Abstract
Among the three Saccharomyces cerevisiae DNA repair epistasis groups, the RAD6 group is the most complicated and least characterized, primarily because it consists of two separate repair pathways: an error-free postreplication repair pathway, and a mutagenesis pathway. The rad6 and rad18 mutants are defective in both pathways, and the rev3 mutant affects only the mutagenesis pathway, but a yeast gene that is involved only in error-free postreplication repair has not been reported. We cloned the MMS2 gene from a yeast genomic library by functional complementation of the mms2-1 mutant [Prakash, L. & Prakash, S. (1977) Genetics 86, 33–55]. MMS2 encodes a 137-amino acid, 15.2-kDa protein with significant sequence homology to a conserved family of ubiquitin-conjugating (Ubc) proteins. However, Mms2 does not appear to possess Ubc activity. Genetic analyses indicate that the mms2 mutation is hypostatic to rad6 and rad18 but is synergistic with the rev3 mutation, and the mms2 mutant is proficient in UV-induced mutagenesis. These phenotypes are reminiscent of a pol30-46 mutant known to be impaired in postreplication repair. The mms2 mutant also displayed a REV3-dependent mutator phenotype, strongly suggesting that the MMS2 gene functions in the error-free postreplication repair pathway, parallel to the REV3 mutagenesis pathway. Furthermore, with respect to UV sensitivity, mms2 was found to be hypostatic to the rad6Δ1–9 mutation, which results in the absence of the first nine amino acids of Rad6. On the basis of these collective results, we propose that the mms2 null mutation and two other allele-specific mutations, rad6Δ1–9 and pol30-46, define the error-free mode of DNA postreplication repair, and that these mutations may enhance both spontaneous and DNA damage-induced mutagenesis.
Ubiquitin (Ub) is a highly conserved 76-residue protein, often found covalently joined to other proteins. Ub conjugation has been shown to participate in many eukaryotic metabolic processes, including ribosome biogenesis (1), mating type regulation (2), cell cycle control (3), DNA repair (4), and other responses (5). Ub is bound to the ubiquitin-activating enzyme (E1), which activates Ub and enables it to bind to the ubiquitin-conjugating enzyme (Ubc or E2). A single cysteine residue in the proper consensus sequence of a Ubc is absolutely required to bind Ub by a thioester bond and attach it to the target molecule. In some instances a third protein, a ubiquitin ligase enzyme (E3), is required to help the Ubc specify the target protein according to an N-terminal rule (for review, see refs. 6 and 7).
The RAD6 gene in Saccharomyces cerevisiae encodes a 172-amino acid, 20-kDa E2 enzyme, Ubc2 (4, 8). Rad6 appears to be a multifunctional protein—-not only do rad6 mutants display a slow-growth phenotype, but they are defective in DNA damage-induced mutagenesis and are sensitive to killing by a variety of DNA-damaging agents, including UV, ionizing radiations, and alkylating agents; as well, rad6 diploids are deficient in sporulation (for review, see ref. 9). It is well established that the Ubc activity of Rad6 is required for all of its functions, because rad6 mutants carrying amino acid substitutions at the sole cysteine residue (C88) displayed phenotypes indistinguishable from the rad6Δ null mutant (10). The N-terminal 15 amino acids are almost identical among Rad6 homologs from S. cerevisiae, Schizosaccharomyces pombe, Drosophila, and humans (8, 11–13). Deletion of the first 9 amino acids from Rad6 (rad6Δ1–9) results in partial sensitivity to UV-induced killing, increased mutagenesis, and the loss of sporulation (14). The Rad6 N terminus is also required for physical interaction with an E3 enzyme Ubr1, and for N-end rule protein degradation (14–16). The C-terminal tail of Rad6 is not required for DNA repair but is essential for sporulation (17); it is also required for in vitro multi-ubiquitination of histones (18). It is thus conceivable that Rad6 possesses both Ubr1-dependent and Ubr1-independent E2 activities, and that these different E2 activities may define its different roles.
Many lines of genetic evidence support the role of Rad6 and Rad18 in both the error-free postreplication repair (PRR) and error-prone mutagenesis pathways (9). When lethal replication-blocking lesions occur within the cell, the stalled replication machinery at the lesion must be resolved to allow for subsequent repair synthesis. Rad6 is targeted to stalled replication machinery, presumably by its interaction with the single-stranded DNA binding protein Rad18 (19). rad18 and rad6 mutants share many phenotypes, including similar levels of sensitivity to killing by a variety of DNA-damaging agents, and defects in UV-induced mutagenesis (9). Indeed, Rad18 and Rad6 belong to the same epistasis group, and they form a stable complex distinct from the Rad6–Ubr1 complex (19, 20). It was thus proposed that the single-stranded DNA-binding and ATP-hydrolytic activities of Rad18 and the E2 activity of Rad6 direct the complex (21) to the stalled replication machinery for initiation of both the PRR and mutagenesis pathways.
Within the yeast Rad6–Rad18 repair pathway, error-prone mutagenesis is mediated by a nonessential DNA polymerase ζ (Polζ), which has been extensively characterized. REV3 encodes the catalytic subunit of Polζ (22). Purified Polζ, consisting of Rev3 and Rev7, has been shown to replicate past a thymine dimer much more efficiently than does Polα (23). Polζ-mediated bypass of replication blocks may result in an elevated number of mutations, because rev3 mutants reduced both spontaneous and DNA damage-induced mutations (for review, see ref. 24). However, because of the lack of mutants defective in PRR but not mutagenesis, it is not clear how the error-free PRR pathway operates. Recently, yeast proliferating cell nuclear antigen (PCNA) (25) and Pol δ (26) have been implicated in the PRR pathway, likely at a late stage of DNA synthesis. We report here the molecular cloning and characterization of a newly identified yeast gene MMS2, encoding a Ubc-like protein. We further demonstrate that the mms2 mutant shares remarkably similar phenotypes with pol30-46 and rad6Δ1–9, and that the mms2 mutation is hypostatic to rad6Δ1–9 but synergistic with rev3. We propose that the above three mutations are all defective in PRR but proficient in mutagenesis.
