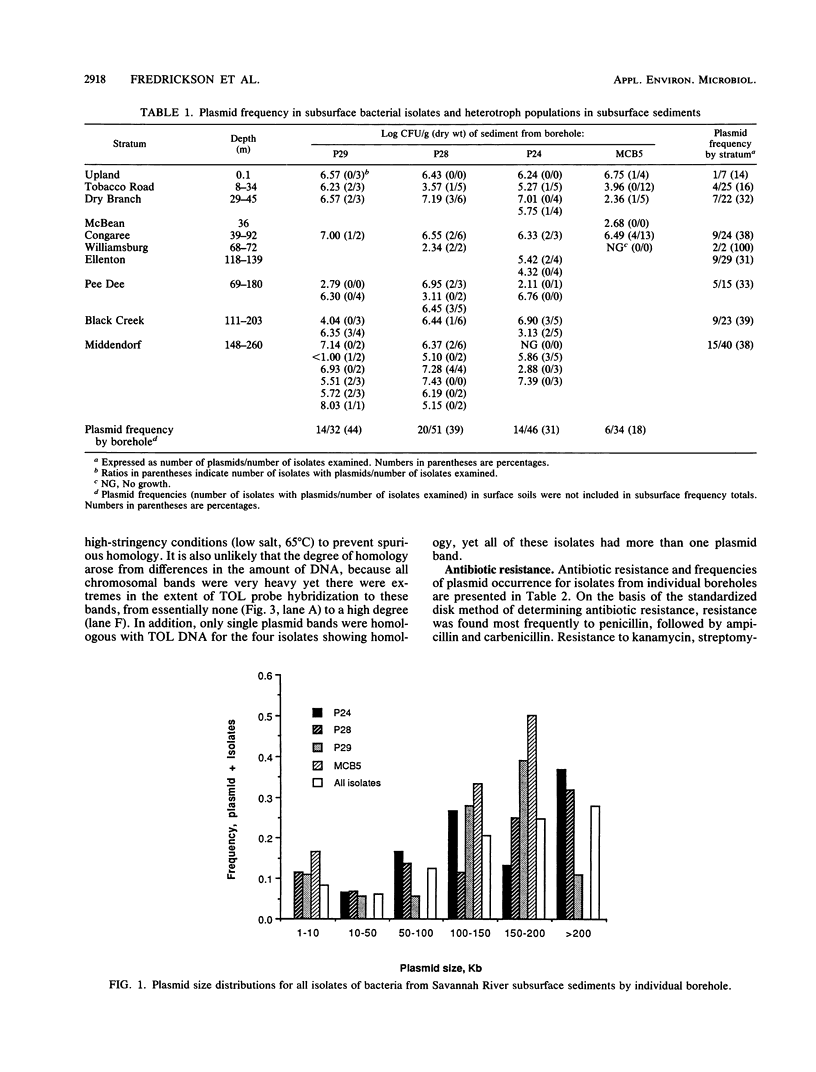
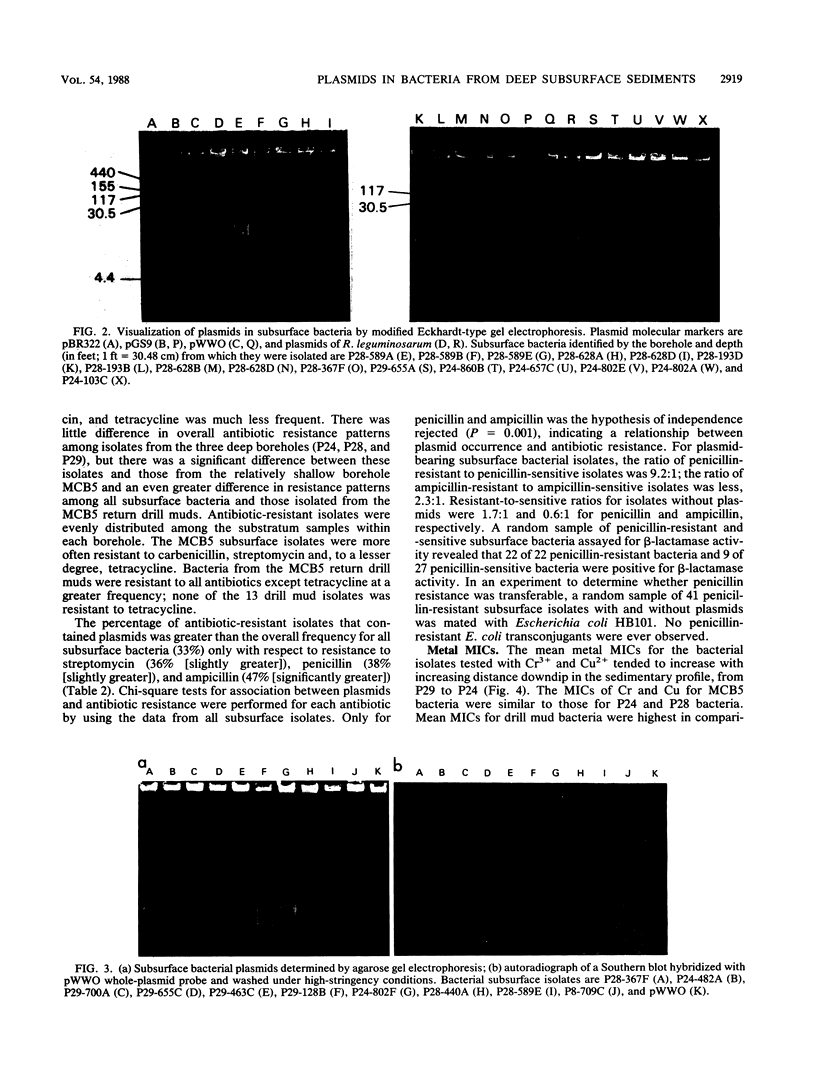
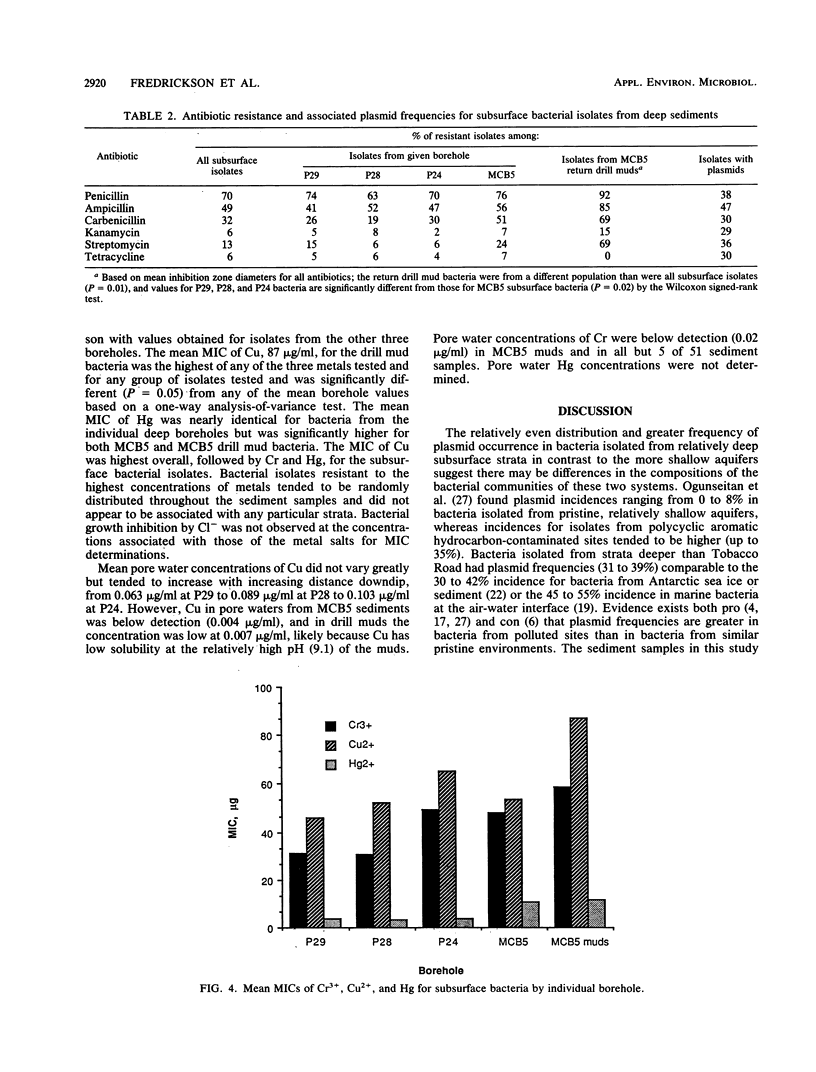
Abstract
Bacteria were isolated from deep terrestrial subsurface sediments underlying the coastal plain of South Carolina. A total of 163 isolates from deep sediments, surface soil, and return drill muds were examined for plasmid DNA content and resistance to the antibiotics penicillin, ampicillin, carbenicillin, streptomycin, kanamycin, and tetracycline. MICs of Cu2+, Cr3+, and Hg2+ for each isolate were also determined. The overall frequency of plasmid occurrence in the subsurface bacteria was 33%. Resistance was most frequent to penicillin (70% of all isolates), ampicillin (49%), and carbenicillin (32%) and was concluded to be related to the concentrations of the individual antibiotics in the disks used for assaying resistance and to the production of low levels of β-lactamase. The frequencies of resistance to penicillin and ampicillin were significantly greater for isolates bearing plasmids than for plasmidless isolates; however, resistance was not transferable to penicillin-sensitive Escherichia coli. Hybridization of subsurface bacterial plasmids and chromosomal DNA with a whole-TOL-plasmid (pWWO) probe revealed some homology of subsurface bacterial plasmid and chromosomal DNAs, indicating a potential for those bacteria to harbor catabolic genes on plasmids or chromosomes. The incidences of antibiotic resistance and MICs of metals for subsurface bacteria were significantly different from those for drill mud bacteria, ruling out the possibility that bacteria from sediments were derived from drill muds.