Abstract
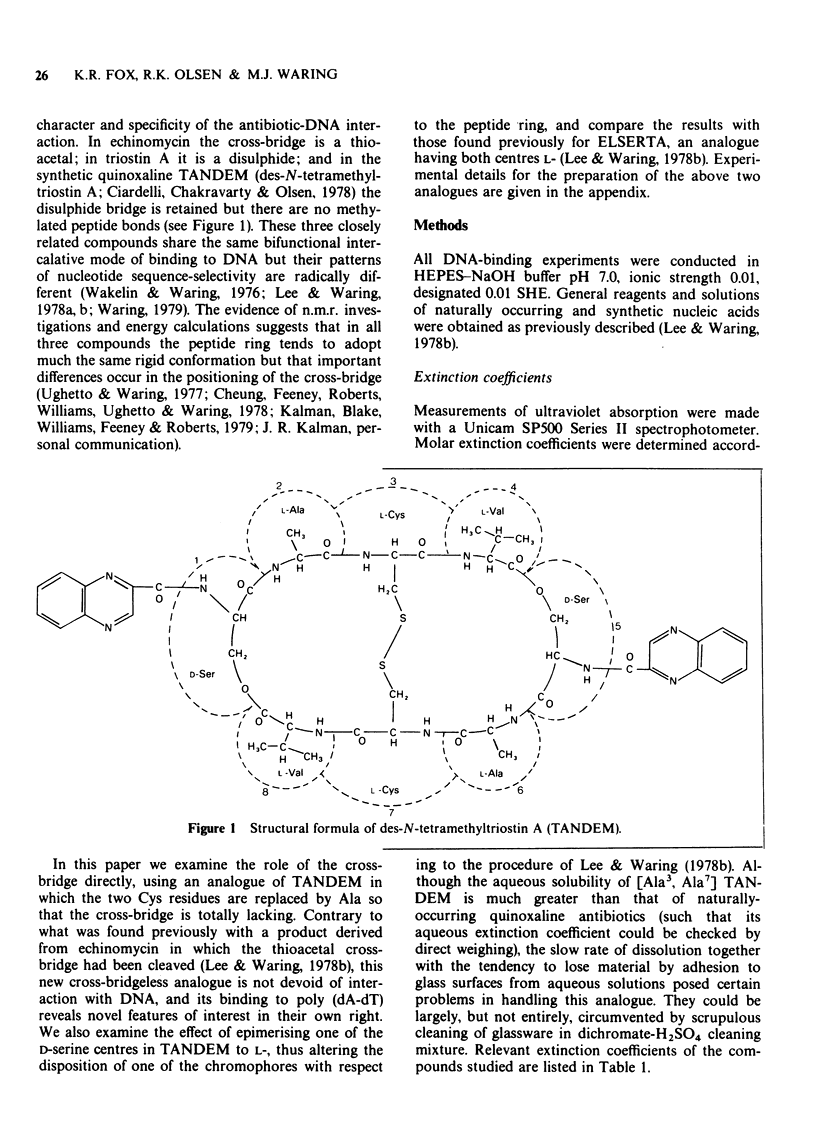
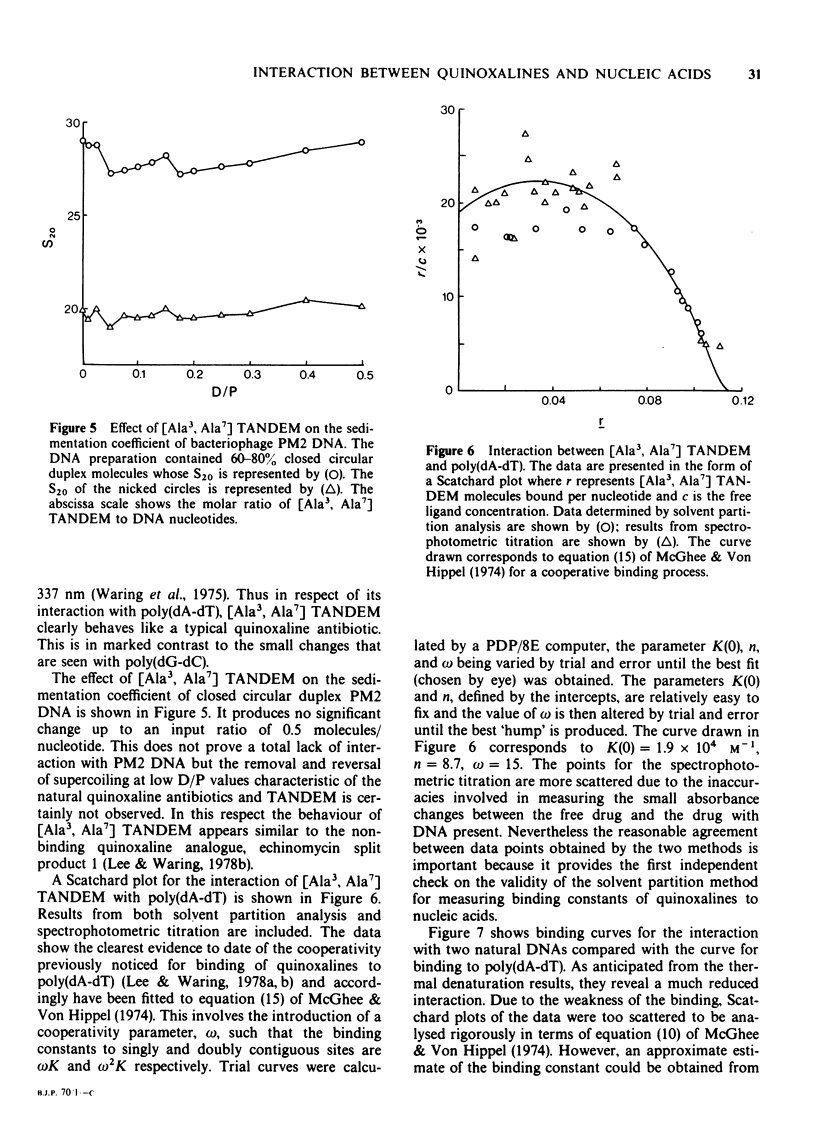
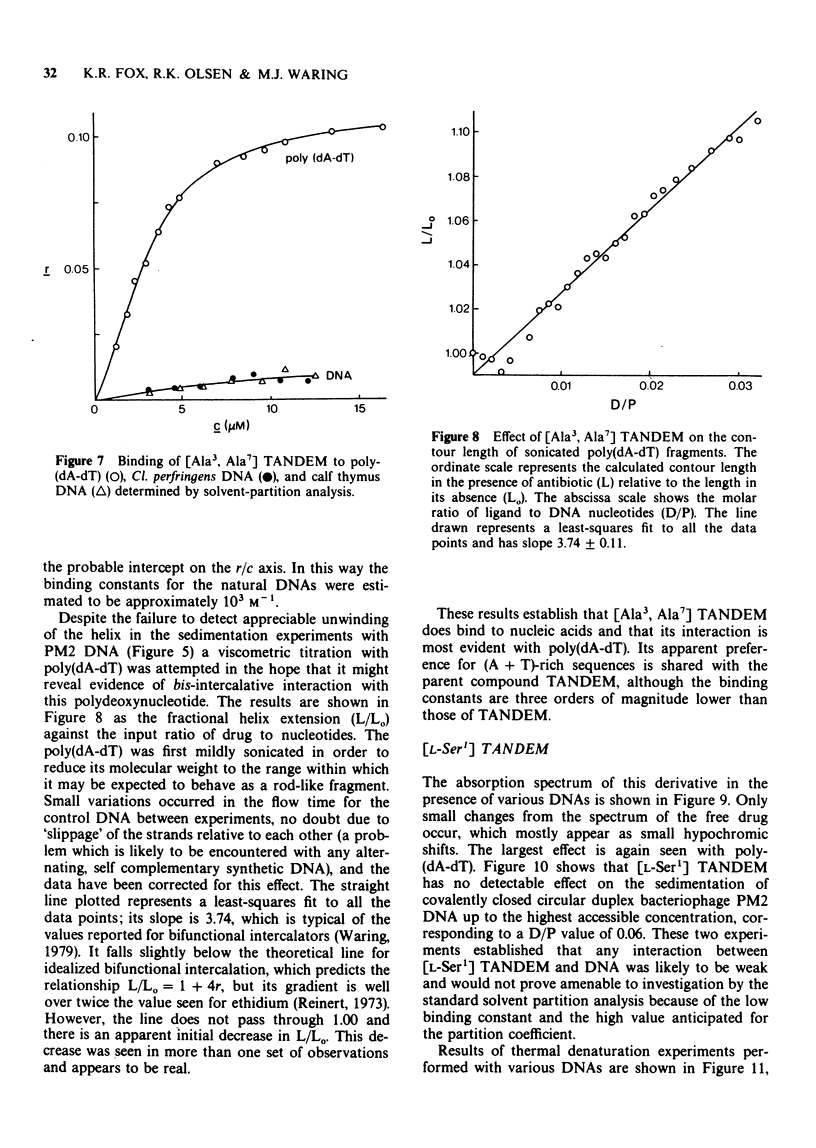
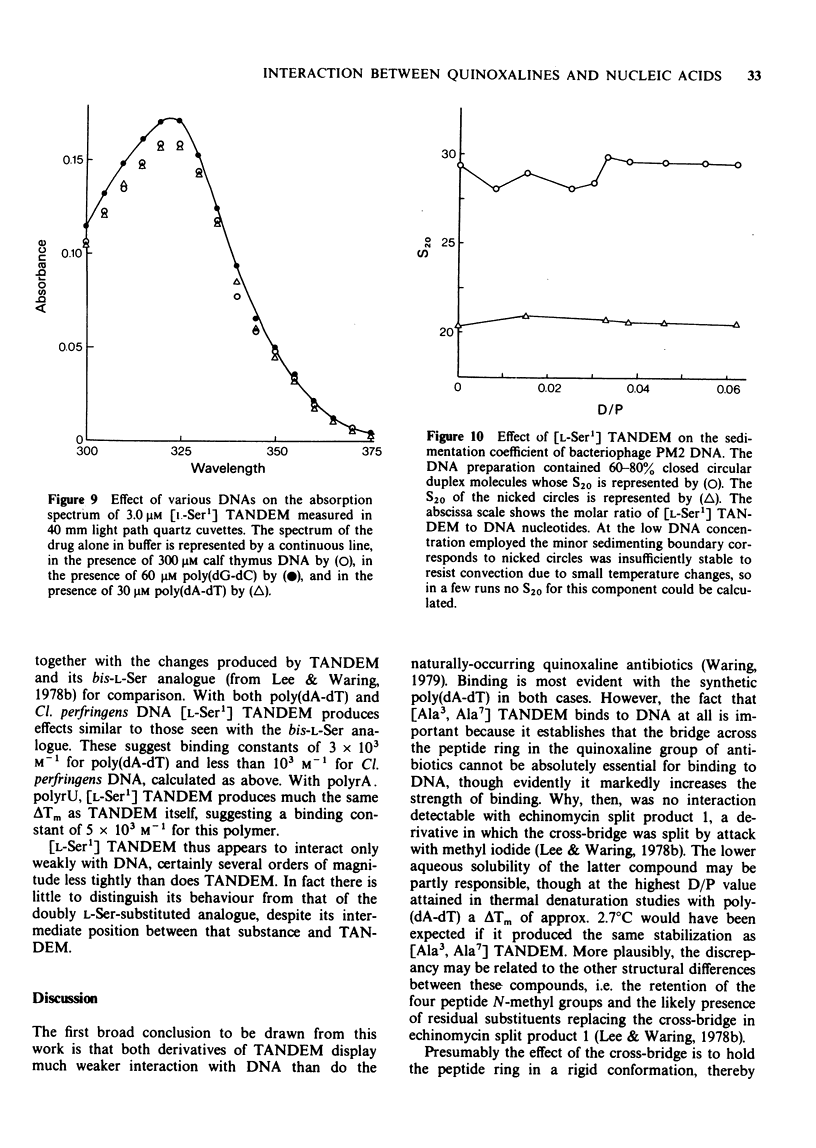
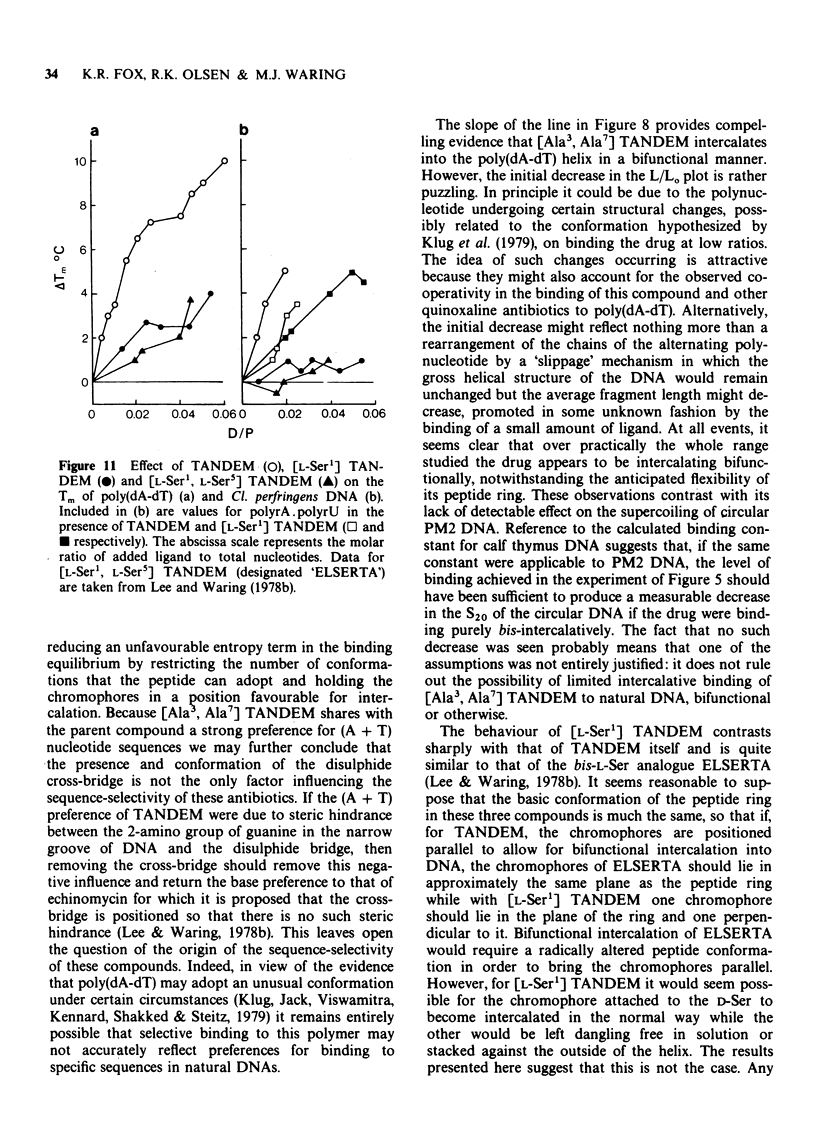
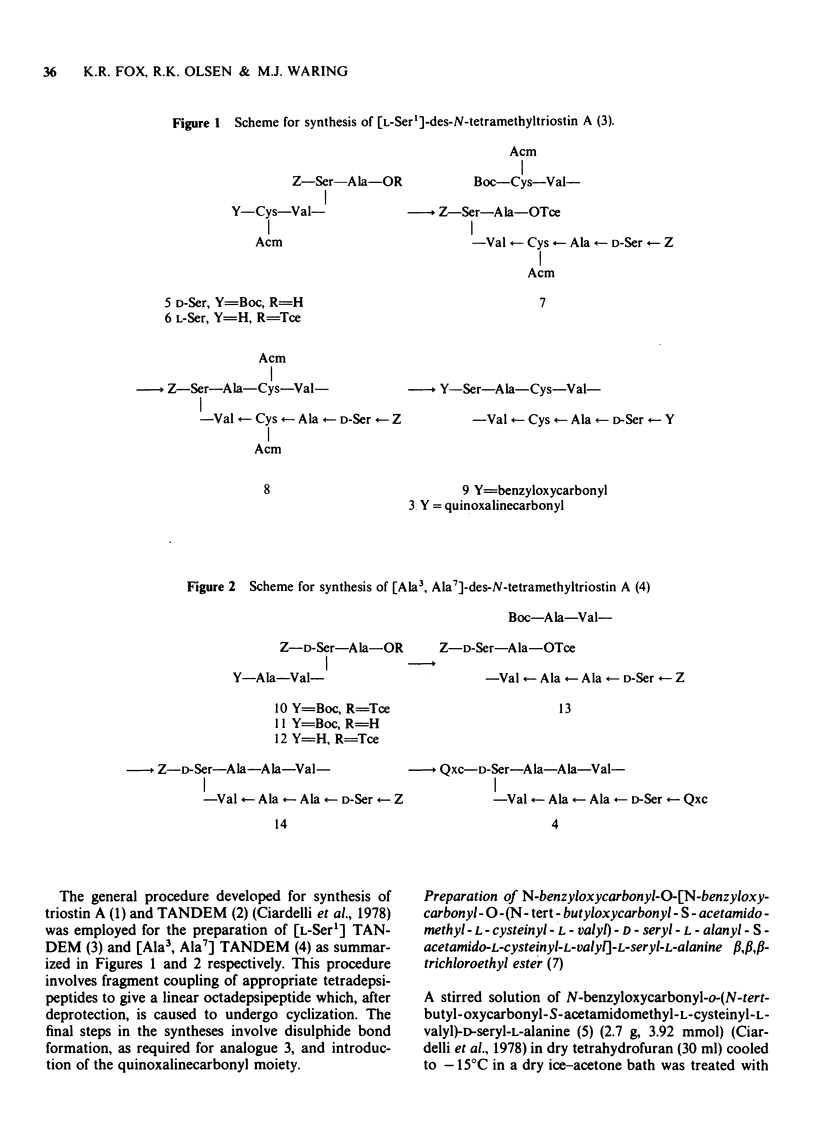
1 [Ala3, Ala7] TANDEM is an analogue of des-N-tetramethyl-triostin A (TANDEM) in which both L-Cys residues of the octapeptide ring are replaced by L-Ala; accordingly it lacks the disulphide cross-bridge which limits the conformational flexibility of TANDEM. 2 In [L-Ser1] TANDEM the configuration of one of the serine residues is inverted, altering the disposition of one of the quinoxaline chromophores with respect to the peptide ring. 3 Both compounds interact weakly but detectably with natural DNAs as judged by spectral shifts and increases in the thermal denaturation ('melting') temperature Tm. They