Abstract
The conserved PAR proteins are localized in asymmetric cortical domains and are required for the polarized localization of cell fate determinants in many organisms. In Caenorhabditis elegans embryos, LET-99 and G protein signaling act downstream of the PARs to regulate spindle positioning and ensure asymmetric division. PAR-3 and PAR-2 localize LET-99 to a posterior cortical band through an unknown mechanism. Here we report that LET-99 asymmetry depends on cortically localized PAR-1 and PAR-4 but not on cytoplasmic polarity effectors. In par-1 and par-4 embryos, LET-99 accumulates at the entire posterior cortex, but remains at low levels at the anterior cortex occupied by PAR-3. Further, PAR-3 and PAR-1 have graded cortical distributions with the highest levels at the anterior and posterior poles, respectively, and the lowest levels of these proteins correlate with high LET-99 accumulation. These results suggest that PAR-3 and PAR-1 inhibit the localization of LET-99 to generate a band pattern. In addition, PAR-1 kinase activity is required for the inhibition of LET-99 localization, and PAR-1 associates with LET-99. Finally, examination of par-1 embryos suggests that the banded pattern of LET-99 is critical for normal posterior spindle displacement and to prevent spindle misorientation caused by cell shape constraints.
INTRODUCTION
Asymmetric cell division, a process that generates two daughter cells with distinct cell fates or functions, is of fundamental importance for normal development (Betschinger and Knoblich, 2004). Two events are coordinated to achieve intrinsically asymmetric cell division. First, cell fate determinants are differentially localized within the cell to create a polarized axis. Second, the mitotic spindle, which determines the position of the cleavage plane, is oriented along the axis of cell polarity so that division results in differential inheritance of determinants.
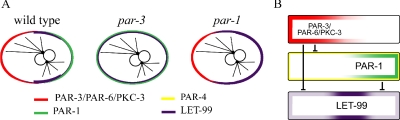
In the nematode Caenorhabditis elegans, a series of asymmetric cell division events occur during early embryo development (reviewed in Schneider and Bowerman, 2003; Cowan and Hyman, 2004; Gönczy and Rose, 2005). The anterior/posterior (A/P) polarity axis is defined at fertilization when the position of the sperm nucleus and its associated centrosome specify the posterior pole of the cell, which is also the future posterior end of the embryo. In response to the sperm cue, several PAR proteins are asymmetrically localized to either the anterior or posterior cortex. The PAR proteins are essential for cell polarization, spindle orientation, and unequal cleavage during the asymmetric division of the one-cell and the P-cell lineage. Genetic and immunolocalization studies have established a PAR-dependent hierarchy that leads to the asymmetric distribution of cell fate determinants (see Figure 1; Kemphues et al., 1988; Morton et al., 1992; Etemad-Moghadam et al., 1995; Guo and Kemphues, 1995; Boyd et al., 1996; Tabuse et al., 1998; Hung and Kemphues, 1999; Schubert et al., 2000; Watts et al., 2000; Cuenca et al., 2003). PAR-3 and -6, both PDZ domain–containing proteins, and an atypical protein kinase, PKC-3, are localized at the anterior cortex. All three are interdependent for anterior localization, which is maintained by posteriorly localized PAR-2. In response to these initial asymmetries, PAR-1, a serine/threonine kinase, is recruited to the posterior cortex and restricts the effectors MEX-5 and -6 (MEX-5/6) to the opposite anterior cytoplasm. MEX-5/6, two nearly identical proteins, are redundantly required for proper asymmetry of several cell fate determinants. In addition to their role in localizing cytoplasmic determinants, MEX-5/6 are required in a positive feedback loop to maintain the normal localization of the PAR proteins. The PAR-4 serine/threonine kinase is uniformly localized to the cortex. Although PAR-4 is not required for polarity establishment, it is required to maintain the normal position of the PAR-3/PAR-2 boundary in the late one-cell embryo, and has been suggested to act downstream of PAR-1 and MEX-5/6 in the generation of cytoplasmic asymmetries.
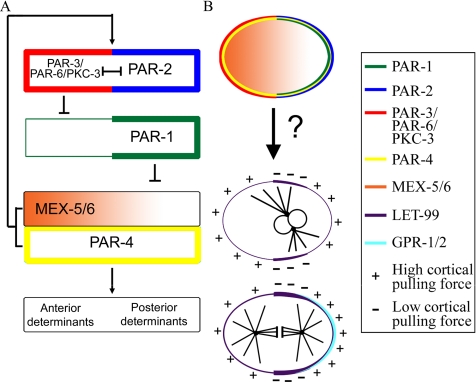
Figure 1.
The PAR proteins regulate the asymmetric localization of cytoplasmic cell fate determinants and the cortical localization of LET-99. (A) The pathway for the localization of the PAR proteins and downstream cell fate determinants: PAR-3/PAR-6/PKC-3 and PAR-2 are at the top of the hierarchy and inhibit each other (|—|) to maintain anterior and posterior PAR domains. PAR-3 inhibits (⊥) the anterior cortical localization of PAR-1, which in turn prevents cytoplasmic MEX-5/6 accumulation at the posterior. Asymmetric MEX-5/6 are required for downstream asymmetries. MEX-5/6 and PAR-4 are also required to maintain the position of the PAR-3–PAR-2 boundary (arrow). (B) Schematic of the one-cell distribution of the components shown in A. PAR-2 and -3 are also required for the asymmetric cortical localization of LET-99 and GPR-1/2, which are proposed to regulate cortical pulling forces that produce nuclear rotation and spindle displacement.
The one-cell embryo exhibits reproducible nuclear and spindle movements that define the division plane such that asymmetrically localized cell fate determinants are segregated into separate daughter cells (reviewed in Cowan and Hyman, 2004; Bellaiche and Gotta, 2005; Gönczy and Rose, 2005). Just after the establishment of cortical PAR domains, the female pronucleus migrates toward the posteriorly located male pronucleus. The pronuclear-centrosomal complex then migrates back to the center of the cell. During centration, the complex rotates to align the two centrosomes with the A/P polarity-axis (Figure 1B). After nuclear envelope breakdown, the spindle shifts posteriorly and undergoes asymmetric anaphase spindle elongation movements, which result in unequal cleavage.
Long astral microtubules and the microtubule motor dynein are required for centration, rotation, and at least some aspects of anaphase spindle positioning (Skop and White, 1998; Gönczy et al., 1999; Severson and Bowerman, 2003; Schmidt et al., 2005; Pecreaux et al., 2006). Moreover, laser-severing experiments indicate that posterior spindle displacement is the result of polarized pulling forces exerted on the mitotic apparatus from the cell cortex (Colombo et al., 2003; Grill et al., 2003). The precise mechanism through which the PAR proteins regulate these cortical forces remains to be determined, but it is known that a heterotrimeric G protein signaling pathway acts downstream of the PARs to transmit polarity cues to the machinery that moves the spindle (Gotta and Ahringer, 2001; Colombo et al., 2003; Gotta et al., 2003; Srinivasan et al., 2003; Tsou et al., 2003a; Afshar et al., 2004, 2005; Couwenbergs et al., 2004). The Gα proteins, GOA-1 and GPA-16, the guanine nucleotide dissociation inhibitors GPR-1/2, and several other positively acting factors are required for generation of cortical pulling forces that drive asymmetric spindle movements. In their absence, no asymmetric metaphase/anaphase movements are observed, and spindle pole separation is greatly attenuated in the one-cell stage; nuclear rotation is abnormal at the two-cell stage as well. The Gα proteins and GPR-1/2 are localized to the cytoplasm, spindle poles and cortex. GPR-1/2 are asymmetrically localized to the posterior cortex during metaphase/anaphase in a polarity dependent manner, which correlates with the higher pulling forces observed during this time.
LET-99, a DEP domain–containing protein, is also required for spindle positioning and is asymmetrically localized in cells of the P lineage (Rose and Kemphues, 1998; Tsou et al., 2002, 2003b). In one-cell embryos, cortical LET-99 is present at highest levels in a posterior band (Figure 1) in response to PAR-3 and -2. In let-99 mutant embryos after pronuclear meeting, exaggerated pronuclear-centrosomal rocking movements occur instead of normal centration and rotation. As a result, the spindle forms at a random orientation in the posterior end of the cell. The spindle continues rocking and orients onto the A/P axis during metaphase, and spindle pole movements are symmetric during anaphase. However, in let-99 embryos in which the eggshell is removed to abolish the oblong shape of the cell, nuclear rotation fails. This is in contrast to the normal rotation of wild-type one-cell embryos made spherical by eggshell removal, showing that LET-99 is essential for intrinsic polarity driven rotation (Tsou et al., 2002). The hyperactive nuclear and spindle rocking movements suggest that let-99 is required to reduce the forces exerted on microtubules from the cortex.
Based on these observations and the LET-99 localization pattern, a LET-99 band model was proposed for spindle movements during asymmetric cell division (Figure 1B; Tsou et al., 2002). In this model, the forces acting on microtubules in the lateral-posterior cortical region enriched for LET-99 are reduced relative to the forces acting from the anterior and posterior-most cortical regions where LET-99 levels are lower. This reduction in force generation is through inhibition of G protein signaling, based on genetic interactions and the observation that LET-99 inhibits the cortical accumulation of GPR and restricts GPR enrichment to the posterior-most cortex (Tsou et al., 2003a). Specifically, the model proposes that during prophase, the inhibition of force generation in the lateral posterior LET-99 domain would lead to a higher net force from the anterior that would pull the centrosome complex away from the posterior and initiate rotation; higher forces at both poles would enhance rotational movement. At metaphase/anaphase when the centrosomes have aligned with the A/P axis, the LET-99 dependent accumulation of GPR at the very posterior, the down-regulation of G protein signaling laterally, or both, would lead to higher net forces acting from the posterior to produce spindle displacement and oscillations. Consistent with this model, the absence of the LET-99 band correlates with defects in rotation in par-2 and -3 embryos. However, as with let-99 embryos, rotation defects occurred in par-3 embryos only when the embryo was made spherical by eggshell removal, showing that in wild type, extrinsic cell shape can compensate for the loss of intrinsic polarity cues (Tsou et al., 2002, 2003b).
