Abstract
The Hog1 mitogen-activated protein kinase (MAPK) plays a central role in stress responses in the human pathogen Candida albicans. Here, we have investigated the MAPK kinase kinase (MAPKKK)-dependent regulation of the pathway. In contrast to the Hog1 pathway in Saccharomyces cerevisiae, which is regulated by three MAPKKKs (Ssk2, Ssk22, and Ste11), our results demonstrate that Hog1 in C. albicans is regulated by a single MAPKKK Ssk2. Deletion of SSK2 results in comparable stress and morphological phenotypes exhibited by hog1Δ cells, and Ssk2 is required for the stress-induced phosphorylation and nuclear accumulation of Hog1, and for Hog1-dependent gene expression. Furthermore, phenotypes associated with deletion of SSK2 can be circumvented by expression of a phosphomimetic mutant of the MAPKK Pbs2, indicating that Ssk2 regulates Hog1 via activation of Pbs2. In S. cerevisiae, the Hog1 pathway is also regulated by the MAPKKK Ste11. However, we can find no connection between Ste11 and the regulation of Hog1 in C. albicans. Furthermore, expression of a chimeric Pbs2 protein containing the Ste11-dependent regulatory region of S. cerevisiae Pbs2, fails to stimulate Ste11-dependent stress signaling in C. albicans. Collectively, our data show that Ssk2 is the sole MAPKKK to relay stress signals to Hog1 in C. albicans and that the MAPK signaling network in C. albicans has diverged significantly from the corresponding network in S. cerevisiae.
INTRODUCTION
Mitogen-activated protein kinase (MAPK) pathways are important signaling modules found in all eukaryotes (for review, see Pearson et al., 2001). Each pathway comprises of three tiers of protein kinases, a MAP kinase, a MAP kinase kinase (MAPKK), and a MAPKK kinase (MAPKKK). The MAPKKK phosphorylates and thereby activates the MAPKK, which subsequently phosphorylates the MAPK on conserved threonine and tyrosine residues located in the activation loop of the catalytic domain. This increases the kinase activity and the nuclear translocation of the MAPK, which results in the MAPK-dependent phosphorylation of cellular substrates, thereby triggering an appropriate cellular response to the stimulus.
One of the best-studied MAPK modules is the high osmolarity glycerol (HOG) pathway in the model yeast Saccharomyces cerevisiae that responds to changes in external osmolarity. Activation of the HOG pathway culminates in the phosphorylation, activation, and nuclear translocation of the Hog1 MAPK. Activated Hog1 mounts a multifaceted response to osmotic stress, which includes the induction or repression of the expression of various genes, regulation of protein translation, adjustments to cell cycle progression, and synthesis of the osmolyte glycerol (for review, see Hohmann, 2002; Saito and Tatebayashi, 2004).
Extracellular hyperosmolarity in S. cerevisiae is largely detected by two independent transmembrane osmosensors, Sln1 and Sho1. Signals from both these sensors converge at the MAPKK Pbs2, which activates the Hog1 MAPK by phosphorylation of conserved threonine and tyrosine residues. The Sln1 osmosensor is a membrane-located histidine kinase that regulates phosphotransfer to a response regulator, Ssk1, via the phosphorelay protein Ypd1 (Maeda et al., 1995; Posas et al., 1996). Sln1 is active under conditions of low osmolarity that results in the phosphorylation and inactivation of Ssk1. However, upon increases in osmolarity, the Sln1 histidine kinase domain is inactivated, resulting in a cessation of the Sln1–Ypd1–Ssk1 phosphorelay and an accumulation of unphosphorylated Ssk1 that can now bind and activate the two redundant MAPKKKs Ssk2 and Ssk22 (Posas and Saito, 1998). When activated the Ssk2/Ssk22 MAPKKKs specifically phosphorylate and activate the Pbs2 MAPKK (Maeda et al., 1995), by binding to an Ssk2/Ssk22-specific docking site in the N-terminal region of Pbs2 (Tatebayashi et al., 2003). The Sho1 osmosensor contains four transmembrane segments and a cytoplasmic Src homology (SH)3 domain that interacts with a proline-rich region on Pbs2. Under conditions of high osmolarity, signal transduction from Sho1 to Pbs2 also requires Cdc42, Ste20, Ste50, and Ste11 (Posas and Saito, 1997; O'Rourke and Herskowitz 1998; Posas et al., 1998, Raitt et al., 2000; Reiser et al., 2000). Cdc42 is a Rho-type small G protein that binds and activates Ste20, a p21-activated protein kinase (PAK) homologue. Ste20 in turn phosphorylates and activates the MAPKKK Ste11, which is present in a complex with the SAM-domain containing protein Ste50. Activated Ste11, in turn phosphorylates the Pbs2 MAPKK. However, in contrast to the specificity of the Ssk2/Ssk22 MAPKKKs for Pbs2, Ste11 can also phosphorylate the MAPKK Ste7 of the pheromone response and the pseudohyphal/invasive growth MAPK pathways. Significantly, there is no nonphysiological cross-talk between these MAPK pathways as the Pbs2 MAPKK and the Sho1 osmosensor act as coscaffolds to direct osmotic stress signals from Ste11 to the Hog1 MAPK (Posas and Saito, 1997; Zarrinpar et al., 2004; Tatebayashi et al., 2006).
The Sln1 and Sho1 osmosensing pathways have been shown to be largely functionally redundant in that both pathways can stimulate the phosphorylation of Pbs2, stimulate the phosphorylation and nuclear translocation of Hog1, and allow growth on high osmolarity media (Maeda et al., 1995; Posas et al., 1998). Consequently, osmotic-stress signaling to Hog1 is prevented only when both pathways are inactivated; for example, only when the Ssk2 and Ssk22 MAPKKKs are deleted in combination with the Ste11 MAPKKK are osmosensitive phenotypes and defects in osmotic stress-induced activation of Pbs2 and Hog1 observed (Posas and Saito, 1997). However, some differences between the two pathways have been identified; for example, the kinetics of Hog1 phosphorylation are different in sln1 and sho1 pathway mutants and only the Sln1 pathway responds to moderate levels of osmotic stress (Maeda et al., 1995; Van Wuytswinkel et al., 2000; O'Rourke and Herskowitz, 2004).
Homologues of S. cerevisiae Hog1 have been studied in the human fungal pathogens Cryptococcus neoformans (Bahn et al., 2005, 2006), Aspergillus fumigatus (Xue et al., 2004; Du et al., 2006), and Candida albicans (San-Jose et al., 1996). Such studies have uncovered interesting differences in the role and regulation of Hog1 in pathogenic fungi compared with that in S. cerevisiae. In C. neoformans, Hog1 responds to diverse stress conditions, and it also has important roles in drug sensitivity, sexual reproduction, and virulence (Bahn et al., 2005). Intriguingly, Hog1 is constitutively phosphorylated under normal growth conditions in the more virulent serotype A, but not the less virulent serotype D, strains. This unusual constitutive phosphorylation of Hog1 is thought to result in cross-talk with signaling cascades regulating C. neoformans virulence factors (Bahn et al., 2005). With regard to the relay of stress signals to Hog1, the response regulator Ssk1 plays a major role (Bahn et al., 2006), and although C. neoformans does not contain a homologue of the S. cerevisiae Sln1 histidine kinase, seven other histidine kinases (Tco1-7) have been identified. Recently, Tco1 and Tco2 have been demonstrated to have overlapping and distinct roles in the regulation of Hog1 (Bahn et al., 2006). In A. fumigatus, the Hog1 homologue SakA is activated in response to oxidative and osmotic stress, and it also negatively regulates conidial germination under nitrogen-limiting conditions (Xue et al., 2004; Du et al., 2006). In contrast to S. cerevisiae, deletion of the Sln1 histidine kinase homologue tscB, in A. fumigatus, is viable, and does not impair stress signaling to Hog1 (Du et al., 2006).
C. albicans HOG1 was originally cloned by functional complementation of the osmosensitive phenotype associated with S. cerevisiae hog1 mutant cells (San-Jose et al., 1996), and it is essential for the virulence of this pathogen (Alonso-Monge et al., 1999; Arana et al., 2007). Subsequent studies revealed that the C. albicans Hog1 MAPK responds to a diverse range of environmental signals, in addition to osmotic stress, including various oxidative stress agents (Alonso-Monge et al., 2003; Smith et al., 2004). Recently, homologues of the S. cerevisiae PBS2, SSK1, and SHO1 genes have been identified in C. albicans, and their potential role in the relay of stress signals to Hog1 examined. Deletion of PBS2 in C. albicans prevents the relay of both osmotic and oxidative stress signals to Hog1, indicating this to be the sole MAPKK that phosphorylates Hog1 in response to these stresses (Arana et al., 2005). In contrast, deletion of SSK1 in C. albicans prevents activation of Hog1 in response to oxidative stress, whereas wild-type levels of Hog1 phosphorylation are induced in ssk1Δ cells after osmotic stress (Chauhan et al., 2003). Furthermore, osmotic stress-induced activation of Hog1 is not impaired in an ssk1sho1 double mutant in C. albicans (Roman et al., 2005). This contrasts with the observation in S. cerevisiae where inactivation of both the Sho1- and Sln1-osmosensing branches prevents the relay of osmotic stress signals to Hog1 (Maeda et al., 1995).
