Abstract
By yeast two-hybrid screening, we found three novel interactors (UNC-95, LIM-8, and LIM-9) for UNC-97/PINCH in Caenorhabditis elegans. All three proteins contain LIM domains that are required for binding. Among the three interactors, LIM-8 and LIM-9 also bind to UNC-96, a component of sarcomeric M-lines. UNC-96 and LIM-8 also bind to the C-terminal portion of a myosin heavy chain (MHC), MHC A, which resides in the middle of thick filaments in the proximity of M-lines. All interactions identified by yeast two-hybrid assays were confirmed by in vitro binding assays using purified proteins. All three novel UNC-97 interactors are expressed in body wall muscle and by antibodies localize to M-lines. Either a decreased or an increased dosage of UNC-96 results in disorganization of thick filaments. Our previous studies showed that UNC-98, a C2H2 Zn finger protein, acts as a linkage between UNC-97, an integrin-associated protein, and MHC A in myosin thick filaments. In this study, we demonstrate another mechanism by which this linkage occurs: from UNC-97 through LIM-8 or LIM-9/UNC-96 to myosin.
INTRODUCTION
Cell-to-substratum attachment plays pivotal roles in morphogenesis during development in many tissues (Hynes, 1994). The transmembrane proteins, the integrins, and their associated proteins inside cells are the main contributors for cell–substratum attachments, and they form a specific structure, the focal adhesion (Petit and Thiery, 2000; Miranti and Brugge, 2002). In muscle cells, this kind of attachment structure, called a “costamere,” is involved in the attachment of muscle cells to basement membrane, and it is important for force transmission, generated during actin–myosin interaction, to the outside of cells (Ervasti, 2003; Samarel, 2005). Integrins and associated proteins that compose the costameres are located at Z-disks of peripheral myofibrils. At focal adhesions and Z-disk costameres, integrins, and associated proteins connect actin cytoskeletal filaments and myofibril thin filaments, respectively. Although some components of focal adhesions are also located at peripheral M-lines (Porter et al., 1992; McDonald et al., 1995), a precise mechanism linking integrins to myofibril thick filaments is not well characterized.
In Caenorhabditis elegans body wall muscle, the actin thin filaments are attached to dense bodies (Z-disk analogues), and the myosin thick filaments are organized around M-lines (Moerman and Williams, 2006). C. elegans muscle cells are an excellent model for functional characterization of costameres. First, all the dense bodies and M-lines are anchored to the cell membrane as revealed by electron microscopy (Waterston, 1988). This means that M-lines are also involved in costamere functions. Second, by forward and reverse genetics approaches, integrins and their associated proteins have been identified to be crucial for the development of C. elegans muscle (Moerman and Williams, 2006). Third, several tools for probing dense bodies or M-lines specifically, such as monoclonal antibodies or translational gfp constructs, are available (Waterston, 1988; Moerman and Williams, 2006). In C. elegans adult muscle, dense bodies, and M-lines are easily detected using the tools mentioned above. By molecular and genetic approaches, we previously identified one molecular linkage from an integrin-associated protein to myosin thick filaments (Miller et al., 2006). UNC-97, a homologue of mammalian PINCH (Hobert et al., 1999), interacts with UNC-98, a C2H2 zinc finger protein (Mercer et al., 2003). All four zinc finger domains of UNC-98 are required for binding to UNC-97. We found that the N-terminal portion of UNC-98, excluding the zinc fingers, binds to the C terminus of myosin heavy chain (MHC) A. From biochemical, cell biological, and genetic evidence, we concluded that UNC-97/UNC-98/MHC A functions as one of the mechanisms to link integrin-associated proteins to myosin thick filaments at M-lines. In our previous report, we suggested that additional mechanisms for anchoring myosin thick filaments at M-lines are likely to exist. Here, we report an additional mechanism that links integrin-associated proteins to myosin thick filaments from a systematic characterization of protein–protein interactions at M-lines by using the yeast two-hybrid approach.
MATERIALS AND METHODS
Worm Strains and Culture
Nematodes were grown at 20°C on NGM agar plates with Escherichia coli strain OP50 (Brenner, 1974). We used Bristol N2 as the wild-type strain and the following mutant strains: VC654 (lim-8 (ok941)); VC209 (lim-9 (gk106));VC349 (lim-9 (gk210)); HE33 (unc-95 (su33)); VC627 (unc-95 (ok893)); GB244 (unc-96 (sf18)); GB247 (sfEx39 [pPD49.78/83-HA-UNC-96(FL), pTG96]).
Yeast Two-Hybrid
The procedures for identifying and testing protein–protein interactions were described previously (Mackinnon et al., 2002). Bait and prey plasmids were made by using pGBDU and pGAD plasmids, and they were transformed into yeast host strain PJ69-4A (James et al., 1996). PAT-3, PAT-4, PAT-6, UNC-96, UNC-97, UNC-98, UNC-112, and UIG-1 plasmids were described previously (Mackinnon et al., 2002; Tsuboi et al., 2002; Lin et al., 2003; Mercer et al., 2003, 2006; Hikita et al., 2005). LIM-8, LIM-9, and UNC-95 plasmids were originally isolated from two-hybrid screening with UIG-1 (HQ, KK; to be described elsewhere). Corresponding cDNA sequences were amplified by using synthesized primers, and they were cloned into the pBluescript KS+ vector. After confirming DNA sequences to be error-free, the cDNA fragments were inserted into pGBDU and pGAD vectors, resulting in bait and prey plasmids.
Glutathione Bead Pull-Down Assay
Plasmids for expression of recombinant proteins were made by cloning of cDNA fragments used in yeast two-hybrid into pGEX-KK plasmids (for glutathione S-transferase [GST] fusions) or pMAL-KK plasmids (for maltose binding protein [MBP] fusions). The procedure for recombinant protein preparation from E. coli was described previously (Mercer et al., 2006). Glutathione bead pull-down assays were carried out according to a previously described method (Mackinnon et al., 2002). Briefly, glutathione agarose beads coated with GST or GST fusion proteins (30 μg) were mixed with MBP or MBP fusions (50 μg), and washed. Samples eluted by Laemmli buffer were separated by 10% SDS-PAGE gels, and they were subjected to Coomassie brilliant blue (CBB) staining.
Far-Western Assay
Total myosin II from wild-type C. elegans was prepared as described previously (Miller et al., 2006). A far-Western assay between myosin II and an MBP fusion of either UNC-96 or LIM-8 was carried out according to a previously described procedure (Mercer et al., 2006). A far-Western assay for interaction between GST-UNC-96 and either MBP-LIM-8 or MBP-LIM-9 was performed similarly except that the blocking step was extended to overnight, and the incubation with MBP or MBP fusions was shortened to 1 h.
Expression Pattern of lim-8 and lim-9 Promoters
Genomic sequences upstream of lim-8 and lim-9 coding sequences were amplified by polymerase chain reaction (PCR) and cloned into pPD95.95 (a gift from A. Fire, Stanford University, Stanford, CA). The promoter–gfp expression plasmids were injected into wild-type strain N2 worms, and several independent transgenic lines were established for each gene. Green fluorescent protein (GFP) fluorescence images of adult body wall and pharyngeal muscles were obtained with a Zeiss Axioskop microscope (Carl Zeiss, Jena, Germany) on Fuji Sensia 100 slide film, scanned, and processed using Adobe Photoshop (Adobe Systems, Mountain View, CA).
