Abstract
Chromosome 8p11–12 is the site of a recurrent breakpoint in a myeloproliferative disorder that involves lymphoid (T- or B-cell), myeloid hyperplasia and eosinophilia, and evolves toward acute leukemia. This multilineage involvement suggests the malignant transformation of a primitive hematopoietic stem cell. In this disorder, the 8p11–12 region is associated with three different partners 6q27, 9q33, and 13q12. We describe here the molecular characterization of the t(8;13) translocation that involves the FGFR1 gene from 8p12, encoding a tyrosine kinase receptor for members of the fibroblast growth factor family, and a gene from 13q12, tentatively named FIM (Fused In Myeloproliferative disorders). FIM is related to DXS6673E, a candidate gene for X-linked mental retardation in Xq13.1; this defines a gene family involved in different human pathologies. The two reciprocal fusion transcripts, FIM/FGFR1 and FGFR1/FIM are expressed in the malignant cells. The FIM/FGFR1 fusion protein contains the FIM putative zinc finger motifs and the catalytic domain of FGFR1. We show that it has a constitutive tyrosine kinase activity.
The investigation of hematopoietic malignant tumors associated with recurrent translocations has provided insights into the mechanisms of transformation of human blood cells and led to the identification of several molecules involved in the regulation of normal hematopoiesis (1).
Recurrent chromosome rearrangements involving the p11–12 region of chromosome 8 are associated with either acute myeloid leukemia (see references in ref. 2) or stem-cell myeloproliferative disorder (MPD). The latter involves proliferation of lymphoblastic T or B cells, myeloid hyperplasia and eosinophilia leading to acute myeloid leukemia (3). This clinicopathological entity, likely to affect a multilineage precursor, is associated with three different translocations in which chromosomal band 8p12 is rearranged with a partner at either 6q27 (4, 5), 9q32–34 (6–11), or 13q12 (12–19). The genes involved in these translocations may play a role in the biology of hematopoietic stem cells.
Previous examination of breakpoints by fluorescent in situ hybridization (FISH) showed that cosmids containing the FGFR1 gene, one of four genes that encode tyrosine kinase receptors for members of the fibroblast growth factor (FGF) family (reviewed in ref. 20), spanned the 8p11–12 breakpoint of all three translocations associated with MPD (21). We report here that the t(8;13) rearrangement is likely to convert FGFR1 into a potent transforming gene involved in leukemogenesis. It results in a chimeric protein made of the FGFR1 tyrosine kinase (22) fused at its N terminus to a portion of a molecule, named FIM, that contains zinc finger motifs.
MATERIALS AND METHODS
Patient Samples and Cell Lines.
Peripheral blood and bone marrow cells were obtained from two patients with hematologic disorders after informed consent. Patient 1 suffered from a multilineage disorder of lymphoblastic T cell non-Hodgkin lymphoma, eosinophilia and myeloid hyperplasia that ended in acute myeloid leukemia.
At relapse, the karyotype from the pleural fluid cells was: 48,XX,t(8;13)(p12;q12),+der(13)t(8;13)(p12;q12),+19[4]/51,idem,+6, +der(8)t(8;13)(p12;q12), +der(13)t(8;13)(p12;q12)[2].
A six-week-old severe combined immunodeficient (SCID) female mouse was inoculated i.p. with pleural fluid cells from this patient. After engraftment (2 months), malignant human cells were collected from abdominal nodes and injected repeatedly in female SCID mice. Passage 13 cells, hereafter designated SCID t(8;13) cells, as well as malignant cells from the patient, were used in the experiments. The karyotype and immunophenotypic analysis of passage number 13 cells were identical to those of the original pleural fluid malignant cells with the exception that only the clone with three derivative 13 chromosomes was present.
Bone marrow cells with a t(8;13) from patient 2, suffering from a B cell acute lymphoblastic leukemia, with involvement of the myeloid lineage (19) (provided by A. Hagemeijer, Center for Human Genetics, University of Leuven, Belgium) were also used.
All cell lines were purchased from the American Type Culture Collection, except IE8 (a gift from T. LeBien, Medical School, University of Minnesota), and SU-DHL-1 (provided by R. Rimokh, Centre L. Bérard, Lyon, France). MDA-MB-134 is a mammary carcinoma cell line with an amplification of the 8p12 region (23).
cDNA Library Screening and DNA Sequence Analysis.