MATERIALS AND METHODS
Yeast Strains and Cell Culture.
The yeast strains used in this study are listed in Table 1. WX17–4a was isolated from MD-2/FY86 diploid segregants to combine mms2-1 with ura3 for library screening. The isogenic rad4Δ, rad6Δ, rad18Δ, rad50Δ, and rev3Δ mutants were described previously (27). The URA3 selectable marker of WXY9221 was removed by selection on a 5-fluoroorotic acid plate (28) to obtain WXY9579. The haploid strain T43 bearing an mms2Δ mutation and its corresponding wild type BY448 were a gift from S. Bacchetti and L. Ma (McMaster University, Hamilton, ON, Canada). Yeast cells were grown at 30°C either in a rich yeast extract/peptone/dextrose (YPD) medium or in a synthetic dextrose (SD) medium supplemented with amino acids and bases at recommended concentrations (29).
Table 1.
Saccharomyces cerevisiae strains
| Strain | Genotype |
|---|---|
| B635 | a cyc1-115 lys2 his1 trp2 |
| MD-2 | B635 with mms2-1 |
| FY86 | α his3-Δ200 ura3-52 leu2-Δ1 GAL+ |
| WX17-4a | a his3-Δ200 ura3-52 lys2 mms2-1 |
| DBY747 | a his3-Δ1 leu2-3,112 trp1-289 ura3-52 |
| SBU | DBY747 with mms2∷URA3 |
| SBL | DBY747 with mms2∷LEU2 |
| WXY9579 | DBY747 with rad50Δ∷hisG |
| SBU50h | DBY747 with rad50Δ∷hisG mms2∷URA3 |
| WXY9394 | DBY747 with rad4Δ∷hisG∷URA3∷hisG |
| SBL4U | DBY747 with rad4Δ∷hisG∷URA3∷hisG mms2∷LEU2 |
| WXY9376 | DBY747 with rad6Δ∷LEU2 |
| SBU6L | DBY747 with rad6Δ∷LEU2 mms2∷URA3 |
| WXY9326 | DBY747 with rad18Δ∷TRP1 |
| SBU18T | DBY747 with rad18Δ∷TRP1 mms2∷URA3 |
| WXY9382 | DBY747 with rev3Δ∷LEU2 |
| SBUr3L | DBY747 with rev3Δ∷LEU2 mms2∷URA3 |
| BY448 | α leu2Δ his3Δura3-52 trp1Δ ade2 lys2 |
| T43 | BY448 with mms2Δ∷TRP1 |
Screening a Genomic Library.
A YCp50-based yeast genomic library (30) was obtained from M. Rose (Princeton University, Princeton, NJ) and used to transform WX17–4a. A two-step screening protocol was followed. First, uracil-independent (Ura+) transformants were obtained on SD − uracil plates. A total of approximately 10,000 Ura+ colonies were then streaked onto YPD and YPD + 0.04% methyl methanesulfonate (MMS). The MMS-resistant clones were subjected to a plasmid cosegregation test and the YCp50-based plasmids were recovered by transforming Escherichia coli with total yeast DNA.
DNA Sequencing and Sequence Analysis.
Nucleotide sequences of the MMS2 open reading frame and its surrounding regions were determined by a dideoxynucleotide chain-terminating method (31) using a T7 DNA polymerase sequencing kit (Pharmacia LKB). The MMS2 sequence (GenBank accession no. U66724) was analyzed to search for intron sequences, and the deduced amino acid sequence was used to perform homology searches and multiple sequence alignments.
The mms2-1 mutant allele was PCR-amplified by using the MD-2 yeast genomic DNA as a template, and was cloned as a 1.1-kb BglII fragment (see Fig. 1A) into the general-purpose plasmid pTZ18R (Pharmacia). The entire mms2-1 sequences of three independent clones were determined.
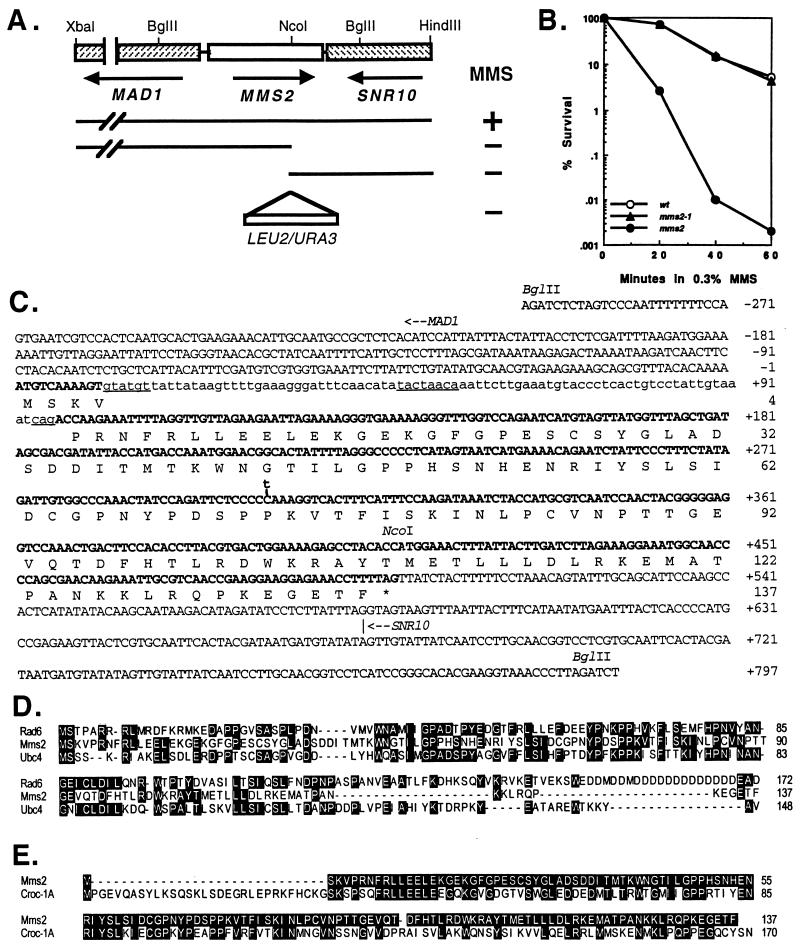
Figure 1.
