Abstract
Drosophila myosin IB (Myo1B) is one of two class I myosins in the Drosophila genome. In the larval and adult midgut enterocyte, Myo1B is present within the microvillus (MV) of the apical brush border (BB) where it forms lateral tethers between the MV membrane and underlying actin filament core. Expression of green fluorescent protein-Myo1B tail domain in the larval gut showed that the tail domain is sufficient for localization of Myo1B to the BB. A Myo1B deletion mutation exhibited normal larval gut physiology with respect to food uptake, clearance, and pH regulation. However, there is a threefold increase in terminal deoxynucleotidyl transferase dUTP nick-end labeling-positive enterocyte nuclei in the Myo1B mutant. Ultrastructural analysis of mutant midgut revealed many perturbations in the BB, including membrane tethering defects, MV vesiculation, and membrane shedding. The apical localization of both singed (fascin) and Dmoesin is impaired. BBs isolated from mutant and control midgut revealed that the loss of Myo1B causes the BB membrane and underlying cytoskeleton to become destabilized. Myo1B mutant larvae also exhibit enhanced sensitivity to oral infection by the bacterial pathogen Pseudomonas entomophila, and severe cytoskeletal defects are observed in the BB of proximal midgut epithelial cells soon after infection. Resistance to P. entomophila infection is restored in Myo1B mutant larvae expressing a Myo1B transgene. These results indicate that Myo1B may play a role in the local midgut response pathway of the Imd innate immune response to Gram-negative bacterial infection.
INTRODUCTION
The class I myosins are a diverse group of motor proteins, found across phylogeny and tissue types. The heavy chain of class I myosins is made up of a single N-terminal motor or head domain that binds both F-actin and ATP; a neck domain consisting of one or more IQ motifs, each of which binds a calmodulin light chain or other members of the calmodulin superfamily; and a C-terminal tail domain of variable length. There are two general subclasses of myosins-I (Myo1s) based on tail domain structure. Short-tailed Myo1s have a single tail homology one (TH-1) domain that is basic in charge and thought to effect interactions with membranes. Long-tailed (or amoeboid type) Myo1s have a TH-1 domain followed by a proline-rich TH-2 domain, and a Src homology–3-containing TH-3 domain (Krendel and Mooseker, 2005). The class I myosins have been implicated in a variety of processes, including cell growth, cell motility, cell adhesion, endocytosis, regulation of actin assembly dynamics, and signal transduction (Krendel and Mooseker, 2005).
The Drosophila genome contains only two class I myosin heavy chain genes, myosin 1A (Myo1A, Myo31DF, CG7438) and myosin 1B (Myo1B, Myo61F, CG9155), both of which are short-tailed (Morgan et al., 1994; Berg et al., 2001). Based on head domain sequence homology, Drosophila Myo1A and B are most closely related to the mammalian Myo1d and Myo1c, respectively (Morgan et al., 1994; Gillespie et al., 2001). Myo1A and B are both expressed in the apical brush-border (BB) domain of the midgut epithelium in late-stage embryos, larvae, and adults (Morgan et al., 1995). In the larval midgut, Myo1A is localized at the base of the apical MV and Myo1B is localized along the full length of the MV (Morgan et al., 1995). Recent studies have shown that these two class I myosins are involved in maintaining left-right symmetry of the visceral organs (Hozumi et al., 2006; Speder et al., 2006). Loss of Myo1A results in a high frequency of reversed symmetry. Hindgut expression of Myo1A may drive handedness in the embryonic gut, whereas expression in the A8 segment of the larval genital imaginal disk may drive handedness of the adult genitalia (for review, see Speder and Noselli, 2006). β-catenin has been identified as a candidate binding protein for Myo1A, giving rise to the hypothesis that this myosin may asymmetrically target polarity determinants to the adherens junction (Speder et al., 2006). Interestingly, overexpression of Myo1B in a wild-type background also causes 100% left-right reversal, suggesting that these two myosins act antagonistically to generate proper left-right asymmetry (Hozumi et al., 2006). Given the limited number of Myo1 genes in Drosophila, compared with other organisms (e.g., there are 8 class I myosins in humans and mice), we used the larval gut as a model system to determine the function of Myo1B in the midgut intestinal epithelial cell where its expression is high and subcellular localization to the BB has been well characterized. We have generated a deletion mutant, and we demonstrate that Myo1B is not an essential gene during larval development. However, Myo1B does have a role in maintaining the highly ordered structure and composition of the BB of the larval midgut epithelium. We also show that larvae lacking Myo1B are hypersensitive to oral infection by the lethal pathogen Pseudomonas entomophila (Vodovar et al., 2005). This increased sensitivity to pathogen infection suggests that Myo1B has an important role in the local Imd innate immune response by midgut enterocytes (Lemaitre and Hoffmann, 2007). This local response in the gut has been shown to be the primary and most effective form of host defense against P. entomophila (Liehl et al., 2006).
MATERIALS AND METHODS
Drosophila Stocks
All fly stocks were maintained under standard culture conditions.
Immunofluorescence
Intact larval gut tissue segments from the proventriculus and attached gastric caeca to the hindgut were dissected and fixed in phosphate-buffered saline (PBS), 4% paraformaldehyde, and 20 mM EDTA for 15 min; rinsed with Tris-buffered saline (TBS; 50 mM Tris and 150 mM NaCl, pH 8.0); and quenched in TBS for 15 min. The gut tissue was permeabilized with 0.2% Triton X-100 in 0.05% Tween 20, TBS (TBS-T) for 20 min and blocked in 25% normal goat serum (NGS) for 1 h. For Myo1B and singed staining, gut tissue was incubated overnight at 4°C in 5% NGS in TBS-T and mouse anti-Myo1B serum (1:400 dilution), or in undiluted anti-singed mouse monoclonal antibody (mAb) supernatant (singed mAb 7C; Cant et al., 1994; Developmental Studies Hybridoma Bank, University of Iowa, Iowa City, IA). For Dmoesin localization, a rabbit polyclonal antisera to Dmoesin (Edwards et al., 1997) was used at 1:1000 (gift from D. Kiehart, Duke University). After TBS-T washes, the samples were incubated in goat anti-mouse or anti-rabbit secondary antibodies (Invitrogen, Carlsbad, CA) at 1:200 in 5% NGS, TBS-T for 1 h and washed. When costained for actin, rhodamine-phalloidin (Invitrogen) was added to the secondary antibody at a concentration of 0.33 μM. Images were obtained using a Zeiss Axiovert 10 microscope (Carl Zeiss, Thornwood, NY) with a Bio-Rad 1024 confocal scanning head (Bio-Rad, Hercules, CA). For comparison of singed and Dmoesin localization, all images of heterozygous and mutant gut tissues were obtained at the same microscope settings. Similarly, to quantify F-actin levels in the BB, each genotype was imaged under identical microscope settings, the resulting images were binarized, and fluorescence intensity was determined using MetaMorph (Molecular Devices, Sunnyvale, CA) imaging software. Fluorescence intensity of the BB was measured in several individual cells per genotype; the intensities were averaged, and the Myo1B mutant and heterozygote fluorescence intensities were tested for statistical significance by t test.
To stain the adult eye and optic lobe, heads were removed, hemisected, and fixed for 1 h in 4% paraformaldehyde in phosphate-buffered saline (PBS); rinsed for 30 min in TBS; and soaked in 12% sucrose in PBS at 4°C for 16 h. Heads were transferred to O.C.T Compound (Tissue-Tek; Ted Pella, Redding, PA) for 20 min in specimen blocks and frozen; 10–12-μm cryosections were cut and adhered to Superfrost/Plus slides (Fisher Scientific, Hampton, NH), allowed to air dry, rinsed with TBS, and blocked with TBS-T + 1% bovine serum albumin for 1 h. Slides were rinsed with TBS and incubated with antibodies and phalloidin as described above except mouse Myo1B antiserum was used at 1:100.