Here, we describe our studies seeking to identify the MAPKKKs that relay stress signals to the Pbs2 MAPKK in C. albicans. We have identified putative homologues of the S. cerevisiae Ssk2,22 and Ste11 MAPKKKs and constructed deletions in these genes. Importantly, our results reveal that the Ssk2 MAPKKK functions alone to relay stress signals to Hog1 in C. albicans. This is in stark contrast with the situation in S. cerevisiae in which the Ssk2, Ssk22, and Ste11 MAPKKKs function in overlapping pathways to relay osmotic stress signals to Hog1. The potential significance of this rewiring of Hog1 signaling components in C. albicans is discussed.
MATERIALS AND METHODS
Strains and Growth Conditions
The strains used in this study are given in Table 1. C. albicans strains were grown in either rich YPD medium or SD minimal medium (Sherman, 1991). All strains were grown at 30°C. Cell morphology was analyzed using a Zeiss axioscope (Carl Zeiss, Jena, Germany).
Table 1.
Strains used in this study
| Strain | Genotype | Reference |
|---|---|---|
| BWP17 | ura3::λ imm434/ura3::λimm434, his1::hisG/his1::hisG, arg4::hisG/arg4::hisG | Wilson et al. (1999) |
| JC47 | hog1::loxP-ARG4-loxP/hog1::loxP-HIS1-loxP | Enjalbert et al. (2006) |
| JC74 | pbs2::loxP-ARG4-loxP/pbs2::loxP-HIS1-loxP | This work |
| JC482 | ssk2::loxP-ARG4-loxP/ssk2::loxP-HIS1-loxP | This work |
| JC524 | ste11::loxP-ARG4-loxP/ste11::loxP-HIS1-loxP | This work |
| JC620 | ssk2::loxP-ARG4-loxP/ssk2::loxP-HIS1-loxP, Cip20-SSK2(URA3,HIS1) | This work |
| JC638 | ste11::loxP-ARG4-loxP/ste11::loxP-HIS1-loxP, Cip20-STE11(URA3,HIS1) | This work |
| JC112 | PBS2-HM:URA3 | This work |
| JC517 | ssk2::loxP-ARG4-loxP/ssk2::loxP-HIS1-loxP, PBS2-HM:URA3 | This work |
| JC507 | ssk2::loxP-ARG4-loxP/ssk2::loxP-HIS1-loxP, PBS2DD-HM:URA3 | This work |
| JC167 | pbs2::loxP-ARG4-ura3-loxP/pbs2::loxP-HIS1-loxP, CaEXP-CaPBS2 (URA3) | This work |
| JC559 | pbs2::loxP-ARG4-ura3-loxP/pbs2::loxP-HIS1-loxP, CaEXP-ScCaPBS2 (URA3) | This work |
| JC613 | ssk2::loxP-ARG4-loxP/ssk2::loxP-HIS1-loxP, CaEXP-ScCaPBS2 (URA3) | This work |
| JC54 | HOG1-YFP-URA3 | This work |
| JC522 | ssk2::loxP-ARG4-loxP/ssk2::loxP-HIS1-loxP, HOG1-YFP-URA3 | This work |
| JC545 | ste11::loxP-ARG4-loxP/ste11::loxP-HIS1-loxP, HOG1-YFP-URA3 | This work |
All strains were constructed in the BWP17 background (ura3::λ imm434/ura3::λimm434, his1::hisG/his1::hisG, arg4::hisG/arg4::hisG).
Strain Construction
Oligonucleotide primers used in this study are listed in Table 2.
Table 2.
Oligonucleotides used in this study
| Oligonucleotide | Sequence 5′–3′ |
|---|---|
| PBS2delF | atggttgaagataaagatatagacttgaatattaataatttgaaaattcacgatgcgccaacacgaacaccaccggtaagccagggttttcccagtcacg |
| PBS2delR | ctgtttctatataatatactgtttataatacagcccaataacctgggcttcatattcatcaatgattattaagaaagcttcctcactaaagggaacaaaagc |
| SSK2delF | gaatacatttcacaactactactgctaatagttcaattggaaaagggcccaattaatttacaatttataattatcttataccagggttttcccagtcacg |
| SSK2delR | gtagaggtccctgtaatggcttgtggtgacatgaacatcagtgtacccgtcctggaattcaaattattatggccatcacgcctcactaaagggaacaaaagc |
| STE11delF | tcatatagaaaccctaatacattagttcgtgtgtatatggtactcaaatctaacaaacagtcatgacagagattaatgatccagggttttcccagtcacg |
| STE11delR | aaacatctctctagattgttcctactacgtcatacaacaatcacgtggtacagtataactttatatatttatcattgtttcctcactaaagggaacaaaagc |
| SSK2BglF | aaggaagatctgtcaattaaattcctattcactcc |
| SSK2BglR | aaggaagatcttaataaaataaggggagaatagagtc |
| STE11BamF | gcgcggatccaaaaccccgaaagttacaccagg |
| STE11BamR | gcgcggatccgattcaaaaagcttagccctccc |
| PBS2MHR | gaattcgctagcttaatgatggtgatgatgctctaagtcctcctcgctgatcaatttttgttcttcagccatggacaaatcttcttcagaaattaacttttgctcctcatgattattaagaaagcttct |
| PBS2PstA | cgtcatctgcagcacgatgcgccaacacgaacacc |
| PBS2PstB | cgtcatctgcagctatatcagcttcaacatcgg |
| PBS2TagR | gaggatccccggggacacgagaaaatattatcatatgtttcagca |
| PBS2DD | ggtaatttagttgccgatttagccaaagataatattggttgtcaa |
| PBS2KDF | gcgcggatccggtttgtttgctaattattctgat |
| PBS2KDR | aaggaagatctttaatgatggtgatgatggtgtaagtcctcc |
| PBS2FLF | gcgcggatccatggttgaagataaagatatagac |
| PBS2FLR | gcgcggatccttaatggtgatgatggtctaagtcctcc |
| 127MUTF | agcactcttaagaacgttctcgacaatc |
| 180MUTR | ccttctgttaggattgagcaattgcgc |
| ScPBS2F | gcgcggatccatggaagacaagtttgctaacctcag |
| ScPBS2R | gcgcggatccaccaccactactaccagaatttgaagag |
Deletion of SSK2, STE11, and PBS2
To delete SSK2, disruption cassettes comprising either the ARG4 or HIS1 gene flanked by loxP sites and 80 base pairs of DNA sequence corresponding to regions 5′ and 3′ of the SSK2 open reading frame (ORF) were generated by polymerase chain reaction (PCR) using the oligonucleotide primers SSK2delF and SSK2delR and the plasmid templates pLAL2 or pLHL2, respectively (Dennison et al., 2005). Disruption cassettes were transformed into C. albicans BWP17 (Wilson et al., 1999) to sequentially disrupt both alleles of SSK2 and generate strain JC482. The same strategy was used to delete STE11 and PBS2 by using the oligonucleotide primers STE11delF and STE11delR, or PBS2delF and PBS2delR, to generate strains JC524 or JC74, respectively. The SSK2 disruption cassettes deleted codons 1-1344 of the 1483 codon ORF of SSK2, the STE11 disruption cassettes deleted codons 6–820 of the 823 codon STE11 ORF, and the PBS2 disruption cassettes deleted codons 28–539 of the 545 codon PBS2 ORF. Gene disruptions were confirmed by PCR. To construct reintegrant control strains, the SSK2 gene plus 550 base pairs of upstream and 200 base pairs of downstream sequences (total 5.2 kb), and the STE11 gene plus 999 base pairs of upstream and 232 base pairs of downstream sequences (total 3.7 kb), were amplified by PCR by using the oligonucleotide primers SSK2BglF and SSK2BglR or STE11BamF and STE11BamR, respectively, and cloned into the BamHI site of CIp20, a derivative of CIp10 (Murad et al., 2000). The resulting plasmids were digested with StuI and introduced into the Ura− null strains.
Tagging and Mutagenesis of PBS2
To chromosomally tag Pbs2 at the C terminus with 6-His residues and two copies of the myc-epitope, the PBS2 gene was amplified by PCR by using the oligonucleotide primers PBS2MHR and PBS2PstA and ligated into CIp-C-ZZ (Blackwell et al., 2003), which had been digested with PstI and NheI to remove the TEV-protein A sequence. The resulting CIp-C-PBS2HM plasmid was linearized by digestion with SgrAI to target integration at the PBS2 locus in C. albicans ssk2Δ cells to generate strain JC517. Chromosomal insertion of the C-terminal 6His-myc tag was confirmed by PCR and DNA sequencing. Mutagenesis of PBS2 to create PBS2S355D,T359D was performed by overlapping PCR using the oligonucleotides PBS2TagR, PBS2DD, and PBS2PstB, and the resulting PCR fragment was digested with PstI and NdeI and cloned into CIp-C-PBS2HM previously digested with the same enzymes to remove the wild-type sequence. The resulting CIp-C-PBS2DDHM plasmid was introduced into ssk2Δ cells as described above to generate strain JC507.