Preparation of Antibodies
For expression of GST or MBP fusions in E. coli, plasmids that contained cDNA sequence for each protein in pGEX-KK or pMAL-KK were used. GST fusions of LIM-8 (amino acids [aa] 451-799 in LIM-8b), LIM-9 (aa 1-280), and UNC-95 (aa 1-269) were purified as described previously (Mercer et al., 2006), and they were supplied to Spring Valley Laboratories (Woodbine, MD) for generation of rabbit polyclonal antibodies (Benian-7 for LIM-8, Benian-9 for LIM-9, and Benian-13 for UNC-95). Polyclonal antibodies were affinity purified by using Affigel columns (Bio-Rad, Hercules, CA) conjugated with MBP fusions of LIM-8 (aa 451-626 in LIM-8b), LIM-9 (aa 1-280), and UNC-95 (aa 1-269) as described previously (Mercer et al., 2003).
Western Blot
Procedures for preparing worm protein lysates and Western blots were described previously (Mercer et al., 2006). The following mutant worm strains were used to make lysates: VC654 for lim-8, VC209 and VC349 for lim-9, and HE33 and VC627 for unc-95. Benian-7 and Benian-9 were used at 1:1000 dilution. Benian-13 was used at 1:200 dilution. Anti-HA antibodies (rabbit) were purchased from Sigma-Aldrich (St. Louis, MO) and used at 1:500 dilution.
Immunostaining
For adult worm immunostaining, worms were fixed as described previously (Nonet et al., 1993). Benian-7, Benian-9, and Benian-13 were used at 1:100 dilution. As markers of dense bodies and M-lines, MH35 (anti-α actinin, 1:200 dilution) and MH42 (anti-UNC-89, 1:200 dilution) were used, respectively. For staining of UNC-97 and MHC A, Benian-16 (1:100 dilution) and 5–6 (1:400 dilution) were used (Miller et al., 2006). Rabbit antibodies were visualized by anti-rabbit antibodies conjugated with Alexa 488 (Invitrogen, Carlsbad, CA), and mouse antibodies were visualized by anti-mouse antibodies conjugated with Alexa 647 (Invitrogen) or with Cy3 (Jackson ImmunoResearch Laboratories, West Grove, PA). Images were captured with a Bio-Rad Radiance model 2100 confocal microscopy system and displayed using LaserSharp2000 or Carl Zeiss LSM 510 confocal microscopy system software.
Overexpression of UNC-96
The cDNA for full-length (aa 1-418) UNC-96, designated “UNC-96(FL)” was inserted into the pKS-HA(Nhe) vector for adding three hemagglutinin (HA) tags. The HA-UNC-96(FL) fragment was cloned into pPD49.78 and pPD49.83 for expression in worms under the control of heat-shock promoters (these vectors were also gifts from A. Fire). Mixtures of pPD49.78/83-HA-UNC-96(FL) with pTG96 (SUR-5::GFP, as a marker) (Yochem et al., 1998) were injected into wild type (N2) for generating transgenic worms [GB247 (sfEx39)]. Transgenic animals were identified by finding animals in which SUR-5::NLS::GFP was expressed in nearly all cells. Several independent lines were examined. Transgenic worms were placed at 30°C for 2 h to induce expression of HA-tagged full-length UNC-96. After an additional 24 h incubation at 20°C, worms were collected and used for preparation of either lysates for Western blots or for fixation by the Nonet method (Nonet et al., 1993). Fixed worms were stained with anti-HA rabbit antibodies (1:500 dilution; Sigma Aldrich) and anti-MHC A (5–6, 1:400 dilution).
RESULTS
Protein Interaction Matrix for UNC-97
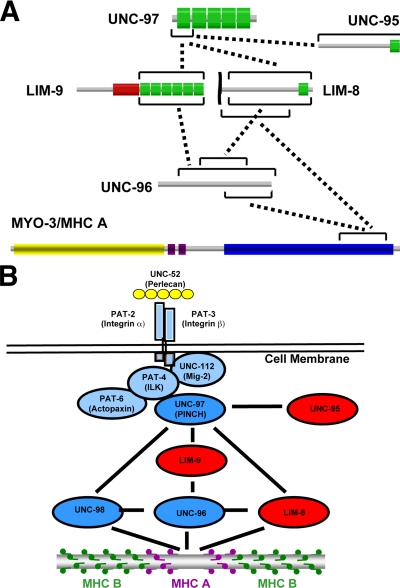
To identify additional proteins that interact with UNC-97, we screened a yeast two-hybrid “bookshelf” that includes the following proteins: known and candidate components of dense bodies and/or M-lines, UNC-112 interactors, and UIG-1 interactors (see citations in Materials and Methods). A list of proteins in the bookshelf and the results of screening are shown in Table 1. Among the interactions, we found that UNC-97 binds to UNC-95 and two new proteins harboring LIM domains, named LIM-8 and LIM-9, respectively. The LIM-8 and LIM-9 proteins also interact with UNC-96, a known M-line component (Mercer et al., 2006). Finally, UNC-96 and LIM-8 showed interaction with the C-terminal portion of MHC A, as we found previously for UNC-98 (Miller et al., 2006). The interactions described above are summarized in Figure 7.
Table 1.
List of proteins in the bookshelf and the results of screening
| Bait side | Prey side |
||||||
|---|---|---|---|---|---|---|---|
| UNC-96 (45-418) | UNC-96 (201-418) | UNC-95 | LIM-9 | LIM-8 | MYO-3/MHC A | Empty vector | |
| UNC-112 | − | − | − | − | − | − | − |
| UNC-97 | − | − | ++ | ++ | ++ | − | − |
| PAT-3 | − | − | − | − | − | − | − |
| PAT-4 | − | − | − | − | − | − | − |
| PAT-6 | − | − | − | − | − | − | − |
| UNC-98 | ++ | ++ | ++ | ++ | ++ | ++ | ++ |
| DIM-1 | − | − | − | − | ++ | − | − |
| ATN-1a | − | − | − | − | − | − | − |
| ATN-1b | − | − | − | − | − | − | − |
| DEB-1 | + | + | ++ | − | ++ | + | + |
| Y71G12B.11a(CeTalin) | − | − | − | − | − | − | − |
| C28H8.6a(Cepaxillin) | − | − | ++ | − | − | − | − |
| KIN-32(CeFAK) | − | − | − | − | − | − | − |
| UIG-1 | − | ++ | ++ | ++ | ++ | ++ | − |
| HUM-6 | − | ++ | ++ | − | − | − | − |
| LIM-9 | + | − | ++ | ++ | ++ | − | − |
| LIM-8 | ++ | − | − | − | ++ | ++ | − |
| MYO-3/MHC A | − | ++ | − | − | − | − | − |
| Prey side | Bait side |
||||||
|---|---|---|---|---|---|---|---|
| UNC-96 (45–418) | UNC-96 (201-418) | UNC-95 | LIM-9 | LIM-8 | MYO-3/MHC A | ||
| UNC-112 | − | − | − | − | − | − | |
| UNC-97 | − | − | − | + | − | − | |
| PAT-3 | − | − | − | − | − | − | |
| PAT-4 | − | − | − | − | − | − | |
| PAT-6 | − | − | − | − | − | − | |
| UNC-98 | − | ++ | − | − | − | − | |
| DIM-1 | − | − | − | − | ++ | − | |
| ATN-1a | − | − | − | − | − | − | |
| ATN-1b | − | − | − | − | − | − | |
| DEB-1 | − | − | − | − | − | − | |
| Y71G12B.11a(CeTalin) | − | − | − | − | − | − | |
| C28H8.6a(Cepaxillin) | − | − | − | − | − | − | |
| KIN-32(CeFAK) | − | − | − | − | − | − | |
| UIG-1 | − | ++ | − | ++ | ++ | − | |
| HUM-6 | − | ++ | − | − | ++ | − | |
| MYO-3/MHC A | − | ++ | − | − | ++ | − | |
| Empty vector | − | − | − | − | − | − | |
Bait and prey plasmids contain full-length cDNA except for the following gene products: PAT-3 (aa 765-809), HUM-6 (aa 1222-2098), and MYO-3/MHC A (aa 1636-1969).