P-element Mutagenesis
P-element P{lacW} l(3)j4A6 (FBti0004954) with background mutations removed (2644-R2/TM3) was obtained from the laboratory of Susan Parkhurst (Fred Hutchinson Cancer Research Center, Seattle, WA). This P-element was mobilized by crossing to the transposase source yw; Δ2-3,Sb/TM6. w; p2644-R2/Δ2-3,Sb flies were backcrossed to w; 2644-R2/TM3 flies to select for excision of the P-element by loss of the white marker; TM3 balanced lines were produced by crossing flies that lost the transposon to TM1/TM3 balanced flies. We screened 1200 P-element excision lines for deletion of the Myo1B gene by Southern blotting and polymerase chain reaction (PCR). Blots were probed with an EcoRI–XmaI fragment from the Myo1B cDNA (nucleotides [nt] 1-2482, probe 1) in Figure 3A. Approximately 150 lines had a band that deviated from the expected size, and they were screened using an Eco47III-AflII probe from the tail region of the Myo1B cDNA (nt 1990–2740, probe 2) in Figure 3A. Five lines showed a loss of the aberrant band detected in the first screen, indicating that a deletion of at least a portion of the Myo1B gene occurred, the best characterized of these affects Myo1B and mtacp1 and is designated Df(3L)920.
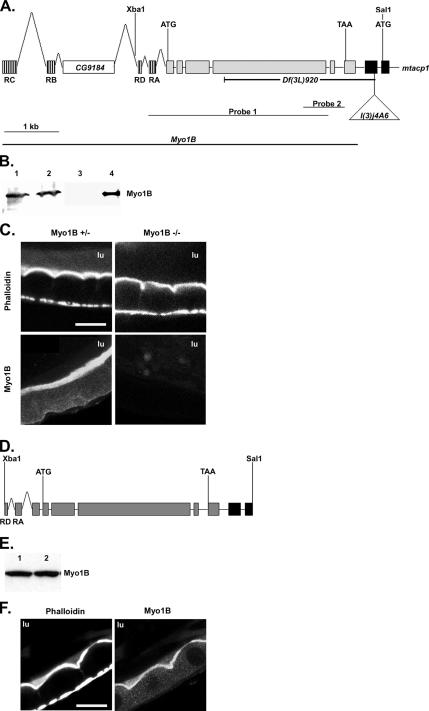
Figure 3.
Creation of a Myo1B deletion mutation. (A) The genomic structure of Myo1B and part of mtacp1 are shown. Transcription start and stop sites are labeled with ATG and TAA, respectively. The position of the selected restriction sites are shown above and the P{lacW}l(3)j4A6 insertion below. The four predicted isoforms of Myo1b use different alternatively spliced first exons, these are shown hashed, only one is used in each allele; the first 2 exons of mtacp1 are filled black. The CG9184 gene is found in the Myo1b intronic sequence. The extent of the Df(3L)920 deficiency is shown below. Probes 1 and 2 represent the Southern probes used to identify deletion mutations in Myo1B. (B) Western blot showing Myo1B expression in P{lacW}I(3)j4A6 and the Df(3L)920 mutation. Lane 1, homozygous P insertion; lane 2, heterozygous P insertion; lane 3, homozygous deletion; lane 4, heterozygous deletion. (C) Myo1B mutant and heterozygous control anterior midgut costained for actin and Myo1B. Loss of Myo1B does not disrupt actin distribution in midgut enterocyte BBs. (D) An Xba1–Sal 1 fragment of genomic DNA was used to rescue the loss of Myo1B in the deletion mutation. This construct contains two of the four alternate first exons. Transcription start and stop sites are represented by ATG and TAA, respectively. (E) Western blot showing expression of P{Myo1B} in the deletion background. Lane 1, P{Myo1B} heterozygote; lane 2, P{Myo1B} homozygote. Myo1B is expressed at equal levels in both genotypes. (F) Anterior midgut of the P{Myo1B} rescue line costained for actin and Myo1B. Transgene-expressed Myo1B localizes to the BB of midgut enterocytes. Bar, 20 μm (C and F). lu, midgut lumen.
Fly Transformation
Transformation was performed using standard methods as described previously (Spradling and Rubin, 1982). Constructs were coinjected into w1118 embryos with the helper plasmid Δ2-3.
mtacp1 Rescue
A full-length cDNA for NADH-ubiquinone oxidoreductase acyl-carrier subunit (mtacp1-RA) was assembled from a partial cDNA (gift of C. Caggese, Istituto di Genetica, Universita di Bari, Italy) extending from nt 1313696 of the genomic sequence (in the 1st exon) to the poly A tail. The 5′-untranslated region (UTR) and the missing portion of the first exon were recovered by PCR from genomic DNA. The two portions of mtacp1-RA were spliced into the pUB-2 transformation vector (gift of Lynn Cooley, Yale University; modified from pCasper3-Up2-RX). w1118 flies were transformed using standard protocols, and stable lines were established. To test rescue of the Df(3L)920 deficiency mutant, the w; P{w+ubmtacp1-RA}/CyO and w; Df(3L)920/Tm3,Ser, P{w+act5CGFP} fly lines were crossed to create w; P{w+ubmtacp1-RA}/CyO; Df(3L)920/TM3,Ser,P{w+act5CGFP}.
Myo1B Rescue
A 14-kilobase (kb) HindIII fragment from genomic cosmid 74B7 (cDROS library; Geneservice, Cambridge, England) containing the Myo1B locus was subcloned into pBluescript (Stratagene, La Jolla, CA). This fragment was further digested with Xba1 and Sal1 to produce a 5.7-kb fragment. The XbaI site is 92 nt 5′ to the mRNA, and the Sal1 site is in the first translated exon of mtacp1. This fragment was cloned into the XbaI/SalI sites of pBluescript, digested with KpnI/NotI, and ligated into the KpnI/NotI sites of pUAST (Brand and Perrimon, 1993), and transformed into w;Df(3L)920/TM3,Sb. Insertions were identified on the Df(3L)920 chromosome, and these lines were crossed to w; P{w+ubmtacp1-RA}/CyO; Df(3L)920/TM3,Ser,P{w+act5CGFP} to create w; P{w+ubmtacp1-RA}/CyO; Df(3L)920,P{w+ UASMyo1B}/TM3,Ser,P{w+act5CGFP}. Unless otherwise indicated, P{Myo1B} refers to flies homozygous for the Myo1B transgene.
Although two other exons have been identified 5′ of Myo61F-RD (Myo61F-RC and -RB), these were not included in our genomic construct as an unrelated gene (CG9184) is found 5′ of the Xba1 site in the intron between Myo61F-RB and -RD.
Green Fluorescent Protein (GFP)-Myo1B Tail Construct
Full-length Myo1B cDNA was used as a PCR template to create the tail fragment used for this construct. PCR (Expand; Roche Diagnostics, Indianapolis, IN) was used to introduce a BglII restriction site at the 5′ end, and an Apa I site at the 3′ end of the amplified cDNA, yielding a 243-amino acid fragment of the Myo1B tail (aa 784-1026). This cDNA sequence was ligated into the polylinker of pEGFP-C1 (Clontech, Palo Alto, CA). The GFP-M1B tail sequence was excised from the vector with NheI and XbaI and ligated into the XbaI site of the pUAST transformation vector. Plasmid DNA was purified using the Maxiprep kit (QIAGEN, Valencia, CA) and sequenced (Keck Biotech, Yale University) to confirm accuracy. The construct was transformed into w1118 following standard protocol, creating w; P{w+UAS GFP Myo1B Tail}. This line was crossed to the GAL4 driver line P{GawB}c355,w1118 (FBst0003750) to induce expression of GFP Myo1B tail domain in the larval gut.
Terminal Deoxynucleotidyl Transferase dUTP Nick-end Labeling (TUNEL) Assay
Early stage third instar guts were processed for the TUNEL assay based on the manufacturer's instructions (Roche Diagnostics). Guts were dissected in 4% paraformaldehyde in PBS and fixed for 20 min at room temperature. The samples were washed three times with PBS and permeabilized for 20 min with 0.2% Triton X-100 in TBS-T. Guts were then rinsed twice with PBS, incubated for 1 h in the TUNEL reagent, and rinsed two more times with PBS. Images were collected using a Zeiss Axiovert 10 microscope (Carl Zeiss) with a Bio-Rad MRC-1024 laser scanning confocal unit (Bio-Rad), and TUNEL-positive nuclei were scored in the Myo1B mutant, Myo1B heterozygote, and the Myo1B rescue line. To maintain consistency, all images used to quantify TUNEL-positive nuclei were taken of the anterior midgut immediately posterior to the proventriculus, with the focal plane focused on the layer of epithelial cell nuclei between the gut lumen and the muscle layer. Means were compared using one-way ANOVA (SPSS Inc., Chicago, IL), and multiple comparisons were analyzed using the Bonferroni test. When costaining for actin, the TUNEL assay was carried out as described above, with the addition of 0.33 μM Alexa 488 phalloidin (Invitrogen) in PBS for 30 min at room temperature after incubation in the TUNEL reagent. Guts were then washed three times with PBS, mounted, and imaged as described above.