To express a chimera of PBS2 in C. albicans made up of the kinase domain of C. albicans PBS2 fused to the N-terminal region of S. cerevisiae PBS2, initially the C. albicans PBS2 kinase domain was amplified by PCR by using the oligonucleotide primers PBS2KDF and PBS2KDR and CIp-C-PBS2HM as a template. The resulting PCR product was digested with BglII and BamHI and ligated into the BamHI site of CaEXP (Care et al., 1999), to generate plasmid CaEXP-CaPBS2kin. The N-terminal region of S. cerevisiae PBS2 contains two CUG (Ser) codons at positions 127 and 180, which would be read as Leu codons in C. albicans. Hence, these were mutated to CUC (Ser) codons by overlapping PCR by using the oligonucleotides 127MUTF, 180MUTR, ScPBS2F, and ScPBS2R. The resulting 950-base pair fragment was digested with BamHI and ligated into CaEXP-CaPBS2kin to generate CaEXP-ScCaPBS2. As a control, the full-length C. albicans PBS2 sequence was amplified by PCR using the oligonucleotide primers CaPBS2FLF and CaPBS2FLR and CIp-C-PBS2HM as a template, and ligated into the BamHI site of CaEXP to generate plasmid CaEXP-CaPBS2. The CaEXP-ScCaPBS2 and CaEXP-CaPBS2 plasmids were linearized at the StuI restriction site in the RSP10 locus and introduced into pbs2Δ and ssk2Δ cells. CaEXP directs the expression of genes under the control of the MET3 promoter, which is repressed in the presence of methionine and cysteine (Care et al., 1999). Hence, strains carrying CaEXP derivatives were grown in minimal SD media lacking methionine and cysteine.
Chromosomal Tagging of HOG1
To chromosomally tag Hog1 with yellow fluorescent protein (YFP) in ssk2Δ and ste11Δ cells, HOG1-specific sequences were added to the universal primer sequences described previously (Gerami-Nejad et al., 2001) to generate the oligonucleotide primers HOG1YFPF and HOG1YFPR. These primers were used in combination with the plasmid template pYFP-URA3 to generate a HOG1-YFP cassette by PCR (Smith et al., 2004). This cassette was introduced into JC482 (ssk2Δ) and JC524 (ste11Δ) cells to create strains JC522 and JC545, respectively. Integration at the HOG1 locus was confirmed by PCR and DNA sequencing.
Stress Sensitivity Tests
For stress sensitivity assays, strains to be tested were grown in liquid culture and spotted onto YPD or SD plates containing the indicated compounds. Plates were incubated at 30°C for 24 h, unless indicated otherwise.
Hog1 Phosphorylation Assays
Cells were grown to mid-exponential phase and exposed to a range of stress conditions for the indicated times. Protein extracts were prepared and phosphorylated Hog1 was detected by Western blot with an anti-phospho-p38 antibody (New England Biolabs, Ipswich, MA) as described previously (Smith et al., 2004). Blots were stripped and total levels of Hog1 were determined by probing with an anti-Hog1 antibody (Santa Cruz Biotechnology, Santa Cruz, CA).
Fluorescence Microscopy
Localization of chromosomally YFP-tagged Hog1 was determined by fluorescence microscopy as described previously (Smith et al., 2004).
RNA Analysis
Preparation and Northern blot analysis of RNA isolated from C. albicans were performed as described previously (Enjalbert et al., 2006). Gene-specific probes were amplified by PCR from genomic DNA by using oligonucleotide primers specific for GPD2, RHR2, and ACT1 (Enjalbert et al., 2006).
Coprecipitation Assays
Exponentially growing wild-type (BWP17) and ssk2Δ (JC482) cells, or wild-type (JC112) and ssk2Δ (JC517) cells expressing chromosomally tagged PBS2-6His-myc, were harvested and snap frozen in liquid nitrogen. Pelleted cells were lysed into lysis buffer (50 mM Tris-HCl, pH 7.5, 250 mM NaCl, 20 mM imidazole, 0.1% NP-40, 50 mM NaF, 2 mM NaF, 1 mM phenylmethylsulfonyl fluoride, 0.07 trypsin inhibition unit/ml aprotinin, 10 μg/ml leupeptin, and 10 μg/ml pepstatin). Pbs2-6His-myc was precipitated from extracts by using nickel-nitrilotriacetic acid (Ni-NTA) agarose (QIAGEN, Dorking, Surrey, United Kingdom). The agarose beads were washed five times with lysis buffer and resuspended in SDS-loading buffer. Proteins were resolved on SDS-polyacrylamide gel electrophoresis (PAGE) and Western blotting was performed using an anti-myc antibody (Sigma Chemical, Poole, Dorset, United Kingdom) to detect precipitation of Pbs2-6His-myc, and coprecipitation of Hog1 was monitored using an anti-Hog1 antibody (Santa Cruz Biotechnology).
RESULTS
C. albicans ssk2Δ Cells Are Phenotypically Comparable to hog1Δ Cells
In S. cerevisiae, Ssk2/22 and Ste11 function in overlapping pathways to relay osmotic stress signals to the Pbs2-Hog1 module (Posas and Saito, 1997). A search of the C. albicans genome database (http://www.candidagenome.org/genome) revealed one locus designated SSK2 (orf19.3775), encoding an 1344-codon open reading frame, with significant similarity to S. cerevisiae SSK2 and SSK22, and one locus designated STE11 (orf19.844), encoding an 823-codon open reading frame, with significant similarity to S. cerevisiae STE11. To avoid confusion, we use the prefixes Ca and Sc to distinguish between the C. albicans and S. cerevisiae SSK2 and STE11 orthologues in this article.
The primary amino acid sequences of ScSte11 and ScSsk2 were aligned with the putative C. albicans orthologues using ClustalW (Thompson et al., 1994). Alignment of the ScSte11and the CaSte11 sequences revealed significant similarity (65%) across the entire length of the protein (our unpublished data). However, whilst alignment of ScSsk1 with CaSsk2 revealed a high degree of similarity, the C-terminal region of ScSsk2, containing the kinase activation loop and subdomains VIII to XI, was absent in the CaSsk2 sequence. To clarify this, we sequenced the CaSSK2 allele in the laboratory strain BWP17 (Wilson et al., 1999), and we discovered that codon 1345, annotated as a STOP codon in the C. albicans genome database, encodes a lysine residue and that the CaSSK2 ORF in the BWP17 background extends to 1483 codons (Supplemental Figure 1). We also found that the stop codon at position 430, which is annotated as separating two exons of CaSSK2 (1-1287 and 1291-4038; http://www.candidagenome.org/genome), encodes a lysine residue.
The extended CaSsk2 sequence in BWP17 contains the kinase activation loop and subdomains VIII to XI. Importantly, Thr1460 in the ScSsk2 kinase activation loop, which has been shown to be important for the activation of the kinase (Posas and Saito, 1998), is conserved in the extended CaSsk2 sequence (Thr1359). Furthermore, a region at the N terminus of ScSsk2 (residues 294-413), which binds to the upstream response regulator ScSsk1 (Posas and Saito, 1998), is also conserved in the CaSsk2 sequence (Supplemental Figure 1).
To examine the function of CaSsk2 and CaSte11 in C. albicans, homozygous null mutants were generated. Each copy of the CaSSK2 allele in this diploid fungus was inactivated by replacing 1344 codons of the 1483-codon ORF with either the HIS1 or ARG4 disruption cassette. Similarly, each CaSTE11 allele was inactivated by replacing the majority of the 823 codon ORF with either the HIS1 or ARG4 disruption cassette (see Materials and Methods). Previous studies have shown that deletion of C. albicans HOG1 results in cells with pleiotropic stress phenotypes (San-Jose et al., 1996; Alonso-Monge et al., 2003; Smith et al., 2004). Hence, to examine the potential role of the CaSsk2 and CaSte11 MAPKKKs in relaying stress signals to Hog1, we compared the sensitivity of C. albicans ssk2Δ, ste11Δ, and hog1Δ cells to a range of stress conditions. Due to the functional redundancy of ScSsk2 and ScSte11 in S. cerevisiae, we were surprised to find that deletion of CaSSK2 resulted in comparable stress-sensitive phenotypes to those exhibited by C. albicans hog1Δ cells. Both hog1Δ and ssk2Δ mutants displayed impaired growth in response to the oxidative stress agents H2O2, tert-butyl hydroperoxide (t-BOOH), and menadione, the osmotic stress agents sorbitol and NaCl, the heavy metals Cd2+ and As3+, the purine analogue caffeine, and the drug staurosporine (Figure 1A). Furthermore, hog1Δ and ssk2Δ cells were equally sensitive to a range of concentrations of osmotic and oxidative stress agents (Supplemental Figure 2), verifying that these mutants display equivalent stress phenotypes. C. albicans hog1Δ and ssk2Δ cells also displayed similar increased resistance, compared with wild-type cells, to the cell wall biogenesis inhibitor calcofluor white (Figure 1A). Hyperactivation of the MAPK Cek1 in C. albicans hog1Δ cells is thought to underlie the resistance to cell wall inhibitors such as calcofluor white (Eisman et al., 2006). Consistent with this, we found that ssk2Δ cells, like hog1Δ cells, display a high basal level of phosphorylation of the Cek1 MAPK (our unpublished data). Importantly, all of the stress-phenotypes exhibited by ssk2Δ cells were reversed upon reintroduction of the wild-type SSK2 gene into the ssk2Δ strain (Figure 1A).
Figure 1.
ssk2Δ and hog1Δ cells display similar phenotypes. (A) Approximately 103 cells, and 10-fold dilutions thereof, of exponentially growing wild-type (Wt-BWP17), hog1Δ (JC47), ssk2Δ (JC482), ssk2Δ+SSK2 (JC620), ste11Δ (JC524) and ste11Δ+STE11 (JC637) strains were spotted onto YPD plates containing the indicated additives: NaCl (0.6 M), sorbitol (1.0 M), t-BOOH (2.0 mM), menadione (0.35 mM), H2O2 (3 mM), CdSO4 (0.4 mM), Na2As3 (2 mM), staurosporine (2.0 μM), caffeine (10 mM), and calcofluor white (CFW; 20 μg/ml). Plates were incubated at 30°C for 24 h. (B) Morphology of wild-type (Wt), hog1Δ, ssk2Δ, and ssk2Δ+SSK2 strains grown in YPD at 30°C. (C) Northern analysis of hyphae-associated transcripts (HWP1 and ECE1) in wild-type (Wt), hog1Δ, and ssk2Δ strains grown in YPD at 30°C. ACT1 was analyzed as a loading control.