In this study, we address several questions regarding the mechanism of LET-99 localization and the role of the LET-99 band pattern: First, which other members of the canonical PAR-dependent pathway for cytoplasmic polarity regulate cortical LET-99 localization? Second, how do anterior and posterior polarity domains result in a band pattern? Third, is localization of LET-99 to the entire posterior domain, instead of a band, compatible with normal nuclear rotation and spindle displacement? Our results show that LET-99 localization is regulated only by the cortical part of the canonical PAR protein pathway and suggest a model in which gradients of PAR-3 and -1 act to inhibit localization of LET-99 at the anterior and posterior poles respectively, potentially through phosphorylation. Further, although nuclear rotation can occur when LET-99 is localized to the entire posterior domain in par-1 embryos, this rotation is easily perturbed by cell shape constraints. Posterior spindle displacement is also defective in the absence of the LET-99 band at metaphase.
MATERIALS AND METHODS
Strains and Maintenance
C. elegans worms were maintained using standard protocols (Brenner, 1974; Church et al., 1995). The following worm strains were used in this study: RL19, let-99(or81) unc-22(e66)/DnT1 (Tsou et al., 2002); N2, wild-type Bristol; BS509, ozDf2/dpy-21(e428) par-4(it33); JJ1244, mex-6(pk440) mex-5(zu199) unc-30(e191)/nT1; KK288, par-1(b274) rol-4(sc8)/DnT1; KK292, par-1(it51) rol-4(sc8)/DnT1; KK434, dpy-21(e428) par-4(it75)/+; KK653, par-3(it71) unc-32(e189)/qC1; EU772, spn-4(or80)/DnT1; TH32, unc-119(ed3) ddIs6[tbg-1::GFP + unc-119(+)]; ruIs32[unc-119(+) pie-1::GFP::H2B]. RL19 was constructed in this lab, the KK strains were kindly provided by D. Morton and K. Kemphues (Cornell University), EU772 by B. Bowerman (University of Oregon), and TH32 by K. Oegema (University of California, San Diego), and the other strains were obtained from the Caenorhabditis Genetics Center. All mutants analyzed bear strong loss-of-function or null alleles, and these were used for all analyses unless otherwise indicated. Strains were grown at 20°C, except TH32, which was maintained at 25°C for optimal transgene expression. Embryo filming was performed at room temperature (23–25°C).
Immunofluorescence and Quantification of Staining Intensities
For determination of the localization of LET-99, PAR-3 and -1, and PKC-3, in situ immunofluorescence staining was performed using standard freeze-crack and methanol fixation protocols as described (Miller and Shakes, 1995; Tsou et al., 2002). Primary antibodies used were as follows: rabbit anti-PAR-1 antibodies (1:100 in PBS, a gift from K. Kemphues; Guo and Kemphues, 1995), mouse monoclonal anti-PAR-3 P4A1 (1:20; purchased from Developmental Studies Hybridoma Bank, University of Iowa, Iowa City, IA; Nance et al., 2003), rabbit anti-PKCζ C-20 (1:50, Santa Cruz Biotechnology, Santa Cruz, CA), rat anti-LET-99 (1:2), and rabbit anti-LET-99 (1:40; Tsou et al., 2002). Fluorescein isothiocyanate (FITC) goat anti-rabbit, FITC goat anti-rat, FITC goat anti-mouse, rhodamine goat anti-rabbit and rhodamine goat anti-mouse antibodies (1:100 in PBS, Sigma, St. Louis, MO) were used as secondary antibodies. Primary and secondary antibodies were preabsorbed with acetone powder from glutathione S-transferase (GST)-expressing bacteria and wild-type worms, respectively. DAPI (4′,6-diamidino-2-phenylindole dihydrochloride) was used to label nuclei for determination of cell cycle stages.
Single-section (0.215 μm) confocal images were taken at the midfocal plane of the embryo with an Olympus FV1000 confocal microscope (Melville, NY), using the same laser power and nonsaturating exposure settings for all specimens. For quantification of cortical staining intensities in one-cell embryos, images were analyzed with IP Lab Images software (Scanalytics, Fairfax, VA). Embryos were divided into segments corresponding to 10% egg length. A 0.5-μm-thick cortical region was highlighted with the segmentation tool, and the average pixel intensity was obtained for each cortical segment, as well as the underlying cytoplasmic region. For most experiments, staining intensities are presented as the ratio between cortical and cytoplasmic values (i.e., relative cortical intensity) to control for variations in staining between replicates. However, for the direct comparison of cortical PAR-1 levels in Figure 6, the absolute fluorescence intensities are given. Whole cell staining intensities were measured by outlining the entire embryo and measuring the total intensity over the whole area. Cortical and whole cell intensity values were compared using the Student's t test (Excel, Microsoft, Redmond, WA). For quantification of cortical LET-99 localization in the P1 cell, an intensity trace for a 0.4-μm-thick line drawn along the cortex of the P1 cell was obtained using ImageJ (http://rsb.info.nih.gov/ij/). For each genotype, the trace region from the cell–cell contact site to the posterior tip of the cell for the upper and lower cortices (0–100% cell length) were separately smoothened using the running average function of SigmaPlot (Systat Software, Point Richmond, CA) and then averaged and plotted using SigmaPlot.
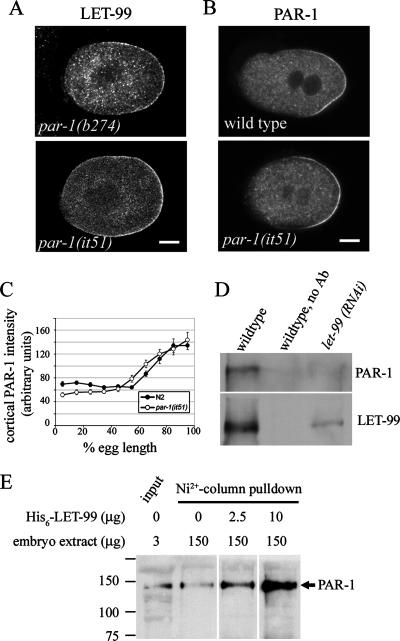
Figure 6.
PAR-1 kinase activity is required for LET-99 localization. (A) Confocal images of LET-99 staining in two par-1 alleles. (B) Confocal images of PAR-1 staining in wild-type and par-1(it51) embryos. (C) Cortical PAR-1 intensities in wild-type and par-1(it51) embryos. Error bars, SEM. (D) Western blots of LET-99 immunoprecipitation products. LET-99 antibodies were used for immunoprecipitation from wild-type or let-99(RNAi) embryos as indicated; a no-primary antibody control from wild-type extracts is also shown. Products were probed with PAR-1 and LET-99 antibodies. (E) Western blots of His6-LET-99-pulldown products. Various amounts of bacterially expressed His6-LET-99 were incubated with embryo extracts and purified with Ni2+-beads. Products were probed with PAR-1 antibodies.
RNA Interference
RNA interference was used to inhibit par-3 for the comparisons in Figure 5. Antisense and sense RNAs were transcribed in vitro (MEGAscript; Ambion, Austin, TX) from a linear DNA template produced by the PCR, using T3 and T7 primers to amplify a partial par-3 cDNA (from K. Kemphues). Double-stranded RNA was prepared as described (Fire et al., 1998), and L4 stage worms were soaked (Tabara et al., 1998) with 1 mg/ml double-strand DNA (dsRNA) for 4 h at 20°C; 24–36 h after recovery the worms were examined for the par-3 phenotype by differential interference contrast (DIC) microscopy and prepared for immunofluorescence. RNAi by feeding was used to inhibit par-1 expression in TH32 worms and to inhibit let-99 for immunoprecipitation experiments. Bacterial strains expressing dsRNA corresponding to par-1 cDNA clone yk1179c04 (from Y. Kohara, National Institute of Genetics, Japan) or a full-length let-99 cDNA (Tsou et al., 2002) were generated, and feeding was performed as described (Timmons and Fire, 1998). Depletion was confirmed by observation of the mutant phenotype or loss of immunofluorescence staining.
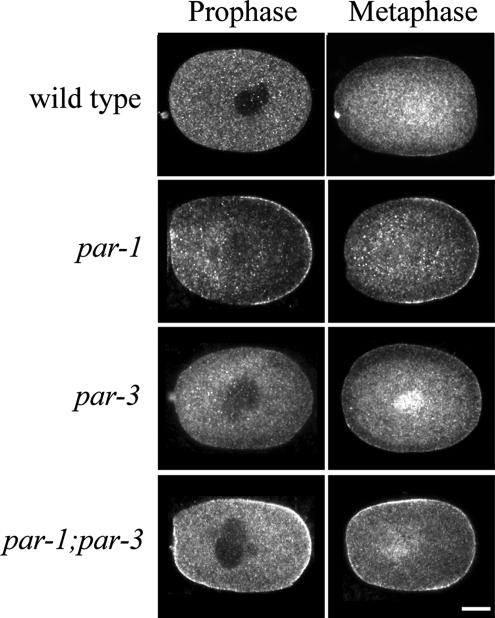
Figure 5.
Cortical LET-99 levels are inhibited by both par-1 and -3. Confocal images of LET-99 staining in N2, par-1, par-3, and par-1;par-3 embryos. Scale bar, 10 μm.