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Balkwill D. L., Ghiorse W. C. Characterization of subsurface bacteria associated with two shallow aquifers in oklahoma. Appl Environ Microbiol. 1985 Sep;50(3):580–588. doi: 10.1128/aem.50.3.580-588.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauer A. W., Kirby W. M., Sherris J. C., Turck M. Antibiotic susceptibility testing by a standardized single disk method. Am J Clin Pathol. 1966 Apr;45(4):493–496. [PubMed] [Google Scholar]
- Baya A. M., Brayton P. R., Brown V. L., Grimes D. J., Russek-Cohen E., Colwell R. R. Coincident plasmids and antimicrobial resistance in marine bacteria isolated from polluted and unpolluted Atlantic Ocean samples. Appl Environ Microbiol. 1986 Jun;51(6):1285–1292. doi: 10.1128/aem.51.6.1285-1292.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bopp L. H., Chakrabarty A. M., Ehrlich H. L. Chromate resistance plasmid in Pseudomonas fluorescens. J Bacteriol. 1983 Sep;155(3):1105–1109. doi: 10.1128/jb.155.3.1105-1109.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burton N. F., Day M. J., Bull A. T. Distribution of bacterial plasmids in clean and polluted sites in a South Wales river. Appl Environ Microbiol. 1982 Nov;44(5):1026–1029. doi: 10.1128/aem.44.5.1026-1029.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell A. Evolutionary significance of accessory DNA elements in bacteria. Annu Rev Microbiol. 1981;35:55–83. doi: 10.1146/annurev.mi.35.100181.000415. [DOI] [PubMed] [Google Scholar]
- Eckhardt T. A rapid method for the identification of plasmid desoxyribonucleic acid in bacteria. Plasmid. 1978 Sep;1(4):584–588. doi: 10.1016/0147-619x(78)90016-1. [DOI] [PubMed] [Google Scholar]