In contrast, C. albicans ste11Δ cells did not demonstrate the stress-sensitive phenotypes exhibited by ssk2Δ and hog1Δ cells, and overall displayed wild-type levels of stress resistance to the compounds tested (Figure 1A). However, we did find that ste11Δ cells are more sensitive than wild-type cells to the cell wall-damaging agent calcofluor white, and this sensitivity can be reversed by reintegration of the wild-type gene (Figure 1A). This phenotype contrasts with that seen with ssk2Δ and hog1Δ cells, which display significant resistance to this agent (Figure 1A).
Previous work has shown that Hog1 also functions as a repressor of morphogenetic switching in C. albicans (Alonso-Monge et al., 1999; Arana et al., 2005; Eisman et al., 2006; Enjalbert et al., 2006). For example, hog1Δ mutant cells form filaments in the absence of any morphogenetic signal (Enjalbert et al., 2006). Significantly, such morphogenetic defects were also observed in ssk2Δ cells but not ste11Δ cells; similar levels of filamentation were observed in hog1Δ and ssk1Δ cells when cultured in YPD, and reintroduction of SSK2 into ssk2Δ cells reversed this morphogenetic defect (Figure 1B). Furthermore, consistent with Hog1 and Ssk2 functioning as negative regulators of filamentation, we found that the hypha-specific genes HWP1 and ECE1 were up-regulated in hog1Δ and ssk2Δ cells, compared with wild-type cells, under noninducing conditions (Figure 1C).
These data, which collectively illustrate that C. albicans hog1Δ and ssk2Δ cells display equivalent phenotypes, indicate that CaSsk2 functions independently of CaSte11 to relay signals to Hog1 in C. albicans. This contrasts with the situation in S. cerevisiae in which ScSsk2 and ScSsk22 function redundantly with the ScSte11 MAPKKK to regulate the Hog1 MAPK (Posas and Saito, 1997).
CaSsk2, but Not CaSte11, Is Required for the Activation of Hog1 in C. albicans
Exposure of C. albicans to a range of stress conditions results in the phosphorylation and nuclear translocation of Hog1 (Alonso-Monge et al., 2003, Smith et al., 2004, Enjalbert et al., 2006). We explored the effect of deleting CaSSK2 and CaSTE11 on each of these processes. To investigate whether CaSsk2 or CaSte11 regulates Hog1 phosphorylation in response to stress, wild-type, ssk2Δ and ste11Δ cells were treated with osmotic stress (0.3 M NaCl), or oxidative stress (5 mM H2O2), and Hog1 phosphorylation was monitored by Western blotting with an antibody that recognizes only the active phosphorylated form of Hog1 (Millar et al., 1995; Smith et al., 2004). Blots were subsequently reprobed with an anti-Hog1 antibody to determine total levels of Hog1. As illustrated in Figure 2A, Hog1 was rapidly phosphorylated in extracts isolated from wild-type cells and ste11Δ cells after exposure to osmotic or oxidative stress. In contrast, phosphorylated Hog1 could not be detected in extracts isolated from ssk2Δ cells after equivalent stress treatments. However, Hog1 phosphorylation was restored in ssk2Δ upon reintegration of the wild-type CaSSK2 gene (Figure 2A). Increased exposure of the Western blots also revealed that CaSsk2 is required for the low basal level of Hog1 phosphorylation seen in wild-type cells under nonstressed conditions (Figure 2B). Importantly, deletion of CaSSK2 does not, however, affect the total cellular levels of Hog1, because equivalent amounts of Hog1 are present in both wild-type and ssk2Δ cells (Figure 2).
Figure 2.
CaSsk2 but not CaSte11 is required for Hog1 phosphorylation. (A) Western blot analysis of whole cell extracts isolated from wild-type (Wt-BWP17), ssk2Δ (JC482), ssk2Δ+SSK2 (JC620), and ste11Δ (JC524) cells after treatment with either 5 mM H2O2 or 0.3 M NaCl for the specified times. Western blots were probed with an anti-phospho-p38 antibody, which only recognizes the phosphorylated, active form of C. albicans Hog1 (Hog1-P). Total levels of Hog1 protein was determined by stripping and reprobing the blot with an anti-Hog1 antibody that recognizes both phosphorylated and unphosphorylated forms of Hog1 (Hog1). (B) Western blot analysis of whole cell extracts isolated from unstressed Wt and ssk2Δ cells.
Studies in S. cerevisiae have uncovered concentration-dependent differences in the activation of the two osmosensing branches (Maeda et al., 1995; Van Wuytswinkel et al., 2000; O'Rourke and Herskowitz, 2004); hence, we also compared the levels of Hog1 phosphorylation in wild-type, ssk2Δ, and ste11Δ cells in response to increased levels of osmotic stress (1 M NaCl). However, even at elevated levels of osmotic stress, no significant activation of Hog1 was observed in extracts isolated from ssk2Δ cells and wild-type levels of Hog1 phosphorylation were detected in extracts isolated from ste11Δ cells (Supplemental Figure 3).
To determine the effect of deleting CaSSK2 or CaSTE11 on the stress-induced nuclear accumulation of Hog1, one copy of the HOG1 gene was chromosomally tagged with YFP in ssk2Δ and ste11Δ cells, and the cellular localization of Hog1-YFP was examined by fluorescence microscopy. Our earlier work validated the use of Hog1-YFP in examining the cellular localization of Hog1 (Smith et al., 2004). As observed previously, Hog1-YFP rapidly accumulated in the nucleus in wild-type cells after osmotic (0.3 M NaCl) and oxidative (5 mM H2O2) stress treatments (Smith et al., 2004). Osmotic and oxidative stress-induced nuclear accumulation of Hog1 was also evident in ste11Δ cells. However, in contrast, no stress-induced nuclear accumulation of Hog1-YFP was observed in ssk2Δ cells (Figure 3).
Figure 3.
CaSsk2 but not CaSte11 is required for Hog1 nuclear localization. The localization of YFP-tagged Hog1 was determined by fluorescence microscopy in wild-type (Wt-JC54), ssk2Δ (JC522), and ste11Δ (JC545) cells before stress and after treatment with either 5 mM H2O2 or 0.3 M NaCl for 10 min (Hog1-YFP). The position of the nuclei in these cells was visualized with 4,6-diamidino-2-phenylindole (DAPI). An autofluorescence control is shown (control).
Because the above-mentioned data implicate CaSsk2, but not CaSte11, in the activation of Hog1, we examined the role of CaSsk2 in the regulation of Hog1-dependent gene expression. Northern analyses were performed to compare RHR2 and GPD2 expression in wild-type, hog1Δ, and ssk2Δ cells after exposure to oxidative (5 mM H2O2) and osmotic (0.3 M NaCl) stress. Consistent with previous studies (Smith et al., 2004; Enjalbert et al., 2006), RHR2 and GPD2 were found to be induced in response to both osmotic and oxidative stress in a Hog1-dependent manner (Figure 4). However, deletion of CaSSK2 impaired the osmotic and oxidative stress-induced expression of RHR2 and GPD2 compared with wild-type cells. Furthermore, quantification of mRNA levels illustrated that the expression profiles of RHR2 and GPD2 were almost identical in hog1Δ and ssk2Δ cells (Figure 4).
Figure 4.
CaSsk2 is required for the activation of Hog1-dependent gene expression. (A) Northern blot analysis of RNA isolated from mid-log cultures of Wt (BWP17), hog1Δ (JC47), and ssk2Δ (JC482) cells treated with either 5 mM H2O2 or 0.3 M NaCl for 0, 10, 30 and 60 min by using probes specific for RHR2, GPD2, and ACT1 (loading control). Fold-induction of the ratios of RHR2 and GPD2 to ACT1 were calculated relative to time 0 for the wild-type strain.
Collectively, these experiments show that CaSsk2 is required for phosphorylation of Hog1, the nuclear accumulation of Hog1, and the induction of Hog1-dependent gene expression in response to osmotic and oxidative stress in C. albicans.
Hog1 Forms a Complex with the MAPKK Pbs2 Independently of CaSsk2 in C. albicans
Pbs2 has recently been identified as the sole MAPKK that relays stress signals to Hog1 in C. albicans (Arana et al., 2005). Similarly, as with Hog1, the levels of Pbs2 were not affected upon deletion of CaSSK2 (Figure 5). We had previously found that Pbs2 forms a complex with Hog1 in C. albicans (our unpublished data); hence, we investigated whether CaSsk2 was necessary for this interaction. Such experiments were performed using wild-type and ssk2Δ strains in which Pbs2 was C-terminally tagged with six histidine residues and two copies of the myc epitope (Pbs2-6His-myc). Pbs2-6His-myc was precipitated from cell extracts using Ni-NTA-agarose, and coprecipitation of Hog1 was assayed by Western blotting using an anti-Hog1 antibody (α-Hog1). As illustrated in Figure 5, Hog1 coprecipitated with Pbs2-6His-myc purified from both wild-type and ssk2Δ cell extracts. Collectively, these results indicate that the loss of Hog1 signaling seen in ssk2Δ cells (Figures 2–4) is not due to a decrease in the stability of the downstream kinases Pbs2 and Hog1, nor is it due to Ssk2 functioning as a scaffold protein to promote interaction of Pbs2 with Hog1 in vivo.
Figure 5.