Immunoprecipitations and Pulldown Assays
Synchronized L1 stage animals were generated and plated onto NGM plates with control or RNAi bacteria strains. Animals were collected at the early adult stage, and the embryos were obtained by hypochlorite treatment (Lewis and Fleming, 1995). The packed embryos were resuspended in lysis buffer (100 mM phosphate buffer, pH 7.2, 120 mM NaCl, 1% NP-40, plus protease inhibitors), frozen with liquid nitrogen, ground to powder with a mortar and pestle, and stored at −80°C for further use. Embryo extract (1 mg) was treated with the cross-linking reagent DSP (1 mM, dithio-bis-succimidylpropionate) on ice for 2 h. The reaction was stopped by adding 25 mM Tris, pH 7.5, for 15 min. Anti-LET-99 antibodies (2 μl) were added to the extract, incubated 4°C overnight, and precipitated with protein A-Sepharose beads (Pharmacia, Piscataway, NJ) using the manufacturer's directions. The products were separated by 8% SDS-PAGE, transferred to membranes, and blotted with anti-PAR-1 (1:4000) and anti-LET-99 antibodies (1:4000).
A full-length let-99 cDNA was cloned into pET28 (Novagen, Madison, WI) such that the His6 tag was at the N-terminus of LET-99. His6-LET-99 fusion protein was expressed in Escherichia coli and purified with Ni2+-NTA agarose (Qiagen, Chatsworth, CA) using the manufacturer's protocols. Increasing amounts of His6-LET-99 protein (0–20 μg) were added to embryonic extracts and incubated at 4°C with rocking overnight. The extracts were then purified over Ni2+-NTA agarose beads that were preblocked with 5% bovine serum albumin (BSA). The beads were washed and subjected to 8% SDS PAGE, and Western blots were probed with anti-PAR-1 antibodies.
Analysis of Nuclear and Spindle Movements
To analyze nuclear rotation and anaphase spindle pole rocking events, one-cell embryos were isolated from adult hermaphrodites, mounted on polylysine-coated slides without flattening (Rose and Kemphues, 1998), and filmed under DIC optics using time-lapse videomicroscopy. For nuclear rotation analyses, the positions of both centrosomes were followed from pronuclear meeting to nuclear envelope breakdown (NEB) at 1-s intervals. The coordinates of both centrosomes were depicted with OpenLab (Improvision, Lexington, MA), and the angles of the centrosomal axis with respect to the A/P axis were calculated using Excel. To generate spherical one-cell embryos, the eggshell was removed from early stage embryos mounted in chitinase and examined by DIC time-lapse videomicroscopy as described (Tsou et al., 2002). Because the effectiveness of chitinase varies, only embryos in which the ratio of the transverse to the long axis was greater than 0.7 by NEB were scored. Generation of spherical P1 cells and introduction of ectopic flat sides were performed as described (Tsou et al., 2003b). For anaphase spindle movement analyses, live imaging of GFP::histone and GFP::γ-tubulin expressing worms was carried out using strain TH32 (Oegema et al., 2001) and an Olympus BX60 epifluorescence microscope. Images were taken every 5 s from pronuclear meeting to the completion of the first cell cycle using OpenLab. Kymographs were generated from the exported image stacks in ImageJ (NIH) using the kymograph plug-in (http://www.embl.de/eamnet/html/kymograph.html).
RESULTS
Cortical LET-99 Asymmetry Is Dependent on Cortical PAR Proteins
The asymmetric cortical localization of LET-99 in the one-cell embryo was previously shown to be dependent on PAR-3 and -2 (Tsou et al., 2002; Figure 1), which are at the top of the pathway that establishes polarity in C. elegans embryos (Figure 1A). In par-3 embryos LET-99 was localized uniformly. LET-99 was weakly localized to the cortex in par-2 embryos, with higher levels in a small posterior cap; in this background, there is a gradient of PAR-3 that expands farther into the posterior in wild type (Etemad-Moghadam et al., 1995). Together these results suggest that PAR-3 inhibits LET-99 localization, but the role of other factors downstream of PAR-3 was not tested. Therefore to gain insight into how the PAR pathway regulates LET-99 localization, we investigated the role of downstream polarity factors in generating LET-99 asymmetry.
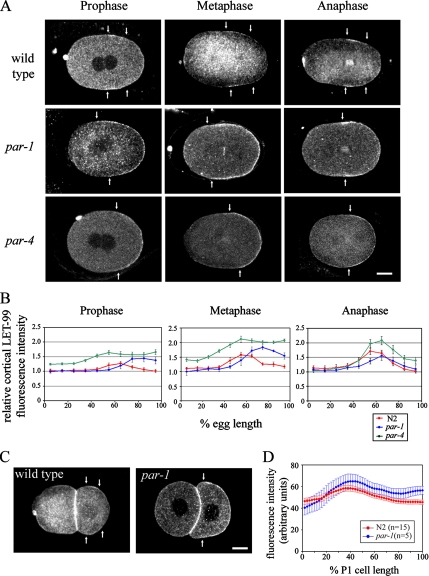
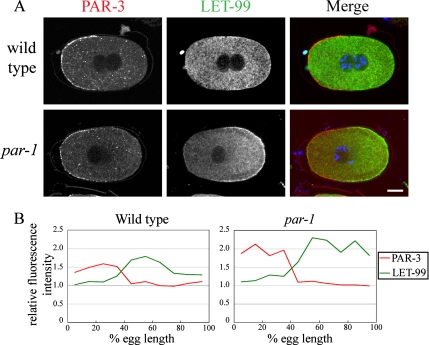
We first tested the requirement for PAR-1 by examining LET-99 localization in par-1 mutant embryos (Kemphues et al., 1988). We found that LET-99 was localized to the entire posterior cortex in par-1 embryos at prophase and metaphase (Figure 2A), instead of in a band pattern as in wild type. To quantify cortical LET-99 levels, we measured cortical LET-99 staining intensity relative to the cytoplasm, across the A/P axis (Figure 2B). In wild-type embryos, cortical LET-99 intensity at the anterior (0% egg length) was similar to that of the cytoplasm and then increased such that the highest levels occur from ∼50 to 70% egg length. Beyond 70% egg length, cortical LET-99 levels gradually decreased again to cytoplasmic levels. The enrichment of cortical LET-99 was detected in wild-type prophase embryos in which the pronuclei had met and became more prominent in metaphase and anaphase embryos. In par-1 mutant embryos, cortical intensities were higher throughout the embryo compared with wild type, and a 1.5-fold and twofold relative intensity increase of posterior cortical LET-99 intensity was observed in prophase and metaphase par-1 embryos, respectively (Figure 2B). In par-1 embryos at anaphase, a weak band pattern was observed, but the band was broader than in wild type, and intensities did not decrease as much at the posterior end (Figure 2B). We conclude that posteriorly localized PAR-1 inhibits LET-99 recruitment to the cortex at the very posterior end of the embryo.
Figure 2.
Cortical LET-99 asymmetry is dependent on par-1 and -4. (A) Confocal images of LET-99 from wild-type and par-1 and -4 embryos at the cell cycle stages indicated. Arrows indicate the boundaries of the LET-99 band in wild-type embryos, and the anterior border of the LET-99 domain in par-1 and -4 embryos. Scale bar, 10 μm. (B) Quantification of cortical LET-99 staining intensities in N2 and par-1 and -4 embryos. Intensities were measured on the cortex from anterior (0% egg length) to posterior and are expressed as a ratio of the cortical staining to underlying cytoplasmic staining. Each line represents the average of 5–10 embryos. Error bars, SEM. Only embryos in late prophase (pronuclear meeting through rotation) were included in prophase measurements. The intensities of cortical LET-99 at the posterior-most cortex in par-1 mutant are higher than wild-type embryos (p < 0.05). (C) Wild-type and par-1 two-cell embryos stained with LET-99. Arrows indicate the boundary of the LET-99 domain. Scale bar, 10 μm. (D) Average linescan intensities for cortical LET-99 measured from the point of cell–cell contact (0% cell length) to the posterior tip (100%) of the P1 cell from wild-type and par-1 embryos. Error bars, SEM.
In par-1 embryos, LET-99 levels were still reduced at the anterior cortex compared with the posterior cortex (Figure 2, A and B). However, the position at which cortical LET-99 levels began to rise was more anterior in par-1 mutant embryos at prophase and metaphase. It was previously shown that the posterior PAR-2 domain expands anteriorly in par-1 mutant embryos, and the PAR-3/6 domain is concomitantly smaller (Cuenca et al., 2003). To determine if the anterior extension of LET-99 accumulation in par-1 mutants correlated with the smaller PAR-3 domain in this background, we double stained embryos with antibodies directed against PAR-3 (Etemad-Moghadam et al., 1995; Nance et al., 2003) and LET-99. In all embryo stages examined in both wild-type and par-1 embryos, the anterior boundary of the LET-99 domain did not overlap with the PAR-3 domain (Figure 3A). Quantification of cortical PAR-3 and LET-99 intensities confirmed that the decrease of PAR-3 staining at midembryo correlated with the increase in LET-99 staining, as in wild type (Figure 3B). Together with the previous findings that cortical LET-99 is uniform in prophase and metaphase par-3 mutants (Tsou et al., 2002), these results suggest that PAR-3 inhibits LET-99 recruitment to the anterior cortex and that this effect is independent of PAR-1 activity.
Figure 3.
PAR-3 and -1 inhibit cortical LET-99 localization. (A) Confocal images of embryos double stained for PAR-3 and LET-99. Scale bar, 10 μm. (B) Traces of cortical PAR-3 and LET-99 intensities from the embryos shown in A.