- Foster T. J. Plasmid-determined resistance to antimicrobial drugs and toxic metal ions in bacteria. Microbiol Rev. 1983 Sep;47(3):361–409. doi: 10.1128/mr.47.3.361-409.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franklin F. C., Lehrbach P. R., Lurz R., Rueckert B., Bagdasarian M., Timmis K. N. Localization and functional analysis of transposon mutations in regulatory genes of the TOL catabolic pathway. J Bacteriol. 1983 May;154(2):676–685. doi: 10.1128/jb.154.2.676-685.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fredrickson J. K., Bezdicek D. F., Brockman F. J., Li S. W. Enumeration of Tn5 mutant bacteria in soil by using a most- probable-number-DNA hybridization procedure and antibiotic resistance. Appl Environ Microbiol. 1988 Feb;54(2):446–453. doi: 10.1128/aem.54.2.446-453.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hada H. S., Sizemore R. K. Incidence of Plasmids in Marine Vibrio spp. Isolated from an Oil Field in the Northwestern Gulf of Mexico. Appl Environ Microbiol. 1981 Jan;41(1):199–202. doi: 10.1128/aem.41.1.199-202.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanson C. W., Martin W. J. Microwave oven for melting laboratory media. J Clin Microbiol. 1978 Apr;7(4):401–402. doi: 10.1128/jcm.7.4.401-402.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harvey R. W., Smith R. L., George L. Effect of organic contamination upon microbial distributions and heterotrophic uptake in a Cape Cod, Mass., aquifer. Appl Environ Microbiol. 1984 Dec;48(6):1197–1202. doi: 10.1128/aem.48.6.1197-1202.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hermansson M., Jones G. W., Kjelleberg S. Frequency of antibiotic and heavy metal resistance, pigmentation, and plasmids in bacteria of the marine air-water interface. Appl Environ Microbiol. 