Abstract
Cilia and basal bodies are essential organelles for a broad spectrum of functions, including the development of left-right asymmetry, kidney function, cerebrospinal fluid transport, generation of photoreceptor outer segments, and hedgehog signaling. Zebrafish fleer (flr) mutants exhibit kidney cysts, randomized left-right asymmetry, hydrocephalus, and rod outer segment defects, suggesting a pleiotropic defect in ciliogenesis. Positional cloning flr identified a tetratricopeptide repeat protein homologous to the Caenorhabditis elegans protein DYF1 that was highly expressed in ciliated cells. flr pronephric cilia were shortened and showed a reduced beat amplitude, and olfactory cilia were absent in mutants. flr cilia exhibited ultrastructural defects in microtubule B-tubules, similar to axonemes that lack tubulin posttranslational modifications (polyglutamylation or polyglycylation). flr cilia showed a dramatic reduction in cilia polyglutamylated tubulin, indicating that flr encodes a novel modulator of tubulin polyglutamylation. We also found that the C. elegans flr homologue, dyf-1, is also required for tubulin polyglutamylation in sensory neuron cilia. Knockdown of zebrafish Ttll6, a tubulin polyglutamylase, specifically eliminated tubulin polyglutamylation and cilia formation in olfactory placodes, similar to flr mutants. These results are the first in vivo evidence that tubulin polyglutamylation is required for vertebrate cilia motility and structure, and, when compromised, results in failed ciliogenesis.
INTRODUCTION
Cilia and basal bodies play important roles in vertebrate development and physiology, as sensors of mechanical, chemical, and osmotic stimuli, or in driving fluid flow in epithelial organs. Recently, the cilium has emerged as a central organelle in an unexpectedly wide spectrum of disease pathologies, including cystic kidney disease, retinal degeneration, organ laterality defects and hydrocephalus (Pazour, 2004). Nonmotile or sensory “9 + 0” cilia present at nerve endings in Caenorhabditis elegans and in mammalian kidney tubules serve to localize receptors and ion channels that function as chemo-, osmo-, or mechanosensors (Perkins et al., 1986; Scholey and Anderson, 2006). Motile, “9 + 2” cilia in epithelia tissues, including the mammalian lung and oviduct, and in the zebrafish pronephros are essential to propel fluid (Ibanez-Tallon et al., 2003; Kramer-Zucker et al., 2005). In early vertebrate development, cilia motility in transient structures such as the embryonic node in mouse or Kupffer's vesicle in zebrafish determines laterality of visceral organs (Nonaka et al., 1998; McGrath et al., 2003; Kramer-Zucker et al., 2005). Mutations in genes affecting ciliogenesis and/or basal body structure have pleiotropic effects on the function of all of these tissues (Kramer-Zucker et al., 2005), indicating a highly conserved, essential function for cilia in organogenesis.
Cilia assembly has been well studied in the flagellate Chlamydomonas reinhardtii and the nematode C. elegans (Rosenbaum and Witman, 2002; Scholey, 2003). Ciliogenesis involves a process of intraflagellar transport (IFT) that mediates the delivery of components such as tubulin, dyneins, and membrane proteins to the cilia tip where new cilia assembly occurs (Rosenbaum and Witman, 2002; Scholey, 2003; Qin et al., 2004). Transport in the anterograde direction is mediated by kinesin II motor proteins, whereas retrograde transport is driven by dynein motor proteins, and it serves to return components to the basal body (Rosenbaum and Witman, 2002; Scholey, 2003). The proteins that comprise the IFT particles (IFT proteins) have been biochemically characterized and shown to be selectively associated with anterograde (complex B) and retrograde (complex A) transport (Rosenbaum and Witman, 2002; Scholey, 2003). Localization of IFT proteins by immunoelectron microscopy, and live studies of fluorescent IFT-fusion proteins indicate that IFT and ciliogenesis occur in a stepwise process. First, particles assemble on basal body transitional fibers where cargo, kinesin motors, and IFT proteins associate. These complexes translocate to the cilia transition zone microtubules, and then to axonemal microtubules where transport of the IFT particle to the cilia tip is initiated (Rosenbaum and Witman, 2002; Scholey, 2003). Core IFT proteins, together with peripheral IFT proteins, link the IFT particles to kinesins or cargo molecules (Lucker et al., 2005) and form the basis of axonemal transport as well as Golgi-to-cilia trafficking of cilia membrane proteins (Follit et al., 2006). Engagement of kinesin and dynein motors on the cilia axoneme is essential for IFT, and it has been shown to be enhanced by a posttranslational modification of axonemal tubulin that adds polyglutamyl side chains to acidic residues in the tubulin C terminus (Gagnon et al., 1996; Wang and Sheetz, 2000; Skiniotis et al., 2004; Vent et al., 2005). These acidic domains are proposed to interact electrostatically with basic residues on kinesins resulting in efficient microtubule based transport of kinesin and dynein-linked cargo molecules (Okada and Hirokawa, 2000; Thorn et al., 2000).
Although progress has been made in the characterization of IFT and its IFT particle proteins, studies of the cilia/basal body proteome indicate that there may be 600 or more proteins involved in cilia structure and assembly (Ostrowski et al., 2002; Andersen et al., 2003; Avidor-Reiss et al., 2004; Li et al., 2004; Blacque et al., 2005; Efimenko et al., 2005; Pazour et al., 2005; Stolc et al., 2005); from this, it is clear that ciliogenesis is a complex process and that many other proteins are involved. For example, in C. elegans amphid cilia, two IFT motors, kinesin-II and osmotic avoidance defective (OSM)-3 kinesin, cooperate to drive two separate anterograde IFT pathways that build distinct cilia segments (Ou et al., 2005). The conserved tetratricopeptide protein DYF-1 has been shown to be required for OSM-3 kinesin to dock onto and move IFT particles, and it has been proposed to act as a cofactor required for homodimeric kinesin OSM-3 motility (Ou et al., 2005). However, the role of DYF-1 in protein interactions that drive ciliogenesis is currently unknown. We have analyzed ciliogenesis in the zebrafish embryo and demonstrated that zebrafish embryos and larvae exhibit multiple different forms of cilia, including 9 + 2 motile, 9 + 0 motile, and immotile sensory cilia (Tsujikawa and Malicki, 2004; Kramer-Zucker et al., 2005). Zebrafish cystic kidney mutants typically exhibit pleiotropic defects in photoreceptor cell morphology and organ situs that can arise from a common defect in ciliogenesis (Kramer-Zucker et al., 2005). Here we describe the positional cloning and characterization of the ENU mutant flr, which exhibits the multiorgan phenotype of cystic kidney, hydrocephalus, retinal degeneration, and left-right asymmetry defects. We find that flr encodes a tetratricopeptide repeat protein homologue of the C. elegans dyf-1 and demonstrate that it is required for axonemal tubulin polyglutamylation. We also show that the tubulin polyglutamylase Ttll6 is required for the formation of zebrafish olfactory cilia, demonstrating for the first time that vertebrate ciliogenesis and function in vivo requires tubulin polyglutamylation.
MATERIALS AND METHODS
Zebrafish Strains and Maintenance
Zebrafish lines for mapping and breeding were maintained according to standard procedures. The flrm477 mutation (Drummond et al., 1998) was maintained in the AB/Tubingen hybrid genetic background. flr heterozygotes were outcrossed to wild-type fish of TL background to generate a map cross. Three heterozygous F1 sibling pairs were incrossed, and in total 725 F2 mutant embryos were used to map the flr locus.
Positional Cloning of fleer
Standard linkage analysis using simple sequence repeat polymorphisms was used for both low- and high-resolution mapping of flr. The initial assignment of flr to chromosome 3 was performed by bulk segregant analysis with a panel of sslp markers spaced every 10 cM over the 25 zebrafish chromosomes. For high-resolution mapping within the interval flanked by z10805 and z22516, 34 Candidate SSR marker primer pairs were generated using the Zebrafish SSR search website (Massachusetts General Hospital, Charlestown, MA; http://danio.mgh.harvard.edu/markers/ssr.html). Genomic sequence in the genetic interval defined by markers z10805 and z63912 was repeatmasked using Repeatmasker (http://www.repeatmasker.org) and analyzed using Genscan (Burge and Karlin, 1997) to predict genes in the interval. Predicted peptides were compared with cilia proteome blast databases (Ostrowski et al., 2002; Avidor-Reiss et al., 2004; Li et al., 2004), using blastp. A high scoring hit to all three databases emerged as the Fleer candidate peptide. A hypothetical flr open reading frame (ORF) was predicted from genomic DNA based on similarity to mouse and human expressed sequence tags. Sequences within the putative 5′ and 3′ Untranslated regions of the flr ORF were used to generate nested polymerase chain reaction (PCR) primers. Reverse transcription (RT)-PCR performed on total RNA from 2.5-d-old zebrafish larvae yielded a 2.4-kb cDNA product whose sequence yielded the complete Fleer predicted polypeptide. The amplified flr cDNA was cloned into pCRII-Topo Dual vector (Invitrogen, Carlsbad, CA). The sequence of the primers used for amplification of flr cDNA is as follows: flrF1 TGTTTTTAAGCTCTGGGAAGCGTCA, flrR1 CCATTGACTGTGATGCACAGAACC, flrF2 GCTCTGGGAAGCGTCATGTATTGA, and flrR2 TGCCCTTCTGTGTCGAATATTTCC. Genbank accession numbers for sequences in this work: fleer EF653429, fleer_tv1 EU124003, ttll6 EU124004.
Morpholino Knockdown of fleer and ttll6
Two antisense morpholinos, flrMoex9d and flrMoex17d, were designed to target the splicing donor sites of exons 9 and 17, respectively. Reverse complement of the exon 9 sequence was used to generate a control morpholino flx9Con. The actual sequence of individual antisense morpholinos is as follows, with the splice donor corresponding site underlined: flrMoex9d 5′ TAGTACACTTACCTCATATTTACAG 3′, flrMoex9Con 5′ GACATTTATACTCCATTCACATGAT 3′, and flrMoex17d 5′ AATTACCTTTTTGTTGTATGGCTC 3′.
