Abstract
Bladder cancer transformation and immortalization require the inactivation of key regulatory genes, including TP53. Genotyping of a large cohort of bladder cancer patients (n = 256) using the TP53 GeneChip showed mutations in 103 cases (40.2%), the majority of them mapping to the DNA-binding core domain. TP53 mutation status was significantly associated with tumor stage (P = 0.0001) and overall survival for patients with advanced disease (P = 0.01). Transcript profiling using oligonucleotide arrays was performed on a subset of these cases (n = 46). Supervised analyses identified genes differentially expressed between invasive bladder tumors with wild-type (n = 24) and mutated TP53 (n = 22). Pathway analyses of top-ranked genes supported the central role of TP53 in the functional network of such gene patterns. A proteomic strategy using reverse phase arrays with protein extracts of bladder cancer cell lines validated the association of identified differentially expressed genes, such as gelsolin, to TP53 status. Immunohistochemistry on tissue microarrays (n = 294) revealed that gelsolin was associated with tumor stage and overall survival, correlating positively with TP53 status in a subset of these patients. This study further reveals that TP53 mutations are frequent events in bladder cancer progression and identified gelsolin related to TP53 status, tumor staging, and clinical outcome by independent high-throughput strategies.
The nuclear protein Tp53 plays an essential role in the regulation of cell cycle and apoptosis, contributing to transformation and malignancy.1 Tp53 is a DNA-binding protein containing transcription, DNA binding, and oligomerization activation domains, functioning as a tumor suppressor.2,3 Mutants of TP53 that frequently occur in a number of different human cancers, including bladder cancer, fail to bind the consensus DNA-binding site and hence cause the loss of tumor suppressor activity.4 Alterations of the TP53 gene occurs both as germline mutations, such as in cancer-prone families with Li-Fraumeni syndrome, or somatic mutations in diverse human malignancies.5
TP53 is one of the proteins better characterized in cancer research with reported targets, regulators, and binding proteins. For example, targets regulated by TP53 include cell-cycle genes, such as p21, and anti-apoptotic genes, such as bax. Regulators of TP53 include ataxia telangiectasia mutated (ATM) and Chk2, whereas Abl1 and the adenomatous polyposis gene (APC) are among known binding TP53 proteins.6,7,8 However, little is known of the differential gene expression patterns of human tumors presenting wild-type TP53 compared with those with a mutant protein. With the advent of microarray technologies, characterization of TP53 sequences and gene expression profiles associated with TP53 status are available in a high-throughput manner. Bladder cancer is one of the tumors in which TP53 is altered with a high frequency, mutation rates being ∼40% in advanced stages of the disease.9,10,11,12 The present study was designed to identify targets that would differentiate patients presenting advanced disease with wild-type versus mutant TP53 (Figure 1). Gelsolin was selected as one on the genes located to chromosome 9q33, a frequently mutated locus in bladder cancer.9,13 Two proteomic approaches were used to evaluate the link of gelsolin with tumor progression and TP53 status. Immunohistochemical analyses on tissue arrays containing well-annotated bladder tumors and known TP53 status served to associate the expression of gelsolin with TP53, tumor stage, and survival. The differential expression of gelsolin among several bladder cancer cell lines of known TP53 alterations was evaluated by custom-made reverse phase arrays.
Figure 1.
Experimental design. TP53 genotyping was performed on 256 bladder tumors using the TP53 sequencing arrays. Gene expression analyses using the U133A array were performed in a subset of 46 bladder tissues to identify targets differentially expressed in bladder cancer regarding their TP53 status (TP53 wild type, n = 24; and TP53 mutated, n = 22). Supervised methods identified 149 probes differentially expressed between those cases with either wild-type or mutated TP53. Two types of validation studies of the association of molecular profiles with TP53 status were performed. Immunohistochemical patterns were analyzed on tissue arrays containing 294 tumors, a subset of them of known TP53 and clinical outcome status. Proteomic reverse phase arrays were also performed on protein extracts of bladder cancer cell lines of known TP53 status.
Materials and Methods
TP53 Sequencing Analyses
DNA Extraction and Tissue Samples
Total DNA was extracted using a nonorganic method (Oncor, Gaithersburg, MD). Macrodissection of OCT-embedded tissue blocks was performed to ensure a minimum of 75% tumor cells.13 DNA quality was evaluated based on 260/280 ratios of absorbances. Specimens were collected under institutional review board approval. These tumors comprised 10 pTa, 32 pT1, 22 pT2, 175 pT3, and 15 pT4 specimens from patients with bladder cancer.
