Abstract
Fission yeast mitogen-activated protein kinase (MAPK) Pmk1p is involved in morphogenesis, cytokinesis, and ion homeostasis as part of the cell integrity pathway, and it becomes activated under multiple stresses, including hyper- or hypotonic conditions, glucose deprivation, cell wall-damaging compounds, and oxidative stress. The only protein phosphatase known to dephosphorylate and inactivate Pmk1p is Pmp1p. We show here that the stress-activated protein kinase (SAPK) pathway and its main effector, Sty1p MAPK, are essential for proper deactivation of Pmk1p under hypertonic stress in a process regulated by Atf1p transcription factor. We demonstrate that tyrosine phosphatases Pyp1p and Pyp2p, and serine/threonine phosphatase Ptc1p, that negatively regulate Sty1p activity and whose expression is dependent on Sty1p-Atf1p function, are involved in Pmk1p dephosphorylation under osmostress. Pyp1p and Ptc1p, in addition to Pmp1p, also control the basal level of MAPK Pmk1p activity in growing cells and associate with, and dephosphorylate Pmk1p both in vitro and in vivo. Our results with Ptc1p provide the first biochemical evidence for a PP2C-type phosphatase acting on more than one MAPK in yeast cells. Importantly, the SAPK-dependent down-regulation of Pmk1p through Pyp1p, Pyp2p, and Ptc1p was not complete, and Pyp1p and Ptc1p phosphatases are able to negatively regulate MAPK Pmk1p activity by an alternative regulatory mechanism. Our data also indicate that Pmk1p phosphorylation oscillates as a function of the cell cycle, peaking at cell separation during cytokinesis, and that Pmp1p phosphatase plays a main role in regulating this process.
INTRODUCTION
In eukaryotic organisms, mitogen-activated protein kinase (MAPK) pathways transduce extracellular signals from hormones, growth factors, cytokines, or environmental stresses to trigger a diverse array of biological responses. Functional MAPK cascades comprise MAPK kinase kinases (MAPKKKs), which phosphorylate and activate MAPK kinases (MAPKKs), which in turn phosphorylate and activate MAPKs (Marshall, 1995; Waskiewicz and Cooper, 1995). Finally, active MAPKs phosphorylate different substrates to promote changes in gene expression that play a critical role in the adjustment of cells to environmental conditions. Three distinct MAPK signaling cascades have been identified so far in the fission yeast Schizosaccharomyces pombe. These include the stress-activated protein kinase (SAPK) pathway, the mating pheromone-responsive pathway, and the cell integrity pathway, whose central elements are MAPKs Sty1p/Spc1p, Spk1p, and Pmk1p/Spm1p, respectively (Toda et al., 1991, 1996; Millar et al., 1995; Shiozaki and Russell, 1995; Zaitsevskaya-Carter and Cooper, 1997).
The key player of the SAPK cascade in S. pombe is MAPK Sty1p, which shows high homology to mammalian p38 kinase and becomes activated by a wide range of stresses (Millar et al., 1995; Shiozaki and Russell, 1995; Degols et al., 1996; Soto et al., 2002) (Figure 1A). Sty1p is directly phosphorylated by MAPKK Wis1p, whereas Wis1p activation is dual, via either MAPKKKs Wak1p (also known as Wis4p or Wik1p) or Win1p (Shieh et al., 1997). Also, a response regulator protein, Mcs4p, associates with Wak1p and probably with Win1p to regulate MAPKKK activity in response to external stimuli (Shieh et al., 1997) (Figure 1A). Expression of many stress responsive genes in fission yeast is controlled by Sty1p through transcription factor Atf1p (Degols et al., 1996; Shiozaki and Russell, 1996; Wilkinson et al., 1996; Chen et al., 2003). In contrast, Pmk1p/Spm1p is a structural homologue to the cell integrity MAPK Mpk1p/Slt2p from Saccharomyces cerevisiae (Toda et al., 1996; Hohmann, 2002), and their activation is similar to the extracellular signal-regulated kinases ERK1/2p (p42/p44) from animal cells that become activated by growth factors, phorbol esters, cytokines, or osmotic stress (Roux and Blenis, 2004). In S. pombe, the involvement of MAPK Pmk1p in the maintenance of cell integrity, cytokinesis, and ion homeostasis derives from phenotypic analyses of mutant strains deleted in genes encoding the main components of the cascade, namely, mkh1+ (encoding MAPKKK Mkh1p) (Sengar et al., 1997), skh1+/pek1+ (encoding MAPKK Skh1p/Pek1p) (Sugiura et al., 1999; Loewith et al., 2000) and pmk1+/spm1+ (encoding MAPK Pmk1p/Spm1p) (Toda et al., 1996; Zaitsevskaya-Carter and Cooper, 1997). Deletion in any of these genes causes morphological changes, and cells display multiseptate phenotype, growth inhibition in response to potassium ions, hypersensitivity to β-1,3-glucanases, and defective vacuole fusion under hypotonic stress (Toda et al., 1996; Sengar et al., 1997; Zaitsevskaya-Carter and Cooper, 1997; Bone et al., 1998; Sugiura et al., 1999; Loewith et al., 2000). MAPK Pmk1p is dually phosphorylated by Pek1p at two conserved threonine and tyrosine residues in positions 186 and 188, respectively (Sugiura et al., 1999; Loewith et al., 2000). Inactive (unphosphorylated) Pek1p binds Pmk1p in the absence of external stimulus and acts as a potent inhibitor of Pmk1p signaling (Sugiura et al., 1999). Although the Mkh1p–Pek1p-Pmk1p cascade was initially described as activatable by high temperatures or sodium chloride only (Toda et al., 1996; Zaitsevskaya-Carter and Cooper, 1997), recent work has shown that Pmk1p phosphorylation is induced by multiple stressing conditions (Madrid et al., 2006). In all cases, the stress-induced activation of Pmk1p was completely dependent on Mkh1p and Pek1p function, suggesting the existence of a linear, nonbranched MAPK cascade. Fluorescence microscopy studies detect Pmk1p into the cytoplasm and nucleus, whereas Mkh1p MAPKKK and Pek1p MAPKK are found exclusively into the cytoplasm. Notably, all three kinases also locate at the septum during cell separation (Madrid et al., 2006). Furthermore, stress treatments or the absence of Mkh1p or Pek1p do not alter the subcellular localization of MAPK Pmk1p, suggesting that its activation occurs at the cytoplasm or septum and that both its active and inactive forms cross the nuclear membrane. At present, the identity of transcription factor(s) regulated by Pmk1p is unknown.
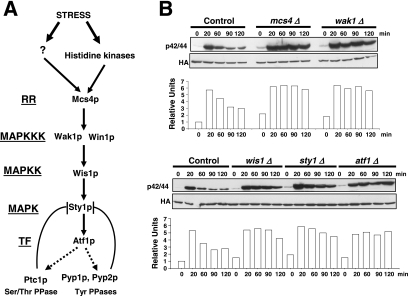
Figure 1.
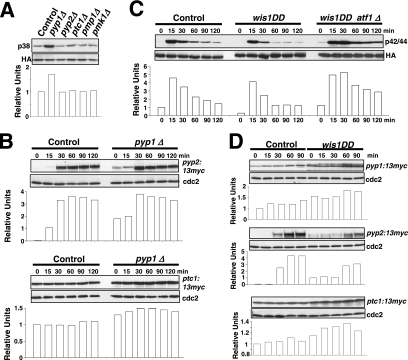
The SAPK pathway controls Pmk1p deactivation in growing cells and under hypertonic stress. (A) Schematic arrangement of the main functional components of the SAPK pathway in the fission yeast. RR, response regulator; TF, transcription factor; →, signal transmission; ■ ■ ■, enhanced transcription; ⊥, phosphatase inhibitory effect. See Introduction for details. (B) Strains MI200 (pmk1-HA6H, Control), MI208 (pmk1-HA6H, mcs4Δ), MI 217 (pmk1-HA6H, wak1Δ), MI210 (pmk1-HA6H, wis1Δ), MI204 (pmk1-HA6H, sty1Δ), and MI211 (pmk1-HA6H, atf1Δ) were grown in YES medium to mid-log phase and treated with 0.6 M KCl. Aliquots were harvested at the times indicated, and Pmk1-HA6H was purified by affinity chromatography. Activated Pmk1p was detected by immunoblotting with anti-phospho-p42/44 and total Pmk1p with anti-HA antibodies.
The duration and magnitude of MAPK activation result from a balance between activation and deactivation, with specific MAPK phosphatases (MKPs) playing a crucial role. In human cells, alterations of this equilibrium cause a number of diseases, such as diabetes or cancer. MKPs are divided into three major categories depending on their specificity to carry out dephosphorylation in tyrosine, serine/threonine, or both tyrosine and threonine residues (dual specificity) (Farooq and Zhou, 2004). Negative regulation of MAPK Sty1p is exerted by tyrosine phosphatases Pyp1p and Pyp2p and by type 2C serine/threonine phosphatases (PP2Cs) Ptc1p and Ptc3p (Hohmann, 2002). Pyp1p is the main phosphatase inactivating Sty1p in growing cells (Degols et al., 1996; Samejima et al., 1997). However, dephosphorylation of Sty1p activated under osmotic or oxidative stresses is carried out by both Pyp1p and Pyp2p (Millar et al., 1995; Shiozaki and Russell, 1995). Interestingly, the basal expression of pyp1+ and the stress-induced expression of pyp1+ and pyp2 + is regulated by Sty1p and Atf1p, forming a negative-feedback loop (Degols et al., 1996; Shiozaki and Russell, 1996; Wilkinson et al., 1996) (Figure 1A). Besides, during heat shock Pyp1p binding to MAPK Sty1p is abolished, resulting in a quick and strong Sty1p activation that is subsequently attenuated by threonine (PP2C) phosphatases Ptc1p and Ptc3p (Nguyen and Shiozaki, 1999). Again, expression of ptc1+ is induced by cadmium, hydrogen peroxide, heat shock, or osmotic stress and regulated by the Sty1p–Atf1p loop (Gaits et al., 1997; Chen et al., 2003).
Currently, Pmp1p is the only phosphatase known to dephosphorylate and inactivate Pmk1p in fission yeast. The corresponding gene pmp1+ was originally isolated as a multicopy suppressor for the chloride hypersensitivity of one strain deleted in ppb1+, which encodes calcineurin (Sugiura et al., 1998). Further analysis indicated that Pmp1p is closely related to dual-specificity phosphatases and that it is able to bind and inactivate Pmk1p in vitro and in vivo, suggesting that it may negatively regulate the Pmk1p MAPK pathway (Sugiura et al., 1998). Microarray analyses have shown no significant changes in pmp1+ transcription during cell cycle or stress treatments (Chen et al., 2003). However, a recent work has demonstrated that the stability of pmp1+ mRNA relies on the function of the RNA binding protein Rnc1p, whose activity is up-regulated by Pmk1p-mediated phosphorylation (Sugiura et al., 2003). Thus, the negative-feedback loop that controls the posttranscriptional stabilization of Pmp1p transcripts by Rnc1p provides a regulatory mechanism for fine-tuning of the Pmk1p pathway.
We described previously the existence of cross-talk between Sty1p and Pmk1p MAPKs in S. pombe (Madrid et al., 2006). Sty1p function is required for correct deactivation of Pmk1p in cells subjected to osmotic upshifts by a mechanism dependent on Atf1p transcriptional activity and on de novo protein synthesis (Madrid et al., 2006). This result strongly suggests that one or several phosphatases whose expression is up-regulated by the SAPK pathway might be responsible for Pmk1p dephosphorylation. In this work, we have focused on this question to disclose that the SAPK-regulated tyrosine phosphatases Pyp1p and Pyp2p, as well as the threonine phosphatase Ptc1p, are able to down-regulate Pmk1p activity in S. pombe depending on the nature of the stimulus.
MATERIALS AND METHODS
Strains and Growth Conditions
S. pombe strains (Table 1) were grown with shaking at 28°C in YES or EMM2 medium with 3% of glucose (Moreno et al., 1991), and supplemented with adenine, leucine, histidine, or uracil (100 mg/l; Sigma-Aldrich Co., St. Louis, MO) depending on their particular requirements. Mutant strains were obtained by standard transformation procedures or by mating and selecting diploids in EMM2 medium without supplements. Spores were obtained in MEL medium (Moreno et al., 1991), purified by glusulase treatment (Soto et al., 2002) and allowed to germinate in EMM2 plus the appropriate requirements. Transformation of yeast strains was performed by the lithium acetate method as described previously (Soto et al., 2002). In experiments performed with cdc25-22 thermosensitive mutant strains, the cells were grown in YES medium to an A600 of 0.2 at 25°C (permissive temperature), shifted to 37°C for 3.5 h, and released from the growth arrest by transfer back to 25°C. In experiments for the expression of a particular gene driven by the thiamine repressible promoter, yeast cultures were grown in EMM2 with or without 5 mg/l thiamine for 12–24 h. Escherichia coli DH5αF′ was used as host to propagate plasmids by growth at 37°C in Luria-Bertani medium plus 50 μg/ml ampicillin.
