Abstract
Inorganic polyphosphate (poly P) is a biopolymer that occurs in all organisms and cells and in many cellular compartments. It is involved in numerous biological phenomena and functions in cellular processes in all organisms. However, even the most fundamental aspects of poly P metabolism are largely unknown. In yeast, large amounts of poly P accumulate in the vacuole during growth. It is neither known how this poly P pool is synthesized nor how it is remobilized from the vacuole to replenish the cytosolic phosphate pool. Here, we report a systematic analysis of the yeast phosphate transporters and their function in poly P metabolism. By using poly P content as a read-out, it was possible to define novel functions of the five phosphate transporters: Pho84, Pho87, Pho89, Pho90, and Pho91, in budding yeast. Most notably, it was found that the low-affinity transporter Pho91 limits poly P accumulation in a strain lacking PHO85. This phenotype was not caused by a regulatory effect on the PHO pathway, but can be attributed to the unexpected localization of Pho91 in the vacuolar membrane. This finding is consistent with the hypothesis that Pho91 serves as a vacuolar phosphate transporter that exports phosphate from the vacuolar lumen to the cytosol.
INTRODUCTION
Phosphate is an essential macronutrient: it is an important component of nucleic acids and phospholipids, represents a source of energy in nucleotides, modulates protein activities, or serves as a signal through phosphorylation of specific amino acid residues, and it can be polymerized to form inorganic polyphosphate (poly P). But even though yeast can store up to 20% of its dry weight as poly P (Kornberg et al., 1999) and furthermore, many pathways are known to be involved in poly P metabolism (Freimoser et al., 2006), it is still not known how this polymer is synthesized in yeast or any higher eukaryote. To learn more about the regulation of the metabolism of phosphate and poly P, we performed a detailed study of the yeast phosphate transporters, the PHO pathway, and poly P levels.
The yeast genome encodes five phosphate transporters (Pho84, Pho87, Pho89, Pho90, and Pho91) that are involved in the intricately regulated phosphate uptake (Wykoff and O'Shea, 2001). Pho84 and Pho89 are two high-affinity transporters that are both regulated by the PHO pathway (Bun-Ya et al., 1991; Martinez and Persson, 1998): Under low-phosphate conditions, the transcription factor Pho4 is dephosphorylated and localized in the nucleus, where it activates transcription of phosphate-regulated genes such as PHO84 and PHO89 (Lenburg and O'Shea, 1996; O'Neill et al., 1996; Springer et al., 2003). In the high-phosphate state, Pho4 is phosphorylated by the cyclin-dependent kinase Pho85, which causes its relocalization to the cytosol and concomitant down-regulation of phosphate responsive genes (Lenburg and O'Shea, 1996; O'Neill et al., 1996; Springer et al., 2003). Despite its strict regulation by the PHO pathway, Pho84 is the most important phosphate transporter in yeast, even in high-phosphate media (Wykoff and O'Shea, 2001). Transcription of PHO84 and PHO89 is not only regulated by the PHO pathway, but for example, also by the SAGA and the SWI-SNF complexes (Lee et al., 2000; Sudarsanam et al., 2000; Bhaumik and Green, 2002) or by shifts to acidic or alkaline pH, the calcineurine pathway, or the cell cycle (Causton et al., 2001; Serrano et al., 2002; Ruiz et al., 2003; Luan and Li, 2004). In the absence of the high-affinity transporters, the low-affinity transporters, Pho87, Pho90, and Pho91, are necessary for phosphate transport, but otherwise exhibit few phenotypes if deleted (Wykoff and O'Shea, 2001). In addition to phosphate uptake, the three low-affinity phosphate transporters are involved in the external sensing of phosphate and the regulation of the phosphate-signaling pathway (Auesukaree et al., 2003; Giots et al., 2003; Pinson et al., 2004).
The PHO pathway has been characterized in great detail by studying secreted acid phosphatase (rAPase) activity, PHO84 transcription, and Pho84 localization, or trehalase activity (Petersson et al., 1999; Auesukaree et al., 2003, 2004; Giots et al., 2003; Pinson et al., 2004; Huang and O'Shea, 2005) and because of its function as a phosphate store, poly P was specifically studied as a potential regulator (Neef and Kladde, 2003; Auesukaree et al., 2004; Thomas and O'Shea, 2005). But the regulation of poly P metabolism by the PHO pathway and by phosphate transporters has not been studied systematically. Here, we used the poly P content as the read-out for a thorough characterization of mutant strains affected in the five phosphate transporters. With poly P content as a distinguishing mark, it was possible to define novel functions of the low-affinity phosphate transporters in phosphate, poly P, and general cell metabolism. Most notably, we suggest that Pho91 is an intracellular phosphate transporter that exports phosphate from the vacuole and thereby regulates intracellular phosphate homeostasis and poly P levels.
MATERIALS AND METHODS
Yeast Strains
All yeast strains used in this study are based on the haploid knock-out (YKO) strains from the Saccharomyces Genome Deletion Project (Winzeler et al., 1999), which were generated in the background of the strain BY4741 (MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0; Brachmann et al., 1998). A detailed listing of all strains used and created in this study is given in Table 1.
Table 1.
