Abstract
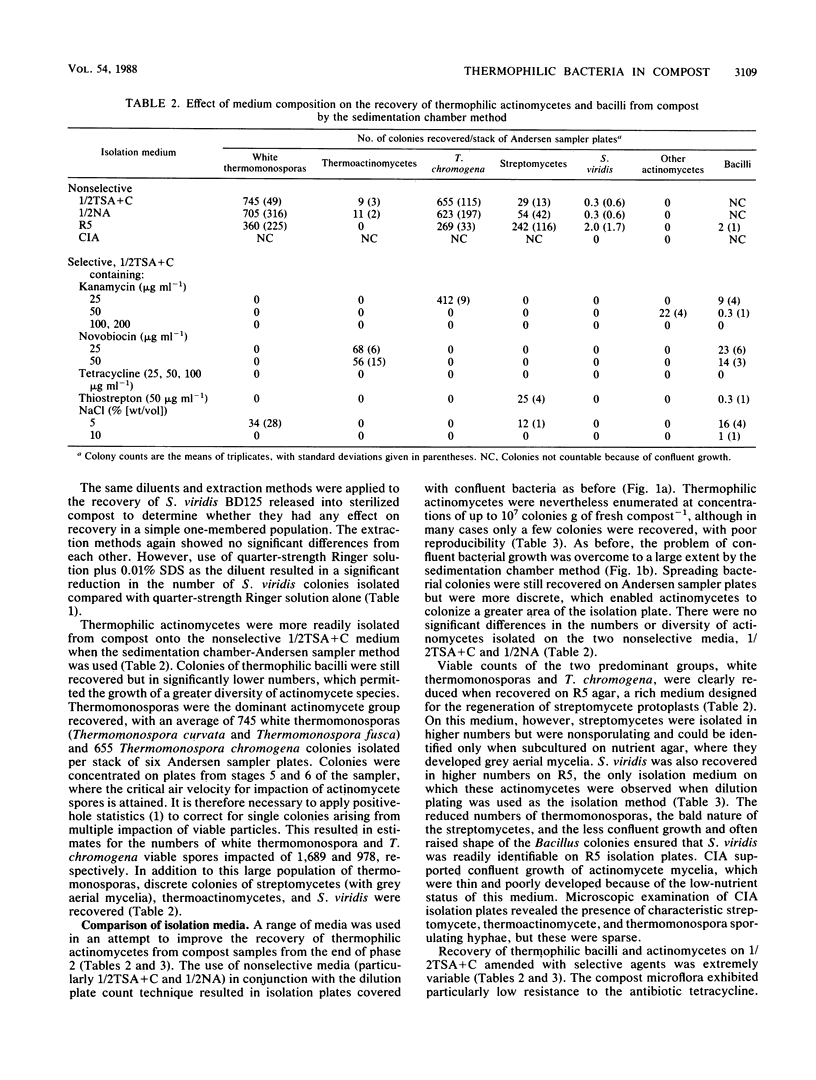
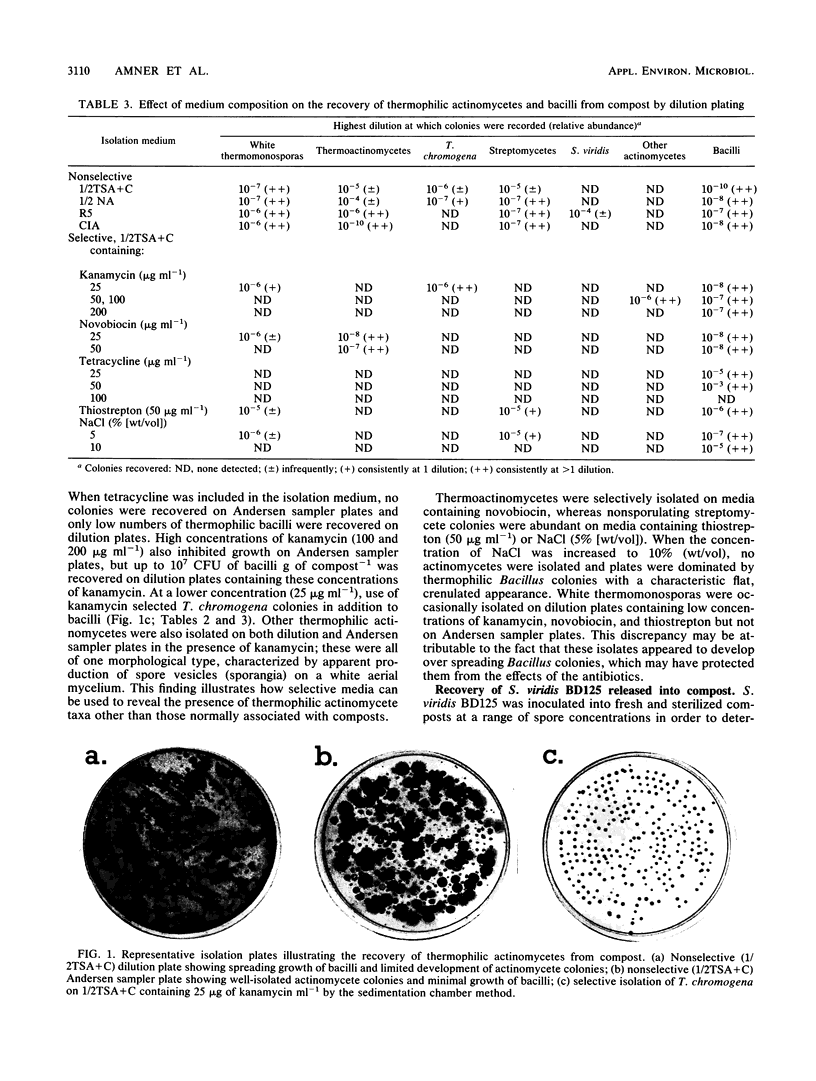
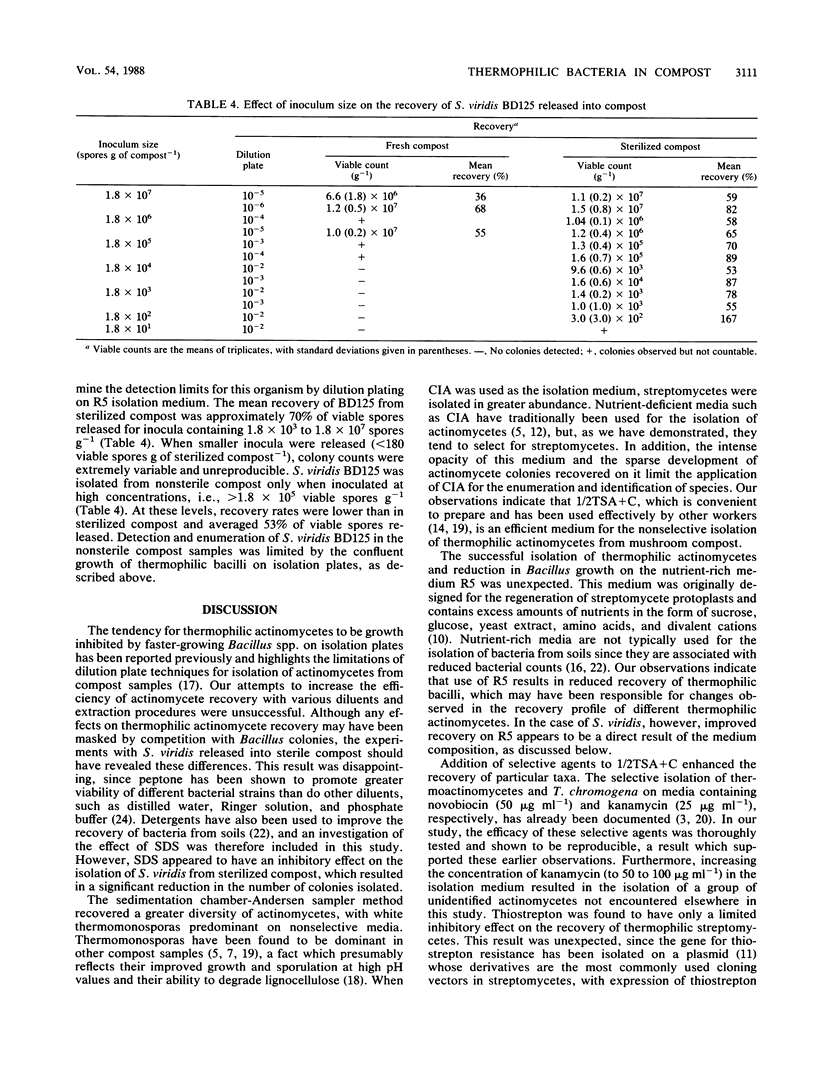
Thermophilic actinomycetes and bacilli were recovered from mushroom compost by conventional dilution plating and sedimentation chamber-Andersen sampler methods. Excessive growth of thermophilic bacilli on dilution plates accounted for the poor recovery and limited diversity of actinomycete colonies, and this result was largely unaffected by the use of modified extraction procedures and diluents. Assessment of the actinomycete population was more successfully achieved by applying the sedimentation chamber method, by using selective media, or both. Background resistance of the compost microflora to selective agents (kanamycin, novobiocin, tetracycline, thiostrepton, and NaCl) was extremely varied, but both actinomycetes and bacilli were particularly sensitive to tetracycline. The selective isolation of Thermoactinomyces spp. and Thermomonospora chromogena by novobiocin and kanamycin, respectively, was shown to be reproducible, and the use of high concentrations of kanamycin resulted in the isolation of a novel group of unidentified thermophilic actinomycetes. Comparison of nonselective nutrient media demonstrated that the nutrient-rich protoplast regeneration medium R5 was surprisingly efficient for actinomycete recovery. This medium was found to be particularly appropriate for the recovery of Saccharomonospora viridis BD125, introduced as spores into both sterile and fresh samples of mushroom compost. This stable pigmented variant of the S. viridis strains indigenous to compost was released at concentrations of up to 107 spores g of compost−1 in order to provide