CaSsk2 is not essential for the interaction of Hog1 with Pbs2. Coprecipitation experiments to examine the association of Hog1 with Pbs2 in wild-type and ssk2Δ cells. Extracts were prepared from mid-log cultures of Wt (BWP17) and ssk2Δ (JC482) cells, and wild-type (JC112) and ssk2Δ (JC517) cells expressing Pbs2-6His-myc. The 6His-myc-tagged Pbs2 protein was purified with Ni-NTA agarose, and coprecipitation of Hog1 was analyzed by Western blotting with an anti-Hog1 antibody (α-Hog1). Blots were reprobed with an anti-myc antibody (α-myc) to allow comparison of the levels of Pbs2-6His-myc precipitated from Wt and ssk2Δ cells.
Phenotypes Associated with Deletion of CaSSK2 Can Be Overcome by Mutating the Pbs2 Phosphorylation Sites to Phosphomimetic Residues
Several protein kinases, including MAPKKs and MAPKs, are activated by phosphorylation between the kinase subdomains VII and VIII (Johnson et al., 1996). Two such phosphorylation sites have been identified in the S. cerevisiae Pbs2 (Maeda et al., 1995) and S. pombe Wis1 MAPKKs (Shiozaki et al., 1998), and significantly both of these sites (Ser355 and Thr359) are conserved in C. albicans Pbs2 (Figure 7). Because the above-mentioned data indicate that CaSsk2 is the MAPKKK that activates the Hog1 pathway in C. albicans, we predicted that mutation of the conserved phosphorylation sites of Pbs2 to phosphomimetic residues (aspartic acid residues) would bypass the requirement for CaSsk2. To test this prediction, ssk2Δ strains were created which expressed either a wild-type PBS2 allele or a PBS2DD allele in which the putative phosphorylation sites, Ser355 and Thr359, were mutated to aspartic acid residues. Both the PBS2 and PBS2DD alleles were expressed from the native PBS2 locus and tagged with a sequence encoding six histidine residues and two copies of the myc epitope located just before the termination codon, to allow expressed mutant proteins to be detected by anti-myc epitope antibodies (see Materials and Methods). Western blotting experiments with lysates isolated from the Pbs2-tagged and Pbs2DD-tagged strains after Ni-NTA enrichment showed that proteins of the expected molecular weight were expressed and that the wild-type and mutant proteins were present at similar levels (our unpublished data). Significantly, expression of the PBS2DD allele, but not the wild-type allele, rescued the stress-sensitive phenotypes and the hyperfilamentation defect associated with deletion of CaSSK2 (Figure 6). These results are consistent with the hypothesis that CaSsk2 functions upstream of Pbs2 in the Hog1 pathway, and, moreover, that it phosphorylates Pbs2 to relay stress signals to Hog1 in C. albicans.
Figure 7.
A Pbs2 chimera containing the S. cerevisiae Pbs2 N-terminal region complements C. albicans pbs2Δ, but not ssk2Δ, phenotypes. (A) Schematic illustration comparing the S. cerevisiae (S.c.) and C. albicans (C.a.) Pbs2 kinase sequences. The phosphorylation sites (P) and the S. cerevisiae Ssk2/22 and Sho1 binding domains are highlighted, and the kinase domain is depicted by a gray box. In the experiments below, full-length CaPBS2 (MET3-CaPBS2) or a chimera in which codons 1-315 of ScPBS2 were fused to codons 160-546 of CaPBS2 (MET3-ScCaPBS2) were tagged with a sequence encoding 2-myc epitopes and 6-His residues and expressed from the MET3 promoter. The junction at which the ScPbs2 sequence was fused to the CaPbs2 sequence to form a ScCaPbs2 chimera is indicated by an arrow. (B) Western blot analysis of Ni-NTA agarose-purified protein extracts isolated from cells expressing Pbs2-6His-myc from the native promoter (JC112), pbs2Δ cells (JC74), or pbs2Δ cells expressing either MET3-CaPBS2 (JC167) or MET3-ScCaPBS2 (JC559). Pbs2 levels were detected using an anti-myc antibody. (C) Approximately 103 cells from exponentially growing pbs2Δ cells or pbs2Δ cells expressing MET3-CaPBS2 or MET3-ScCaPBS2 were spotted onto SD plates lacking methionine and cysteine and containing either 0.6 M KCl, 3.5 mM H2O2, 0.5 mM CdSO4, 8 mM caffeine, and 2 mM Na2As3. In addition, ssk2Δ cells and ssk2Δ cells expressing MET3-ScCaPBS2 were spotted onto the same plates containing 0.6 M KCl. (D) Western blot analysis of whole-cell extracts isolated from pbs2Δ cells, pbs2Δ cells expressing MET3-CaPBS2 or MET3-ScCaPBS2, and ssk2Δ cells expressing MET3-ScCaPBS2 after the indicated treatments. Phosphorylated Hog1 (Hog1-P) and total levels of Hog1 were detected as described in Figure 2.
Figure 6.
Expression of a phosphomimetic mutant of Pbs2 rescues phenotypes associated with deletion of CaSSK2. (A) Approximately 103 cells of exponentially growing Wt (BWP17), ssk2Δ (JC482), or ssk2Δ strains expressing either Pbs2-6His-myc (JC517) or Pbs2DD-6His-myc (JC507) were spotted onto YPD plates containing 1.2 M sorbitol, 0.6 M KCl, 300 mM menadione, 3.5 mM H2O2, 0.5 mM CdSO4, and 2 mM Na2As3. Plates were incubated at 30°C for 24 h. (B) Morphology of ssk2Δ cells expressing either Pbs2-6His-myc or Pbs2DD-6His-myc grown in YPD liquid media at 30°C.
A Pbs2 Chimera Containing the S. cerevisiae Pbs2 N-Terminal Region Complements C. albicans pbs2Δ- but not ssk2Δ-associated Phenotypes
In S. cerevisiae, the ScSsk2 and ScSte11 MAPKKKs both relay osmotic stress signals to the Pbs2 MAPKK. However, our data indicate that CaSsk2 alone, and not CaSte11, relays stress signals to Pbs2 in C. albicans. Extensive analysis of the S. cerevisiae Pbs2 MAPKK has identified distinct regions in the N terminus of the protein that are required for the relay of signals via the Ssk2 or Ste11 MAPKKKs (Tatebayashi et al., 2003). Notably, as illustrated by the schematic diagram in Figure 7A, the N-terminal region of C. albicans Pbs2 is significantly shorter than that of S. cerevisiae Pbs2. In addition, strong homology between the two proteins is restricted to the C terminus, which contains the kinase domain. The lack of homology between S. cerevisiae and C. albicans Pbs2 proteins in the N-terminal regulatory region may underlie the differences in MAPKKK regulation of these kinases. To investigate this, we examined whether expression of ScPBS2 in C. albicans would permit CaSte11-dependent signaling to Hog1. Unfortunately, however, expression of ScPBS2 in C. albicans did not complement the osmotic stress phenotypes associated with deletion of CaPBS2 (our unpublished observations). Although the reasons for this are unknown, ScPBS2 contains five CUG (Ser) codons that would be translated as Leu codons in C. albicans. Subsequently, we tested whether the N-terminal domain of ScPbs2 could function in C. albicans. To this end, we created a construct that expresses a chimera of the S. cerevisiae and C. albicans Pbs2 proteins, composed of residues 1–315 of ScPbs2 fused to residues 160-546 of CaPbs2, which essentially replaces the N-terminal region of CaPbs2 with the equivalent region from ScPbs2. In addition, the two CUG (Ser) codons present in the N-terminal region of S. cerevisiae PBS2 were mutated to CUC (Ser) codons to ensure the correct sequence was maintained upon expression in C. albicans (see Materials and Methods). This construct was expressed from the MET3 promoter (Care et al., 1999) and tagged with a sequence encoding 2-myc epitopes and 6-His residues to allow detection (MET-ScCaPBS2). As a control a similar construct expressing the full-length CaPBS2 sequence was created (MET-CaPBS2). These constructs were targeted to the RPS10 locus in C. albicans pbs2Δ cells and expression induced by growing in defined medium lacking methionine and cysteine (Care et al., 1999).
Western blotting analysis revealed that proteins of the expected molecular weight were expressed from the MET-ScCaPBS2 and the MET-CaPBS2 constructs (Figure 7B). Although the levels of wild-type Pbs2 expressed from the MET3 promoter were similar to those from the endogenous promoter, the ScCaPbs2 chimera was expressed at significantly lower levels (Figure 7B). Nonetheless, expression of MET-ScCaPBS2 largely rescued the stress sensitive phenotypes associated with deletion of PBS2 in C. albicans (Figure 7C). This was dependent on the presence of the ScPbs2 N-terminal regulatory domain, as expression of the CaPbs2 kinase domain-encoding fragment (residues 160-546) did not rescue the osmosensitive phenotype of pbs2Δ cells (our unpublished observations). Furthermore, Hog1 phosphorylation in response to osmotic and oxidative stress was restored in pbs2Δ cells upon expression of MET-ScCaPBS2 (Figure 7D), although, consistent with the sensitivity data, not quite to wild-type levels. Collectively, these results illustrate that the ScCaPbs2 chimera can complement deletion of wild-type Pbs2 in C. albicans to relay osmotic and oxidative stress signals to Hog1. This suggests that ScCaPbs2 can be phosphorylated by CaSsk2, because this MAPKKK regulates both osmotic and oxidative stress signal transmission to Hog1 in C. albicans (Figures 2–4). However, because ScCaPbs2 also contains the sequences necessary for Ste11 regulation of Pbs2 in S. cerevisiae, we investigated whether expression of the ScCaPbs2 chimera in C. albicans ssk2Δ cells would allow for CaSsk2-independent activation of Hog1 in response to osmotic stress. As illustrated in Figure 7, C and D, this did not occur, because expression of ScCaPbs2 in ssk2Δ cells failed to restore osmotic stress-induced Hog1 phosphorylation or resistance to osmotic stress. This was not due to increased instability of the chimera in ssk2Δ cells (our unpublished data). These results indicate that although the ScCaPbs2 chimera contains both S. cerevisiae Ssk2- and Ste11-dependent regulatory regions, the chimeric protein can only function in the presence of CaSsk2. Thus, these data suggest that the Sho1–Ste11 pathway in C. albicans, unlike the analogous pathway in S. cerevisiae, may not respond to changes in osmolarity.
