Abstract
Thymocytes are positively or negatively selected depending on interactions between their T cell receptors (TCR) and peptides presented by major histocompatibility complex molecules. We have previously shown that apoptosis of thymocytes from an αβ TCR-transgenic mouse (F5), induced by antigenic peptide, can be inhibited specifically by an antagonist peptide variant in an in vitro culture model. We have now extended these experiments by demonstrating that the antagonist peptide can inhibit natural negative selection of maturing thymocytes, induced by endogenously expressed antigen, in fetal thymic organ cultures (FTOC). This inhibition resulted in the rescue and maturation of thymocytes that would otherwise have been deleted. Mature T cells generated in these cultures were able to respond to antigen by producing limited quantities of interferon-γ, but unlike T cells from control FTOC, they required exogenous interleukin-2 to generate cytolytic effector cells. Interestingly, the antagonist peptide also accelerated the development of F5 thymocytes in the absence of the negatively selecting ligand. These data suggest that the developmental fate of a thymocyte may be determined by the recognition of multiple distinct peptide ligands during thymic selection. Alterations in the profiles of selecting peptides presented in the thymus would thus have profound effects on the size and autoreactive potential of the T cell repertoire generated.
T lymphocytes are positively or negatively selected in the thymus as a result of interactions between their T cell receptors (TCR) and thymic peptide—major histocompatibility complexes (MHC) (1–11). Much of the recent work on thymic selection was influenced by experiments that examined T cell responses to peptide analogs, produced from the antigenic peptide by substitution of amino acid residues involved in interaction with the TCR. Such peptide analogs can generate qualitatively different T cell responses compared with those produced by the antigenic peptide (12). In particular, some analogs were shown to act as TCR antagonists and inhibit T cell responses to the antigenic peptide (13). Several studies have shown that antagonist peptides are capable of positively selecting (9, 10), negatively selecting (14, 15), or blocking positive selection (16) of thymocytes. However, other studies have found that the ability of a peptide to select thymocytes positively or negatively does not always correlate with its properties as a TCR antagonist (10, 11, 17, 18).
We have previously reported an analog peptide (19) that was capable of inhibiting antigen-specific lysis of target cells by cytotoxic T lymphocyte (CTL) from αβ TCR (F5)-transgenic mice (20, 21). Furthermore, this peptide was shown to inhibit apoptosis of transgenic thymocytes (19) in an in vitro suspension culture assay (22). Because the suspension culture could not support the development of immature thymocytes, we could not determine whether this inhibition would result in the rescue and complete maturation of thymocytes. In the present study, we have extended these experiments by examining the effect of the antagonist peptide on the selection of transgenic thymocytes in fetal thymic organ cultures (FTOC) from mice doubly transgenic for the F5 TCR and the influenza nucleoprotein, which express the antigenic NP68 peptide endogenously (23). We report that the antagonist peptide can rescue thymocytes from negative selection in such FTOC and that this results in the production of mature T cells, which are partially responsive to the antigenic peptide. In addition, we show that the antagonist peptide accelerates the positive selection of thymocytes in single transgenic F5 FTOC. Taken together, these data provide further evidence suggesting that interactions with multiple peptides can influence the selection of an individual thymocyte.
MATERIALS AND METHODS
Mice.
Mice transgenic for the αβ TCR from the F5 cytotoxic T cell clone (21) and for a truncated influenza virus A/NT/60/68 nucleoprotein (encoding the NP366–374 peptide epitope) (23) were generated in our laboratory as previously reported. Mice deficient in Rag-1 gene expression, generated by homologous recombination, were obtained from Eugenia Spanopoulou (24).
Reagents.
Three nonamer peptides were used: NP68 [from the nucleoprotein of influenza virus A/NT/60/68 (NP366–374); ASNENMDAM], NP34 [from the nucleoprotein of influenza virus A/PR/8/34 (NP366–374); ASNENMETM], and the control peptide GAG [from the gag protein of the SF2 strain of HIV (390–398); SQVTNPANI]. The peptides were synthesized on an Applied Biosystems 430A peptide synthesizer.
Fetal Thymic Organ Culture.
