.
3.2. Pax3 and Pax3-FKHR mRNA levels do not decrease throughout early myogenesis
The expression levels of proteins can be regulated on the transcriptional, translational, or the post-translational level. To determine if the observed differences in the expression of protein of Pax3 and Pax3-FKHR resulted from differences in their transcript levels, we analyzed mRNA for Pax3 and Pax3-FKHR throughout the first twenty-four hours of differentiation. Primary myoblasts or primary myoblasts were stably transduced with Pax3 or Pax3-FKHR as described above, cytoplasmic poly(A)+ mRNA was isolated from undifferentiated primary myoblasts and myoblasts differentiated for 8, 12, 18, and 24 hours, and a constant amount of mRNA was used for real-time one-step reverse transcriptase PCR (RT-PCR) analysis to determine the levels of endogenous Pax3, ectopic Pax3, or ectopic Pax3-FKHR mRNA. These time points of differentiation correlate to Pax3 protein expression levels of 100%, 60%, 20% and 5%, respectively (Figure 2), and would be expected to result in significantly different levels of mRNA expression. In order to amplify endogenous Pax3 mRNA we utilized primers specific for exon 6 and exon 7 of Pax3 (Figure 3A). In order to specifically amplify ectopic Pax3 or Pax3-FKHR mRNA we utilized primers specific for the 3′-end of Pax3 or FKHR, respectively, and a primer specific for GFP (Figure 3A). Therefore, only mRNA that derives from virally introduced DNA will be amplified. Real-time RT-PCR was also performed in parallel reactions using primers specific for β-actin to control for mRNA levels. A gel electrophoretic analysis of the reactions demonstrated single DNA bands with sizes that were consistent with the predicted PCR products for the mature spliced mRNA (data not shown) confirming the purity of the isolated mRNA and the specificity of the PCR primers used in the real-time RT-PCR analysis.
The real-time one-step RT-PCR analysis demonstrated very similar amplification curves and threshold cycle numbers (CT) for all of the time points of differentiation for Pax3, either endogenous or ectopically expressed, and Pax3-FKHR (data not shown). We used the 2-ΔΔCT method [29] to quantify the levels of mRNA for each of the time points of differentiation relative to the undifferentiated cells, as described in the Materials and Methods. If the protein expression levels of Pax3 and Pax3-FKHR were being regulated on a transcriptional level we would expect to see a decrease in mRNA transcript as differentiation progresses consistent with the decrease in levels of protein. However, in contrast to the results for Pax3 protein expression levels, we observed a moderate increase in the relative levels of mRNA for endogenous Pax3 and no statistical difference in the relative levels of ectopic Pax3 mRNA throughout the first twenty-four hours of differentiation (Figure 3B). In addition, we observed moderate changes in the relative levels of mRNA for ectopic Pax3-FKHR (Figure 3B). Therefore, because the changes in mRNA level do not correlate with the observed changes in protein expression, we conclude that the differences in the protein levels for Pax3 and Pax3-FKHR during myogenic differentiation are not regulated on the transcriptional level.
3.3. Pax3-FKHR is more stable than Pax3 at twenty-four hours of differentiation
We performed an in vivo pulse-chase labeling experiment to determine if the differences in Pax3 and Pax3-FKHR protein expression at twenty-four hours of differentiation resulted from changes in protein translation or protein stability. The levels of radioactively labeled endogenous Pax3 were below the limit of detection for the assay (data not shown). However, our results demonstrate that there is no significant difference between the expression of endogenous and ectopic Pax3 protein or mRNA levels (Figures 1 - 3), indicating that the mechanism is similar for Pax3 regardless of whether it derives from an endogenous or ectopic DNA source. Therefore, primary myoblasts stably expressing either Pax3 or Pax3-FKHR were differentiated for twenty-two and a half hours, starved of methionine and cysteine for thirty minutes, and pulsed with [35S]-methionine and [35S]-cysteine for one hour, allowing the non-radioactive chase to be initiated at twenty-four hours of differentiation. Total cell extracts were made at the indicated times after the initiation of the chase and Pax3 or Pax3-FKHR were immunoprecipitated from 400μg of total cell extracts and visualized by SDS-PAGE. The amount of labeled Pax3 and Pax3-FKHR were quantified by densitometry and normalized for the amount of total cell extract used for the immunoprecipitation.