Table 1.
S. pombe strains used in this study
| Strain | Genotype | Source/reference |
|---|---|---|
| MI200 | h+ade6-M216 pmk1-HA6H::ura4+leu1-32 ura4D-18 | Madrid et al. (2006) |
| MI201 | h−ade6-M210 pmk1-HA6H::ura4+leu1-32 ura4D-18 | Madrid et al. (2006) |
| TP319-13c | h−pmk1::ura4+leu1-32 ura4D-18 | T. Toda |
| PPG148 | h−cdc25-22 ura4D-18 | S. Moreno |
| JM1368 | h−ade6-M216 his7-336 mcs4::ura4+leu 1-32 ura4-D18 | J. B. Millar |
| JM1478 | h−ade6-M216 his7-366 wak1::ura4+ leu 1-32 ura4-D18 | J. B. Millar |
| TK102 | h−his1-102 wis1::his1+leu 1-32 ura4-D18 | T. Kato |
| TK107 | h−sty1::ura4+leu 1-32 ura4-D18 | T. Kato |
| MI102 | h+ade6-M216 pmk1::KanR leu1-32 ura4D-18 | This work |
| MI103 | h+ade6-M216 atf1::ura4+leu1-32 ura4D-18 | This work |
| MI104 | h+ade6-M216 pmp1::KanR leu1-32 ura4D-18 | This work |
| MI105 | h+ade6-M216 pyp1::KanR leu1-32 ura4D-18 | This work |
| MI106 | h+ade6-M216 pyp2::KanR leu1-32 ura4D-18 | This work |
| MI107 | h−sty1::ura4+pmk1::KanR leu1-32 ura4D-18 | This work |
| JM1032 | h−ade 6-704 pyp2::LEU2 leu1-32 ura4D-18 | J. B. Millar |
| MI110 | h+ade6-M216 ptc1::KanR leu1-32 ura4D-18 | This work |
| MI113 | h+ade6-M216 ptc2::KanR leu1-32 ura4D-18 | This work |
| MI114 | h+ade6-M216 ptc3::HygR leu1-32 ura4D-18 | This work |
| MI108 | h−pyp1::KanR pmk1::ura4+leu1-32 ura4D-18 | This work |
| MI109 | h−pyp1::KanR sty1::ura4+leu 1-32 ura4-D18 | This work |
| MI111 | h−ptc1::KanR pmk1::ura4+leu1-32 ura4D-18 | This work |
| MI112 | h−ptc1::KanR sty1::ura4+leu 1-32 ura4-D18 | This work |
| MI115 | h−pmp1::KanR pmk1::ura4+leu 1-32 ura4-D18 | This work |
| MI116 | h−pmp1::KanR sty1::ura4+leu 1-32 ura4-D18 | This work |
| MI117 | h−pyp2::KanR pmk1::ura4+leu 1-32 ura4-D18 | This work |
| MI118 | h−pyp2::KanR sty1::ura4+leu 1-32 ura4-D18 | This work |
| MI208 | h−ade6-M216 his7-336 mcs4::ura4+pmk1-HA6H::ura4+leu 1-32 ura4-D18 | This work |
| MI217 | h−ade6-M216 his7-336 wak1::ura4+pmk1-HA6H::ura4+leu 1-32 ura4-D18 | This work |
| MI210 | h−his1-102 wis1::his1+pmk1-HA6H::ura4+leu 1-32 ura4-D18 | This work |
| MI204 | h+ade−sty1::ura4+pmk1-HA6H::ura4+leu1-32 ura4D-18 | Madrid et al. (2006) |
| MI211 | h+ade6-M216 atf1::ura4+pmk1-HA6H::ura4+leu1-32 ura4D-18 | Madrid et al. (2006) |
| MI212 | h+ade6-M216 pmp1::KanR pmk1-HA6H::ura4+leu1-32 ura4D-18 | This work |
| MI213 | h+ade6-M216 pyp1::KanR pmk1-HA6H::ura4+leu1-32 ura4D-18 | This work |
| MI214 | h+ade6-M216 pyp2::KanR pmk1-HA6H::ura4+leu1-32 ura4D-18 | This work |
| MI216 | h+ade6-M216 ptc1::KanR pmk1-HA6H::ura4+leu1-32 ura4D-18 | This work |
| MI218 | h+ade6-M216 ptc2::KanR pmk1-HA6H::ura4+leu1-32 ura4D-18 | This work |
| MI219 | h+ade6-M216 ptc3::HygR pmk1-HA6H::ura4+leu1-32 ura4D-18 | This work |
| MI215 | h−ade 6-704 pyp2::LEU2 nmt1:pyp1::KanR pmk1-HA6H::ura4+leu1-32 ura4D-18 | This work |
| MI107 | h+sty1::ura4+pmk1::KanR leu1-32 ura4D-18 | This work |
| MI220 | h+pyp1::KanR sty1::ura4+pmk1-HA6H::ura4+leu1-32 ura4D-18 | This work |
| MI221 | h+pyp2::KanR sty1::ura4+pmk1-HA6H::ura4+leu1-32 ura4D-18 | This work |
| MI222 | h+ptc1::KanR sty1::ura4+pmk1-HA6H::ura4+leu1-32 ura4D-18 | This work |
| MI223 | h+pyp1::KanR ptc1::KanR pmk1-HA6H::ura4+leu1-32 ura4D-18 | This work |
| MI301 | h−pmk1::ura4+pmk1-GFP::leu1+leu1-32 ura4D-18 | Madrid et al. (2006) |
| MI311 | h+ade−sty1::ura4+pmk1-GFP::leu1+leu1-32 ura4D-18 | This work |
| MI312 | h+ade6-M216 atf1::KanR pmk1-GFP::leu1+leu1-32 ura4D-18 | This work |
| MI313 | h+ade6-M216 pmp1::KanR pmk1-GFP::leu1+leu1-32 ura4D-18 | This work |
| MI314 | h+ade6-M216 pyp1::KanR pmk1-GFP::leu1+leu1-32 ura4D-18 | This work |
| MI315 | h+ade6-M216 pyp2::KanR pmk1-GFP::leu1+leu1-32 ura4D-18 | This work |
| MI500 | h+ade6-M216 sty1-HA6H::ura4+nmt41:GST-pyp1::KanR leu1-32 ura4D-18 | This work |
| MI501 | h+ade6-M216 sty1-HA6H::ura4+nmt41:GST-pyp2::KanR leu1-32 ura4D-18 | This work |
| MI502 | h+ade6-M216 pmk1-HA6H::ura4+nmt41:GST-pyp1::KanR leu1-32 ura4D-18 | This work |
| MI503 | h+ade6-M216 pmk1-HA6H::ura4+nmt41:GST-pyp2::KanR leu1-32 ura4D-18 | This work |
| MI505 | h−ade6-M216 pmk1-HA6H::ura4+ptc1-GST::KanR leu1-32 ura4D-18 | This work |
| MI506 | h−ade6-M216 pmk1-HA6H::ura4+pyp1-GST::KanR leu1-32 ura4D-18 | This work |
| JM1521 | h+ade6-M216 his7-366 sty1-HA6H::ura4+leu1-32 ura4D-18 | J. B. Millar |
| MI600 | h−cdc25-22 pmk1-HA6H::ura4+ura4D-18 | This work |
| MI601 | h−cdc25-22 pmk1::KanR ura4D-18 | This work |
| MI602 | h−cdc25-22 pmp1::KanR pmk1-HA6H::ura4+leu1-32 ura4D-18 | This work |
| MI701 | h+ade6-M216 pyp1-13myc::KanR leu1-32 ura4D-18 | This work |
| MI702 | h+pyp2-13myc::ura4+leu1-32 ura4D-18 | J. B. Millar |
| MI703 | h+ade6-M216 ptc1-13myc::KanR leu1-32 ura4D-18 | This work |
| MI704 | h+pyp2-13myc::ura4+pyp1::KanR leu1-32 ura4D-18 | This work |
| MI705 | h+ade6-M216 ptc1-13myc::KanR pyp1::HygR leu1-32 ura4D-18 | This work |
| MI706 | h−sty1::ura4+pyp1-13myc::KanR leu1-32 ura4D-18 | This work |
| MI707 | h−sty1::ura4+pyp2-13myc::KanR leu1-32 ura4D-18 | This work |
| MI708 | h+sty1::ura4+ptc1-13myc::KanR leu1-32 ura4D-18 | This work |
| 2119 | h−his7-336 wis1DD-12myc::ura4+leu 1-32 ura4-D18 | M. A. Rodriguez-Gabriel |
| MI709 | h−his7-336 wis1DD-12myc::ura4+pmk1-HA6H::ura4+leu 1-32 ura4-D18 | This work |
| MI710 | h−his7-336 wis1DD-12myc::ura4+pyp1-13myc::KanR leu 1-32 ura4-D18 | This work |
| MI711 | h−his7-336 wis1DD-12myc::ura4+pyp2-13myc::KanR leu 1-32 ura4-D18 | This work |
| MI712 | h−his7-336 wis1DD-12myc::ura4+ptc1-13myc::KanR leu 1-32 ura4-D18 | This work |
| MI713 | h−wis1DD-12myc::ura4+pmk1-HA6H::ura4+atf1::ura4+leu 1-32 ura4-D18 | This work |
| MI100 | h+ade6-M216 his7-366 pmk1::KanR sty1-HA6H::ura4+leu1-32 ura4D-18 | Madrid et al. (2006) |
| MI1000 | h+ade6-M216 his7-366 pmp1::KanR sty1-HA6H::ura4+leu1-32 ura4D-18 | This work |
| MI1001 | h+ade6-M216 his7-366 pyp1::KanR sty1-HA6H::ura4+leu1-32 ura4D-18 | This work |
| MI1002 | h+ade6-M216 his7-366 pyp2::KanR sty1-HA6H::ura4+leu1-32 ura4D-18 | This work |
| MI1003 | h+ade6-M216 his7-366 ptc1::KanR sty1-HA6H::ura4+leu1-32 ura4D-18 | This work |
Plasmid Constructs
The complete pmp1+, pyp1+, pyp2+, and ptc1+ open reading frames (ORFs) were amplified by polymerase chain reaction (PCR) by using genomic S. pombe DNA as template and the following oligonucleotide pairs: PMP1F-5 (TATATCCCGGGAATGTCTCAAAAACTACC, SmaI site is underlined) and PMP1F-3 (TATATTCTAGATCAAGAAGCATCATTACT, XbaI site is underlined), PYP1F-5 (TATATCCCGGGAATGAATTTTTCAAACGG, SmaI site is underlined) and PYP1F-3 (TATATCCATGGTCATGTTAAAACCGGGA, NcoI site is underlined), PYP2F-5 (TATATCCCGGGAATGCTCCATCTTCTGTCT, SmaI site is underlined) and PYP2F-3 (TATATTCTAGATTAAGTCATCAAGGGCTT, XbaI site is underlined), PTC1F-5 (TATATCCCGGGAATGAAGGGAAGCCATCC, SmaI site is underlined) and PTC1F-3 (TATATTCTAGACTAATAATAGTCATTACTG, XbaI site is underlined). The resulting DNA fragments were digested with SmaI and XbaI or NcoI, and cloned into plasmid pGEX-KG, which allows the expression in E. coli of the corresponding proteins as glutathione S-transferase (GST) fusions at their N terminus (Guan and Dixon, 1991). The resulting plasmids (pGEX-pmp1, pGEX-pyp1, pGEX-pyp2, and pGEX-ptc1) were transformed into E. coli DH5αF′, and the protein fusions were purified with glutathione-Sepharose beads as indicated below. During coprecipitation experiments, the expression plasmid pDS472a (Forsburg and Sherman, 1997) was used to express the GST tag under the control of the strong thiamine repressible promoter in S. pombe cells.