F5 and F5/NP fetal thymic lobes were isolated from day 15 embryos, obtained from timed matings of F5/Rag-1−/− females with Rag-1−/− and NP/Rag-1−/− males, respectively. The fetal thymic lobes were transferred onto Nucleopore polycarbonate filters (Costar) and cultured at 37°C, 5% CO2 in RPMI 1640 (GIBCO) supplemented with 10% heat-inactivated fetal calf serum, 2 mM l-glutamine, and antibiotics (RP-10 medium) for 10 days, unless stated otherwise. The lobes were either cultured in medium alone or sister lobes were divided between medium containing 10 μM NP34 or 10 μM GAG peptides. The medium was changed every 24 h during the culture period. Thymocytes were harvested for analysis by gently disrupting the thymic lobes manually in 1.5-ml Eppendorf tubes. In some cases, thymocytes from several thymic lobes were pooled and cultured in flat-bottom 96-well plates with 0.5 × 105 irradiated (3500 rad) Rag-1−/− splenocytes in RP-10 medium with 50 μM 2-mercaptoethanol, with or without 1 μM NP68. Where stated, 10 units/ml interleukin-2 (IL-2) (Genzyme) was added to the cultures. Supernatants were removed on day 2 of culture for quantification of interferon (IFN)-γ, or effector CTL were harvested on day 4 of culture.
Flow Cytometry.
For flow cytometric analysis, between 105 and 106 cells were stained with various combinations of monoclonal antibodies (mAbs). The following mAbs and second layer reagents were used: FITC-conjugated YTS169.4 [anti-CD8α (25)], biotin-conjugated KT11 [anti-Vβ11 (26)], biotin-conjugated YBM5.10 [anti-HSA (27)], phycoerythrin-conjugated anti-CD4 (Sigma), and streptavidin-RED 670 (GIBCO). For analysis of cell cycle, cells were stained with 7-aminoactinomycin D (7AAD) (Sigma) in PBS containing 0.3% saponin (Sigma), 2% fetal calf serum, and 0.1% sodium azide. Stained cells were analyzed on a FACScan flow cytometer (Becton Dickinson) using Cellquest software (Becton Dickinson).
IFN-γ ELISA.
The amount of IFN-γ in the culture supernatants was estimated using the Interest-γ ELISA kit (Genzyme) and a recombinant IFN-γ standard.
CTL Assays.
EL4 cells were labeled with 50 μCi of 51Cr-sodium chromate in RP-10 medium for 1–2 h at 37°C at a concentration of 107 cells/ml, either alone or in the presence of 10 μM NP68. Cells were then washed three times in warm RP-10 medium and resuspended at 2 × 104 cells/ml in RP-10 medium; 100-μl volumes were then plated out onto a V-bottom 96-well plate containing 100 μl of serial dilutions of thymocyte effector cells. After brief centrifugation, the plates were maintained at 37°C with 5% CO2 for 5 h, after which plates were centrifuged again and 25 μl removed and spotted onto glass fiber Spot-on filtermats (Wallac, Milton Keynes, UK). Filtermats were analyzed on a 1205 Betaplate counter (Wallac). Percent specific lysis was determined as follows: % specific lysis = [(experimental release − spontaneous release)/(maximum release − spontaneous release)] × 100. In all experiments, spontaneous release was <20%.
RESULTS
An Antagonist Peptide Inhibits Negative Selection by an Endogenously Expressed Antigenic Epitope.
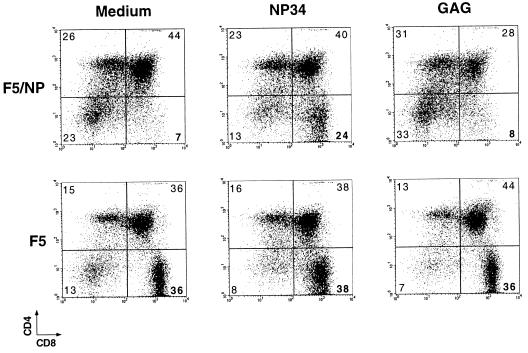
The F5 αβ TCR recognizes a nonamer peptide from influenza virus nucleoprotein (NP68) in the context of MHC class I H-2Db (28). In F5/NP68/Rag-1−/− double transgenic mice (23, 29), which express the NP68 peptide epitope endogenously, this results in the deletion of thymocytes in the transition stage between the CD8+CD4+TCRint and CD8+CD4+TCRhi subsets. Despite apparent interaction with the antigenic peptide, as judged by expression of activation markers and TCR down-regulation, a few thymocytes do mature in these mice and exit into the periphery. These T cells are unresponsive to antigen in vitro and are only capable of differentiating into effector CTL in the presence of exogenous IL-2 (23). Negative selection of transgenic thymocytes was also observed in FTOC of day 15 thymic lobes from F5/NP68/Rag-1−/− (F5/NP) double transgenic mice. After 10 days in vitro, these FTOCs contained a relatively small percentage of CD8+CD4− (CD8 SP) thymocytes, expressing low levels of the CD8 molecule, when compared with the discrete population of CD8 SP thymocytes generated in F5/Rag-1−/− (F5) FTOC (Fig. 1).
Figure 1.
