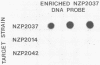
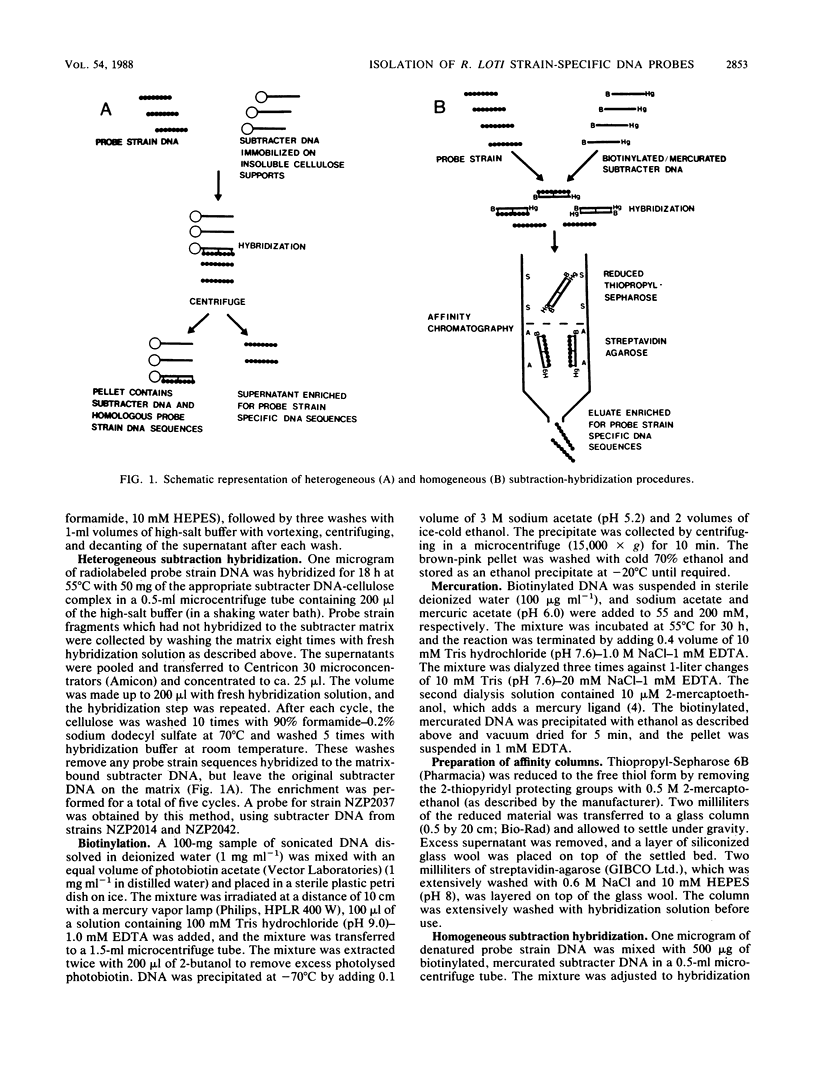
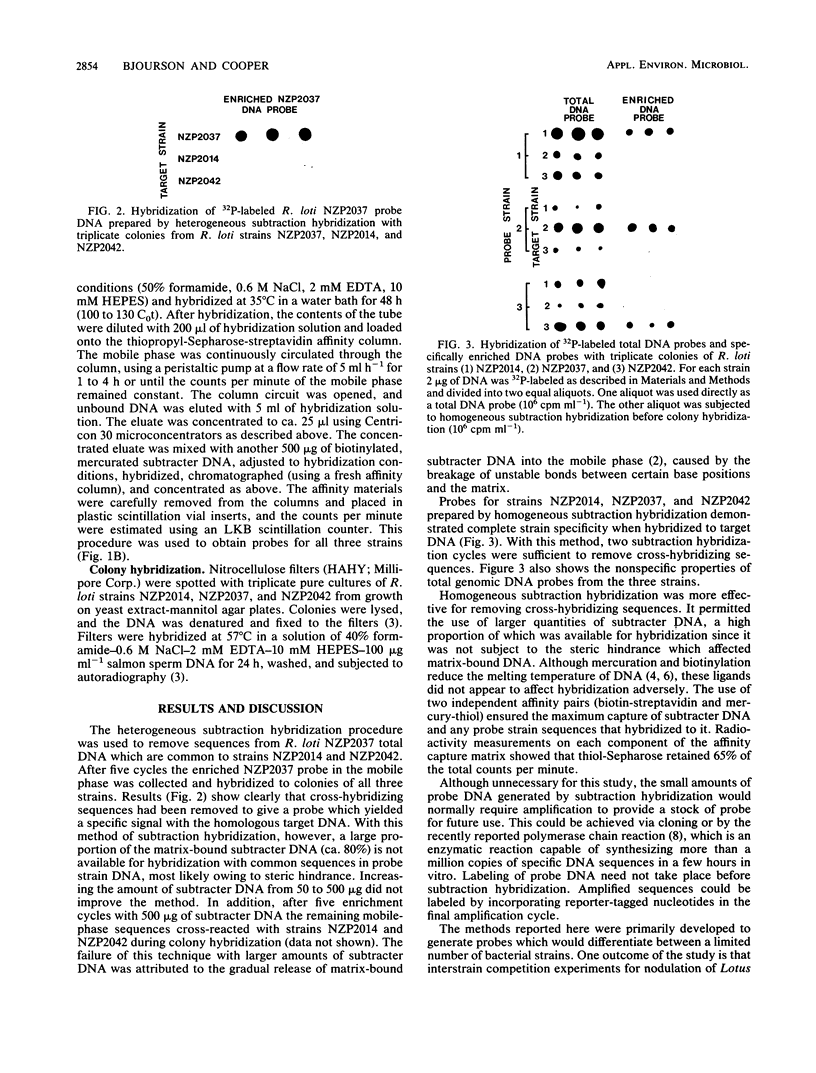
Abstract
Mixed-phase (heterogeneous) and single-phase (homogeneous) DNA subtraction-hybridization methods were used to isolate specific DNA probes for closely related Rhizobium loti strains. In the heterogeneous method, DNA from the prospective probe strain was repeatedly hybridized to a mixture of DNA from cross-hybridizing strains (subtracter DNA) which was immobilized on an epoxy-activated cellulose matrix. Probe strain sequences which shared homology with the matrix-bound subtracter DNA hybridized to it, leaving unique probe strain sequences in the mobile phase. In the homogeneous method, probe strain sequences were hybridized in solution to biotinylated, mercurated subtracter DNA. Biotinylated, mercurated subtracer DNA and probe strain sequences hybridized to it were removed by two-step affinity chromatography on streptavidin-agarose and thiol-Sepharose. The specificity of the sequences remaining after subtraction hybridization by both methods was assessed and compared by colony hybridization with R. loti strains. Both methods allowed the rapid isolation of strain-specific DNA fragments which were suitable for use as probes.
Full text
PDF

Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bünemann H., Westhoff P., Herrmann R. G. Immobilization of denatured DNA to macroporous supports: I. Efficiency of different coupling procedures. Nucleic Acids Res. 1982 Nov 25;10(22):7163–7180. doi: 10.1093/nar/10.22.7163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper J. E., Bjourson A. J., Thompson J. K. Identification of lotus rhizobia by direct DNA hybridization of crushed root nodules. Appl Environ Microbiol. 