TP53 oligonucleotide array assay (GeneChip p53; Affymetrix, Santa Clara, CA). Purified DNA (100 ng) was subjected to multiplex-polymerase chain reactions (PCRs) amplifying exons 2 to11 simultaneously, using reagents supplied by the manufacturer (Affymetrix). Apart from the DNA, each PCR reaction contained 10 U of AmpliTaq Gold, PCR buffer II, 2.5 mmol/L MgCl2, 5 μl of the primer set, and 0.2 mmol/L each dNTP. The reaction was performed in a final volume of 100 μl. The PCR profile consisted of an initial heating at 95°C for 10 minutes, followed by 35 cycles of 95°C for 30 seconds, 60°C for 30 seconds, and 72°C for 45 seconds, with a final extension step at 72°C for 10 minutes. Forty-five μl of the PCR product was then fragmented by the addition of 0.25 U of fragmentation reagent (DNase I in 10 mmol/L Tris-HCl, pH 7.5, 10 mmol/L CaCl2, 10 mmol/L MgCl2, and 500 ml/L glycerol) along with 2.5 U of calf intestine alkaline phosphatase, 0.4 mmol/L ethylenediaminetetraacetic acid, and 0.5 mol/L Tris-acetate, and incubation at 25°C for 15 minutes, followed by heat inactivation at 95°C for 10 minutes. For labeling, 50 μl of the fragmented DNA was incubated at 37°C for 45 minutes with 10 μmol/L fluorescein-N6-dideoxy-ATP, 25 U of terminal transferase, and TdTase buffer in a total volume of 100 μl, followed by heat inactivation at 95°C for 10 minutes. The sample was hybridized to the chip in a volume of 0.5 ml containing 6× sodium chloride/sodium phosphate/EDTA (SSPE) buffer, 0.5 ml/L Triton X-100, 1 mg of acetylated bovine serum albumin, 2 nmol/L control oligonucleotide, and the labeled DNA sample. Hybridization was done in an oven with constant agitation at 45°C for 30 minutes. The chip was then washed on the wash station four times with 3× SSPE containing 0.05 ml/L Triton X-100. After washing, GeneChips were read using a confocal laser scanner, and data were aligned and analyzed. A reference from the control DNA supplied was also analyzed. This reference belonged to the same PCR round and was measured on the same batch of chips.14,15
Gene Profiling Using U133A GeneChips
Tissue Samples and RNA Extraction
Tumors belonging to patients with invasive bladder cancer (pT2+) were obtained by cystectomy or cystoprostatectomy at Memorial Sloan-Kettering Cancer Center. Specimens were collected under institutional review board approval of this institution. Macrodissection of OCT-embedded tissue blocks was performed to ensure a minimum of 75% tumor cells. Because of the high heterogeneity of muscle-invasive bladder tumors, this conservative cutoff of 75% would guarantee that tumor subpopulations would be representative enough to identify targets associated with TP53 status in cancer cells. Total RNA was extracted using TRIzol (Life Technologies, Rockville, MD) and purification with RNeasy columns (Qiagen, Valencia, CA). RNA quality was evaluated based on 260/280 ratios of absorbances and by gel analysis using an Agilent 2100 BioAnalyzer (Agilent Technologies, Palo Alto, CA).13 Selection of cases for oligonucleotide arrays focused on balancing numbers of cases with wild-type (n = 24) and mutant TP53 (n = 22), covering the most frequent TP53 mutations in the DNA-binding core domain in cases displaying all advanced disease (PT2+).
Labeling and Hybridization
Complementary DNA of the analyzed specimens was synthesized from 1.5 μg of total RNA using a T7-promoter- tagged oligo-dT primer. RNA target was synthesized by in vitro transcription and labeled with biotinylated nucleotides (Enzo Biochem, Farmingdale, NY). Labeled target was hybridized on GeneChip test 3 arrays (Affymetrix) to assess the quality of the sample before hybridizing onto the human genome U133A arrays including 22,283 probes representing known genes and expressed sequence tags (Affymetrix), as previously reported.13
GeneChip Analysis
Scanned image files were visually inspected for artifacts and analyzed using Affymetrix Microarray Suite 5.0 (MAS 5.0). Expression values of each array were multiplicatively scaled to have an average expression of 500 at least across the central 95% of all genes on the array. Signal was used as the primary measure of expression level, and detection was retained as a complementary measure.13
Immunohistochemical Analyses
Cell Lines, Tissue Arrays, and Immunohistochemistry
Cytospins of bladder cancer cell lines were obtained after centrifugation at low speed, 800 rpm, for 5 minutes.16 Four different bladder cancer microarrays were constructed in the Division of Molecular Pathology and used in this study. These arrays included a total of 294 primary transitional cell carcinomas (TCCs) of the bladder, belonging to patients recruited at Memorial Sloan-Kettering Cancer Center under institutional review board-approved protocols. A total of 93 non-muscle-invasive and 201 invasive TCC tumors were analyzed in these microarrays. These tumors corresponded to 34 grade 1, 69 grade 2, and 191 grade 3 lesions. One of these tissue microarrays comprised a cohort of four non-muscle-invasive lesions and 91 invasive tumors with annotated follow-up and known status of TP53. This array allowed clinical outcome assessment and evaluation of the associations of novel markers with TP53. Protein expression patterns of gelsolin were assessed at the microanatomical level on these tissue microarrays by immunohistochemistry using standard avidin-biotin immunoperoxidase procedures. Western blot assays were performed to address the specificity of the antibodies under study. We used a mouse monoclonal antibody against TP53 (1801) at 1:500 dilution (Calbiochem, San Diego, CA) and gelsolin at 1/1000 (Sigma, St. Louis, MO) on formalin-fixed/paraffin-embedded sections. The avidin-biotin immunoperoxidase technique was the immunohistochemical method applied. For specific epitopes on paraffin sections, we used antigen retrieval methods (0.01% citric acid for 15 minutes under microwave treatment) before incubation with primary antibodies or antiserum overnight at 4°C. Secondary antibodies were biotinylated horse anti-mouse or goat anti-rabbit antibodies (Vector Laboratories, Peterborough, UK), which were used at 1:500 or 1:1000 dilution, respectively. Diaminobenzidine was used as the final chromogen and hematoxylin as the nuclear counterstain. Two independent pathologists (C.C.-C. and N.B.), blinded to the TP53 or clinical status of the samples, reviewed immunohistochemical stainings.
Statistical Analysis
All TCCs (n = 294) were used for the analysis of association among gelsolin with clinicopathological variables and the expression patterns of TP53. The consensus value of the representative cores from each tumor sample arrayed was used for statistical analyses. The association of the expression of the selected targets with histopathological stage and tumor grade was evaluated using the nonparametric Wilcoxon-Mann-Whitney and Kruskall-Wallis tests. There is no consensus on the cutoffs of the immunohistochemical expression of the other markers, and thus they were analyzed as continuous variables.17 Survival analyses were performed taking the cutoffs of 20% for TP53 and 5% for gelsolin.
The associations of the markers identified in the DNA microarray analysis to outcome were also evaluated at the protein level using a subset of 95 TCCs of the bladder cases for which follow up was available. Overall-survival time was defined as the years elapsed between transurethral resection or cystectomy and death from disease (or the last follow-up date). Patients who were alive at the last follow-up or lost to follow-up were censored. For survival analysis, the association of marker expression levels with overall survival was analyzed using the Wald test, and the log-rank test was used to examine their relationship when different cutoffs were applied.17 Survival curves were plotted using the standard Kaplan-Meier methodology. Associations among gelsolin with TP53 were analyzed using Kendall’s τ b-test.17 Statistical analyses were performed using the SPSS statistical package (version 10.0).