The antagonist peptide NP34 rescues F5 thymocytes from negative selection in F5/NP FTOC. Fetal thymic lobes were isolated from day 15 F5/NP68/Rag-1−/− (F5/NP) and F5/Rag-1−/− (F5) embryos and were cultured alone or in the presence of 10 μM NP34 or 10 μM GAG peptides. Thymocytes were harvested at day 10 of culture and stained for CD4 and CD8 molecules. The percentage of cells in each quadrant region is indicated in the dot plots.
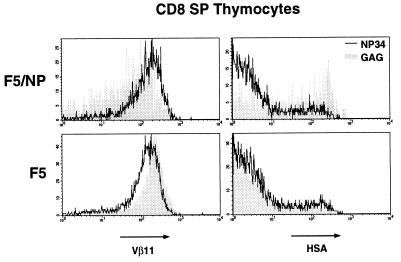
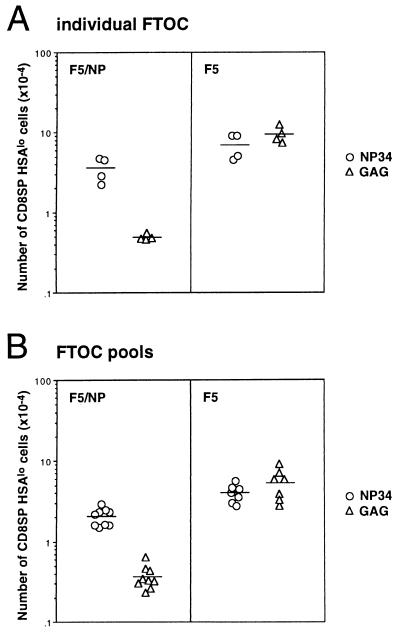
We have previously reported that the NP34 antagonist peptide can inhibit apoptosis of F5 thymocytes, in response to NP68, in an in vitro assay that used the down-regulation of the CD8 and CD4 coreceptor molecules as a sensitive measure of cell death (19). It is important to note that such coreceptor down-modulation has been indirectly implicated in positive selection as well as apoptosis (30, 31). We decided to analyze the effects of the NP34 peptide in the FTOC described above to determine whether inhibition of negative selection by peptide analogs could also occur in more physiological conditions and, more importantly, whether such inhibition would result in the rescue and development of mature thymocytes. The percentage of CD8 SP thymocytes from F5/NP thymic lobes cultured with 10 μM NP34 increased from 7–8% to 24% when compared with thymic lobes cultured alone or with 10 μM of the control GAG peptide (Fig. 1). Although NP34 had no significant effect on the percentage of CD8 SP thymocytes generated in F5 thymic lobes, this population expressed a wider range of CD8 levels than the thymocytes from control thymic lobes (Fig. 1). The control peptide GAG had no measurable effect on thymocyte development in either F5/NP or F5 FTOC. Furthermore, the CD8 SP thymocytes in NP34-treated F5/NP FTOC expressed higher TCR levels than those from GAG-treated FTOC and contained a larger proportion of HSAlo mature thymocytes (Fig. 2). Although NP34 had no effect on the proportion of mature thymocytes in the CD8 SP population from F5 FTOC, these thymocytes expressed lower levels of TCR than those from GAG-treated F5 FTOC (Fig. 2). Inhibition of negative selection in F5/NP FTOC by NP34 was also evident when absolute numbers of mature HSAlo CD8 SP thymocytes were calculated from individual thymic lobes (a 7-fold increase over GAG-treated FTOC) or from FTOC pools (a 6-fold increase) (Fig. 3). There was no significant difference in the numbers of mature thymocytes generated from F5 FTOC treated with NP34 or the control GAG peptide.
Figure 2.
CD8+CD4− thymocytes from NP34-treated FTOC have higher levels of TCR expression and contain a greater proportion of HSAlo mature thymocytes than cells from control FTOC. Histograms show the expression of Vβ11 and HSA molecules on CD8+CD4− (CD8 SP) thymocytes from FTOC treated with NP34 (solid line) or GAG (shaded plots) peptides, as in Fig. 1.
Figure 3.
The antagonist peptide NP34 stimulates the production of an increased number of mature F5 thymocytes in F5/NP FTOC. F5/NP and F5 FTOC were cultured in the presence of 10 μM NP34 (○) or 10 μM GAG (▵) peptides for 10 days. Thymocytes were harvested and stained for CD4, CD8, Vβ11, and HSA molecules. (A) Each symbol in the graph represents the absolute number of CD8+CD4−HSAlo thymocytes generated from an individual FTOC. Similar data were obtained from analysis of individual thymic lobes in six independent experiments. (B) Data from analysis of FTOC pools in nine independent experiments is shown. Each symbol represents the absolute number of CD8+CD4−HSAlo thymocytes per thymic lobe from a pool of 12–39 F5/NP and 5–20 F5 FTOC.