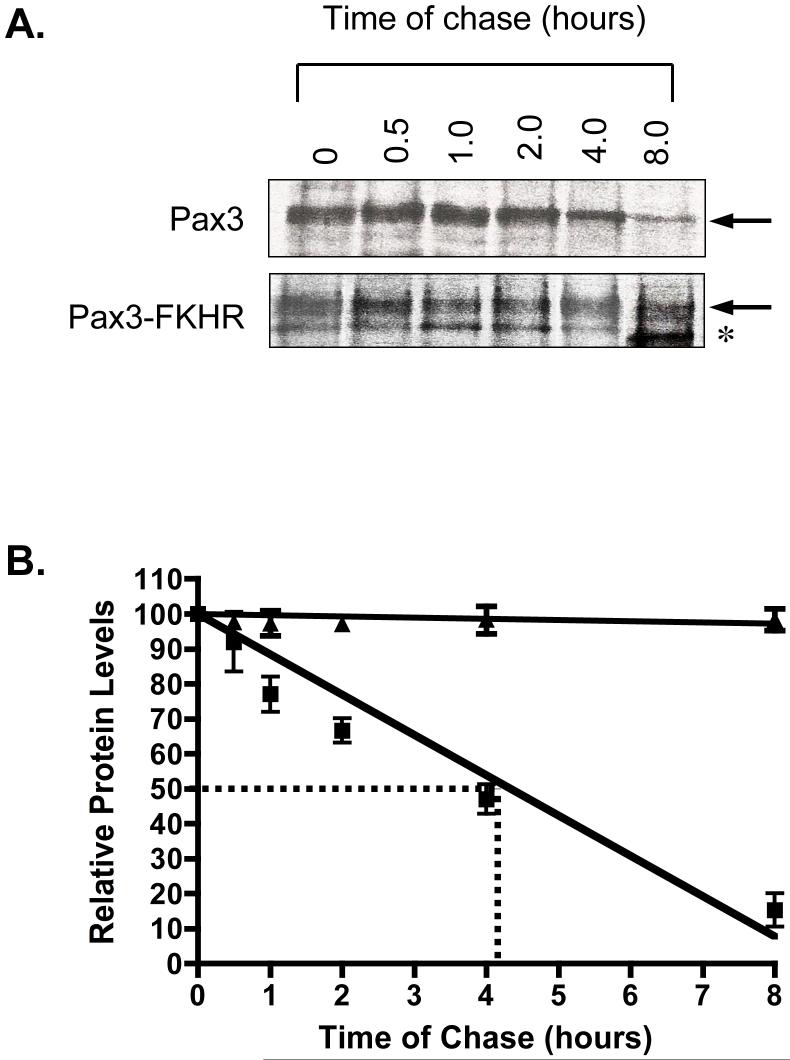
A representative pulse-chase analysis is illustrated in Figure 4A and demonstrates that we were able to immunoprecipitate radiolabeled Pax3 and Pax3-FKHR at the initiation of the chase. This result demonstrates the efficient translation of both proteins at twenty-four hours of differentiation. We observed a consistent decrease in the amount of radiolabeled Pax3 over the time of the chase with no apparent change in the amount of radiolabeled Pax3-FKHR (Figure 4A). A quantification of these results by densitometry demonstrated no significant change in the levels of radiolabeled Pax3-FKHR over the eight-hour chase, indicating that the half-life of Pax3-FKHR is significantly greater than eight hours. In contrast, we determined that Pax3 has a half-life of approximately four hours (Figure 4B). Therefore, we conclude that Pax3-FKHR has significantly greater post-translational protein stability than wild type Pax3, which accounts for the observed differences in protein expression levels between the two proteins during early myogenic differentiation.