A cDNA library [Lambda Uni-Zap XR vector, oligo (dT)-primed, Stratagene] was constructed with mRNA extracted from SCID t(8;13) cells by using a messenger RNA isolation kit (Stratagene). Human placenta [oligo (dT)-and random primed] and skeletal muscle [oligo-(dT) primed] libraries in Lambda Uni-Zap XR vector were purchased from Stratagene. For screening, 105 plaques were plated per 150-mm dish and transferred to nitrocellulose (Schleicher & Schuell) membranes. Approximately 106 plaque forming units were screened with each probe: OL9, a 500-bp BglII-EcoRI fragment from pFLG16 FGFR1 cDNA corresponding to the portion encoding the second tyrosine kinase subdomain and the 3′ untranslated region of FGFR1; 20.2T3, a 200-bp BglI-EcoRI fragment from cDNA clone 20.2; 28PP2, a 536-bp EcoRI-PstI fragment from cDNA clone 28; FIM#3T3RR1, a 507-bp EcoRI-EcoRI fragment from cDNA clone FIM#3; FIM#10.1a, a 344-bp PstI-PstI fragment from cDNA clone FIM#10.1. Probes OL9, 20.2T3 and FIM#10.1a were successively used to screen the SCID cDNA library; probes 28PP2 and FIM#3T3RR1 were used to screen the skeletal muscle and placenta libraries, respectively.
Double-stranded DNA sequencing was done at Génome Express (Grenoble, France) on an automated sequencer (Applied Biosystems 373). Comparisons with GenBank and dbEST entries were done by using blastn, comparisons with protein databases (EMBL and SwissProt) using blastx, tblastx. Protein alignment was performed using geneworks 2.4 (IntelliGenetics).
FISH Analysis.
Two-color FISH experiments were done on metaphase chromosome spreads from normal human male lymphocytes and from SCID t(8;13) as described (24), with the following modifications: cosmid and plasmid DNA probes were labeled by nick-translation with 16-dUTP biotin (BRL/Life Technologies) and digoxigenin-11-dUTP (Boehringer Mannheim), respectively. Plasmid probe (300 ng) in combination with two biotinylated cosmid probes (150 ng for each) were incubated on metaphase spreads and washed at low stringency (50% formamide/2× standard saline citrate and 2× standard saline citrate at 42°C). Immunofluorescence detection and image processing were done as described (24).
Southern and Northern Analyses.
Genomic DNA samples were extracted and BamHI digests analyzed by Southern blot hybridization according to standard procedures (25). Probes 27.1a and 27.1b, corresponding to 545-bp PstI-PstI and 785-bp PstI-XhoI fragments from cDNA clone 27.1, respectively, were used. The multiple tissue Northern blots (CLONTECH, no. 7759–1 and 7760–1) were hybridized according to the manufacturer’s instructions by using probe 27.1a.
Analysis of Gene Expression by RT-PCR.
Total RNAs were extracted from frozen patient samples (25). Reverse transcription (RT)–PCR reaction was performed after standard procedures by using Superscript reverse transcriptase (GIBCO/BRL), and random hexamers. PCR reactions were done in a Perkin–Elmer/Cetus apparatus by using an equivalent of 500 ng of reversed transcribed RNA and 50 pmol of each oligonucleotide as primers. The first cycle was denaturation at 95°C for 5 min, annealing at 60°C for 2 min, and synthesis at 72°C for 3 min. The next 30 cycles were run using the same conditions except that the denaturation step was 1 min. For the last cycle, synthesis time was 7 min. The oligonucleotide primers (Fig. 1) used for gene expression analysis were either FGFR1-specific primers (nucleotides 1,132–1,151 and 1,227–1,208, sense and antisense primers FA and FB, respectively, PCR product of 96 bp) or FIM-specific primers (nucleotides 2,824–2,843 and 2,989–2,968, sense and antisense primers X1 and 1R, respectively, PCR product of 176 bp). Two primer pairs were used for detection of fusion transcripts: FIM-specific sense primer X1 and FGFR1 exon 9-specific antisense primer (nucleotides 1,316–1,297, F9 primer, PCR product of 155 bp) or FGFR1 exon 8-specific sense primer (nucleotides 1,295–1,316, FC primer), and FIM-specific antisense primer 1R (PCR product of 155 bp). The human β-actin (nucleotides 2,106–2,128 and 2,707–2,685, sense and antisense, respectively, PCR product of 506 bp), was used as control for RT and PCR efficiency. The amplified products were run in 2.5% agarose gels in 1× Tris borate buffer (0.5× 45 mM Tris–borate/1 mM EDTA) and stained with ethidium bromide (25). PCR products were also cloned in the pUC18 vector by using the Sure Clone Ligation kit (Pharmacia). Nucleotide sequences were obtained by using the T7Sequencing kit (Pharmacia) and the forward and reverse sequencing primers for pUC18. Products were analyzed on 5% polyacrylamide/urea sequencing gels.