Yeast strains that were used or created in the course of this study
| Strain | Genotype | Reference |
|---|---|---|
| EY57 | MATa ade2-1 trp1-1 can1-100 leu2-3,112 his3-11,15 ura3 | Schwob and Nasmyth (1993); Wykoff and O'Shea (2001) |
| EY916 | MATa pho84Δ::HIS3 pho87Δ::CgHIS3 pho89Δ::CgHIS3 pho90Δ::CgHIS3 pho91Δ::KlURA3 | Wykoff and O'Shea (2001) |
| EY917 | MATa pho84Δ::HIS3 pho87Δ::CgHIS3 pho89Δ::CgHIS3 pho90Δ::CgHIS3 pho91Δ ADE2 | Wykoff and O'Shea (2001) |
| EY918 | MATa pho87Δ::CgHIS3 pho89Δ::CgHIS3 pho90Δ::CgHIS3 pho91Δ::KlURA3 ADE2 | Wykoff and O'Shea (2001) |
| EY919 | MATa pho84Δ::HIS3 pho89Δ::CgHIS3 pho90Δ::CgHIS3 pho91Δ::KlURA3 ADE2 | Wykoff and O'Shea (2001) |
| EY920 | MATa pho84Δ::HIS3 pho87Δ::CgHIS3 pho89Δ::CgHIS3 pho91Δ::KlURA3 ADE2 | Wykoff and O'Shea (2001) |
| EY921 | MATa pho84Δ::HIS3 pho87Δ::CgHIS3 pho89Δ::CgHIS3 pho90Δ::CgHIS3 ADE2 | Wykoff and O'Shea (2001) |
| EY922 | MATa pho84Δ::HIS3 pho87Δ::CgHIS3 pho90Δ::CgHIS3 pho91Δ::KlURA3 ADE2 | Wykoff and O'Shea (2001) |
| BY4741 | MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 | Brachmann et al. (1998) |
| pho84Δ | MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 pho84Δ::kanMX4 | Winzeler et al. (1999) |
| pho85Δ | MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 pho85Δ::kanMX4 | Winzeler et al. (1999) |
| pho87Δ | MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 pho87Δ::kanMX4 | Winzeler et al. (1999) |
| pho89Δ | MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 pho89Δ::kanMX4 | Winzeler et al. (1999) |
| pho90Δ | MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 pho90Δ::kanMX4 | Winzeler et al. (1999) |
| pho91Δ | MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 pho91Δ::kanMX4 | Winzeler et al. (1999) |
| FFSc228 (pho85ΔNAT) | MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 pho85Δ::natMX4 | This study |
| FFSc370 (pho85Δ pho84Δ) | MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 pho85Δ::natMX4 pho84Δ::kanMX4 | This study |
| FFSc256 (pho85Δ pho87Δ) | MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 pho85Δ::natMX4 pho87Δ::kanMX4 | This study |
| FFSc271 (pho85Δ pho89Δ) | MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 pho85Δ::natMX4 pho89Δ::kanMX4 | This study |
| FFSc269 (pho85Δ pho90Δ) | MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 pho85Δ::natMX4 pho90Δ::kanMX4 | This study |
| FFSc266 (pho85Δ pho91Δ) | MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 pho85Δ::natMX4 pho91Δ::kanMX4 | This study |
| FFSc519 (PTEF GFP-PHO90) | MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 natNT2-PTEF-yeGFP-PHO90 | This study |
| FFSc486 (PTEF GFP-PHO91) | MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 natNT2-PTEF-yeGFP-PHO91 | This study |
| FFSc501 (PTEF GFP-PHO91 pho85Δ) | MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 pho85Δ::kanMX4 natNT2- PTEF-yeGFP-PHO91 | This study |
| FFSc528 (PTEFPHO91) | MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 kanMX4-PTEF-PHO91 | This study |
| FFSc634 (PCYC1 GFP-PHO91) | MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 natNT2-PCYC1-yeGFP-PHO91 | This study |
| FFSc482 (PADH GFP-PHO91) | MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 natNT2-PADH1-yeGFP-PHO91 | This study |
| FFSc528 (PTEFPHO91 pho85Δ) | MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 pho85Δ::natMX4 kanMX4-PTEF-PHO91 | This study |
| FFSc412 (PHO84-GFP) | MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 PHO84-yeGFP:HIS3MX6 | This study |
| FFSc609 (PHO84-GFP pho87Δ) | MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 pho87Δ::kanMX4 PHO84-yeGFP:HIS3MX6 | This study |
| FFSc610 (PHO84-GFP pho90Δ) | MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 pho90Δ::kanMX4 PHO84-yeGFP:HIS3MX6 | This study |
| FFSc611 (PHO84-GFP pho91Δ) | MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 pho91Δ::kanMX4 PHO84-yeGFP:HIS3MX6 | This study |
| FFSc562 (PTEF-PHO87-GFP) | MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 natNT2- PTEF-yeGFP-PHO87 | This study |
The “EY-strains” were derived from the strain EY57, which corresponds to the strain K699 (Schwob and Nasmyth, 1993).
Cultivation and Handling of Yeast Strains
Strains were routinely grown at 30°C in YPD medium (10 g/l yeast extract, 20 g/l peptone, 20 g/l glucose), with the optional addition of 200 mg/l G418 (PAA Laboratories Gmbh, Pasching, Austria), 300 mg/l hygromycin (Apollo Scientific, Bredbury, United Kingdom) or 100 mg/l clonNat (Werner BioAgents, Jena, Germany). Low-orthophosphate YPD medium (YPD-Pi) was prepared by precipitating free phosphate with ammonium as described previously (Kaneko et al., 1982; Werner et al., 2005). The strains EY57 and EY916-EY922 were a gift from Dr. E. O'Shea (Harvard University) and Dr. D. Wykoff (Villanova University) and were maintained on YPGal medium (YPD medium containing 20 g/l galactose instead of glucose; Wykoff and O'Shea, 2001). Strains harboring plasmids were kept on SC medium lacking the corresponding amino acid (0.68 g/l yeast nitrogen base, complete supplement dropout mix [both either from Qbiogene, Carlsbad, CA, or Formedium, Norfolk, United Kingdom], 20 g/l glucose). For poly P analysis, strains were always precultured for 3 d, and fresh medium was inoculated to an OD600 = 1. Cells were harvested at different time points, dissolved in 1 M H2SO4, and stored at −20°C until extraction.
Manipulation of Yeast Strains
Plasmids were transformed into yeast by the method of Gietz et al. (1992). The kanamycin marker in the YKO strains was replaced with either the nourseothricine (natMX4, clonNat) or the hygromycin (hphMX4) resistance gene by transformation with Not I-digested plasmids pAG25 (natMX4) or pAG32 (hphMX4; Goldstein and McCusker, 1999), respectively. To inactivate two genes, these markers were amplified with flanking up- and downstream genomic sequences (with the “A” and “D” primers). Double deletion strains were created by transforming existing single YKO strains with these PCR products following the high-efficiency yeast transformation protocol (Gietz and Woods, 2002). Some double deletions were also created by crossing a bait strain to selected single YKO strains (Tong et al., 2001) or by crossing and tetrad dissection according to standard procedures (Kaiser et al., 1994). All strains showed almost identical phenotypes irrespective of the method used for their generation. All double deletion strains were verified by PCR with the “A,” “B,” and kanB primers. The yeast phosphate transporters Pho87, Pho90, and Pho91 were endogenously tagged with green fluorescent protein (GFP) at the N-terminus by the PCR-based epitope-tagging strategy (Janke et al., 2004). The sequences encoding the GFP and/or the different promoters were amplified with the S1 and S4 primer pair from the plasmids pYM-N9, pYM-N13, pYM-N17, pYM-N18, and pYM-N21 (for the ADH, CYC1, GPD, and TEF promoters, respectively; Janke et al., 2004). In the resulting strains the endogenous promoter is replaced with a ADH, CYC1, GPD, or TEF promoter, and the GFP tag is fused to the N-terminus of the Pho87, Pho90, or Pho91. The correct fusion of the N-terminal GFP with Pho91 was also verified by sequencing the region of the junction of these two parts. Pho84 was tagged by integrating a PCR fragment of the GFP tag and a HIS-selection marker (amplified with the S2 and S3 primers from the plasmid pYM28) at the C-terminus.
DNA Manipulations
Yeast genomic DNA was isolated following the smash-and-grab protocol (Rose et al., 1990). Modified versions of the plasmids pRS416-ADH, pRS416-TEF, and pRS426-GPD (Mumberg et al., 1995) were made by the introduction of the sequence for one hemagglutinin (HA) tag. The novel plasmids (pRS416-ADH-HA, pRS416-TEF-HA, and pRS426-GPD-HA) allow C-terminal HA tag fusions by cloning using the XmaI restriction site. The gene encoding PHO84 was cloned into the above mentioned vectors with the restriction sites EcoRI and XmaI that were included in the primers used for amplification. The insert in the resulting construct (pRS416-ADH-HA-PHO84, pRS416-TEF-HA-PHO84, and pRS426-GPD-HA-PHO84) were verified by sequencing.