Figure 4.
Pulse-chase analysis of Pax3 and Pax3-FKHR at twenty-four hours of differentiation. A. Representative pulse-chase analysis of Pax3 and Pax3-FKHR from primary myoblasts induced to differentiate for twenty-four hours. After twenty-four hours of differentiation pulse-chase analysis was performed to determine the stability of the proteins, as described in the Materials and Methods. B. The protein levels of Pax3 and Pax3-FKHR observed in panel A were quantified by densitometric analysis and normalized for the amount of total cell lysate used for the immunoprecipitation. The normalized values of Pax3 (squares) and Pax3-FKHR (triangles) for each time point were plotted relative to the level of radiolabeled protein present at the 0 hour time point, which was given a relative value of 100%. The dotted line indicates the half-life for Pax3 as determined from the graph. The errors bars indicate the standard deviation from the mean for the average of three independent determinations.
3.4. Differentiation capability of primary myoblasts expressing Pax3 and Pax3-FKHR
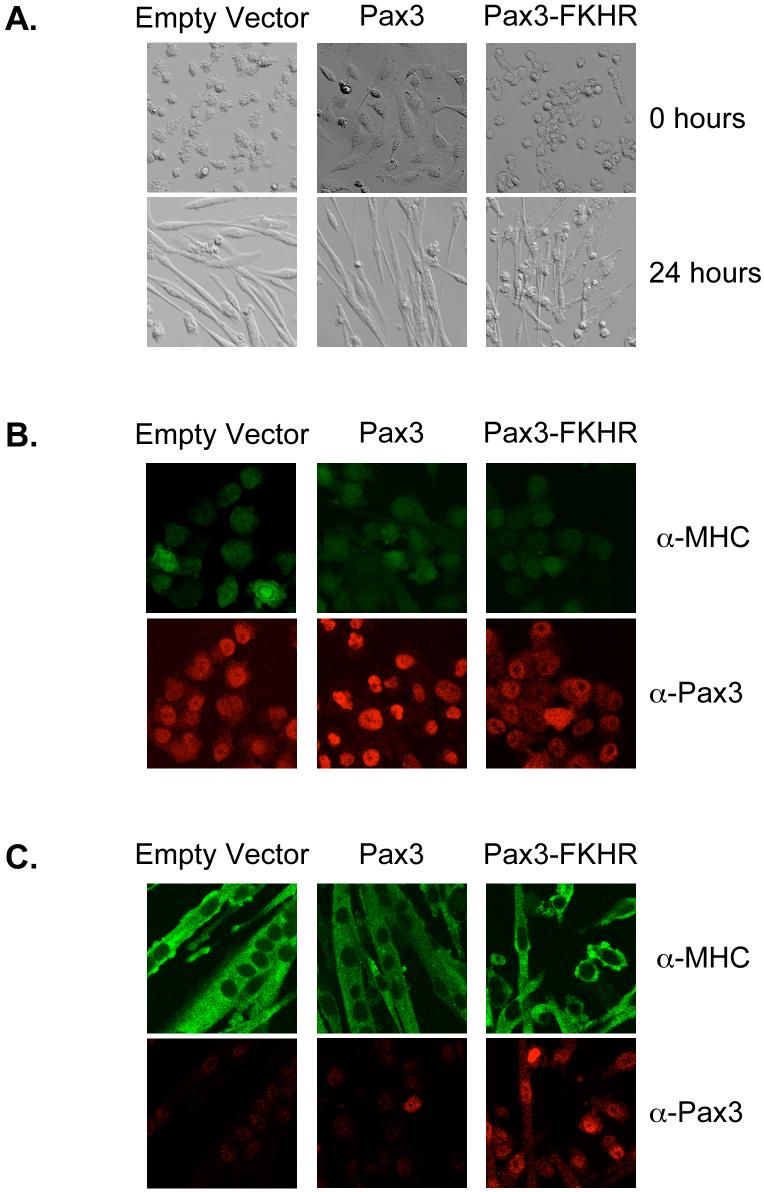
Finally, it has previously been reported that the ectopic expression of Pax3 and Pax3-FKHR inhibited the differentiation of C2C12 myoblasts [30]. Therefore, it would be expected that the loss of Pax3 would therefore allow differentiation to progress while the aberrant persistent expression of Pax3-FKHR might affect the differentiation capability of the mouse primary myoblasts. As demonstrated in Figure 5A and consistent with the morphology of undifferentiated primary myoblasts [25], proliferating primary myoblasts stably transduced with either the empty vector or Pax3-FKHR are present as small round cells. Proliferating cells expressing Pax3 appeared flatter and more elongated, which may result from the ability of Pax3 to activate the expression of early myogenic factors [10]. Immunofluorescence of the proliferating cells confirmed the expression of Pax3 and Pax3-FKHR and that these proteins were present primarily as nuclear proteins (Figure 5B, bottom panels), consistent with previous reports [4, 5, 28]. As expected for proliferating primary myoblasts, only minimal levels of non-specific MHC staining were observed (Figure 5B, top panels).
Figure 5.
Differentiation capability of primary myoblasts expressing Pax3 or Pax3-FKHR. A) Phase contrast microscopy of proliferating (top panels) or differentiated (bottom panels) primary myoblasts stably transduced with empty vector, Pax3, or Pax3-FKHR. Cells are shown at a 40X magnification. Co-immunofluorescence of proliferating (B) and cells differentiated for twenty-four hours (C) to examine the expression of Pax3 (red) and Myosin Heavy Chain (green).