We next asked if cortical LET-99 asymmetry requires PAR-4 (Kemphues et al., 1988; Watts et al., 2000). In par-4 embryos during prophase and metaphase, LET-99 was localized to the entire posterior cortex instead of a band (Figure 2A). Quantification of cortical LET-99 intensity in par-4 embryos showed that cortical LET-99 staining starts to increase from ∼60–70% egg length to the posterior end, and the cortical levels of LET-99 at the posterior during prophase and metaphase are slightly higher than in the band region in wild-type embryos (Figure 2B). The more posterior position at which LET-99 levels increase is consistent with the finding that PAR-3 expands toward the posterior in par-4 embryos (Hung and Kemphues, 1999). The results suggest that PAR-4, like PAR-1, is required to inhibit LET-99 localization at the posterior cortex. As in par-1 embryos, a LET-99 band pattern became apparent again during anaphase in par-4 embryos, suggesting that a PAR independent mechanism can generate a band late in the cell cycle.
We also examined LET-99 localization in mex-5; mex-6 double mutant embryos, as MEX-5/6 are required for nearly all cytoplasmic asymmetries downstream of the PAR proteins (Schubert et al., 2000). MEX-5/6 also play a feedback role in PAR asymmetry, such that many embryos have an expanded PAR-3 domain, and some embryos do not establish anterior versus posterior PAR domains (Cuenca et al., 2003; Cheeks et al., 2004). We therefore double-labeled mex-5;mex-6 embryos with LET-99 and PKC-3 to mark the anterior PAR domain. The overall staining intensity of LET-99 was greatly reduced in mex-5;mex-6 mutant embryos, and thus scoring of the cortical pattern in prophase embryos was difficult. However, in metaphase embryos, where cortical LET-99 was more apparent, a band was visible in 10/10 mex-5;mex-6 embryos. In these embryos, the band did not overlap with the PKC-3 domain just as in wild type, but the LET-99 band was more posteriorly positioned when the PKC-3 domain was larger than in wild type (5 of 10 embryos had larger PKC-3 domains; Supplementary Figure S1A). In contrast, no par-1, -4, or -3 metaphase embryos examined exhibited a banded pattern of LET-99 (this report; Tsou et al., 2002) Thus, we conclude that MEX-5/6 are not required for formation of a cortical LET-99 band downstream of PAR-3 and -1 in metaphase one-cell embryos. However, MEX-5/6 are required for normal levels of LET-99. Quantification of whole cell staining intensities confirmed that mex-5;mex-6 one-cell embryos had significantly lower levels of LET-99 than wild-type embryos (Supplementary Figure S1C). The levels of LET-99 were also slightly decreased in embryos mutant for SPN-4, which is required for the normal expression of several factors downstream of MEX-5/6 (Supplementary Figure S1). Together these results suggest that the cortical PAR pathway regulates the pattern of LET-99, and that in addition, a mechanism involving MEX-5/6 and SPN-4 regulates the levels of LET-99 in the embryo.
The presence of a LET-99 band in the P1 cell but not the AB cell suggests the PAR proteins function to localize LET-99 in this cell as well (Tsou et al., 2002). Because par-1 mutants reestablish PAR-3 and -2 domains in the P1 cell (Etemad-Moghadam et al., 1995), this background allowed us to address the role of PAR proteins in the localization of LET-99 in the P1 cell. par-1 mutant embryos stained for LET-99 showed a pattern similar to that seen in P0. The anterior domain exhibited lower staining intensities (Figure 2, C and D), suggesting that PAR-3 inhibits LET-99 cortical localization in the anterior. In addition, the entire posterior domain had higher levels of cortical LET-99 than in wild-type; however, a gradient of LET-99 was still present. This observation suggests that at the two-cell stage, another factor may cooperate with PAR-1 to regulate the LET-99 pattern. Nonetheless, the overall regulation of LET-99 by PAR-3 and -1 appears similar to that in P0.
PAR-3 and -1 Inhibit LET-99 Localization to Generate a Cortical Band
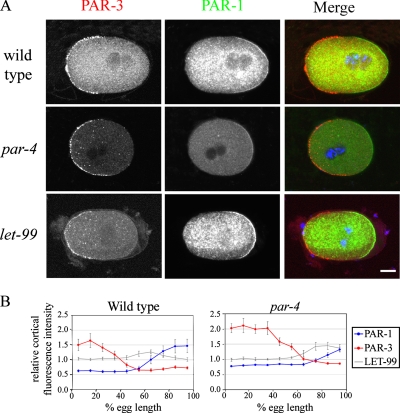
The results above indicate that only the cortical parts of the polarity pathway regulate LET-99 localization and point to a role for PAR-3 and -1 in inhibiting cortical LET-99 localization. To gain more insight into possible mechanisms by which PAR proteins regulate LET-99 localization, we analyzed the distribution of PAR-3 and -1 in more detail. In particular, because the LET-99 band appears to overlap with the posterior PAR domain, we reasoned that PAR-1 should be present in a gradient with the highest PAR-1 levels at the posterior, such that LET-99 localization is only inhibited at the posterior-most cortex. The quantification of prophase staining intensities of embryos double-labeled for PAR-3 and -1 is shown in Figure 4(n = 9); similar results were obtained for metaphase and anaphase embryos (n = 11). PAR-1 staining intensity increased from 50 to 75% egg length, and then reached a plateau (Figure 4B). Furthermore, there was a gap between the PAR-1 and -3 domains in wild-type embryos at all stages, similar to what was reported for PAR-3 and -2 (Munro et al., 2004). The gap between PAR-3 and -1 staining correlates with the region where LET-99 cortical levels rise, although the highest levels of LET-99 overlap with the PAR-1 domain. The staining intensity of LET-99 then decreases as PAR-1 levels increase at the posterior (Figure 4B). In contrast, the gap between the PAR-1 and -3 domains was still present in let-99 mutant embryos (n = 8). These observations suggest that low levels of both PAR-3 and -1 are required for LET-99 to accumulate to high levels at the cortex.
Figure 4.
A gap is present between the PAR-3 and -1 domains. (A) Confocal images of N2, par-4, and let-99 embryos double-stained with PAR-3 and -1. Scale bar, 10 μm. (B) Averaged cortical intensities of PAR-3 and -1 in prophase wild-type (n = 10) and par-4 (n = 9) embryos. The graphs are superimposed with the average LET-99 cortical intensities from Figure 2. Error bars, SEM.
If both PAR-3 and -1 inhibit cortical LET-99 localization, embryos lacking both proteins should have high levels of LET-99 at the cortex. To test this, we compared LET-99 staining in wild-type, par-3, par-1 and par-3;par-1 embryos. As expected, LET-99 was highly enriched at the entire cortex in par-1;par-3 double mutant embryos at prophase and metaphase (n = 19; Figure 5). The staining intensity of LET-99 in the double mutant appeared comparable to the level of posteriorly mislocalized LET-99 in par-1 mutants and higher at prophase than in par-3 single mutants. The latter observation is consistent with the model that PAR-1, which is uniformly cortical in par-3 mutants, inhibits LET-99 localization to the cortex. All together, these observations support the model that both proteins inhibit cortical localization of LET-99 and thus act together to generate an asymmetric cortical LET-99 band.
If the cortical PAR-1 gradient correlates with LET-99 localization, then the lack of a LET-99 band in par-4 embryos could be due to defects in PAR-1 localization. The PAR-3/PAR-6 domain expands in par-4 mutant embryos during mitosis (Hung and Kemphues, 1999), and PAR-1 is localized to a smaller posterior domain (D. Morton and K. Kemphues, personal communication). However, the levels of PAR-1 have not been measured. Quantification of PAR-1 intensity in par-4 embryos showed that the PAR-1 gradient is shifted toward the posterior as expected (Figure 4B). In addition PAR-1 intensity did not increase to the same extent in par-4 embryos as observed in control embryos. Thus, the absence of the LET-99 band in par-4 embryos may be a consequence of lower levels of PAR-1. These observations and the fact that LET-99 overlaps with the PAR-1 gradient in wild type (Figure 4B) suggest that a certain threshold of PAR-1 is required to inhibit LET-99 localization at the posterior cortex.
PAR-1 Kinase Activity Is Required for LET-99 Localization, and PAR-1 and LET-99 Associate In Vivo
High levels of PAR-1 could inhibit LET-99 accumulation at the posterior cortex through its activity as a kinase or through a direct competition for the same cortical sites. To distinguish between these possibilities, we examined LET-99 localization in par-1(it51) mutants, which have a mutation in a conserved residue of the kinase domain that is predicted to eliminate kinase activity (Guo and Kemphues, 1995). In these embryos, mutant PAR-1 localized to the posterior cortex (Guo and Kemphues, 1995) in a gradient similar to wild-type PAR-1 (Figure 6C). LET-99 was mislocalized to the entire posterior domain in it51 embryos, just as in par-1(b274) embryos (cf. Figure 6A to Figure 1). These observations suggest that PAR-1 kinase activity is required to inhibit cortical LET-99 localization.
PAR-1 could phosphorylate LET-99 directly or regulate it indirectly through PAR-4 or an unknown intermediate. We have not detected modified forms of LET-99 that would allow us to determine if LET-99 is phosphorylated in a PAR-dependent manner. However, if LET-99 is a substrate of PAR-1, we reasoned that they may interact, albeit transiently. Immunoprecipitations from embryonic extracts were carried out using an anti-LET-99 antibody. Western blotting showed that PAR-1 coimmunoprecipitated with LET-99 in extracts from wild-type embryos, but not in extracts from let-99(RNAi) embryos (Figure 6D). In addition, recombinant His6-LET-99 associated with PAR-1 in embryonic extracts. His6-LET-99 was added to embryonic extracts, repurified by nickel column chromatography, and the products examined by Western blotting. PAR-1 copurified with LET-99 in this experiment, and the amount of PAR-1 was dependent on the input of His-tagged LET-99 (Figure 6E). These observations along with the analysis of par-1(it51) lead to the model that PAR-1 phosphorylates LET-99 to inhibit accumulation at the cortex.