Reverse-Phase Arrays
Bladder Cancer Cell Lines
Nine bladder cancer cell lines were obtained from the American Type Culture Collection (Rockville, MD), grown, and collected under standard tissue culture protocols as previously reported.16 These cell lines were derived from TCCs of the bladder of early stage (RT4), low grade (5637), invasive (T24, J82, UM-UC-3, HT-1376, and HT-1197), and metastatic bladder tumors (TCCSUP), as well as a squamous cell carcinoma cell line (ScaBER). Bladder cancer cell lines were wild type for TP53 (RT4) or presented mutations in TP53 at the following exons: 4 (UM-UC-3, ScaBER), 5 (T24), 7 (HT-1376), 8 (5637 and J82), 10 (TCCSUP), and 11 (HT-1197).16
Protein Lysate Preparation
The bladder cancer cell lines were cultured, and protein extracts were prepared from them as previously described.16 In brief, cells were collected by scraping and washed three times with ice-cold phosphate-buffered saline. The resulting pellets were lysed in buffer containing 9 mol/L urea (Sigma), 4% 3-[(3-cholamidopropyl) dimethylammonio]-1-propanesulfonate (CHAPS; Calbiochem), 2%, pH 8.0 to 10.5, Pharmalyte (Amersham Pharmacia Biotech, Piscataway, NJ), and 65 mmol/L dithiothreitol (Amersham Pharmacia Biotech). After lysis, the samples were centrifuged briefly, and the supernatants were stored at −80°C.
Protein Lysate Array Design and Production
Arrays were prepared on nitrocellulose-coated glass slides (FAST Slides; Schleicher & Schuell, Keene, NH) by using a pin-in-ring format GMS 417 arrayer (Affymetrix) with four 500-μm-diameter pins. Because the samples were viscous, the pin-in-ring format was used to avoid problems because of clogging of quills. Five twofold serial dilutions were made from each lysate. Four 384-well microtiter plates (Genetix, New Milton, UK) were used to array 180 spots (plus eight spatial registration marks for use in image processing) on a 21 × 35-mm area of nitrocellulose membrane. The first dilution (fourfold) was made with buffer containing 5 mol/L urea, 2% Pharmalyte, pH 8 to 10.5, and 65 mmol/L dithiothreitol. The remaining dilutions were then made with buffer containing 6 mol/L urea, 1% CHAPS, 2% Pharmalyte, pH 8 to 10.5, and 65 mmol/L dithiothreitol. Hence, only the lysate concentration changed along each dilution series. The urea concentration was thus kept at 6 mol/L and the CHAPS concentration at 2%, to keep proteins in their denatured forms. To avoid evaporation in the microtiter plate during spotting, humidity in the array chamber was kept at 70 to 90% with a Vicks ultrasonic humidifier (Kaz, Hudson, NY).18
Detection of Specific and Total Protein on Microarrays
Each array was incubated with a specific primary antibody, which was detected by using the catalyzed signal amplification system (DAKO, Carpinteria, CA). Briefly, each slide was washed manually with deionized water to remove urea. Then, in an Autostainer universal staining system (DAKO), it was blocked with I-block (Tropix, Bedford, MA) and incubated with primary and secondary antibodies. Also in the Autostainer, it was then incubated with streptavidin-biotin complex, biotinyl tyramide (for amplification) for 15 minutes, streptavidin-peroxidase for 15 minutes, and 3,3′-diaminobenzidine tetrahydrochloride chromogen for 5 minutes. Between steps, the slide was washed with catalyzed signal amplification buffer. The signal was scanned with a Perfection 1200S scanner (Epson America, Long Beach, CA) with 256-shade gray scale at 600 dots per inch. For detection of total protein, arrays were stained with SYPRO ruby protein blot stain (Molecular Probes, Eugene, OR) and scanned with a FluorImager SI (Amersham Pharmacia Biotech) at 100-μm resolution. Gelsolin expression was quantified at a 1/1000 dilution using a mouse monoclonal antibody (Sigma), whereas mutated TP53 was measured using a mouse monoclonal at 1/500 (Calbiochem, Darmstadt, Germany). Spot images were converted to raw pixel values by a modified version of the p-scan (Peak Quantification with Statistical Comparative Analysis) software.18,19
Western Blotting
Murine monoclonal antibodies were screened for specificity by Western blotting with 20 μg of lysate protein per lane. Western blotting of gelsolin was performed at a 1/500 dilution using a mouse monoclonal antibody (Sigma). The running buffer contained 62.5 mmol/L Tris-HCl, pH 6.8, 2% sodium dodecyl sulfate, 10% glycerol, and 2.5% 2-mercaptoethanol. We used a 4 to 15% sodium dodecyl sulfate-polyacrylamide linear gradient gel (Tris·HCl Ready Gel; Bio-Rad, Hercules, CA), secondary alkaline phosphatase-conjugated goat anti-mouse antibody, and the chemiluminescent immunoblot detection system (ECL, Amersham).
Results
TP53 Mutation Status Is Associated with Bladder Cancer Progression and Overall Survival in Patients with Invasive Bladder Cancer
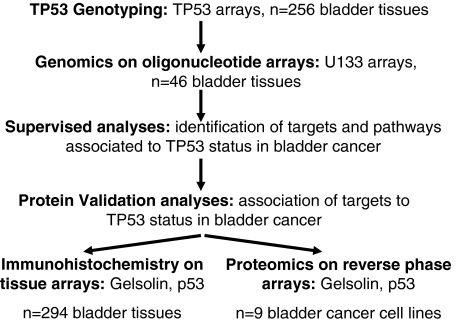
Of the 256 bladder tumors under study, 153 cases (59.8%) presented a wild-type TP53, whereas 103 cases were found to have TP53 mutations (40.2%). The distribution of TP53 mutations and their association with bladder cancer stage is illustrated in Figure 2A. TP53 mutations were more frequently observed in tumors with advanced disease stages than in those classified as non-muscle-invasive bladder tumor lesions. TP53 mutation status was found to be significantly associated with tumor stage (Kruskall-Wallis, P = 0.001). The association of TP53 status with clinical outcome was analyzed on those patients with invasive disease for which annotated follow-up was available. Cases with mutated TP53 displayed a median survival of 12.0 months [SE,: 2.0; 95% confidence interval (CI), 8.1 to 15.9]. However, those cases presenting a wild-type status lived for 51.0 months (SE,: 10.8; 95% CI, 29.8 to 72.2). The survival of patients with bladder cancer and mutant TP53 was significantly shorter than those presenting a wild-type TP53 (log-rank, P = 0.01) (Figure 2B). Thus, TP53 mutation status was further confirmed to be associated with tumor progression and clinical outcome in patients with invasive bladder tumors.