After twenty-four hours of differentiation the cells stably transduced with either the empty vector or Pax3 have elongated, fused, and formed multi-nucleated myotubes (Figure 5A, bottom panels, and 5C). Consistent with our Western blot analyses (Figures 1 and 2), immunofluorescent staining of these cells demonstrate the significant loss of the expression of Pax3 (Figure 5C, bottom panels). In addition, we observed a strong cytoplasmic staining for MHC (Figure 5C, top panels) demonstrating efficient differentiation has occurred in these cells. In contrast, cells stably transduced with Pax3-FKHR have flattened and show some elongation but are present primarily as mononucleated myoblasts (Figure 5A, bottom panel, and 5C). This result is consistent with previous reports [30] and indicates that the presence of Pax3-FKHR inhibits the ability of primary myoblasts to terminally differentiate. However, despite the inability of myoblasts expressing the fusion protein to fuse and form multinucleated myotubes, MHC is still being expressed in the cytoplasm (Figure 5C, top panels), indicating that the block to terminal differentiation may occur at the level of fusion and not through the inhibition of the expression of late myogenic markers. The results for the control cells and cells stably transduced with Pax3 or Pax3-FKHR were observed multiple times at a variety of cell densities (data not shown) indicating that the differentiation capability of these cells was reproducible and independent of the cell density.
4. Discussion
The oncogenic fusion protein Pax3-FKHR, which results from the t(2;13)(q35;q14) chromosomal translocation, is one of the hallmarks of the childhood skeletal muscle tumor ARMS. To better understand some of the underlying mechanisms of how the fusion protein contributes to the development of ARMS, it is necessary to determine its biological actions relative to each of its constituent parts (i.e. - wild type Pax3 and/or FKHR). Along these lines, the protein expression levels of FKHR and the effects of myogenic differentiation on these levels have previously been characterized [21, 22]. In contrast, similar studies have yet to be performed on wild-type Pax3 or the Pax3-FKHR fusion protein. Knowledge of the effects of myogenic differentiation on the protein expression levels of Pax3 and how fusion of FKHR to Pax3 affects these levels would provide important insights into the nature of ARMS. Therefore, we demonstrate for the first time that Pax3-FKHR has a significantly greater post-translational stability relative to wild type Pax3, which results in the aberrant persistence of its expression throughout myogenic differentiation and may contribute to the inability of Pax3-FKHR expressing cells from achieving terminal differentiation.