1987 Oct;53(10):2338–2342. doi: 10.1128/aem.53.10.2338-2342.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelch W. J., Lee J. S. Antibiotic resistance patterns of gram-negative bacteria isolated from environmental sources. Appl Environ Microbiol. 1978 Sep;36(3):450–456. doi: 10.1128/aem.36.3.450-456.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobori H., Sullivan C. W., Shizuya H. Bacterial plasmids in antarctic natural microbial assemblages. Appl Environ Microbiol. 1984 Sep;48(3):515–518. doi: 10.1128/aem.48.3.515-518.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ladd T. I., Ventullo R. M., Wallis P. M., Costerton J. W. Heterotrophic activity and biodegradation of labile and refractory compounds by groundwater and stream microbial populations. Appl Environ Microbiol. 1982 Aug;44(2):321–329. doi: 10.1128/aem.44.2.321-329.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinclair J. L., Ghiorse W. C. Distribution of protozoa in subsurface sediments of a pristine groundwater study site in oklahoma. Appl Environ Microbiol. 1987 May;53(5):1157–1163. doi: 10.1128/aem.53.5.1157-1163.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Summers A. O., Jacoby G. A. Plasmid-determined resistance to boron and chromium compounds in Pseudomonas aeruginosa. Antimicrob Agents Chemother. 1978 Apr;13(4):637–640. doi: 10.1128/aac.13.4.637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timoney J. F., Port J., Giles J., Spanier J. Heavy-metal and antibiotic resistance in the bacterial flora of sediments of New York Bight. Appl Environ Microbiol. 1978 Sep;36(3):465–472. doi: 10.1128/aem.36.3.465-472.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]