1987 Jul;53(7):1705–1707. doi: 10.1128/aem.53.7.1705-1707.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dale R. M., Ward D. C. Mercurated polynucleotides: new probes for hybridization and selective polymer fractionation. Biochemistry. 1975 Jun 3;14(11):2458–2469. doi: 10.1021/bi00682a028. [DOI] [PubMed] [Google Scholar]
- Langer P. R., Waldrop A. A., Ward D. C. Enzymatic synthesis of biotin-labeled polynucleotides: novel nucleic acid affinity probes. Proc Natl Acad Sci U S A. 1981 Nov;78(11):6633–6637. doi: 10.1073/pnas.78.11.6633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saiki R. K., Gelfand D. H., Stoffel S., Scharf S. J., Higuchi R., Horn G. T., Mullis K. B., Erlich H. A. Primer-directed enzymatic amplification of DNA with a thermostable DNA polymerase. Science. 1988 Jan 29;239(4839):487–491. doi: 10.1126/science.2448875. [DOI] [PubMed] [Google Scholar]
- Scott M. R., Westphal K. H., Rigby P. W. Activation of mouse genes in transformed cells. Cell. 1983 Sep;34(2):557–567. doi: 10.1016/0092-8674(83)90388-4. [DOI] [PubMed] [Google Scholar]
- Welcher A. A., Torres A. R., Ward D. C. Selective enrichment of specific DNA, cDNA and RNA sequences using biotinylated probes, avidin and copper-chelate agarose. Nucleic Acids Res. 1986 Dec 22;14(24):10027–10044. doi: 10.1093/nar/14.24.10027. [DOI] [PMC free article] [PubMed] [Google Scholar]