The Antagonist Peptide Affects the Kinetics of Thymocyte Development in F5 FTOC.
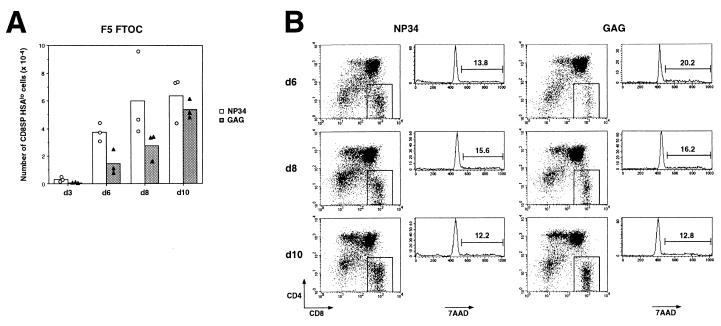
The generation of mature thymocytes was analyzed in a time-course experiment. Significant numbers of mature thymocytes could be detected by day 6 of NP34-treated F5/NP FTOC, whereas only a small number of mature thymocytes was found in control F5/NP FTOC at any of the time points analyzed (data not shown). Interestingly, although there was no difference in the final numbers of mature thymocytes produced in either NP34- or GAG-treated F5 FTOC on day 10 of culture, significantly greater numbers were found consistently in NP34-treated FTOC on days 6 and 8 of the time course (Fig. 4A). It is important to note that the effect of NP34 cannot be explained by proliferation of mature thymocytes in response to this peptide, as there was no increase in the percentage of cycling mature thymocytes in NP34-treated FTOC when compared with GAG-treated FTOC on day 6, 8, or 10 of the time course, as judged by 7AAD incorporation (Fig. 4B). In addition, mature thymocytes did not express the IL-2 receptor on day 10 of culture in any of the FTOC (data not shown). These data suggest that the antagonist peptide NP34 not only modulates selection of F5 thymocytes by the endogenously expressed antigenic peptide in F5/NP FTOC but also by the endogenous positively selecting ligands in F5 FTOC.
Figure 4.
NP34 accelerates the production of mature thymocytes in F5 FTOC. F5 FTOC were cultured in the presence of 10 μM NP34 (circles and open bars) or 10 μM GAG (triangles and shaded bars) peptides for 3, 6, 8, or 10 days. (A) Thymocytes were harvested at each time point and stained with anti-CD4, anti-CD8, and anti-HSA mAbs. Each symbol represents absolute number of CD8+CD4−HSAlo thymocytes generated from an individual FTOC, and bars represent the mean of these numbers. Similar data were obtained from two independent time course experiments. (B) Thymocytes harvested from NP34-treated (Left) and GAG-treated (Right) FTOC at days 6, 8, and 10 were stained with anti-CD4, anti-CD8, and 7AAD. Histograms show the 7AAD profiles of the CD8 SP thymocytes included in the gate shown in the dot plots. The percentage of cycling thymocytes is indicated above the marker in the histograms.
Thymocytes Rescued from Negative Selection Are Capable of Differentiating into IFN-γ-Producing Effector Cells but Require IL-2 for CTL Activity.
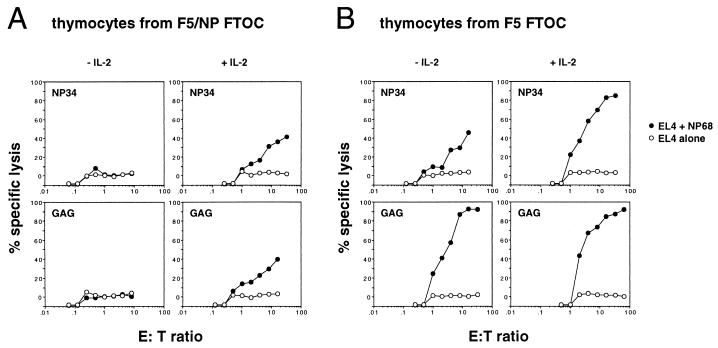
We next examined whether the mature thymocytes rescued from deletion in F5/NP FTOC were capable of responding to the antigenic peptide and generating effector T cells. Thymocytes from NP34-treated FTOC responded by producing (IFN-γ) when stimulated with NP68 presented by splenic antigen-presenting cell (APC) (Table 1), although they produced much less IFN-γ than thymocytes from F5 FTOC. In contrast, thymocytes from control GAG-treated F5/NP cultures were incapable of producing detectable amounts of IFN-γ in any of the experiments. On the other hand, thymocytes from NP34- and GAG-treated F5/NP FTOC were incapable of generating cytotoxic effector cells in vitro unless exogenous IL-2 was added to the cultures (Fig. 5A). Exogenous IL-2 was not necessary for the production of effector CTL from either NP34- or GAG-treated F5 FTOC (Fig. 5B).