The conclusion that Pax3-FKHR is more stable than Pax3 and that this persistent expression may inhibit myoblast differentiation is based on the following observations. (i) Pax3 protein levels decrease significantly in the first twenty-four hours of myogenic differentiation. In contrast, the protein expression levels of Pax3-FKHR remain relatively unchanged (Figures 1 and 2); (ii) These differences in protein expression do not result from differences on the transcriptional level since changes in mRNA levels for Pax3 do not correlate with the decrease in Pax3 protein levels (Figure 3); (iii) These differences in protein expression also do not result from changes in translation since we observed no differences in the level of radioactively labeled Pax3 and Pax3-FKHR at twenty-four hours of differentiation (Figure 4A, zero time point); (iv) The half-life of Pax3-FKHR (≫8 hours) is significantly greater than the half-life of wild-type Pax3 (≈ 4 hours) (Figure 4); and (v) Primary myoblasts that have a persistent expression of Pax3-FKHR are unable to fuse and form multi-nucleated myotubes (Figure 5).
Wild-type Pax3 is a myogenic transcription factor that regulates the expression of early myogenic genes [10, 31, 32] and as such it is essential that its expression be tightly regulated for the successful progression through myogenic differentiation. A previous report demonstrated that the enforced expression of Pax3 inhibited myogenic differentiation of C2C12 myoblasts and that this inhibition may be required to suppress differentiation of migrating limb muscle progenitors [30]. This model would therefore predict that in order for efficient myogenic differentiation to occur there must be a concomitant decrease in the expression of Pax3. In the present report we demonstrate that Pax3 is expressed in proliferating mouse primary myoblasts (Figures 1 and 2). More importantly, and consistent with the prediction stated above, we found that Pax3 protein expression levels decrease nearly 95% in differentiated primary myoblasts and that this decrease resulted from the post-translational regulation of Pax3 protein stability. Therefore, taken together with previous literature reports that demonstrate Pax3 inhibits myogenesis in an immortalized cell line, our results are the first to indicate that the post-translationally regulated loss of Pax3 protein expression during early myogenic differentiation may be required for the non-immortalized primary myoblasts to achieve terminal differentiation.
It was previously reported that like Pax3, Pax3-FKHR is also able to inhibit the terminal differentiation of C2C12 myoblasts [30]. However, unlike Pax3, whose expression decreases significantly by twenty-four hours of differentiation, the enhanced post-translational stability of Pax3-FKHR results in its aberrant persistence of expression throughout early myogenic differentiation. In fact, we have demonstrated that primary myoblasts that contain a persistent expression of Pax3-FKHR are unable to terminally differentiate (Figure 5). We believe that this inability to differentiate is a direct result of the expression of Pax3-FKHR since primary myoblasts stably transduced with either Pax3 or an empty vector negative control are capable of forming multi-nucleated myotubes. The only difference between these three cell lines is the expression of Pax3-FKHR and as such the inability of these myoblasts to differentiate must result from the expression of Pax3-KFHR. Therefore, combined with its enhanced transcriptional activity and its inability to be repressed by the co-repressor hDaxx, the enhanced stability of Pax3-FKHR provides yet one more mechanism by which the deregulation of normal Pax3 or FKHR functions may contribute to the development of ARMS.