Figure 2.
A: Distribution of wild-type and mutated TP53 status along bladder cancer progression. An increased altered TP53 was observed in advanced disease compared with non-muscle-invasive bladder lesions. TP53 genotype status was found significantly associated with tumor stage (Kruskall-Wallis, P = 0.001). B: Association of TP53 status with clinical outcome. Overall survival of patients with bladder cancer and mutant TP53 was significantly shorter than those presenting a wild-type TP53 genotype (log-rank, P = 0.01).
TP53 Mutations in the Core-Binding Domain Are Frequently Present in Bladder Tumors
Analyses of the distribution of the TP53 mutations along the functional domains of TP53 revealed that the majority were mapped to the DNA-binding core. Percent rates of mutations within each TP53 exon are summarized in Table 1. In brief, they affected exons 4 (10.6%), 5 (21.8%), 6 (12.1%), 7 (10.6%), 8 (38.0%), and 9 (0.1%). Eight bladder cancer cases presented more than one mutation in the TP53 gene. The codons most frequently mutated included 285 (n = 12), 213 (n = 6), and 248 (n = 6). Detailed information regarding exon location, base changes, and amino acid residues mutated in the cases analyzed in this series is included as Supplemental Table 1 available at http://ajp.amjpathol.org.
Table 1.
Summary of the Most Frequent TP53 Mutations in the Bladder Tumors
| Exon | Codon | Base change | Amino acid change | Number of cases |
|---|---|---|---|---|
| 8 | 285 | G>A | E>K | 12 |
| 4 | 36 | G>A | Silent | 6 |
| 7 | 248 | G>A | R>Q | 5 |
| 4 | 125 | G>A | Silent | 4 |
| 6 | 220 | A>G | Y>C | 4 |
| 6 | 213 | C>T | R>stop | 3 |
| 8 | 273 | C>T | R>C | 3 |
| 8 | 274 | G>T | V>F | 3 |
| 5 | 132 | G>T | K>N | 2 |
| 5 | 141 | G>T | C>F | 2 |
| 5 | 175 | G>A | R>H | 2 |
| 5 | 179 | A>R | H>R | 2 |
| 6 | 192 | C>T | Q>stop | 2 |
| 6 | 213 | C>T | R>W | 2 |
| 7 | 237 | G>A | M>I | 2 |
| 8 | 271 | G>A | E>K | 2 |
| 8 | 276 | C>G | A>G | 2 |
| 8 | 279 | G>A | G>E | 2 |
| 8 | 298 | G>T | E>stop | 2 |
The table refers to some of the most frequent mutations with at least two individuals presenting a specific genetic alteration in exon, codon, base change, and amino acid changes and the number of cases presenting each specific mutation. Expanded information is provided in Supplemental Table 1 available at http://ajp.amjpathol.org.
Gene Expression Analyses Identify Molecular Targets Associated with TP53 Status
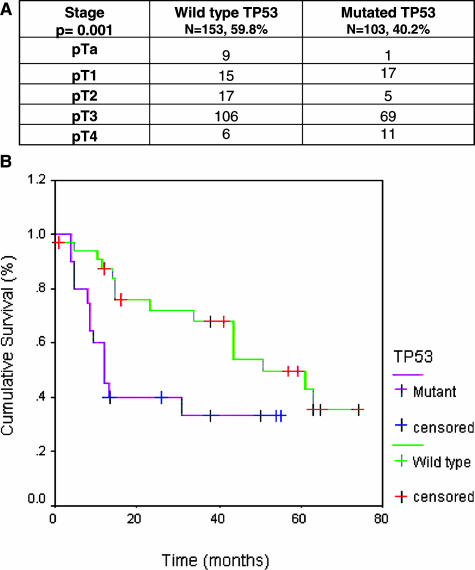
Transcript profiling studies using the U133A oligonucleotide array were performed on a subset of the bladder specimens sequenced for TP53 using the GeneChip array (TP53 wt, n = 24; TP53 mutated, n = 22). t-Test analyses revealed 149 probes differentially expressed between these groups with P values lower than 0.002 at a false discovery rate of 0.02. The top 30 differentially expressed genes are summarized in Table 2. An expanded version provides the information regarding all these 149 probes in Supplemental Table 2 available at http://ajp.amjpathol.org. Further unsupervised pathway analyses using the Ingenuity software revealed the central relevance of TP53 in these networks (Figure 3), supporting the experimental design of the present study. Functional annotation of top-ranked genes differentially expressed between these groups revealed that molecular pathways associated with TP53 status included not only cell cycle and apoptotic signaling but also adhesion and angiogenesis networks.
Table 2.