Table 1.
IFN-γ production by thymocytes from FTOC in response to stimulation with antigenic peptide
| FTOC | Treatment | IFN-γ (pg/ml)* |
|---|---|---|
| F5/NP | NP34 | 167 ± 8 |
| F5/NP | GAG | — |
| F5 | NP34 | 1574 ± 26 |
| F5 | GAG | 1770 ± 34 |
Data show IFN-γ produced by thymocytes, from pools of 12 NP34- or GAG-treated FTOC, after stimulation for 2 days with APC and 1 μM NP68. Data shown are mean values and standard deviations of triplicate wells. IFN-γ was not detected (<20 pg/ml) in cultures of thymocytes from GAG-treated F5/NP FTOC (−) or when thymocytes were cultured with APC in the absence of NP68. Similar data were obtained from five independent experiments.
Figure 5.
Effector CTL can only be generated from F5/NP FTOC in the presence of exogenous IL-2. F5/NP (A) and F5 (B) FTOC were cultured in the presence of 10 μM NP34 or GAG peptides for 10 days. Thymocytes were harvested, pooled from 26 (F5/NP) or 15 (F5) FTOC, and cultured with APC and 1 μM NP68 in the presence or absence of 10 units/ml IL-2. The graphs show the % specific lysis of EL4 target cells, loaded (•) or not (○) with NP68, by CTL generated from these cultures. Each point represents the mean of duplicate wells. The data are representative of four independent experiments.
DISCUSSION
In this study, we demonstrate that an antagonist peptide NP34 can inhibit deletion of TCR transgenic thymocytes by the endogenously expressed antigenic NP68 peptide in FTOC. This inhibition resulted in the rescue and maturation of thymocytes in these cultures. Furthermore, NP34 modified positive selection of transgenic thymocytes on endogenous ligands in single transgenic F5 FTOC, accelerating the production of mature T cells.
Several studies have addressed the ability of agonist peptides to positively select TCR transgenic thymocytes. Although all these studies identified strong agonist peptide ligands that were capable of inducing the maturation of phenotypically mature T cells (10, 11, 32), the significance of such selection was called into question in one of these studies because the thymocytes selected on an agonist peptide were unable to respond to the antigenic peptide (32). A more recent study demonstrated that agonist peptides could select transgenic T cells and that although the selected T cells no longer responded to the selecting peptide, they could proliferate in response to a peptide that was a stronger agonist (17). This suggests that T cells selected on unique peptide ligands are tolerant to these ligands but may be responsive to peptides that are significantly more potent agonists (17, 21, 33). In light of this work, it was important to establish whether or not the T cells rescued from deletion in our experiments were functional.
Although the majority of thymocytes are deleted by the endogenously expressed NP68 peptide in F5/NP mice in vivo (23) and in F5/NP FTOC (see above), a small number of T cells escape this deletion and mature. These cells are unresponsive to APC presenting the antigenic peptide and require exogenous IL-2 to proliferate or differentiate into effector CTL. In contrast to the cells that mature in control F5/NP FTOC, thymocytes rescued by NP34 from deletion in F5/NP FTOC were able to produce IFN-γ in response to the antigenic peptide. However, the magnitude of this response was lower than that of thymocytes from single transgenic F5 FTOC. In addition, the rescued thymocytes required exogenous IL-2 to generate CTL. Taken together, these results demonstrate that the phenotypically mature thymocytes rescued from negative selection in F5/NP FTOC were able to respond to the antigenic peptide, although some effector functions remained compromised. Mature thymocytes from both F5/NP and F5 FTOC treated with NP34 expressed lower levels of CD8 and TCR than cells from control F5 FTOC. The reason for this down-regulation is unclear, but it could in part explain why these thymocytes were less responsive to antigenic peptide than control thymocytes (34). A recent study has shown that nondeleting concentrations of agonist peptide can lead to the generation of mature thymocytes expressing CD8 αα homodimers (35). This was not the case in the experiments described above, as CD8 SP thymocytes from both F5 and F5/NP FTOC expressed the heterodimeric CD8αβ coreceptor (data not shown).