Gene Disruption and Epitope Tagging
The pmk1+, pmp1+, atf1+, pyp1+, pyp2+, ptc1+, ptc2+, and ptc3+ null mutants were obtained by entire deletion of the corresponding coding sequences and their replacement with the KanMX6 (KanR) cassette by PCR-mediated strategy by using plasmid pFA6a-kanMX6 as template (Bahler et al., 1998). Alternatively, gene deletion was performed using plasmid pCR2.1.hph conferring hygromycin B resistance (Sato et al., 2005). Plasmid pFA6a-kanMX6-P41nmt1-GST (Bahler et al., 1998) was used to obtain strains expressing N-terminal, GST-tagged versions of Pyp1p and Pyp2p under the control of the medium-strength thiamine repressible promoter. To construct strains expressing C-terminal 13-myc or GST tagged versions of either pyp1+, pyp2+, or ptc1+, we used plasmids pFA6a-13Myc-kanMX6 and pFA6a-GST-kanMX6, respectively (Bahler et al., 1998). Primer sequences used in each case are available upon request. The Pmk1-HA6H–tagged strains were obtained after transformation with integrative plasmid pIH-pmk1-ura (Madrid et al., 2006) previously digested with BstxI at the unique site within the pmk1+ coding region. Strains expressing a C-terminal–tagged version of Pmk1p fused to the green fluorescent protein (GFP) were obtained by transforming with integrative plasmid pIL-pmk1-GFP (Madrid et al., 2006) digested at the unique NruI site within leu1+ and selecting for leucine prototrophy. In all cases, the correct construction of strains was verified by PCR, Western blot. or both (see below).
Stress Treatments
To investigate Pmk1p activation under hypertonic stress most experiments were made using log phase cell cultures (A600 of 0.5) growing at 28°C in YES medium and the addition of 0.6 M KCl. At different times, the cells from 30 ml of culture were harvested by centrifugation at 4°C, washed with cold phosphate-buffered saline buffer, and the yeast pellets immediately frozen in liquid nitrogen for analysis.
Purification and Detection of Activated Pmk1-HA6H and Sty1-HA6H
Cell homogenates were prepared under native conditions by using chilled acid-washed glass beads and lysis buffer (10% glycerol, 50 mM Tris-HCl, pH 7.5, 150 mM NaCl, 0.1% Nonidet NP-40, plus specific protease and phosphatase inhibitor cocktails for fungal and yeast extracts; Sigma Chemical). The lysates were cleared by centrifugation at 16,000 × g for 15 min and HA6H-tagged Pmk1p or Sty1p were purified by using nickel-nitrilotriacetic acid (Ni2+-NTA)-agarose beads (QIAGEN, Hilden, Germany) (Madrid et al., 2004). The purified proteins were resolved in 10% SDS-polyacrylamide gel electrophoresis (PAGE) gels, transferred to nitrocellulose filters (GE Healthcare, Little Chalfont, Buckinghamshire, United Kingdom), and incubated with either monoclonal mouse anti-hemagglutinin (HA) (clone 12CA5; Roche Molecular Biochemicals, Basel, Switzerland), polyclonal rabbit anti-phospho-p42/44 antibodies (Cell Signaling Technology, Danvers, MA) (Madrid et al., 2006), or monoclonal mouse anti-phospho-p38 antibodies (Cell Signaling Technology) (Soto et al., 2002). Alternatively, we used a mouse monoclonal anti-phosphotyrosine antibody (PY99; Santa Cruz Biotechnology, Santa Cruz, CA). In this case, cell extracts were prepared as described by Shiozaki and Russell (1997) and incubated with Ni2+-NTA-agarose beads. After being processed as described above, immunoreactive bands were revealed with anti-mouse or anti-rabbit horseradish peroxidase (HRP)-conjugated secondary antibodies (Sigma Chemical) and the SuperSignal System (Pierce Chemical, Rockford, IL). Densitometric quantification of Western blot signals was performed using Molecular Analyst Software (Bio-Rad, Hercules, CA).
Detection of myc-tagged Fusions
Cell homogenates were prepared under native conditions as indicated above. The cleared lysates were resolved in 8% SDS-PAGE gels, transferred to nitrocellulose filters, and incubated with monoclonal mouse anti-c-myc antibody (clone 9E10; Roche Molecular Biochemicals). A polyclonal rabbit anti-Cdc2 antibody (PSTAIR; Upstate Biotechnology, Lake Placid, NY) was used as loading control. To detect immunoreactive bands, anti-mouse or anti-rabbit HRP-conjugated secondary antibodies (Sigma Chemical) and the SuperSignal System (Pierce Chemical) were used.
Purification and Detection of GST Fusions in E. coli and S. pombe
Purification of GST or GST-fused versions of phosphatases Pmp1p, Pyp1p, Pyp2p, and Ptc1p in E. coli DH5α was performed according to Millar et al. (1992). Briefly, the expression of the corresponding GST fusion was induced by adding 0.4 mM isopropyl β-d-thiogalactoside to mid-log phase bacterial cultures. GST-fused proteins were precipitated from lysate supernatants by incubation at 4°C with glutathione-Sepharose (GE Healthcare). Sepharose-bound proteins were then eluted by incubating 1 h at 4°C with 10% glycerol, 100 mM KCl, 5 mM MgCl2, 0.1 mM ZnCl2, 0.1 mM EDTA, 2 mM dithiothreitol (DTT), 10 mM HEPES, and 10 mM reduced glutathione, pH 9.0. To purify GST or GST fusions in S. pombe, cells were grown in 1 l of EMM2 medium without thiamine to induce expression of GST, GST-Pyp1, or GST-Pyp2. The same medium was used to grow strains expressing Pyp1-GST or Ptc1-GST fusions under control of its own promoter. Yeast cells were recovered by filtration, washed, resuspended in 4 ml of lysis buffer (10% glycerol, 20 mM Tris-HCl, pH 8.2, 100 mM NaCl, 10 mM MgCl2, 2 mM EDTA, 0.05% Nonidet NP-40, plus specific protease and phosphatase inhibitors), and disrupted with chilled acid-washed glass beads. The lysates were cleared by centrifugation at 15,000 × g for 25 min and incubated for 3 h with 1 ml of glutathione-Sepharose beads previously equilibrated in lysis buffer. After exhaustive washing with the same buffer, the bound proteins were eluted in the presence of reduced glutathione and concentrated by trichloroacetic acid (TCA) precipitation.
Coprecipitation Experiments
To detect interaction of GST-fusions with Pmk1-HA6H or Sty1-HA6H, the TCA precipitates were resolved by SDS-PAGE, transferred to nitrocellulose filters, and hybridized separately with anti-HA, anti-phospho-p44/42, or a polyclonal goat anti-GST antibody conjugated to HRP (GE Healthcare). The immunoreactive bands were revealed as indicated above.
Pmk1p Dephosphorylation In Vitro
Activated Pmk1-HA6H was purified with Ni2+-NTA-agarose beads (see above) from strain MI200 growing to mid-log phase in YES medium and subjected to stress with 0.6 M KCl for 15 min. The beads were washed twice and resuspended in phosphatase buffer containing 1 mM EDTA, 2 mM DTT, 50 mM imidazole, pH 7.2. Aliquots of Pmk1-HA6H were incubated at 30°C for 60 min in phosphatase buffer with 5 μg of either GST-Pmp1, GST-Pyp1, or GST-Pyp2, with or without 30 mM sodium orthovanadate. Phosphatase assays with GST-Ptc1 were performed by incubating activated Pmk1-HA6H in solution A (50 mM Tris-HCl, pH 7.0, 0.1 mM EGTA, 1 mg/ml bovine serum albumin, and 10 mM glutathione) with 20 mM MgCl2 or 1 mM EDTA at 30°C for 45 min (Nguyen and Shiozaki, 1999). The reactions were stopped by adding sample buffer, subjected to SDS-PAGE, and analyzed by Western blot analysis with anti-p42/44, anti-HA, and anti-GST antibodies as described above.
Fluorescence Microscopy
Images were taken on a Leica DM 4000B fluorescence microscope with a 100× objective, captured with a cooled Leica DC 300F camera and IM50 software, and then imported and processed with Adobe PhotoShop 6.0 (Adobe Systems, Mountain View, CA). To localize Pmk1-GFP fusion, small aliquots (10 μl) from cells growing in YES medium were loaded onto poly-l-lysine–coated slides or fixed with formaldehyde as described previously (Alfa et al., 1993). Calcofluor white was used for cell wall/septum staining as described previously (Alfa et al., 1993). A minimum of 200 cells per strain was analyzed.
β-1,3-Glucanase Sensitivity
Resistance to β-1,3-glucanase was assayed in different mutants by the method of Loewith et al. (2000) with some modifications. Cells were grown in YES medium to an A600 of 0.6, washed with 10 mM Tris-HCl, pH 7.5, 1 mM EDTA, and 1 mM β-mercaptoethanol, and incubated with vigorous shaking at 30°C in the same buffer supplemented with 100 μg/ml Zymolyase 20-T (Seikagaku, Tokyo, Japan). Samples were taken every 15 min, and cell lysis was monitored by measuring A600 decay.
Plate Assay of Stress Sensitivity for Growth
Wild-type and mutant strains of S. pombe were grown in YES liquid medium to log phase. Appropriate dilutions were spotted per duplicate on YES solid media or in the same medium supplemented with different concentrations of MgCl2. The plates were incubated at 28°C for 3 d.
Reproducibility of Results
All experiments were repeated at least three times with similar results. Representative results are shown.
RESULTS
SAPK Controls Pmk1p Deactivation during Growth and Hypertonic Stress
Sty1p MAPK is essential for proper Pmk1p deactivation after hypertonic stress in a process regulated by transcription factor Atf1p (Madrid et al., 2006). We performed a comparative analysis of Pmk1p activation/deactivation in salt-stressed S. pombe strains expressing an HA6H-tagged version of Pmk1p MAPK and deleted in different components of the SAPK pathway (Figure 1A) by using a p42/44 antibody, which detects dual phosphorylation at threonine and tyrosine. Pmk1p phosphorylation was evident in control cells after 20 min of hypertonic treatment, followed by a deactivation trend (Figure 1B). However, strains deleted in mcs4+ (response regulator), wak1+ (MAPKKK), wis1+ (MAPKK), sty1+ (MAPK), or atf1+ (transcription factor) showed a comparatively maintained stress-induced Pmk1p activation (Figure 1B). Identical results were obtained when these strains were subjected to nonsaline hypertonic stress (1 M sorbitol; data not shown). Also, a moderate but reproducible increase in basal Pmk1 phosphorylation was evident in the above-mentioned mutants compared with control cells (Figure 1B). These data confirm that the SAPK pathway transduces hypertonic stress signals via Sty1p-Atf1p to promote Pmk1p dephosphorylation, and they suggest that such pathway negatively regulates Pmk1p activity in growing cells.
Role of Tyrosine Phosphatases Pyp1p and Pyp2p as Negative Regulators of Pmk1p Activity
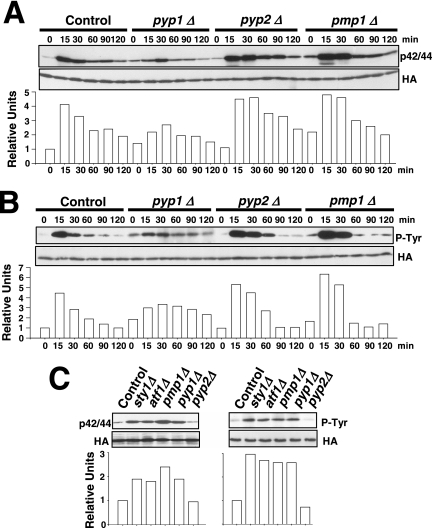
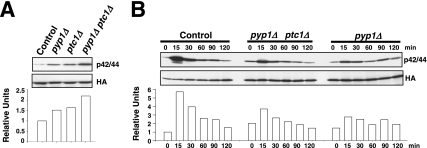
The above-mentioned results support that one or more protein phosphatases induced through the SAPK pathway might be responsible for Pmk1p deactivation in growing cells and under hypertonic stress. Thus, we first focused our attention on tyrosine phosphatases Pyp1p and Pyp2p, which account for Sty1p dephosphorylation during normal growth conditions (Pyp1p) and under osmostress (Pyp1p and Pyp2p) (Millar et al., 1995; Degols et al., 1996; Shiozaki and Russell, 1996). In fact, Sty1p–Atf1p function regulates during osmostress the basal expression of pyp1+ and the induced level of pyp1+ (moderate) and pyp2+ (strong) mRNAs (Degols et al., 1996; Shiozaki and Russell, 1996; Wilkinson et al., 1996, Chen et al., 2003). We determined Pmk1p activation/deactivation under hypertonic stress in control cells, and in cells lacking Pyp1p, Pyp2p, or Pmp1p, which is the only known protein phosphatase able to down-regulate Pmk1p activation in S. pombe (Sugiura et al., 1998). In pyp1Δ strain MI213, basal Pmk1p phosphorylation was higher than in control cells, whereas under osmostress the activation was significantly lower (Figure 2A). On the contrary, pyp2Δ cells showed unchanged basal level of Pmk1p activity, but the deletion prompted a marked phosphorylation and a kinetics of Pmk1p deactivation slower than in wild-type cells (Figure 2A). Except for a clear increase in the basal level of Pmk1p activity, pmp1+ deletion did not seem to affect significantly the rate of Pmk1p dephosphorylation under osmostress, although the overall level of Pmk1 phosphorylation was higher at all times (Figure 2A). We also analyzed Pmk1p phosphorylation under osmostress in control, pyp1Δ, pyp2Δ, and pmp1Δ strains by using specific anti-phosphotyrosine antibody (pY99) instead of anti-p42/44 antibody. The pattern of tyrosine phosphorylation of Pmk1p in these mutants was very similar to that observed with the antibody detecting dual phosphorylation at threonine and tyrosine (Figure 2B). To examine further the involvement of Pyp1p and Pmp1p phosphatases in regulating the basal level of activated Pmk1p, we analyzed this character in unstressed growing cells from different mutants. The p42/44 and pY99 levels in sty1Δ, atf1Δ, pyp1Δ, and pmp1Δ strains were higher than in control or pyp2+-deleted cells (Figure 2C). Together, these results strongly suggest that both Pmp1p and Pyp1p phosphatases are responsible for Pmk1p tyrosine dephosphorylation under normal growth conditions, whereas Pyp2p is involved in Pmk1p down-regulation during hypertonic stress. However, additional phosphatases might be involved in this process, because Pmk1p dephosphorylation can be still recorded in the absence of Pyp2p. In contrast, the unexpected Pmk1 hypoactivation observed in pyp1Δ cells could result from Sty1 hyperactivation (see below).