Quantification of Poly P and Total Phosphate Content
Poly P was extracted, purified, and quantified as previously described (Werner et al., 2005). In short, 1 OD600 equivalent of cells was harvested and pelleted. The supernatant was removed, 50 μl 1 M H2SO4 was added, and the suspension was neutralized with 50 μl 2 M NaOH and 100 μl Tris-malate buffer (1 M Tris, 0.5 M malate, pH 7.5, 6% neutral red solution [0.1% neutral red in 70% ethanol]). Cell fragments were removed by centrifugation. After addition of 600 μl 6 M NaI, the extracts were applied to Qiagen PCR purification columns (Chatsworth, CA). The columns were washed twice with wash buffer (10 mM Tris buffer, pH 7.5, 50% ethanol, 1 mM EDTA, and 100 mM NaCl). Poly P was eluted in 100 μl H2O, and 50 μl of this eluate was specifically digested with Saccharomyces cerevisiae exopolyphosphatase. To quantify the released phosphate, 86 μl of 28 mM ammonium heptamolybdate (in 2.1 M H2SO4) and 64 μl of 1.6 mM malachite green (in 0.35% polyvinyl alcohol) were added. The malachite green solution was measured in a BioTek PowerWaveTM XS microplate spectrophotometer (BioTek Instruments, Winooski, VT) at 600 nm and compared with that of phosphate standards (5–100 μM Pi; Cogan et al., 1999). To quantify total phosphate content, 0.5 OD600 equivalent of cells were pelleted, resuspended in 200 μl 1 M H2SO4, and heated in a boiling water bath for 20 min. Released phosphate was quantified as described above.
Acid Phosphatase Assay
Acid Phosphatase (rAPase) activity was assayed according to Huang and O'Shea (2005) in 50 μl cell suspension by adding 200 μl p-nitrophenyl-phosphate (20 mM in 100 mM sodium acetate, pH 4.2). After 5–15 min reaction time at 28°C, 200 μl of 10% ice-cold trichloroacetic acid and 400 μl sodium carbonate solution (2 M) were added, and the OD420 was measured. The measurements were normalized with the cell density (OD600), the reaction time, and the sample volume (in ml). rAPase activity (Miller units) was obtained by multiplying the normalized OD420 values with a factor of 1000.
β-Galactosidase Reporter Assay
The PHO84-lacZ and the ACT1-lacZ reporter plasmids have been described earlier (Hughes et al., 2001) and were a gift from Dr. S. Fields (University of Washington). The β-galactosidase reporter assay was performed similar to the protocols of Möckli and Auerbach (2004) and Miller (1972). To 10 μl of a yeast culture, 500 μl Z-buffer (60 mM Na2HPO4, 40 mM NaH2PO4, 10 mM KCl, 1 mM MgSO4, 50 mM β-mercaptoethanol, pH 7), 10 μl CHCl3, and 15 μl SDS (0.2%) were added. The cells were briefly vortexed and incubated for 5 min at 28°C, and the reaction was started by the addition of 100 μl ONPG solution (4 mg/ml O-nitrophenyl β-d-galactopyranoside in Z-buffer). As soon as the solution turned yellow, the reaction was stopped by the addition of 250 μl of 2 M Na2CO3 solution. The cells were spun down, and absorption in the supernatant was measured at 420 nm. The values were normalized with the reaction time, the cell density (OD600), and the sample volume (in ml). LacZ activity (Miller units) was calculated by multiplication with 1000. For each strain three independent transformants were measured, and all experiments were performed at least twice.
FM4-64 Staining and Confocal Microscopy
Vacuoles were stained with FM4-64 as described (Vida and Emr, 1995). In short, yeast cultures were grown in YPD or YPD-Pi to an OD600 ≈ 0.5–2, and 3 OD600 units were concentrated in 100 μl YPD (or YPD-Pi) and stained at 30°C for 15–20 min with 60 μM FM4-64 (8 mM stock solution in DMSO). Cells were pelleted (700 × g, 3 min), resuspended in 5 ml YPD (or YPD-Pi), and shaken at 30°C for 45–60 min, and then the cells were washed three times with 1 ml PBS (4 mM KH2PO4, 16 mM Na2HPO4, 8.7 mM NaCl, pH 7.3) or TBS (50 mM Tris, 150 mM NaCl, pH 7.5, for cells grown in YPD-Pi). Cells were then applied to agarose-coated microscope slides (Hailey et al., 2002) and observed with a confocal laser scanning microscope (Leica DM IRBE and Leica TCS SP laser; Leica, Unterentfelden, Switzerland) using an ArKr laser at 488 and 568 nm for excitation. Pictures of the stained samples and their controls were not processed digitally except for overlay of different channels and contrast adjustments.
RESULTS
Pho84 Is the Most Important Phosphate Transporter for poly P Metabolism
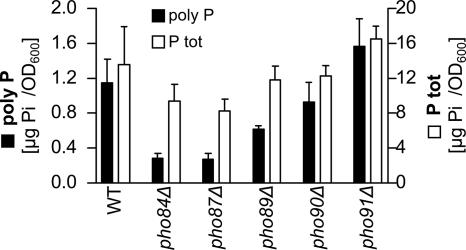
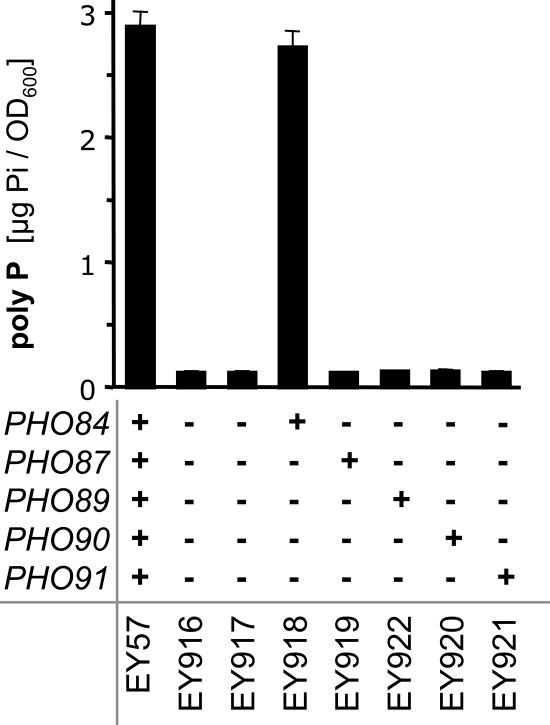
First, we quantified poly P and total phosphate in strains lacking either of the five phosphate transporters: Pho84, Pho87, Pho89, Pho90, or Pho91. Poly P levels were most strongly affected by the deletion of PHO84 and PHO87 and less so by the inactivation of PHO89 (Figure 1). The total phosphate content was reduced to a lesser extent in these three strains (Figure 1). In contrast, strains lacking either Pho90 or Pho91 did not have strongly reduced poly P levels, and the pho91Δ strain even showed a slight but not significant increase in poly P (Figure 1). To further characterize the effect of each phosphate transporter on poly P levels in yeast, the mutant strains expressing only one of the five transporters (Wykoff and O'Shea, 2001) were tested. The strain that contains Pho84 but lacks all other phosphate transporters (EY918) had poly P levels similar to the wild-type strain (EY57; Figure 2). However, none of the other phosphate transporters could support measurable amounts of poly P by itself (Figure 2). This suggested that Pho84 is the most important transporter for the maintenance of normal poly P levels.