Morpholino oligonucleotides (oligos) were diluted to 0.5 mM in 100 mM KCl, 10 mM HEPES, 0.1% phenol red (Sigma-Aldrich, St. Louis, MO), and 4.6 nl was injected into two- to four-cell–stage wild-type TuAB embryos by using a nanoliter 2000 microinjector (WPI, Sarasota, FL). To verify the morpholino induced mRNA splicing defects, nested RT-PCR was performed on total RNA extracted from individual morpholino-injected embryos. Sequences of the primers located in flanking exons are listed here, with the numbers referring to exons and letters F and R denote forward and reverse, respectively: flrEx4F 5′ GCAGGCTGCAATCAAATATGGAGA 3′, flrEx9F 5′ TTCCTCCTTCAGCAAAACCCCTTC 3′, flrEx16F 5′ AGGCAGAGGAACTGATGAGGAAA 3′, flrEx16F2 5′ TCTTTCATTTGTGCATTGTCAACC 3′, flrEx12R 5′ TGTTTCGTAACCTTGCGGAGCTGT 3′, flrEx11R 5′ GAGCTGTTTGGCATGTGATCATGG 3′, flrEx19R 5′ TCATACGTGACGGTGTTCTTGC 3′, and flrEx18R 5′ TCACAGTGCTCAAGGAACTGAATGC 3′.
Antisense morpholino to the exon 10 splice donor site (ttll6MoEx10d) of the zebrafish ttll6 gene were designed based on cDNA sequence to the ttll6 C terminus (GenBank accession no. CT667061) and corresponding ttll6 genomic sequence in bac BX001014.6. PCR primers located within flanking exons 9 and 11 were used to amplify and sequence the morphant mRNA to assay induced mRNA splicing defects. Sequence of the morpholino and RT-PCR oligos are as follows: ttll6MoEx10d: GCAACTGAATGACTTACTGAGTTTG, ttll6Ex9F GACGGAGGAGAAATATGAAAAGT, ttll6Ex9F2 CCAACACAGCAGCTCTCTTTTCCA, ttll6Ex11R2 TCACGCTGGACAGCTCAAGCATT, and ttll6Ex11R TCCGCTCAAGAGAAACAGGT.
In Situ Hybridization
Whole mount in situ hybridization was performed as described previously (Thisse and Thisse, 1998). Digoxygenin-labeled antisense RNA probe (1.1 kb) was synthesized by in vitro transcription reaction by using the T7 polymerase (Ambion, Austin, TX) and HindIII-linearized flr_tv1-TopoII clone as template. The stained embryos were dehydrated, cleared with benzyl benzoate:benzyl alcohol (2:1), and photographed with a Spot digital camera (National Diagnostics, Atlanta, GA) mounted on a Leica MZ12 stereomicroscope (Leica Microsystems, Deerfield, IL).
Histology
Zebrafish larvae were fixed for histology with 4% formaldehyde in phosphate-buffered saline (PBS), overnight at 4°C. The fixed specimens were dehydrated, embedded in JB-4 (Polysciences, Warrington, PA), and sectioned at 5–7 μm. Slides were stained with methylene blue/azure II (Humphrey and Pittman, 1974), and they were examined using a Nikon E800 microscope (Nikon, Tokyo, Japan).
Antibody Staining and Immunofluorescence
Whole zebrafish embryos or larvae were fixed for immunolabeling in Dent's fixative (80% methanol:20% dimethyl sulfoxide) at 4°C overnight or for 4 h at room temperature in Prefer fixative (Anatech, Battle Creek, MI). Before antibody labeling, the fixed specimens were rehydrated, washed with PBS containing 0.5% Tween 20 (PBST), and blocked with 10% normal goat serum (NGS) in PBST.
The C. elegans cultures were grown on agar plates by using standard methods (Sulston and Hodgkin, 1988). Adult worms from wild-type N2, dyf-1 SP1205, and osm-3 strains were prepared for immunohistochemistry as per the procedure of Nonet et al. (1997). Briefly, worms were suspended for 1 h in modified Bouin's fixative (0.75 ml of saturated picric, 0.25 ml of 37% Formalin, 0.05 ml of acetic acid, 0.25 ml of methanol, and 0.01 ml of β-mercaptoethanol). While suspended in the fix, the worms were permeabilized by repeat freeze-thaw in liquid nitrogen. After an additional hour of incubation, the worms were washed with BTB (25 mM Na-borate, 0.5% Triton X-100, and 2% β-mercaptoethanol) for a total of 3 h. After a brief wash in BT (25 mM Na-borate and 0.5% Triton X-100) buffer, the worms were stored in PBST (PBS containing 0.1% Tween 20) for immunohistochemistry.
Polyclonal rabbit antibody to Fleer was raised against a 13-amino acid synthetic peptide (YYHMQDFTNAAEC) epitope located at the start of first tetratricopeptide repeat (TPR) domain in the Fleer N terminus (Cocalico Biologicals, Reamstown, PA). The antibody was affinity purified on antigen peptide-conjugated beads (Pierce Chemical, Rockford, IL).
Primary antibodies were diluted in PBST containing 5% NGS at following concentrations. rabbit anti-Fleer (1:400) mouse anti-acetylated tubulin, monoclonal antibody (mAb) 6-11B-1 (1:750; Sigma-Aldrich); mouse anti-glutamylated tubulin, mAb GT335 (1:400; gift from C. Janke); mouse anti β-tubulin, mAb 3F3-G2 (1:100; Seven Hills Bioreagents, Cincinnati, OH); mouse anti-OSM-5 (1:350; gift from B. Yoder); and rabbit anti-OSM10 (1:200; gift from Anne Hart). Antibody incubations were performed overnight at 4°C followed by washing five times, 15 min each with PBST, blocked with 10% NGS in PBST for 1 h at room temperature (RT), and detected using the secondary antibodies Alexa 546 anti-mouse (1:1000) or Alexa 488 anti-mouse (1:750). Double immunostaining using two antibodies from mouse, mAb GT335 and mAb 6–11B-1, incorporated a blocking step to prevent secondary antibody cross-reactivity (Supplemental Figure S2; Negoescu et al., 1994). mAb GT335 was applied first and detected with the secondary A546 anti-mouse antibody. This binary complex was stabilized by 1-h fixation with 4% formaldehyde at RT, followed by a brief rinse and blocking with 10% normal mouse serum (2 h at 4°C). Nonspecifically bound normal mouse serum was removed by extensive washing with PBST (5 × 15 min), and unconjugated mouse F(ab) fragments (1:20) were applied overnight at 4°C to eliminate recognition of the first mouse primary antibody with the later secondary anti-mouse antibody. After washes with PBST, sequential labeling with the second primary (mAb 6–11B-1) and its detection with Alexa 488 anti-mouse was carried out as for the single antibody labeling. Immunolabeled specimens were dehydrated in methanol, cleared with 2:1 benzyl benzoate:benzyl alcohol, and examined with a Zeiss LSM5 Pascal-confocal microscope. Two-color confocal z-series images were acquired using sequential laser excitation. For measurements of cilia length, z-series images were converted into a single plane projection and analyzed using Image J software (LSM reader).
Electron Microscopy
Embryos were prepared for electron microscopy as described previously (Drummond et al., 1998).
High-Speed Video Microscopy
The 54-h phenylthiourea (PTU)-treated embryos were put in E3 egg water containing 40 mM 2,3-butanedione monoxime (BDM; Sigma-Aldrich) for 5 min to stop the heart beat, and then they were changed to 20 mM BDM-containing egg water for observation. The embryos were analyzed with Nomarski optics by using a 40×/0.55 water immersion lens on a Zeiss Axioplan microscope (Carl Zeiss, Jena, Germany) equipped with a high-speed Photron FastCAM-PCI500 videocamera (Photron, Tokyo, Japan). Images of beating cilia were acquired at 250 frames/s by using Photron FastCAM version 1.2.0.7 (Photron). Image processing was done using Photoshop (Adobe Systems, Mountain View, CA), and movies were compiled in Graphic converter version 4.5.2 (Lemke Software, Peine, Germany) and Quicktime (Apple, Cupertino, CA).
RESULTS
fleer Encodes a Novel Tetratricopeptide Repeat Protein Related to C. elegans dyf1
The fleerm477 mutant (flrm477) was originally isolated in the Boston large-scale zebrafish mutagenesis screen (Drummond et al., 1998), and it belongs to a class of mutants exhibiting ventral axis curvature, kidney cysts, hydrocephalus, and retinal degeneration (Figure 1). Randomization of heart looping is also observed in flr heterozygote crosses (data not shown). Because recent evidence suggests that defects in ciliogenesis or cilia function can account for this specific set of phenotypes (Kramer-Zucker et al., 2005), we undertook the isolation of the flr gene by positional cloning to discover a potentially novel gene involved in ciliogenesis. Bulk segregant analysis mapped the flrm477 locus to chromosome 3 (Figure 2A). Subsequent genotyping of individual embryos with a panel of CA repeat SSR markers generated from sequence in the region narrowed the interval to a 0.24-cM interval between z10805 (0.17 cM) and z63912 (0.07 cM) (Figure 2A). Additional markers in the flr interval showed no recombination. Sequenced bacs in the interval were analyzed using Genscan (Burge and Karlin, 1997), and predicted proteins were compared with cilia proteome databases to generate candidate genes for the flr mutation. BAC zC76L23, identified by the zero recombination marker s246.3, harbored a novel predicted gene that encoded a protein with multiple TPRs; a motif often detected in proteins associated with cilia. In addition, this evolutionarily conserved flr candidate gene was represented in three independent sequence databases of the cilia proteome (Ostrowski et al., 2002; Avidor-Reiss et al., 2004; Li et al., 2004). A 2.3-kb ORF of the flr candidate was amplified from whole embryo RNA by RT-PCR. Comparison of flr candidate sequence from wild type and mutants identified a T798A nonsense mutation at the beginning of exon 8 that introduced a stop codon at the end of exon 7 (Figure 2B). Homozygosity of the T798A mutation in the genomic sequence of flr mutants, including the single rare recombinant, genetically confirmed that the candidate TPR gene encoded flr.