also raise the Tm of poly rA . poly rU. 4 Binding isotherms determined by solvent partition analysis with [Ala3, Ala7] TANDEM yield association constants of about 10(3) M-1 for its interaction with natural DNAs. A Scatchard plot for binding to poly(dA-dT) determined by solvent partition and spectrophotometric methods shows marked evidence of cooperativity with an intrinsic association constant 1.9 x 10(4) M-1, 8.7 nucleotides per binding site, and cooperativity parameter 15. 5 Binding of [Ala3, Ala7] TANDEM to short rod-like fragments of poly(dA-dT) increases their contour length by almost the theoretical amount expected for an ideal process of bifunctional intercalation. 6 No effect of either compound on the winding of the DNA helix could be detected in sedimentation experiments with closed circular duplex PM2 DNA. 7 It is concluded that the cross-bridge of TANDEM greatly stabilizes its binding to DNA, most probably via entropic factors, but is not the only structural feature that influences its AT sequence-selectivity. The consequences of epimerising one of the D-Ser residues appear as disastrous as epimerising both. 8 The experimental details for the synthesis of [Ala3, Ala7] TANDEM and [L-Ser1] TANDEM are given in an appendix to this paper.
Full text
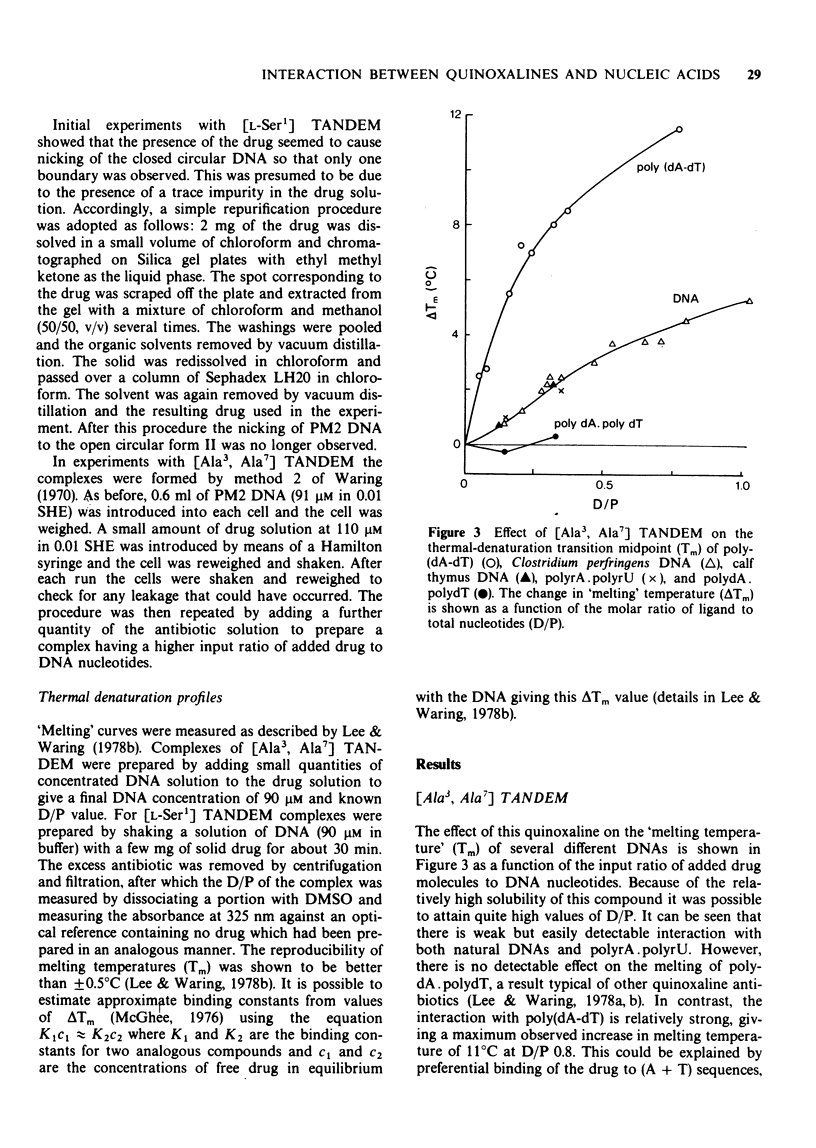
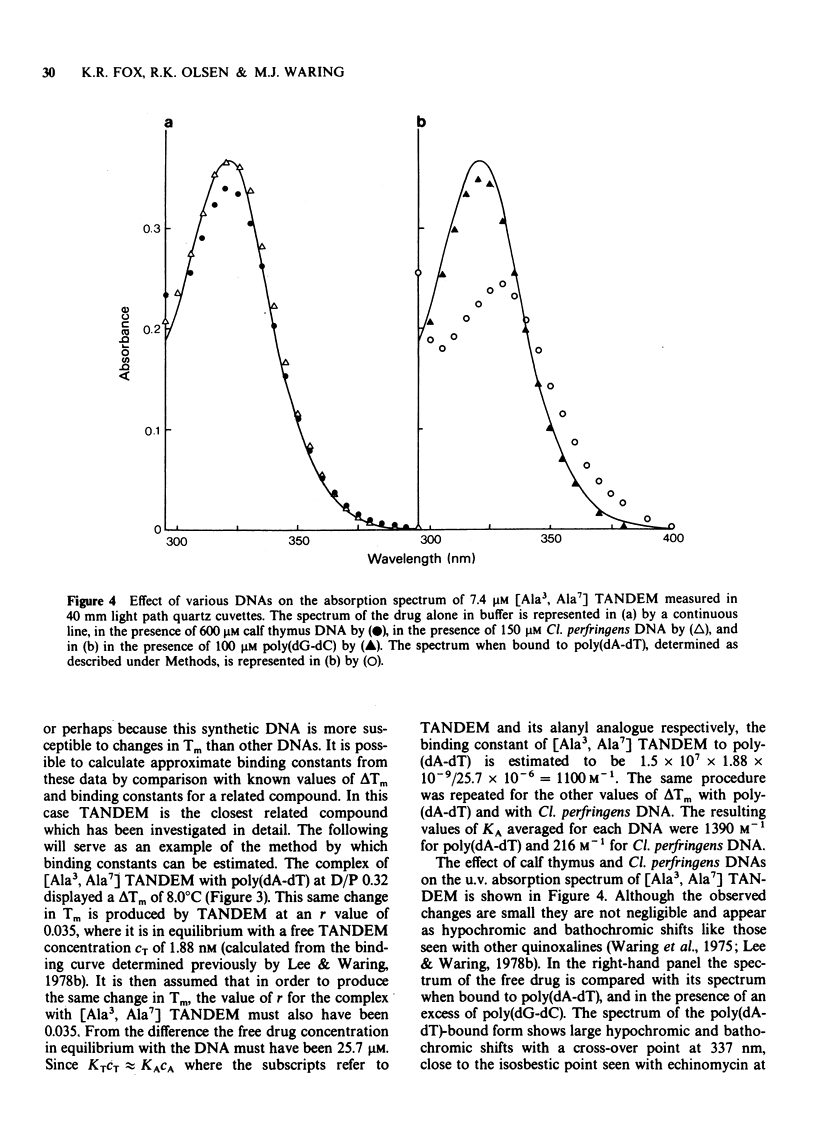
PDF

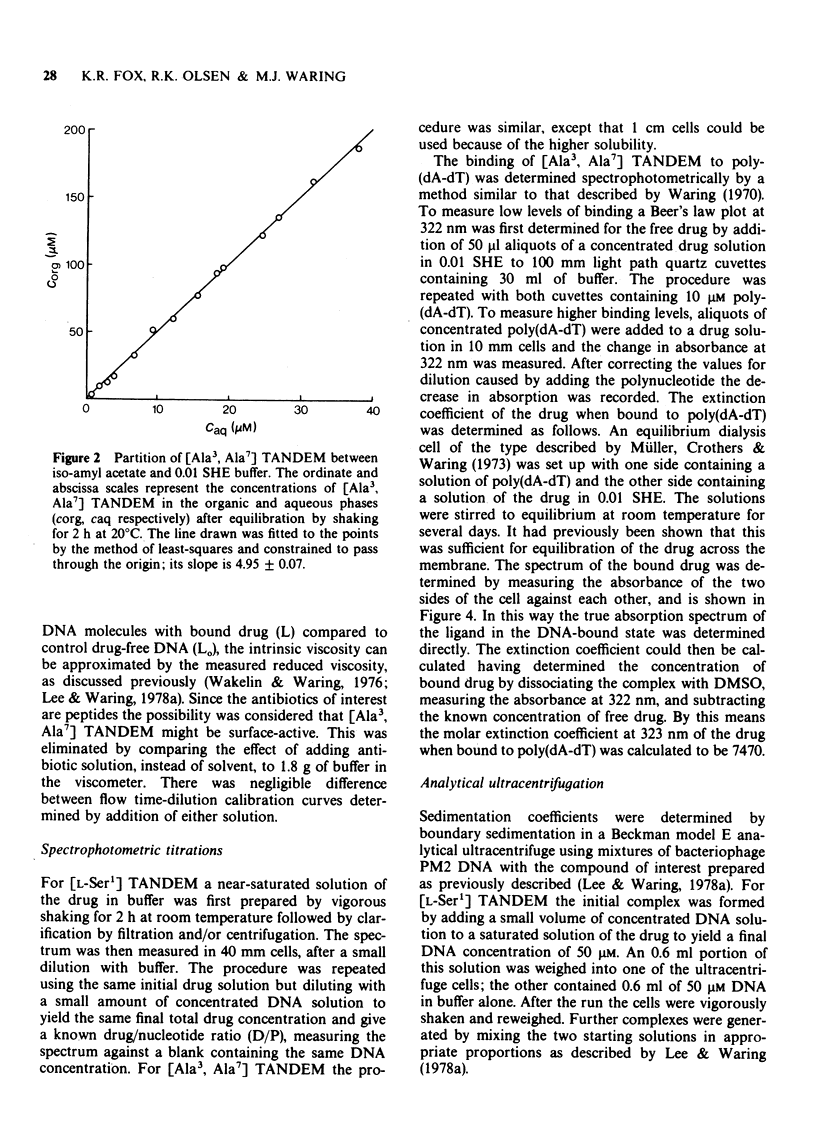





Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Arnott S., Fuller W., Hodgson A., Prutton I. Molecular conformations and structure transitions of RNA complementary helices and their possible biological significance. Nature. 1968 Nov 9;220(5167):561–564. doi: 10.1038/220561a0. [DOI] [PubMed] [Google Scholar]