LIM-8 (aa 567-1004), LIM-9 (aa 251-655), UNC-96 (aa 45-418 or aa 201-418).
++, strong growth on the selection media; +, means growth on the selection media; and −, means no growth on the selection media.
Figure 7.
Summary of interactions and working model: Multiple protein clusters link muscle focal adhesions to thick filaments. (A) Summary of protein–protein interactions that were found in this study. Each protein is represented schematically with the indicated domains. Green boxes show LIM domains, a red box shows a PET domain, purple boxes show IQ repeats, a blue box shows the coiled coil region, and a yellow box shows the myosin head region. According to the results of domain mapping, minimum interacting regions are indicated by black bars. (B) Multiple protein clusters link cell adhesion structures to thick filaments. In C. elegans muscle, myofilaments are located close to the surface and anchored by dense bodies and M-lines to the muscle cell membrane. At these attachment structures, UNC-52/Perlecan is located in the ECM. Inside the muscle cell, the cytoplasmic tail of integrin (PAT-3/PAT-2) is associated with a four-protein complex, including UNC-97 (Norman et al., 2007). Among the UNC-97 interactors is UNC-98 (a C2H2 zinc finger protein) that also binds to the C-terminal tail of MHC A, suggesting a protein linkage from integrin-associated proteins to thick filaments (Miller et al., 2006). In this study, we found that UNC-97 binds to two independent LIM domain proteins (LIM-8 and LIM-9), and these two LIM domain proteins bind to UNC-96. UNC-96 has been shown to be an M-line component and to interact with UNC-98 (Mercer et al., 2006). We show that UNC-96 and LIM-8 can also bind to the C terminus of MHC A. Together, we propose that the UNC-97/LIM-8/LIM-9/UNC-96 protein complex functions as one more pathway that links integrin-associated proteins to thick filaments.
Mapping of Domains Essential for the Interactions
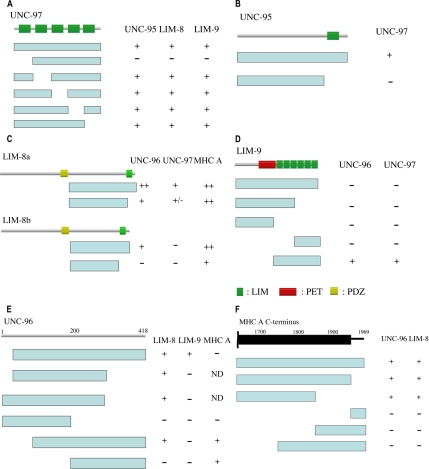
We found that UNC-97 interacts with UNC-95, LIM-8, and LIM-9. UNC-97 is composed of five LIM domains (Hobert et al., 1999). We examined which LIM domain is essential for binding by using deletion constructs lacking single LIM domains. Removal of the first LIM domain abolished binding of UNC-97 to all three interactors (Figure 1A), suggesting that the first LIM domain is essential for binding. A similar requirement was found previously for the interaction of UNC-97 with PAT-4 (Mackinnon et al., 2002).
Figure 1.
Domain mapping for each interacting pair of proteins by the yeast two-hybrid system. The columns contain the results of testing for interaction. ++, strong growth; +, growth; +/−, poor growth; and −, no growth on the selection media. ND, not done. In A–D, the top lines represent schematics of the indicated proteins in which green bars denote LIM domains, yellow bars denote PDZ domains, and a red box denotes a PET domain. (A) Mapping of binding sites in UNC-97 for UNC-95, LIM-8, and LIM-9. Blue bars represent deletion constructs of UNC-97 as bait. Prey plasmids of UNC-95, LIM-8, and LIM-9 were tested for interaction with UNC-97 deletion series baits. (B) Mapping of the binding site in UNC-95 for UNC-97. Blue bars represent deletion constructs of UNC-95 as prey. Bait plasmids of UNC-97 were examined for interaction with UNC-95 deletion series preys. (C) Mapping of the binding site in LIM-8 for UNC-96, UNC-97, and MHC A. LIM-8a contains 1004 amino acid residues, and LIM-8b contains 868 amino acid residues. Blue bars represent deletion constructs of LIM-8 as bait for UNC-96 and MHC A, and prey for UNC-97. Prey plasmids of UNC-96 and MHC A, and the bait plasmid of UNC-97 were examined for interaction with LIM-8 deletion series baits and preys. (D) Mapping of the binding site in LIM-9 for UNC-96 and UNC-97. Blue bars represent deletion constructs of LIM-9 as bait for UNC-96 and prey for UNC-97. Prey plasmids of UNC-96 and a bait plasmid of UNC-97 were examined for interaction with LIM-9 deletion series baits and preys. (E) Mapping of the binding site in UNC-96 for LIM-8, LIM-9, and MHC A. Top schematic is the UNC-96 protein structure with amino acids numbers displayed. Blue bars represent deletion constructs of UNC-96 as bait for MHC A and prey for LIM-8 and LIM-9. Prey plasmids of MHC A and bait plasmids of LIM-8 and LIM-9 were examined for interaction with UNC-96 deletion series baits and preys. Because some part of UNC-96 can activate transcription in yeast cells, two deletion constructs as baits could not be used to examine binding to MHC A (shown as ND). (F) Mapping of the binding site in MHC A for UNC-96 and LIM-8. Top schematic denotes the C-terminal half of MHC A, in which the black bar represents the coiled-coil domain, the black line the nonhelical tail, and amino acids numbers are shown. Blue bars represent deletion constructs of MHC A as prey. Bait plasmids of UNC-96 and LIM-8 were examined for interaction with MHC A deletion series preys.
UNC-95 has one LIM domain at its C terminus (Broday et al., 2004). We tested UNC-95 two-hybrid constructs with and without this LIM domain for interaction with UNC-97. In UNC-95, the region upstream of the LIM domain did not show interaction with UNC-97 (Figure 1B), suggesting that the LIM domain of UNC-95 is important for binding to UNC-97.