Summary of the Top 30 Differentially Expressed Genes between Invasive Bladder Tumors with Wild-Type and Mutant TP53
| Probe ID | P value | FC | Symbol | Gene description | Location |
|---|---|---|---|---|---|
| 205909_at | 9.4369 × 10−08 | −2.08 | POLE2 | Polymerase (DNA-directed), ε2 | 14q21 |
| 207828_s_at | 3.7682 × 10−07 | −3.88 | CENPF | Centromere protein F, 350/400 ka (mitosin) | 1q32 |
| 213951_s_at | 4.6765 × 10−07 | −2.09 | HUMGT198 | GT198, complete ORF | 17q12 |
| 204531_s_at | 1.4276 × 10−06 | −2.50 | BRCA1 | Breast cancer 1, early onset | 17q21 |
| 219363_s_at | 2.744 × 10−06 | −1.53 | CGI-12 | CGI-12 protein | 8q22.1 |
| 203063_at | 3.5733 × 10−06 | 1.92 | PPM1F | Protein phosphatase 1F | 22q11.22 |
| 201417_at | 4.1024 × 10−06 | −2.86 | SOX4 | SRY (sex determining region Y)-box 4 | 6p22.3 |
| 204170_s_at | 5.124 × 10−06 | −3.26 | CKS2 | CDC28 protein kinase regulatory subunit 2 | 9q22 |
| 201303_at | 6.8435 × 10−06 | −1.62 | DDX48 | DEAD (Asp-Glu-Ala-Asp) box polypeptide 48 | 17q25.3 |
| 208694_at | 7.5283 × 10−06 | −1.66 | PRKDC | Protein kinase, DNA-activated, polypeptide | 8q11 |
| 203755_at | 7.5301 × 10−06 | −2.92 | BUB1B | Budding uninhibited by benzimidazoles 1β | 15q15 |
| 204023_at | 7.6062 × 10−06 | −1.88 | RFC4 | Replication factor C (activator 1) 4, 37 kDa | 3q27 |
| 220840_s_at | 9.1669 × 10−06 | −2.48 | FLJ10706 | Hypothetical protein FLJ10706 | 1q23.3 |
| 222233_s_at | 9.6983 × 10−06 | −1.78 | DCLRE1C | DNA cross-link repair 1C | 10p13 |
| 200696_s_at | 9.8959 × 10−06 | 2.54 | GSN | Gelsolin | 9q33 |
| 222039_at | 1.0428 × 10−05 | −2.97 | LOC146909 | Hypothetical protein LOC146909 | 17q21.31 |
| 221521_s_at | 1.3368 × 10−05 | −4.22 | LOC51659 | HSPC037 protein | 16q24.1 |
| 214051_at | 1.3675 × 10−05 | −1.75 | MGC39900 | Hypothetical protein MGC39900 | Xq22.2 |
| 200020_at | 1.5754 × 10−05 | −1.24 | TARDBP | TAR DNA-binding protein | 1p36.22 |
| 210766_s_at | 1.5886 × 10−05 | −1.67 | CSE1L | CSE1 chromosome segregation 1-like | 20q13 |
| 201088_at | 1.7071 × 10−05 | −2.15 | KPNA2 | Karyopherin α2 | 17q23.1 |
| 217943_s_at | 1.7774 × 10−05 | 1.66 | FLJ10350 | Hypothetical protein FLJ10350 | 1p34.3 |
| 206102_at | 1.7818 × 10−05 | −3.06 | KIAA0186 | KIAA0186 gene product | 20p11.21 |
| 219167_at | 1.8871 × 10−05 | 2.43 | RIS | Ras family member Ris | 15q11.2 |
| 206050_s_at | 2.2264 × 10−05 | 1.54 | RNH | Ribonuclease/angiogenin inhibitor | 11p15.5 |
| 202440_s_at | 2.3849 × 10−05 | 1.79 | ST5 | Suppression of tumorigenicity 5 | 11p15 |
| 205883_at | 2.3961 × 10−05 | 2.63 | ZNF145 | Zinc finger protein 145 | 11q23.1 |
| 219306_at | 2.5219 × 10−05 | −2.34 | KNSL7 | Kinesin-like 7 | 3p21.32 |
| 204108_at | 2.5651 × 10−05 | −1.67 | NFYA | Nuclear transcription factor Y, α | 6p21.3 |
| 218883_s_at | 2.7391 × 10−05 | −2.84 | KLIP1 | KSHV latent nuclear antigen-interacting protein 1 | 4q35.1 |
A positive fold change (FC) indicates an increased expression in tumors with wild-type TP53 compared with mutated cases. A negative fold change (FC) indicates an increased expression in tumors with mutant TP53 compared with wild-type cases. Expanded information of the 149 probes is provided in Supplemental Table 2 available at http://ajp.amjpathol.org.
Figure 3.
Gene profiles associated with TP53 status. Pathway analyses revealed the central relevance of TP53, highlighted with a blue arrow, in the signaling networks of the genes differentially expressed between cases with wild-type and mutant TP53, supporting the experimental design by which they were identified. Overexpressed genes in patients with wild-type TP53 are highlighted in red, and underexpressed probes in these cases are highlighted in green. Fold changes are indicated in parentheses with positive and negative signs, respectively.
Gelsolin, a Target Identified as Related to TP53 Status, Is Associated with Bladder Cancer Progression and Clinical Outcome
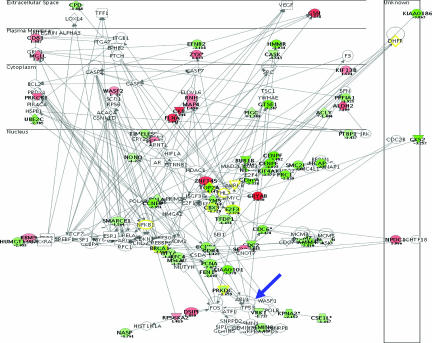
Validation strategies of the clinical relevance of top-ranked targets differentially expressed between cases with wild-type and mutant TP53 included clinicopathological correlations using immunohistochemistry on an independent series of patients with bladder cancer. Gelsolin expression was analyzed on four tissue microarrays containing 294 bladder tumor specimens and found to be associated with tumor stage and tumor grade (both, P < 0.001). Representative staining profiles of gelsolin along disease progression states are illustrated in Figure 4. Clinical follow-up data and mutation status in 95 cases allowed the evaluation of gelsolin protein expression patterns with overall survival and TP53. Patients with low gelsolin expression showed shorter survival than those with high protein levels of gelsolin (P = 0.03) (Figure 4C). Moreover, lower protein expression of gelsolin was observed in patients with mutated TP53 (P < 0.001). Thus, protein expression patterns of gelsolin were associated not only with tumor progression and clinical outcome but also with TP53 status.