Figure 2.
Tyrosine phosphatases Pyp1p and Pyp2p are negative regulators of Pmk1p activation during growth and under hypertonic stress. (A) Strains MI200 (pmk1-HA6H, Control), MI213 (pmk1-HA6H, pyp1Δ), MI214 (pmk1-HA6H, pyp2Δ), and MI212 (pmk1-HA6H, pmp1Δ) were grown in YES medium to mid-log phase and treated with 0.6 M KCl. At timed intervals, Pmk1-HA6H was purified by affinity chromatography under native conditions. Activated and total Pmk1p was detected by immunoblotting with anti-phospho-p42/44 or anti-HA antibodies, respectively. (B) The same experiment described in A, except that Pmk1p phosphorylation was detected by immunoblotting with anti-phosphotyrosine (PY99) antibody. (C) Strains MI200 (pmk1-HA6H, Control), MI204 (pmk1-HA6H, sty1Δ), MI211 (pmk1-HA6H, aft1Δ), MI212 (pmk1-HA6H, pmp1Δ), MI213 (pmk1-HA6H, pyp1Δ), and MI214 (pmk1-HA6H, pyp2Δ) were grown in YES medium to mid-log phase, and Pmk1p basal activity was determined with anti-phospho-p42/44 or anti-phosphotyrosine antibodies.
Pmk1p localizes in both cytoplasm and nucleus, as well as in the mitotic spindle and septum during cytokinesis, whereas Pmp1p, Pyp1p, and Pyp2p are mostly restricted to the cytoplasm (Gaits and Russell, 1999; Madrid et al., 2006). To clarify whether the subcellular localization of Pmk1p was altered in the absence of these protein phosphatases or by lack of components of the SAPK pathway, we performed fluorescence microscopy observations in strains expressing a C-terminal–tagged version of Pmk1 fused to GFP in sty1Δ, atf1Δ, pmp1Δ, pyp1Δ, or pyp2Δ backgrounds. No noticeable effect was observed on the distribution of Pmk1-GFP at the cell nucleus, cytoplasm, and septum (data not shown).
Tyrosine Phosphatases Pyp1p and Pyp2p Associate with, and Dephosphorylate Pmk1p
If Pmk1p were a substrate for Pyp1p and Pyp2p, these protein tyrosine phosphatases should be able to bind such MAPK to some extent. To test this, we expressed N-terminal GST-tagged versions of Pyp1p and Pyp2p under the control of the medium-strength thiamine repressible promoter nmt41 in strains MI502 and MI503, which contain genomic versions of Pmk1p fused to HA6H epitope at its C terminus. The Sty1p-tagged strains MI500 and MI501 were used as a positive control for binding to Pyp1p and Pyp2p, respectively. After growth of the cells in minimal medium with or without thiamine, GST-Pyp1p and GST-Pyp2p fusions were purified by affinity chromatography with glutathione-Sepharose beads. Finally, the bound proteins were subjected to Western blot analysis with anti-HA antibodies. Figure 3A shows that purification of both GST-Pyp1p and GST-Pyp2p fusions from cultures growing in the absence of thiamine allowed to detect Sty1p and Pmk1p with anti-HA antibodies, whereas in negative controls or in cultures grown in the presence of thiamine these MAPKs did not copurify with GST alone. These results suggest that Pyp1p and Pyp2p associate with Pmk1p. To ensure that the above-mentioned interactions were not due to overexpression of the phosphatases, we performed a coprecipitation experiment by using strain MI506, which expresses a C-terminal GST-tagged version of Pyp1p under the regulation of its own promoter in a Pmk1-HA background. Approximately equal amounts of GST (negative control) and Pyp1-GST fusions were purified from cell extracts with glutathione-Sepharose beads. As shown in Figure 3B, Pmk1-HA was exclusively detected after Pyp1-GST purification, indicating that most likely Pmk1 and Pyp1 interact in vivo.
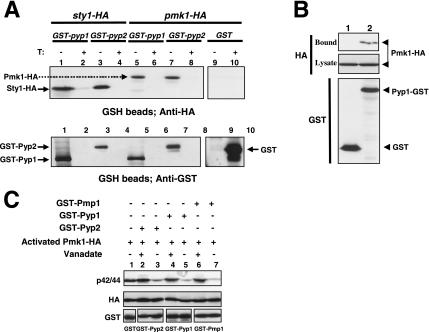
Figure 3.
Pyp1p and Pyp2p associate with, and dephosphorylate Pmk1p. (A) Strains MI500 (sty1-HA6H, nmt1:GST-pyp1, lanes 1 and 2), MI501 (sty1-HA6H, nmt1:GST-pyp2, lanes 3 and 4), MI502 (pmk1-HA6H, nmt1:GST-pyp1, lanes 5 and 6), MI503 (pmk1-HA6H, nmt1:GST-pyp2, lanes 7 and 8), and MI200 expressing unfused GST from plasmid pDS472a (lanes 9 and 10) were grown in EMM2 medium in the presence (+T) or absence (−T) of thiamine for 16 h. GST, GST-Pyp1, and GST-Pyp2 fusions were purified with glutathione-Sepharose beads, resolved by SDS-PAGE, transferred to nitrocellulose membranes, and immunoblotted with anti-HA or anti-GST antibodies. (B) Pyp1p and Pmk1p associate in vivo. An MI200 transformant expressing unfused GST from plasmid pDS472a (lane 1), and strain MI502 (pmk1-HA6H, pyp1-GST) expressing a genomic version of Pyp1p fused to GST at its C terminus (lane 2) were grown in EMM2 medium in the absence of thiamine for 16 h. GST and Pyp1-GST fusions were purified with glutathione-Sepharose beads, resolved by SDS-PAGE, transferred to nitrocellulose membranes, and immunoblotted with anti-HA or anti-GST antibodies. (C) Pyp1p and Pyp2p dephosphorylate Pmk1p in vitro. Activated Pmk1-HA6H was purified with Ni2+-NTA-agarose beads from strain MI200 after treatment with 0.6 M KCl for 15 min. The beads were incubated at 30°C for 60 min in phosphatase buffer with 5 μg of either GST (lane 1), GST-Pyp2 (lanes 2 and 3), GST-Pyp1 (lanes 4 and 5), or GST-Pmp1 (lanes 6 and 7) in the presence (even lanes) or absence (odd lanes) of 30 mM sodium orthovanadate. The samples were analyzed by SDS-PAGE and immunoblotted with anti-p42/44, anti-HA, and anti-GST antibodies.
Further demonstration of Pmk1p dephosphorylation by Pyp1p and Pyp2p was obtained by in vitro assays. To accomplish this, Pyp1p, Pyp2p, and Pmp1p phosphatases fused to GST at their N terminus were expressed in E. coli and purified by affinity chromatography. The protein fusions were then incubated with salt-activated Pmk1-HA6H purified from strain MI200. Results from Western blot analyses with anti-p42/44 antibodies showed that active Pmk1p was efficiently dephosphorylated in vitro by each of the three phosphatases (Figure 3C). Moreover, incubation of the purified proteins with the strong inhibitor of protein tyrosine phosphatases and dual specificity phosphatases, sodium orthovanadate, greatly prevented Pmk1 dephosphorylation (Figure 3C).
Threonine Phosphatase Ptc1p Attenuates Basal and Activated Pmk1p under Osmostress
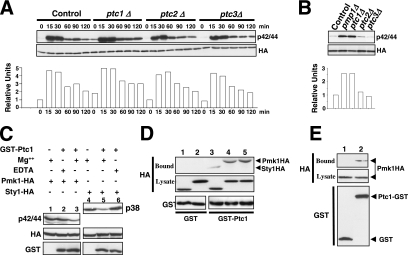
We focused on PP2C protein phosphatases Ptc1p, Ptc2p, and Ptc3p as potential candidates to negatively modulate Pmk1p activity because of the possible contribution of other phosphatases to Pmk1p deactivation (Figure 2, A and B). Threonine phosphatases Ptc1p and Ptc3p are involved in Sty1p down-regulation, and the induced expression of ptc1+ under stress is regulated by Sty1p-Atf1p (Gaits et al., 1997; Nguyen and Shiozaki, 1999). We found no significant change in the phosphorylation pattern of Pmk1p in ptc2Δ or ptc3Δ cells from either untreated or osmotic-stressed growing cultures compared with control cells (Figure 4A). However, deletion of ptc1+ resulted in increased basal level of Pmk1 activity and slower deactivation kinetics under osmostress than in wild-type cells (Figure 4A). This suggests that Ptc1p is involved in Pmk1p inactivation in both growing and osmostressed cells. Comparative analyses in control and mutant strains showed that deletion of either ptc1+ or pmp1+ induced a clear increase in Pmk1p phosphorylation in growing cells (Figure 4B).
Figure 4.
Ptc1p associates with, and dephosphorylates, Pmk1p. (A) Strains MI200 (pmk1-HA6H, Control), MI216 (pmk1-HA6H, ptc1Δ), MI218 (pmk1-HA6H, ptc2Δ), and MI219 (pmk1-HA6H, ptc3Δ) were grown in YES medium to mid-log phase and treated with 0.6 M KCl. At different times Pmk1-HA6H was purified by affinity chromatography, and either activated or total Pmk1p was detected by immunoblotting with anti-phospho-p42/44 or anti-HA antibodies, respectively. (B) Strains MI200 (pmk1-HA6H, Control), MI212 (pmk1-HA6H, pmp1Δ), MI216 (pmk1-HA6H, ptc1Δ), MI218 (pmk1-HA6H, ptc2Δ), and MI219 (pmk1-HA6H, ptc3Δ) were grown in YES medium to mid-log phase, and Pmk1p basal activity was determined by using anti-phospho-p42/44 antibody as described above. (C) Pmk1p dephosphorylation in vitro. Activated Pmk1-HA6H or Sty1-HA6H was purified with Ni2+-NTA-agarose beads from strains MI200 (Pmk1p; lanes 1–3) or JM1521 (Sty1p; lanes 4–6) after treatment with 0.6 M KCl for 15 min. The beads were incubated at 30°C for 45 min in phosphatase buffer with 10 μg of GST-Ptc1 (lanes 2, 3, 5, and 6) in the presence of 10 mM MgCl2 (lanes 3 and 5) or 10 mM EDTA (lanes 2 and 6). The samples were analyzed by SDS-PAGE and immunoblotted with anti-p42/44, anti-HA, and anti-GST antibodies. (D) Pmk1p and Ptc1p interact in vitro. Cell lysates from exponentially growing strains JM1521 (Sty1-HA6H; lanes 1 and 3), MI200 (Pmk1-HA6H; lanes 2 and 4), and strain MI200 subjected to a 15-min treatment with 0.6 M KCl (lane 5) were incubated each with 15 μg of bacterially purified GST (lanes 1 and 2) or GST-Ptc1 (lanes 3–5) bound to glutathione-Sepharose beads. The beads were washed extensively, and the binding of Pmk1p or Sty1p fusions was detected by inmunoblotting with monoclonal anti-HA antibodies. (E) Pmk1p and Ptc1p interact in vivo. An MI200 transformant expressing unfused GST from plasmid pDS472a (lane 1) and strain MI505 (pmk1-HA6H, ptc1-GST) expressing a genomic version of Ptc1p fused to GST at its C terminus (lane 2) were grown in EMM2 medium in the absence of thiamine for 16 h. GST and Pyp1-GST fusions were purified with glutathione-Sepharose beads, resolved by SDS-PAGE, transferred to nitrocellulose membranes, and immunoblotted with anti-HA or anti-GST antibodies.