- Cohen G., Eisenberg H. Conformation studies on the sodium and cesium salts of calf thymus deoxyribonucleic acid (DNA). Biopolymers. 1966 Apr-May;4(4):429–440. doi: 10.1002/bip.1966.360040404. [DOI] [PubMed] [Google Scholar]
- Crothers D. M., Zimm B. H. Viscosity and sedimentation of the DNA from bacteriophages T2 and T7 and the relation to molecular weight. J Mol Biol. 1965 Jul;12(3):525–536. doi: 10.1016/s0022-2836(65)80310-2. [DOI] [PubMed] [Google Scholar]
- Dell A., Williams D. H., Morris H. R., Smith G. A., Feeney J., Roberts G. C. Structure revision of the antibiotic echinomycin. J Am Chem Soc. 1975 Apr 30;97(9):2497–2502. doi: 10.1021/ja00842a029. [DOI] [PubMed] [Google Scholar]
- Klug A., Jack A., Viswamitra M. A., Kennard O., Shakked Z., Steitz T. A. A hypothesis on a specific sequence-dependent conformation of DNA and its relation to the binding of the lac-repressor protein. J Mol Biol. 1979 Jul 15;131(4):669–680. doi: 10.1016/0022-2836(79)90196-7. [DOI] [PubMed] [Google Scholar]
- Lee J. S., Waring M. J. Bifunctional intercalation and sequence specificity in the binding of quinomycin and triostin antibiotics to deoxyribonucleic acid. Biochem J. 1978 Jul 1;173(1):115–128. doi: 10.1042/bj1730115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J. S., Waring M. J. Interaction between synthetic analogues of quinoxaline antibiotics and nucleic acids. Changes in mechanism and specificity related to structural alterations. Biochem J. 1978 Jul 1;173(1):129–144. doi: 10.1042/bj1730129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin D. G., Mizsak S. A., Biles C., Stewart J. C., Bacynsky L., Meulman P. A. Structure of quinomycin antibiotics. J Antibiot (Tokyo) 1975 Apr;28(4):332–336. doi: 10.7164/antibiotics.28.332. [DOI] [PubMed] [Google Scholar]