Figure 4.
Gelsolin validation analyses by means of immunohistochemistry on tissue arrays. Representative stainings of gelsolin in non-muscle-invasive (A) and invasive (B) bladder tumors. C: Gelsolin protein expression patterns were significantly associated with overall survival in the 95 cases for which follow-up was available.
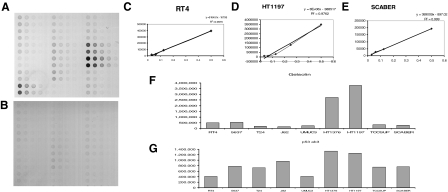
Gelsolin is differentially expressed among bladder cancer cell lines. An independent in vitro validation strategy of the association of gelsolin with TP53 status was undertaken using a proteomic approach. Reverse-phase protein lysate microarrays were optimized to compare gelsolin expression levels among bladder cancer cell lines representing different stages along bladder cancer progression and of known TP53 status. Each protein array included serial dilutions of the protein extracts of the nine bladder cancer cell lines under study (180 spots in all). Figure 5 illustrates the design of the arrays incubated with mouse anti-human-gelsolin antibodies. As a negative control, a duplicated slide was incubated with mouse secondary antibodies, avoiding primary antibody targeting incubation and displaying minimal to no background staining (Figure 5B). The antibody-antigen binding properties were assessed through dilution curves obtained for gelsolin and TP53 proteins expression, as shown for independent series of RT4, HT1197, or ScaBER (Figure 5, C–E). The experimental variability could be evaluated by two independent protein extracts obtained on separate days for each bladder cancer cell line. To assess reproducibility of protein profiles, each protein extract was printed in duplicate. Thus, reproducibility could be assessed by four-replicated spots for each cell line. The average of such measurements correlated with Western blotting analyses of the cell lines under study (data not shown). Figure 5F shows the averaged quantitative differential expression of gelsolin among duplicated experimental and spotted protein extracts of the cell lines under study. Interestingly, bladder cancer cell lines harboring either wild-type or mutated TP53 showed differential protein expression of gelsolin. The average of gelsolin protein measurements correlated with the differential protein expression of mutated TP53 as measured by Ab-3 antibody (Figure 5G). More specifically, HT-1376 and HT-1197 cells, known to have mutations at exons 7 and 11, respectively, were shown to have higher levels of gelsolin and mutant TP53 when measured by means of reverse-phase protein arrays.
Figure 5.
Gelsolin validation analyses by means of reverse phase arrays on protein extracts of bladder cancer cell lines. A: Design of the arrays incubated with mouse anti-human gelsolin. B: Negative control incubating a mouse secondary antibody and omitting the primary antibody. C–E: Dilution curves obtained for gelsolin for several bladder cancer cell lines, RT4, HT1197, or ScaBER, as shown. F: Averaged quantitative differential expression of gelsolin among duplicated experimental and spotted protein extracts of the cell lines under study. G: Averaged quantitative differential expression of mutant TP53 among duplicated experimental and spotted protein extracts of the cell lines under study.
Discussion
TP53 has been reported to play a critical role in initiation and progression of human bladder cancer. The present report was designed to identify critical targets and pathways differentiating wild-type and mutant TP53 phenotypes in bladder tumors. The undertaken approach further refines the nature, clinical relevance, and prognostic abilities of detecting TP53 alterations in invasive bladder cancer. The TP53 sequencing exercise represents an expanded analysis of previous studies from our group on the predictive role of TP53 in clinical outcome,6,9,10,11,12,13,14,15,16 as well as its association with bladder cancer progression. Similarly to previous reports,6,10,14,15 the majority of TP53 mutations were found in the DNA-binding region.
The novelty of the present report relies on the identification and validation of novel targets associated with TP53 status from gene profiles of invasive bladder tumors. Furthermore, independent pathway analyses linking the functional annotation available for these targets reveal the relevance of TP53 as a central node involved in the regulation of a large number of genes at late stages in bladder cancer progression. This observation supports the rationale for our experimental design, in which gene profiles of tumors with different TP53 status are compared using supervised methods and reveals the influence of this gene in the progression of the disease. The use of protein expression patterns of gelsolin, a differentially expressed gene identified in this study, complements the molecular classification of the patients studied according to their TP53 status in combination with tumor stage and clinical outcome. Moreover, validation analyses at the protein level using immunohistochemistry on tissue microarrays have revealed gelsolin as a prognostic marker of overall survival. An independent proteomic approach, by means of custom-made reverse phase protein arrays, has served to confirm a differential protein expression of gelsolin among cell lines derived from early and late stages of bladder tumors. Using this strategy, gelsolin expression could be linked to specific TP53 mutations as well.
The TP53 sequencing technology applied using the GeneChip p53 from Affymetrix has limitations that need to be considered in the interpretation of the results. Although correlation with conventional sequencing has been described,14 and this GeneChip may detect the majority of TP53 mutations,15 it does not cover the full TP53 exons and has not been designed to detect long deletions or insertions. However, the U133A array covers certain known genes and expressed sequence tags. Further versions of the arrays will progressively provide more comprehensive views of the targets associated with TP53 status in parallel with the innovations and development of the technology. It is important to be aware that these two considerations might not allow detection of mutations of low penetrance or discovery of relevant unknown genes.