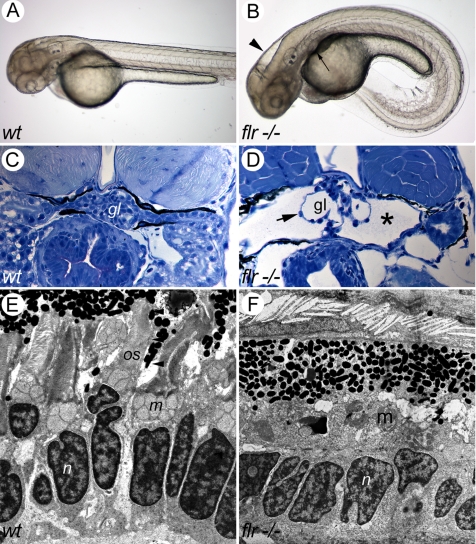
Figure 1.
The fleer mutant phenotype. (A) Wild-type embryo at 56 hpf. (B) flr mutant embryo exhibiting ventral axis curvature, hydrocephalus (arrowhead), and kidney cysts (arrow). (C) Histological section of normal pronephric kidney at 72 hpf (gl, glomerulus). (D) flr mutant kidney exhibiting cystic distension (asterisk) and expanded capillary loops (arrow; gl, glomerulus). (E) Electron micrograph of the photoreceptor cell layer of a 84-hpf larva showing rod cells and their outer segments (os) beneath the retinal pigmented epithelium. (n, nucleus; m, mitochondria). (F) flr mutant retina specifically lack rod outer segments.
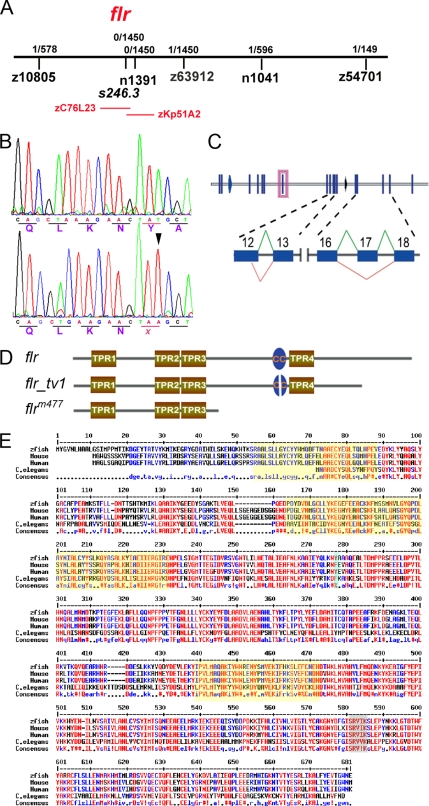
Figure 2.
Positional cloning and characterization of fleer mRNAs. (A) flr is located on chromosome 3, tightly linked to flanking SSR markers z10805 (1/578 meioses) and z63912 (1/1450 meioses). BAC zC76L23, located within the flr interval, contained a predicted TPR protein that is present in multiple cilia proteome databases and thus a potential flr candidate. (B) cDNA sequence of the flr candidate from WT and flr−/− embryos identified a T (798) > A nonsense mutation (arrowhead) that replaces a wild-type tyrosine (266) with an ochre stop codon. (C) The flr gene contains 19 exons which encode two mRNA isoforms. In flr_tv1, an upstream splice donor is used in exon 12 and exon 17 is omitted. (D) Fleer and Fleer_tv1 are predicted to contain four TPR motifs and a coiled-coil domain (CC). (E) Alignment of predicted Fleer related proteins from invertebrate and vertebrate species demonstrate evolutionary conservation. Red indicates identity, and yellow shading outlines conserved TPR domains.
Amplification of flr cDNA by RT-PCR using wild-type larvae RNA and sequencing of the cloned products revealed that the flr gene encodes two splice variants. The principal splice variant contains all 19 flr exons, whereas the sequence of a second flr transcript variant indicated alternative splicing and deletion of exon 17 coupled with use of an alternate splice donor site in exon 12 (Figure 2C). As a result of alternate splicing, the flr transcript variant (flr_tv1) mRNA contains a frame shift after exon 16, introducing 23 unique codons before a stop codon in exon 18. Use of an alternate exon 12 donor is predicted to result in an in-frame deletion of four amino acids (TKQV). Overall, flr_tv1 would be predicted to encode a protein truncated at the C terminus by 94 amino acids (Figure 2D). Prominent structural features of the flr polypeptide, including the four TPR motifs and a coiled-coil region, are shared between the two flr isoforms (Figure 2D). The nonsense point mutation in the flr mutant would truncate the Fleer polypeptide at amino acid 266 after the first three TPR domains (Figure 2D). The protein encoded by flr displayed a high degree of evolutionary conservation across predicted invertebrate and vertebrate homologues (Figure 2E). Interestingly, the C. elegans flr homolog has been recently shown to be mutated in the dye-filling mutant dyf-1. dyf-1 mutants exhibit cilia defects in chemosensory neurons, further suggesting a conserved role for flr in ciliogenesis.
Phenocopy of flr by Morpholino Knockdown
Antisense oligo-mediated knockdown of the predicted TPR repeat containing protein phenocopied the entire range of defects characteristic of the ENU mutant flrm477 (Figure 3, A and B). Wild-type embryos injected with morpholinos targeting splice donor sites in either exon 9 (Figure 3, C and D) or exon 17 (Figure 3, E and F) developed the axis curvature and pronephric cysts characteristic of flr mutants. Embryos injected with a control morpholino were normal in appearance. Molecular analysis of RT-PCR products from morpholino-injected embryos revealed misplicing events consistent with a flr loss of function phenotype. In exon 9 morphants, inclusion of intron 10 in the altered mRNA introduced a premature termination codon (Figure 3, G and H). In exon 17 morphants (Figure 3, G and I), the targeted exon was eliminated. Complete deletion of exon 17 would be expected to yield a polypeptide resembling flr_tv1; however, a single base frame shift at the splice acceptor site in exon 18 introduced a stop codon, prematurely truncating the morphant polypeptide. These results suggest that the C-terminal 114 amino acids of Fleer encoded by exons 17–19 are essential for its function and that protein encoded by the flr_tv1 splice variant may be nonfunctional.
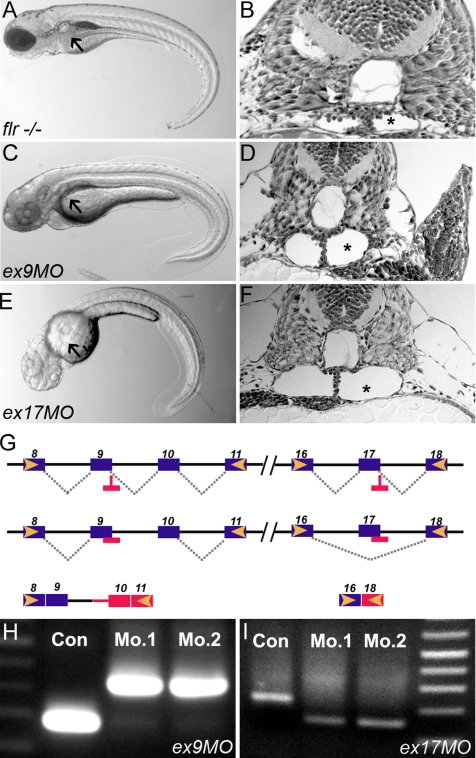
Figure 3.
Antisense morpholinos targeting the fleer candidate phenocopy the flrm477 mutant. (A) flr−/− embryos exhibit axis curvature and bilateral pronephric cysts (arrows). (B) Histological section of flr mutant showing tangential section of pronephric cyst (asterisk). (C) Embryos injected with morpholinos targeting the splice donor sites in flr exon 9 exhibit kidney cysts and axis curvature identical to flrm477. (D) Histology of the pronephric region in flr exon 9 morphant showing cyst formation. (E) Exon 17 morphants show cystic distention (asterisk) within the pronephros. (G) Summary of molecular defects caused by morpholino targeting of splice donor sites at exon 9 and 17. (H) RT-PCR analysis using primers flanking the exon 9 show an increase in amplicon size due to inclusion of intron 9 in the altered mRNA. (I) RT-PCR analysis using primers flanking the exon 17 shows a decrease in amplicon size due to exon 17 skipping. Analysis of two different single morphant larvae for each morpholino demonstrates reproducibility of flr knockdown.
flr Is Expressed in Ciliated Organs and Tissues
flr mRNA was first detected in two-cell–stage zebrafish embryos by nonquantitative RT-PCR, indicating that flr is maternally expressed (data not shown). By in situ hybridization, flr expression was first detected in Kupffer's vesicle of seven somite embryos (Figure 4A) and the lateral mesoderm of 11 somite embryos (Figure 4B). By 24–30 h postfertilization (hpf), widespread expression of flr was evident within the central nervous system and the pronephros (Figure 4C). In the pronephros, flr expression paralleled the development of multiciliated cells (Figure 4D; Liu et al., 2007). During later stages of larval development (54 h), flr expression was detected throughout the pronephros, whereas expression intensified in the olfactory placode (Figure 4E) and lateral line organs (Figure 4F). To examine the subcellular localization of the Fleer protein, we raised an antibody against an N-terminal Fleer peptide (see Materials and Methods). Fleer protein was detected in pronephric cilia (Figure 4G) and colocalized with acetylated tubulin (Figure 4H). Fleer also colocalized with acetylated tubulin in olfactory placode cilia (Figure 4, J–L). Cilia of the brain ventricle and lateral line organs were also positive for Fleer protein (data not shown). These findings show that flr mRNA is expressed in many, if not all, ciliated epithelial cells and that Fleer protein is localized to multiple types of cilia axonemes, suggesting that Fleer plays a general role in ciliogenesis.
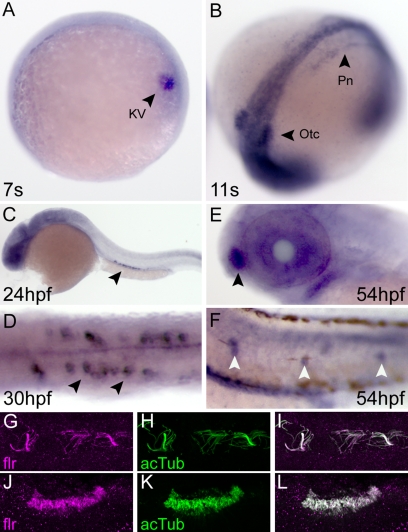
Figure 4.