At present the exact mechanism by which Pax3-FKHR obtains its enhanced post-translational stability relative to Pax3 is not known. It is possible that the fusion of FKHR to Pax3 may sterically hinder post-translational modifications that are required to regulate the degradation of the wild-type proteins. Alternatively, the fusion of Pax3 to FKHR, or vice versa, may add sites of post-translational modification that result in increasing the stability of Pax3-FKHR. Although it has been reported that post-translational modifications of FKHR, in particular phosphorylation, regulate its cellular localization and protein stability [24, 33], it is not known if these sites present in the Pax3-FKHR are phosphorylated and if these events can modify the stability of the fusion protein. Experiments are presently being performed to determine how and if phosphorylation of Pax3 and Pax3-FKHR regulate their protein stability.
Acknowledgements
We thank Dr. Gerard Grosveld, St. Jude Children’s Research Hospital, for kindly providing us with laboratory and office space and allowing us access to reagents and supplies during the aftermath of Hurricane Katrina. We also thank San-San Ng, Louisiana State University Health Sciences Center, New Orleans, for her assistance with the real-time one-step PCR analysis. This work was supported, in part, by grant number 1-P20-RR020152-01 from the National Center for Research Resources (NCRR), a component of the National Institutes of Health (NIH), grant number LEQSF(2004-07)-RD-A-19 from the Louisiana Board of Regents (BoRSF), and the Louisiana Cancer Research Consortium (LCRC). Its contents are solely the responsibility of the authors and do not necessarily represent the official views of NCRR, NIH, BoRSF, or the LCRC.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- [1].Barr FG. The role of chimeric paired box transcription factors in the pathogenesis of pediatric rhabdomysarcoma. Cancer Res. 1999;59:1711s–1715s. [PubMed] [Google Scholar]
- [2].Galili N, Davis RJ, Fredericks WJ, Mukhopadhyay S, Rauscher FJ, 3rd, Emanuel BS, Rovera G, Barr FG. Fusion of a fork head domain gene to PAX3 in the solid tumour alveolar rhabdomyosarcoma. Nat Genet. 1993;5:230–5. doi: 10.1038/ng1193-230. [DOI] [PubMed] [Google Scholar]
- [3].Shapiro DN, Sublett JE, Li B, Downing JR, Naeve CW. Fusion of PAX3 to a member of the forkhead family of transcription factors in human alveolar rhabdomyosarcoma. Cancer Res. 1993;53:5108–12. [PubMed] [Google Scholar]
- [4].Hollenbach AD, Sublett JE, McPherson CJ, Grosveld G. The Pax3-FKHR oncoprotein is unresponsive to the Pax3-associated repressor hDaxx. Embo J. 1999;18:3702–11. doi: 10.1093/emboj/18.13.3702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Hollenbach AD, McPherson CJ, Lagutina I, Grosveld G. The EF-hand calcium-binding protein calmyrin inhibits the transcriptional and DNA-binding activity of Pax3. Biochim Biophys Acta. 2002;1574:321–8. doi: 10.1016/s0167-4781(02)00230-0. [DOI] [PubMed] [Google Scholar]
- [6].Stuart ET, Kioussi C, Gruss P. Mammalian Pax genes. Annu Rev Genet. 1994;28:219–36. doi: 10.1146/annurev.ge.28.120194.001251. [DOI] [PubMed] [Google Scholar]
- [7].Bober E, Franz T, Arnold HH, Gruss P, Tremblay P. Pax-3 is required for the development of limb muscles: a possible role for the migration of dermomyotomal muscle progenitor cells. Development. 1994;120:603–12. doi: 10.1242/dev.120.3.603. [DOI] [PubMed] [Google Scholar]