Physical characterization of the S. cerevisiae MMS2 gene. (A) Mapping and disruption of the MMS2 gene. A subclone containing the 3-kb XbaI–HindIII fragment enables (+) the mms2-1 mutant to grow on YPD plates containing 0.4% MMS. Further deletions to the NcoI site from either end abolished (−) the MMS2 function. Either a URA3 or a LEU2 fragment was inserted at the NcoI site to construct the mms2∷URA3 and mms2∷LEU2 disruption cassettes. (B) Killing of DBY747 (wt), WX17–4a (mms2-1), and SBU (mms2) in a liquid culture containing 0.3% MMS. (C) The nucleotide and deduced amino acid sequences of the MMS2 gene (GenBank accession no. U66724). Exons are in boldface. Lowercase indicates intron sequences. Consensus sequences within the intron are underlined. The translation initiation site for MAD1 and the transcriptional termination site for SNR10 are indicated with an arrow for direction. A C-to-T transition found in the mms2-1 mutation at nucleotide 303 is marked. (D) Amino acid sequence alignments of Mms2 with two yeast Ubc proteins, Rad6 (Ubc2) and Ubc4. Residues shared by two or more proteins are highlighted. (E) Amino acid sequence alignment of Mms2 with Croc1. Residues in Croc1 identical to Mms2 are highlighted.
Plasmid Manipulation.
A series of deletions was made within the library clone YCpM2 insert, and the resulting plasmids were used to transform WX17–4a to map the MMS2 gene. The MMS2 gene was subcloned as a 3-kb XbaI–HindIII fragment from YCpM2 into YCplac33 (32) to form YCp-MMS2, and into pTZ18R to form pTZ-MMS2. A linker was inserted into the unique NcoI site of pTZ-MMS2 to convert it to a BamHI site, which was then used to clone either the 1.8-kb LEU2 gene from YDp-L (33) or the 1.2-kb URA3 gene from YDp-U (33), resulting in pmms2∷LEU2 and pmms2∷URA3, respectively (Fig. 1A).
Plasmid pSCW-rad6Δ1–9 was constructed by PCR amplification of the RAD6 gene with a mutation primer 5′-TCGAATTCAAGTCCACACCAGCTAG-3′, where an EcoRI site was created, followed by a mutated translation initiation codon (AAG instead of ATG), and a 3′ end specific primer to generate a BglII site 3′ to the RAD6 translation stop codon. The EcoRI–BglII fragment was inserted into the EcoRI–BamHI sites of pSCW231 (14) to place the rad6Δ1–9 under the control of an ADH1 promoter. This mutation results in translation initiation of the rad6 gene at the second ATG, which encodes the 10th amino acid of Rad6 (14). It has been determined that the pSCW-rad6Δ1–9 transformants will produce >10-fold more Rad6 protein than do wild-type cells (14).
Cell Killing and Mutagenesis Assays.
MMS-induced liquid killing was performed at 30°C in YPD as previously described (27). For UV treatment, cells were plated at different dilutions and then exposed to 254-nm UV light, either in a UV crosslinker (Fisher model FB-UVXL-1000 at ≈2,400 μW/cm2) or with a UV lamp (Ultraviolet Products model UVGL-25 at 40 μW/cm2) at given doses in the dark. Cells were plated in duplicate either on YPD to score cell survival or on SD − tryptophan to score tryptophan-independent (Trp+) revertants. DBY747 bears a trp1-289 amber mutation that can be reverted to Trp+ by several different mutational events. The plates were incubated at 30°C in a dark chamber for 4 days to prevent photoactivation.
Spontaneous Trp+ reversion rates of DBY747 derivatives were measured by a modified Luria and Delbruck fluctuation test as described (34). An overnight yeast culture was used to inoculate five tubes, each containing 10 ml of fresh YPD, to a final titer of 20 cells per ml. Incubation was continued until a titer of 2 × 108 cells per ml was reached. Cells were collected, washed, resuspended, and plated. Each set of experiments contained five independent cultures of each strain; each culture was plated onto YPD in duplicate to score total survivors, and onto SD − tryptophan plates to score Trp+ revertants. Spontaneous mutation rates (number of revertants per cell per generation) were calculated as previously described (35).
RESULTS
MMS2 Encodes a Ubc-like Protein.
The mms2-1 mutant was originally isolated by Prakash and Prakash (36) by its enhanced sensitivity to MMS. The MMS2 gene was cloned by screening a single-copy yeast genomic library for the functional complementation of the mms2-1 MMS-sensitive phenotype. The initial MMS2 clone, YCpM2, contains a 10-kb insert. By a combination of deletion mapping and DNA sequencing, a small MMS2 open reading frame was discovered that conferred an MMS-resistant phenotype in the mms2-1 strain (Fig. 1A). The MMS2 gene is located on chromosome 7 and resides between MAD1 and SNR10 (Fig. 1 A and C). It encodes a predicted 137-amino acid, 15.5-kDa protein with a single intron. The splicing sequences located within S. cerevisiae introns are highly conserved. We confirmed that the first exon encoding three amino acids is required for MMS2 function, because MMS2 clones using the second ATG codon at nucleotide 218 as the translational start were unable to complement the mms2-1 mutation, whereas an intronless MMS2 clone is able to complement the MMS-sensitive phenotype of mms2-1 (data not shown).