Finally, quantification of whole cell staining intensities at specific stages showed that the overall LET-99 levels did not change significantly with cell cycle progression in wild-type or par-1 embryos (Supplementary Figure S1C). In addition, the overall staining intensities of LET-99 in par-1 embryos compared with wild-type were similar, although par-1 embryos had slightly higher levels of LET-99 at metaphase. These observations suggest that the accumulation of cortical LET-99 in the band pattern in wild-type embryos and at the posterior cortex in par-1 embryos results from regulation of cortical versus cytoplasmic localization, rather than to changes in protein expression levels.
Nuclear Rotation in par-1 Embryos Is Sensitive to Changes in Cell Shape
If the cortical band pattern of LET-99 is important for rotation, then the presence of a posterior domain of high LET-99 levels in par-1 and -4 embryos should correlate with defects in spindle orientation. Previous studies revealed only weak defects in par-1 two-cell rotation, whereas par-4 mutants appeared to have normal spindle orientation in both the one- and two-cell stages (Kemphues et al., 1988; Watts et al., 2000). However, it has also been shown that cell shape asymmetries can rescue spindle positioning in par-3 embryos, where both polarity and LET-99 localization are disrupted (Tsou et al., 2003b). Similarly, nuclear rotation fails completely in let-99 embryos made spherical by digestion of the eggshell (Tsou et al., 2003b). We therefore reexamined nuclear rotation in par-1 and -4 mutant embryos in more detail.
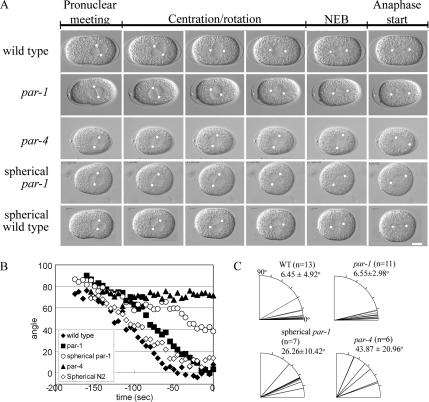
In all intact par-1 mutant embryos examined, the pronuclear-centrosomal complex centered and underwent rotation with similar kinetics to wild-type embryos (n = 13; representative embryos are shown in Figure 7, A and B; Supplementary Movies 1 and 2). In all par-1 embryos the pronuclear-centrosome complex rotated to within 15° of a line drawn on the A/P axis before NEB as in wild type (Figure 7C). On the other hand, in par-4 mutant embryos, the pronuclear-centrosomal complex did not rotate fully before NEB (n = 7, Supplementary Movie 3). After NEB in par-4 mutant embryos, the spindle gradually oriented onto the A/P axis (Figure 7A) and showed asymmetric spindle pole oscillations (Supplementary Movie 3). Therefore, nuclear rotation is defective in par-4 mutant embryos in which LET-99 is localized to the posterior cap instead of a discrete band.
Figure 7.
Defective nuclear rotation in par-1 and -4 embryos. (A) DIC images from time-lapse movies showing spindle orientation events in one-cell embryos. White dots indicate the location of centrosomes. Scale bar, 10 μm. (B) Plots of the angles between the A/P axis (0°) and a line drawn through the centrosomes, for each embryo shown in A from pronuclear meeting to NEB (t = 0). (C) Quantification of the extent of nuclear-centrosome rotation at NEB in indicated genetic backgrounds and manipulations. Each line represents a single embryo, with 0° indicating complete rotation of the centrosomes onto the A/P axis. The average rotation angle ± SD is also given.
The difference between par-1 and par-4 phenotypes could be due to the difference in the size of the LET-99 posterior cap (compare Figure 2, A and B) or because par-4 embryos are more spherical than par-1 mutant embryos, or both. Thus, we examined par-1 embryos that were made spherical by chitinase digestion of the eggshell. In such embryos, nuclear rotation was not completed during prophase; instead, at NEB the spindle formed at an angle to the long axis in most embryos, but then oriented onto the long axis as in par-4 embryos (Figure 7A; Supplementary Movie 4). These results suggest that a banded pattern of LET-99 is necessary to ensure efficient nuclear rotation onto the A/P axis before spindle formation in response to intrinsic polarity cues.
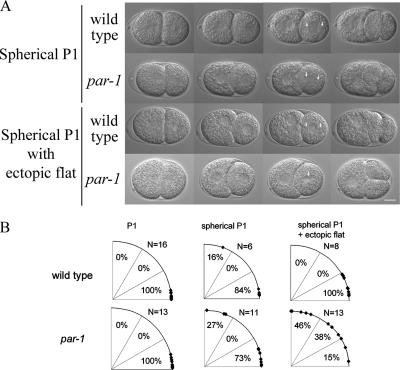
The LET-99 band pattern is reestablished in the wild-type P1 cell (Tsou et al., 2002; Figure 2A), which undergoes nuclear rotation to orient the mitotic spindle onto the A/P axis. We previously showed that the nuclear rotation that occurs in two-cell par-3 embryos (which have uniform cortical LET-99) is induced by cell shape asymmetries such as flat cell contact regions, and rotation does not occur in spherical cells (Tsou et al., 2003b). Conversely, wild-type embryos showed robust rotation even in the presence of ectopically induced flat surfaces. Thus, it was not surprising that nuclear rotation occurred normally in the P1 cell of par-1 embryos (Figure 8B). However, we reasoned that rotation in the P1 cell of par-1 embryos could be sensitive to cell shape effects. To test this, we UV-irradiated the AB cell (Tsou et al., 2003b), which results in loss of the flat cell contact region and a more spherical P1 cell at the time of rotation. As previously reported, in wild-type embryos with spherical P1 cells, the nuclear centrosome complex rotated during prophase onto the A/P axis (Figure 8A; Supplementary Movie 5) so that by the time of NEB, 5 of 6 wild-type spindles were aligned within 30° of the A/P axis (Figure 8B). In par-1 embryos with spherical P1 cells, nuclear rotation also occurred in most embryos (Figure 8; Supplementary Movie 6). However, when an ectopic flat side was introduced on top of the P1 cell by pressure of a coverslip, the nuclear-centrosomal complex oriented toward the ectopic flat side in 6 of 13 par-1 P1 cells during prophase; after NEB, the elongating spindle reoriented parallel to the coverslip (Figure 8, A and B; Supplementary Movie 7). In contrast, in 8 of 8 wild-type embryos filmed under the same flattening conditions, the nucleus rotated onto the A/P axis with normal timing (Supplementary Movie 8). We conclude that par-1 embryos are sensitive to changes in cell shape, unlike wild-type embryos. Together, the par-4 and -1 results support the model that a cortical LET-99 band is required for efficient nuclear rotation in response to intrinsic polarity cues and importantly, to prevent rotation according to cell shape asymmetries. In wild-type embryos cell shape asymmetries can compensate for the lack of a band during rotation in the P0 and P1 cells, but we speculate that cell shape asymmetries would interfere with spindle orientation during later divisions when more cell shape constraints are present.
Figure 8.
P1 nuclear rotation is perturbed by cell shape changes in par-1 embryos. (A) DIC images from time-lapse movies taken from wild-type and par-1 embryos with different manipulations. (B) Quantification of the extent of nuclear-centrosome rotation at NEB in untreated and manipulated P1 cells.
par-1 Embryos Exhibit Anterior Spindle Displacement and Oscillations of the Anterior Spindle Pole
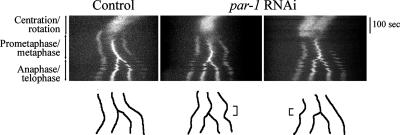
Based on the LET-99 band model, alterations in posterior spindle pole displacement and oscillations are predicted when LET-99 is mislocalized (Tsou et al., 2002). par-1 mutants were previously described as having a reduction in the overall posterior displacement of the spindle, but individual spindle pole movements were not tracked in detail (Kemphues et al., 1988). We therefore traced the displacement of the spindle and of each spindle pole using both DIC time-lapse microscopy and kymograph analysis of embryos expressing green fluorescent protein (GFP)::γ-tubulin and GFP::histone (Oegema et al., 2001; Figure 9). As previously described, in wild-type embryos, the entire spindle shifted posteriorly before spindle pole separation began (n = 11), and anaphase elongation was asymmetric, with the posterior spindle pole moving faster during early anaphase (9 of 11; Figure 9 shows a representative kymograph; 2 of 11 showed symmetric spindle pole elongation during anaphase). Posterior spindle pole oscillations began on average 133 s after NEB, and the maximum amplitudes of the posterior spindle pole oscillations were higher than those of the anterior spindle pole (Table 1). In all par-1 embryos, the spindle showed some initial posterior displacement as in wild type, but then the entire spindle shifted more anteriorly during metaphase (10 of 16) or the anterior spindle pole moved faster during early anaphase (6 of 16; Figure 9). These movements resulted in the final spindle position being less posterior on average than in wild type, and the extent of spindle pole separation was also reduced in par-1 embryos compared with wild type (Table 1). Finally, spindle pole oscillations in par-1 embryos began earlier than in wild type (on average 102 s after NEB), and the maximum amplitude of the anterior spindle pole was larger than that of the posterior spindle pole (Table 1). The anterior-directed spindle movements and the more vigorous anterior spindle pole rocking suggest that cortical forces are higher at the anterior in par-1 embryos. These observations support the model that LET-99 inhibits force generation and that the band pattern is important for normal posterior spindle displacement.