- [8].Goulding M, Lumsden A, Paquette AJ. Regulation of Pax-3 expression in the dermomyotome and its role in muscle development. Development. 1994;120:957–71. doi: 10.1242/dev.120.4.957. [DOI] [PubMed] [Google Scholar]
- [9].Tajbakhsh S, Rocancourt D, Cossu G, Buckingham M. Redefining the genetic hierarchies controlling skeletal myogenesis: Pax-3 and Myf-5 act upstream of MyoD. Cell. 1997;89:127–38. doi: 10.1016/s0092-8674(00)80189-0. [DOI] [PubMed] [Google Scholar]
- [10].Maroto M, Reshef R, Munsterberg AE, Koester S, Goulding M, Lassar AB. Ectopic Pax-3 activates MyoD and Myf-5 expression in embryonic mesoderm and neural tissue. Cell. 1997;89:139–48. doi: 10.1016/s0092-8674(00)80190-7. [DOI] [PubMed] [Google Scholar]
- [11].Anderson MJ, Viars CS, Czekay S, Cavenee WK, Arden KC. Cloning and characterization of three human forkhead genes that comprise an FKHR-like gene subfamily. Genomics. 1998;47:187–99. doi: 10.1006/geno.1997.5122. [DOI] [PubMed] [Google Scholar]
- [12].Gajiwala KS, Burley SK. Winged helix proteins. Curr Opin Struct Biol. 2000;10:110–6. doi: 10.1016/s0959-440x(99)00057-3. [DOI] [PubMed] [Google Scholar]
- [13].Modur V, Nagarajan R, Evers BM, Milbrandt J. FOXO proteins regulate tumor necrosis factor-related apoptosis inducing ligand expression. Implications for PTEN mutation in prostate cancer. J Biol Chem. 2002;277:47928–37. doi: 10.1074/jbc.M207509200. [DOI] [PubMed] [Google Scholar]
- [14].Dijkers PF, Medema RH, Pals C, Banerji L, Thomas NS, Lam EW, Burgering BM, Raaijmakers JA, Lammers JW, Koenderman L, Coffer PJ. Forkhead transcription factor FKHR-L1 modulates cytokine-dependent transcriptional regulation of p27(KIP1) Mol Cell Biol. 2000;20:9138–48. doi: 10.1128/mcb.20.24.9138-9148.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Medema RH, Kops GJ, Bos JL, Burgering BM. AFX-like Forkhead transcription factors mediate cell-cycle regulation by Ras and PKB through p27kip1. Nature. 2000;404:782–7. doi: 10.1038/35008115. [DOI] [PubMed] [Google Scholar]
- [16].Hall RK, Yamasaki T, Kucera T, Waltner-Law M, O’Brien R, Granner DK. Regulation of phosphoenolpyruvate carboxykinase and insulin-like growth factor-binding protein-1 gene expression by insulin. The role of winged helix/forkhead proteins. J Biol Chem. 2000;275:30169–75. doi: 10.1074/jbc.M004898200. [DOI] [PubMed] [Google Scholar]
- [17].Schmoll D, Walker KS, Alessi DR, Grempler R, Burchell A, Guo S, Walther R, Unterman TG. Regulation of glucose-6-phosphatase gene expression by protein kinase Balpha and the forkhead transcription factor FKHR. Evidence for insulin response unit dependent and -independent effects of insulin on promoter activity. J Biol Chem. 2000;275:36324–33. doi: 10.1074/jbc.M003616200. [DOI] [PubMed] [Google Scholar]
- [18].Daitoku H, Yamagata K, Matsuzaki H, Hatta M, Fukamizu A. Regulation of PGC-1 promoter activity by protein kinase B and the forkhead transcription factor FKHR. Diabetes. 2003;52:642–9. doi: 10.2337/diabetes.52.3.642. [DOI] [PubMed] [Google Scholar]
- [19].Nakae J, Kitamura T, Kitamura Y, Biggs WH, 3rd, Arden KC, Accili D. The forkhead transcription factor Foxo1 regulates adipocyte differentiation. Dev Cell. 2003;4:119–29. doi: 10.1016/s1534-5807(02)00401-x. [DOI] [PubMed] [Google Scholar]