Some evidence favors that Ptc1p down-regulation of Pmk1p activity occurs by direct interaction. First, a purified GST-Ptc1 fusion expressed in E. coli was able to dephosphorylate in vitro both activated Pmk1-HA6H and Sty1-HA6H to a similar extent in the presence of Mg2+ ions required for Ptc1p activity (Nguyen and Shiozaki, 1999) (Figure 4C, lanes 3 and 5). Second, when GST or GST-Ptc1 bound to glutathione-Sepharose beads was incubated with cell extracts from Pmk1-HA and Sty1-HA strains MI200 and JM1521, respectively, Pmk1-HA and Sty1-HA coprecipitated with GST-Ptc1 beads but not with GST beads after extensive washes (Figure 4D). Hence, similar to Sty1p, Ptc1p seems to interact in vitro with Pmk1p. Moreover, this association is unaffected by the activation status of Pmk1p (Figure 4D). Finally, we constructed a S. pombe strain expressing a C-terminal GST-tagged version of Ptc1p under the control of its own promoter in a Pmk1-HA6H background (strain MI505). As Figure 4E shows, Pmk1p-HA6H copurified with GST-Ptc1p but not with GST alone. This result strongly supports that Ptc1p and Pmk1p associate in vivo. Similar to what happens in the absence of phosphatases Pyp1p, Pyp2p, or Pmp1p, the subcellular localization of Pmk1p was unaffected by deletion of Ptc1p (data not shown).
Defective Pmk1p Activation under Saline Stress in pyp1Δ Cells Is Due to Sty1p Hyperactivation
The above-mentioned results strongly suggest that the SAPK-regulated phosphatases Pyp1p, Pyp2p, and Ptc1p deactivate Pmk1p, either in growing cells (Pyp1p and Ptc1p) or under osmostress (Pyp2p and Ptc1p). However, the defective Pmk1p activation in pyp1Δ cells subjected to osmostress was an intriguing result (Figure 2A). As indicated in Figure 5A, basal Sty1p phosphorylation measured by immunoblotting with anti-phospho-p38 antibody increased significantly in pyp1Δ cells, but not in the absence of Pyp2p, Ptc1p or Pmp1p. A reasonable interpretation for this is that, because the expression of pyp2+ and ptc1+ genes during osmostress depends on Sty1p-Atf1p (Degols et al., 1996; Shiozaki and Russell, 1996; Wilkinson et al., 1996, Chen et al., 2003), the hyperactivation of Sty1p in pyp1Δ cells should entail increased expression and synthesis of the two phosphatases, which would in turn lead to the decreased Pmk1p activation observed in the absence of Pyp1p. To examine this hypothesis, we undertook three different approaches. First, we constructed control and pyp1Δ strains expressing C-terminal 13myc-tagged versions of Pyp2p or Ptc1p phosphatases, and we determined Pyp2-13myc and Ptc1-13myc protein levels in cultures subjected to osmostress. As Figure 5B (top) shows, Pyp1p deletion prompted a clear increase in Pyp2-13myc levels in growing cells (0 min) and 15–30 min after the stress treatment with 0.6 M KCl compared with control cells. Pyp1p deletion also induced a modest but reproducible increase in Ptc1-13myc protein levels (Figure 5B, bottom). Second, we analyzed the osmostress-induced Pmk1p activation/deactivation in control cells and in strain MI709, expressing a hyperactive version of Wis1p MAPKK (wis1DD), which prompts the constitutive activation of Sty1p (Shiozaki et al., 1998). As shown in Figure 5C, Pmk1p activation in the wis1DD mutant was lower than in control cells. Moreover, Pyp1p and Ptc1p protein levels were enhanced in the mutant relative to control cells upon osmostress (Figure 5D), whereas Pyp2p protein levels were detected in growing and 15-min salt-stressed wis1DD cells (Figure 5D). Third, additional deletion of atf1+ gene in a wis1DD background rescued the defective Pmk1p activation observed in stressed cultures from the wis1DD mutant (Figure 5C). As a whole, these data support the notion that an increased synthesis of phosphatases Pyp2p and Ptc1p (due to up-regulation of the Sty1p-Atf1p loop) causes defective Pmk1p activation under osmostress in pyp1Δ cells. Also, they confirm that Pyp2p and Ptc1p negatively modulate Pmk1p activity in such conditions.
Figure 5.
Sty1p hyperactivation promotes defective Pmk1p activation under saline stress. (A) Sty1p basal activity in different mutants. Strains JM1521 (sty1-HA6H, Control), MI1001 (sty1-HA6H, pyp1Δ), MI1002 (sty1-HA6H, pyp2Δ), MI1003 (sty1-HA6H, ptc1Δ), MI1000 (sty1-HA6H, pmp1Δ), and MI100 (sty1-HA6H, pmk1Δ) were grown in YES medium to mid-log phase, and Sty1-HA6H was purified by affinity chromatography. Activated or total Sty1p was detected by immunoblotting with anti-phospho-p38 and anti-HA antibodies, respectively. (B) Increased Pyp2p and Ptc1p synthesis in pyp1Δ cells. Strains MI702 (pyp2-13myc, Control) and MI704 (pyp2-13myc, pyp1Δ) (top), MI703 (ptc1-13myc, Control), and MI705 (ptc1-13myc, pyp1Δ) (bottom) were grown in YES medium to mid-log phase and treated with 0.6 M KCl. Aliquots were harvested at the times indicated, and total extracts were obtained. Pyp2-13myc and Ptc1p-myc fusions were detected by immunoblotting with anti-c-myc antibody. Anti-Cdc2 antibody was used as loading control. (C) Pmk1p activation in wis1DD cells. Strains MI200 (pmk1-HA6H, Control), MI709 (pmk1-HA6H, wis1DD), and MI713 (pmk1-HA6H, wis1DD, atf1Δ) were grown in YES medium to mid-log phase and treated with 0.6 M KCl. Aliquots were harvested at the times indicated and Pmk1-HA6H was purified by affinity chromatography. Activated Pmk1p was detected by immunoblotting with anti-phospho-p42/44 and total Pmk1p with anti-HA antibodies. (D) Increased Pyp1p, Pyp2p and Ptc1p synthesis in wis1DD cells. Strains MI701 (pyp1-13myc, Control) and MI710 (pyp1-13myc, wis1DD) (top), MI702 (pyp2-13myc, Control) and MI711 (pyp2-13myc, wis1DD) (middle), MI703 (ptc1-13myc, Control) and MI712 (ptc1-13myc, wis1DD) (bottom) were grown in YES medium to mid-log phase and treated with 0.6 M KCl. Aliquots were harvested at the times indicated, and total extracts were prepared. Pyp1-myc, Pyp2-13myc, and Ptc1p-myc fusions were detected by immunoblotting with anti-c-myc antibody. Anti-Cdc2 antibody was used as loading control.
An apparently contradictory result is the increased basal level in Pmk1p phosphorylation in growing pyp1Δ cells (Figure 2, A–C). In this mutant, Sty1p hyperactivation promotes increased Pyp2p and Ptc1p protein levels (Figure 5B), which should account for Pmk1 deactivation. Considering that both Pyp1p and Ptc1p phosphatases down-regulate Pmk1p activity in growing cells, one possible explanation would be that their effect on Pmk1p is additive, and thus that the basal Pmk1 phosphorylation observed in pyp1Δ cells results from lack of negative regulation by Pyp1p plus increased MAPK dephosphorylation due to enhanced Ptc1p protein levels. Coincident with this suggestion, simultaneous deletion of pyp1+ and ptc1+ induced an increase in basal Pmk1p phosphorylation stronger than in single-deleted cells (Figure 6A). Moreover, partial reversion of the Pmk1p hypoactivation phenotype was observed by simultaneous deletion of ptc1+ and pyp1+ compared with pyp1Δ cells subjected to osmostress (Figure 6B; for example, compare Pmk1p phosphorylation after 15 min in control, pyp1Δ ptc1Δ, and pyp1Δ cells).
Figure 6.
Synergistic down-regulation of Pmk1p activity by Pyp1p and Ptc1p phosphatases. (A) Pmk1p basal activity in phosphatase-null mutants. Pmk1-HA6H was purified by affinity chromatography from strains MI200 (pmk1-HA6H, Control), MI213 (pmk1-HA6H, pyp1Δ), MI216 (pmk1-HA6H, ptc1Δ), and MI223 (pmk1-HA6H, pyp1Δ, ptc1Δ). Activated or total Pmk1p was detected by immunoblotting with anti-phospho-p42/44 or anti-HA antibodies, respectively. (B) Pmk1p activation in phosphatase-null mutants subjected to a saline stress. Strains MI200 (pmk1-HA6H, Control), MI223 (pmk1-HA6H, pyp1Δ, ptc1Δ), and MI213 (pmk1-HA6H, pyp1Δ) were grown in YES medium to mid-log phase and treated with 0.6 M KCl. At different times Pmk1-HA6H was purified by affinity chromatography, and either activated or total Pmk1p was detected by immunoblotting as described above.
Pyp1p and Ptc1p Phosphatases Negatively Modulate Pmk1p Phosphorylation under Osmostress Both in a Sty1p-dependent and -independent Manner
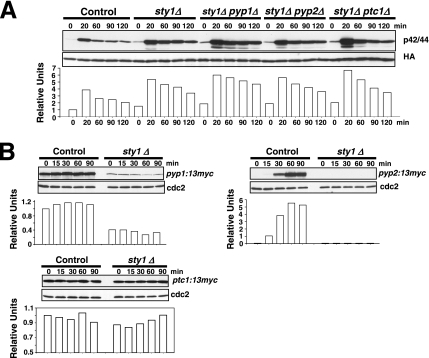
If Pmk1p down-regulation by Pyp1p, Pyp2p, and Ptc1p were completely dependent on the maintenance of appropriate phosphatase levels under transcriptional control by Sty1p, the defective Pmk1 deactivation observed in sty1Δ cells under osmostress should be unaffected by simultaneous deletion of Sty1p and any of the a above-mentioned phosphatases. Our results show that such prediction is correct in some cases. For example, the rate of Pmk1p activation/deactivation in osmostressed cells was virtually identical in sty1Δ and sty1Δ pyp2Δ mutants (Figure 7A). However, double-deleted sty1Δ pyp1Δ cells subjected to osmostress displayed more Pmk1p activity than the sty1Δ mutant (Figure 7A). Importantly, Sty1p deletion was able to suppress the defective Pmk1 activation observed in pyp1Δ cells (Figure 2, A and B), supporting previous results suggesting that this effect is due to increased phosphatase synthesis by Sty1p hyperactivation. Similar to sty1Δ pyp1Δ cells, the overall Pmk1p phosphorylation in sty1Δ ptc1Δ osmostressed cells was higher than in single sty1Δ mutant (Figure 7A). The most logical scenario to explain these results is that, unlike for pyp2+, the expression of both pyp1+ and ptc1+ is not fully dependent on transcriptional control by Sty1p-Atf1p. We tested this interpretation by analyzing Pyp1-13myc, Pyp2-13myc, and Ptc1-13myc protein levels in control and sty1Δ cultures under osmostress. As shown in Figure 7B, moderate Pyp1-13myc protein levels were detected in sty1Δ cells compared with control cells. On the contrary, Pyp2-13myc fusion was virtually undetectable in the absence of Sty1p, whereas the level of Ptc1-13myc protein under osmostress was largely unaffected by the MAPK deletion (Figure 7B). These results imply that Pmk1p deactivation by phosphatases Pyp1p and Ptc1p may be relatively (Pyp1p) or largely (Ptc1p) independent on the transcriptional control by Sty1p.
Figure 7.