Two pieces of evidence strongly suggest that the cloned MMS2 gene is allelic to mms2-1. First, the cloned MMS2 was used to disrupt the chromosomal MMS2 gene and the mutant became sensitive to MMS; this mms2 mutation was unable to complement the mms2-1 mutation in a diploid. Secondly, the mms2-1 allele from MD-2 was cloned by PCR amplification and a C-to-T transition at nucleotide 303, which results in a single amino acid substitution, P73L (Fig. 1C), was identified from all three independent mms2-1 clones. mms2-1 in a single-copy plasmid was unable to complement MMS sensitivity of the mms2 mutant (data not shown).
The C-terminal two-thirds of the deduced Mms2 shares significant homology with almost all known Ubc proteins, with P values ranging from 10−11 to 10−3. The middle third of Mms2 shares up to 40% identity and 60% similarity with some Ubc proteins (Fig. 1D), suggesting an evolutionary conservation. Like Ubcs, the deduced Mms2 is rich in proline residues (12/137), particularly in the middle one-third region (8/41), indicating that it may form a globular protein. Despite the high overall degree of homology between Mms2 and Ubcs, Mms2 lacks a critical consensus sequence that all Ubc proteins display around their active Cys residue (Fig. 1D). The absence of this consensus sequence suggests that Mms2 does not possess direct Ub-conjugating ability. Partially purified Mms2 from an E. coli expression system was unable to bind Ub and lacked E2 activity (V. Chau, Wayne State University, personal communication). As a matter of fact, Mms2 has even stronger homology (P = 10−39) with Croc1, a recently identified transactivator of the c-fos enhancer (37), which also lacks E2 activity (38). The two proteins share 49% amino acid identity throughout the entire length of Mms2 (Fig. 1E).
The mms2 Null Mutant Is Sensitive to Both MMS and UV.
The original mms2-1 mutant was characterized as sensitive to MMS- but not to UV-induced killing (36). We found that the mms2-1 mutant is sensitive to MMS by a plate assay, but not in a liquid assay (Fig. 1 A and B) and that the mms2-1 mutation results in only partial loss of function, since a single copy of mms2-1 was unable to rescue the mms2 mutant from killing by MMS, yet mms2-1 in a multicopy plasmid was able to complement the mms2 mutant (data not shown). It should be noted that P73 mutated in mms2-1 is conserved in all the corresponding Ubcs as well as in Croc1 (Fig. 1 D and E), and that it is adjacent to another highly conserved proline residue, which, when mutated (e.g., P64S in Rad6 and P71S in Ubc3), results in temperature-sensitive mutants (39). In contrast to the mms2-1 mutant, the mms2 mutant was sensitive to MMS in both plate and liquid assays (Fig. 1B). In addition, compared with an isogenic wild-type strain, the mms2 mutant is more sensitive to UV (see Fig. 2) and a UV-mimetic agent, 4-nitroquinoline 1-oxide (data not shown). Thus the role of the MMS2 gene appears not to be limited to the protection of cells from DNA methylation damage. Our mms2 disruption mutant is likely a complete loss-of-function mutant because it was as sensitive to MMS and UV as was the mms2Δ strain T43, in which the entire MMS2 gene is deleted (data not shown).
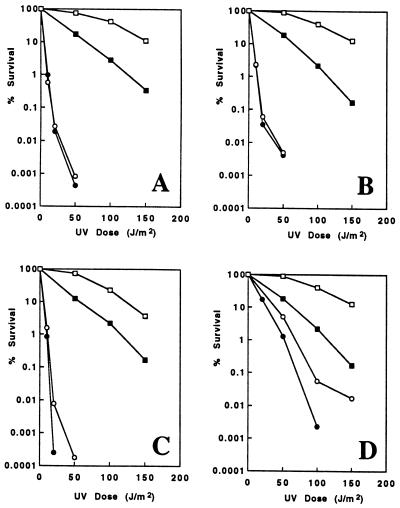
Figure 2.
Epistatic analyses of mms2 with radiation repair pathway mutations with respect to UV sensitivity. (A–D) □, DBY747 (wt); ▪, SBU (mms2). (A) mms2 and rad6. ○, WXY9376 (rad6Δ); •, SBU6L (rad6Δ mms2). (B) mms2 and rad18Δ. ○, WXY9326 (rad18Δ); •, SBU18T (rad18Δ mms2). (C) mms2 and rad4Δ. ○, WXY9394 (rad4Δ); •, SBL4U (rad4Δ mms2). (D) mms2 and rad50Δ. ○, WXY9579 (rad50Δ); •, SBU50h (rad50Δ mms2). The results are an average of two to four independent experiments.
MMS2 Belongs to the RAD6 Epistasis Group.
The relative level of MMS and UV sensitivity of the mms2 null mutant, as well as the Ubc-like sequence of the Mms2 protein, led us to speculate that MMS2 may function in the RAD6 pathway. To test this hypothesis, we created mms2 rad6Δ and mms2 rad18Δ double mutants and found that, as expected, the double mutants were no more sensitive to UV (Fig. 2 A and B) and MMS (data not shown) than were the respective rad single mutants. Thus rad6 and rad18 are epistatic to mms2, indicating that MMS2 belongs to the RAD6 group. We also performed epistatic analysis of mms2 with nucleotide excision repair (rad4Δ) and recombination repair (rad50Δ) mutations. The mms2 rad4Δ (Fig. 2C) and mms2 rad50Δ (Fig. 2D) double mutants were found to be more sensitive to UV than either of the corresponding single mutants, and the killing effects appeared to be additive. Thus the MMS2 gene does not belong to the RAD3 or RAD52 epistasis groups; it is specific for the RAD6 pathway. However, unlike rad6, rad18, and other rev mutants, mms2 does not impair UV-induced mutagenesis, and the mutation frequency before and after low-dose UV-treatment actually increased to a certain degree (Table 2). This result indicates that MMS2 does not affect the mutagenesis pathway.
Table 2.