In WormBase, LIM-8 has been predicted to have three alternative isoforms, and all three contain one PDZ and one LIM domain. These three isoforms have different initiator methionines. The region for interaction with UNC-97, UNC-96, and MHC A is C-terminal of the PDZ domain, because we used this region for bookshelf screening. In this region, isoforms a and c contain the same exon structure, and they are extended compared with isoform b (an addition of 181 amino acid residues). We prepared two-hybrid plasmids harboring fragments downstream of the PDZ domain of LIM-8a and LIM-8b and corresponding fragments lacking the C-terminal LIM domain. Both LIM-8a and LIM-8b could bind to UNC-96, but constructs lacking the LIM domain showed weak binding to UNC-96 (Figure 1C). For the interaction with UNC-97, LIM-8a could bind to UNC-97, but LIM-8b did not. Deletion of the LIM domain from LIM-8a almost abolished binding to UNC-97. For the interaction with MHC A, both LIM-8a and LIM-8b could bind. Even in the absence of the C-terminal LIM domain, binding to MHC A occurred. Together, the C-terminal LIM domain of LIM-8 is essential for interaction with UNC-96 and UNC-97, and an isoform a/c-specific extension is required for binding to UNC-97. The isoform a/c-specific extension contains 181 amino acid residues. In this sequence, serine, arginine, and tyrosine are found more frequently than in an average C. elegans protein, and serine residues are usually found in pairs (5 times).
In WormBase, six isoforms of LIM-9 are predicted. Among them, we could confirm only one isoform by PCR generation and sequencing of a cDNA that encodes 655 amino acid residues and contains one PET domain and six LIM domains. This confirmed isoform, which we designate isoform “g” (GenBank accession no. EF443133), starts from the predicted isoform “f” ATG site and terminates with some extension beyond the predicted isoform “f.” Despite the different number of LIM domains and the presence of the PET domain, the closest homologue for LIM-9 in mammalian cells is FHL (4 and a half LIM domain) protein (Chu et al., 2000). When human FHL2 was used as a query in a BLAST search against C. elegans proteins, the highest scoring match was to LIM-9 (over score value 800), and the matching scores to other LIM domain proteins were less than score value 300. In the Drosophila genome sequence, we found that the closest homologue for LIM-9 is limpet, which contains a PET domain followed by five LIM domains.
We prepared deletion constructs as shown in Figure 1D to test interaction of LIM-9 with UNC-96 and UNC-97. One construct harboring only the six LIM domains could bind to UNC-96 and UNC-97 (Figure 1D), suggesting that the LIM domains are essential and sufficient for interaction with UNC-96 and UNC-97. Surprisingly, a full-length construct of LIM-9 did not show interaction to UNC-96 or UNC-97, suggesting that the region N-terminal of the LIM domains may inhibit interaction between the LIM domains of LIM-9 and UNC-96/UNC-97.
UNC-96 has been reported to interact with UNC-98 (Mercer et al., 2006). We found that UNC-96 also binds to LIM-8, LIM-9 and MYO-3/MHC A. Using deletion constructs for UNC-96, we mapped the regions that are essential for the interactions (Figure 1E). LIM-8 could bind to various constructs of UNC-96 as long as these constructs contained a central portion of UNC-96 (aa 101-300), suggesting that a central portion of UNC-96 is essential for binding to LIM-8. In the case of LIM-9, only one construct of UNC-96 (aa 45-418) showed interaction with LIM-9. MYO-3/MHC A interacts with the C-terminal half of UNC-96, the same region of UNC-96 that interacts with UNC-98 (Mercer et al., 2006).
We have reported that UNC-98 can bind to the C-terminal 200 amino acid residues of MHC A, but not those of other MHCs (Miller et al., 2006). We found that UNC-96 and LIM-8 also could bind to MHC A, but not to other MHCs (MHC B, MHC C, and MHC D) (data not shown). Using the same set of deletion derivatives of MHC A C terminus, we determined the minimum region for interaction with UNC-96 and LIM-8 (Figure 1F). In contrast to UNC-98, the C-terminal 200 amino acid resides (aa 1771-1969) of MHC A did not bind to UNC-96 or LIM-8, and instead, a more internal 200 amino acid residues (aa 1636-1870) of the coiled-coil domain could bind to UNC-96 or LIM-8.
Minimum regions for the binding of each pair of proteins are summarized in Figure 7A.
In Vitro Binding Assays Using Purified Proteins
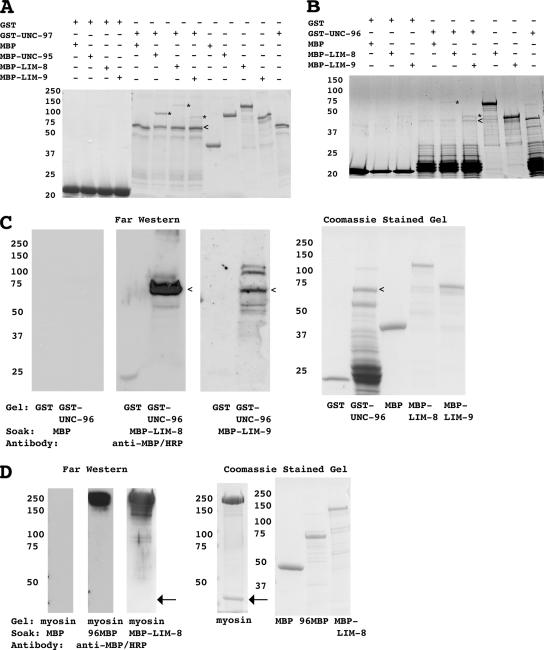
To confirm protein–protein interactions identified by the yeast two-hybrid system, we carried out in vitro glutathione bead pull-downs or far-Western assays by using purified proteins. We prepared GST fusion proteins for UNC-97 (full length) and UNC-96 (aa 45-418), and MBP fusion proteins for UNC-95 (full length), LIM-8 (aa 401-1004 in isoform a), and LIM-9 (aa 281-655). After incubating GST-UNC-97 either with MBP, MBP-UNC-95, MBP-LIM-8, or MBP-LIM-9, GST fusion proteins were pulled out of solution with glutathione agarose. In those pellets, the presence of any MBP fusion proteins was examined by CBB staining. As shown in Figure 2A, this assay demonstrates that UNC-97 interacts with UNC-95, LIM-8, and LIM-9 in vitro. Similarly, when MBP-LIM-8 and MBP-LIM-9 were incubated with GST-UNC-96, MBP-LIM-8 and MBP-LIM-9 were pelleted (Figure 2B). Despite our best efforts, the GST-UNC-96 had multiple bands; thus, we performed a far-Western assay to show interaction between GST-UNC-96 and either MBP-LIM-8 or MBP-LIM-9. As shown in Figure 2C, MBP-LIM-8 or MBP-LIM-9 binds on a far-Western to a band corresponding to the size expected for GST-UNC-96. Thus, by two independent assays, UNC-96 interacts with both LIM-8 and LIM-9 in vitro.
Figure 2.