Immunoblot Analysis
Protein samples were transferred to Hybond nitrocellulose (GE Healthcare, Chalfont St. Giles, United Kingdom), processed, and detected according to the manufacturer's instructions (ECL detection system; GE Healthcare) using horseradish peroxidase-conjugated secondary antibodies (Roche Diagnostics). Blots were probed with anti-Drosophila Myo1B mouse antiserum (Morgan et al., 1995) at 1:1000, with anti-GFP polyclonal antibody at 1 mg/ml, with anti-singed mAb supernatant at 1:20, or with anti-Dmoesin sera at 1:1000.
Gut pH and Food Clearance
Gut pH and food clearance were assessed by feeding pH indicator dyes (bromophenol blue and phenol red) or fast green in yeast paste (Dubreuil et al., 1998). The color of the pH indicators was photographed immediately after dissection in glycerol with a Nikon E800 microscope (Nikon USA, Melville, NY) equipped with a Spot charge-coupled device camera (Diagnostic Instruments, Sterling Heights, MI), courtesy of Weimin Zhong (Yale University). No differences in either gut pH regulation or food clearance rates were observed in Myo1B mutant larvae.
BB Preparation
Third instar guts were dissected in divalent cation-free insect media (Ashburner, 1989) and rinsed in ice-cold BB homogenization buffer (10 mM imidazole, pH 7.2, 4 mM K-EDTA, 1 mM K-EGTA, 2 mM dithiothreitol [DTT], 1 mM Pefabloc (Centerchem, Norwalk, CT), and 1.3 μM phalloidin; Morgan et al., 1995). Guts were then transferred to a microfuge tube containing the same buffer and homogenized with 10 passes of a plastic conical pestle (Kimble/Kontes, Vineland, NJ). The homogenate was spun at 900 × g, and the pellet was resuspended with a 25-gauge needle in BB stabilization buffer (1 mM EGTA, 75 mM KCl, 20 mM MgCl2, 1 mM DTT, 1 mM Pefabloc, 16 μg/ml phalloidin, and 10 mM imidazole, pH 7.2; Mooseker and Tilney, 1975; Morgan et al., 1995) and incubated on ice for 5 min. The isolated BBs were then analyzed by differential interference contrast (DIC) light microscopy (LM) or fixed and processed for transmission electron microscopy (TEM).
Histology
Drosophila larvae were fixed in Carnoy's fixative (60% ethanol, 30% chloroform, and 10% acetic acid) for 1 h at room temperature. Larvae were dehydrated in an ethanol series (80–100%) and then incubated twice in 100% xylenes. All steps were carried out for 10 min each. Larvae were infiltrated with a 1:1 mixture of xylenes and Paraplast Plus (Fisher, Houston, TX) for 40 min at 60°C, followed by a second infiltration step with 100% Paraplast Plus for 20 min at 60°C, and then they were embedded in fresh Paraplast Plus. Blocks were serially sectioned and stained with hematoxylin and eosin (Research Histology, Yale University School of Medicine). Images (35-mm film images) were taken using an Olympus (Center Valley, PA) SZX9 stereomicroscope.
Electron Microscopy
Whole gut or BB preparations were fixed in 2% glutaraldehyde, 0.2% tannic acid, 20 mM EGTA, in 0.1 M sodium phosphate buffer, pH 7.0, for 1 h on ice. The samples were postfixed in 1% OsO4 in 0.1 M sodium phosphate buffer, pH 6.0, for 1 h on ice, and stained overnight at 4°C with 1% uranyl acetate. The specimens were dehydrated in an ethanol series (30–100%) and embedded in EmBed 812 (Electron Microscopy Sciences, Hatfield, PA) following established protocol. Imaging was done on a JEOL 1230 TEM (JEOL, Tokyo, Japan), and digital images were recorded with a Hamamatsu ORCA-HR digital camera (Hamamatsu, Hamamatsu City, Japan).
Bacterial Infection
P. entomophila was grown for 24 h in Luria-Bertani media supplemented with 50 μg/ml rifampicin (Sigma Aldrich, St. Louis, MO) at 29°C. Fifty milliliters of bacterial culture (OD600 ∼ 1.1) was pelleted and mixed with an equal amount of crushed banana (∼100 μl of each; Vodovar et al., 2005). Larvae were added to the mixture in a 24-well plate and allowed to eat for 30 min at room temperature. The larvae were then transferred to fresh apple juice plates and incubated at 29°C. Larvae were scored for viability for 48 h after oral infection.
RESULTS
Myo1B Is Expressed throughout the Larval Midgut
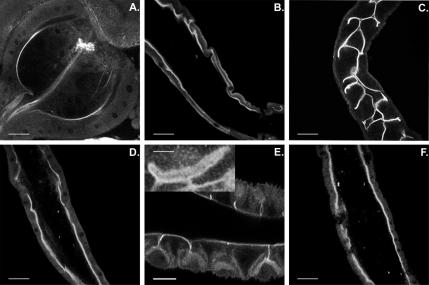
Previous work (Morgan et al., 1995) showed that Myo1B is expressed in the apical BB domain of enterocytes in the developing embryo, larvae, and adults, as well as in the egg chamber of adult females. In the larval midgut enterocyte, based on immuno-electron microscopic analysis, Myo1B is restricted to the MV of the BB, and it is not found in the subapical terminal web domain (Morgan et al., 1995). Confocal microscopy (Figure 1) of the midgut showed that Myo1B localized to the BB of enterocytes in the many distinct regions of the midgut. Expression of Myo1B in the BB remains consistently high throughout the length of the midgut (Figure 1, B, D, and F). Myo1B is also expressed in the BB of the specialized secretory structures of the alimentary system, including the proventriculus (Figure 1A), gastric ceaca (Figure 1C), cuprophilic cells (Figure 1E), and Malphigian tubules (unpublished data), where, as in gut, it is concentrated at the apical domain of the epithelium. Myo1B is also expressed in adult brain and eyes. In the optic lobe, Myo1B exhibits relatively high levels of expression in the inner chiasm, although there is also significant expression in the lamina and medulla (Supplemental Figure 1, A and B). Myo1B is also expressed in photoreceptors but in contrast to its colocalization with MV actin in the midgut BB, Myo1B is largely absent from the actin-rich rhabdomere MV, and it is concentrated at the base of MV (Supplemental Figure 1, C and D).
Figure 1.
Myo1B localizes to the enterocyte BB throughout the length of the larval midgut. (A) Proventriculus. (B) Anterior midgut. (C) Gastric caeca. (D) Middle midgut. (E) Cuprophilic cell region of middle midgut. Inset shows high-magnification image of a single cuprophilic cell. These specialized cells secrete acid in this region of the midgut. (F) Posterior midgut. Bar, 30 μm (A, B, D, and F), 20 μm (C), and 10 μm (E). Inset (E), 5 μm.
The Tail Domain Is Sufficient for Localization of Myo1B In Vivo
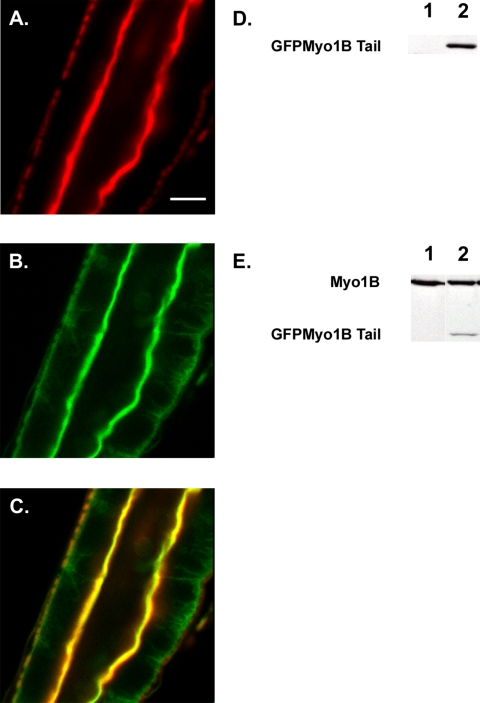
Myo1B, like the vertebrate enterocyte class I myosin, Myo1a (Mooseker and Cheney, 1995), forms lateral bridges linking the MV actin core to the plasma membrane (Morgan et al., 1995). Our laboratory has shown that the tail domain of vertebrate Myo1a is necessary and sufficient to localize Myo1a to the MV membrane (Tyska and Mooseker, 2002, 2004). To test the potential role of the Myo1B tail domain in localization to the BB, expression of GFP-tagged Myo1Btail domain under the control of the GAL4/UAS binary expression system in wild-type larvae was examined. Costaining of tail-expressing guts with phalloidin confirmed that GFP Myo1B tail domain localized specifically to the BB domain of midgut enterocytes (Figure 2).