The differences observed in the antigen-specific CTL and IFN-γ response by mature thymocytes rescued in NP34-treated F5/NP FTOC may have been due to differences in sensitivity of the assays used to detect these responses. Alternatively, these results can be explained by postulating that certain “resting thresholds” (17, 36) are set in thymocytes maturing in the presence of both NP34 and the endogenously expressed NP68. It is possible that these new “resting thresholds” are such that the mature thymocytes can only respond to antigenic peptide by production of IFN-γ. Different thresholds have been described for distinct effector responses by T cell clones against single TCR ligands (37). In the case described in the present report, induction of CTL and IFN-γ production by the rescued thymocytes may be dependent on the affinity and avidity of the stimulating peptide required to surpass the imprinted threshold for both responses.
The accelerated production of mature thymocytes in NP34-treated F5 FTOC could be due to NP34 antagonizing the negative selection of F5 thymocytes by putative endogenous ligands. However, it is also possible that NP34 directly increased the efficiency of positive selection in early stages of the cultures, as the absolute number of mature CD8 SP thymocytes produced in these and control FTOC were the same at the end of the culture period. It seems that the absolute number of cells produced in either culture was finally determined by the number of precursors originally present in the fetal thymic lobes. It is possible that NP34 also accelerated the positive selection of thymocytes in FTOC using thymic lobes from double transgenic (F5/NP) mice. However, it is unlikely that this alone would result in the increased numbers of mature thymocytes generated in NP34-treated F5/NP FTOC because the endogenously expressed NP68 peptide could interact with and delete F5 thymocytes at any stage in the culture. In some cases, thymocytes have been reported to mature directly from CD8−CD4− cells without going through the CD8+CD4+ stage (38). It is important to note that in both NP34 and GAG-treated F5 and F5/NP FTOC, the appearance of CD8+CD4+ thymocytes preceded that of mature CD8 SP thymocytes and the absolute numbers of CD8+CD4+ thymocytes declined as the numbers of mature thymocytes increased (data not shown). This suggests that NP34 did not induce the selection of F5 thymocytes before the CD8+CD4+ stage, although this possibility cannot be discounted.
It has been well established that a given TCR can interact with multiple peptides, in some cases with ones that share little or no amino acid sequence homology (39–42). In addition, multiple potential endogenous peptide ligands have been found by database searches (43, 44). In the work described here, a single peptide was shown to influence the selection of thymocytes by both a negatively selecting peptide and by unidentified positively selecting ligands. Recent studies have identified several self-peptides that were capable of interacting with and positively selecting TCR transgenic thymocytes (45, 46). These experiments suggest that multiple peptides may be involved in the processes of positive and negative selection and that the ability of a given peptide to modulate selection of thymocytes by another peptide may be of physiological importance in thymocyte maturation (47). In addition, they indicate that such interactions may determine the degree of self-reactivity exhibited by the peripheral T cell repertoire and the consequent benefits or dangers to the immune system.
Acknowledgments
We thank Dr. Kyuhei Tomonari for kindly providing us with the KT11 antibody and Dr. Eugenia Spanopoulou for the gift of Rag-1−/− mice and Trisha Norton and Mauro Tolaini for technical help. In addition, we thank Dr. Hugh Brady, Dr. Rose Zamoyska, and Dr. Gitta Stockinger for critical reading of the manuscript. This work was supported by the Medical Research Council. O.W. is supported by a fellowship from the Leukaemia Research Fund; A.W. is supported by a grant from the Boehringer Ingelheim Fonds.
Footnotes
This paper was submitted directly (Track II) to the Proceedings Office.
Abbreviations: CTL, cytotoxic T lymphocyte; FTOC, fetal thymic organ culture; IFN-γ, interferon-γ; IL-2, interleukin-2; MHC, major histocompatibility complex; NP, nucleoprotein; Rag, recombination activating gene; TCR, T cell receptor; 7AAD, 7-aminoactinomycin D; APC, antigen-presenting cells.