Figure 1.
Cloning of the FIM/FGFR1 fusion cDNA involved in the t(8;13) translocation breakpoint and of wild-type FIM. Long-range restriction map of FGFR1 cDNA is shown in green at the top. FIM cDNA is shown in blue. Probes generated for 5′-cDNA walking are indicated under the respective partial cDNA clones (blue arrows). Useful restriction enzyme sites are indicated: BI: BglI; BII; BglII; E: EcoRI; H: HindIII; P: PstI; S: SacI; and X: XhoI. The FGFR1 (FA, FB, FC, and F9) and FIM (X1, 1R) oligonucleotide primers used in PCR experiments are indicated by arrowheads. Probes used for Southern and Northern experiments are indicated by open boxes under the respective cDNAs. Red double arrows indicate the position of the breakpoint.
To determine the relative levels of chimeric and wild-type transcripts, four competitive PCR reactions were performed by using various combinations of sense and antisense FIM and FGFR1 oligonucleotides located near the breakpoint, and by using RT-PCR products from SCID t(8;13) and normal breast (control) RNAs as follows: FGFR1 (FA, F9) and FGFR1/FIM, (FIM primer 1R) leading to four PCR products of 180 and 186 bp corresponding to FGFR1 cDNA, and of 202 and 208 bp corresponding to the FGFR1/FIM fusion gene on the der(8) chromosome; FGFR1 (FA, F9), and FIM/FGFR1 (FIM primer X1) leading to three PCR products of 180 and 186 bp (FGFR1 gene) and 155 bp [FIM/FGFR1 fusion gene on chromosome der(13)]; FIM (X1, 1R) and FIM/FGFR1 (FGFR1 primer F9) leading to two PCR products of 176 and 155 bp for the FIM and FIM/FGFR1 cDNAs, respectively; FIM (X1, 1R) and FGFR1/FIM (FGFR1 primer FA) leading to three PCR products of 176 bp (FIM gene), and 202 and 208 bp [FGFR1/FIM fusion gene on chromosome der(8)]. The PCR products were electrophoresed in 2.5% agarose gels and quantified by spectrophotometry using bio image software (Millipore).
Analysis of FIM/FGFR1 and FGFR1/FIM fusion sequences at the Genomic Level.
Using the above described X1/F9 and FA/1R primer pairs for FIM/FGFR1 and FGFR1/FIM, PCR amplification of the genomic SCID t(8;13) DNA generated fusion fragments of 2.2 kb and 600 bp, respectively. Cloned PCR products were sequenced.
Sequencing of FGFR1 Intron 8.
A 1.2-kb PCR product was amplified from the genomic SCID t(8;13) DNA by using primers located in exon 8 (FA) and exon 9 (F9) of the FGFR1 gene. The amplified fragment was cloned and sequenced. In addition, part of the 134.8 cosmid, specific for the FGFR1 gene (23) was sequenced by using FGFR1 primer FA.
Autokinase Activity and Tyrosine Phosphorylation Analysis of the FIM/FGFR1 Chimeric Protein.