Figure 2.
The tail domain of Myo1B is sufficient for localization. (A) Middle midgut actin visualized with Alexa 568 phalloidin. (B) GFP Myo1B Tail visualized with anti-GFP. (C) Overlay of A and B showing that the tail domain of Myo1B is sufficient for localization to the BB domain of the midgut enterocyte. (D) Western blot probed with anti-GFP to verify expression of GFP Myo1B Tail. Lane 1, WT; lane 2, tail-expressing line. (E) Western blot probed with anti-Myo1B to verify expression of GFP Myo1B tail. Lane 1, WT; lane 2, tail-expressing line. Bar, 20 μm (A–C).
Generation of a Myo1B Deletion Mutation
To study the loss of Myo1B function, we used the P-element P{lacW}l(3)j4A6, which is located 391 base pairs 3′ of Myo1B (Figure 3A). Larvae homozygous and heterozygous for the P{lacW}l(3)j4A6 insertion have normal Myo1B protein expression (Figure 3B, lanes 1 and 2). We mobilized P{lacW}l(3)j4A6 and screened for excisions using standard techniques. The resulting excision lines were screened for deletion of the 3′ end of the Myo1B gene by Southern blotting; yielding five lines with deletions in Myo1B. The best characterized of these deletions (#920) removes most of the Myo1B gene and the untranslated first exon of the mtacp-1 gene 183 base pairs 3′ of the Myo1B gene. Because this deletion affects two adjacent loci, we have named it Df(3L)920. Homozygous Df(3L)920 larva have no detectable Myo1B protein expression, compared with heterozygous control (Figure 3B, lanes 3 and 4), and they are first instar lethal. We removed the contribution of the loss of mtacp1 function by rescue of the deletion. We used cDNA for one of the two isoforms (mtacp1-RA) identified (Ragone et al., 1999) under the control of the ubiquitin promoter, creating a fly line with the genotype w P{w+ubmtacp-1}/Cyo; 920/TM3,Ser, P{w+act5CGFP}. Expression of P{w+ubmtacp-1} restored larval viability, with pupation occurring at near wild-type levels. This fly line is referred to as the Myo1B mutant. Histological examination of longitudinally oriented serial sections of whole mount heterozygous and mutant third instars revealed no obvious alterations in the morphology of larval organs (Supplemental Figures 2 and 3).
Phalloidin staining of first instar gut tissue from Myo1B mutant larvae revealed no obvious morphological differences at the light microscopic level in BB organization in comparison to heterozygous control larvae (Figure 3C). Quantification of phalloidin staining showed that there is not a statistically significant difference between the amount of F-actin in the BB of midgut enterocytes of mutant and heterozygous larvae. Full-length genomic DNA was used to create the P{Myo1B}/P{mtacp1} double rescue line used as a control for all experiments (Figure 3D). Although the Myo1B sequence was cloned into the pUAST expression vector, Myo1B protein is expressed at wild-type levels (Figure 3E) in the absence of a GAL4 driver. PCR was used to confirm that the deletion was still present; allowing us to conclude that the endogenous promoter is contained within our genomic sequence. Dissected guts were costained for both actin and Myo1B, and transgene expressed Myo1B localized specifically to the apical BB of the enterocyte (Figure 3F). Expression of P{Myo1B} alone in the deletion background does not rescue the deletion, showing that the larval lethality is due to the loss of mtacp-1. Larvae expressing transgenes for both mtacp1 and Myo1B also pupate at near wild-type levels, although very few adults eclose. Because of this, we restricted our phenotypic analyses to larvae. These results, combined with the results from P{mtacp-1} rescue suggest that Myo1B is not an essential gene during embryonic and larval development.
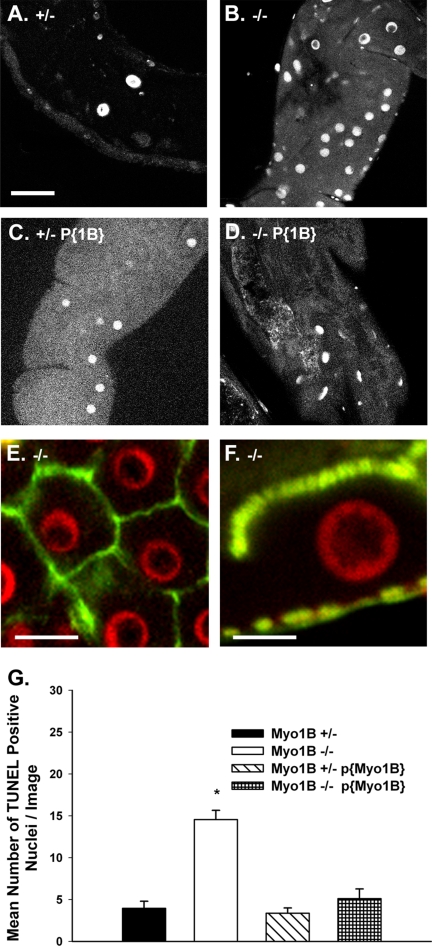
Myo1B Mutant Midgut Epithelium Has Elevated Numbers of TUNEL-positive Nuclei
As noted in Materials and Methods, no discernible differences in gut pH regulation or food clearance were observed in larvae lacking Myo1B. However, TUNEL staining of Myo1B mutant gut revealed that the number of TUNEL-positive nuclei is greater in the mutant midgut than in heterozygous control gut. Confocal microscopy and quantification of images taken of the proximal region of the anterior midgut showed that Myo1B mutant guts had a near threefold increase in the mean number of apoptotic nuclei per image as compared with Myo1B heterozygote and P{Myo1B} guts (Figure 4, A–D). The number of apoptotic nuclei is consistently higher in the mutant compared with heterozygous control in many regions of the larval midgut, including the gastric cecum, anterior midgut, and the copper cell region (data not shown). To verify that the TUNEL-positive nuclei were indeed midgut epithelial cells and not nuclei of invading hemocytes, Myo1B mutant midgut tissue was double stained with phalloidin to visualize the basolateral (Figure 4E) and apical domains (Figure 4F) of the enterocytes; this clearly shows that the TUNEL-positive nuclei are in enterocytes. Expression of full-length Myo1B in the mutant background restores the number of TUNEL-positive nuclei to the level found in the Myo1B heterozygous control. The mean number of TUNEL-positive nuclei in the Myo1B mutant is significantly greater compared with the other genotypes used for the experiment (Figure 4G). Results from one-way ANOVA gave a p value of 0.000, and the Bonferroni multiple comparison test showed that the Myo1B mutant has significantly more TUNEL-positive nuclei compared with the other three genotypes. There was no significant difference between the Myo1B heterozygote, P{Myo1B} homozygous rescue, and P{Myo1B} heterozygous rescue lines (Figure 4G).
Figure 4.
Myo1B mutant larvae have increased numbers of TUNEL-positive nuclei. All TUNEL assay images were taken from same region of the anterior midgut, on the focal plane of the enterocyte nuclei. (A) Myo1B heterozygous control. (B) Myo1B mutant. (C) P{Myo1B} heterozygous (D) P{Myo1B} homozygous. (E) En face confocal image of Myo1B mutant midgut shows that TUNEL-positive nuclei are contained within the junctional boundaries of the midgut enterocytes. (F) Longitudinal section through Myo1B mutant midgut enterocyte. TUNEL staining is specific to enterocyte nuclei. The apical BB domain is at the top of the image, and the gut muscle layer is at the bottom. (G) Quantification of images showed that the loss of Myo1B resulted in a statistically significant increase in the number of TUNEL-positive nuclei per image. Means were compared using one-way ANOVA, p = 0.000. Bonferroni multiple comparison test showed that the Myo1B mutant was significantly different from the other three genotypes. SEM represented by error bars. Bar, 30 μm (A–D), 20 μm (E), and 10 μm (F).