UV-induced mutagenesis of S. cerevisiae strains
| Strain (genotype) | UV dose, J/m2 | Viability, % | Reversion frequency* per 107 viable cells |
|---|---|---|---|
| DBY747 | 0 | 100 | 4.8 ± 0.8 |
| (wild type) | 2 | 100 | 20.2 ± 3.4 |
| 4 | 100 | 29.2 ± 2.6 | |
| 6 | 96 | 36.0 ± 8.4 | |
| 10 | 92 | 65.8 ± 1.3 | |
| SBU | 0 | 100 | 18.6 ± 0.4 |
| (mms2) | 2 | 93 | 45.5 ± 8.0 |
| 4 | 92 | 65.0 ± 5.0 | |
| 6 | 82 | 72.5 ± 3.6 | |
| 10 | 80 | 119.2 ± 24.4 |
DBY747 and SBU are isogenic strains carrying the trp1-289 mutation. Trp+ revertants were scored as the mean ± SD of at least two independent experiments.
MMS2 and REV3 Mutagenesis.
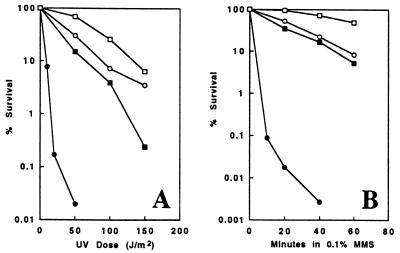
To investigate the role of MMS2 within the RAD6 pathway, we measured UV- and MMS-induced killing of the mms2 rev3Δ double mutant. To our surprise, while each of the mms2 and rev3Δ single mutants was only moderately sensitive to the DNA-damaging agents, the mms2 rev3Δ double mutant was extremely sensitive to both UV (Fig. 3A) and MMS (Fig. 3B); the levels of mms2 rev3Δ sensitivity to killing by UV and MMS were actually comparable to those of an isogenic rad18Δ single mutant (Fig. 2B, Fig. 3, and data not shown). The effect of the two mutations was clearly synergistic, because the fractions of mms2 rev3Δ surviving after the highest dose of UV (50 J/m2) or 40-min treatment with 0.1% MMS were 1,000-fold lower than expected if the mms2 and rev3Δ effects were simply additive (Fig. 3). This result led us to speculate that MMS2 represents a repair pathway that is an alternative to REV3 mutagenesis –namely, the error-free PRR pathway.
Figure 3.
mms2 is synergistic to rev3 with respect to both UV (A) and MMS (B) sensitivity. □, DBY747 (wild type); ▪, SBU (mms2); ○, WXY9382 (rev3Δ); •, SBUr3L (rev3Δmms2). The results are an average of two independent experiments.
We further predicted that if the mms2 mutation affects only error-free PRR, it may increase the potential for mutagenesis by channeling lesions to the error-prone pathway. The spontaneous mutation rates of mms2 and rev3Δ single mutants and the mms2 rev3Δ double mutant were measured. The mms2 single mutant increased the spontaneous mutation rate by >30-fold at the trp1-289 allele, and this increase is completely dependent on the functional REV3 gene, because the spontaneous mutation rate of the mms2 rev3Δ double mutant is as low as the rev3Δ single mutant (Table 3). This result is consistent with a role for MMS2 in PRR apart from REV3 mutagenesis.
Table 3.
Spontaneous mutation rates of S. cerevisiae strains
| Strain | Key alleles | Mutation rate* × 109 | Relative rate† |
|---|---|---|---|
| DBY747 | Wild type | 7.0 ± 4.4 | 1 |
| SBU | mms2 | 216.6 ± 50.0 | 31.0 |
| WXY9382 | rev3Δ | 8.7 ± 5.0 | 1.2 |
| SBUr3L | mms2 rev3Δ | 8.7 ± 5.0 | 1.2 |
All strains are isogenic and carry the revertable trp1-289 amber mutation. Rates are expressed as number of revertants per cell per generation. The results shown are the mean ± SD of five (DBY747 and SBU) or three (WXY9382 and SBUr3L) sets of experiments.
Relative to the wild-type strain.
mms2 Is Hypostatic to rad6Δ1–9.
Some of the mms2 mutant phenotypes, including the intermediate level of UV sensitivity and increased mutagenesis, are remarkably similar to the rad6 mutant with an N-terminal deletion, rad6Δ1–9, described by the Prakash laboratories (14). We reasoned that the rad6Δ1–9 mutation may also affect the error-free PRR but not the mutagenesis pathway. A critical experiment would be to see whether rad6Δ1–9 and mms2 mutations show an epistatic interaction. We constructed a rad6Δ1–9 allele in a pSCW231 plasmid as previously described (14). The resulting plasmid, pSCW-rad6Δ1–9, was used to transform rad6Δ and mms2 rad6Δ mutants to create rad6Δ1–9 and mms2 rad6Δ1–9 strains, respectively. While rad6Δ1–9 partially rescued rad6 sensitivity (cf. Fig. 2A and Fig. 4A), epistatic analysis (Fig. 4) showed that the mms2 rad6Δ1–9 double mutant was as sensitive to killing by both UV (Fig. 4A) and MMS (Fig. 4B) as was the rad6Δ1–9 single mutant. Thus, mms2 not only is hypostatic to rad6Δ it is also hypostatic to the rad6Δ1–9 deletion with respect to both UV and MMS sensitivity.
Figure 4.
mms2 is hypostatic to rad6Δ1–9 with respect to both UV (A) and MMS (B) sensitivity. □, DBY747 (wild type); ▪, SBU (mms2); ○, WXY9376/pSCW-rad6Δ1–9 (rad6Δ1–9); •, SBU6L/pSCW-rad6Δ1–9 (mms2 rad6Δ1–9). The results are an average of two independent experiments.