Expression of fleer mRNA and protein in ciliated epithelial cells. (A) Whole mount in situ hybridization in seven somite-stage embryos demonstrated flr expression in Kupffer's vesicle cells (arrowhead; KV). (B) At the 11-somite stage, flr expression is detected in the intermediate mesoderm that will form the pronephros (arrowhead; Pn) and the otic vesicle (arrowhead; Otc). (C) At 24 hpf, pronephric expression persists in differentiated epithelial cells (arrowhead). (D) flr expression is highest at 30 hpf in multiciliated cells of the pronephros (arrowheads) (E) At 54 hpf, flr is expressed in the olfactory placode (arrowhead). (F) Lateral line organs are also positive for flr mRNA at 54 hpf (white arrowheads). (G–L) Colocalization of Fleer and acetylated tubulin in cilia. Whole mount double immunofluorescence with anti-Fleer (G) and anti-acetylated tubulin (H) in lumenal cilia of the pronephros. (I) Merge of G and H. Expression of Fleer (J) and acetylated tubulin (K) in cilia that ring the olfactory placode. (L) Merge of J and K.
fleer Loss of Function Affects Cilia Elongation
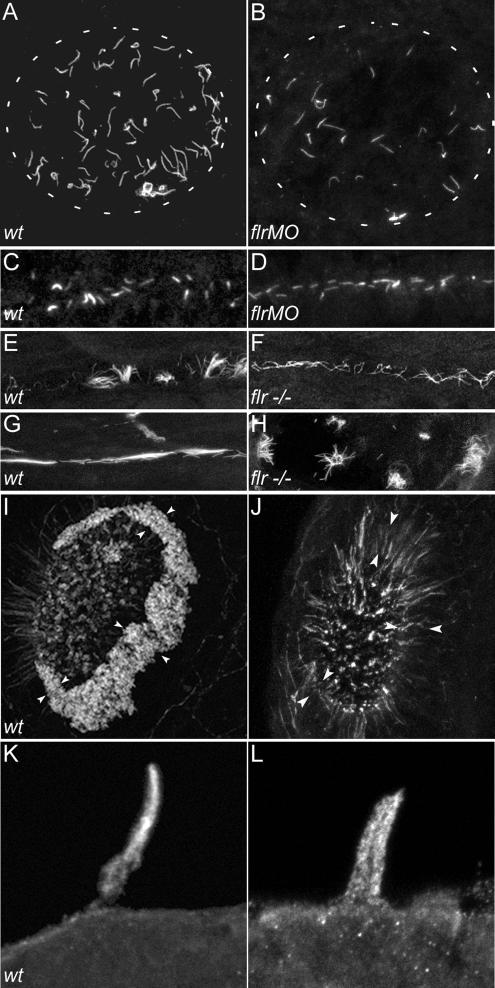
To directly test the role of flr in ciliogenesis, we visualized cilia in whole mount-fixed embryos by immunohistochemistry with antibodies to acetylated tubulin or β-tubulin. Motile cilia in Kupffer's vesicle generate a directional fluid flow thought to constitute a signal determining organ laterality (Bisgrove et al., 2005; Kramer-Zucker et al., 2005). By immunocytochemistry, cilia in Kupffer's vesicle of wild-type embryos were 4.5 ± 1.3 μm (n = 86) in length and arranged radially (Figure 5A). flr morphant cilia were shortened (3.1 ± 1.5 μm; n = 16; Figure 5B) to ∼70% of control length.
Figure 5.
flr loss of function differentially affects cilia in different organs. Cilia were visualized in wild-type and flr mutant or morphant larvae stained with an antibody to acetylated tubulin. (A) Kupffer's vesicle contains radially arranged cilia of uniform length in wild-type embryos. White dashed outline represents the borders of Kupffer's vesicle in A and B. (B) flr morphant embryos exhibit severe shortening or absence of cilia preferentially at the periphery of Kupffer's vesicle. (C) In the presumptive pronephric duct at the 10-somite stage, short cilia were apparent in both wild-type (C) and flr morphant (D) embryos. (E) Later, at 48 hpf, single cilia and multiciliated cells are observed in wild-type embryos, whereas in the flr mutant pronephros, (F) cilia were shorter and multiciliated cells were not observed. (G) In 88-hpf wild-type larvae, multiciliated cells project dense, brightly staining bundles of cilia into the narrow lumen of the pronephros. (H) flr mutant pronephroi at 88 hpf contain dispersed bundles of cilia that are nonuniform in length. (I) The dense ring of cilia on the epithelium surrounding the olfactory placode (arrowheads) of 60-hpf wild-type larvae was absent in 60h flr mutant larvae (J). Residual staining for acetylated tubulin in flr mutant olfactory placodes was associated with intracellular projections of olfactory neurons. (K) Lateral line organ cilia project from the skin of a 96-hpf larva. (L) flr mutant lateral line organ cilia at 96 hpf are shorter and slightly dispersed.
In the pronephros, motile cilia drive fluid flow, and they are essential to prevent fluid backup and kidney cyst formation (Kramer-Zucker et al., 2005). Cilia are present either singly on transporting epithelia or in groups of 15–20 on multiciliated cells (Liu et al., 2007). In 10 somite-stage embryos, single short cilia ranging in length from 1 to 2 μm were observed in the forming pronephric nephron of both wild-type (Figure 5C) and flr morphant (Figure 5D) embryos (Table 1). Elongation of cilia occurred during pronephric development, and by 48 hpf, wild-type cilia length was dependent on position within the pronephric nephron (Table 1 and Figure 5E). Cilia length in the anterior, proximal nephron was roughly twice as long as cilia in the posterior pronephros (Table 1). flr mutant cilia were consistently shorter compared with wild-type cilia in equivalent nephron segments, attaining only 60–80% of the length of their normal counterparts. (Figure 5F and Table 1). Although multiciliated cells were observed in the wild-type pronephros at 48 hpf (Figure 5G), they were not apparent in flr mutant pronephroi until much later at 88 hpf (Figure 5H). At this stage, multiciliated cells in flr mutants seemed disorganized, and the individual cilia in each multiciliated tuft were shortened (Figure 5H).
Table 1.
Cilia length (micrometers) in wild-type and fleer mutant larvae depends on position in the pronephros
| Anterior | Midsegment | Posterior | |
|---|---|---|---|
| wt | |||
| 14 somites | 1.3 ± 0.4 (n = 28) | 1.7 ± 0.3 (n = 34) | 1.1 ± 0.2 (n = 11) |
| 48 hpf | 11.5 ± 1.7 (n = 34) | 8.5 ± 1.8 (n = 27) | 5.5 ± 1 (n = 15) |
| Fleer−/− | |||
| 14 somites | 1.5 ± 0.3 (n = 68) | 2.0 ± 0.5 (n = 52) | 1.0 ± 0.2 (n = 18) |
| 48 hpf | 7.2 ± 1.6 (n = 115) | 6.6 ± 1.3 (n = 79) | 3.5 ± 1.3 (n = 18) |
Cilia lengths were measured from confocal z-series projections (see Materials and Methods) by using ImageJ software. At early stages of development (14 hpf), cilia were similar in length in wild-type and flr embryos. At 48 hpf, cilia were consistently shorter in flr mutants. Data represents the mean cilia length ± SD; n equals the number of individual cilia measured. At least three different embryos were examined for each data point.
Additional cilia defects were observed in the olfactory placode and lateral line organs. The dense ring of cilia visible on the epithelial cells of the wild-type olfactory placode at 54 h (Figure 5I) was almost entirely absent in flr mutants (Figure 5J). The mechanosensory kinocilia of the lateral line hair cells in flr larvae were consistently shorter (Figure 5L; 15.2 ± 3.5 μm, n = 8) than control kinocilia (Figure 5K; 17.5 ± 3.6 μm, n = 10).
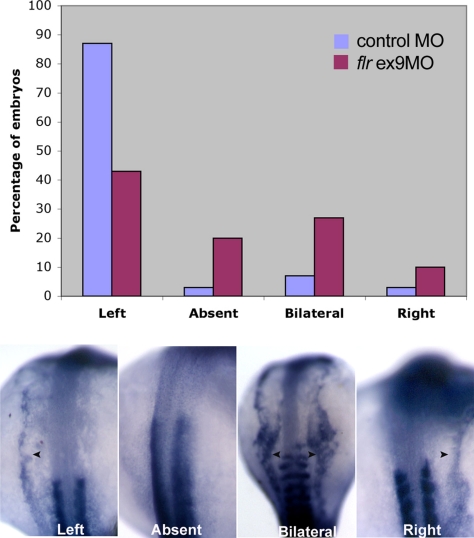
Defects seen in flr morphant cilia in Kupffer's vesicle would suggest that these embryos would show randomization of embryonic left-right asymmetry (Bisgrove et al., 2005; Kramer-Zucker et al., 2005). We used the laterality marker southpaw to assess left-right Asymmetry in flr exon 9 morphants, because the uniformity of phenotype in morphants would be expected to give a greater number of affected embryos than a clutch of embryos from an intercross of flrm477 heterozygotes. Approximately 90% of wild-type embryos showed normal left-sided expression of southpaw at the 18 somite stage, whereas <50% of flr morphants showed normal left-sided expression (Figure 6). The remaining flr morphants showed either an absence of southpaw expression, bilateral expression, or right-sided expression. These results are consistent with previous studies that point to an essential role of cilia in Kupffer's vesicle, the equivalent of the mouse embryonic node, and fluid movement in generating a signal that breaks left-right symmetry (Bisgrove et al., 2005; Kramer-Zucker et al., 2005).
Figure 6.
fleer loss of function results in defects of left-right asymmetry. In situ hybridization of wild-type and flr morphant embryos with southpaw probe. Nearly, all wild-type embryos showed normal left-sided expression at the 18-somite stage. flr exon 9 morphants exhibited randomization of the left-right axis with 44% left-sided expression, 20% absent expression, 26% bilateral expression, and 10% right-sided expression (top). Examples of southpaw expression patterns (bottom) showing normal left-sided expression or altered absent, bilateral, or right-sided expression.