Midgut BB Ultrastructure Is Disrupted by the Loss of Myo1B
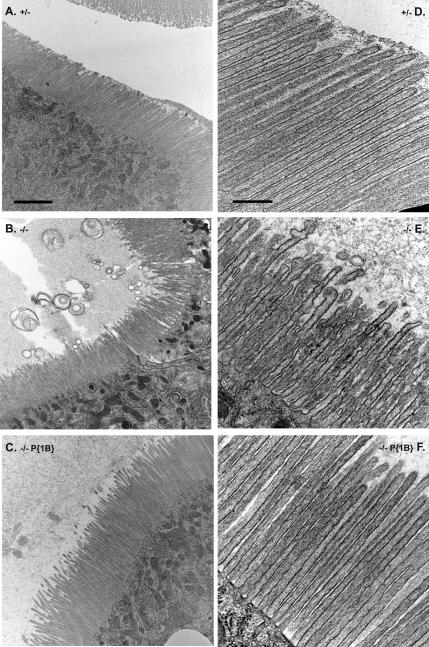
The results of whole gut TEM analysis revealed that the loss of Myo1B causes structural defects in the larval midgut BB in larvae raised under standard laboratory conditions, compared with Myo1B heterozygous control (Figure 5, A and D). Many defects were observed in the midgut epithelium of the Myo1B mutant (Figure 5, B and E), including MV vesiculation, membrane-tethering defects, and a general disorganized appearance. These defects were present in all sampled regions of the midgut, with varying degrees of severity depending on the region. The large, bullet shaped enterocytes of the anterior midgut generally contained the most severe BB defects, whereas defects were less severe to absent in the posterior midgut, where MV are much shorter and less densely arrayed. Expression of P{Myo1B} in the deletion background rescues the observed defects (Figure 5, C and F).
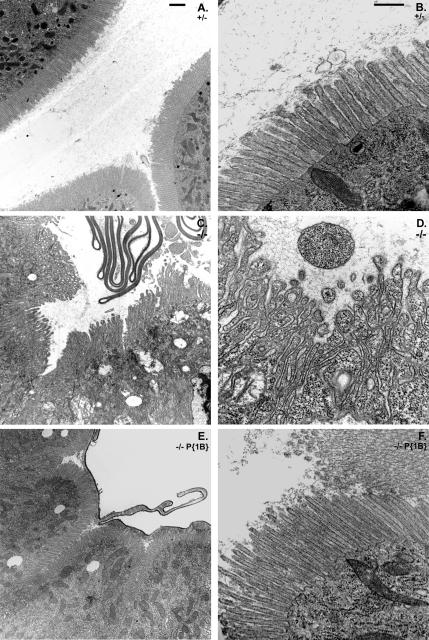
Figure 5.
Loss of Myo1B disrupts BB structure in the larval gut. (A) Low-magnification TEM of the anterior midgut shows that Myo1B heterozygote control has ordered, normal looking MV. (B) Low-magnification TEM of Myo1B mutant anterior midgut. MV are disorganized in appearance, and they are shedding membrane into the lumen of the gut. (C) Low-magnification TEM of P{Myo1B} rescue line anterior midgut. MV are ordered and normal in appearance. (D) High-magnification TEM of Myo1B heterozygote control. MV are densely arrayed and have a uniform morphology. (E) High-magnification TEM of Myo1B mutant anterior midgut; note numerous herniations on MV, vesiculation of MV tips, and general disorganized appearance. (F) High-magnification TEM of P{Myo1B} rescue line. Proper MV ultrastructure is restored by Myo1B expression. Bar, 2 μm (A–C) and 500 nm (D–F).
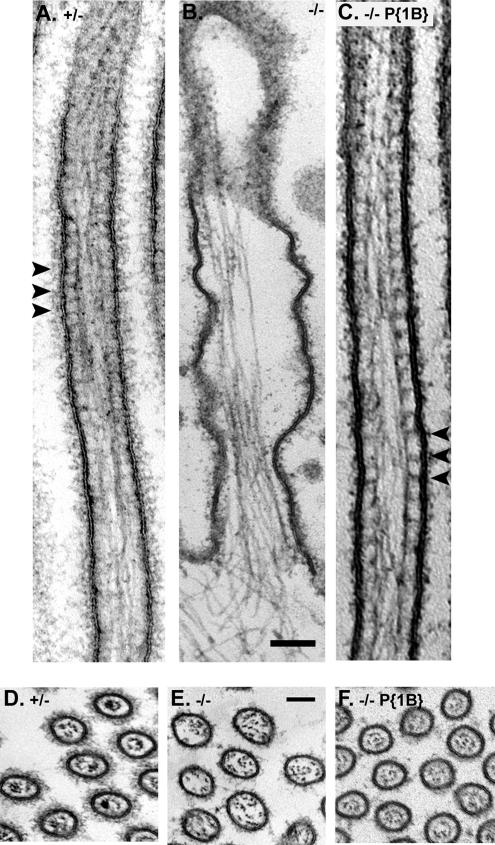
To directly test the contributions of Myo1B to the structural integrity of the BB membrane–cytoskeleton, the response of the midgut enterocytes to mechanical lysis under hypotonic conditions was examined. We have previously shown that intact, isolated BBs with well-ordered arrays of MV can be obtained by homogenization of larval midgut tissue using the same hypotonic lysis buffer used for isolation of vertebrate BBs (Bonfanti et al., 1992; Morgan et al., 1995). Light microscopy and low-magnification TEM examination of homogenates from Myo1B heterozygous larvae revealed numerous intact BBs with “fans” of MV of uniform length (Figure 6, A and D). In contrast, homogenates from Myo1B mutant midgut preparations contained far fewer isolated BBs. Moreover, those cellular fragments recognizable as derived from the BB domain exhibited MV that were less densely packed, more irregular in length, and they were characterized by membrane sloughing from the underlying actin cytoskeleton (Figure 6, B and E). Expression of P{Myo1B} in the deletion background rescues the observed defects in isolated BBs (Figure 6, C and F). Ultrastructural examination of these BB preparations revealed additional defects in BBs derived from the Myo1B mutant larvae. As predicted from previous studies, the lateral tethers linking the MV core to the membrane are absent in the Myo1B mutant, but they are present in the heterozygous control and in the P{Myo1B} rescue line (Figure 7, A–C). Cross-sections through Myo1B mutant MV showed that the central actin bundles of the MV were less densely packed than in control BBs, with some of the actin filaments resting against the outer membrane, instead of being tightly packed in the center of the MV, as seen in heterozygous control and P{Myo1B} rescue. (Figure 7, D–F). These observations suggest that Myo1B makes an essential contribution to both the stability of the BB membrane and to the structural integrity of the MV actin core.
Figure 6.
Loss of Myo1B causes numerous defects in isolated BBs. (A) DIC LM image of Myo1B heterozygote isolated BB. MV remain uniform in length and proper organization is preserved. (B) DIC image of Myo1B mutant isolated BB. Mechanical disruption of mutant guts under hypotonic conditions disrupts MV morphology. (C) DIC image of P{Myo1B} rescue line. Expression of Myo1B restores MV morphology. (D) Low-magnification TEM of Myo1B heterozygote isolated BBs shows that MV retain structure after release from the cell by mechanical lysis in hypotonic buffer. (E) Low-magnification TEM of isolated Myo1B mutant BBs. MV have numerous defects including fusion, vesiculation, and loss of membrane. (F) Low-magnification TEM of P{Myo1B} rescue line. Bar, 2 μm (A–C) and 1 μm (D–F).
Figure 7.
Myo1B forms the cross bridges between the MV actin bundle and the cell membrane. (A) Myo1B heterozygote MV. Cross-bridges are visible between the actin bundle and the cell membrane. (B) Myo1B mutant MV lack cross bridges between the central actin bundle and the cell membrane, causing the cell membrane to detach from the MV core. (C) MV cross bridges and BB structural integrity are restored in the P{Myo1B} rescue line. (D) Cross section of Myo1B heterozygote MV, note that the actin bundle is densely packed in the center of the MV. (E) Cross section of Myo1B mutant MV. The central actin bundle is less densely packed than seen in heterozygote control and P{Myo1B} rescue lines. (F) Cross section of P{Myo1B} rescue MV shows that expression of Myo1B restores proper actin bundle organization. Bar (A–F), 100 nm.