DISCUSSION
Several lines of direct and indirect evidence support the assertion that the MMS2 gene belongs to the RAD6 epistasis group. First, the mms2 null mutant is sensitive to killing by both MMS and UV at the level characteristic of a RAD6-pathway mutant. This is in contrast to the rad3-pathway mutants, which are especially sensitive to UV and chemicals that produce structurally distorting lesions, but are marginally sensitive to MMS; and to the rad52-pathway mutants, which are extremely sensitive to ionizing radiations and MMS, but less sensitive to UV compared with mutants belonging to the other two pathways (9, 24, 27). Second, the deduced Mms2 is homologous to all Ubcs, indicating its possible involvement in ubiquitination, an activity required for the RAD6 DNA repair pathway. Finally, the mms2 mutation is hypostatic to both rad6 and rad18, but is additive to rad4 and rad50, suggesting that it is defective in the RAD6 pathway. Within the RAD6 pathway, mms2 is hypostatic to the N-terminal deletion rad6Δ1–9 but is synergistic with the rev3 mutation. These seemingly controversial results are reconcilable, taking into account the unique ability of RAD6 to control two parallel subpathways, namely, error-free PRR and mutagenesis. While the mutagenesis pathway is known to be mediated by REV3, encoding the catalytic subunit of Polζ, as well as other genes such as REV1 (40, 41) and REV7 (23), little is known about the error-free PRR pathway. Our declaration that MMS2 functions in an error-free PRR pathway alternative to the error-prone pathway is based on the combined evidence that MMS2 belongs to the RAD6 epistasis group, that the mms2 mutant does not affect REV3-mediated spontaneous and UV-induced mutagenesis, and that the mms2 mutation is synergistic with the rev3 mutation. Our analysis of the rad6Δ1–9 mutation, along with the results of others (14), also demonstrated that, like MMS2, the N terminus of Rad6 is required for error-free PRR but not for mutagenesis. The DNA repair phenotypes of mms2 are also strikingly similar to the phenotype of the pol30-46 mutant, including the level of UV sensitivity, epistasis to rad6 and rad18, and synergism with rev3 (25). It is interesting to note that a pol3-3 (polδ) mutant at a restrictive temperature impairs PRR (26) and that a pol3-13 mutant has been characterized as having a defect in the RAD6 pathway (42). Very recently, an S. cerevisiae RAD30 gene, encoding an E. coli DinB and UmuC homolog, was reported to function in the error-free PRR pathway (43). However, unlike mms2, the rad30 mutation does not seem to increase spontaneous mutation rate, nor does it show a synergistic interaction with rev3 with respect to UV sensitivity (43). Nevertheless, it will be of great interest to extend epistatic analysis to encompass mms2, rad6Δ1–9, pol30-46, pol3-13, and rad30 to determine whether they do in fact belong to the same epistasis subgroup.
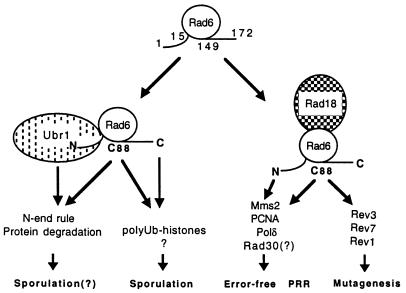
Although the mms2 single mutant is only moderately sensitive to MMS and UV, we argue that the mms2 mutation, and probably rad6Δ1–9 and pol30-46 as well, may be completely defective in the error-free PRR pathway. This argument is derived from results demonstrating that the mms2 rev3 and pol30-46 rev3 double mutants are as sensitive to killing by UV as their respective rad18 single mutants. In other words, simultaneous defects in both error-free PRR (mms2 or pol30-46) and mutagenesis (rev3) branches are equivalent to a mutation (rad18) known to be defective in both pathways. Furthermore, we have shown that the increased spontaneous mutation rate observed in the mms2 mutant is completely dependent on the functional REV3, indicating that when error-free PRR is dysfunctional, all the spontaneous lesions could be channeled to the mutagenesis pathway. A working model is presented in Fig. 5 to illustrate our current understanding of the RAD6 DNA repair pathway. PCNA and Polδ likely function in the late stage of PRR for DNA synthesis. Considering that Ubcs may form either a heterodimer (44, 45) or a homodimer (46), and that Ubc-like proteins are similar to Ubcs not only in sequence but also in predicted secondary and tertiary structure (38), it is possible that Mms2 physically interacts with Rad6 to direct the Rad6–Rad18 complex to PRR, and that this interaction may require the Rad6 N terminus. The N terminus of Rad6 is also essential for its interaction with Ubr1 and for sporulation (14, 15). Indeed, we found that mms2 mutant strains are partially defective in both N-end-rule protein degradation and sporulation (unpublished observations), suggesting that Mms2 may physically interact with Rad6.
Figure 5.
Schematic diagram of possible Rad6-mediated metabolic pathways. Rad6 forms distinct complexes with either Ubr1 or Rad18; the Rad6–Rad18 complex is proposed to be responsible for DNA repair (19–21). The DNA repair component consists of an N-terminal-dependent error-free PRR pathway and an N-terminal-independent error-prone mutagenesis pathway. Mms2, PCNA, Polδ, and Rad30 are proposed to participate in the error-free PRR, whereas Rev1, -3, and -7 are responsible for mutagenesis.
We have consistently observed a >30-fold increase in spontaneous mutation rate of the mms2 mutant. This is compared with a fewfold increase in other DNA repair mutants (e.g., rad1Δ, rad6Δ, rad18Δ, apn1Δ, and mgt1Δ; data not shown), and an approximately 20-fold increase with the msh2 DNA mismatch repair mutant (47), in an otherwise isogenic background. Hence, mms2 is probably one of the most prominent mutators reported in budding yeast. The rad6Δ1–9 (14) and pol30-46 (25) mutations also appear to increase the spontaneous mutation frequency, albeit to a lesser degree than mms2, a difference due either to experimental systems, including strain backgrounds, different reversion alleles, and experimental protocols, or to a possibility that rad6Δ1–9 and pol30-46 mutations affect other cellular processes in addition to error-free PRR.