Cilia Length Defects Are Accompanied by Motility Defects in flr Mutant Embryos
Structural differences in individual cilia and the disorganization of multiciliated cell tufts in the pronephros suggested that cilia motility might also be affected by the flr mutation. Imaging of pronephric cilia by high-speed video microscopy revealed that normal pronephric cilia beat synchronously at 20 Hz (Supplemental Movie 1). In contrast, the majority of single cilia in flr mutants were immotile, and the few motile cilia that were observed displayed a severely reduced beat amplitude (Supplemental Movie 2). Interestingly, although motility was severely impaired, the beat frequency in mutant cilia was not different from wild-type cilia (Supplemental Movie 2). These data suggest that a principal defect in flr mutant cilia may be the proper assembly or engagement of dynein arm motors and not the regulation or initiation of cilia beat frequency per se.
flr Mutant Cilia Exhibit Ultrastructural Defects in the B-Tubule of Microtubule Doublets
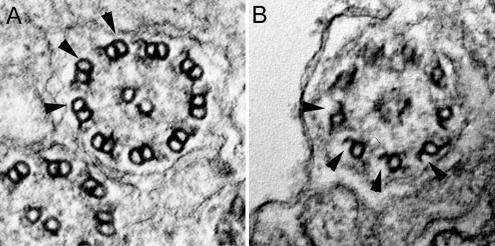
To assess whether the motility defects we observed in flr mutant cilia were due to defects in dynein arm assembly or alternatively, might be reflected changes in the structure of cilia microtubules themselves, we pursued ultrastructural analysis of flr cilia. Single pronephric cilia from 2.5 days postfertilization (dpf) wild-type zebrafish exhibit a typical “9 + 2” structure in cross section, with well formed microtubule doublets and both inner and outer dynein arm motors (Figure 7A). In contrast, flr mutant cilia exhibited a gap or discontinuity in the outer aspect of the microtubule doublet B-tubule (Figure 7B). Discontinuities were observed in B-tubules of all flr cilia examined however in some cases not all 9 doublets were affected.
Figure 7.
fleer loss of function causes structural defects in the ciliary axoneme. (A) Ultrastructure of normal single pronephric cilia showing 9 + 2 microtubule architecture at 56 hpf and intact B-tubules of the microtubule doublets (arrowheads). (B) flr mutant cilia in cross section show a specific loss of structure in the outer aspect of the B-tubules (arrowheads), with gaps in microtubules and accumulation of some electron-dense material between the doublets and the cilia outer membrane.
Defects in Axonemal Tubulin Polyglutamylation in fleer Mutants
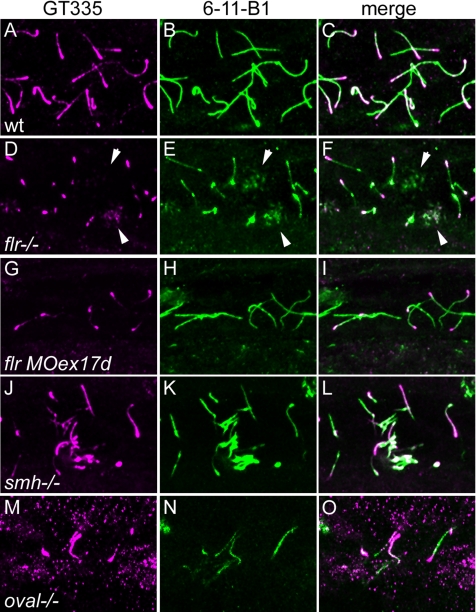
Previous studies have shown that mutations in β-tubulin that prevent posttranslational modifications of tubulin, i.e., polyglutamylation and/or polyglycylation, result in structural defects in cilia doublet B-tubules, similar to what we observe in flr mutants (Redeker et al., 2005). Also, cytoplasmic injection of antibody against polyglutamylated tubulin has been shown to reduce cilia beat amplitude without affecting beat frequency (Gagnon et al., 1996; Vent et al., 2005). The similarity of these defects to the flr phenotype prompted us to assess whether axonemal distribution of glutamylated tubulins was altered by the flr mutation. Confocal analysis of 2.5-dpf wild-type embryos, sequentially immunolabeled with the glutamylated tubulin-specific antibody GT335 and anti-acetylated tubulin, indicated that microtubules in wild-type cilia are polyglutamylated, with polyglutamylation strongest at the basal body and progressively reduced toward the cilia tip (Figure 8A). As has been shown previously, acetylated tubulin is uniformly present throughout the length of the cilium (Figure 8, B and C). flr mutant cilia showed a strong reduction in the level of polyglutamylated tubulin in axonemes (Figure 8D), whereas polyglutamylated tubulin remained present in the basal body region of cells with single cilia (Figure 8D). Cells with multiple short cilia (multiciliated cells) were also detected by acetylated tubulin staining (Figure 8E), and these showed an even more marked absence of tubulin polyglutamylation (Figure 8D) in flr mutants. A similar reduction of tubulin polyglutamylation occurred in flr morphant cilia (Figure 8, G–I). The reduction in tubulin polyglutamylation in flr mutants was specific to cilia, because neuronal microtubule polyglutamylation was not affected in mutants (data not shown). To control for the reduction in pronephric cilia length in flr mutants, we calculated the percentage of cilia length (anti-acetylated tubulin immunostaining) that exhibited polyglutamylated tubulin (GT335 immunostaining). Even compensating for slightly shortened flr cilia, polyglutamylated tubulin extended to no >34% of axoneme length in flr mutants compared with 80–90% of the length of control cilia. To test whether the reduction in tubulin polyglutamylation was simply a by-product of cilia dysfunction or perhaps an effect of epithelial cystic distension, we assayed tubulin polyglutamylation in the cystic mutant schmalhans (Brand et al., 1996). schmalhans mutant cilia showed a normal pattern of tubulin polyglutamylation, with strong polyglutamylation signal near the basal body tapering toward the cilia tip (Figure 8, J–L). To assess whether loss of tubulin polyglutamylation was a general effect of mutations in TPR proteins, we examined GT335 staining in cilia of the zebrafish mutant oval (Tsujikawa and Malicki, 2004) that carries a mutation in ift88, another TPR protein required for ciliogenesis. Although oval mutant cilia were shortened, the extent of tubulin polyglutamylation resembled wild-type (Figure 8, L–M), with GT335 staining extending ∼75% of axoneme length. These results demonstrate that the flr gene product is essential for proper tubulin polyglutamylation, and they further suggest that a failure in posttranslational modification of tubulin may account for the observed defects in flr cilia structure and motility.
Figure 8.
fleer is required for axonemal tubulin polyglutamylation. Tubulin polyglutamylation and acetylation was visualized in pronephric cilia by sequential labeling with antibodies mAb GT335 and mAb 6-11B-1. (A) In wild-type pronephric cilia at 56 hpf, polyglutamylated tubulin is distributed in a decreasing gradient from the basal body to the cilia tip. (B) Acetylated tubulin immunostaining of wild-type cilia reveals the total length of the cilia. (C) Merge of A and B. (D) In flr mutant pronephric cilia, polyglutamylated tubulin is restricted to the basal body region and severely reduced in axonemes. (E) Acetylated tubulin staining could still be observed in axonemes devoid of the polygutamylated tubulin. (F) Merge of D and E shows that polyglutamylated tubulin is found only in the proximal segment of flr cilia. (G) flr exon 17 morphants also show a dramatic reduction in polyglutamylated tubulin, whereas overall cilia length (H) is only slightly affected. (I) Merge of G and H shows restriction of the polyglutamylated tubulin to the basal body region in flr morphants. (J) The cystic mutant schmalhans shows normal tubulin polyglutamylation and cilia length (K). (L) Merge of J and K. (M) The TPR protein mutant oval/ift88 shows polyglutamylation extending to the tips of cilia. (N) Cilia are shortened in oval/ift88 mutants; however, the merged anti-polyglutamylated tubulin/acetylated tubulin image (O) shows a relatively normal pattern of polyglutamylation.
DYF-1 Is Required for Axonemal Tubulin Polyglutamylation in C. elegans Sensory Cilia
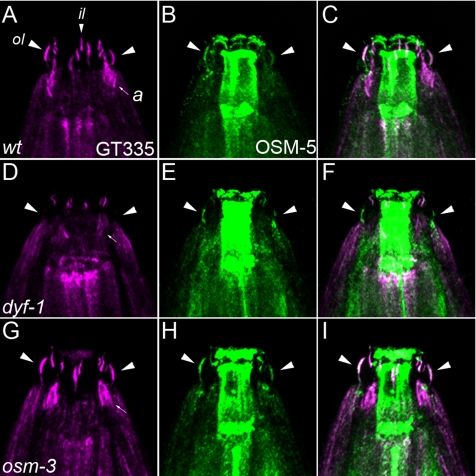
The flr homologue in C. elegans is dyf-1, a gene required for sensory cilia outer segment formation in amphid neurons. dyf-1 mutants exhibit arrested motility of the homodimeric kinesin OSM-3 in the cilia midsegment and reduced motility of other, kinesin-II–dependent IFT particles. The high degree of protein sequence conservation between Fleer and DYF-1 prompted us to assess whether the level of axonemal tubulin polyglutamylation differed in wild-type and dyf-1 mutant hermaphrodites. Double staining with antipolyglutamylated tubulin antibody GT335 (Figure 9A) and anti-OSM-5 (Figure 9B) identified multiple cilia subtypes in wild-type hermaphrodites, including outer labial cilia, inner labial cilia, and amphid cilia (Figure 9A). In outer labial cilia where anti-OSM-5 reliably reported the overall cilia structure (Figure 9B), GT335 and OSM-5 colocalized along the entire length of the axonemes (Figure 9C). Strikingly, dyf-1 mutant outer labial cilia were devoid of GT335 staining (Figure 9D). GT335 staining in amphid cilia and inner labial cilia in dyf-1 mutants was variable and less affected overall than in outer labial cilia (Figure 9D). The loss of polyglutamylation in outer labial cilia was not due to a simple absence of cilia, because anti-OSM-5 clearly identified outer labial axonemes in dyf-1 mutants (Figure 9E). A merged image of GT335 and OSM-5 double stained dyf-1 mutants shows a clear absence of polyglutamylation in outer labial cilia (Figure 9F). (For animated three-dimensional confocal images of GT335 staining in wild-type and dyf-1 mutants, see Supplemental Movie 3 and 4, respectively.) Previous studies have linked the dyf-1 mutant phenotype to the phenotype of osm-3 mutants on the basis of a similar loss of amphid cilia singlet microtubule outer segments (Ou et al., 2005). In addition, OSM-3 kinesin motility is arrested in dyf-1 mutants (Ou et al., 2005). osm-3 mutant cilia did not show a reduction in tubulin polyglutamylation in outer or inner labial cilia (Figure 9G), and they exhibited an expected reduction in GT335 staining in amphid cilia (Figure 9, G–I) corresponding to their shortened length. The results show that Fleer and DYF-1 have a conserved function in regulating axonemal tubulin polyglutamylation and that this function is distinct from the function of OSM-3 kinesin, despite the similarity in dyf-1 and osm-3 phenotypes.