Figure 1.
Poly P (■, left y-axis) and total phosphate (P tot, □, right y-axis) levels in single deletion strains of the five yeast phosphate transporters. The six strains were grown in YPD medium, and samples were taken at different time points. The figure shows only the measurements of the samples harvested after 4 h (late exponential phase). The bars represent the mean of four poly P extractions and quantifications, and the SD is indicated.
Figure 2.
Poly P content in yeast strains containing none or only one of the five phosphate transporters. The strains were precultured in YPGal medium, and poly P experiments were performed in YPD medium. The figure shows average values and standard deviations of three independent samples that were harvested 2 h after the transfer to YPD medium.
The PHO Pathway Affects poly P Levels by Regulating PHO84, and Low-Affinity Phosphate Transporters Limit poly P Accumulation in a pho85Δ Strain
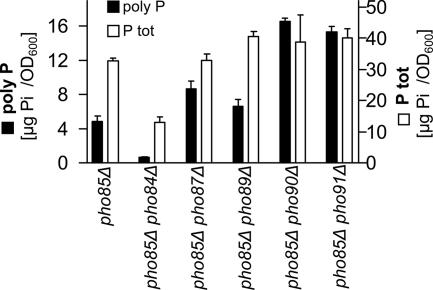
It has been shown previously that inactivation of PHO85 causes hyper-accumulation of poly P (McDonald et al., 2001). It is also known that the deletion of PHO85 results in the constitutive up-regulation of the PHO pathway and that PHO84 is one of the most strongly up-regulated genes (Ogawa et al., 2000). Therefore, we tested if the poly P hyperaccumulation phenotype of the pho85Δ strains correlated with up-regulation of PHO84. Poly P content in the double deletion strain of PHO84 and PHO85 was strongly reduced and lower than in the strain lacking only PHO84 (Figure 3). We therefore conclude that up-regulation of Pho84 is essential for poly P hyperaccumulation in the pho85Δ strain. In addition to PHO84, we also tested the influence of the other four phosphate transporters in a pho85Δ strain background. Because these transporters, except Pho87, contribute only minimally to phosphate uptake and showed a weaker poly P phenotype, it was expected that these four double deletion strains would have the same poly P phenotype as the strain with an inactivated PHO85 gene. Unexpectedly, the deletion of the different phosphate transporters in the strain lacking PHO85 had a much stronger effect on poly P levels than that observed in the single deletions of the five transporters (Figure 3). The poly P levels in the double deletions strains of PHO85 and PHO89 were little affected, but the deletion of PHO87, PHO90, or PHO91 in the pho85Δ strain background caused an increase of poly P levels (Figure 3). The pho85Δ pho87Δ strain usually showed slightly increased poly P levels by a factor of 1.5–2, while the double deletion of PHO85 with either PHO90 or PHO91 caused 3 - 4 times higher poly P content as compared with the pho85Δ strain (Figure 3). Contrary to the poly P levels, total phosphate content in these strains changed less. Consequently, the pho85Δ pho87Δ, pho85Δ pho90Δ, and the pho85Δ pho91Δ strains stored more of their total phosphate as poly P (Figure 3). From these results we conclude that the low-affinity phosphate transporters suppress poly P levels in cells with a constitutively up-regulated PHO pathway, i.e., in PHO85 mutant cells.
Figure 3.
Low-affinity phosphate transporters limit poly P accumulation in a pho85Δ strain background. Poly P content (■) and total phosphate levels (P tot, □) in double deletion strains of PHO85 and PHO84, PHO87, PHO89, PHO90, or PHO91 were determined in quadruplet samples (harvested at the 4-h time point from cultures grown in YPD), and the standard deviations are indicated.
Increased poly P Levels in Double Deletion Strains Are Not Due to the Up-Regulation of the PHO Pathway or to Posttranslational Effects on Pho84
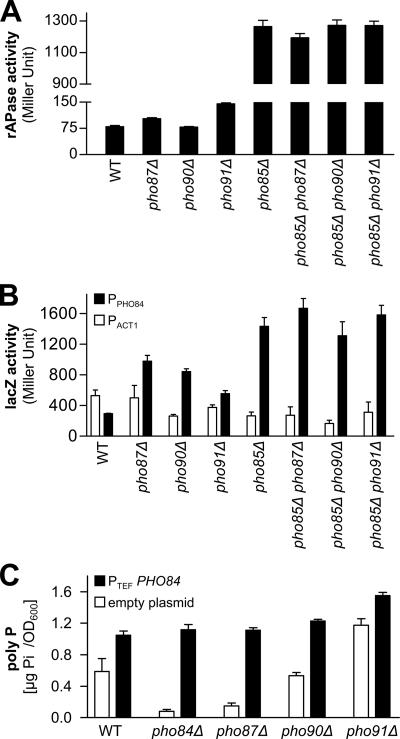
The single deletion strains of PHO90 and PHO91 were least affected in their poly P accumulation, and in the pho85Δ background all three low-affinity phosphate transporters (Pho87, Pho90, and Pho91) limited poly P levels (Figure 1). Therefore, we further characterized their effect on the metabolism of phosphate and poly P. The inhibitory effect of Pho87, Pho90, and Pho91 on poly P levels in the pho85Δ strain could have been caused by a repression of the PHO pathway and Pho84 levels or localization. As a measure for the activation of the PHO pathway, we determined rAPase activity in the single and double deletion strains that were used for the poly P measurements. In comparison to wild type, and as shown previously (Huang and O'Shea, 2005), rAPase activity was strongly increased in the pho85Δ strain, but only insignificantly higher in the pho87Δ and pho91Δ strains (Figure 4A). The double deletion strains of PHO90 or PHO91 with PHO85 secreted very similar rAPase levels as the pho85Δ strain and only the pho85Δ pho87Δ double deletion strain exhibited minimally reduced rAPase activity (Figure 4A). Neither did the double deletion of PHO85 and PHO87, PHO90 or PHO91 significantly alter transcription from the PHO84 promoter as shown by β-galactosidase activities in a LacZ-reporter assay (determined with the actin [ACT1] promoter as a reference; Figure 4B). However, the pho87Δ and pho90Δ and to a lesser extent also the pho91Δ single deletion strains showed strongly up-regulated transcription from the PHO84 promoter, even though these strains did not accumulate more poly P. This suggests that the low-affinity phosphate transporters influence PHO84 transcription by still unknown mechanism. Because Pho84 is regulated posttranslationally by internalization and degradation in the vacuole, it was tested if Pho91 represses the effect of PHO84 overexpression or affects Pho84 localization at the plasma membrane. The overexpression of PHO84 in a pho91Δ strain did not cause a stronger poly P increase (compared with the strain with the empty plasmid) than in the wild type (Figure 4C). Overexpression of PHO84 in the other two single deletion strains or in the pho84Δ strain restored wild-type poly P levels (Figure 4C). Finally, Pho84 localization at the plasma membrane was not abolished by the deletion of PHO87, PHO90, or PHO91. Pho84-GFP could be detected at the plasma membrane, inside the vacuole and presumably within the endoplasmic reticulum in cells grown in low-phosphate and in normal YPD medium, albeit the latter resulted in a much weaker signal in all compartments. However, Pho84 was slightly less frequently and less strongly internalized in the vacuole in the strains lacking PHO90 or PHO91 compared with the wild-type and the pho87Δ strain (Figure 5). On the other hand, the deletion of PHO87 had no apparent effect on the localization of Pho84-GFP under the conditions tested. Based on the rAPase measurements, the reporter gene assays and Pho84 localization, it was concluded that the low-affinity phosphate transporters Pho87, Pho90, and Pho91 only lightly affect the PHO pathway, PHO84 expression, and PHO84 function. This small effect on the PHO pathway seemed unlikely to account for the large poly P increase that was observed in the pho85Δ strain upon the deletion of PHO90 or PHO91.