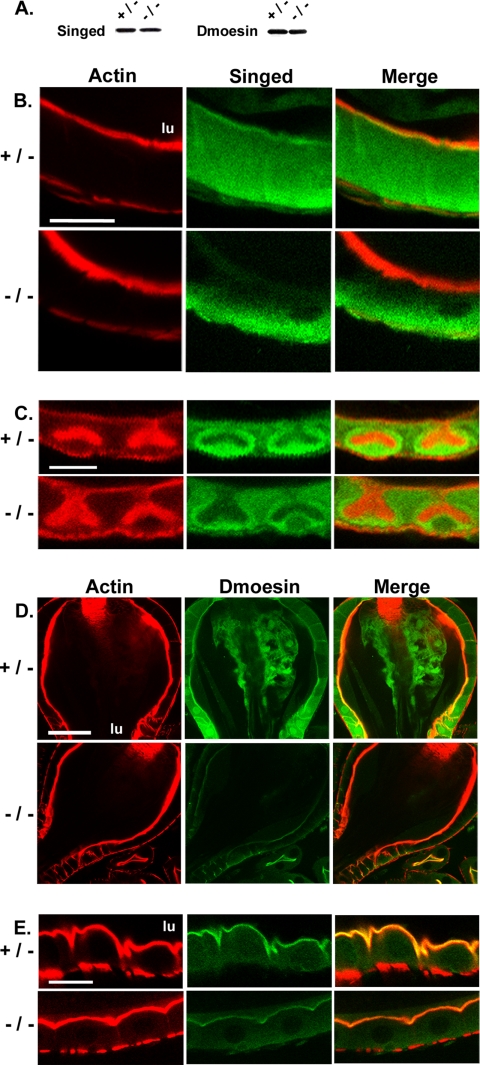
Myo1b Is Required for Proper Localization of the Cytoskeletal Components Singed and Dmoesin in Specific Regions of the Larval Gut
Immunoblot analysis of isolated gut tissue from heterozygote and mutant larvae revealed comparable levels of expression (Figure 8A) of both singed, the Drosophila homologue of the actin-bundling protein fascin (Cant et al., 1994), and Dmoesin, the sole member of the ezrin–radixin–moesin family of actin-membrane linkers in Drosophila (Fievet et al., 2007). In the heterozygote, singed localizes specifically to the base of BB microvilli in the proventriculus (Figure 8B), surrounds the actin-rich invaginations of the cuprophillic cells (Figure 8C), and it has a diffuse cytoplasmic distribution in the other regions of the midgut (data not shown). Immunofluorescence of Myo1B mutant gut revealed that singed localization is completely lost from the proventriculus BB, and it is greatly reduced from the area surrounding the actin-rich cuprophilic cell invaginations (Figure 8, B and C).
Figure 8.
Loss of Myo1B affects the localization of both singed and Dmoesin in the larval gut. (A) Singed and Dmoesin are expressed at comparable levels in Myo1B heterozygote and mutant whole gut homogenates as compared by Western blot. (B) Loss of Myo1B disrupts singed localization in the proventriculus. Apical BB staining seen in the heterozygote is completely lost in the mutant. (C) Loss of Myo1B disrupts singed localization in cuprophilic cells. The strong staining seen surrounding the cuprophilic cell invagination in the heterozygote is greatly reduced in the Myo1B mutant. (D) Loss of Myo1B reduces the amount of Dmoesin localized to the proventriculus. (E) Loss of Myo1B reduces the amount of Dmoesin that is localized to the apical BB of the anterior midgut. Bar, 20 μm (B), 10 μm (C), 30 μm (D), and 15 μm (E). lu, midgut lumen.
Loss of Myo1B also impacts the localization of the actin-membrane linker, Dmoesin. In the Myo1B heterozygote, Dmoesin is tightly localized to the apical domain of epithelial cells in the proventriculus (Figure 8D), anterior midgut (Figure 8E), and along the length of the midgut (data not shown). In the Myo1B mutant gut, there is a dramatic reduction in the level of apically localized Dmoesin in the proventriculus (Figure 8D) and anterior midgut (Figure 8E), but not as severe a reduction in the other regions of the midgut (data not shown).
Myo1B Mutant Larvae Are Hypersensitive to Oral Infection by P. entomophila
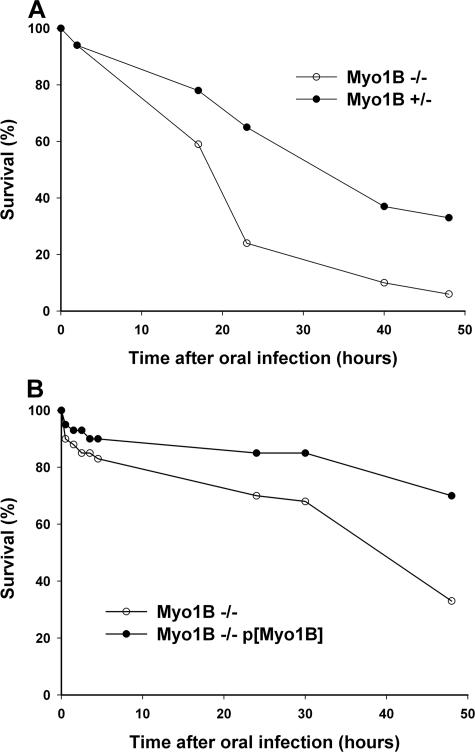
Although Myo1B does not seem to play an essential role in the absorptive functions of the gut, at least under laboratory conditions, the destabilization of the BB domain may impair the barrier properties of the gut in response to environmental challenge. To test this possibility, we examined the response of control and Myo1B mutant larvae to oral infection by P. entomophila, which has been shown to be lethal at high doses of infection (Vodovar et al., 2005). The response to this pathogen is of particular interest, because these bacteria induce significant (presumably toxin-mediated) disruption of the midgut epithelium in addition to activation of the Drosophila innate immune system that has been observed in response to other pathogens (Vodovar et al., 2005). Myo1B mutant larvae exhibit increased sensitivity to oral infection by P. entomophila compared with heterozygous larvae. Mutant larvae died at a faster rate, and in greater numbers, than either heterozygous control or Myo1B transgene rescue larvae (Figure 9). Ultrastructural analysis of proximal midgut from infected larvae showed that regions of the Myo1B mutant gut had severe BB defects as early as 2 h post infection (Figure 10, C and D). Both heterozygous control and Myo1B transgene rescue larvae had near normal BB morphology at the same time point (Figure 10, A and B, E and F).
Figure 9.
Loss of Myo1B causes increased sensitivity to infection by P. entomophila. (A) Survival curve comparing orally infected Myo1B mutant and heterozygous control first instars. Myo1B mutant larvae die at a faster rate and in greater overall numbers. (B) Survival curve comparing Myo1B mutant and P{Myo1B} rescue first instars. Expression of P{Myo1B} in the deletion background rescues the hypersensitivity to infection.
Figure 10.
Oral infection with P. entomophila causes a rapid destabilization of the BB in the Myo1B mutant gut. (A) Low-magnification TEM of Myo1B heterozygote 2 h after infection. The BB is intact and cells seems normal. (B) High-magnification TEM of Myo1B heterozygote control 2 h postinfection. (C) Low-magnification TEM of first instar Myo1B mutant gut 2 h after infection with P. entomophila. MV are fused, elongated, and shedding cytoplasm-filled membrane into the lumen. (D) High-magnification TEM of Myo1B mutant midgut 2 h after infection showing severe cytoskeletal damage induced by P. entomophila infection. MV are elongated, fused, and are destabilized. Also note that the MV are filled with cytoplasm, and they are detaching from the enterocyte. (E) Low-magnification TEM of P{Myo1B} rescue gut 2 h after infection. The BB is intact and cells appear normal. (F) High-magnification TEM of P{Myo1B} rescue 2 h post-infection. Bar, 2 μm (A, C, and E) and 500 nm (B, D, and F).