In vitro binding of each interacting pair using purified proteins. In each subfigure, the column of numbers indicates the positions of molecular weight markers in kilodaltons. In A and B, a + means included in assay and a − means not included in assay. (A) Glutathione bead pull down using GST-UNC-97 and MBP-UNC-95, MBP-LIM-8, and MBP-LIM-9. GST or GST-UNC-97–coated glutathione agarose beads were mixed with MBP, MBP-UNC-95, MBP-LIM-8, or MBP-LIM-9. After several washes with buffer, proteins associated with glutathione agarose were eluted by Laemmli buffer. These samples were separated on a 10% SDS-PAGE gel and stained with CBB. The < shows the position of GST-UNC-97. The * shows the positions of MBP-UNC-95, MBP-LIM-8, MBP-LIM-9. (B) Glutathione bead pull down using GST-UNC-96 and MBP-LIM-8 and MBP-LIM-9. GST or GST-UNC-96–coated glutathione agarose beads were mixed with MBP, MBP-LIM-8, or MBP-LIM-9. After several washes with buffer, proteins associated with glutathione agarose were eluted with Laemmli buffer. Eluted samples were separated on a 10% SDS-PAGE gel and stained with CBB. The < shows the position of GST-UNC-96. The * shows the position of MBP-LIM-8 or MBP-LIM-9. (C) Far-Western assay showing interaction between UNC-96 and LIM-8 and between UNC-96 and LIM-9. GST and GST-UNC-96 were separated on 10% SDS-PAGE and transferred to a nitrocellulose membrane. The membrane was incubated with MBP or MBP-LIM-8 or MBP-LIM-9, and after washing, bound proteins were visualized by using horseradish peroxidase (HRP)-conjugated anti-MBP antibodies and enhanced chemiluminescence (ECL). The far-Western column shows the ECL film of the anti-MBP antibody reaction. Coomassie staining shows CBB staining of proteins used in this assay. The < shows the position of GST-UNC-96. (D) Far-Western assay showing interaction between purified myosin and UNC-96 and LIM-8. Total myosin II purified from C. elegans was separated on a 7.5% SDS-PAGE gel and transferred to a nitrocellulose membrane. The membrane was incubated with MBP or 96MBP or MBP-LIM-8, and after washing, bound proteins were visualized by using HRP-conjugated anti-MBP antibodies and ECL. The arrows indicate the position of actin that is found in a small amount in this purified myosin preparation.
We used a far-Western assay to confirm interactions between myosin and either UNC-96 or LIM-8. (We attempted a glutathione pull-down assay to demonstrate binding between GST-MHC A and an MBP-UNC-96 [aa 201-418]. Unfortunately, because MBP-UNC-96 [aa 201-418] could bind to GST itself, we could not proceed with this approach.) We prepared myosin II from nematodes and MBP fusions for UNC-96 (aa 201-418) and LIM-8 (aa 401-1004 in isoform a). The myosin was separated on a gel, blotted to a membrane, and incubated with either MBP or 96MBP (MBP-UNC-96 [aa 201-418]) or MBP-LIM-8 (Figure 2D). After incubation with 96MBP or MBP-LIM-8, anti-MBP antibodies recognize a band at a size corresponding to myosin II (200 kDa). The myosin preparation also contained a small amount of actin (43 kDa, indicated with an arrow in Figure 2D), but neither 96MBP nor MBP-LIM-8 interacted with actin in this assay. In addition, MBP alone did not show interaction with purified myosin II.
Expression Pattern of lim-8 and lim-9 Promoters
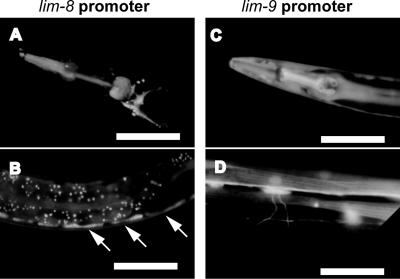
To begin to characterize the newly identified LIM domain proteins, LIM-8 and LIM-9, we investigated the expression pattern of the lim-8 and lim-9 promoters. In WormBase, multiple isoforms of lim-8 and lim-9 are listed as predictions. For each gene, we used a single genomic sequence that is likely to act as a promoter for all the isoforms. We fused 3.7 kb of sequence upstream of the initiator methionine to GFP for lim-8, and 4.5 kb of sequence upstream of the initiator methionine was fused to GFP for lim-9. Transgenic worms harboring lim-8 or lim-9 promoter gfp fusions showed GFP fluorescence in the pharyngeal and body wall muscles (Figure 3), suggesting these two new LIM domain proteins function in muscle cells. The lim-8 promoter gfp fusion was also expressed in vulva, spermathecae, anal sphincter and depressor muscles, head neurons, gonadal sheath, and excretory canal. lim-8 expression was higher in larvae than that in adults, and the actual expression in the adult stage was very weak. In the case of lim-9 expression, GFP fluorescence was also detected in some neuronal processes (Figure 3D), vulva, spermathecae, anal sphincter and depressor muscles, gonadal sheath, and excretory canal.
Figure 3.
lim-8 and lim-9 expression patterns by using promoter-gfp fusions. Approximately 3–4 kb of genomic sequence upstream of the predicted initiation methionines of lim-8 or lim-9 were fused to gfp and used to create transgenic worms. (A and B) lim-8 promoter::gfp expression in the pharynx of adult (A) and in body wall muscle of larvae (B). Arrows indicate positions of body wall muscle cells. (C and D) lim-9 promoter::gfp expression in pharynx (C) and in body wall muscle and neurons (D), all in adults. White bars, 100 μm (A–C) or 50 μm (D).
Localization of Three New UNC-97 Interactors in Adult Body Wall Muscle
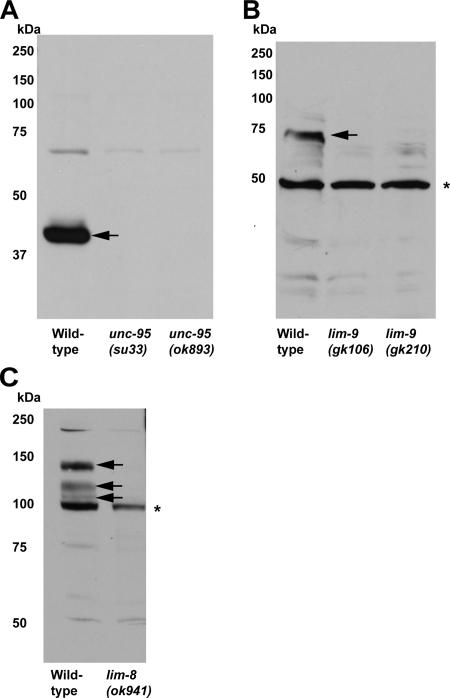
Because both gfp translational fusions (Hobert et al., 1999) and antibodies (Miller et al., 2006) localize UNC-97 to M-line and dense bodies, we wondered whether the new UNC-97 interactors are also localized to those structures. For this purpose, rabbit polyclonal antibodies were generated to UNC-95, LIM-8, and LIM-9. In Western blot analysis with these antibodies, we detected bands corresponding to the predicted size of each protein in wild-type worm lysates (Figure 4). Such bands were eliminated or shifted to smaller size in knockout mutant lysates (Figure 4, marked with arrows), showing that those proteins are derived from each gene locus.
Figure 4.
Western blot analysis of anti-UNC-95, anti-LIM-8, and anti-LIM-9 antibodies. Each antibody detects a protein of predicted size from wild-type C. elegans lysates (indicated with arrows), and the proteins are missing or in truncated form from mutant lysates. Asterisks denote nonspecific bands that also appeared in wild-type and mutant lysates. (A) Anti-UNC-95 antibodies. (B) Anti-LIM-9 antibodies. (C) Anti-LIM-8 antibodies.