Figure 4.
Deletion of PHO87, PHO90, or PHO91 in the pho85Δ background does not significantly affect the PHO pathway. (A) rAPase activity in single or double deletion strains was determined as a measure for the activity of the PHO pathway. All strains were precultured in YPD medium (3d), inoculated into fresh YPD medium, and harvested after 4 h. Four measurements were taken, and average values with their SD are represented. (B) PHO84 expression was assayed with LacZ-reporter constructs. β-Galactosidase activity is shown for the ACT1 promoter (□) and the PHO84 promoter (■). All strains were precultured for 2 d in SC-URA medium, transferred to low-phosphate SC-URA medium (100 μM Pi, 1d), and then inoculated into fresh YPD medium. Three independent transformants were assayed (at the 4-h time point), and the average values and standard deviations are shown. (C) Overexpression of PHO84 from the plasmid pRS416-TEF-HA-PHO84 in the wild-type and the deletion strains of PHO84, PHO87, PHO90, and PHO91. The poly P content in strains that were transformed with the empty plasmid pRS416-TEF-HA (□) or with the plasmid containing PHO84 (pRS416-TEF-HA-PHO84) (■). All strains were precultured in SC-URA medium and were measured after growing for 4 h in YPD medium. The bars represent average values of four extractions, and quantifications and the SD is represented.
Figure 5.
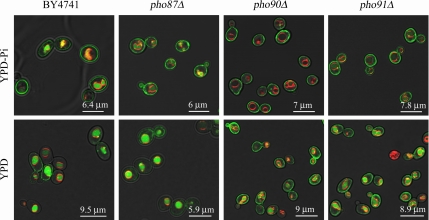
Pho84 localization at the plasma membrane is not abolished by the deletion of PHO87, PHO90, or PHO91. Localization of C-terminal GFP fusions of Pho84 in the BY4741, pho87Δ, pho90Δ, and pho91Δ strain backgrounds by confocal laser scanning microscopy. The four strains were either grown in YPD-Pi or in normal YPD medium, and vacuolar membranes were specifically stained with the dye FM4-64. Fluorescence intensity of the different mutant strains and of the cells from YPD and YPD-Pi medium cannot be compared. The laser power level was adjusted in order to visualize the Pho84-GFP fusion protein in strains with weak signal (in YPD medium, particularly in the BY4741 and the pho87Δ strain).
The Low-Affinity Transporter Pho91 Is Localized in the Vacuolar Membrane
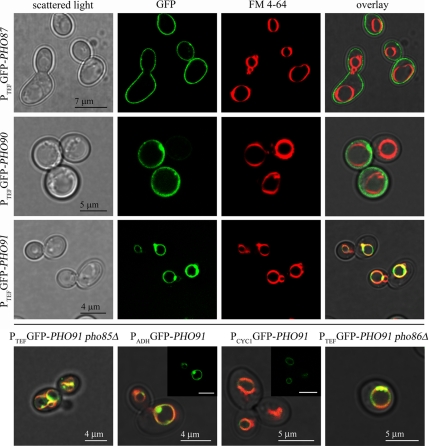
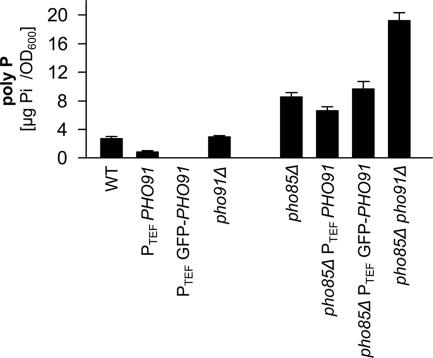
Because the low-affinity phosphate transporters Pho87, Pho90, and Pho91 affected poly P metabolism and phosphate allocation within the cell without clearly affecting the PHO pathway, we hypothesized that either of these transporters could be involved in intracellular phosphate transport. To test if any of the low-affinity phosphate transporters functions as an intracellular phosphate transporter and thereby influences poly P metabolism, we determined the localization of these three proteins by overexpressing N-terminal GFP fusions under the control of the strong ADH, the TEF and the GPD promoters, respectively. This approach was chosen because we could only observe extremely faint staining with the C-terminal GFP fusions created by Huh et al. (2003). In agreement with the assumed function as a part of a low-affinity phosphate uptake and/or sensing system, Pho87 and Pho90 localized to the plasma membrane independent of the promoter that was used (Figure 6). In contrast, Pho91 showed a distinct localization in intracellular membranes, which colocalized with FM4-64 staining (Figure 6). This colocalization of the FM4-64 signal with the staining from the GFP-Pho91 fusion protein placed the Pho91 transporter in the vacuolar membrane. GFP-Pho91 was always detected in the vacuole, irrespective of the promoter used and even when expressed from the weak CYC1 promoter (Figure 6). Furthermore, the vacuolar localization of Pho91 was independent of Pho86 (Figure 6), Pho87 or Pho90 (not shown). We therefore concluded that Pho91 is a vacuolar phosphate transporter and that the vacuolar localization was not an artifact of overexpression. In addition to the signal from the vacuolar periphery, we also observed bright fluorescence from clusters in or at the vacuolar membrane, mostly between adjacent vacuoles (Figure 6). These clusters became more abundant as the yeast cultures approached stationary phase and were less frequently found in cells grown in SC medium compared with YPD. Brightly stained clusters were also more abundant in a pho85Δ background (Figure 6). However, despite the numerous clusters at interfaces between adjacent vacuoles or vesicles in the pho85Δ strain, the GFP-Pho91 signal was still detected from the entire vacuolar periphery. These clusters could also have been an artifact from overexpression because they were less frequent if GFP-Pho91 was expressed from the weak CYC1 promoter. To test if the N-terminally GFP-tagged Pho91 protein was still functional, poly P was quantified in the wild-type and pho85Δ strains with and without the GFP tag (Figure 7). The poly P content in the PHO85 deletion strain containing the GFP-tagged Pho91 transporter increased only slightly, which indicated a functional fusion protein (Figure 7). To the contrary, the promoter exchange alone and introduction of the GFP-Pho91 fusion protein in the wild type rather reduced poly P levels (Figure 7), supposedly due to the presumptive overexpression of PHO91. This effect was much more apparent in the wild-type background (Figure 7). From these localization results it was thus concluded that active Pho91 is localized in the vacuolar membrane.