Pyp1p and Ptc1p phosphatases can negatively modulate Pmk1p phosphorylation under osmotic stress in a Sty1p-independent manner. (A) Strains MI200 (pmk1-HA6H, Control), MI204 (pmk1-HA6H, sty1Δ), MI220 (pmk1-HA6H, sty1Δ, pyp1Δ), MI221 (pmk1-HA6H, sty1Δ, pyp2Δ), and MI222 (pmk1-HA6H, sty1Δ, ptc1Δ) were grown in YES medium to mid-log phase and treated with 0.6 M KCl. At timed intervals, Pmk1-HA6H was purified by affinity chromatography under native conditions, and activated or total Pmk1p was detected by immunoblotting with anti-phospho-p42/44 or anti-HA antibodies, respectively. (B) Synthesis of Pyp1p and Ptc1p phosphatases in sty1Δ cells. Strains MI701 (pyp1-13myc, Control) and MI706 (pyp1-13myc, sty1Δ) (left), MI702 (pyp2-13myc, Control) and MI707 (pyp2-13myc, sty1Δ) (middle), MI703 (ptc1-13myc, Control) and MI708 (ptc1-13myc, sty1Δ) (right) were grown in YES medium to mid-log phase and treated with 0.6 M KCl. Aliquots were harvested at the times indicated, and total extracts prepared. Pyp1-myc, Pyp2-13myc, and Ptc1p-myc fusions were detected by immunoblotting with anti-c-myc antibody. Anti-Cdc2 antibody was used as loading control.
Differential Regulation of MAPK Functions by Pyp1p, Ptc1p, and Pmp1p Phosphatases
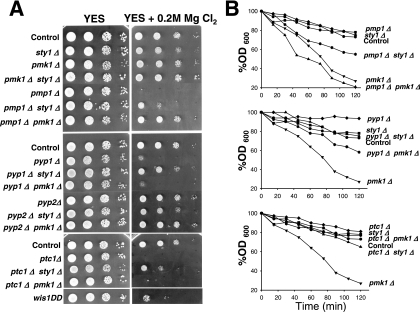
In S. pombe, the phosphorylation state of Pmk1p has a dramatic effect on chloride homeostasis. Deletion of pmp1+, and consequently Pmk1p hyperactivation, leads to strong sensitivity to this anion (Sugiura et al., 1998). We analyzed the ability of strains deficient in different elements of the SAPK and Pmk1p pathways to grow in rich medium supplemented with MgCl2. Strains lacking sty1+, pmk1+, and the double-deleted pmk1Δ sty1Δ strain did not show significant growth changes in this medium (Figure 8A). As expected, the chloride sensitivity of the pmp1+-deleted mutant was rescued by additional deletion of pmk1+, indicating that Pmk1p hyperactivation is responsible for such behavior (Figure 8A). Some tolerance was also observed in the sty1Δ pmp1Δ double mutant (Figure 8A). Because Pmp1p does not seem to down-regulate Sty1p activity (Sugiura et al., 1998; this work), this observation suggests that, in addition to Pmk1p, basal Sty1p activity contributes to the hypersensitivity of pmp1Δ cells against chloride ions. Notably, deletion of pyp1+ or ptc1+, but not of pyp2+, resulted in a strong growth inhibition (Figure 8A). Nevertheless, because chloride sensitivity in pyp1Δ and ptc1Δ cells might be also due to Sty1p hyperactivation, we analyzed the sensitivity of pmk1Δ pyp1Δ, sty1Δ pyp1Δ, pmk1Δ ptc1Δ, and sty1Δ ptc1Δ double mutants. Consistently, pmk1Δ pyp1Δ and pmk1Δ ptc1Δ double mutants were as sensitive as single pyp1Δ and ptc1Δ mutants, respectively, whereas deletion of sty1+ alleviated the chloride sensitivity of pyp1Δ and ptc1Δ mutants (Figure 8A). Moreover, the constitutive activation of Sty1p in a wis1DD strain brought about increased chloride sensitivity. These results indicate that Sty1p hyperactivation, rather than Pmk1p hyperactivation, accounts for the chloride sensitivity in the absence of Pyp1p or Ptc1p phosphatases.
Figure 8.
Differential regulation of MAPK functions by Pyp1p, Ptc1p, and Pmp1p. (A) Chloride sensitivity assays. Strains MI200 (Control), TK107 (sty1Δ), MI102 (pmk1Δ), MI107 (pmk1Δ sty1Δ), MI104 (pmp1Δ), MI116 (pmp1Δ sty1Δ), MI115 (pmp1Δ pmk1Δ), MI105 (pyp1Δ), MI109 (pyp1Δ sty1Δ), MI108 (pyp1Δ pmk1Δ), MI106 (pyp2Δ), MI118 (pyp2Δ sty1Δ), MI117 (pyp2Δ pmk1Δ), MI110 (ptc1Δ), MI112 (ptc1Δ sty1Δ), MI111 (ptc1Δ pmk1Δ), and 2119 (wis1DD) were grown in YES medium to an A600 of 0.5. Samples containing 105, 104, 103 or 102 cells were spotted onto YES plates supplemented with 0.2 M MgCl2 and incubated for 3 d at 28°C before being photographed. (B) Cell wall integrity assays. Strains MI200 (Control), TK107 (sty1Δ), MI102 (pmk1Δ), MI104 (pmp1Δ), MI116 (pmp1Δ sty1Δ), MI115 (pmp1Δ pmk1Δ), MI105 (pyp1Δ), MI109 (pyp1Δ sty1Δ), MI108 (pyp1Δ pmk1Δ), MI110 (ptc1Δ), MI112 (ptc1Δ sty1Δ), and MI111 (ptc1Δ pmk1Δ) were grown in YES medium to an A600 of 0.5, and the cells treated at 30°C with 100 μg/ml Zymolyase 20-T. Cell lysis was monitored by measuring A600 decay at different incubation times. Results represent the mean value of three independent experiments.
Another feature of pmk1+-deleted cells is their sensitivity to β-1,3-glucanase, indicative of cell wall defects (Toda et al., 1996; Zaitsevskaya-Carter and Cooper, 1997). We also analyzed β-1,3-glucanase sensitivity in exponentially growing cultures of strains deleted in elements of the MAPK cascade. The strain lacking Pmk1p, displayed the expected hypersensitivity to β-1,3-glucanase (Figure 8B). Deletion of pmp1+ did not affect cell sensitivity against glucanase, whereas pmk1Δ pmp1Δ cells were as sensitive to the lytic enzyme as single pmk1Δ mutant (Figure 8B, top). However, sty1Δ pmp1Δ cells were more sensitive to the treatment than sty1Δ or pmp1Δ strains (Figure 8B, top), indicating that Sty1p activity contributes to the maintenance of cell wall structure in the absence of Pmp1p. Deletion of pyp1+, and less noticeably that of ptc1+, resulted in moderately increased resistance against the lytic enzyme (Figure 8B, middle and bottom). Unexpectedly, pmk1Δ pyp1Δ and pmk1Δ ptc1Δ cells were less sensitive to glucanase treatment than pmk1Δ cells, suggesting that Sty1p hyperactivation partially suppress the sensitive phenotype. In this context, the glucanase-resistant trait of pyp1Δ cells (and ptc1Δ cells) was clearly suppressed by simultaneous deletion in sty1+ (Figure 8B, middle and bottom). Together, these data suggest that although Pyp1p and Ptc1p can dephosphorylate both Sty1p and Pmk1p MAPKs, their role in the maintenance of the cell wall structure is mainly mediated through the regulation of Sty1p activity.
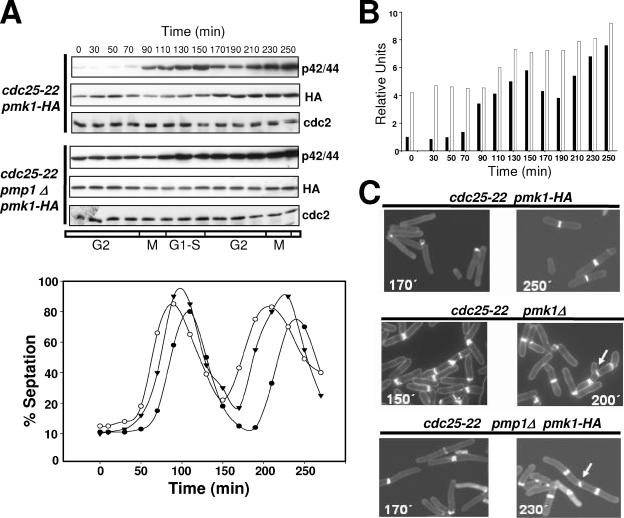
Cell Cycle-dependent Activation of Pmk1p
Microarray analysis has shown that pmk1+ is periodically expressed during the cell cycle, with increased transcript levels during mitosis and G1 phase (Rustici et al., 2004). We explored Pmk1p levels and its phosphorylation state during the cell cycle by introducing the Pmk1-HA6H fusion into a cdc25-22 strain. Cells from this mutant (strain MI600) were grown at 25°C to log phase, shifted to 37°C for 3.5 h to synchronize the cells in G2, and then returned to 25°C. Figure 9A indicates that the level of HA6H-tagged Pmk1p varied slightly through the cell cycle, with a small increase during G2 transition followed by a very low decrease at M and G1-S phases (Figure 9A). However, Pmk1p phosphorylation clearly changed during the cell cycle, increasing during M phase and reaching its maximum during cytokinesis (Figure 9, A and B). Notably, deletion of phosphatase Pmp1p altered the activation state of Pmk1p along the cell cycle. As seen in Figure 9, A and B, phosphorylation level of p42/44 in G2-arrested cells from strain MI602 (cdc25-22 pmp1Δ Pmk1-HA6H) was higher than in wild-type cells, increased moderately during cell separation, and did not decrease thereafter. A quantitative analysis of the effect of pmk1+ or pmp1+ deletion in the completion of cytokinesis indicated that whereas the septation index reached a value of 0–2% in control cells at the point of maximal cell separation, the separation was partially defective in pmk1Δ and pmp1Δ strains, with ∼20% cells unable to complete cytokinesis (Figure 9A). The consequence of this defect in pmk1Δ and pmp1Δ cells is a cumulative multiseptate phenotype during the second division cycle (Figure 9C, multiseptate cells marked with arrows), which is coincident with the morphological feature previously described for both mutants (Toda et al., 1996; Zaitsevskaya-Carter and Cooper, 1997; Sugiura et al., 1998).
Figure 9.
Influence of Pmp1p activity on the cell cycle-dependent activation of Pmk1p. (A) Pmk1p activates periodically during the cell cycle. Top, cells from strains MI600 (cdc25-22, pmk1-HA6H) and MI602 (cdc25-22, pmp1Δ, pmk1-HA6H) were grown to an A600 of 0.3 at 25°C, shifted to 37°C for 3.5 h, and then released from the growth arrest by transfer back to 25°C. Aliquots were taken at different time intervals, and Pmk1-HA6H was purified by affinity chromatography. Activated or total Pmk1p were detected by immunoblotting with anti-phospho-p42/44 or anti-HA antibodies, respectively, and anti-Cdc2 antibody was used as loading control. Bottom, septation index of strains MI600 (filled circles), MI601 (cdc25-22, pmk1Δ; open circles), and MI602 (filled triangles). (B) Quantitative analysis of Pmk1p activity during cell cycle in strains MI600 (filled bars) and MI602 (empty bars) obtained from data in A. (C) Defective cell separation in pmk1Δ and pmp1Δ cells. Cells from strains MI600 (Control), MI601, and MI602 were taken at the times shown during cell cycle experiments, stained with Calcofluor white, and observed by fluorescence microscopy. Arrows indicate cells with multiseptate phenotype and defective cell separation.
DISCUSSION
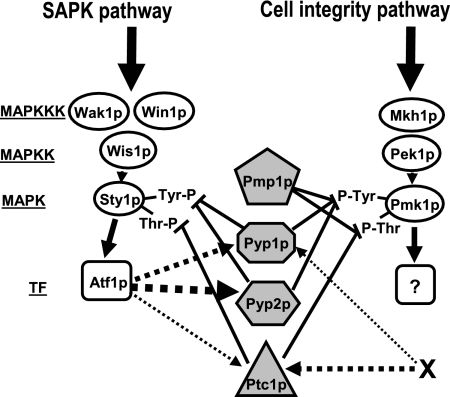
In S. pombe, protein tyrosine phosphatases Pyp1p and Pyp2p, and serine/threonine phosphatase Ptc1p negatively control the stimulation of MAPK Sty1p (Millar et al., 1995; Shiozaki and Russell, 1995; Degols et al., 1996; Samejima et al., 1997; Nguyen and Shiozaki, 1999). In this work we provide evidence for a role of these phosphatases in the down-regulation of the key element of the cell integrity pathway MAPK Pmk1p. The only MAPK phosphatase previously known to dephosphorylate Pmk1p was dual specificity phosphatase Pmp1p (Sugiura et al., 1998). We demonstrate here that Pyp1p, Pyp2p, and Ptc1p phosphatases associate in vivo with Pmk1p and that they are able to dephosphorylate activated Pmk1p. The contribution of these protein phosphatases to Pmk1p inactivation seems dependent on the physiological state of the yeast cells. Analyses of MAPK activity in different null mutants indicate that Pyp1p and Ptc1p control both the basal activity level of Pmk1p and its dephosphorylation during adaptation to osmostress, whereas Pyp2p mainly limits Pmk1p hyperactivation during such adaptation. Thus, Pmk1p activation is negatively regulated by at least two different dephosphorylation mechanisms, one mediated by Pmp1p and another by the effectors of the SAPK pathway Pyp1p, Pyp2p, and Ptc1p (Figure 10).