- McGhee J. D. Theoretical calculations of the helix-coil transition of DNA in the presence of large, cooperatively binding ligands. Biopolymers. 1976 Jul;15(7):1345–1375. doi: 10.1002/bip.1976.360150710. [DOI] [PubMed] [Google Scholar]
- McGhee J. D., von Hippel P. H. Theoretical aspects of DNA-protein interactions: co-operative and non-co-operative binding of large ligands to a one-dimensional homogeneous lattice. J Mol Biol. 1974 Jun 25;86(2):469–489. doi: 10.1016/0022-2836(74)90031-x. [DOI] [PubMed] [Google Scholar]
- Müller W., Crothers D. M., Waring M. J. A non-intercalating proflavine derivative. Eur J Biochem. 1973 Nov 1;39(1):223–234. doi: 10.1111/j.1432-1033.1973.tb03120.x. [DOI] [PubMed] [Google Scholar]
- Reinert K. E. DNA stiffening and elongation caused by the binding of ethidium bromide. Biochim Biophys Acta. 1973 Aug 24;319(2):135–139. doi: 10.1016/0005-2787(73)90004-x. [DOI] [PubMed] [Google Scholar]
- Sato K., Shiratori O., Katagiri K. The mode of action of quinoxaline antibiotics. Interaction of quinomycin A with deoxyribonucleic acid. J Antibiot (Tokyo) 1967 Sep;20(5):270–276. [PubMed] [Google Scholar]
- Ughetto G., Waring M. J. Conformation of the deoxyribonucleic acid-binding peptide antibiotic echinomycin based on energy calculations. Mol Pharmacol. 1977 May;13(3):579–584. [PubMed] [Google Scholar]
- Ward D. C., Reich E., Goldberg I. H. Base specificity in the interaction of polynucleotides with antibiotic drugs. Science. 1965 Sep 10;149(3689):1259–1263. doi: 10.1126/science.149.3689.1259. [DOI] [PubMed] [Google Scholar]
- Waring M. J., Wakelin L. P. Echinomycin: a bifunctional intercalating antibiotic. Nature. 1974 Dec 20;252(5485):653–657. doi: 10.1038/252653a0. [DOI] [PubMed] [Google Scholar]
- Waring M. J., Wakelin L. P., Lee J. S. A solvent-partition method for measuring the binding of drugs to DNA. Application to the quinoxaline antibiotics echinomycin and triostin A. Biochim Biophys Acta. 1975 Oct 1;407(2):200–212. doi: 10.1016/0005-2787(75)90285-3. [DOI] [PubMed] [Google Scholar]
- Waring M., Makoff A. Breakdown of pulse-labeled ribonucleic acid and polysomes in Bacillus megaterium: actions of streptolydigin, echinomycin, and triostins. Mol Pharmacol. 1974 Mar;10(2):214–224. [PubMed] [Google Scholar]
- Waring M. Variation of the supercoils in closed circular DNA by binding of antibiotics and drugs: evidence for molecular models involving intercalation. J Mol Biol. 1970 Dec 14;54(2):247–279. doi: 10.1016/0022-2836(70)90429-8. [DOI] [PubMed] [Google Scholar]