Figure 6.
The low-affinity transporter Pho91 is localized in the vacuolar membrane. Localization of N-terminal GFP fusions of Pho87, Pho90, and Pho91 by confocal laser scanning microscopy. The N-terminal GFP tag, a selection marker (natMX4) and the TEF (strong), ADH (intermediate), or CYC1 (weak) promoter were integrated into the genome by homologous recombination. Cells were grown in YPD, and as a specific marker for the vacuolar membrane, the dye FM4-64 was used. The bottom row shows only the overlay pictures of the cells expressing GFP-PHO91 from the TEF promoter in the pho85Δ or the pho86Δ background or in the wild type under the control of the ADH and the CYC1 promoters. For the latter two images the GFP channel is shown as an inlay (with the same scale bar).
Figure 7.
N-terminally GFP-tagged Pho91 is functional. The poly P content of wild-type and pho85Δ strains that express either untagged or the GFP-tagged Pho91 under the control of the TEF promoter was compared with the poly P levels of wild type, pho91Δ, pho85Δ, and pho85Δ pho91Δ. Yeast cultures were grown in YPD medium and quadruplet samples were harvested after 4 h. The bars represent average values of four poly P extractions, and quantifications and the SD is represented.
DISCUSSION
Poly P levels in yeast depend on many cellular pathways and functions such as energy metabolism, vesicle trafficking, the vtc complex, and vacuolar functions (Ogawa et al., 2000; Freimoser et al., 2006). Because of the obvious importance of phosphate uptake for the synthesis of poly P, we chose the phosphate transporters and the PHO pathway as a starting point to disentangle this complex regulation of the poly P metabolism in yeast.
To test if the five yeast phosphate transporters differentially affect poly P metabolism, we compared poly P levels in the single deletion strains of the five transporters and in strains that contain only one of all five transporters. In all media tested, the inactivation of PHO84 caused the lowest poly P total, roughly half that of the wild-type strain, and only Pho84 (and none of the other transporters) conferred a normal poly P state if present as the only phosphate transporter. Thus, Pho84 is the most important phosphate transporter for poly P metabolism; irrespective of the growth medium, growth stage, or pH of the medium (not shown).
Next, we analyzed the poly P state after deletion of either one of the five phosphate transporters in the background of a pho85Δ strain. It had previously been shown that constitutive up-regulation of the PHO pathway, by the deletion of PHO85, causes strongly increased poly P levels (McDonald et al., 2001). This increase was completely abolished by the additional deletion of PHO84, which suggested that up-regulation of PHO84 was required for poly P hyperaccumulation in the pho85Δ strain. However, deletion of many other genes that are also involved in poly P metabolism, for example, the VTC genes, also abolishes or diminishes poly P hyperaccumulation in the pho85Δ strain (data not shown). Surprisingly, the pho84Δ pho85Δ double deletion strain contained less poly P than the pho84Δ single deletion strain. This suggests that some of the (numerous) effects of a PHO85 deletion reduce poly P levels but that this phenotype is masked by the drastically increased phosphate uptake due to the up-regulation of PHO84. If Pho84 is indeed the determining transporter for poly P metabolism, the removal of any one of the other transporters (PHO87, PHO89, PHO90, or PHO91) in the pho85Δ strain should not suppress the poly P hyperaccumulation caused by the deletion of PHO85. Unexpectedly, combination of the PHO87, PHO90, or PHO91 inactivation with the PHO85 deletion strongly increased poly P hyperaccumulation compared with the pho85Δ strain. Although deletion of PHO90 and PHO91 did not have a significant effect on poly P content in the single deletion strains, it caused strong and highly significant poly P hyperaccumulation in the pho85Δ background. Interestingly, the double deletion strains also showed an altered phosphate allocation: The total phosphate content increased much less than the poly P content and consequently the proportion of phosphate fixed as poly P was ∼2–4 times higher than in the single deletion strains. Next, we attempted to further characterize the involvement of all three low-affinity transporters in phosphate and poly P metabolism.
The double deletion strains (pho85Δ pho87Δ, pho85Δ pho90Δ, and pho85Δ pho91Δ) had neither a strongly altered rAPase activity nor did they up-regulate PHO84. In addition, Pho84 localization at the plasma membrane, and internalization in the vacuole was not affected by PHO87 and only slightly dependent on PHO90 or PHO91. The pho90Δ and pho91Δ strains showed slightly less internalized Pho84 than in the wild type or in the strain lacking PHO87. This suggests that the increased poly P hyperaccumulation was not caused by a general up-regulation of the PHO pathway. In contrast, Pho90 and Pho91 affected the relative poly P content and must therefore be involved in intracellular phosphate allocation. However, a triple deletion of PHO85, PHO90, and PHO91 did not result in a further increased poly P content compared with the pho85Δ pho90Δ and the pho85Δ pho91Δ double deletion strains (not shown). Consequently, Pho90 and Pho91 do not perform redundant functions and must exert their effect on poly P metabolism by different mechanisms. We therefore conclude that Pho90 and Pho91 act independently in the regulation of phosphate metabolism or in intracellular phosphate transport.
Within the context of the global analysis of all yeast proteins Huh et al. (2003) localized Pho90 and Pho91 to the ER, and Pho87 could not be localized definitively. Therefore, there is little experimental evidence to support the widely held view that Pho87, Pho90, and Pho91 function in phosphate uptake from the environment across the plasma membrane (Wykoff and O'Shea, 2001; Auesukaree et al., 2003; Giots et al., 2003; Pinson et al., 2004). Because our poly P data could be explained by an intracellular localization and functioning of either Pho87, Pho90, or Pho91, localization of these low-affinity phosphate transporters was specifically addressed.
Although Pho87 and Pho90 were indeed clearly localized to the plasma membrane, the GFP-Pho91 fusion protein revealed a clear and unmistakable localization in the vacuolar membrane. It is therefore suggested that Pho91 serves as a vacuolar phosphate transporter that exports Pi from the vacuolar lumen to the cytosol. According to this model, a deletion of PHO91 blocks phosphate export from the vacuole, which could in turn impair degradation of the vacuolar poly P pool. A pho85Δ pho91Δ strain would thus not take up more phosphate compared with the pho85Δ single deletion strain, but more phosphate would be “trapped” in the form of poly P because of the reduced poly P degradation and phosphate export. This hypothesis also implies that the vacuolar poly P pool does not exert a strong negative feedback regulation on poly P synthesis in strains lacking PHO91. In contrast to Pho91, the observation that Pho90 limits poly P accumulation cannot be explained by its action as a phosphate transporter per se, because it was clearly localized in the plasma membrane, where it would be involved in phosphate uptake. At present, it is hypothesized that Pho90 must have a regulatory or sensory function that is involved in controlling phosphate allocation. Indeed, Pho87 or Pho90 and Pho91 have been suggested to serve as components of an external phosphate sensor (Giots et al., 2003; Pinson et al., 2004). However, more detailed studies will be necessary to identify the exact function of Pho87 and Pho90 in poly P metabolism.