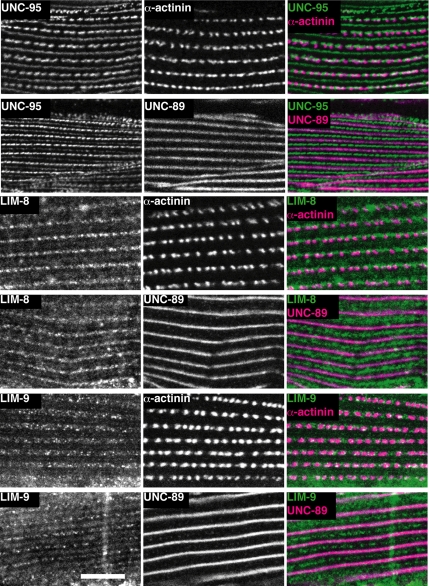
Immunostaining with each the three antibodies showed staining of body wall muscle in a striated pattern that is characteristic of myofibrils (Figure 5). We determined precise localization of each protein in body wall muscle by costaining with marker antibodies for dense bodies (α-actinin) and M-lines (UNC-89). By this method, UNC-95 was localized at dense bodies and M-lines, in agreement with the previous localization of UNC-95 by using a translational gfp fusion (Broday et al., 2004). LIM-8 and LIM-9 localized to the M-line as marked by UNC-89 (see merged images in Figure 5). However, LIM-8 and LIM-9 appeared as discontinuous dots rather than continuous lines. Similar discontinuous M-line localization has been reported for UNC-96 (Mercer et al., 2006). The LIM-8 and LIM-9 were not clearly colocalized with α-actinin staining; instead, they showed staining around and between dense bodies in the I-band. Because our affinity-purified antibodies to LIM-9 and LIM-8, by Western, reacted to additional proteins not affected in the mutants (indicated by * in Figure 4, B and C), we worried that the staining patterns might reflect these reactivities. However, this does not seem to be the case: Preabsorption of each of these antibodies to the immunogens eliminated muscle staining (data not shown). In summary, all three newly identified UNC-97 interactors were at least partially localized at M-lines, suggesting that we have identified three new members of an UNC-97 interacting complex at M-lines.
Figure 5.
Localization of UNC-95, LIM-8, and LIM-9 in adult body wall muscle cells. Localization of UNC-95, LIM-8, and LIM-9 are shown together with antibodies to markers for dense bodies (α-actinin) and M-lines (UNC-89). The left column shows UNC-95, LIM-8, or LIM-9 localization. The center column shows dense body and M-line marker localization. The right column shows merged images of left and center columns. Rabbit antibodies were visualized by anti-rabbit IgG conjugated with Alexa 488, and mouse antibodies were visualized by anti-mouse IgG conjugated with Alexa 647. White bar, 10 μm.
Precise Dosage of UNC-96 Is Important for the Organization of Thick Filaments
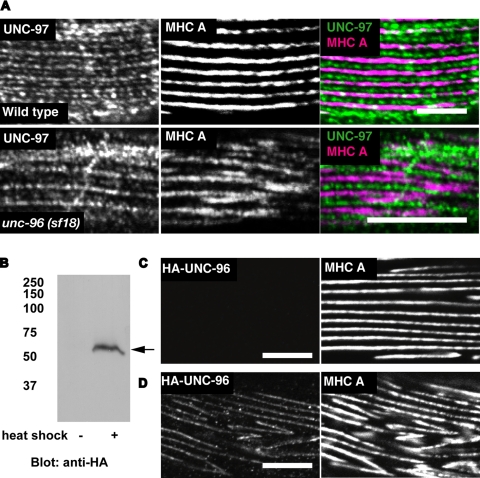
Genetic, protein interaction, and localization data suggest that a key function of UNC-96 is to regulate thick filament organization (Mercer et al., 2006). To further explore this model, we carried out two experiments. One experiment is the costaining of an unc-96 loss of function mutant with anti-UNC-97 and anti-MHC A antibodies. In unc-96 (sf18) mutant worm muscle cells, UNC-97 is localized at dense bodies and M-lines (Figure 6A) similar to those in wild-type animals (Miller et al., 2006). However, MHC A staining is disorganized, as reported previously (Figure 6A). The other experiment is overexpression of UNC-96 full-length protein (including the MHC A binding portion). HA-tagged UNC-96 was expressed in adults under the control of a heat shock promoter in a wild-type genetic background. By Western blot, we confirmed that overexpression of UNC-96 had occurred after heat-shock induction (Figure 6B). In worms overexpressing UNC-96, the pattern of MHC A localization was disorganized (Figure 6D) compared with worms not overexpressing HA-tagged full-length UNC-96 (Figure 6C). Both loss of function (sf18 mutant) and gain of function (overexpression of HA-tagged full-length protein) showed disorganization of MHC A staining, suggesting that a precise level of UNC-96 is important for the organization of thick filaments.
Figure 6.
Either a decrease or an increase of UNC-96 results in disorganization of thick filaments. (A) Wild-type or unc-96 (sf18) worms were stained with anti-UNC-97 and anti-MHC A antibodies. (B–D) represent experiments using lines of worms carrying extrachromosomal arrays of plasmids consisting of HA-tagged UNC-96 full-length cDNA under the control of a heat-shock promoter. (B) Laemmli soluble proteins were prepared from these transgenic animals after they were subjected to heat shock (+), or without heat shock (−). Proteins (100 μg) were separated by 10% SDS-PAGE, transferred to a membrane and reacted with anti-HA antibodies. An arrow indicates a protein in the heat shock (+) lane that is expected for HA-UNC-96. (C and D) Worms were stained with anti-HA (rabbit) and anti-MHC A (mouse monoclonal). Results of antibody staining after heat shock on worms lacking (C) or harboring the extrachromosomal array (D). In A, C, and D, rabbit antibodies were visualized by anti-rabbit IgG conjugated with Alexa 488, and mouse antibodies were visualized by anti-mouse IgG conjugated with Cy3. White bars, 10 μm.
DISCUSSION
Multiple Proteins Link Membrane Attachment Proteins to Myosin Thick Filaments
We found three novel interactors of UNC-97/PINCH, one of the proteins associated with integrin that is crucial for cell substratum attachment in C. elegans muscle (Norman et al., 2007). Two of the three UNC-97 interactors, LIM-8 and LIM-9, could also bind to UNC-96, a known M-line component. Finally, UNC-96 and LIM-8 could bind to myosin heavy chain A (MHC A) that is localized to the middle of thick filaments in the proximity of M-lines. Together, we hypothesize that LIM-8 or LIM-9/UNC-96 proteins function as molecular linkers from UNC-97, an integrin-associated peripheral protein, to MHC A, one of the components of thick filaments (Figure 7B). The fact that both UNC-97 and LIM-9 (PINCH and FHLs, respectively) have homologues in mammalian skeletal muscle suggests that similar interactions might exist in mammalian muscle. Indeed, FHL2 is located primarily to Z-disks and also to M-lines in heart muscles (Scholl et al., 2000), and FHL3 is localized to Z-disks in skeletal muscles (Samson et al., 2004). PINCH1 is reported to be expressed in cardiac muscle and to be localized at costameres in cultured cardiac muscle cells (Chen et al., 2005).