References
- 1.Sha W C, Nelson C A, Newberry R D, Kranz D M, Russell J H, Loh D Y. Nature (London) 1988;336:73–76. doi: 10.1038/336073a0. [DOI] [PubMed] [Google Scholar]
- 2.Kisielow P, Bluthmann H, Staerz U D, Steinmetz M, von Boehmer H. Nature (London) 1988;333:742–746. doi: 10.1038/333742a0. [DOI] [PubMed] [Google Scholar]
- 3.Kisielow P, Teh H S, Bluthmann H, von Boehmer H. Nature (London) 1988;335:730–733. doi: 10.1038/335730a0. [DOI] [PubMed] [Google Scholar]
- 4.Teh H S, Kisielow P, Scott B, Kishi H, Uematsu Y, Bluthmann H, von Boehmer H. Nature (London) 1988;335:229–233. doi: 10.1038/335229a0. [DOI] [PubMed] [Google Scholar]
- 5.Scott B, Bluthmann H, Teh H S, von Boehmer H. Nature (London) 1989;338:591–593. doi: 10.1038/338591a0. [DOI] [PubMed] [Google Scholar]
- 6.Murphy K M, Heimberger A B, Loh D Y. Science. 1990;250:1720–1723. doi: 10.1126/science.2125367. [DOI] [PubMed] [Google Scholar]
- 7.Ashton-Rickardt P G, Van Kaer L, Schumacher T N M, Ploegh H L, Tonegawa S. Cell. 1993;73:1041–1049. doi: 10.1016/0092-8674(93)90281-t. [DOI] [PubMed] [Google Scholar]
- 8.Hogquist K A, Gavin M A, Bevan M J. J Exp Med. 1993;177:1469–1473. doi: 10.1084/jem.177.5.1469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hogquist K A, Jameson S C, Heath W R, Howard J L, Bevan M J, Carbone F R. Cell. 1994;76:17–27. doi: 10.1016/0092-8674(94)90169-4. [DOI] [PubMed] [Google Scholar]
- 10.Ashton-Rickardt P G, Bandeira A, Delaney J R, Van Kaer L, Pircher H P, Zinkernagel R M, Tonegawa S. Cell. 1994;76:651–663. doi: 10.1016/0092-8674(94)90505-3. [DOI] [PubMed] [Google Scholar]
- 11.Sebzda E, Wallace V A, Mayer J, Yeung R S M, Mak T W, Ohashi P S. Science. 1994;263:1615–1618. doi: 10.1126/science.8128249. [DOI] [PubMed] [Google Scholar]
- 12.Evavold B D, Sloan-Lancaster J, Allen P M. Immunol Today. 1993;14:602–609. doi: 10.1016/0167-5699(93)90200-5. [DOI] [PubMed] [Google Scholar]
- 13.De Magistris M T, Alexander J, Coggeshall M, Altman A, Gaeta F C A, Grey H M, Sette A. Cell. 1992;68:625–634. doi: 10.1016/0092-8674(92)90139-4. [DOI] [PubMed] [Google Scholar]
- 14.Page D M, Alexander J, Snoke K, Appella E, Sette A, Hedrick S M, Grey H M. Proc Natl Acad Sci USA. 1994;91:4057–4061. doi: 10.1073/pnas.91.9.4057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Vasquez N J, Kane L P, Hedrick S M. Immunity. 1994;1:45–56. doi: 10.1016/1074-7613(94)90008-6. [DOI] [PubMed] [Google Scholar]
- 16.Spain L M, Jorgensen J L, Davis M M, Berg L J. J Immunol. 1994;152:1709–1717. [PubMed] [Google Scholar]
- 17.Sebzda E, Kündig T M, Thomson C T, Aoki A, Mak S-Y, Mayer J P, Zamborelli T, Nathenson S G, Ohashi P S. J Exp Med. 1996;183:1093–1104. doi: 10.1084/jem.183.3.1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pawlowski T J, Singleton M D, Loh D Y, Berg R, Staerz U D. Eur J Immunol. 1996;26:851–857. doi: 10.1002/eji.1830260419. [DOI] [PubMed] [Google Scholar]
- 19.Williams O, Tanaka Y, Bix M, Murdjeva M, Littman D R, Kioussis D. Eur J Immunol. 1996;26:532–538. doi: 10.1002/eji.1830260305. [DOI] [PubMed] [Google Scholar]
- 20.Mamalaki C, Norton T, Tanaka Y, Townsend A R, Chandler P, Simpson E, Kioussis D. Proc Natl Acad Sci USA. 1992;89:11342–11346. doi: 10.1073/pnas.89.23.11342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mamalaki C, Elliott J, Norton T, Yannoutsos N, Townsend A R, Chandler P, Simpson E, Kioussis D. Dev Immunol. 1993;3:159–174. doi: 10.1155/1993/98015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tanaka Y, Mamalaki C, Stockinger B, Kioussis D. Eur J Immunol. 1993;23:2614–2621. doi: 10.1002/eji.1830231035. [DOI] [PubMed] [Google Scholar]