In summary, we have shown that among the five described yeast phosphate transporters, Pho84 is the most important protein for poly P metabolism. In the absence of Pho84, poly P levels were reduced by at least 50%. Surprisingly, the low-affinity transporters Pho87, Pho90, and Pho91 negatively regulate poly P levels, which was most apparent in a pho85Δ background with a constitutively up-regulated PHO pathway. We could not detect a direct regulatory influence of Pho91 on the PHO pathway or on Pho84. Instead, it was found that Pho91 localizes to the vacuolar membrane. We therefore suggest that Pho91 is an intracellular phosphate transporter that affects poly P levels by regulating intracellular phosphate allocation and organellar phosphate levels.
ACKNOWLEGMENTS
We thank N. Amrhein and M. Aebi for helpful and motivating discussions and S. Zeeman and his group for providing a plate reader. Two anonymous reviewers are acknowledged for constructive criticism, helpful suggestions, and very fast reviews. E. O'Shea and D. Wykoff are given thanks for providing yeast strains and S. Fields for plasmids. This work was supported by the Swiss National Science Foundation (Grant 3100A0-112083/1) and ETH Zurich.
Abbreviations used:
- GFP
green fluorescent protein
- HA
hemagglutinin
- poly P
inorganic polyphosphate
- rAPase
acid phosphatase
- SC medium
synthetic complete medium
- YPD
yeast peptone dextrose.
Footnotes
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E07-05-0457) on September 5, 2007.
REFERENCES
- Auesukaree C., Homma T., Kaneko Y., Harashima S. Transcriptional regulation of phosphate-responsive genes in low-affinity phosphate-transporter-defective mutants in Saccharomyces cerevisiae. Biochem. Biophys. Res. Commun. 2003;306:843–850. doi: 10.1016/s0006-291x(03)01068-4. [DOI] [PubMed] [Google Scholar]
- Auesukaree C., Homma T., Tochio H., Shirakawa M., Kaneko Y., Harashima S. Intracellular phosphate serves as a signal for the regulation of the PHO pathway in Saccharomyces cerevisiae. J. Biol. Chem. 2004;279:17289–17294. doi: 10.1074/jbc.M312202200. [DOI] [PubMed] [Google Scholar]
- Bhaumik S. R., Green M. R. Differential requirement of SAGA components for recruitment of TATA-box-binding protein to promoters in vivo. Mol. Cell. Biol. 2002;22:7365–7371. doi: 10.1128/MCB.22.21.7365-7371.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brachmann C. B., Davies A., Cost G. J., Caputo E., Li J., Hieter P., Boeke J. D. Designer deletion strains derived from Saccharomyces cerevisiae S288C: a useful set of strains and plasmids for PCR-mediated gene disruption and other applications. Yeast. 1998;14:115–132. doi: 10.1002/(SICI)1097-0061(19980130)14:2<115::AID-YEA204>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- Bun-Ya M., Nishimura M., Harashima S., Oshima Y. The PHO84 gene of Saccharomyces cerevisiae encodes an inorganic phosphate transporter. Mol. Cell. Biol. 1991;11:3229–3238. doi: 10.1128/mcb.11.6.3229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Causton H. C., Ren B., Koh S. S., Harbison C. T., Kanin E., Jennings E. G., Lee T. I., True H. L., Lander E. S., Young R. A. Remodeling of yeast genome expression in response to environmental changes. Mol. Biol. Cell. 2001;12:323–337. doi: 10.1091/mbc.12.2.323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cogan E. B., Birrell G. B., Griffith O. H. A robotics-based automated assay for inorganic and organic phosphates. Anal. Biochem. 1999;271:29–35. doi: 10.1006/abio.1999.4100. [DOI] [PubMed] [Google Scholar]
- Freimoser F. M., Hürlimann H. C., Jakob C. A., Werner T. P., Amrhein N. Systematic screening of polyphosphate (poly P) levels in yeast mutant cells reveals strong interdependence with primary metabolism. Genome Biol. 2006;7:R109. doi: 10.1186/gb-2006-7-11-r109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gietz D., St. Jean A., Woods R. A., Schiestl R. H. Improved method for high efficiency transformation of intact yeast cells. Nucleic Acids Res. 1992;20:1425. doi: 10.1093/nar/20.6.1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gietz R. D., Woods R. A. Transformation of yeast by lithium acetate/single-stranded carrier DNA/polyethylene glycol method. Methods Enzymol. 2002;350:87–96. doi: 10.1016/s0076-6879(02)50957-5. [DOI] [PubMed] [Google Scholar]
- Giots F., Donaton M. C., Thevelein J. M. Inorganic phosphate is sensed by specific phosphate carriers and acts in concert with glucose as a nutrient signal for activation of the protein kinase A pathway in the yeast Saccharomyces cerevisiae. Mol. Microbiol. 2003;47:1163–1181. doi: 10.1046/j.1365-2958.2003.03365.x. [DOI] [PubMed] [Google Scholar]
- Goldstein A. L., McCusker J. H. Three new dominant drug resistance cassettes for gene disruption in Saccharomyces cerevisiae. Yeast. 1999;15:1541–1553. doi: 10.1002/(SICI)1097-0061(199910)15:14<1541::AID-YEA476>3.0.CO;2-K. [DOI] [PubMed] [Google Scholar]
- Hailey D. W., Davis T. N., Muller E.G.D. Fluorescence resonance energy transfer using color variants of green fluorescent protein. Methods Enzymol. 2002;351:34–49. doi: 10.1016/s0076-6879(02)51840-1. [DOI] [PubMed] [Google Scholar]
- Huang S., O'Shea E. K. A systematic high-throughput screen of a yeast deletion collection for mutants defective in PHO5 regulation. Genetics. 2005;169:1859–1871. doi: 10.1534/genetics.104.038695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes R. E., Lo R. S., Davis C., Strand A. D., Neal C. L., Olson J. M., Fields S. Altered transcription in yeast expressing expanded polyglutamine. Proc. Natl. Acad. Sci. USA. 2001;98:13201–13206. doi: 10.1073/pnas.191498198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huh W. K., Falvo J. V., Gerke L. C., Carroll A. S., Howson R. W., Weissman J. S., O'Shea E. K. Global analysis of protein localization in budding yeast. Nature. 2003;425:686–691. doi: 10.1038/nature02026. [DOI] [PubMed] [Google Scholar]
- Janke C., et al. A versatile toolbox for PCR-based tagging of yeast genes: new fluorescent proteins, more markers and promoter substitution cassettes. Yeast. 2004;21:947–962. doi: 10.1002/yea.1142. [DOI] [PubMed] [Google Scholar]
- Kaiser C., Michaelis S., Mitchell A. Methods in Yeast Genetics. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1994. [Google Scholar]
- Kaneko Y., Tohe A., Oshima Y. Identification of the genetic locus for the structural gene and a new regulatory gene for the synthesis of repressible alkaline phosphatase in Saccharomyces cerevisiae. Mol. Cell. Biol. 1982;2:127–137. doi: 10.1128/mcb.2.2.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kornberg A., Rao N. N., Ault-Riché D. Inorganic polyphosphate: a molecule of many functions. Annu. Rev. Biochem. 1999;68:89–125. doi: 10.1146/annurev.biochem.68.1.89. [DOI] [PubMed] [Google Scholar]