NIH 3T3 cells were transiently transfected with a reconstructed full length FIM/FGFR1 cDNA inserted into the pcDNA3 expression vector (Invitrogen) or with the empty vector. 24 hours after transfection and after a 2 hr serum starvation, cells were stimulated or not with FGF1 in the presence of heparin. The FGFR1 expressing cell line NFlg 26 (22) was used as a positive control. Cell lysates and immunoprecipitation using an antibody directed against a FGFR1 C-terminal peptide (anti-C-FGFR1) (C15, Santa Cruz Biotechnology) were done as described (26). Half of each immunoprecipitate was challenged for autokinase activity in the presence of [γ32P]ATP and 5 mM MnCl2 (27) and analyzed by electrophoresis and autoradiography. The phosphorylation on tyrosine and the level of expression were analyzed by immunoblotting, with an anti-phosphotyrosine antibody (4G10, Upstate Biotechnology, Lake Placid, NY) and the anti-C-FGFR1 antibody, respectively (26, 28), using the second half of the samples.
RESULTS
Isolation of cDNA Clones Spanning the Translocation Breakpoint.
To isolate clones specific of the translocation (Fig. 1), we constructed a cDNA library by using mRNA from patient 1 cells engrafted in SCID mouse [SCID t(8;13)]. The first round of screening was done with a FGFR1 cDNA probe (OL9). Clone named 20.2 was first isolated and characterized. It contained FGFR1 sequences (from exon 9 to the 3′ end of the coding sequence) and a stretch of unknown sequence in its by 5′ portion. Several rounds of cDNA cloning using various probes and libraries were done to obtain the FIM/FGFR1 fusion cDNA and subsequently the wild-type FIM cDNA: by 5′-cDNA walking starting from the 20.2T3 probe derived from the 5′-part of the 20.2 chimeric clone, other partially overlapping cDNA clones were identified from SCID t(8;13) (clones 28, 27.1 and FIM#39.5), skeletal muscle (clone FIM#3), and placental (clone FIM#10.1) human cDNA libraries. This led to the identification of the chimeric FIM/FGFR1 cDNA (clones 20.2 and 28), and of the wild-type FIM cDNA (clones 27.1, and FIM#3, 10.1, and 39.5).
FIM Is the Gene Affected by the Translocation at 13q12.
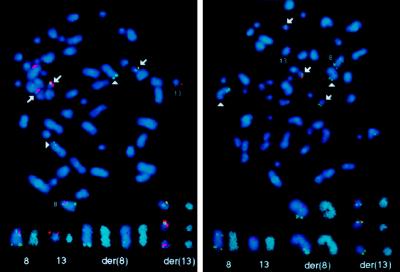
To verify that the cDNA clones (Fig. 1) contained sequences derived from chromosome 13, they were digoxigenin-labeled (red signals) and used as probes in FISH experiments. FISH results obtained with the chimeric FIM/FGFR1 (clone 28) and wild-type FIM (clone FIM#3) cDNAs are shown in Fig. 2. The probes were cohybridized with cosmids labeled with biotin (shown in green) and localized either telomeric to FGFR1 on 8p12 or on 8qter. On normal metaphase chromosomes, probe 28 gave signals on both 8p12 and 13q12 regions and the wild-type FIM cDNA hybridized to band 13q12 (data not shown). On SCID t(8;13) cells, the chimeric cDNA gave signals on normal chromosomes 8 and 13, and mainly on the der (13) (Fig. 2 Left), confirming the 8pter-3′FGFR1-5′-8cen orientation of the FGFR1 gene. This shows that the FIM/FGFR1 cDNA originates from the der(13) fusion gene. The FIM cDNA hybridized to the normal 13, all the der(8) and der(13) chromosomes (Fig. 2 right panel), confirming its position and involvement in the t(8;13)(p12;q12). The same results were obtained with cells from patient 2 presenting with a B-acute lymphoblastic leukemia associated with a t(8;13) (not shown).
Figure 2.
Mapping of the chimeric FIM/FGFR1 and FIM cDNAs. FISH analyses were done on metaphase spreads from SCID t(8;13) cells. These cells contain two der(8) and three der(13). FISH analyses done using the FIM/FGFR1 (clone 28) and wild-type FIM (clone FIM#3) cDNAs are presented in the Left and Right panels, respectively. Normal chromosomes 8 and 13 are indicated, and arrows point to der(13), arrowheads to der(8). Isolated chromosomes counterstained in blue with 4′,6-diamidino-2-phenylindole dihydrochloride and image processed with a sharpen filter to increase the chromosome banding resolution are shown at bottom (at right in each pair).
FIM Is Rearranged in t(8;13)(p12;q12).