Figure 10.
Proposed regulatory links between the SAPK and the cell integrity pathways by protein phosphatases (shaded figures) during growth and osmostress. Pyp1p, Pyp2p, and Ptc1p protein phosphatases are negative regulators of MAPK Sty1p and MAPK Pmk1 activities, whereas Pmp1 only dephosphorylates MAPK Pmk1p (⊥, inhibition). The expression/synthesis of the shared phosphatases is in turn under Sty1p/Atf1p-dependent and Sty1p/Atf1p-independent (X-dependent) regulation. The size of the discontinuous arrows indicates the relative transcriptional reliance. The question mark shows that the corresponding transcription factor is unknown. For more details on abbreviations and symbols, see Figure 1.
Work with budding yeast S. cerevisiae has shown that tyrosine phosphatases Ptp2p and Ptp3p (homologues to Pyp1p and Pyp2p, respectively) regulate the basal stimulation of Hog1p and Slt2p MAPKs (which are Sty1p and Pmk1p orthologues, respectively) (Jacoby et al., 1997; Mattison et al., 1999). In contrast, our results suggest that Pyp1p is the only tyrosine phosphatase able to down-regulate the basal level of both Pmk1p and Sty1p in S. pombe growing cells. Also, the budding yeast PP2C phosphatase Ptc1p regulates endoplasmic reticulum inheritance by modulating Slt2p activity, and the level of phosphorylated Slt2p is elevated in ptc1Δ cells (Du et al., 2006). Our findings in fission yeast strongly suggest that Ptc1p controls Pmk1p activity by direct association, because Ptc1p copurifies with Pmk1p and dephosphorylates the active protein kinase both in vivo and in vitro. This result provides the first direct biochemical evidence on the action of a PP2C type phosphatase upon an ERK-type MAPK in yeast cells.
It has been recently reported that the hyperosmotic shock induces in budding yeast a delayed transient phosphorylation of Slt2p, which is not observed in strains deleted in members of the HOG pathway (García-Rodriguez et al., 2005). In striking contrast, we show that deletion of members of the SAPK pathway in fission yeast raises both the basal and the osmostress-induced Pmk1 phosphorylation. In fact, a constitutive Sty1p hyperactivation prompted by either pyp1+ deletion or the presence of wis1DD hyperactive allele caused a significant decrease in Pmk1p phosphorylation. Previous work has shown that the basal expression of pyp1+ and the stress-induced expression of pyp1+, pyp2+, and ptc1 + is triggered by the Sty1p–Atf1p pathway through a negative feedback loop (Degols et al., 1996; Shiozaki and Russell, 1996; Wilkinson et al., 1996, Gaits et al., 1997). Our detailed analysis of protein levels confirmed that the Sty1p-dependent increased expression and synthesis of phosphatases Pyp1p, Pyp2p, and Ptc1p is the main responsible for Pmk1p down-regulation under osmostress. The question remains as to why these phosphatases are needed to inactivate Pmk1p under osmostress. A possible explanation might be related to Pmp1p availability under osmostress. It has been described that the mRNA levels of pmp1+ in S. pombe decrease shortly after osmotic stress, to recover slowly thereafter (Chen et al., 2003). This effect would allow Pyp1p, Pyp2p, or Ptc1p to effectively down-regulate activated Pmk1p, because under osmostress they are rapidly and strongly induced through the Sty1p–Atf1p pathway.
An important result from this work is that phosphatases Pyp1p and Ptc1p can negatively regulate Pmk1p activity in a manner independent on the transcriptional control of the Sty1p-Atf1p. This is supported by Pyp1p and Ptc1p synthesis in the absence of Sty1p and by the defective dephosphorylation observed in sty1Δ pyp1Δ and sty1Δ ptc1Δ double mutants compared with single mutants. This was particularly evident in the case of Ptc1p. High protein levels of this phosphatase were detected in sty1Δ cells under osmotic stress, although the induced expression of the ptc1+gene is fully dependent on the Sty1p–Atf1p pathway (Gaits et al., 1997). Thus, the SAPK-independent regulation of Ptc1p and Pyp1p synthesis reveals a novel mechanism for down-regulation of Pmk1p and Sty1p activities in fission yeast.
In addition to its role in down-regulating Pmk1p phosphorylation under osmostress, we analyzed the biological relevance of Pyp1/2p and Ptc1p phosphatases in the modulation of other cellular responses regulated by Pmk1p. One example is the control of chloride homeostasis. In this process calcineurin phosphatase and the Pmk1p MAPK pathway play antagonistic roles (Sugiura et al., 1998). Most likely, calcineurin deactivates one substrate phosphorylatable by Pmk1p and involved in Cl− transport so that under Pmk1p hyperactivation (caused for instance by pmp1+ deletion) the phosphorylated component would accumulate, thus preventing Cl− transport and leading to cell toxicity and death (Sugiura et al., 2002). Surprisingly, the results presented in this work demonstrate that, in the absence of pyp1+or ptc1+, the hyperactivation of Sty1p, and not that of Pmk1p, is the main responsible for the increase in cell sensitivity to Cl−. Hence, Pyp1p, Ptc1p, and Pmp1p phosphatases control chloride homeostasis in S. pombe by differential inactivation of Sty1p (Pyp1p and Ptc1p) and Pmk1p (Pmp1p). Pmk1p activity is also essential for the control of cell wall structure in S. pombe. Accordingly, yeast mutants devoid of Pyp1p and Ptc1p displayed increased resistance to cell wall digestion by Zymolyase, which is a feature opposed to the hypersensitive phenotype described for pmk1Δ cells (Toda et al., 1996; Zaitsevskaya-Carter and Cooper, 1997). However, the analysis of cell wall sensitivity in double mutants indicates that the increased resistance to Zymolyase in the absence of pyp1+ or ptc1+ is due to Sty1p hyperactivation. Hence, both Pmk1p and Sty1p are involved in the control of cell wall structure. It would be interesting to perform a detailed comparative analysis of the cell wall composition in pmk1+, sty1+, pmp1+, pyp1+, ptc1+, and pyp2+ single and double null mutants to establish the role of the Pmk1p and Sty1p pathways in morphogenesis.
The subcellular localization of Pmk1p seems unaffected by deletion of Pyp1p, Pyp2p, Ptc1p, and Pmp1p phosphatases, and it is also independent of its phosphorylation state. Because all these phosphatases are mainly cytoplasmic proteins (Gaits and Russell, 1999; Madrid et al., 2006), deactivation of Pmk1p probably occurs at the cytoplasm and/or the septum, and the activated Pmk1p found at the nucleus must shift to the cytoplasm to be dephosphorylated. This to-and-fro mechanism for Pmk1p deactivation in S. pombe differs clearly from that of S. cerevisiae, where phosphatases Ptp2p and Msg5p are nuclear proteins and only phosphatase Ptp3p localizes at the cytoplasm, with Ptp2p acting as a nuclear anchor for Hog1 (Mattison and Ota, 2000; Martín et al., 2005).
The absence of Mkh1p, Pek1p, or Pmk1p causes multiseptation in cells plus thickened cell walls (Toda et al., 1996; Sengar et al., 1997; Zaitsevskaya-Carter and Cooper, 1997; Loewith et al., 2000), and the three proteins localize at the septum during cell separation (Madrid et al., 2006). Another finding in our study is that Pmk1p phosphorylation varies periodically during the cell cycle, with maximum activity during cytokinesis. These results suggest the existence of a signal that specifically activates the Pmk1p pathway to control septum formation and/or dissolution during cell separation in a localized manner. Moreover, Pmp1p phosphatase activity is essential for a tight control of Pmk1p activity during cell separation, and its absence not only disrupts the periodic deactivation of the MAPK but originates a multiseptate phenotype like that of pmk1Δ cells (Sugiura et al., 1998; and our results). Therefore, the cycling of Pmk1p activity along the cell cycle in S. pombe seems critical to ensure the efficient completion of cytokinesis. This is clearly distinct from what happens in the budding yeast, where Slt2p is activated periodically through the cell cycle but peaks in G1 in coincidence with bud emergence and not during cell separation (Levin, 2005). Nevertheless, some points of this control in the fission yeast remain unknown. One refers to the potential role, if any, of Pyp1p or Ptc1p in the deactivation of Pmk1p during the cell cycle. We could not perform an analysis of Pmk1p activation along the cell cycle in a cdc25-22 pyp1Δ background because of the difficulty to synchronize cell cultures due to the Sty1p hyperactivation that follows Pyp1p deletion. However, the absence of a multiseptate phenotype in pyp1Δ and ptc1Δ cells suggests that, contrary to Pmp1p, the activity of these phosphatases is most likely irrelevant to the regulation of Pmk1p activity during cell separation. One attractive possibility would be that Pmp1p was the only phosphatase responsible for the inactivation of the Pmk1p pool localized at the septum. However, we have been unable to visualize Pmp1p at the septum, although this might result from a very quick and transient localization.
In summary, our results allow to conclude that negative regulation of Pmk1p activity in S. pombe is more complex than originally thought. In addition to the previously characterized role of dual specificity phosphatase Pmp1p, the MAPK phosphatases Pyp1p, Pyp2p, and Ptc1p integrate SAPK-dependent or -independent signals that promote Pmk1p down-regulation. The identification of SAPK-independent elements controlling Pmk1p inactivation by Ptc1p and Pyp1p phosphatases and the meaning of their apparent redundancy are currently unanswered questions. Also, the elucidation of mechanisms determining how the specificity of action against Sty1p and Pmk1p is attained deserves further work.
ACKNOWLEDGMENTS
We thank P. Pérez for helpful discussions and comments; A. Durán (University of Salamanca, Spain), T. Kato (ERATO, Kyoto, Japan), S. Moreno (University of Salamanca, Spain), J. B. Millar (University of Warwick, United Kingdom), P. Nurse (Rockefeller University, NY), M. A. Rodriguez-Gabriel (Complutense University, Madrid, Spain), and T. Toda (London Research Institute, United Kingdom) for kind supply of yeast strains; and F. Garro for technical assistance. M.M. and A.N. are predoctoral fellows from Ministerio de Educación, Cultura y Deporte (Formación de Profesorado Universitario) and from Fundación Séneca (Spain), respectively. This work was supported by grants BFU2005-01401/BMC from Ministerio de Educación y Ciencia (to J.C.) and 00475/PI/04 from Fundación Séneca (Región de Murcia), Spain.
Abbreviations used:
- GFP
green fluorescent protein
- GST
glutathione S-transferase
- HA
hemagglutinin
- HA6H
epitope comprising hemagglutinin antigen plus six histidine residues
- MAPK
mitogen-activated protein kinase
- MAPKK
mitogen-activated protein kinase kinase
- MAPKKK
mitogen-activated protein kinase kinase kinase
- SAPK
stress-activated protein kinase.
Footnotes
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E07-05-0484) on August 29, 2007.