- Lee T. I., Causton H. C., Holstege F. C., Shen W. C., Hannett N., Jennings E. G., Winston F., Green M. R., Young R. A. Redundant roles for the TFIID and SAGA complexes in global transcription. Nature. 2000;405:701–704. doi: 10.1038/35015104. [DOI] [PubMed] [Google Scholar]
- Lenburg M. E., O'Shea E. K. Signaling phosphate starvation. Trends Biochem. Sci. 1996;21:383–387. [PubMed] [Google Scholar]
- Luan Y., Li H. Model-based methods for identifying periodically expressed genes based on time course microarray gene expression data. Bioinformatics. 2004;20:332–339. doi: 10.1093/bioinformatics/btg413. [DOI] [PubMed] [Google Scholar]
- Martinez P., Persson B. L. Identification, cloning and characterization of a derepressible Na+-coupled phosphate transporter in Saccharomyces cerevisiae. Mol. Gen. Genet. 1998;258:628–638. doi: 10.1007/s004380050776. [DOI] [PubMed] [Google Scholar]
- McDonald A. E., Niere J. O., Plaxton W. C. Phosphite disrupts the acclimation of Saccharomyces cerevisiae to phosphate starvation. Can. J. Microbiol. 2001;47:969–978. doi: 10.1139/w01-099. [DOI] [PubMed] [Google Scholar]
- Miller J. H. Assay of beta-galactosidase. In: Miller J. H., editor. Experiments in Molecular Genetics. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1972. pp. 352–355. [Google Scholar]
- Möckli N., Auerbach D. Quantitative beta-galactosidase assay suitable for high-throughput applications in the yeast two-hybrid system. Biotechniques. 2004;36:872–876. doi: 10.2144/04365PT03. [DOI] [PubMed] [Google Scholar]
- Mumberg D., Muller R., Funk M. Yeast vectors for the controlled expression of heterologous proteins in different genetic backgrounds. Gene. 1995;156:119–122. doi: 10.1016/0378-1119(95)00037-7. [DOI] [PubMed] [Google Scholar]
- NCRR. NCRR Yeast Resource Center > Fluorescence Microscopy > Fluorescence Resonance Energy Transfer (FRET) [Accessed January 2007]. http://depts.washington.edu/yeastrc/pages/FRET_1.html.
- Neef D. W., Kladde M. P. Polyphosphate loss promotes SNF/SWI- and Gcn5-dependent mitotic induction of PHO5. Mol. Cell. Biol. 2003;23:3788–3797. doi: 10.1128/MCB.23.11.3788-3797.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Neill E. M., Kaffman A., Jolly E. R., O'Shea E. K. Regulation of PHO4 nuclear localization by the PHO80-PHO85 cyclin-CDK complex. Science. 1996;271:209–212. doi: 10.1126/science.271.5246.209. [DOI] [PubMed] [Google Scholar]
- Ogawa N., DeRisi J., Brown P. O. New components of a system for phosphate accumulation and polyphosphate metabolism in Saccharomyces cerevisiae revealed by genomic expression analysis. Mol. Biol. Cell. 2000;11:4309–4321. doi: 10.1091/mbc.11.12.4309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersson J., Pattison J., Kruckeberg A. L., Berden J. A., Persson B. L. Intracellular localization of an active green fluorescent protein-tagged Pho84 phosphate permease in Saccharomyces cerevisiae. FEBS Lett. 1999;462:37–42. doi: 10.1016/s0014-5793(99)01471-4. [DOI] [PubMed] [Google Scholar]
- Pinson B., Merle M., Franconi J. M., Daignan-Fornier B. Low affinity orthophosphate carriers regulate PHO gene expression independently of internal orthophosphate concentration in Saccharomyces cerevisiae. J. Biol. Chem. 2004;279:35273–35280. doi: 10.1074/jbc.M405398200. [DOI] [PubMed] [Google Scholar]
- Rose M. D., Winston F., Hieter P. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1990. Methods in Yeast Genetics: A Laboratory Course Manual. [Google Scholar]
- Ruiz A., Yenush L., Arino J. Regulation of ENA1 Na+-ATPase gene expression by the Ppz1 protein phosphatase is mediated by the calcineurin pathway. Eukaryot. Cell. 2003;2:937–948. doi: 10.1128/EC.2.5.937-948.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwob E., Nasmyth K. CLB5 and CLB6, a new pair of B cyclins involved in DNA replication in Saccharomyces cerevisiae. Genes Dev. 1993;7:1160–1175. doi: 10.1101/gad.7.7a.1160. [DOI] [PubMed] [Google Scholar]
- Serrano R., Ruiz A., Bernal D., Chambers J. R., Arino J. The transcriptional response to alkaline pH in Saccharomyces cerevisiae: evidence for calcium-mediated signalling. Mol. Microbiol. 2002;46:1319–1333. doi: 10.1046/j.1365-2958.2002.03246.x. [DOI] [PubMed] [Google Scholar]
- SGD. Saccharomyces Genome Deletion Project (primers) [Accessed April 2005]; http://www-sequence.stanford.edu/group/yeast_deletion_project/Deletion_primers_PCR_sizes.txt.
- Springer M., Wykoff D. D., Miller N., O'Shea E. K. Partially phosphorylated Pho4 activates transcription of a subset of phosphate-responsive genes. PLoS Biol. 2003;1:261–270. doi: 10.1371/journal.pbio.0000028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sudarsanam P., Iyer V. R., Brown P. O., Winston F. Whole-genome expression analysis of snf/swi mutants of Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA. 2000;97:3364–3369. doi: 10.1073/pnas.050407197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas M. R., O'Shea E. K. An intracellular phosphate buffer filters transient fluctuations in extracellular phosphate levels. Proc. Natl. Acad. Sci. USA. 2005;102:9565–9570. doi: 10.1073/pnas.0501122102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong A. H., et al. Systematic genetic analysis with ordered arrays of yeast deletion mutants. Science. 2001;294:2364–2368. doi: 10.1126/science.1065810. [DOI] [PubMed] [Google Scholar]
- Vida T. A., Emr S. D. A new vital stain for visualizing vacuolar membrane dynamics and endocytosis in yeast. J. Cell Biol. 1995;128:779–792. doi: 10.1083/jcb.128.5.779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werner T. P., Amrhein N., Freimoser F. M. Novel method for the quantification of inorganic polyphosphate (iPoP) in Saccharomyces cerevisiae shows dependence of iPoP content on the growth phase. Arch. Microbiol. 2005;184:129–136. doi: 10.1007/s00203-005-0031-2. [DOI] [PubMed] [Google Scholar]
- Winzeler E. A., et al. Functional characterization of the S. cerevisiae genome by gene deletion and parallel analysis. Science. 1999;285:901–906. doi: 10.1126/science.285.5429.901. [DOI] [PubMed] [Google Scholar]
- Wykoff D. D., O'Shea E. K. Phosphate transport and sensing in Saccharomyces cerevisiae. Genetics. 2001;159:1491–1499. doi: 10.1093/genetics/159.4.1491. [DOI] [PMC free article] [PubMed] [Google Scholar]