Figure 9.
Metaphase and anaphase spindle movements in wild-type and par-1 embryos. Representative kymograph analyses of spindle movements in embryos expressing GFP::γ-tubulin to label centrosomes and GFP::histone to label DNA. Images were recorded every 5 s from pronuclear meeting to telophase and kymographs were generated from a line drawn through middle of the embryo along the A/P axis. During centration/rotation the centrosome and DNA signals appear as one bright area. The centrosomes become visible at prometaphase (two outer fluorescent signals); the chromosomes become visible as a bright line between the poles when aligned at metaphase and then separate into two masses during anaphase. Line traces of the paths of the spindle poles and chromosomes are shown below each kymograph. Brackets indicate anterior displacement during metaphase in a par-1 embryo (middle panels) and anterior spindle pole movement during anaphase in another par-1 embryo (right).
Table 1.
Anaphase spindle movements are defective in par-1 mutant embryos
| Wild type (n = 7) |
par-1 (n = 10) |
|||||
|---|---|---|---|---|---|---|
| Anterior pole | Posterior pole | Anterior pole | Posterior pole | |||
| Position at NEB (%EL) | 33.8 ± 2.1 | 65.0 ± 3.8 | 32.7 ± 2.0 | 65.7 ± 4.8 | ||
| Position at anaphase rocking (%EL) | 39.0 ± 1.4 | 79.3 ± 1.6 | 36.1 ± 12.6a | 73.1 ± 3.2a | ||
| Max rocking amplitude (μm) | 5.29 ± 0.74 | 6.63 ± 0.64 | 7.45 ± 1.74a | 6.83 ± 2.74 | ||
| Pole separation at NEB (μm) | 14.43 ± 0.94 | 14.07 ± 0.96 | ||||
| Pole separation at anaphase rocking (μm) | 21.37 ± 1.22 | 20.11 ± 0.80a | ||||
All measurements were made from tracking the centrosomes/spindle poles in DIC time-lapse microscopy movies. Values are means ± SD.
a Values are significantly different from wild type (p < 0.05).
DISCUSSION
In many polarized cell types, the PAR proteins localize to cortical domains that then specify the asymmetric localization of downstream components. Much progress has been made in understanding the regulation of PAR asymmetry and that of cytoplasmic cell fate determinants (for review see Golden, 2000; Schneider and Bowerman, 2003; Betschinger and Knoblich, 2004; Cowan and Hyman, 2004; Macara, 2004; Gönczy and Rose, 2005; Nance, 2005). However, the pathways by which PAR proteins regulate cortical proteins involved in spindle positioning are less well studied. In this report, we present evidence that the cortical components of the PAR pathway regulate the localization pattern of LET-99 independent of effects on cytoplasmic polarity, which suggests that the PAR proteins may act directly on LET-99. We also provide further evidence that the LET-99 band pattern is important for transmitting polarity cues to the machinery that positions nuclei and spindles.
PAR-3 and -1 Inhibit LET-99 Localization to Generate a Cortical LET-99 Band
Of the proteins that show asymmetric localization in early C. elegans embryos, LET-99 is unique in being enriched in a cortical band encircling the cell, rather than to either the anterior or posterior cortex. In this report we show that the highest levels of LET-99 staining correlate with a gap between the graded distributions of PAR-3 and -1 in wild-type embryos, although the cortical band of LET-99 extends into the PAR-1 domain as well. Quantification of staining intensities shows that cortical LET-99 levels are higher at the anterior in par-3 embryos, higher at the posterior cortex in par-1 embryos (Figure 10A) and high overall in par-1;par-3 double mutants. These observations suggest a simple mechanism for formation of the LET-99 band: The presence of high levels of cortical PAR-3 inhibits LET-99 accumulation at the anterior cortex (independently of PAR-1), and high levels of PAR-1 inhibit LET-99 accumulation at the posterior-most cortex (Figure 10B). It would appear that a threshold of PAR-1 levels are required for complete inhibition at the cortex, because the LET-99 and PAR-1 domains overlap. This model also explains the previous observation that LET-99 levels are uniform in par-3 mutants (Tsou et al., 2002), because PAR-1 is not present at high enough levels at the posterior in par-3 mutants to completely inhibit LET-99 accumulation at the posterior-most cortex (Figure 10A). Our results are most consistent with the model that PAR-4 acts upstream of PAR-1 in LET-99 localization, because the levels of PAR-1 at the posterior cortex are lower in par-4 embryos than in wild type and because PAR-1 associates with LET-99. However, even if PAR-4 acts downstream of or parallel to PAR-1, the gradient of PAR-1 would be the essential factor that results in the LET-99 band, because PAR-4 is uniformly localized.
Figure 10.
Model. (A) Summary of LET-99 localization patterns in wild-type and par-3 and -1 embryos. (B) Model for the formation of the cortical LET-99 band by PAR-3 and -1. Both PAR-3/PAR-6/PKC-3 and PAR-1 inhibit the recruitment of LET-99 onto the cortex. Because PAR-3 and -1 are both present in gradients with the highest levels at the poles, inhibition of LET-99 cortical localization results in a band pattern. PAR-4 is uniformly localized but is proposed to act via promoting high levels of PAR-1 at the very posterior.
How might the PARs regulate LET-99 localization? Because only the cortical part of the pathway is required and the effect of PAR-3 on LET-99 is independent of PAR-1 and -4, we speculate that LET-99 could be a direct target of PAR-3/PAR-6/PKC-3 and of PAR-1 or -4. PAR-1 and -4, and PKC-3 are kinases, and there are several precedents for protein localization being regulated by PAR-associated phosphorylation in other organisms (Nagai-Tamai et al., 2002; Riechmann et al., 2002; Benton and St. Johnston, 2003; Hurov et al., 2004; Suzuki et al., 2004; Betschinger et al., 2005). In C. elegans embryos, PAR-2 was recently shown to be phosphorylated by PKC-3, which results in exclusion of PAR-2 from anterior cortex (Hao et al., 2006). These observations and our findings that PAR-1 associates with LET-99 and that a predicted PAR-1 kinase-dead mutant fails to localize LET-99 are all consistent with the model that the cortical localization of LET-99 is regulated through phosphorylation by PAR-1 and potentially PKC-3. Testing this model will be an active area of future study.
Although the PAR inhibition model can explain the establishment of the LET-99 pattern, a band “reappears” during anaphase in par-1, -3, and -4 mutants. In these cases, the position of the band correlates with that of the mitotic spindle, suggesting that the spindle may somehow feedback on LET-99 localization at this stage (this report; Tsou et al., 2002). Indeed, a recent report on a role for LET-99 in cytokinesis showed that the position of the LET-99 band can be changed by experimentally moving the spindle during late anaphase but not at earlier stages (Bringmann et al., 2007).
The Role of the LET-99 Band in Spindle-positioning Events
The defects observed in let-99 mutant embryos suggest that LET-99 has roles in centration, rotation, and posterior spindle displacement (Rose and Kemphues, 1998; Tsou et al., 2002). The cortical LET-99 band has been proposed to function in these processes by inhibiting G protein signaling, and thus force generation, on astral microtubules contacting the lateral-posterior cortex enriched for LET-99 (Figure 1; Tsou et al., 2002, 2003a). Centration and rotation occur simultaneously in most wild-type embryos and thus have often been grouped together when describing effects of mutants like let-99 on spindle positioning. However, recent modeling experiments suggest that centration of the pronuclear complex could be mediated in part by cytoplasmic pulling forces that are length dependent and thus would not require asymmetries at the cortex (Kimura and Onami, 2005). Consistent with this model, we have seen no correlation between the degree of centration and the boundary of either the LET-99 domain or the PAR-3 domain in par-1 and various mutants (this report; Tsou et al., 2002; Cuenca et al., 2003), and the centration defects of let-99 can be rescued in double mutant combinations (Rose and Kemphues, 1998; Tsou et al., 2003b). These observations are consistent with the view that in wild-type embryos, LET-99 is required to down-regulate GPR-1/2 and thus cortical forces, either generally or at the posterior cortex, to allow centration; however, LET-99 is probably not part of the centration mechanism itself.
Unlike centration, the cortical localization patterns of the PARs and LET-99 do appear important for nuclear rotation (Tsou et al., 2002). However, it was not surprising that par-1 embryos undergo rotation, given that anterior cortical LET-99 levels are still low in this background (Figure 10), and thus anterior cortical pulling forces are predicted to be high. Interestingly, however, par-1 embryos did exhibit rotation delays when made spherical. These observations confirm earlier findings that in wild-type embryos extrinsic cell shape acts redundantly with intrinsic polarity mechanisms to orient the spindle both in P0 and P1 (Tsou et al., 2002, 2003b). Further, our findings suggest that when cell shapes are perturbed, the banded pattern of LET-99 results in more efficient rotation than that produced by a posterior domain of LET-99. Cell-shape–dependent rotation could be mediated by differential forces produced when microtubules contact the cortex at different angles in an oval shaped embryo, as we previously suggested (Tsou et al., 2003b). Alternatively, the length-dependent-pulling force mechanism mentioned above (Kimura and Onami, 2005) provides another model for robust centration and rotation in one-cell par-1 and par-3 embryos in the absence of the LET-99 band: In an oval embryo, the microtubules projecting toward the anterior are longer than those projecting laterally or posteriorly and could thus result in forces pulling the centrosome anteriorly even in the absence of polarity or a LET-99 band (Figure 10A). However, this asymmetry of microtubule length, and thus force generation would be lost in spherical embryos.