Figure 9.
Loss of tubulin polyglutamylation in dyf-1 C. elegans mutant cilia. Tubulin polyglutamylation in C. elegans hermaphrodite sensory cilia was detected with the mAb GT335 (magenta) and overall cilia structure was detected with anti-OSM-5 antibody (green). Anti-OSM-5 staining also gave nonspecific labeling of the pharynx, unrelated to cilia. (A) GT335 immunoreactivity identifies polyglutamylated tubulin in multiple wild-type hermaphrodite cilia: outer labial (large arrowheads; ol), inner labial (small arrowheads; il) and amphid cilia (small arrow; a). (B) OSM-5 positive outer labial cilia (arrowheads). (C) Merge of A and B; arrowheads denote outer labial cilia positive for both GT335 and OSM-5. (D) Polyglutamylated tubulin in dyf-1 mutant hermaphrodites is reduced. Large arrowheads, position of outer labial cilia; small arrow, amphid cilia. (E) OSM-5–positive outer labial cilia in dyf-1 mutants. (F) Merge of D and E; dyf-1 outer labial cilia are negative for GT335. (G) Polyglutamylated tubulin is unaffected in osm-3 mutant hermaphrodite cilia. Large arrowheads, outer labial cilia. (H) OSM-5 positive outer labial cilia are present in osm-3 mutants. (I) Merge of G and H; GT335 and OSM-5 colocalize in osm-3 mutant outer labial cilia (large arrowheads).
The Tubulin Polyglutamylase Ttll6 Is Required for Olfactory Epithelia Ciliogenesis
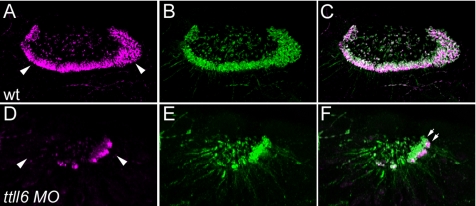
The enzymes catalyzing tubulin polyglutamylation are related to the tubulin tyrosine ligase family of proteins, and they are encoded by the tubulin tyrosine ligase-like (TTLL) genes (Janke et al., 2005; van Dijk et al., 2007). At least 13 different ttll genes are present in the zebrafish genome (data not shown). Our results would predict that knockdown of ttll genes might result in a partial phenocopy of flr, depending on where and when ttll genes were expressed. Our results also show that motile cilia in the olfactory placodes are the most sensitive to flr loss of function (Figure 5J), suggesting that they could be highly sensitive to loss of tubulin polyglutamylation activity. Consistent with this idea, we found that olfactory cilia are polyglutamylated along most of their length (Figure 10, A and B). We then tested zebrafish ttll candidate genes by gene knockdown for an effect on cilia structure or motility. We initially focused on ttll1 and ttll6, because they had been previously implicated in cilia and basal body polyglutamylation (Janke et al., 2005; van Dijk et al., 2007). ttll6 knockdown by targeting with a splice donor blocking morpholino to exon 10 (Supplemental Figure S1), produced a strong loss of function phenotype and nearly eliminated olfactory cilia (Figure 10, C and D), similar to the elimination of olfactory cilia seen in flr mutant larvae (Figure 5J). Also, like flr mutant cilia in the kidney, residual olfactory cilia in ttll6 morphants showed a relative absence of tubulin polyglutamylation in the axoneme and concentration of polyglutamylation at the basal body (Figure 10F), whereas control olfactory cilia were more uniformly polyglutamylated (Figure 10C). Sequencing of ttll6 morphant mRNA revealed that splicing failure occurred at the targeted exon donor 10, leading to inclusion of intron 10. This resulted in the introduction of 10 missense codons followed by a stop codon (Supplemental Figure S1A). The morphant ttll6 mRNA would be predicted to encode a truncated protein lacking the 231 C-terminal amino acids of Ttll6. The results indicate that tubulin polyglutamylation is essential for ciliogenesis and that ttll6 is the polyglutamylase that acts in the olfactory placode.
Figure 10.
Knockdown of ttll6 polyglutamylase prevents cilia formation and polyglutamylation in the olfactory placode. (A) Olfactory cilia are polyglutamylated. GT335 immunoreactivity is strong in olfactory multiciliated cells (arrowheads). (B) Acetylated tubulin staining identifies all olfactory cilia. (C) Merge of A and B; olfactory cilia are uniformly polyglutamylated. (D) Knockdown of ttll6 polyglutamylase severely reduces GT335 immunoreactivity in the position of olfactory cilia (arrowheads). (E) ttll6 knockdown severely reduces cilia number and length as indicated by immunostaining for acetylated tubulin. (F) Merge of D and E; in a small tuft of remnant cilia, polyglutamylation of tubulin is restricted to the basal body region of cilia, and it does not extend to the axonemes (double arrows).
DISCUSSION
We have demonstrated that the zebrafish flr gene encodes a novel tetratricopeptide repeat protein that regulates cilia structure and axonemal tubulin polyglutamylation. Tubulin polyglutamylation involves the addition of polyglutamyl side chains to glutamate residues present in an evolutionarily conserved C-terminal acidic motif on both α- and β-tubulin (Kann et al., 2003). Tubulin polyglutamylation is mediated by members of the TTLL family of enzymes (Janke et al., 2005; van Dijk et al., 2007), and this posttranslational modification enhances the binding of microtubule-associated proteins to microtubules, and it regulates the processivity of kinesin-based transport (Westermann and Weber, 2003). In Tetrahymena, mutants lacking C-terminal polyglutamylation/polyglycylation sites in tubulin, the B-subfiber of cilia axonemes is unstable, leading to structural discontinuity of the outer doublets and disruption of intraflagellar transport (Redeker et al., 2005). Cilia motility is also compromised by antibodies that bind polyglutamyl tubulin, suggesting that efficient engagement or processivity of dynein motor arms requires tubulin polyglutamylation (Gagnon et al., 1996; Vent et al., 2005). Loss of tubulin polyglutamylation in flr mutant cilia was not an indirect consequence of cystic dilation and was specific to loss of the Fleer protein and not a general effect of TPR protein loss of function. Moreover we show that flr function is conserved in C. elegans where its homologue dyf-1 plays a similar role in modulating axonemal tubulin polyglutamylation. We found that the tubulin glutamylase ttll6 is essential for ciliogenesis in the zebrafish olfactory epithelium and that morpholino knockdown of ttll6 phenocopies the flr olfactory cilia phenotype. These results point to a novel role for flr/dyf-1 in modulating an essential enzyme activity that is required for axonemal structural stability, microtubule-based transport, and cilia motility.
The C. elegans Fleer homologue DYF-1 is transported by IFT in neuronal cilia, suggesting that Fleer/DYF-1 interacts directly or indirectly with microtubule motors (Ou et al., 2005). Evidence has also been presented suggesting that DYF-1 may regulate kinesin motor activity. C. elegans dyf-1 mutants lack the most distal, singlet microtubule segments of neurosensory cilia and seem similar in this regard to osm-3 mutants that harbor mutations in a homodimeric kinesin (Ou et al., 2005). In dyf-1 mutants, transport processes dependent on the multimeric kinesin kinesin II continue at reduced velocity; however, motility of the homodimeric kinesin OSM-3 is arrested (Ou et al., 2005). This finding has lead to the idea that multiple anterograde motors function in amphid cilia and that OSM-3 is specifically required for IFT in the singlet microtubule outer segments. DYF-1 could function as a specific activator of OSM-3 (Ou et al., 2005; Imanishi et al., 2006). In light of our results, an alternative view could be that a reduction in axonemal tubulin polyglutamylation could lead to a loss of OSM-3 motility in dyf-1 mutants. In support of this idea, in vitro studies of kinesin motility have shown that removal of the tubulin C terminus containing polyglutamylation sites does not prevent binding of kinesin motors to microtubules, but most potently reduces the processivity of kinesin movement (Thorn et al., 2000; Wang and Sheetz, 2000). Basic residues in the kinesin neck coiled-coil have been proposed to interact electrostatically with acidic tubulin C termini to maintain proximity of kinesin heads to microtubules, facilitating reengagement and processive, hand-overhand kinesin movement (Okada and Hirokawa, 2000; Thorn et al., 2000). Although our results clearly show an absence of polyglutamylation in outer labial cilia, the extent of polyglutamylation in amphid cilia, where OSM-3 motility has been measured in vivo (Ou et al., 2005), was more difficult to assess given the variable staining intensity we saw with GT335 antibody in dyf-1 mutants and the organization of amphid cilia in bundles with staggered basal body positions. Nonetheless, the lack of polyglutamylation in outer labial cilia in dyf-1 mutants suggests that a polyglutamylation defect may exist in amphid cilia and that this could contribute to defects in kinesin motility.
OSM-3 and Kinesin II are affected differently in dyf-1 mutants; OSM-3 motility is arrested while Kinesin II motility persists (Ou et al., 2005). Although further experiments will be required to clearly link polyglutamylation to the behavior of OSM-3 kinesin in amphid neuron cilia, our results raise the possibility that OSM-3 and Kinesin II may be differentially sensitive to a reduction in tubulin polyglutamylation. The idea that different classes of kinesins may have different requirements for tubulin polyglutamylation is supported by recent studies of the ROSA22 mouse mutant that lacks the PGs1 subunit of tubulin polyglutamylase. These mutant mice show a substantial reduction of polyglutamylated tubulin in neurons and a selective impairment of KIF1A cargo trafficking, whereas KIF3A and KIF5 transport is unaltered (Ikegami et al., 2007). Differential sensitivity of IFT in different cell types to loss of polyglutamylation may also help account for the high sensitivity of zebrafish olfactory cilia and kidney multiciliated cells to loss of flr function compared with other cilia. It may be that distinct kinesin motors function in olfactory and kidney multiciliated cells and these kinesins may be more sensitive to loss of tubulin polyglutamylation.