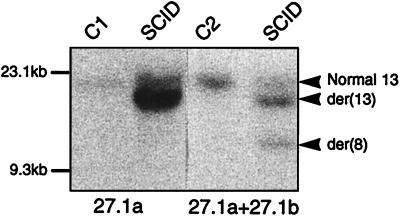
The breakpoints on chromosomes 8 and 13 were detected by Southern blot analysis of DNA from SCID t(8;13) cells. Using the 5′ FIM probe 27.1a subcloned from the wild-type clone 27.1, an aberrant 20-kb BamHI band was detected that corresponds to the der(13) fusion gene in addition to the normal 22-kb fragment (Fig. 3, lane 2). The strong intensity of this aberrant-sized junction fragment reflects the presence of three der(13) chromosomes. The rearranged segment was also revealed by using a 3′-FGFR1 genomic probe (data not shown). Hybridization with the 5′ EcoRI-XhoI fragment from FIM cDNA (clone 27.1, Fig. 1) spanning the breakpoint, showed two rearranged BamHI fragments of 13- and 20-kb, respectively (Fig. 3, lane 4) as compared with control DNA (Fig. 3, lane 3). This probe recognized the two reciprocal products of the t(8;13). Mouse DNA was not detected in SCID t(8;13) cells.
Figure 3.
FIM rearrangement in t(8;13) cells. Southern blot analysis of BamHI-digested DNAs from SCID t(8;13) and controls (C1, mammary carcinoma MDA-MB-134 cells; C2, normal lymphocytes). Hybridizations were done with the FIM cDNA 27.1a probe (two first lanes) and 27.1a plus 27.1b. Positions of the normal and the two rearranged fragments are indicated (arrowheads) on the right. The sizes of HindIII-digested λ DNA fragments are indicated on the left.
Analysis of the FIM Gene and Evidence for a Gene Family.
Sequence analysis and restriction mapping of 12 overlapping 5′ FIM cDNAs clones showed that all were derived from the same gene. The reconstituted wild-type FIM cDNA gene resulted in a 5,008-bp sequence terminated by a poly(A) stretch. The translation initiation site was identified by the presence of a Kozak consensus sequence (GGCATGG) (29), preceded by stop codons in all three reading frames and followed by an ORF of 4,137 bp. In the 3′ noncoding region, two polyadenylation signals were located upstream of the poly(A) tail at positions 4,357 and 4,987. A cDNA variant of the FIM gene, probably the result of an alternative splicing, was also identified and contained a 261-bp deletion [87 amino acids (aa)] from nucleotide 492 to nucleotide 753 (starting from the ATG).
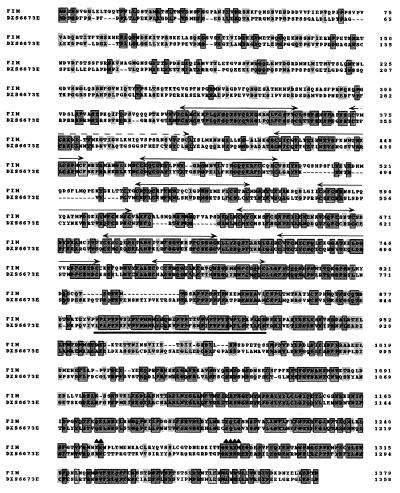
The deduced FIM protein sequence is 1,379 aa long, and is largely hydrophilic (30, 31). The FIM sequence showed several motifs: a N-terminal cysteine-rich region containing nine repeats (from aa 331 to 796) with the consensus sequence C-X2-C-X18–24-F/Y-C-X3-C, which may correspond to putative zing finger motifs (32), a highly hydrophobic, proline-rich stretch (aa 887 to 928), and a putative bipartite nuclear localization signal from aa 1,250–1,284 (33) (Fig. 4). Database searches showed that DXS6673E gene, a candidate for X-linked mental retardation at Xq13.1 (34), has similarity with FIM. DXS6673E encodes a hydrophilic molecule with 10 zinc finger like motifs and a stretch of hydrophobic, proline-rich residues similar to that of FIM. Comparison of the deduced amino acid sequence of FIM with the DXS6673E protein revealed 51, 92, and 57% similarities, for the N-terminal region, the proline-rich stretch, and the C-terminal region, respectively (Fig. 4).