Novel molecular associations have been identified to TP53 status in the present study. Gelsolin was selected for clinical validation, and its expression was associated with tumor staging and overall survival at the protein level by means of immunohistochemistry on an independent set of bladder tumors. Thus, gene expression profiling and analysis of protein expression on tissue microarrays provided complementary information. The loss of gelsolin expression in invasive tumors, as compared with non-muscle-invasive lesions, is consistent with the tumor suppressor role of gelsolin in bladder cancer.20,21,22,23 However, this is the first report describing the association of gelsolin with tumor staging, survival, and TP53 status using two independent sets of clinical material together with in vitro studies using bladder cancer cell lines with wild-type and mutant TP53. Gelsolin is an actin-regulatory protein coded by chromosome 9q33, a region reported as frequently deleted in bladder cancer.24 This observation led us to select this target for further validation analyses. Hemidesmosomes are organized around a core of actin filaments that appear early during cell adhesion. Gelsolin is one of the proteins surrounding the actin core described above.24 Alteration of actin polymerization and/or remodeling plays pivotal role in regulating the morphological and phenotypic events of a malignant cell.24 The actin remodeling is the result not only of the activation of oncogenic actin signaling pathways (eg, Ras in bladder cancer) but also of the inactivation of several important actin-binding proteins that have tumor suppressive functions (eg, Gelsolin).9,24 Accumulation of evidence exists to indicate the tumor suppressive functions of actin regulatory proteins. Gene expression analysis suggests that actin alterations are progressive and that distinctive actin remodeling profiles are associated with different stages of bladder cancer development and progression. These patterns can be used as markers for cancer staging and prognostic indication. The detection of specific types of actin signaling pathway alterations suggests potential targeted preventive or therapeutic intervention with specific actin signaling pathway blockers, thereby providing an actin-based paradigm for individualized monitoring and intervention of human bladder cancer.
In our study, neoplastic cellular populations were obtained after hematoxylin and eosin examination of bladder cancer samples assuming that at least 75% pure tumor cells were included. This strategy may imply certain variability in neoplastic populations between cases varying from 75 to 100% because of the tissue heterogeneity in muscle-invasive T2+ tumors and may potentially affect the results observed. In this regard, such tumor subpopulation variation would not skew defining the TP53 status of the advanced cases under study because of the PCR-based, probe-specific sequencing strategy of the TP53 Affymetrix array. Potential differences in percent rates of neoplastic and nonneoplastic cells would eventually vary the transcript profiling intensities. This is an issue affecting any transcript profiling analysis unless laser microdissection is performed ensuring 95 to 100% neoplastic populations. Aware of this common limitation affecting any quantitative high-throughput profiling of nonlaser-microdissected neoplastic nucleic acids, data analyses were undertaken estimating false-positive rates so that only highly significant changes were considered for further validation analyses. As an example, gelsolin appeared the 15th in the list of top ranked probes, with a P value of 9.9 × 10−6 and was shown to correlate with TP53 status using independent methodologies on human cells and clinical specimens. Thus, although the differential percentage of tumor cells from which nucleic acids were extracted among cases could potentially affect the transcript results, the data analyses undertaken provide an estimated reliability on the differentially expressed probes identified to be associated with TP53 status.
In summary, our study revealed that TP53 mutations are frequent events in bladder cancer and provide prognostic information. Sequencing TP53 arrays represent a high-throughput means for assessing TP53 status as a predictor of overall survival in patients with bladder tumors. Gene profiling identified novel molecular associations to TP53 status, which in turn seem to provide information that is of clinical utility, mainly for the stratification of patients with advanced bladder cancer. This is of clinical relevance because the criteria to determine patient outcome are incompletely defined, and new biological determinants are needed for selecting patients who harbor tumors more prone to display a more aggressive clinical behavior and who would benefit from tailored intervention. Evaluation of protein profiles of bladder cancer cell lines and clinical specimens rendered complementary information at dissecting gelsolin as a TP53-related molecule. Thus, in vitro and clinical material provided complementary information in showing the relevance of gelsolin and TP53 in bladder cancer progression.
Supplementary Material
Acknowledgments
We thank all members of the laboratories of Drs. Cordon-Cardo, Petricoin, and Sánchez-Carbayo for their technical support and constructive suggestions in the preparation of the manuscript; and the Tissue Procurement Core, especially Cora Mariano, at Memorial Sloan-Kettering Cancer Center for their support in facilitating the procurement of the tumor specimens and clinical follow-up of the bladder cancer cases analyzed in this study.
Footnotes
Address reprint requests to Marta Sanchez-Carbayo, Ph.D., Group Leader, Tumor Markers Group-208A, Spanish National Cancer Center, Melchor Fernandez Almagro, 3, Madrid 28029, Spain; or Carlos Cordon-Cardo, M.D., Ph.D., Division of Molecular Pathology, Memorial Sloan-Kettering Cancer Center, 1275 York Ave., New York, NY 10021. E-mail: mscarbayo@cnio.es and cordon-c@mskcc.org.
Supplemental material for this article can be found on http:// ajp.amjpathol.org.