DISCUSSION
In this study, we present several lines of evidence that Hog1 is regulated by a single MAPKKK, Ssk2, in the human fungal pathogen C. albicans. This is strikingly different from the situation in the model yeast S. cerevisiae in which the Hog1 MAPK is regulated by three functionally redundant MAPKKKs Ste11, Ssk2, and Ssk22 (Posas and Saito, 1997). However, in C. albicans we find that deletion of the homologue of S. cerevisiae SSK2 and SSK22 results in 1) equivalent stress-related phenotypes as exhibited by hog1Δ cells (Figure 1 and Supplemental Figure 2); 2) the hyperfilamentous growth defect seen in hog1Δ mutants (Figure 1); 3) loss of detectable Hog1 phosphorylation, or nuclear accumulation of Hog1, in response to osmotic or oxidative stress (Figures 2 and 3); and 4) loss of activation of Hog1-dependent gene expression (Figure 4). Significantly, the stress sensitivities and morphological defects exhibited by C. albicans ssk2Δ cells can be rescued by expression of a mutant version of the Pbs2 MAPKK, in which the conserved MAPKKK phosphorylation sites (Ser355 and Thr359) have been mutated to phosphomimetic aspartic acid residues (Figure 6), strongly supporting the proposal that the function of CaSsk2 is to phosphorylate Pbs2. In contrast to the phenotypes exhibited by ssk2Δ cells, wild-type levels of stress-induced phosphorylation and nuclear accumulation of Hog1 are seen in C. albicans ste11Δ cells (Figures 2 and 3), and ste11Δ cells do not display any overlapping stress-sensitive phenotypes exhibited by hog1Δ cells (Figure 1).
What is the basis for this rewiring of MAPK signaling components in C. albicans compared with S. cerevisiae? In S. cerevisiae, the molecular mechanisms underlying the regulation of Hog1 by the different MAPKKKs are well characterized. The Ssk2 MAPKKK constitutively binds to a novel docking site on the Pbs2 MAPK, which directs Ssk2 specificity (Tatebayashi et al., 2003). This docking site is found between residues 46 and 56 of S. cerevisiae Pbs2, and it is largely conserved in the C. albicans Pbs2 sequence (Figure 7A). Four amino acids were identified in ScPbs2 to be essential for activation by ScSsk2 (Tatebayashi et al., 2003), two of which (Ala52 and Ala56) are identical in the CaPbs2 sequence, and two of which (Arg53 and Val54) are substituted with functionally conserved amino acids. However, the regions of S. cerevisiae Pbs2 previously shown to be required for signaling from the Sho1–Ste11 branch (residues 55-107, which includes the proline-rich motif that binds to the Sho1-SH3 domain; Tatebayashi et al., 2003), or the binding of Ste11 (residues 2–162; Zarrinpar et al., 2004) are less conserved in the C. albicans Pbs2 sequence. It was possible that the lack of sequence homology between the N-terminal regulatory regions of the S. cerevisiae and C. albicans Pbs2 MAPKKs may underlie the differences in MAPKKK regulation of Hog1 in these fungi. However, expression of a chimeric Pbs2 protein containing the S. cerevisiae N-terminal regulatory domain of Pbs2 facilitated Ssk2-dependent, but not Ste11-dependent, signaling to Hog1 in C. albicans (Figure 7). Although we cannot rule out the possibility that the chimera may influence the function of the N-terminal regulatory domain, this result suggests that the Sho1–Ste11 pathway in C. albicans is not involved in osmosensing and signaling.
Consistent with these conclusions, a recent study revealed that deletion of SHO1 in C. albicans does not impair the relay of osmotic stress signals to the Hog1 MAPK (Roman et al., 2005). This is not due to functional redundancy of the Sho1 and two-component osmosensing pathways, as seen in S. cerevisiae, because osmotic stress-induced activation of Hog1 was also seen in sho1Δssk1Δ cells (Roman et al., 2005). Instead, this study revealed Sho1 to play a major role in the control of the Cek1 MAPK and to regulate cell wall biogenesis and morphogenesis (Roman et al., 2005). In this study, we find that ste11Δ cells, similar to sho1Δ cells, are sensitive to the cell wall inhibitor calcofluor white. This is consistent with a model in which Sho1 relays signals to the Cek1 MAPK via the Ste11 MAPKKK in C. albicans. Similar findings have been previously documented in S. cerevisiae in which Sho1 regulates the filamentous growth MAPK Kss1 (the homologue of C. albicans Cek1) in combination with the signaling mucin Msb2 (O'Rourke and Herskowitz, 1998; Cullen et al., 2000, 2004). Thus, these data suggest that although Sho1–Ste11 regulation of the Kss1/Cek1 MAPKs is conserved between S. cerevisiae and C. albicans, the osmosensing function of Sho1 in the regulation of Hog1 seems to be restricted to S. cerevisiae. In this regard, it is also relevant that the Sho1 homologue in the filamentous fungus Aspergillus nidulans seems not to respond to changes in osmolarity (Furukawa et al., 2005), and the fission yeast Schizosaccharomyces pombe does not contain an apparent Sho1 homologue.
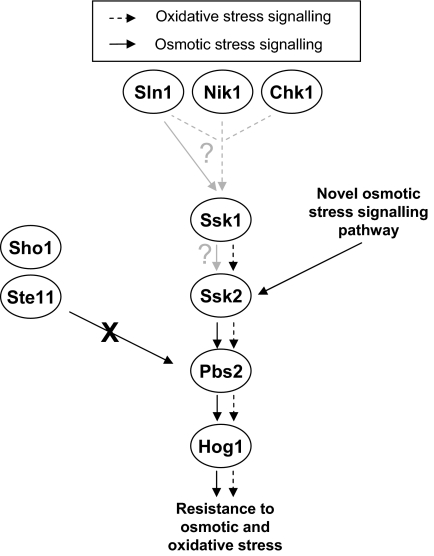
What relays stress signals to the CaSsk2 MAPKKK in C. albicans? Figure 8 illustrates the known stress signaling components of the C. albicans Hog1 pathway. Intriguingly, deletion or mutation of the two-component response regulator Ssk1 impairs activation of Hog1 in response to oxidative stress but not osmotic stress (Chauhan et al., 2003; Menon et al., 2006). Hence the relay of oxidative stress signals to the Ssk2 MAPKKK in C. albicans seems to be two-component dependent. This begs the question as to which of the three histidine kinases in C. albicans, Chk1, Sln1, or Nik1, functions as a peroxide sensor? The most likely candidate would seem to be Chk1, because this kinase shows significant similarity to the peroxide-sensing histidine kinases Mak2 and Mak3 in S. pombe (Buck et al., 2001; Quinn et al., 2002). Indeed, chk1 cells do show increased sensitivity to oxidative stress (Li et al., 2004) and to killing by human neutrophils in vitro (Torosantucci et al., 2002). Nonetheless, deletion of CHK1 does not impair oxidative stress-induced activation of Hog1 (Li et al., 2004). Furthermore, wild-type levels of Hog1 phosphorylation in response to oxidative stress are seen in double chk1sln1 and chk1nik1 histidine kinase mutants (Roman et al., 2005). Hence, it remains unclear how oxidative stress signals are relayed to CaSsk2 via the Ssk1 response regulator. Similarly, it is not known how osmotic stress signals are relayed to the Hog1 pathway. As discussed above, osmotic stress regulation of Hog1 still occurs in cells in which the Sho1 and two-component pathways have been inactivated (Roman et al., 2005). It was suggested that a homologue of the S. cerevisiae Msb2 mucin, which has been proposed to function in parallel with Sho1 to relay osmotic stress signals to Pbs2 via the MAPKKK3 Ste11 (O'Rourke and Herskowitz, 2002), may be responsible for the osmotic stress activation of Hog1 seen in sho1Δssk1Δ C. albicans cells (Roman et al., 2005). However, based on our work, we think this unlikely as we find that the Ssk2 MAPKKK, and not Ste11, regulates Hog1 in response to osmotic stress. Moreover, recent studies indicate that the primary function of S. cerevisiae Msb2 may not be in the regulation of the Hog1 MAPK pathway but rather in the regulation of the Kss1 invasive growth and cell wall integrity pathway (Cullen et al., 2004). Instead, the integral membrane protein Opy2 seems to function with Sho1 for Hog1 pathway activation (Wu et al., 2006). In the fission yeast, S. pombe, two-component signal transduction is used to specifically relay oxidative signals to the stress responsive MAPK, Sty1, and no Sho1 pathway has been described in this organism. Hence, it is possible that C. albicans and S. pombe may use a similar, but as yet, uncharacterized osmosensor (Figure 8).
Figure 8.