- [20].Hu MC, Lee DF, Xia W, Golfman LS, Ou-Yang F, Yang JY, Zou Y, Bao S, Hanada N, Saso H, Kobayashi R, Hung MC. IkappaB kinase promotes tumorigenesis through inhibition of forkhead FOXO3a. Cell. 2004;117:225–37. doi: 10.1016/s0092-8674(04)00302-2. [DOI] [PubMed] [Google Scholar]
- [21].Nishiyama T, Kii I, Kudo A. Inactivation of Rho/ROCK signaling is crucial for the nuclear accumulation of FKHR and myoblast fusion. J Biol Chem. 2004;279:47311–9. doi: 10.1074/jbc.M403546200. [DOI] [PubMed] [Google Scholar]
- [22].Bois PR, Grosveld GC. FKHR (FOXO1a) is required for myotube fusion of primary mouse myoblasts. Embo J. 2003;22:1147–57. doi: 10.1093/emboj/cdg116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Nakae J, Barr V, Accili D. Differential regulation of gene expression by insulin and IGF-1 receptors correlates with phosphorylation of a single amino acid residue in the forkhead transcription factor FKHR. Embo J. 2000;19:989–96. doi: 10.1093/emboj/19.5.989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Matsuzaki H, Daitoku H, Hatta M, Tanaka K, Fukamizu A. Insulin-induced phosphorylation of FKHR (Foxo1) targets to proteasomal degradation. Proc Natl Acad Sci U S A. 2003;100:11285–90. doi: 10.1073/pnas.1934283100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Rando TA, Blau HM. Primary mouse myoblast purification, characterization, and transplantation for cell-mediated gene therapy. J Cell Biol. 1994;125:1275–87. doi: 10.1083/jcb.125.6.1275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Laker C, Meyer J, Schopen A, Friel J, Heberlein C, Ostertag W, Stocking C. Host cis-mediated extinction of a retrovirus permissive for expression in embryonal stem cells during differentiation. J Virol. 1998;72:339–48. doi: 10.1128/jvi.72.1.339-348.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Swift S, Lorens J, Achacoso P, Nolan GP. In: Current Protocols in Immunology. Coligan JE, Kruisbeek AM, Margulies DH, Shevach EM, Strober W, editors. John Wiley and Sons; Boston, MA: 1999. pp. 10–17. [Google Scholar]
- [28].Lam PY, Sublett JE, Hollenbach AD, Roussel MF. The oncogenic potential of the Pax3-FKHR fusion protein requires the Pax3 homeodomain recognition helix but not the Pax3 paired-box DNA binding domain. Mol Cell Biol. 1999;19:594–601. doi: 10.1128/mcb.19.1.594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–8. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- [30].Epstein JA, Lam P, Jepeal L, Maas RL, Shapiro DN. Pax3 inhibits myogenic differentiation of cultured myoblast cells. J Biol Chem. 1995;270:11719–22. doi: 10.1074/jbc.270.20.11719. [DOI] [PubMed] [Google Scholar]
- [31].Ridgeway AG, Skerjanc IS. Pax3 is essential for skeletal myogenesis and the expression of Six1 and Eya2. J Biol Chem. 2001;276:19033–9. doi: 10.1074/jbc.M011491200. [DOI] [PubMed] [Google Scholar]
- [32].Epstein JA, Shapiro DN, Cheng J, Lam PY, Maas RL. Pax3 modulates expression of the c-Met receptor during limb muscle development. Proc Natl Acad Sci U S A. 1996;93:4213–4218. doi: 10.1073/pnas.93.9.4213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Rena G, Woods YL, Prescott AR, Peggie M, Unterman TG, Williams MR, Cohen P. Two novel phosphorylation sites on FKHR that are critical for its nuclear exclusion. Embo J. 2002;21:2263–71. doi: 10.1093/emboj/21.9.2263. [DOI] [PMC free article] [PubMed] [Google Scholar]