Genetic instability of the human genome contributes to cancer and other diseases (24, 48). Like other DNA repair pathways (24), the DNA PRR and mutagenesis pathways also appear to be highly conserved from yeast to human. For example, two human Rad6 homologs have been identified (13); inactivation of HR6B from mice results in phenotypes parallel to the yeast rad6 mutant (49). In addition, a hREV3 cDNA encoding a putative human Polζ has been identified (50). The human MMS2 homolog, CROC1, initially isolated by its ability to transactivate the c-fos enhancer element (37) was also found to be down-regulated in human colon carcinoma cells upon chemical-induced differentiation (38) and to be up-regulated when simian virus 40-transformed human embryonic kidney cells became immortal (L. Ma, S.B., C. Lavery, S. Lin, W.X., and S. Bacchetti, unpublished results), suggesting that CROC1 may play a role in tumorigenesis and carcinogenesis. We have also isolated a cDNA encoding another MMS2 homolog that is highly related but distinct from CROC1 (unpublished data). Hence, understanding the role of MMS2 in error-free PRR and its implication in REV3 mutagenesis may prove to be relevant to public health.
Acknowledgments
We thank many laboratories for providing plasmids, Dr. M. Rose for the yeast genomic library, Drs. S. Bacchetti and L. Ma for a yeast mms2Δ strain, Dr. V. Chau for sharing unpublished data, and Dr. M. J. Ellison for valuable discussion. We also thank Dr. J. Game and an anonymous reviewer for their valuable comments. This work was supported by Medical Research Council of Canada operating grant MT-12633 and National Cancer Institute of Canada operating grant NCIC7412 to W.X. W.X. is a Research Scientist of the National Cancer Institute of Canada, and S.B. is supported by a University of Saskatchewan Graduate Fellowship.
Footnotes
This paper was submitted directly (Track II) to the Proceedings Office.
Abbreviations: MMS, methyl methanesulfonate; PCNA, proliferating cell nuclear antigen; Pol, DNA polymerase; PRR, postreplication repair; Ub, ubiquitin; Ubc, ubiquitin-conjugating enzyme.
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. U66724).
References
- 1.Finley D, Bartel B, Varshavsky A. Nature (London) 1989;338:394–401. doi: 10.1038/338394a0. [DOI] [PubMed] [Google Scholar]
- 2.Chen P, Johnson P, Sommer T, Jentsch S, Hochstrasser M. Cell. 1993;74:357–369. doi: 10.1016/0092-8674(93)90426-q. [DOI] [PubMed] [Google Scholar]
- 3.Gobel M G, Yochem J, Jentsch S, McGrath J P, Varshavsky A, Byers B. Science. 1988;241:1331–1335. doi: 10.1126/science.2842867. [DOI] [PubMed] [Google Scholar]
- 4.Jentsch S, McGrath J P, Varshavsky A. Nature (London) 1987;329:131–134. doi: 10.1038/329131a0. [DOI] [PubMed] [Google Scholar]
- 5.Finley D, Ozkaynak E, Varshavsky A. Cell. 1987;48:1035–1046. doi: 10.1016/0092-8674(87)90711-2. [DOI] [PubMed] [Google Scholar]
- 6.Finley D, Chau V. Annu Rev Cell Biol. 1991;7:25–69. doi: 10.1146/annurev.cb.07.110191.000325. [DOI] [PubMed] [Google Scholar]
- 7.Varshavsky A. Proc Natl Acad Sci USA. 1996;93:12142–12149. doi: 10.1073/pnas.93.22.12142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Reynolds P, Weber S, Prakash L. Proc Natl Acad Sci USA. 1985;82:168–172. doi: 10.1073/pnas.82.1.168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Prakash S, Sung P, Prakash L. Annu Rev Genet. 1993;27:33–70. doi: 10.1146/annurev.ge.27.120193.000341. [DOI] [PubMed] [Google Scholar]
- 10.Sung P, Prakash S, Prakash L. Proc Natl Acad Sci USA. 1990;87:2695–2699. doi: 10.1073/pnas.87.7.2695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Reynolds P, Koken M H M, Hoeijmakers J H J, Prakash S, Prakash L. EMBO J. 1990;9:1423–1430. doi: 10.1002/j.1460-2075.1990.tb08258.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Koken M H M, Reynolds P, Bootsma D, Hoeijmakers J H J, Prakash S, Prakash L. Proc Natl Acad Sci USA. 1990;88:3832–3836. doi: 10.1073/pnas.88.9.3832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Koken M H M, Reynolds P, Jaspers-Dekker I, Prakash L, Prakash S, Bootsma D, Hoeijmakers J H J. Proc Natl Acad Sci USA. 1990;88:8865–8869. doi: 10.1073/pnas.88.20.8865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Watkins J F, Sung P, Prakash S, Prakash L. Genes Dev. 1993;7:250–261. doi: 10.1101/gad.7.2.250. [DOI] [PubMed] [Google Scholar]
- 15.Dohmen R J, Madura K, Bartel B, Varshavsky A. Proc Natl Acad Sci USA. 1991;88:7351–7355. doi: 10.1073/pnas.88.16.7351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sung P, Berleth E, Pickart C, Prakash S, Prakash L. EMBO J. 1991;10:2187–2193. doi: 10.1002/j.1460-2075.1991.tb07754.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Morrison A, Miller E J, Prakash L. Mol Cell Biol. 1988;8:1179–1185. doi: 10.1128/mcb.8.3.1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sung P, Prakash S, Prakash L. Genes Dev. 1988;2:1476–1485. doi: 10.1101/gad.2.11.1476. [DOI] [PubMed] [Google Scholar]