Why generate a band pattern of LET-99 if a posterior LET-99 domain can support rotation, especially in combination with cell shape cues? One possible explanation stems from the observation that ectopic cell shape constraints do not alter wild-type rotation but do disrupt rotation in par-1 embryos. Thus, the importance of a LET-99 band may be to ensure efficient nuclear rotation under conditions that alter cell shape, such as compaction within the uterus or cell contact with neighboring cells later in development. Another, not mutually exclusive, explanation for the LET-99 band pattern is that it is critical for metaphase/anaphase spindle movements. LET-99 inhibits the localization of GPR to the cortex, and so the presence of a posterior domain of LET-99 at metaphase is predicted to prevent high levels of GPR from accumulating at the posterior-most cortex (Figure 1). The altered spindle orientation movements observed in par-1 embryos are consistent with this view. In particular, par-1 embryos showed an anterior displacement of the spindle during metaphase or anaphase, as predicted by the model that high levels of LET-99 inhibit force generation. Surprisingly, the timing of this shift varied and all embryos exhibited some posterior displacement. We speculate that the initial posterior shift and variability in phenotype is due to posterior GPR enrichment that occurs independently of LET-99. In wild-type embryos, LET-99 is not present at the anterior cortex where GPR levels are also low (Colombo et al., 2003; Gotta et al., 2003; Tsou et al., 2003a); thus, the presence of PAR-3 or other proteins at the anterior in par-1 embryos may produce a gradient of GPR by early metaphase even when LET-99 is mislocalized. As cortical GPR levels rise with cell cycle progression (Colombo et al., 2003; Tsou et al., 2003a), LET-99 inhibition of further accumulation at the posterior may then allow free GPR to accumulate at the anterior and cause anterior displacement and more vigorous oscillations of the anterior spindle pole.
In summary, these studies provide evidence that the PAR-3/PAR-6/PKC-3 complex and PAR-1 regulate LET-99 localization. We speculate that this is a direct consequence of LET-99 phosphorylation by the PKC-3 and -1 kinases, which would cause LET-99's dissociation from cortical anchors. This work also provides additional evidence that LET-99 is a key downstream component of the PAR pathway and supports the LET-99 band model for spindle positioning. The PAR proteins and GPR-1/2–related proteins are conserved in spindle positioning, and LET-99 shows similarity to the DEP1 family of vertebrate proteins. Although many DEP domain–containing proteins play roles in G protein pathways (Sato et al., 2006), the roles of DEP1 proteins remain to be determined. Further study of LET-99 and its interactions with the PAR proteins and G protein signaling should yield insight into the function of this family of proteins.
Supplementary Material
ACKNOWLEDGMENTS
We thank Bruce Bowerman, and Karen Oegema for strains, Y. Kohara for cDNAs, and Diane Morton and Ken Kemphues for reagents and sharing unpublished results. Other strains were provided by the Caenorhabditis Genetics Center (funded by the National Institutes of Health [NIH] National Center for Research Resources), and the PAR-3 mAb was obtained from the Developmental Studies Hybridoma Bank (under the auspices of the National Institute of Child Health and Human Development and maintained by The University of Iowa, Department of Biological Sciences, Iowa City, IA 52242). We are also grateful to Frank McNally and Geraldine Seydoux for critical reading of the manuscript, as well as members of the Rose, Scholey, McNally, and Kaplan labs for discussions. This work was supported by National Institutes of Health R01 award GM68744 (L.R.).
Footnotes
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E07-02-0105) on August 29, 2007.

 The online version of this article contains supplemental material at MBC Online (http://www.molbiolcell.org).
The online version of this article contains supplemental material at MBC Online (http://www.molbiolcell.org).
REFERENCES
- Afshar K., Willard F. S., Colombo K., Johnston C. A., McCudden C. R., Siderovski D. P., Gönczy P. RIC-8 is required for GPR-1/2-dependent Gα function during asymmetric division of C. elegans embryos. Cell. 2004;119:219–230. doi: 10.1016/j.cell.2004.09.026. [DOI] [PubMed] [Google Scholar]
- Afshar K., Willard F. S., Colombo K., Siderovski D. P., Gönczy P. Cortical localization of the Gα protein GPA-16 requires RIC-8 function during C. elegans asymmetric cell division. Development. 2005;132:4449–4459. doi: 10.1242/dev.02039. [DOI] [PubMed] [Google Scholar]
- Bellaiche Y., Gotta M. Heterotrimeric G proteins and regulation of size asymmetry during cell division. Curr. Opin. Cell Biol. 2005;17:658–663. doi: 10.1016/j.ceb.2005.10.002. [DOI] [PubMed] [Google Scholar]
- Benton R., St. Johnston D. Drosophila PAR-1 and 14-3-3 inhibit Bazooka/PAR-3 to establish complementary cortical domains in polarized cells. Cell. 2003;115:691–704. doi: 10.1016/s0092-8674(03)00938-3. [DOI] [PubMed] [Google Scholar]
- Betschinger J., Eisenhaber F., Knoblich J. A. Phosphorylation-induced autoinhibition regulates the cytoskeletal protein Lethal (2) giant larvae. Curr. Biol. 2005;15:276–282. doi: 10.1016/j.cub.2005.01.012. [DOI] [PubMed] [Google Scholar]
- Betschinger J., Knoblich J. A. Dare to be different: asymmetric cell division in Drosophila, C. elegans and vertebrates. Curr. Biol. 2004;14:R674–R685. doi: 10.1016/j.cub.2004.08.017. [DOI] [PubMed] [Google Scholar]
- Boyd L., Guo S., Levitan D., Stinchcomb D., Kemphues K. PAR-2 is asymmetrically distributed and promotes association of P granules and PAR-1 with the cortex in C. elegans embryos. Development. 1996;122:3075–3084. doi: 10.1242/dev.122.10.3075. [DOI] [PubMed] [Google Scholar]
- Brenner S. The genetics of Caenorhabditis elegans. Genetics. 1974;77:71–94. doi: 10.1093/genetics/77.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bringmann H., Cowan C. R., Kong J., Hyman A. A. LET-99, GOA-1/GPA-16, and GPR-1/2 are required for aster-positioned cytokinesis. Curr. Biol. 2007;17:185–191. doi: 10.1016/j.cub.2006.11.070. [DOI] [PubMed] [Google Scholar]
- Cheeks R. J., Canman J. C., Gabriel W. N., Meyer N., Strome S., Goldstein B. C. elegans PAR proteins function by mobilizing and stabilizing asymmetrically localized protein complexes. Curr. Biol. 2004;14:851–862. doi: 10.1016/j.cub.2004.05.022. [DOI] [PubMed] [Google Scholar]
- Church D., Guan K., Lambie E. Three genes of the MAP kinase cascade, mek-2, mpk-1/sur-1 and let-60 ras, are required for meiotic cell cycle progression in Caenorhabditis elegans. Development. 1995;121:2525–2535. doi: 10.1242/dev.121.8.2525. [DOI] [PubMed] [Google Scholar]
- Colombo K., Grill S. W., Kimple R. J., Willard F. S., Siderovski D. P., Gönczy P. Translation of polarity cues into asymmetric spindle positioning in Caenorhabditis elegans embryos. Science. 2003;300:1957–1961. doi: 10.1126/science.1084146. [DOI] [PubMed] [Google Scholar]
- Couwenbergs C., Spilker A. C., Gotta M. Control of embryonic spindle positioning and Gα activity by C. elegans RIC-8. Curr. Biol. 2004;14:1871–1876. doi: 10.1016/j.cub.2004.09.059. [DOI] [PubMed] [Google Scholar]
- Cowan C. R., Hyman A. A. Asymmetric cell division in C. elegans: cortical polarity and spindle positioning. Annu. Rev. Cell Dev. Biol. 2004;20:427–453. doi: 10.1146/annurev.cellbio.19.111301.113823. [DOI] [PubMed] [Google Scholar]
- Cuenca A. A., Schetter A., Aceto D., Kemphues K., Seydoux G. Polarization of the C. elegans zygote proceeds via distinct establishment and maintenance phases. Development. 2003;130:1255–1265. doi: 10.1242/dev.00284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Etemad-Moghadam B., Guo S., Kemphues K. J. Asymmetrically distributed PAR–3 protein contributes to cell polarity and spindle alignment in early C. elegans embryos. Cell. 1995;83:743–752. doi: 10.1016/0092-8674(95)90187-6. [DOI] [PubMed] [Google Scholar]
- Fire A., Xu S., Montgomery M. K., Kostas S. A., Driver S. E., Mello C. C. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature. 1998;391:806–811. doi: 10.1038/35888. [DOI] [PubMed] [Google Scholar]
- Golden A. Cytoplasmic flow and the establishment of polarity in C. elegans 1-cell embryos. Curr. Opin. Genet. Dev. 2000;10:414–420. doi: 10.1016/s0959-437x(00)00106-4. [DOI] [PubMed] [Google Scholar]
- Gönczy P., Pichler S., Kirkham M., Hyman A. A. Cytoplasmic dynein is required for distinct aspects of MTOC positioning, including centrosome separation, in the one cell stage Caenorhabditis elegans embryo. J. Cell Biol. 1999;147:135–150. doi: 10.1083/jcb.147.1.135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- T.C.e.R. Community: WormBook. Gönczy P., Rose L. S. Asymmetric cell division and axis formation in the embryo. WormBook. 2005 doi: 10.1895/wormbook.1.30.1. doi/ 10.1895/wormbook.1.30.1, http://www.wormbook.org. [DOI] [PMC free article] [PubMed]