All TTLL enzymes contain a conserved core “TTL domain” (Supplemental Figure S1; van Dijk et al., 2007) that is required for polyglutamylation activity. Nonconserved domains are dispensable for enzyme activity, but they are required for normal localization of TTLL proteins (van Dijk et al., 2007). In both Tetrahymena and mammalian cells, TTLL6 is localized to basal bodies and cilia (Janke et al., 2005; van Dijk et al., 2007). The C-terminal truncated TTLL6 proteins localize to basal bodies, but cilia localization is lost (Janke et al., 2005; van Dijk et al., 2007). Our zebrafish ttll6 knockdown mRNA would be predicted to encode the N-terminal 505 amino acids of Ttll6, a domain sufficient for polyglutamylase activity (Janke et al., 2005; van Dijk et al., 2007), but lack the C terminus (Supplemental Figure S1B) required for cilia localization. Morpholino knockdown of ttll6 had strongest effects on olfactory cilia. Most cilia were missing, and interestingly, in remaining cilia, only basal bodies were polyglutamylated. This finding is consistent with the idea that the Ttll6 protein encoded by the morphant mRNA may be enzymatically active but defective in transport to the ciliary axoneme. In contrast to its strong effect on olfactory cilia, ttll6 knockdown had little effect on kidney cilia (data not shown). In the mouse kidney, all nine TTLL polyglutamylases have been shown to be expressed, suggesting that a high degree of functional redundancy may exist (van Dijk et al., 2007). Further experiments will be required to determine how many of the zebrafish Ttll polyglutamylases are expressed in the pronephros and whether a similar degree of functional redundancy can explain the lack of effect of ttll6 knockdown in the larval kidney.
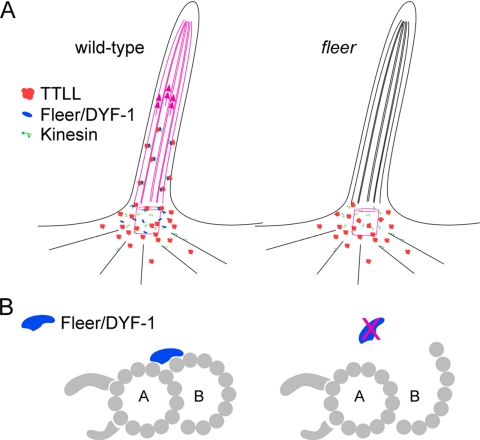
The Fleer protein lacks a TTL domain (van Dijk et al., 2007) and is therefore unlikely to function as a tubulin polyglutamylase. It is more likely that Fleer acts as a regulator of TTLL enzyme activity or plays a role in TTLL enzyme localization or transport. Our results on flr together with the data on TTLL enzyme localization suggest that Fleer/DYF-1 could act as a cargo adaptor protein that links TTLL polyglutamylase enzymes to anterograde intraflagellar transport. In the absence of Fleer, polyglutamylation of basal body tubulin would be maintained, but failure to tether TTLL enzymes to IFT would result in lack of polyglutamylated cilia tubulin leading to B-subfiber instability, impaired dynein motility, and the arrest of polyglutamylation-dependent kinesin motility (Figure 11A). This hypothetical model also raises the possibility that coordination of different kinesin motility in IFT may be, at least in part, achieved indirectly by the action of a primary Kinesin:Fleer/DYF-1:TTLL complex laying down a polyglutamylated tubulin track for a secondary kinesin (OSM-3/Kif17) to follow. As an alternative model, Fleer could act as a structural element of cilia axonemes that stabilizes the junction of cilia B-subfiber protofilaments with the A-subfiber partition (Figure 11B). The outer aspect of cilia B-subfibers are the most sensitive to detergent extraction (Witman et al., 1972), suggesting that these protofilaments may require stabilization by accessory proteins. However, this model does not readily explain how tubulin polyglutamylation would be generally reduced in flr mutant cilia. Also, the active intraflagellar transport of the Fleer homologue DYF-1 in C. elegans (Ou et al., 2005) argues for a more dynamic role for these proteins. Biochemical analysis of fleer-interacting proteins will be required to better understand its function in cilia.
Figure 11.
Hypothetical models of Fleer/DYF-1 function. (A) Fleer/DYF-1 could act as an adaptor protein that links TTLL enzymes to kinesins and intraflagellar transport. Anterograde transport of TTLL enzymes favors tubulin polyglutamylation all along the length of the axoneme (red microtubules). In the absence of Fleer/DYF-1, tubulin is polyglutamylated only in the basal body/basal region of the cilia by local diffusion of TTLL enzymes and distal microtubule doublets are not polyglutamylated (black microtubules). (B) Alternatively, Fleer/DYF-1 could play a structural role in maintaining the connection between B-tubule microtubule protofilaments and the A-subfiber of cilia doublets. In the absence of Fleer/DYF-1, structural defects in cilia microtubule doublets could interfere with intraflagellar transport.
Although our studies have focused solely on the role of the flr/dyf-1 and ttll6 genes on axonemal structure and function, tubulin polyglutamylation could affect other cellular processes in zebrafish larval organs. For example, pigment granule movement in fish melanophores can be blocked by injection of the anti-glutamyl tubulin GT335 antibody (Klotz et al., 1999). Interestingly, we have shown previously that basolateral protein trafficking is altered in flr mutants (Drummond et al., 1998), suggesting that tubulin glutamylation could also contribute to cytoplasmic microtubule-based vesicle transport. However, we have not been able to detect glutamylation of cytoplasmic microtubules in pronephric epithelial cells, and further studies will be required to evaluate the role of polyglutamylated tubulin in this context.
Supplementary Material
ACKNOWLEDGMENTS
We thank Eric Stone of the Massachusetts General Hospital (MGH) fish facility for aquaculture support, Anne Hart of the MGH C. elegans functional genomics core facility for expertise on the dyf-1 mutant and for OSM-10 antibody, Brad Yoder of University of Alabama for OSM-5 antibody and helpful suggestions, Susan Dutcher of Washington University for a key insight on tubulin polyglutamylation, Steven Potter of the University of Cincinnati for the anti-β tubulin antibody, and Carsten Janke for GT335 antibody. This work was supported by National Institutes of Health grants DK-53093 and DK-54711 (to I.A.D.). N.P. was supported by National Research Service Award training grant T32-DK007540-20, and S.M. was supported by fellowship 126a2f from the Polycystic Kidney Disease Foundation.
Footnotes
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E07-06-0537) on August 29, 2007.

 The online version of this article contains supplemental material at MBC Online (http://www.molbiolcell.org).
The online version of this article contains supplemental material at MBC Online (http://www.molbiolcell.org).
REFERENCES
- Andersen J. S., Wilkinson C. J., Mayor T., Mortensen P., Nigg E. A., Mann M. Proteomic characterization of the human centrosome by protein correlation profiling. Nature. 2003;426:570–574. doi: 10.1038/nature02166. [DOI] [PubMed] [Google Scholar]
- Avidor-Reiss T., Maer A. M., Koundakjian E., Polyanovsky A., Keil T., Subramaniam S., Zuker C. S. Decoding cilia function: defining specialized genes required for compartmentalized cilia biogenesis. Cell. 2004;117:527–539. doi: 10.1016/s0092-8674(04)00412-x. [DOI] [PubMed] [Google Scholar]
- Bisgrove B. W., Snarr B. S., Emrazian A., Yost H. J. Polaris and Polycystin-2 in dorsal forerunner cells and Kupffer's vesicle are required for specification of the zebrafish left-right axis. Dev. Biol. 2005;287:274–288. doi: 10.1016/j.ydbio.2005.08.047. [DOI] [PubMed] [Google Scholar]
- Blacque O. E., et al. Functional genomics of the cilium, a sensory organelle. Curr. Biol. 2005;15:935–941. doi: 10.1016/j.cub.2005.04.059. [DOI] [PubMed] [Google Scholar]
- Brand M., et al. Mutations affecting development of the midline and general body shape during zebrafish embryogenesis. Development. 1996;123:129–142. doi: 10.1242/dev.123.1.129. [DOI] [PubMed] [Google Scholar]
- Burge C., Karlin S. Prediction of complete gene structures in human genomic DNA. J. Mol. Biol. 1997;268:78–94. doi: 10.1006/jmbi.1997.0951. [DOI] [PubMed] [Google Scholar]
- Drummond I. A., et al. Early development of the zebrafish pronephros and analysis of mutations affecting pronephric function. Development. 1998;125:4655–4667. doi: 10.1242/dev.125.23.4655. [DOI] [PubMed] [Google Scholar]
- Efimenko E., Bubb K., Mak H. Y., Holzman T., Leroux M. R., Ruvkun G., Thomas J. H., Swoboda P. Analysis of xbx genes in C. elegans. Development. 2005;132:1923–1934. doi: 10.1242/dev.01775. [DOI] [PubMed] [Google Scholar]
- Follit J. A., Tuft R. A., Fogarty K. E., Pazour G. J. The intraflagellar transport protein IFT20 is associated with the Golgi complex and is required for cilia assembly. Mol. Biol. Cell. 2006;17:3781–3792. doi: 10.1091/mbc.E06-02-0133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gagnon C., White D., Cosson J., Huitorel P., Edde B., Desbruyeres E., Paturle-Lafanechere L., Multigner L., Job D., Cibert C. The polyglutamylated lateral chain of alpha-tubulin plays a key role in flagellar motility. J. Cell Sci. 1996;109:1545–1553. doi: 10.1242/jcs.109.6.1545. [DOI] [PubMed] [Google Scholar]
- Humphrey C., Pittman F. A simple methylene blue-azure II-basic fuchsin stain for epoxy-embedded tissue sections. Stain Technol. 1974;49:9–14. doi: 10.3109/10520297409116929. [DOI] [PubMed] [Google Scholar]