information for future experiments on the release and recovery of genetically manipulated strains. The detection limits for this strain were in the region of 102 g−1 from sterilized compost but only 105 g−1 from nonsterile compost. These figures correspond to mean recovery efficiencies of approximately 70% (sterilized compost) and 53% (fresh compost) of viable spores released. Further improvements in the detection and recovery of S. viridis strains released into compost should be achieved by the introduction of selectable markers developed from this information on the antibiotic resistance profile of the indigenous compost microflora.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- ANDERSEN A. A. New sampler for the collection, sizing, and enumeration of viable airborne particles. J Bacteriol. 1958 Nov;76(5):471–484. doi: 10.1128/jb.76.5.471-484.1958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brayton P. R., Tamplin M. L., Huq A., Colwell R. R. Enumeration of Vibrio cholerae O1 in Bangladesh waters by fluorescent-antibody direct viable count. Appl Environ Microbiol. 1987 Dec;53(12):2862–2865. doi: 10.1128/aem.53.12.2862-2865.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cross T. Thermophilic actinomycetes. J Appl Bacteriol. 1968 Mar;31(1):36–53. doi: 10.1111/j.1365-2672.1968.tb00339.x. [DOI] [PubMed] [Google Scholar]
- Fredrickson J. K., Bezdicek D. F., Brockman F. J., Li S. W. Enumeration of Tn5 mutant bacteria in soil by using a most- probable-number-DNA hybridization procedure and antibiotic resistance. Appl Environ Microbiol. 1988 Feb;54(2):446–453. doi: 10.1128/aem.54.2.446-453.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanson C. W., Martin W. J. Microwave oven for melting laboratory media. J Clin Microbiol. 