- Gotta M., Ahringer J. Distinct roles for Gα and Gβγ in regulating spindle position and orientation in Caenorhabditis elegans embryos. Nat. Cell Biol. 2001;3:297–300. doi: 10.1038/35060092. [DOI] [PubMed] [Google Scholar]
- Gotta M., Dong Y., Peterson Y. K., Lanier S. M., Ahringer J. Asymmetrically distributed C. elegans homologs of AGS3/PINS control spindle position in the early embryo. Curr. Biol. 2003;13:1029–1037. doi: 10.1016/s0960-9822(03)00371-3. [DOI] [PubMed] [Google Scholar]
- Grill S. W., Howard J., Schaffer E., Stelzer E. H., Hyman A. A. The distribution of active force generators controls mitotic spindle position. Science. 2003;301:518–521. doi: 10.1126/science.1086560. [DOI] [PubMed] [Google Scholar]
- Guo S., Kemphues K. J. par-1, a gene required for establishing polarity in C. elegans embryos, encodes a putative Ser/Thr kinase that is asymmetrically distributed. Cell. 1995;81:611–620. doi: 10.1016/0092-8674(95)90082-9. [DOI] [PubMed] [Google Scholar]
- Hao Y., Boyd L., Seydoux G. Stabilization of cell polarity by the C. elegans RING protein PAR-2. Dev. Cell. 2006;10:199–208. doi: 10.1016/j.devcel.2005.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hung T. J., Kemphues K. J. PAR-6 is a conserved PDZ domain-containing protein that colocalizes with PAR-3 in Caenorhabditis elegans embryos. Development. 1999;126:127–135. doi: 10.1242/dev.126.1.127. [DOI] [PubMed] [Google Scholar]
- Hurov J. B., Watkins J. L., Piwnica-Worms H. Atypical PKC phosphorylates PAR-1 kinases to regulate localization and activity. Curr. Biol. 2004;14:736–741. doi: 10.1016/j.cub.2004.04.007. [DOI] [PubMed] [Google Scholar]
- Kemphues K. J., Priess J. R., Morton D. G., Cheng N. S. Identification of genes required for cytoplasmic localization in early C. elegans embryos. Cell. 1988;52:311–320. doi: 10.1016/s0092-8674(88)80024-2. [DOI] [PubMed] [Google Scholar]
- Kimura A., Onami S. Computer simulations and image processing reveal length-dependent pulling force as the primary mechanism for C. elegans male pronuclear migration. Dev. Cell. 2005;8:765–775. doi: 10.1016/j.devcel.2005.03.007. [DOI] [PubMed] [Google Scholar]
- Lewis J. A., Fleming J. T. Basic culture methods of Caenorhabditis elegans. In: Epstein H. F., Shakes D. C., editors. Modern Biological Analysis of an Organism. San Diego: Academic Press; 1995. pp. 3–29. [Google Scholar]
- Macara I. G. Parsing the polarity code. Nat. Rev. Mol. Cell Biol. 2004;5:220–231. doi: 10.1038/nrm1332. [DOI] [PubMed] [Google Scholar]
- Miller D. M., Shakes D. C. Immunofluorescence microscopy in Caenorhabditis elegans. In: Epstein H. F., Shakes D. C., editors. Modern Biological Analysis of an Organism. San Diego: Academic Press; 1995. pp. 365–394. [Google Scholar]
- Morton D. G., Roos J. M., Kemphues K. J. par-4, a gene required for cytoplasmic localization and determination of specific cell types in Caenorhabditis elegans embryogenesis. Genetics. 1992;130:771–790. doi: 10.1093/genetics/130.4.771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munro E., Nance J., Priess J. R. Cortical flows powered by asymmetrical contraction transport PAR proteins to establish and maintain anterior-posterior polarity in the early C. elegans embryo. Dev. Cell. 2004;7:413–424. doi: 10.1016/j.devcel.2004.08.001. [DOI] [PubMed] [Google Scholar]
- Nagai-Tamai Y., Mizuno K., Hirose T., Suzuki A., Ohno S. Regulated protein-protein interaction between aPKC and PAR-3 plays an essential role in the polarization of epithelial cells. Genes Cells. 2002;7:1161–1171. doi: 10.1046/j.1365-2443.2002.00590.x. [DOI] [PubMed] [Google Scholar]
- Nance J. PAR proteins and the establishment of cell polarity during C. elegans development. BioEssays. 2005;27:126–135. doi: 10.1002/bies.20175. [DOI] [PubMed] [Google Scholar]
- Nance J., Munro E. M., Priess J. R. C. elegans PAR-3 and PAR-6 are required for apicobasal asymmetries associated with cell adhesion and gastrulation. Development. 2003;130:5339–5350. doi: 10.1242/dev.00735. [DOI] [PubMed] [Google Scholar]
- Oegema K., Desai A., Rybina S., Kirkham M., Hyman A. A. Functional analysis of kinetochore assembly in Caenorhabditis elegans. J. Cell Biol. 2001;153:1209–1226. doi: 10.1083/jcb.153.6.1209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pecreaux J., Roper J.-C., Kruse K., Julicher F., Hyman A. A., Grill S. W., Howard J. Spindle oscillations during asymmetric cell division require a threshold number of active cortical force generators. Curr. Biol. 2006;16:2111–2122. doi: 10.1016/j.cub.2006.09.030. [DOI] [PubMed] [Google Scholar]
- Riechmann V., Gutierrez G. J., Filardo P., Nebreda A. R., Ephrussi A. Par-1 regulates stability of the posterior determinant Oskar by phosphorylation. Nat. Cell Biol. 2002;4:337–342. doi: 10.1038/ncb782. [DOI] [PubMed] [Google Scholar]
- Rose L. S., Kemphues K. The let-99 gene is required for proper spindle orientation during cleavage of the C. elegans embryo. Development. 1998;125:1337–1346. doi: 10.1242/dev.125.7.1337. [DOI] [PubMed] [Google Scholar]
- Sato M., Blumer J. B., Simon V., Lanier S. M. Accessory proteins for G proteins: partners in signaling. Annu. Rev. Pharmacol. Toxicol. 2006;46:151–187. doi: 10.1146/annurev.pharmtox.46.120604.141115. [DOI] [PubMed] [Google Scholar]
- Schmidt D. J., Rose D. J., Saxton W. M., Strome S. Functional analysis of cytoplasmic dynein heavy chain in Caenorhabditis elegans with fast-acting temperature-sensitive mutations. Mol. Biol. Cell. 2005;16:1200–1212. doi: 10.1091/mbc.E04-06-0523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider S. Q., Bowerman B. Cell polarity and the cytoskeleton in the Caenorhabditis elegans zygote. Annu. Rev. Genet. 2003;37:221–249. doi: 10.1146/annurev.genet.37.110801.142443. [DOI] [PubMed] [Google Scholar]
- Schubert C. M., Lin R., de Vries C. J., Plasterk R.H.A., Priess J. R. MEX-5 and MEX-6 function to establish soma/germline asymmetry in early C. elegans embryos. Mol. Cell. 2000;5:671–682. doi: 10.1016/s1097-2765(00)80246-4. [DOI] [PubMed] [Google Scholar]
- Severson A. F., Bowerman B. Myosin and the PAR proteins polarize microfilament-dependent forces that shape and position mitotic spindles in Caenorhabditis elegans. J. Cell Biol. 2003;161:21–26. doi: 10.1083/jcb.200210171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skop A. R., White J. G. The dynactin complex is required for cleavage plane specification in early Caenorhabditis elegans embryos. Curr. Biol. 1998;8:1110–1117. doi: 10.1016/s0960-9822(98)70465-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srinivasan D. G., Fisk R. M., Xu H., van den Heuvel S. A complex of LIN-5 and GPR proteins regulates G protein signaling and spindle function in C. elegans. Genes Dev. 2003;17:1225–1239. doi: 10.1101/gad.1081203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki A., et al. aPKC acts upstream of PAR-1b in both the establishment and maintenance of mammalian epithelial polarity. Curr. Biol. 2004;14:1425–1435. doi: 10.1016/j.cub.2004.08.021. [DOI] [PubMed] [Google Scholar]
- Tabara H., Grishok A., Mello C. C. RNAi in C. elegans: soaking in the genome sequence. Science. 1998;282:430–431. doi: 10.1126/science.282.5388.430. [DOI] [PubMed] [Google Scholar]
- Tabuse Y., Izumi Y., Piano F., Kemphues K., Miwa J., Ohno S. Atypical protein kinase C cooperates with PAR-3 to establish embryonic polarity in Caenorhabditis elegans. Development. 1998;125:3607–3614. doi: 10.1242/dev.125.18.3607. [DOI] [PubMed] [Google Scholar]
- Timmons L., Fire A. Specific interference by ingested dsRNA. Nature. 1998;395:854. doi: 10.1038/27579. [DOI] [PubMed] [Google Scholar]
- Tsou M. F., Hayashi A., DeBella L. R., McGrath G., Rose L. S. LET-99 determines spindle position and is asymmetrically enriched in response to PAR polarity cues in C. elegans embryos. Development. 2002;129:4469–4481. doi: 10.1242/dev.129.19.4469. [DOI] [PubMed] [Google Scholar]
- Tsou M. F., Hayashi A., Rose L. S. LET-99 opposes Gα/GPR signaling to generate asymmetry for spindle positioning in response to PAR and MES-1/SRC-1 signaling. Development. 2003a;130:5717–5730. doi: 10.1242/dev.00790. [DOI] [PubMed] [Google Scholar]
- Tsou M.F.B., Ku W., Hayashi A., Rose L. S. PAR-dependent and geometry-dependent mechanisms of spindle positioning. J. Cell Biol. 2003b;160:845–855. doi: 10.1083/jcb.200209079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watts J. L., Morton D. G., Bestman J., Kemphues K. J. The C. elegans par-4 gene encodes a putative serine-threonine kinase required for establishing embryonic asymmetry. Development. 2000;127:1467–1475. doi: 10.1242/dev.127.7.1467. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.