DISCUSSION
The results presented here demonstrate a role for Myo1B in maintaining the structural integrity of the BB domain in the larval midgut enterocyte and in providing resistance against oral infection by bacterial pathogens. Although Drosophila Myo1B is structurally most similar to vertebrate Myo1c, it seems to be the functional homologue of vertebrate Myo1a, because both of these myosins form lateral links that tether the MV membrane to the underlying actin core in the gut epithelium. A subset of the consequences of loss of Myo1B are reminiscent of the perturbations of the BB domain observed in the Myo1a knockout (KO) mouse (Tyska et al., 2005), although there are distinct differences between the two organisms. On the whole, the mild alterations in BB structure in the Myo1B mutant enterocyte, including irregular MV morphology and membrane shedding were less dramatic than that observed in the Myo1a KO mouse. For example, in the mouse KO, many microvilli have a vesiculated, sausage-link appearance resulting from severing of the MV core actin filaments by villin (Tyska et al., 2005). This is presumably due to elevated intramicrovillar Ca2+ resulting from impaired Ca2+ buffering due to the absence of Myo1a-associated calmodulin light chains. In contrast, Quail, the Drosophila homologue of villin (note that Quail lacks filament severing and capping activity; Matova et al., 1999), is not expressed in midgut (Mahajan-Miklos and Cooley, 1994), although other members of the gelsolin family of actin capping and severing proteins may be expressed based on the detection of villin immunogens in the midgut of Manduca larvae (Bonfanti et al., 1992). It will be important in future studies to determine whether the Myo1B-associated light chains (presumably calmodulin) play a similar role to mouse Myo1a in contributing to cytosolic Ca2+ homeostasis. For example, Ca2+ cytotoxicity could contribute to the elevated apoptosis observed in the Myo1B mutant.
Although the perturbations of the midgut BB are relatively mild in the Myo1B mutant compared with the Myo1a KO mouse BB, Myo1B may play an even more critical role than mouse Myo1a in stabilizing the BB membrane cytoskeleton, based on the fragility of the Myo1B mutant BB in response to mechanical lysis. In contrast to the mouse Myo1a KO, few, if any normal looking BBs can be recovered from the Myo1B mutant gut. This destabilization may be due to the additive effects of the loss of actin-membrane tethering by both Myo1B and Dmoesin, together with the diminished filament bundling activity by singed. It is also possible that due to the small number of class I myosins expressed in the Drosophila enterocyte, Myo1B may have a larger repertoire of functions than Myo1a in the vertebrate enterocyte, in which at least three other Myo1s are also expressed (Myo1b, c, and e), whereas in Drosophila, Myo1A is the only other Myo1 present—a myosin that has been shown to have antagonistic functions with Myo1B in determination of left-right asymmetry. Indeed, in the mouse Myo1a KO enterocyte, myosin Ic is ectopically recruited to the BB from the basolateral domain, effecting partial rescue of the loss of lipid-raft–associated membrane proteins, including sucrase isomaltase (Tyska and Mooseker, 2004; Tyska et al., 2005). In contrast, Drosophila Myo1A remains in the subapical domain in the Myo1B mutant (unpublished observation).
The reduced apical localization of Dmoesin in the Myo1B mutant midgut is of particular interest given the critical roles Dmoesin plays in establishment of epithelial cell polarity (Speck et al., 2003). The absence of Myo1B mechanochemical linkages to the membrane could have an indirect impact on Dmoesin-membrane interactions due to altered biophysical and/or compositional changes in the BB membrane. Conversely, there may be direct interactions between these two membrane-actin linker proteins. One important question for future studies will be to determine whether loss of Myo1B results in reduced levels of active, phosphorylated Dmoesin (Fievet et al., 2007).
In contrast to Myo1B and Dmoesin, singed does not exhibit tight localization to the BB domain in most regions of the gut. Nevertheless, singed could play a key role in MV actin bundle structure, a role that could be modulated by Myo1B actin-membrane interactions. Singed is required for actin bundle assembly in nurse cells during oogenesis, yet singed exhibits diffuse cytoplasmic localization (Cant et al., 1994) rather than bundle association characteristic of other components such as Quail that also contribute to bundle formation. Moreover, the loss of apical localization in epithelial cells of the proventriculus and diminishment of association with invaginations of the cuprophilic cells in the Myo1B mutant indicate that Myo1B motor linkages between filaments and the membrane can also contribute to the localization/recruitment of singed to actin bundles.
In contrast to the vertebrate BB membrane, the physiological roles of the enterocyte in nutrient absorption and digestion in the Drosophila midgut are not well understood at the molecular level. However, the presumed absorptive and secretory functions required for lumenal nutrient digestion and nutrient uptake are not discernibly affected in the absence of Myo1B given that larval growth is not severely impaired and the complex regulation of lumenal pH along the length of the midgut is normal. However, loss of Myo1B does cause a threefold increase in the number of apoptotic enterocytes, suggesting that there is an elevated level of stress in these cells. Similar results were seen in the Myo1a KO mouse (Tyska et al., 2005). Thus, although both Myo1B mutant larvae and Myo1a KO mice exhibit normal growth rates, indicative of adequate nutrient absorption under ideal laboratory feeding and living environments, the increased cellular stress in these mutants may have significant consequences under less than ideal and more realistic environments.
In vertebrates, the intestinal mucosa plays essential roles in the barrier functions of the gut in response to mucosal injury, inflammation, and to pathogens (Turner, 2006). The role of the Drosophila midgut epithelium in providing a similar barrier is less clear, but recent work has highlighted the importance of the Drosophila midgut epithelium during the innate immune response (for review, see Lemaitre and Hoffmann, 2007). In contrast to the vertebrate gut, the lumen of the fly midgut is lined by a tube of extracellular matrix, the peritrophic membrane, which serves as a physical barrier between the epithelium and potential fungal and bacterial pathogens. However, this barrier is permeable to digested nutrients and presumably any toxins released by ingested pathogens. Indeed, the recent studies of Lamaitre and colleagues (Vodovar et al., 2005; Liehl et al., 2006) on the response to oral infection by the lethal bacterial pathogen P. entomophila have provided convincing evidence for a localized response by the midgut epithelium in addition to the systemic innate immune response after pathogen infection.
P. entomophila is one of the few Drosophila pathogens identified that is lethal after oral infection.(Vodovar et al., 2005). Oral infection of P. entomophila elicits a local immune response in the gut that includes secretion of the antibacterial peptide diptericin as well activation of the systemic Imd immune response pathway (Liehl et al., 2006). One of the consequences of P. entomophila infection is disruption of the BB domain at relatively late stages of infection, followed by an increase in Myo1B gene expression (Vodovar et al., 2005), suggesting that the midgut epithelium may play an important role in host resistance to infection and the resultant response to toxins released by P. entomophila into the gut lumen. This possibility is underscored by the observations reported here that Myo1B mutant larvae are hypersensitive to P. entomophila infection, and these larvae exhibit more rapid disruptions of the midgut epithelium after oral infection with P. entomophila, compared with Myo1B heterozygous larvae.
The molecular basis for the role of Myo1B in conferring resistance to infection by P. entomophila is not known. The simplest possibility is that destabilization of the BB domain by loss of Myo1B membrane tethering makes the BB membrane more susceptible to damage by P. entomophila secreted toxins (e.g., lipases; Vodovar et al., 2006). There may also be compositional changes in the BB membrane if Myo1B is involved in the transport to and/or retention of proteins in the BB membrane, as has been demonstrated for retention of lipid raft proteins by vertebrate Myo1a (Tyska and Mooseker, 2004; Tyska et al., 2005). Another possibility is that Myo1B plays an active role in secretion of antibacterial peptides such as diptericin. In this regard, it will be of interest to see whether the Myo1B mutants show increased sensitivity to the secreted P. entomophila metalloprotease, AprA (whose presumed targets may include diptericin), which in the absence of bacteria is sufficient to elicit the local gut immune response after oral ingestion (Liehl et al., 2006). Although the basis for the role of Myo1B in conferring pathogen resistance is unknown, the results presented here demonstrate a novel role for this class I myosin in the barrier functions of the midgut epithelium that is quite distinct from its role, together with Myo1A, in determining right left visceral symmetry.
Supplementary Material
ACKNOWLEDGMENTS
We thank the members of the Mooseker laboratory for numerous discussions and Drs. Steven Ness and Marc Schwartz for technical support during early phases of this project. We especially thank Dr. Bruno Lamaitre (Centre de Genetique Molecular, Centre National de la Recherche Scientifique, Gif-sur-Yvette, France) for providing the P. entomophila strain and for advice and input on these studies. P.S.H. also thanks D. Hegan for helpful discussions and support. This work was supported by National Institutes of Health grants DK-25387 (to M.S.M.), DK-55389 (to Jon Morrow, Yale School of Medicine), and GM-52857 (to L.G.T.) and a research grant from the Crohns and Colitis Foundation of America (to M.S.M.).