References
- Levine AJ. The p53 tumor-suppressor gene. N Engl J Med. 1992;326:1350–1352. doi: 10.1056/NEJM199205143262008. [DOI] [PubMed] [Google Scholar]
- Levine AJ. P53, the cellular gatekeeper for growth and division. Cell. 1997;88:323–331. doi: 10.1016/s0092-8674(00)81871-1. [DOI] [PubMed] [Google Scholar]
- Sengupta S, Harris CC. p53: traffic cop at the crossroads of DNA repair and recombination. Nat Rev Mol Cell Biol. 2005;6:44–55. doi: 10.1038/nrm1546. [DOI] [PubMed] [Google Scholar]
- Wolff EM, Liang G, Jones PA. Mechanisms of disease: genetic and epigenetic alterations that drive bladder cancer. Nat Clin Pract Urol. 2005;2:502–510. doi: 10.1038/ncpuro0318. [DOI] [PubMed] [Google Scholar]
- Soussi T, Lozano G. p53 mutation heterogeneity in cancer. Biochem Biophys Res Commun. 2005;331:834–842. doi: 10.1016/j.bbrc.2005.03.190. [DOI] [PubMed] [Google Scholar]
- Cordon-Cardo C, Dalbagni G, Saez GT, Oliva MR, Zhang ZF, Rosai J, Reuter VE, Pellicer A. p53 mutations in human bladder cancer: genotypic versus phenotypic patterns. Int J Cancer. 1994;56:347–353. doi: 10.1002/ijc.2910560309. [DOI] [PubMed] [Google Scholar]
- Markl IDC, Jones PA. Presence and location of TP53 mutation determines pattern of CDKN2A/ARF pathway inactivation in bladder cancer. Cancer Res. 1998;58:5348–5353. [PubMed] [Google Scholar]
- Zhao R, Gish K, Murphy M, Yin Y, Notterman D, Hoffman WH, Tom E, Mack DH, Levine AJ. Analysis of p53-regulated gene expression patterns using oligonucleotide arrays. Genes Dev. 2000;14:981–993. [PMC free article] [PubMed] [Google Scholar]
- Dalbagni G, Presti J, Reuter V, Fair WR, Cordon-Cardo C. Genetic alterations in bladder cancer. Lancet. 1993;342:469–471. doi: 10.1016/0140-6736(93)91595-d. [DOI] [PubMed] [Google Scholar]
- Lianes P, Orlow I, Zhang ZF, Oliva MR, Sarkis AS, Reuter VE, Cordon-Cardo C. Altered patterns of MDM2 and TP53 expression in human bladder cancer. J Natl Cancer Inst. 1994;86:1325–1330. doi: 10.1093/jnci/86.17.1325. [DOI] [PubMed] [Google Scholar]
- Cordon-Cardo C, Sheinfeld J, Dalbagni G. Genetic studies and molecular markers of bladder cancer. Semin Surg Oncol. 1997;13:319–327. doi: 10.1002/(sici)1098-2388(199709/10)13:5<319::aid-ssu5>3.0.co;2-g. [DOI] [PubMed] [Google Scholar]
- Cordon-Cardo C. p53 and RB: simple interesting correlates or tumor markers of critical predictive nature? J Clin Oncol. 2004;22:975–977. doi: 10.1200/JCO.2004.12.994. [DOI] [PubMed] [Google Scholar]
- Sanchez-Carbayo M, Socci ND, Lozano J, Saint F, Cordon-Cardo C. Defining molecular profiles of poor outcome in patients with invasive bladder cancer using oligonucleotide microarrays. J Clin Oncol. 2006;24:778–789. doi: 10.1200/JCO.2005.03.2375. [DOI] [PubMed] [Google Scholar]
- Wikman FP, Lu ML, Thykjaer T, Olesen SH, Andersen LD, Cordon-Cardo C, Orntoft TF. Evaluation of the performance of a p53 sequencing microarray chip using 140 previously sequenced bladder tumor samples. Clin Chem. 2000;46:1555–1561. [PubMed] [Google Scholar]
- Lu ML, Wikman F, Orntoft TF, Charytonowicz E, Rabbani F, Zhang Z, Dalbagni G, Pohar KS, Yu G, Cordon-Cardo C. Impact of alterations affecting the p53 pathway in bladder cancer on clinical outcome, assessed by conventional and array-based methods. Clin Cancer Res. 2002;8:171–179. [PubMed] [Google Scholar]
- Sanchez-Carbayo M, Socci ND, Charytonowicz E, Lu M, Prystowsky M, Childs G, Cordon-Cardo C. Molecular profiling of bladder cancer using cDNA microarrays: defining histogenesis and biological phenotypes. Cancer Res. 2002;62:6973–6980. [PubMed] [Google Scholar]
- Dawson-Saunders B, Trapp RG. Norwalk: Appleton and Lange,; Basic and Clinical Biostatistics, (ed 2.) 1994 [Google Scholar]
- Carlisle AJ, Prabhu VV, Elkahloun A, Hudson J, Trent JM, Linehan WM, Williams ED, Emmert-Buck MR, Liotta LA, Munson PJ, Krizman DB. Development of a prostate cDNA microarray and statistical gene expression analysis package. Mol Carcinog. 2000;28:12–22. [PubMed] [Google Scholar]
- Nishizuka S, Charboneau L, Young L, Major S, Reinhold WC, Waltham M, Kouros-Mehr H, Bussey KJ, Lee JK, Espina V, Munson PJ, Petricoin E, III, Liotta LA, Weinstein JN. Proteomic profiling of the NCI-60 cancer cell lines using new high-density reverse-phase lysate microarrays. Proc Natl Acad Sci USA. 2003;100:14229–14234. doi: 10.1073/pnas.2331323100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka M, Mullauer L, Ogiso Y, Fujita H, Moriya S, Furuuchi K, Harabayashi T, Shinohara N, Koyanagi T, Kuzumaki N. Gelsolin: a candidate for suppressor of human bladder cancer. Cancer Res. 1995;55:3228–3232. [PubMed] [Google Scholar]
- Sakai N, Ohtsu M, Fujita H, Koike T, Kuzumaki N. Enhancement of G2 checkpoint function by gelsolin transfection in human cancer cells. Exp Cell Res. 1999;251:224–233. doi: 10.1006/excr.1999.4552. [DOI] [PubMed] [Google Scholar]
- Celis A, Rasmussen HH, Celis P, Basse B, Lauridsen JB, Ratz G, Hein B, Ostergaard M, Wolf H, Orntoft T, Celis JE. Short-term culturing of low-grade superficial bladder transitional cell carcinomas leads to changes in the expression levels of several proteins involved in key cellular activities. Electrophoresis. 1999;20:355–361. doi: 10.1002/(SICI)1522-2683(19990201)20:2<355::AID-ELPS355>3.0.CO;2-N. [DOI] [PubMed] [Google Scholar]
- Rao J, Seligson D, Visapaa H, Horvath S, Eeva M, Michel K, Pantuck A, Belldegrun A, Palotie A. Tissue microarray analysis of cytoskeletal actin-associated biomarkers gelsolin and E-cadherin in urothelial carcinoma. Cancer. 2002;95:1247–1257. doi: 10.1002/cncr.10823. [DOI] [PubMed] [Google Scholar]
- Spinardi L, Rietdorf J, Nitsch L, Bono M, Tacchetti C, Way M, Marchisio PC. A dynamic podosome-like structure of epithelial cells. Exp Cell Res. 2004;295:360–374. doi: 10.1016/j.yexcr.2004.01.007. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.