Previously, we reported that UNC-98, another component of M-lines, functions as a bridge from UNC-97 to myosin thick filaments, and we suggested that additional protein linkages to myosin thick filaments might exist. This was based on the observation that loss of function mutation in unc-98 did not result in severe defects in thick filament organization compared with loss of function mutation in MHC A (Miller et al., 2006). Our current study indicates that two additional pathways involve LIM-8, LIM-9, and UNC-96 and that these pathways work in parallel and are redundant to the pathway that includes UNC-98 (Figure 7B). However, because unc-98; unc-96 (RNAi) or unc-96; unc-98 (RNAi) animals do not show enhanced defects in organization of thick filaments (Mercer et al., 2006), there must be still other pathways for linking integrin-associated proteins to thick filaments. One possibility is that UNC-95 provides this linkage, because we show that UNC-95 also binds to UNC-97 (Figure 7B). However, we have found that a triple mutant containing unc-95; unc-96; unc-98(RNAi) did not have a more severe defect in thick filament organization compared with each single mutant (data not shown). Additional UNC-97 binding partners, or other integrin-associated proteins like PAT-6 may also function for linking to thick filaments.
Currently, we know three protein complexes (UNC-98, LIM-8, and LIM-9/UNC-96) function in connecting thick filaments to cell-substratum attachment structures. Because these attachment structures are used for transmission of force generated from actomyosin interaction to the outside of muscle cells, multiple and redundant pathways seem plausible.
The three described pathways might be independent and parallel. Our examination of muscle structure by polarized light or staining with anti-MHC A antibodies failed to reveal any myofibril defects in knockout mutants for either LIM-8 or LIM-9. RNA interference (RNAi) by injection for either gene, at least in wild-type animals, also failed to reveal any muscle phenotypes. Nevertheless, in an RNAi hypersensitive background (rrf-3), we did observe that ∼30% of the F1 progeny from parents injected by lim-8 double-stranded RNA showed paralysis as L2 or L3 larvae (data not shown). This developmental period corresponds to the period in which we observed that the lim-8 promoter was most active (Figure 3B).
Many LIM Domain Proteins Associate with Integrins
There are many proteins containing the LIM domain localized at muscle focal adhesion structures, such as UNC-97, UNC-95, LIM-8, and LIM-9. We have defined a protein interaction matrix among these LIM domain proteins and determined which LIM domains are required for these interactions (Figures 1 and 7A). The LIM domain is well known as a protein–protein interaction module (Kadrmas and Beckerle, 2004). However, our results indicate that not all LIM domains are functionally equivalent. The UNC-97 protein is composed of five LIM domains, but only the first LIM domain is responsible for binding to LIM-8, LIM-9, UNC-95 (our results), and PAT-4 (Mackinnon et al., 2002), suggesting preference of this LIM domain (Figure 1A). In LIM-9, this protein has 6 LIM domains, and we showed that all 6 LIM domains are required for binding to UNC-97 and UNC-96 (Figure 1D), suggesting a binding site that either simply spans these 6 LIM domains or is created by some sort of three-dimensional (3D) structure generated by self-association of these LIM domains. Binding specificities of single LIM domains or complex structures composed of multiple LIM domains could overcome nonspecific binding and ensure formation of precise protein associations that are essential for cell–substratum attachment.
Our finding that four different proteins (LIM-9, LIM-8, UNC-95, and PAT-4) all interact with the first LIM domain of UNC-97 might seem difficult to explain. One possibility is that individual UNC-97 polypeptides might bind through their first LIM domains in a mutually exclusive manner to any one of the four proteins. A variation of this idea is that a two-dimensional or 3D lattice might be formed by invoking UNC-97 multimerizes and that each UNC-97 polypeptide of the multimer could be free to interact with a different protein. Evidence that UNC-97 may multimerize comes from our two-hybrid result that UNC-97 preys were obtained upon screening with UNC-97 as bait (Qadota and Moerman, unpublished data). We have not yet confirmed this possible dimerization by other methods.
UNC-96 and LIM-8 Are Novel MHC A Binding Proteins
We have shown that UNC-96 and LIM-8, each of which are components of M-lines, interact with the C-terminal region of MHC A (Figure 1, C, E, and F). Previously, we reported that UNC-98, another component of M-lines, also interacts with the C-terminal region of MHC A (Miller et al., 2006). More precise mapping of the binding sites in MHC A revealed that UNC-98 binds to aa 1771-1969 of MHC A (Miller et al., 2006), whereas UNC-96 or LIM-8 bind to aa 1636–1870 of MHC A (Figure 1F). This slightly different requirement in the C terminus of MHC A may suggest simultaneous and redundant interaction of UNC-98, UNC-96, and LIM-8 to MHC A for linking thick filaments. However, we also reported that UNC-98 could bind to UNC-96 (Mercer et al., 2006), and here we show that LIM-8 can bind to UNC-96, suggesting that these proteins form a complex. Because we now know that each protein can link to MHC A, it is also possible that interaction with MHC A occurs through a LIM-8/UNC-96/UNC-98 complex.
We now know that in C. elegans striated muscle, at the M-line, there are three myosin binding proteins, UNC-98, UNC-96, and LIM-8. For two of these proteins, UNC-98 (Miller et al., 2006) and UNC-96 (this study), thick filament organization is sensitive to their protein levels: either loss of function or overexpression results in disorganized thick filaments. However, LIM-8 does not show this property: either loss of function (noted above) or overexpression (data not shown) does not affect myofibrillar organization. Perhaps this is why the existence of lim-8 had not been revealed in Unc genetic screens.
ACKNOWLEDGMENTS
We thank Bob Barstead (Oklahoma Medical Research Foundation, Oklahoma City, OK) for RB2, a random primed C. elegans cDNA library; Andy Fire for C. elegans expression plasmids; and Krishna Bhat (University of Texas Medical Branch, Galveston, TX) for the use of a confocal microscope system. We also thank Noriko Mishima for technical assistance. Some strains used in this work were provided by the Caenorhabditis Genetics Center, which is supported by the National Center for Research Resources of the National Institutes of Health. This work was supported by grants from the National Institutes of Health to G.M.B. (AR051466 and AR052133) and from grants-in-aid for scientific research from the Ministry of Education, Culture, Sports, Science and Technology of Japan (to K.K.).
Footnotes
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E07-03-0278) on August 29, 2007.