- 23.Mamalaki C, Murdjeva M, Tolaini M, Norton T, Chandler P, Townsend A, Simpson E, Kioussis D. Dev Immunol. 1996;4:299–315. doi: 10.1155/1995/54219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Spanopoulou E, Roman C A J, Corcoran L M, Schlissel M S, Silver D P, Nemazee D, Nussenzweig M C, Shinton S A, Hardy R R, Baltimore D. Genes Dev. 1994;8:1030–1042. doi: 10.1101/gad.8.9.1030. [DOI] [PubMed] [Google Scholar]
- 25.Cobbold S P, Jayasuriya A, Nash A, Prospero T D, Waldmann H. Nature (London) 1984;312:548–551. doi: 10.1038/312548a0. [DOI] [PubMed] [Google Scholar]
- 26.Tomonari K, Lovering E. Immunogenetics. 1988;28:445–451. doi: 10.1007/BF00355377. [DOI] [PubMed] [Google Scholar]
- 27.Watt S M, Gilmore D J, Davis J M, Clark M R, Waldmann H. Mol Cell Probes. 1987;1:297–326. doi: 10.1016/0890-8508(87)90013-2. [DOI] [PubMed] [Google Scholar]
- 28.Townsend A R M, McMichael A J, Carter N P, Huddleston J A, Brownlee G G. Cell. 1984;39:13–25. doi: 10.1016/0092-8674(84)90187-9. [DOI] [PubMed] [Google Scholar]
- 29.Wack A, Ladyman H M, Williams O, Roderick K, Ritter M A, Kioussis D. Int Immunol. 1996;8:1537–1548. doi: 10.1093/intimm/8.10.1537. [DOI] [PubMed] [Google Scholar]
- 30.Lucas B, Germain R N. Immunity. 1996;5:461–477. doi: 10.1016/s1074-7613(00)80502-6. [DOI] [PubMed] [Google Scholar]
- 31.Groves T, Parsons M, Miyamoto N G, Guidos C J. J Immunol. 1997;158:65–75. [PubMed] [Google Scholar]
- 32.Hogquist K A, Jameson S C, Bevan M J. Immunity. 1995;3:79–86. doi: 10.1016/1074-7613(95)90160-4. [DOI] [PubMed] [Google Scholar]
- 33.Kawai K, Ohashi P S. Nature (London) 1995;374:68–69. doi: 10.1038/374068a0. [DOI] [PubMed] [Google Scholar]
- 34.Jameson S C, Hogquist K A, Bevan M J. Nature (London) 1994;369:750–752. doi: 10.1038/369750a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chidgey A, Boyd R. Int Immunol. 1997;9:1527–1536. doi: 10.1093/intimm/9.10.1527. [DOI] [PubMed] [Google Scholar]
- 36.Grossman Z, Singer A. Proc Natl Acad Sci USA. 1996;93:14747–14752. doi: 10.1073/pnas.93.25.14747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Racioppi L, Ronchese F, Matis L A, Germain R N. J Exp Med. 1993;177:1047–1060. doi: 10.1084/jem.177.4.1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Liu C-P, Kappler J W, Marrack P. J Exp Med. 1996;184:1619–1630. doi: 10.1084/jem.184.5.1619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bhardwaj V, Kumar V, Geysen H M, Sercarz E E. J Immunol. 1993;151:5000–5010. [PubMed] [Google Scholar]
- 40.Nanda N K, Arzoo K K, Geysen H M, Sette A, Sercarz E E. J Exp Med. 1995;182:531–539. doi: 10.1084/jem.182.2.531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hagerty D T, Allen P M. J Immunol. 1995;155:2993–3001. [PubMed] [Google Scholar]
- 42.Quaratino S, Thorpe C J, Travers P J, Londei M. Proc Natl Acad Sci USA. 1995;92:10398–10402. doi: 10.1073/pnas.92.22.10398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wucherpfennig K W, Strominger J L. Cell. 1995;80:695–705. doi: 10.1016/0092-8674(95)90348-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Evavold B D, Sloan-Lancaster J, Wilson K J, Rothbard J B, Allen P M. Immunity. 1995;2:655–663. doi: 10.1016/1074-7613(95)90010-1. [DOI] [PubMed] [Google Scholar]
- 45.Hogquist K A, Tomlinson A J, Kieper W C, McGargill M A, Hart M C, Naylor S, Jameson S C. Immunity. 1997;6:389–399. doi: 10.1016/s1074-7613(00)80282-4. [DOI] [PubMed] [Google Scholar]
- 46.Hu Q, Bazemore-Walker C R, Girao C, Opferman J T, Sun J, Shabanowitz J, Hunt D F, Ashton-Rickardt P G. Immunity. 1997;7:221–231. doi: 10.1016/s1074-7613(00)80525-7. [DOI] [PubMed] [Google Scholar]
- 47.Williams O, Tanaka Y, Tarazona R, Kioussis D. Immunol Today. 1997;18:121–126. doi: 10.1016/s0167-5699(97)01029-3. [DOI] [PubMed] [Google Scholar]