- Ibanez-Tallon I., Heintz N., Omran H. To beat or not to beat: roles of cilia in development and disease. Hum. Mol. Genet. 2003;1:R27–R35. doi: 10.1093/hmg/ddg061. [DOI] [PubMed] [Google Scholar]
- Ikegami K., et al. Loss of alpha-tubulin polyglutamylation in ROSA22 mice is associated with abnormal targeting of KIF1A and modulated synaptic function. Proc. Natl. Acad. Sci. USA. 2007;104:3213–3218. doi: 10.1073/pnas.0611547104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imanishi M., Endres N. F., Gennerich A., Vale R. D. Autoinhibition regulates the motility of the C. elegans intraflagellar transport motor OSM-3. J. Cell Biol. 2006;174:931–937. doi: 10.1083/jcb.200605179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janke C., et al. Tubulin polyglutamylase enzymes are members of the TTL domain protein family. Science. 2005;308:1758–1762. doi: 10.1126/science.1113010. [DOI] [PubMed] [Google Scholar]
- Kann M. L., Soues S., Levilliers N., Fouquet J. P. Glutamylated tubulin: diversity of expression and distribution of isoforms. Cell Motil. Cytoskeleton. 2003;55:14–25. doi: 10.1002/cm.10107. [DOI] [PubMed] [Google Scholar]
- Klotz A., Rutberg M., Denoulet P., Wallin M. Polyglutamylation of atlantic cod tubulin: immunochemical localization and possible role in pigment granule transport. Cell Motil. Cytoskeleton. 1999;44:263–273. doi: 10.1002/(SICI)1097-0169(199912)44:4<263::AID-CM4>3.0.CO;2-V. [DOI] [PubMed] [Google Scholar]
- Kramer-Zucker A. G., Olale F., Haycraft C. J., Yoder B. K., Schier A. F., Drummond I. A. Cilia-driven fluid flow in the zebrafish pronephros, brain and Kupffer's vesicle is required for normal organogenesis. Development. 2005;132:1907–1921. doi: 10.1242/dev.01772. [DOI] [PubMed] [Google Scholar]
- Li J. B., et al. Comparative genomics identifies a flagellar and basal body proteome that includes the BBS5 human disease gene. Cell. 2004;117:541–552. doi: 10.1016/s0092-8674(04)00450-7. [DOI] [PubMed] [Google Scholar]
- Liu Y., Pathak N., Kramer-Zucker A., Drummond I. A. Notch signaling controls the differentiation of transporting epithelia and multiciliated cells in the zebrafish pronephros. Development. 2007;134:1111–1122. doi: 10.1242/dev.02806. [DOI] [PubMed] [Google Scholar]
- Lucker B. F., Behal R. H., Qin H., Siron L. C., Taggart W. D., Rosenbaum J. L., Cole D. G. Characterization of the intraflagellar transport complex B core: direct interaction of the IFT81 and IFT74/72 subunits. J. Biol. Chem. 2005;280:27688–27696. doi: 10.1074/jbc.M505062200. [DOI] [PubMed] [Google Scholar]
- McGrath J., Somlo S., Makova S., Tian X., Brueckner M. Two populations of node monocilia initiate left-right asymmetry in the mouse. Cell. 2003;114:61–73. doi: 10.1016/s0092-8674(03)00511-7. [DOI] [PubMed] [Google Scholar]
- Negoescu A., Labat-Moleur F., Lorimier P., Lamarcq L., Guillermet C., Chambaz E., Brambilla E. F(ab) secondary antibodies: a general method for double immunolabeling with primary antisera from the same species. Efficiency control by chemiluminescence. J. Histochem. Cytochem. 1994;42:433–437. doi: 10.1177/42.3.7508473. [DOI] [PubMed] [Google Scholar]
- Nonaka S., Tanaka Y., Okada Y., Takeda S., Harada A., Kanai Y., Kido M., Hirokawa N. Randomization of left-right asymmetry due to loss of nodal cilia generating leftward flow of extraembryonic fluid in mice lacking KIF3B motor protein. Cell. 1998;95:829–837. doi: 10.1016/s0092-8674(00)81705-5. [DOI] [PubMed] [Google Scholar]
- Nonet M. L., Staunton J. E., Kilgard M. P., Fergestad T., Hartwieg E., Horvitz H. R., Jorgensen E. M., Meyer B. J. Caenorhabditis elegans rab-3 mutant synapses exhibit impaired function and are partially depleted of vesicles. J. Neurosci. 1997;17:8061–8073. doi: 10.1523/JNEUROSCI.17-21-08061.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okada Y., Hirokawa N. Mechanism of the single-headed processivity: diffusional anchoring between the K-loop of kinesin and the C terminus of tubulin. Proc. Natl. Acad. Sci. USA. 2000;97:640–645. doi: 10.1073/pnas.97.2.640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ostrowski L. E., Blackburn K., Radde K. M., Moyer M. B., Schlatzer D. M., Moseley A., Boucher R. C. A proteomic analysis of human cilia: identification of novel components. Mol Cell Proteomics. 2002;1:451–465. doi: 10.1074/mcp.m200037-mcp200. [DOI] [PubMed] [Google Scholar]
- Ou G., Blacque O. E., Snow J. J., Leroux M. R., Scholey J. M. Functional coordination of intraflagellar transport motors. Nature. 2005;436:583–587. doi: 10.1038/nature03818. [DOI] [PubMed] [Google Scholar]
- Pazour G. J. Intraflagellar transport and cilia-dependent renal disease: the ciliary hypothesis of polycystic kidney disease. J. Am. Soc. Nephrol. 2004;15:2528–2536. doi: 10.1097/01.ASN.0000141055.57643.E0. [DOI] [PubMed] [Google Scholar]
- Pazour G. J., Agrin N., Leszyk J., Witman G. B. Proteomic analysis of a eukaryotic cilium. J. Cell Biol. 2005;170:103–113. doi: 10.1083/jcb.200504008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perkins L. A., Hedgecock E. M., Thomson J. N., Culotti J. G. Mutant sensory cilia in the nematode Caenorhabditis elegans. Dev. Biol. 1986;117:456–487. doi: 10.1016/0012-1606(86)90314-3. [DOI] [PubMed] [Google Scholar]
- Qin H., Diener D. R., Geimer S., Cole D. G., Rosenbaum J. L. Intraflagellar transport (IFT) cargo: IFT transports flagellar precursors to the tip and turnover products to the cell body. J. Cell Biol. 2004;164:255–266. doi: 10.1083/jcb.200308132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Redeker V., Levilliers N., Vinolo E., Rossier J., Jaillard D., Burnette D., Gaertig J., Bre M. H. Mutations of tubulin glycylation sites reveal cross-talk between the C termini of α- and β-tubulin and affect the ciliary matrix in Tetrahymena. J. Biol. Chem. 2005;280:596–606. doi: 10.1074/jbc.M408324200. [DOI] [PubMed] [Google Scholar]
- Rosenbaum J. L., Witman G. B. Intraflagellar transport. Nat. Rev. Mol. Cell Biol. 2002;3:813–825. doi: 10.1038/nrm952. [DOI] [PubMed] [Google Scholar]
- Scholey J. M. Intraflagellar transport. Annu. Rev. Cell. Dev. Biol. 2003;19:423–443. doi: 10.1146/annurev.cellbio.19.111401.091318. [DOI] [PubMed] [Google Scholar]
- Scholey J. M., Anderson K. V. Intraflagellar transport and cilium-based signaling. Cell. 2006;125:439–442. doi: 10.1016/j.cell.2006.04.013. [DOI] [PubMed] [Google Scholar]
- Skiniotis G., Cochran J. C., Muller J., Mandelkow E., Gilbert S. P., Hoenger A. Modulation of kinesin binding by the C-termini of tubulin. EMBO J. 2004;23:989–999. doi: 10.1038/sj.emboj.7600118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stolc V., Samanta M. P., Tongprasit W., Marshall W. F. Genome-wide transcriptional analysis of flagellar regeneration in Chlamydomonas reinhardtii identifies orthologs of ciliary disease genes. Proc. Natl. Acad. Sci. USA. 2005;102:3703–3707. doi: 10.1073/pnas.0408358102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sulston J., Hodgkin J. Methods in the Nematode Caenorhabditis elegans. In: Wood W. B., editor. The Nematode Caenorhabditis elegans. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1988. pp. 587–606. [Google Scholar]
- Thisse C., Thisse B. High resolution whole-mount in situ hybridization. Zebrafish Sci. Monit. 1998;5:8–9. [Google Scholar]
- Thorn K. S., Ubersax J. A., Vale R. D. Engineering the processive run length of the kinesin motor. J. Cell Biol. 2000;151:1093–1100. doi: 10.1083/jcb.151.5.1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsujikawa M., Malicki J. Intraflagellar transport genes are essential for differentiation and survival of vertebrate sensory neurons. Neuron. 2004;42:703–716. doi: 10.1016/s0896-6273(04)00268-5. [DOI] [PubMed] [Google Scholar]
- van Dijk J., Rogowski K., Miro J., Lacroix B., Edde B., Janke C. A targeted multienzyme mechanism for selective microtubule polyglutamylation. Mol. Cell. 2007;26:437–448. doi: 10.1016/j.molcel.2007.04.012. [DOI] [PubMed] [Google Scholar]
- Vent J., Wyatt T. A., Smith D. D., Banerjee A., Luduena R. F., Sisson J. H., Hallworth R. Direct involvement of the isotype-specific C-terminus of beta tubulin in ciliary beating. J. Cell Sci. 2005;118:4333–4341. doi: 10.1242/jcs.02550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z., Sheetz M. P. The C-terminus of tubulin increases cytoplasmic dynein and kinesin processivity. Biophys. J. 2000;78:1955–1964. doi: 10.1016/S0006-3495(00)76743-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westermann S., Weber K. Post-translational modifications regulate microtubule function. Nat. Rev. Mol. Cell Biol. 2003;4:938–947. doi: 10.1038/nrm1260. [DOI] [PubMed] [Google Scholar]
- Witman G. B., Carlson K., Berliner J., Rosenbaum J. L. Chlamydomonas flagella. I. Isolation and electrophoretic analysis of microtubules, matrix, membranes, and mastigonemes. J. Cell Biol. 1972;54:507–539. doi: 10.1083/jcb.54.3.507. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.