REFERENCES
- Brenner S. The genetics of Caenorhabditis elegans. Genetics. 1974;77:71–94. doi: 10.1093/genetics/77.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broday L., Kolotuev I., Didier C., Bhoumik A., Podbilewicz B., Ronai Z. The LIM domain protein UNC-95 is required for the assembly of muscle attachment structures and is regulated by the RING finger protein RNF-5 in C. elegans. J. Cell Biol. 2004;165:857–867. doi: 10.1083/jcb.200401133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H., Huang X. N., Yan W., Chen K., Guo L., Tummalapali L., Dedhar S., St-Arnaud R., Wu C., Sepulveda J. L. Role of the integrin-linked kinase/PINCH1/alpha-parvin complex in cardiac myocyte hypertrophy. Lab. Invest. 2005;85:1342–1356. doi: 10.1038/labinvest.3700345. [DOI] [PubMed] [Google Scholar]
- Chu P. H., Ruiz-Lozano P., Zhou Q., Cai C., Chen J. Expression patterns of FHL/SLIM family members suggest important functional roles in skeletal muscle and cardiovascular system. Mech. Dev. 2000;95:259–265. doi: 10.1016/s0925-4773(00)00341-5. [DOI] [PubMed] [Google Scholar]
- Ervasti J. M. Costameres: the Achilles' heel of Herculean muscle. J. Biol. Chem. 2003;278:13591–13594. doi: 10.1074/jbc.R200021200. [DOI] [PubMed] [Google Scholar]
- Hikita T., Qadota H., Tsuboi D., Taya S., Moerman D. G., Kaibuchi K. Identification of a novel Cdc42 GEF that is localized to the PAT-3-mediated adhesive structure. Biochem. Biophys. Res. Commun. 2005;335:139–145. doi: 10.1016/j.bbrc.2005.07.068. [DOI] [PubMed] [Google Scholar]
- Hobert O., Moerman D. G., Clark K. A., Beckerle M. C., Ruvkun G. A conserved LIM protein that affects muscular adherens junction integrity and mechanosensory function in Caenorhabditis elegans. J. Cell Biol. 1999;144:45–57. doi: 10.1083/jcb.144.1.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hynes R. O. Genetic analyses of cell-matrix interactions in development. Curr. Opin. Genet. Dev. 1994;4:569–574. doi: 10.1016/0959-437x(94)90074-d. [DOI] [PubMed] [Google Scholar]
- James P., Halladay J., Craig E. A. Genomic libraries and a host strain designed for highly efficient two-hybrid selection in yeast. Genetics. 1996;144:1425–1436. doi: 10.1093/genetics/144.4.1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadrmas J. L., Beckerle M. C. The LIM domain: from the cytoskeleton to the nucleus. Nat. Rev. Mol. Cell Biol. 2004;5:920–931. doi: 10.1038/nrm1499. [DOI] [PubMed] [Google Scholar]
- Lin X., Qadota H., Moerman D. G., Williams B. D. C. elegans PAT-6/actopaxin plays a critical role in the assembly of integrin adhesion complexes in vivo. Curr. Biol. 2003;13:922–932. doi: 10.1016/s0960-9822(03)00372-5. [DOI] [PubMed] [Google Scholar]
- Mackinnon A. C., Qadota H., Norman K. R., Moerman D. G., Williams B. D. C. elegans PAT-4/ILK functions as an adaptor protein within integrin adhesion complexes. Curr. Biol. 2002;12:787–797. doi: 10.1016/s0960-9822(02)00810-2. [DOI] [PubMed] [Google Scholar]
- McDonald K. A., Lakonishok M., Horwitz A. F. Alpha v and alpha 3 integrin subunits are associated with myofibrils during myofibrillogenesis. J. Cell Sci. 1995;108:975–983. doi: 10.1242/jcs.108.3.975. [DOI] [PubMed] [Google Scholar]
- Mercer K. B., Flaherty D. B., Miller R. K., Qadota H., Tinley T. L., Moerman D. G., Benian G. M. Caenorhabditis elegans UNC-98, a C2H2 Zn finger protein, is a novel partner of UNC-97/PINCH in muscle adhesion complexes. Mol. Biol. Cell. 2003;14:2492–2507. doi: 10.1091/mbc.E02-10-0676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mercer K. B., Miller R. K., Tinley T. L., Sheth S., Qadota H., Benian G. M. Caenorhabditis elegans UNC-96 is a new component of M-lines that interacts with UNC-98 and paramyosin and is required in adult muscle for assembly and/or maintenance of thick filaments. Mol. Biol. Cell. 2006;17:3832–3847. doi: 10.1091/mbc.E06-02-0144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller R. K., Qadota H., Landsverk M. L., Mercer K. B., Epstein H. F., Benian G. M. UNC-98 links an integrin-associated complex to thick filaments in Caenorhabditis elegans muscle. J. Cell Biol. 2006;175:853–859. doi: 10.1083/jcb.200608043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miranti C. K., Brugge J. S. Sensing the environment: a historical perspective on integrin signal transduction. Nat. Cell Biol. 2002;4:E83–E90. doi: 10.1038/ncb0402-e83. [DOI] [PubMed] [Google Scholar]
- Moerman D. G., Williams B. D. WormBook , editor. Sarcomere assembly in C. elegans muscle. The C. elegans Research Community. 2006 doi: 10.1895/wormbook.1.81.1. doi/ 10.1895/wormbook.1891.1881.1891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nonet M. L., Grundahl K., Meyer B. J., Rand J. B. Synaptic function is impaired but not eliminated in C. elegans mutants lacking synaptotagmin. Cell. 1993;73:1291–1305. doi: 10.1016/0092-8674(93)90357-v. [DOI] [PubMed] [Google Scholar]
- Norman K. R., Cordes S., Qadota H., Rahmani P., Moerman D. G. UNC-97/PINCH is involved in the assembly of integrin cell adhesion complexes in Caenorhabditis elegans body wall muscle. Dev. Biol. 2007;309:45–55. doi: 10.1016/j.ydbio.2007.06.014. [DOI] [PubMed] [Google Scholar]
- Petit V., Thiery J. P. Focal adhesions: structure and dynamics. Biol. Cell. 2000;92:477–494. doi: 10.1016/s0248-4900(00)01101-1. [DOI] [PubMed] [Google Scholar]
- Porter G. A., Dmytrenko G. M., Winkelmann J. C., Bloch R. J. Dystrophin colocalizes with beta-spectrin in distinct subsarcolemmal domains in mammalian skeletal muscle. J. Cell Biol. 1992;117:997–1005. doi: 10.1083/jcb.117.5.997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samarel A. M. Costameres, focal adhesions, and cardiomyocyte mechanotransduction. Am. J. Physiol. 2005;289:H2291–H2301. doi: 10.1152/ajpheart.00749.2005. [DOI] [PubMed] [Google Scholar]
- Samson T., Smyth N., Janetzky S., Wendler O., Muller J. M., Schule R., von der Mark H., von der Mark K., Wixler V. The LIM-only proteins FHL2 and FHL3 interact with α- and β-subunits of the muscle α7β1 integrin receptor. J. Biol. Chem. 2004;279:28641–28652. doi: 10.1074/jbc.M312894200. [DOI] [PubMed] [Google Scholar]
- Scholl F. A., McLoughlin P., Ehler E., de Giovanni C., Schafer B. W. DRAL is a p53-responsive gene whose four and a half LIM domain protein product induces apoptosis. J. Cell Biol. 2000;151:495–506. doi: 10.1083/jcb.151.3.495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuboi D., Qadota H., Kasuya K., Amano M., Kaibuchi K. Isolation of the interacting molecules with GEX-3 by a novel functional screening. Biochem. Biophys. Res. Commun. 2002;292:697–701. doi: 10.1006/bbrc.2002.6717. [DOI] [PubMed] [Google Scholar]
- Waterston R. H. Muscle. In: Wood W. B., editor. The Nematode Caenorhabditis elegans. Cold Harbor, NY: Cold Spring Harbor Laboratory Press; 1988. pp. 281–335. [Google Scholar]
- Yochem J., Gu T., Han M. A new marker for mosaic analysis in Caenorhabditis elegans indicates a fusion between hyp6 and hyp7, two major components of the hypodermis. Genetics. 1998;149:1323–1334. doi: 10.1093/genetics/149.3.1323. [DOI] [PMC free article] [PubMed] [Google Scholar]