1978 Apr;7(4):401–402. doi: 10.1128/jcm.7.4.401-402.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holben William E., Jansson Janet K., Chelm Barry K., Tiedje James M. DNA Probe Method for the Detection of Specific Microorganisms in the Soil Bacterial Community. Appl Environ Microbiol. 1988 Mar;54(3):703–711. doi: 10.1128/aem.54.3.703-711.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kieser T., Hopwood D. A., Wright H. M., Thompson C. J. pIJ101, a multi-copy broad host-range Streptomyces plasmid: functional analysis and development of DNA cloning vectors. Mol Gen Genet. 1982;185(2):223–228. doi: 10.1007/BF00330791. [DOI] [PubMed] [Google Scholar]
- Kleyn J. G., Wetzler T. F. The microbiology of spent mushroom compost and its dust. Can J Microbiol. 1981 Aug;27(8):748–753. doi: 10.1139/m81-116. [DOI] [PubMed] [Google Scholar]
- Lacey J. Actinomycetes in soils, composts and fodders. Soc Appl Bacteriol Symp Ser. 1973 Jan;2:231–251. [PubMed] [Google Scholar]
- Lacey J., Dutkiewicz J. Isolation of Actinomycetes and fungi from mouldy hay using a sedimentation chamber. J Appl Bacteriol. 1976 Oct;41(2):315–319. doi: 10.1111/j.1365-2672.1976.tb00636.x. [DOI] [PubMed] [Google Scholar]
- Roszak D. B., Colwell R. R. Metabolic activity of bacterial cells enumerated by direct viable count. Appl Environ Microbiol. 1987 Dec;53(12):2889–2893. doi: 10.1128/aem.53.12.2889-2893.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strom P. F. Identification of thermophilic bacteria in solid-waste composting. Appl Environ Microbiol. 1985 Oct;50(4):906–913. doi: 10.1128/aem.50.4.906-913.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]