Model depicting the relay of osmotic and oxidative stress signals to the Hog1 MAPK in C. albicans. Oxidative stress signaling to Hog1 requires the Ssk1 response regulator (Chauhan et al., 2003), the Ssk2 MAPKKK (this study), and the Pbs2 MAPKK (Arana et al., 2005). However, which histidine kinase(s) regulates Ssk1 in response to oxidative stress is unclear (Li et al., 2004; Roman et al., 2005). Although the relay of osmotic stress signals to Hog1 also requires Ssk2 (this study) and Pbs2 (Arana et al., 2005), Hog1 is activated in response to osmotic stress in ssk1 mutant cells (Chauhan et al., 2003). This, together with the finding that the Sho1–Ste11 pathway does not transmit osmotic stress signals to the Pbs2 MAPKK (Roman et al., 2005; this study) indicates the presence of a novel osmotic-stress signaling pathway in C. albicans. Such a pathway may function in parallel with the Ssk1 response regulator to relay osmotic stress signals to Hog1.
A growing body of evidence is demonstrating that the role and regulation of the Hog1 stress-responsive MAPK pathway in the pathogenic yeast C. albicans has diverged significantly from analogous pathways in the model yeasts S. cerevisiae and S. pombe. For example, C. albicans Hog1 has a distinct activation profile that has likely evolved to survive the particular environmental niches found in the human host (Smith et al., 2004). Furthermore, the role of the stress-responsive MAPK pathways in mediating core transcriptional responses to stress is different in C. albicans (Enjalbert et al., 2006), S. cerevisiae (Gasch et al., 2000; Causton et al., 2001) and S. pombe (Chen et al., 2003). Indeed, a systematic analysis of the Hog1/Sty1-regulated orthologues that exist in C. albicans, S. cerevisiae, and S. pombe revealed only one gene (C. albicans IPF5981, S. cerevisiae YML128C, and S. pombe SPBC365.12) to be commonly regulated by these pathways in all three yeast (Enjalbert et al., 2006). In addition, work from this study and others has also uncovered significant differences in the regulation of the C. albicans Hog1 MAPK compared with analogous pathways in S. cerevisiae and S. pombe (Chauhan et al., 2003; Arana et al., 2005; Roman et al., 2005, Menon et al., 2006). The Hog1 MAPK pathway plays an important role in C. albicans virulence (Alonso-Monge et al., 1999; Arana et al., 2007) thus an important goal is to delineate how the specialization of the role and regulation of Hog1 promotes survival in the human host.
Supplementary Material
ACKNOWLEDGMENTS
We thank Brian Morgan, Elizabeth Veal, and Simon Whitehall for discussions and comments on the manuscript. We also thank Al Brown for valuable help and advice. We are grateful to Cheryl Gale and Judith Berman (University of Minnesota, Minneapolis, MN) for the kind gift of YFP cassettes. This work was funded by the BBSRC and the Wellcome Trust.
Footnotes
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E07-06-0581) on September 5, 2007.
 The online version of this article contains supplemental material at MBC Online (http://www.molbiolcell.org).
The online version of this article contains supplemental material at MBC Online (http://www.molbiolcell.org).
REFERENCES
- Alonso-Monge R., Navarro-Garcia F., Molero G., Diez-Orejas R., Gustin M., Pla J., Sanchez M., Nombela C. Role of the mitogen-activated protein kinase Hog1p in morphogenesis and virulence of Candida albicans. J. Bacteriol. 1999;181:3058–3068. doi: 10.1128/jb.181.10.3058-3068.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alonso-Monge R., Navarro-Garcia F., Roman E., Negredo A. I., Eisman B., Nombela C., Pla J. The Hog1 mitogen-activated protein kinase is essential in the oxidative stress response and chlamydospore formation in Candida albicans. Eukaryot. Cell. 2003;2:351–361. doi: 10.1128/EC.2.2.351-361.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arana D. M., Alonso-Monge R., Du C., Calderone R., Pla J. Differential susceptibility of mitogen-activated protein kinase pathway mutants to oxidative-mediated killing by phagocytes in the fungal pathogen Candida albicans. Cell. Microbiol. 2007;9:1647–1659. doi: 10.1111/j.1462-5822.2007.00898.x. [DOI] [PubMed] [Google Scholar]
- Arana D. M., Nombela C., Alonso-Monge R., Pla J. The Pbs2 MAP kinase kinase is essential for the oxidative-stress response in the fungal pathogen Candida albicans. Microbiology. 2005;151:1033–1049. doi: 10.1099/mic.0.27723-0. [DOI] [PubMed] [Google Scholar]
- Bahn Y. S., Kojima K., Cox G. M., Heitman J. Specialization of the HOG pathway and its impact on differentiation and virulence of Cryptococcus neoformans. Mol. Biol. Cell. 2005;16:2285–2300. doi: 10.1091/mbc.E04-11-0987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bahn Y. S., Kojima K., Cox G. M., Heitman J. A unique fungal two-component system regulates stress responses, drug sensitivity, sexual development, and virulence of Cryptococcus neoformans. Mol. Biol. Cell. 2006;17:3122–3135. doi: 10.1091/mbc.E06-02-0113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blackwell C., Russell C. L., Argimon S., Brown A. J., Brown J. D. Protein A-tagging for purification of native macromolecular complexes from Candida albicans. Yeast. 2003;20:1235–1241. doi: 10.1002/yea.1036. [DOI] [PubMed] [Google Scholar]
- Buck V., Quinn J., Soto Pino T., Martin H., Saldanha J., Makino K., Morgan B. A., Millar J. B. Peroxide sensors for the fission yeast stress-activated mitogen-activated protein kinase pathway. Mol. Biol. Cell. 2001;12:407–419. doi: 10.1091/mbc.12.2.407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Care R. S., Trevethick J., Binley K. M., Sudbery P. E. The MET3 promoter: a new tool for Candida albicans molecular genetics. Mol. Microbiol. 1999;34:792–798. doi: 10.1046/j.1365-2958.1999.01641.x. [DOI] [PubMed] [Google Scholar]
- Causton H. C., Ren B., Koh S. S., Harbison C. T., Kanin E., Jennings E. G., Lee T. I., True H. L., Lander E. S., Young R. A. Remodeling of yeast genome expression in response to environmental changes. Mol. Biol. Cell. 2001;12:323–337. doi: 10.1091/mbc.12.2.323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chauhan N., Inglis D., Roman E., Pla J., Li D., Calera J. A., Calderone R. Candida albicans response regulator gene SSK1 regulates a subset of genes whose functions are associated with cell wall biosynthesis and adaptation to oxidative stress. Eukaryot. Cell. 2003;2:1018–1024. doi: 10.1128/EC.2.5.1018-1024.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen D., Toone W. M., Mata J., Lyne R., Burns G., Kivinen K., Brazma A., Jones N., Bahler J. Global transcriptional responses of fission yeast to environmental stress. Mol. Biol. Cell. 2003;14:214–229. doi: 10.1091/mbc.E02-08-0499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cullen P. J., Sabbagh W., Jr, Graham E., Irick M. M., van Olden E. K., Neal C., Delrow J., Bardwell L., Sprague G. F., Jr A signaling mucin at the head of the Cdc42- and MAPK-dependent filamentous growth pathway in yeast. Genes Dev. 2004;18:1695–1708. doi: 10.1101/gad.1178604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cullen P. J., Schultz J., Horecka J., Stevenson B. J., Jigami Y., Sprague G. F., Jr Defects in protein glycosylation cause SHO1-dependent activation of a STE12 signaling pathway in yeast. Genetics. 2000;155:1005–1018. doi: 10.1093/genetics/155.3.1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dennison P.M.J., Ramsdale M., Manson C. L., Brown A.J.P. Gene disruption in Candida albicans using a synthetic, codon-optimised Cre loxP system. Fungal Genet. Biol. 2005;42:737–748. doi: 10.1016/j.fgb.2005.05.006. [DOI] [PubMed] [Google Scholar]
- Du C., Sarfati J., Latge J. P., Calderone R. The role of the sakA (Hog1) and tcsB (sln1) genes in the oxidant adaptation of Aspergillus fumigatus. Med. Mycol. 2006;44:211–218. doi: 10.1080/13693780500338886. [DOI] [PubMed] [Google Scholar]
- Eisman B., Alonso-Monge R., Roman E., Arana D., Nombela C., Pla J. The Cek1 and Hog1 mitogen-activated protein kinases play complementary roles in cell wall biogenesis and chlamydospore formation in the fungal pathogen Candida albicans. Eukaryot. Cell. 2006;5:347–358. doi: 10.1128/EC.5.2.347-358.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enjalbert B., Smith D. A., Cornell M. J., Alam I., Nicholls S., Brown A. J., Quinn J. Role of the Hog1 stress-activated protein kinase in the global transcriptional response to stress in the fungal pathogen Candida albicans. Mol. Biol. Cell. 2006;17:1018–1032. doi: 10.1091/mbc.E05-06-0501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furukawa K., Hoshi Y., Maeda T., Nakajima T., Abe K. Aspergillus nidulans HOG pathway is activated only by two-component signalling pathway in response to osmotic stress. Mol. Microbiol. 2005;56:1246–1261. doi: 10.1111/j.1365-2958.2005.04605.x. [DOI] [PubMed] [Google Scholar]