REFERENCES
- Alfa C., Fantes P., Hyams J., Mcleod M., Warbrick E. New York: Cold Spring Harbor Laboratory Press; 1993. Experiments with Fission Yeast: A Laboratory Course Manual. [Google Scholar]
- Bahler J., Wu J. Q., Longtine M. S., Shah N. G., McKenzie A., 3rd, Steever A. B., Wach A., Philippsen P., Pringle J. R. Heterologous modules for efficient and versatile PCR-based gene targeting in Schizosaccharomyces pombe. Yeast. 1998;14:943–951. doi: 10.1002/(SICI)1097-0061(199807)14:10<943::AID-YEA292>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- Bone N., Millar J. B., Toda T., Armstrong J. Regulated vacuole fusion and fission in Schizosaccharomyces pombe: an osmotic response dependent on MAP kinases. Curr. Biol. 1998;8:135–144. doi: 10.1016/s0960-9822(98)00060-8. [DOI] [PubMed] [Google Scholar]
- Chen D., Toone W. M., Mata J., Lyne R., Burns G., Kivinen K., Brazma A., Jones N., Bähler J. Global transcriptional responses of fission yeast to environmental stress. Mol. Biol. Cell. 2003;14:214–229. doi: 10.1091/mbc.E02-08-0499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Degols G., Shiozaki K., Russell P. Activation and regulation of the Spc1 stress-activated protein kinase in Schizosaccharomyces pombe. Mol. Cell. Biol. 1996;16:2870–2877. doi: 10.1128/mcb.16.6.2870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du Y., Walker L., Novick P., Ferro-Novick S. Ptc1p regulates cortical ER inheritance via Slt2p. EMBO J. 2006;25:4413–4422. doi: 10.1038/sj.emboj.7601319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farooq A., Zhou M.-M. Structure and regulation of MAPK phosphatases. Cell Signal. 2004;16:769–779. doi: 10.1016/j.cellsig.2003.12.008. [DOI] [PubMed] [Google Scholar]
- Forsburg S. L., Sherman D. A. General purpose tagging vectors for fission yeast. Gene. 1997;191:191–195. doi: 10.1016/s0378-1119(97)00058-9. [DOI] [PubMed] [Google Scholar]
- Gaits F., Shiozaki K., Russell P. Protein phosphatase 2C acts independently of stress-activated kinase cascade to regulate the stress response in fission yeast. J. Biol. Chem. 1997;272:17873–17879. doi: 10.1074/jbc.272.28.17873. [DOI] [PubMed] [Google Scholar]
- Gaits F., Russell P. Active nucleocytoplasmic shuttling required for function and regulation of stress-activated kinase Spc1/Sty1 in fission yeast. Mol. Biol. Cell. 1999;10:1395–1407. doi: 10.1091/mbc.10.5.1395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Rodríguez L. J., Valle R., Durán A., Roncero C. Cell integrity signaling activation in response to hyperosmotic shock in yeast. FEBS Lett. 2005;579:6186–6190. doi: 10.1016/j.febslet.2005.10.001. [DOI] [PubMed] [Google Scholar]
- Guan K. L., Dixon J. E. Eukaryotic proteins expressed in Escherichia coli: an improved thrombin cleavage and purification procedure of fusion proteins with glutathione S-transferase. Anal. Biochem. 1991;192:262–267. doi: 10.1016/0003-2697(91)90534-z. [DOI] [PubMed] [Google Scholar]
- Hohmann S. Osmotic stress signaling and osmoadaptation in yeasts. Microbiol. Mol. Biol. Rev. 2002;66:300–372. doi: 10.1128/MMBR.66.2.300-372.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacoby T., Flanagan H., Faykin A., Seto A. G., Mattison C., Ota I. Two protein-tyrosine phosphatases inactivate the osmotic stress response pathway in yeast by targeting the mitogen-activated protein kinase, Hog1. J. Biol. Chem. 1997;272:17749–17755. doi: 10.1074/jbc.272.28.17749. [DOI] [PubMed] [Google Scholar]
- Levin D. E. Cell wall integrity signaling in Saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev. 2005;69:262–291. doi: 10.1128/MMBR.69.2.262-291.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loewith R., Hubberstey A., Young D. Skh1, the MEK component of the mkh1 signaling pathway in Schizosaccharomyces pombe. J. Cell Sci. 2000;113:153–160. doi: 10.1242/jcs.113.1.153. [DOI] [PubMed] [Google Scholar]
- Madrid M., Soto T., Franco A., Paredes V., Vicente J., Hidalgo E., Gacto M., Cansado J. A cooperative role for Atf1 and Pap1 in the detoxification of the oxidative stress induced by glucose deprivation in Schizosaccharomyces pombe. J. Biol. Chem. 2004;279:41594–41602. doi: 10.1074/jbc.M405509200. [DOI] [PubMed] [Google Scholar]
- Madrid M., Soto T., Khong H. K., Franco A., Vicente J., Pérez P., Gacto M., Cansado J. Stress-induced response, localization, and regulation of the Pmk1 cell integrity pathway in Schizosaccharomyces pombe. J. Biol. Chem. 2006;281:2033–2043. doi: 10.1074/jbc.M506467200. [DOI] [PubMed] [Google Scholar]
- Marshall C. J. Specificity of receptor tyrosine kinase signaling: transient versus sustained extracellular signal-regulated kinase activation. Cell. 1995;80:179–185. doi: 10.1016/0092-8674(95)90401-8. [DOI] [PubMed] [Google Scholar]
- Martín H., Flández M., Nombela C., Molina M. Protein phosphatases in MAPK signalling: we keep learning from yeast. Mol. Microbiol. 2005;58:6–16. doi: 10.1111/j.1365-2958.2005.04822.x. [DOI] [PubMed] [Google Scholar]
- Mattison C. P., Ota I. M. Two protein tyrosine phosphatases, Ptp2 and Ptp3, modulate the subcellular localization of the Hog1 MAP kinase in yeast. Genes Dev. 2000;14:1229–1235. [PMC free article] [PubMed] [Google Scholar]
- Mattison C. P., Spencer S. S., Kresge K. A., Lee J., Ota I. M. Differential regulation of the cell wall integrity mitogen-activated protein kinase pathway in budding yeast by the protein tyrosine phosphatases Ptp2 and Ptp3. Mol. Cell. Biol. 1999;19:7651–7660. doi: 10.1128/mcb.19.11.7651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millar J. B., Russell P., Dixon J. E., Guan K. L. Negative regulation of mitosis by two functionally overlapping PTPases in fission yeast. EMBO J. 1992;11:4943–4952. doi: 10.1002/j.1460-2075.1992.tb05601.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millar J. B., Buck V., Wilkinson M. G. Pyp1 and Pyp2 PTPases dephosphorylate an osmosensing MAP kinase controlling cell size at division in fission yeast. Genes Dev. 1995;9:2117–2130. doi: 10.1101/gad.9.17.2117. [DOI] [PubMed] [Google Scholar]
- Moreno S., Klar A., Nurse P. Methods in Enzymology. Vol. 283. New York: Academic Press, Inc.; 1991. Molecular genetic analysis of fission yeast Schizosaccharomyces pombe; pp. 506–520. [DOI] [PubMed] [Google Scholar]
- Nguyen A. N., Shiozaki K. Heat-shock-induced activation of stress MAP kinase is regulated by threonine- and tyrosine-specific phosphatases. Genes Dev. 1999;13:1653–1663. doi: 10.1101/gad.13.13.1653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roux P. P., Blenis J. ERK and p38 MAPK-activated protein kinases: a family of protein kinases with diverse biological functions. Microbiol. Mol. Biol. Rev. 2004;68:320–344. doi: 10.1128/MMBR.68.2.320-344.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rustici G., Mata J., Kivinen K., Lio P., Penkett C. J., Burns G., Hayles J., Brazma A., Nurse P., Bahler J. Periodic gene expression program of the fission yeast cell cycle. Nat. Genet. 2004;36:809–817. doi: 10.1038/ng1377. [DOI] [PubMed] [Google Scholar]
- Samejima I., Mackie S., Fantes P. A. Multiple modes of activation of the stress-responsive MAP kinase pathway in fission yeast. EMBO J. 1997;16:6162–6170. doi: 10.1093/emboj/16.20.6162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato M., Dhut S., Toda T. New drug-resistant cassettes for gene disruption and epitope tagging in Schizosaccharomyces pombe. Yeast. 2005;22:583–591. doi: 10.1002/yea.1233. [DOI] [PubMed] [Google Scholar]
- Sengar A. S., Markley N. A., Marini N. J., Young D. Mkh1, a MEK kinase required for cell wall integrity and proper response to osmotic and temperature stress in Schizosaccharomyces pombe. Mol. Cell. Biol. 1997;17:3508–3519. doi: 10.1128/mcb.17.7.3508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shieh J. C., Wilkinson M. G., Buck V., Morgan B. A., Makino K., Millar J. B. The Mcs4 response regulator coordinately controls the stress-activated Wak1-Wis1-Sty1 MAP kinase pathway and fission yeast cell cycle. Genes Dev. 1997;11:1008–1022. doi: 10.1101/gad.11.8.1008. [DOI] [PubMed] [Google Scholar]
- Shiozaki K., Russell P. Cell-cycle control linked to extracellular environment by MAP kinase pathway in fission yeast. Nature. 1995;378:739–743. doi: 10.1038/378739a0. [DOI] [PubMed] [Google Scholar]
- Shiozaki K., Russell P. Conjugation, meiosis, and the osmotic stress response are regulated by Spc1 kinase through Atf1 transcription factor in fission yeast. Genes Dev. 1996;10:2276–2288. doi: 10.1101/gad.10.18.2276. [DOI] [PubMed] [Google Scholar]
- Shiozaki K., Russell P. Methods in Enzymology. Vol. 283. New York: Academic Press, Inc.; 1997. Stress-activated protein kinase pathway in cell cycle control of fission yeast; pp. 506–520. [DOI] [PubMed] [Google Scholar]
- Shiozaki K., Shiozaki M., Russell P. Heat Stress activates fission yeast Spc1/Sty1 MAPK by a MEKK-independent mechanism. Mol. Biol. Cell. 1998;9:1339–1349. doi: 10.1091/mbc.9.6.1339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soto T., Beltrán F. F., Paredes V., Madrid M., Millar J.B.A., Vicente-Soler J., Cansado J., Gacto M. Cold induces stress-activated protein kinase-mediated response in the fission yeast Schizosaccharomyces pombe. Eur. J. Biochem. 2002;269:1–10. doi: 10.1046/j.1432-1033.2002.03214.x. [DOI] [PubMed] [Google Scholar]
- Sugiura R., Kita A., Shimizu Y., Shuntoh H., Sio S. O., Kuno T. Feedback regulation of MAPK signalling by an RNA-binding protein. Nature. 2003;424:961–965. doi: 10.1038/nature01907. [DOI] [PubMed] [Google Scholar]
- Sugiura R., Sio S. O., Shuntoh H., Kuno T. Calcineurin phosphatase in signal transduction: lessons from fission yeast. Genes Cells. 2002;7:619–627. doi: 10.1046/j.1365-2443.2002.00557.x. [DOI] [PubMed] [Google Scholar]
- Sugiura R., Toda T., Dhut S., Shuntoh H., Kuno T. The MAPK kinase Pek1 acts as a phosphorylation-dependent molecular switch. Nature. 1999;399:479–483. doi: 10.1038/20951. [DOI] [PubMed] [Google Scholar]
- Sugiura R., Toda T., Shuntoh H., Yanagida M., Kuno T. pmp1+, a suppressor of calcineurin deficiency, encodes a novel MAP kinase phosphatase in fission yeast. EMBO J. 1998;17:140–148. doi: 10.1093/emboj/17.1.140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toda T., Dhut S., Superti-Furga G., Gotoh Y., Nishida E., Sugiura R., Kuno T. The fission yeast pmk1+ gene encodes a novel mitogen-activated protein kinase homolog which regulates cell integrity and functions coordinately with the protein kinase C pathway. Mol. Cell. Biol. 1996;16:6752–6764. doi: 10.1128/mcb.16.12.6752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toda T., Shimanuki M., Yanagida M. Fission yeast genes that confer resistance to staurosporine encode an AP-1-like transcription factor and a protein kinase related to the mammalian ERK1/MAP2 and budding yeast FUS3 and KSS1 kinases. Genes Dev. 1991;5:60–73. doi: 10.1101/gad.5.1.60. [DOI] [PubMed] [Google Scholar]
- Waskiewicz A. J., Cooper J. A. Mitogen and stress response pathways: MAP kinase cascades and phosphatase regulation in mammals and yeast. Curr. Opin. Cell Biol. 1995;7:798–805. doi: 10.1016/0955-0674(95)80063-8. [DOI] [PubMed] [Google Scholar]
- Wilkinson M. G., Samuels M., Takeda T., Toone W. M., Shieh J. C., Toda T., Millar J. B., Jones N. The Atf1 transcription factor is a target for the Sty1 stress-activated MAP kinase pathway in fission yeast. Genes Dev. 1996;10:2289–2301. doi: 10.1101/gad.10.18.2289. [DOI] [PubMed] [Google Scholar]
- Zaitsevskaya-Carter T., Cooper J. A. Spm1, a stress-activated MAP kinase that regulates morphogenesis in S. pombe. EMBO J. 1997;16:1318–1331. doi: 10.1093/emboj/16.6.1318. [DOI] [PMC free article] [PubMed] [Google Scholar]