Abbreviations used:
- BB
brush border
- GFP
green fluorescent protein
- MV
microvillus/i/ar
- Myo1B
myosin IB
- TUNEL
terminal deoxynucleotidyl transferase dUTP nick-end labeling.
Footnotes
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E07-02-0191) on September 12, 2007.
 The online version of this article contains supplemental material at MBC Online (http://www.molbiolcell.org).
The online version of this article contains supplemental material at MBC Online (http://www.molbiolcell.org).
REFERENCES
- Ashburner M. Drosophila: A Laboratory Manual. Cold Spring, NY: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- Berg J. S., Powell B. C., Cheney R. E. A millennial myosin census. Mol. Biol. Cell. 2001;12:780–794. doi: 10.1091/mbc.12.4.780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonfanti P., Colombo A., Heintzelman M. B., Mooseker M. S., Camatini M. The molecular architecture of an insect midgut brush border cytoskeleton. Eur. J. Cell Biol. 1992;57:298–307. [PubMed] [Google Scholar]
- Brand A. H., Perrimon N. Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development. 1993;118:401–415. doi: 10.1242/dev.118.2.401. [DOI] [PubMed] [Google Scholar]
- Cant K., Knowles B. A., Mooseker M. S., Cooley L. Drosophila singed, a fascin homolog, is required for actin bundle formation during oogenesis and bristle extension. J. Cell Biol. 1994;125:369–380. doi: 10.1083/jcb.125.2.369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubreuil R. R., Frankel J., Wang P., Howrylak J., Kappil M., Grushko T. A. Mutations of alpha spectrin and labial block cuprophilic cell differentiation and acid secretion in the middle midgut of Drosophila larvae. Dev. Biol. 1998;194:1–11. doi: 10.1006/dbio.1997.8821. [DOI] [PubMed] [Google Scholar]
- Edwards K. A., Demsky M., Montague R. A., Weymouth N., Kiehart D. P. GFP-moesin illuminates actin cytoskeleton dynamics in living tissue and demonstrates cell shape changes during morphogenesis in Drosophila. Dev. Biol. 1997;191:103–117. doi: 10.1006/dbio.1997.8707. [DOI] [PubMed] [Google Scholar]
- Fievet B., Louvard D., Arpin M. ERM proteins in epithelial cell organization and functions. Biochim. Biophys. Acta. 2007;1773:653–660. doi: 10.1016/j.bbamcr.2006.06.013. [DOI] [PubMed] [Google Scholar]
- Gillespie P. G., et al. Myosin-I nomenclature. J. Cell Biol. 2001;155:703–704. doi: 10.1083/jcb.200110032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hozumi S., et al. An unconventional myosin in Drosophila reverses the default handedness in visceral organs. Nature. 2006;440:798–802. doi: 10.1038/nature04625. [DOI] [PubMed] [Google Scholar]
- Krendel M., Mooseker M. S. Myosins: tails (and heads) of functional diversity. Physiology. 2005;20:239–251. doi: 10.1152/physiol.00014.2005. [DOI] [PubMed] [Google Scholar]
- Lemaitre B., Hoffmann J. The host defense of Drosophila melanogaster. Annu. Rev. Immunol. 2007;25:697–743. doi: 10.1146/annurev.immunol.25.022106.141615. [DOI] [PubMed] [Google Scholar]
- Liehl P., Blight M., Vodovar N., Boccard F., Lemaitre B. Prevalence of local immune response against oral infection in a Drosophila/Pseudomonas infection model. PLoS Pathol. 2006;2:e56. doi: 10.1371/journal.ppat.0020056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahajan-Miklos S., Cooley L. The villin-like protein encoded by the Drosophila quail gene is required for actin bundle assembly during oogenesis. Cell. 1994;78:291–301. doi: 10.1016/0092-8674(94)90298-4. [DOI] [PubMed] [Google Scholar]
- Matova N., Mahajan-Miklos S., Mooseker M. S., Cooley L. Drosophila quail, a villin-related protein, bundles actin filaments in apoptotic nurse cells. Development. 1999;126:5645–5657. doi: 10.1242/dev.126.24.5645. [DOI] [PubMed] [Google Scholar]
- Mooseker M. S., Cheney R. E. Unconventional myosins. Annu. Rev. Cell Dev. Biol. 1995;11:633–675. doi: 10.1146/annurev.cb.11.110195.003221. [DOI] [PubMed] [Google Scholar]
- Mooseker M. S., Tilney L. G. Organization of an actin filament-membrane complex. Filament polarity and membrane attachment in the microvilli of intestinal epithelial cells. J. Cell Biol. 1975;67:725–743. doi: 10.1083/jcb.67.3.725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan N. S., Heintzelman M. B., Mooseker M. S. Characterization of myosin-IA and myosin-IB, two unconventional myosins associated with the Drosophila brush border cytoskeleton. Dev. Biol. 1995;172:51–71. doi: 10.1006/dbio.1995.0005. [DOI] [PubMed] [Google Scholar]
- Morgan N. S., Skovronsky D. M., Artavanis-Tsakonas S., Mooseker M. S. The molecular cloning and characterization of Drosophila melanogaster myosin-IA and myosin-IB. J. Mol. Biol. 1994;239:347–356. doi: 10.1006/jmbi.1994.1376. [DOI] [PubMed] [Google Scholar]
- Ragone G., Caizzi R., Moschetti R., Barsanti P., De Pinto V., Caggese C. The Drosophila melanogaster gene for the NADH:ubiquinone oxidoreductase acyl carrier protein: developmental expression analysis and evidence for alternatively spliced forms. Mol. Gen. Genet. 1999;261:690–697. doi: 10.1007/s004380050012. [DOI] [PubMed] [Google Scholar]
- Speck O., Hughes S. C., Noren N. K., Kulikauskas R. M., Fehon R. G. Moesin functions antagonistically to the Rho pathway to maintain epithelial integrity. Nature. 2003;421:83–87. doi: 10.1038/nature01295. [DOI] [PubMed] [Google Scholar]
- Speder P., Adam G., Noselli S. Type ID unconventional myosin controls left-right asymmetry in Drosophila. Nature. 2006;440:803–807. doi: 10.1038/nature04623. [DOI] [PubMed] [Google Scholar]
- Speder P., Noselli S. Left-right asymmetry: class I myosins show the direction. Curr. Opin. Cell Biol. 2006;19:82–87. doi: 10.1016/j.ceb.2006.12.006. [DOI] [PubMed] [Google Scholar]
- Spradling A. C., Rubin G. M. Transposition of cloned P elements into Drosophila germ line chromosomes. Science. 1982;218:341–347. doi: 10.1126/science.6289435. [DOI] [PubMed] [Google Scholar]
- Turner J. R. Molecular basis of epithelial barrier regulation: from basic mechanisms to clinical application. Am. J. Pathol. 2006;169:1901–1909. doi: 10.2353/ajpath.2006.060681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyska M. J., Mackey A. T., Huang J. D., Copeland N. G., Jenkins N. A., Mooseker M. S. Myosin-1a is critical for normal brush border structure and composition. Mol. Biol. Cell. 2005;16:2443–2457. doi: 10.1091/mbc.E04-12-1116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyska M. J., Mooseker M. S. MYO1A (brush border myosin I) dynamics in the brush border of LLC-PK1-CL4 cells. Biophys. J. 2002;82:1869–1883. doi: 10.1016/S0006-3495(02)75537-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyska M. J., Mooseker M. S. A role for myosin-1A in the localization of a brush border disaccharidase. J. Cell Biol. 2004;165:395–405. doi: 10.1083/jcb.200310031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vodovar N., et al. Complete genome sequence of the entomopathogenic and metabolically versatile soil bacterium Pseudomonas entomophila. Nat. Biotechnol. 2006;24:673–679. doi: 10.1038/nbt1212. [DOI] [PubMed] [Google Scholar]
- Vodovar N., Vinals M., Liehl P., Basset A., Degrouard J., Spellman P., Boccard F., Lemaitre B. Drosophila host defense after oral infection by an entomopathogenic Pseudomonas species. Proc. Natl. Acad. Sci. USA. 2005;102:11414–11419. doi: 10.1073/pnas.0502240102. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.