- Gasch A. P., Spellman P. T., Kao C. M., Carmel-Harel O., Eisen M. B., Storz G., Botstein D., Brown P. O. Genomic expression programs in the response of yeast cells to environmental changes. Mol. Biol. Cell. 2000;11:4241–4257. doi: 10.1091/mbc.11.12.4241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerami-Nejad M., Berman J., Gale C. A. Cassettes for PCR-mediated construction of green, yellow, and cyan fluorescent protein fusions in Candida albicans. Yeast. 2001;18:859–864. doi: 10.1002/yea.738. [DOI] [PubMed] [Google Scholar]
- Hohmann S. Osmotic stress signaling and osmoadaptation in yeasts. Microbiol. Mol. Biol. Rev. 2002;66:300–372. doi: 10.1128/MMBR.66.2.300-372.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson L. N., Noble M. E., Owen D. J. Active and inactive protein kinases: structural basis for regulation. Cell. 1996;85:149–158. doi: 10.1016/s0092-8674(00)81092-2. [DOI] [PubMed] [Google Scholar]
- Li D., Gurkovska V., Sheridan M., Calderone R., Chauhan N. Studies on the regulation of the two-component histidine kinase gene CHK1 in Candida albicans using the heterologous lacZ reporter gene. Microbiology. 2004;150:3305–3313. doi: 10.1099/mic.0.27237-0. [DOI] [PubMed] [Google Scholar]
- Maeda T., Takekawa M., Saito H. Activation of yeast PBS2 MAPKK by MAPKKKs or by binding of an SH3-containing osmosensor. Science. 1995;269:554–558. doi: 10.1126/science.7624781. [DOI] [PubMed] [Google Scholar]
- Menon V., Li D., Chauhan N., Rajnarayanan R., Dubrovska A., West A. H., Calderone R. Functional studies of the Ssk1p response regulator protein of Candida albicans as determined by phenotypic analysis of receiver domain point mutants. Mol. Microbiol. 2006;62:997–1013. doi: 10.1111/j.1365-2958.2006.05438.x. [DOI] [PubMed] [Google Scholar]
- Millar J. B., Buck V., Wilkinson M. G. Pyp1 and Pyp2 PTPases dephosphorylate an osmosensing MAP kinase controlling cell size at division in fission yeast. Genes Dev. 1995;9:2117–2130. doi: 10.1101/gad.9.17.2117. [DOI] [PubMed] [Google Scholar]
- Murad A. M., Lee P. R., Broadbent I. D., Barelle C. J., Brown A. J. CIp10, an efficient and convenient integrating vector for Candida albicans. Yeast. 2000;16:325–327. doi: 10.1002/1097-0061(20000315)16:4<325::AID-YEA538>3.0.CO;2-#. [DOI] [PubMed] [Google Scholar]
- O'Rourke S. M., Herskowitz I. The Hog1 MAPK prevents cross talk between the HOG and pheromone response MAPK pathways in Saccharomyces cerevisiae. Genes Dev. 1998;12:2874–2886. doi: 10.1101/gad.12.18.2874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Rourke S. M., Herskowitz I. A third osmosensing branch in Saccharomyces cerevisiae requires the Msb2 protein and functions in parallel with the Sho1 branch. Mol. Cell Biol. 2002;22:4739–4749. doi: 10.1128/MCB.22.13.4739-4749.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Rourke S. M., Herskowitz I. Unique and redundant roles for HOG MAPK pathway components as revealed by whole-genome expression analysis. Mol. Biol. Cell. 2004;15:532–542. doi: 10.1091/mbc.E03-07-0521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearson G., Robinson F., Beers Gibson T., Xu B. E., Karandikar M., Berman K., Cobb M. H. Mitogen-activated protein (MAP) kinase pathways: regulation and physiological functions. Endocr. Rev. 2001;22:153–183. doi: 10.1210/edrv.22.2.0428. [DOI] [PubMed] [Google Scholar]
- Posas F., Saito H. Osmotic activation of the HOG MAPK pathway via Ste11p MAPKKK: scaffold role of Pbs2p MAPKK. Science. 1997;276:1702–1705. doi: 10.1126/science.276.5319.1702. [DOI] [PubMed] [Google Scholar]
- Posas F., Saito H. Activation of the yeast SSK2 MAP kinase kinase kinase by the SSK1 two-component response regulator. EMBO J. 1998;17:1385–1394. doi: 10.1093/emboj/17.5.1385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Posas F., Witten E. A., Saito H. Requirement of STE50 for osmostress-induced activation of the STE11 mitogen-activated protein kinase kinase kinase in the high-osmolarity glycerol response pathway. Mol. Cell Biol. 1998;18:5788–5796. doi: 10.1128/mcb.18.10.5788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Posas F., Wurgler-Murphy S. M., Maeda T., Witten E. A., Thai T. C., Saito H. Yeast HOG1 MAP kinase cascade is regulated by a multistep phosphorelay mechanism in the SLN1-YPD1-SSK1 “two-component” osmosensor. Cell. 1996;86:865–875. doi: 10.1016/s0092-8674(00)80162-2. [DOI] [PubMed] [Google Scholar]
- Quinn J., Findlay V. J., Dawson K., Millar J. B., Jones N., Morgan B. A., Toone W. M. Distinct regulatory proteins control the graded transcriptional response to increasing H2O2 levels in fission yeast Schizosaccharomyces pombe. Mol. Biol. Cell. 2002;13:805–816. doi: 10.1091/mbc.01-06-0288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raitt D. C., Posas F., Saito H. Yeast Cdc42 GTPase and Ste20 PAK-like kinase regulate Sho1-dependent activation of the Hog1 MAPK pathway. EMBO J. 2000;19:4623–4631. doi: 10.1093/emboj/19.17.4623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiser V., Salah S. M., Ammerer G. Polarized localization of yeast Pbs2 depends on osmostress, the membrane protein Sho1 and Cdc42. Nat. Cell Biol. 2000;2:620–627. doi: 10.1038/35023568. [DOI] [PubMed] [Google Scholar]
- Roman E., Nombela C., Pla J. The Sho1 adaptor protein links oxidative stress to morphogenesis and cell wall biosynthesis in the fungal pathogen Candida albicans. Mol. Cell Biol. 2005;25:10611–10627. doi: 10.1128/MCB.25.23.10611-10627.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito H., Tatebayashi K. Regulation of the osmoregulatory HOG MAPK cascade in yeast. J. Biochem. 2004;136:267–272. doi: 10.1093/jb/mvh135. [DOI] [PubMed] [Google Scholar]
- San-Jose C., Monge R. A., Perez-Diaz R., Pla J., Nombela C. The mitogen-activated protein kinase homolog HOG1 gene controls glycerol accumulation in the pathogenic fungus Candida albicans. J. Bacteriol. 1996;178:5850–5852. doi: 10.1128/jb.178.19.5850-5852.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherman F. Getting started with yeast. Methods Enzymol. 1991;194:3–21. doi: 10.1016/0076-6879(91)94004-v. [DOI] [PubMed] [Google Scholar]
- Shiozaki K., Shiozaki M., Russell P. Heat stress activates fission yeast Spc1/StyI MAPK by a MEKK-independent mechanism. Mol. Biol. Cell. 1998;9:1339–1349. doi: 10.1091/mbc.9.6.1339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith D. A., Nicholls S., Morgan B. A., Brown A. J., Quinn J. A conserved stress-activated protein kinase regulates a core stress response in the human pathogen Candida albicans. Mol. Biol. Cell. 2004;15:4179–4190. doi: 10.1091/mbc.E04-03-0181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatebayashi K., Takekawa M., Saito H. A docking site determining specificity of Pbs2 MAPKK for Ssk2/Ssk22 MAPKKKs in the yeast HOG pathway. EMBO J. 2003;22:3624–3634. doi: 10.1093/emboj/cdg353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatebayashi K., Yamamoto K., Tanaka K., Tomida T., Maruoka T., Kasukawa E., Saito H. Adaptor functions of Cdc42, Ste50, and Sho1 in the yeast osmoregulatory HOG MAPK pathway. EMBO J. 2006;25:3033–3044. doi: 10.1038/sj.emboj.7601192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson J. D., Higgins D. G., Gibson T. J. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position–specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torosantucci A., Chiani P., De Bernardis F., Cassone A., Calera J. A., Calderone R. Deletion of the two-component histidine kinase gene (CHK1) of Candida albicans contributes to enhanced growth inhibition and killing by human neutrophils in vitro. Infect. Immun. 2002;70:985–987. doi: 10.1128/iai.70.2.985-987.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Wuytswinkel O., Reiser V., Siderius M., Kelders M. C., Ammerer G., Ruis H., Mager W. H. Response of Saccharomyces cerevisiae to severe osmotic stress: evidence for a novel activation mechanism of the HOG MAP kinase pathway. Mol. Microbiol. 2000;37:382–397. doi: 10.1046/j.1365-2958.2000.02002.x. [DOI] [PubMed] [Google Scholar]
- Wilson R. B., Davis D., Mitchell A. P. Rapid hypothesis testing with Candida albicans through gene disruption with short homology regions. J. Bacteriol. 1999;181:1868–1874. doi: 10.1128/jb.181.6.1868-1874.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu C., Jansen G., Zhang J., Thomas D. Y., Whiteway M. Adaptor protein Ste50p links the Ste11p MEKK to the HOG pathway through plasma membrane association. Genes Dev. 2006;20:734–746. doi: 10.1101/gad.1375706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue T., Nguyen C. K., Romans A., May G. S. A mitogen-activated protein kinase that senses nitrogen regulates conidial germination and growth in Aspergillus fumigatus. Eukaryot. Cell. 2004;3:557–560. doi: 10.1128/EC.3.2.557-560.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zarrinpar A., Bhattacharyya R. P., Nittler M. P., Lim W. A. Sho1 and Pbs2 act as coscaffolds linking components in the yeast high osmolarity MAP kinase pathway. Mol. Cell. 2004;14:825–832. doi: 10.1016/j.molcel.2004.06.011. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.