- 19.Bailly V, Lamb J, Sung P, Prakash S, Prakash L. Genes Dev. 1994;8:811–820. doi: 10.1101/gad.8.7.811. [DOI] [PubMed] [Google Scholar]
- 20.Bailly V, Prakash S, Prakash L. Mol Cell Biol. 1997;17:4536–4543. doi: 10.1128/mcb.17.8.4536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bailly V, Lauder S, Prakash S, Prakash L. J Biol Chem. 1997;272:23360–23365. doi: 10.1074/jbc.272.37.23360. [DOI] [PubMed] [Google Scholar]
- 22.Morrison A, Christensen R B, Alley J, Beck A K, Bernstine E G, Lemontt J F, Lawrence C W. J Bacteriol. 1989;171:5659–5667. doi: 10.1128/jb.171.10.5659-5667.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nelson J R, Lawrence C W, Hinkle D C. Science. 1996;272:1646–1649. doi: 10.1126/science.272.5268.1646. [DOI] [PubMed] [Google Scholar]
- 24.Friedberg E C, Walker G, Siede W. DNA Repair and Mutagenesis. Washington, DC: Am. Soc. Microbiol.; 1995. [Google Scholar]
- 25.Torres-Ramos C A, Yoder B L, Burgers P M J, Prakash S, Prakash L. Proc Natl Acad Sci USA. 1996;93:9676–9681. doi: 10.1073/pnas.93.18.9676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Torres-Ramos C A, Prakash S, Prakash L. J Biol Chem. 1997;272:25445–25448. doi: 10.1074/jbc.272.41.25445. [DOI] [PubMed] [Google Scholar]
- 27.Xiao W, Chow B L, Rathgeber L. Curr Genet. 1996;30:461–468. doi: 10.1007/s002940050157. [DOI] [PubMed] [Google Scholar]
- 28.Boeke J D, Trueheart J, Natsoulis G, Fink G R. Methods Enzymol. 1987;154:164–175. doi: 10.1016/0076-6879(87)54076-9. [DOI] [PubMed] [Google Scholar]
- 29.Sherman F, Fink G R, Hicks J. Methods in Yeast Genetics. Plainview, NY: Cold Spring Harbor Lab. Press; 1983. [Google Scholar]
- 30.Rose M D, Novick P, Thomas J H, Botstein D, Fink G R. Gene. 1987;60:237–243. doi: 10.1016/0378-1119(87)90232-0. [DOI] [PubMed] [Google Scholar]
- 31.Sanger E, Nicklen S, Coulson A R. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gietz R D, Sugino A. Gene. 1988;74:527–534. doi: 10.1016/0378-1119(88)90185-0. [DOI] [PubMed] [Google Scholar]
- 33.Berben G, Dumont V, Bolle P-A, Hilger F. Yeast. 1991;7:475–477. doi: 10.1002/yea.320070506. [DOI] [PubMed] [Google Scholar]
- 34.Von Borstel R C. Methods Cell Biol. 1978;20:1–24. doi: 10.1016/s0091-679x(08)62005-1. [DOI] [PubMed] [Google Scholar]
- 35.Williamson M S, Game J C, Fogel S. Genetics. 1985;110:609–646. doi: 10.1093/genetics/110.4.609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Prakash L, Prakash S. Genetics. 1977;86:33–55. doi: 10.1093/genetics/86.1.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rothofsky M L, Lin S L. Gene. 1997;195:141–149. doi: 10.1016/s0378-1119(97)00097-8. [DOI] [PubMed] [Google Scholar]
- 38.Sancho E, Vila M R, Sanchez-Pulido L, Lozano J J, Paciucci R, Nadal M, Fox M, Harvey C, Bercovich B, Loukili N, Ciechanover A, Lin S L, Sanz F, Estivill X, Valencia A, Thomson T M. Mol Cell Biol. 1998;18:576–589. doi: 10.1128/mcb.18.1.576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ellison K S, Gwozd T, Prendergast J A, Paterson M C, Ellison M J. J Biol Chem. 1991;266:24116–24120. [PubMed] [Google Scholar]
- 40.Larimer F W, Perry J R, Hardigree A A. J Bacteriol. 1989;171:230–237. doi: 10.1128/jb.171.1.230-237.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nelson J R, Lawrence C W, Hinkle D C. Nature (London) 1996;382:729–731. doi: 10.1038/382729a0. [DOI] [PubMed] [Google Scholar]
- 42.Giot L, Chanet R, Simon M, Facca C, Faye G. Genetics. 1997;146:1239–1251. doi: 10.1093/genetics/146.4.1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.McDonald J P, Levine A S, Woodgate R. Genetics. 1997;147:1557–1568. doi: 10.1093/genetics/147.4.1557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Silver E T, Gwozd T J, Ptak C, Goebl M, Ellison M J. EMBO J. 1992;11:3091–3098. doi: 10.1002/j.1460-2075.1992.tb05381.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chen P, Johnson P, Sommer T, Jentsch S, Hochstrasser M. Cell. 1993;74:357–369. doi: 10.1016/0092-8674(93)90426-q. [DOI] [PubMed] [Google Scholar]
- 46.Ptak C, Prendergast, Hodgins R, Kay C M, Chau V, Ellison M J. J Biol Chem. 1994;269:26539–26545. [PubMed] [Google Scholar]
- 47.Xiao W, Rathgeber L, Fontanie T, Bawa S. Carcinogenesis. 1995;16:1933–1939. doi: 10.1093/carcin/16.8.1933. [DOI] [PubMed] [Google Scholar]
- 48.Lengauer C, Kinzler K W, Vogelstein B. Nature (London) 1997;386:623–627. doi: 10.1038/386623a0. [DOI] [PubMed] [Google Scholar]
- 49.Roest H P, van Klaveren J, de Wit J, van Gurp C G, Koken M H M, Vermey M, van Roijen J H, Hoogerbrugge J W, Vreeburg J T M, Baarends W M, Bootsma D, Grootegoed J A, Hoeijmakers J H J. Cell. 1996;86:799–810. doi: 10.1016/s0092-8674(00)80154-3. [DOI] [PubMed] [Google Scholar]
- 50.Xiao, W., Lechler, T., Chow, B. L., Fontanie, T., Agustus, M., Carter, K. C. & Wei, Y.-F. (1998) Carcinogenesis, in press. [DOI] [PubMed]