Figure 4.
Amino acid sequence alignments of the putative FIM and DXS6673E (34) proteins. Gray boxes represent identities and gray letters conservative substitutions. Double arrows represent the cysteine-rich motifs with the consensus C-X2-C-X18–24-F/Y-C-X3-C sequence, which may correspond to putative zing finger motifs whereas the dotted double arrow shows a nonconsensus motif in the FIM sequence. The stretch of hydrophobic, proline-rich residues is underlined. ▴, Potential bipartite nuclear localization signal.
Sequence identities were also found between several sequence stretches of FIM and 18 human expressed sequence tags. Similarities were observed between 28 other human expressed sequence tags and different portions of the FIM gene, five of them have been mapped between markers D3S3564 and D3S1588 on chromosome arm 3p (35). Searches also showed significant similarities between the 3′ end of the FIM cDNA and six unmapped murine expressed sequence tags (data not shown).
Expression of Wild-Type and Chimeric mRNAs.
Northern blot analysis of human tissues showed that the FIM gene is ubiquitously expressed as two main transcripts of 5.0 and 7.5 kb (Fig. 5a). Additional transcripts were observed in some tissues. FIM expression was found in normal hematopoietic cells, in different hematopoietic cell lines, and the acute leukemias studied by RT-PCR (data not shown). FGFR1 expression was also found in normal and tumor cells of the hematopoietic system, in agreement with previous studies (36, 37). RT-PCR with primer sets designed to amplify the various transcripts was used to analyze the expression of the two reciprocal fusion transcripts in the patient 1 and SCID t(8;13) cells. Same results were obtained for the two samples, and data from SCID t(8;13) are presented in Fig. 5b (Upper Right). Reverse transcribed mRNAs yielded two variant FGFR1 products of 180 and 186 bp, a 176-bp FIM product, a 155-bp FIM/FGFR1 product expressed from the chromosome der(13), and two products of 202 and 208 bp for the FGFR1/FIM fusion gene on der(8). The two reciprocal chimeric transcripts were also amplified from RNA of patient 2 with a t(8;13) (data not shown) but not from normal cell RNAs (Fig. 5b Lower Right). Competitive PCR showed a predominance of chimeric FIM/FGFR1 and wild-type FIM transcripts in the leukemic sample (Fig. 5b Upper Right).
Figure 5.
Expression of FIM and of the fusion transcripts. (a) Northern blot analysis of the expression of FIM. Northern blots with indicated human adult poly(A)+ RNAs were hybridized with the FIM 27.1a probe. β-actin was used as control. (b) RT-PCR products obtained from SCID t(8;13) and normal breast (Upper and Lower Right, respectively) RNAs using FGFR1 and FIM primers located near the translocation, without (four first lanes) or with (five last lanes) oligonucleotide competition. (Right) Primers used were: lane 1, FA and F9; lane 2, X1 and 1R; lane 3, FA and 1R; lane 4, X1 and F9; lane 5, FA, F9, and 1R; lane 6, FA, F9, and X1; lane 7, X1, 1R, and FA; lane 8, F9, X1, and 1R; and lane 9, FA, F9, X1, and 1R. The two variant products of the wild-type FGFR1 gene of 180 and 186 bp (FA and F9 primers) are seen in lanes 1, 5, 6, and 9. β-actin primers were used to estimate the efficiency of the PCR reactions (Left). The respective chromosomal positions and the transcripts identified in each reaction are indicated at the top and on the right, respectively.
Characterization of the FIM/FGFR1 Fusion Junction.
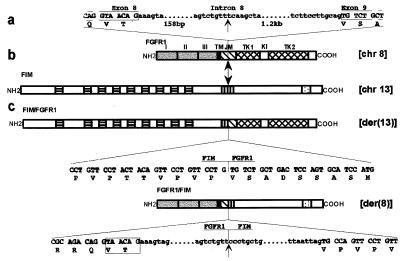
Sequence analysis of chimeric cDNAs (clones 20.2 and 28) confirmed that they code for an in frame fusion of the FIM N-terminal region with the FGFR1 intracellular domain, and correspond to the der(13) fusion gene. Sequencing of PCR products from both the patient 1 and SCID t(8;13) confirmed the presence of the two reciprocal fusion cDNAs whose breakpoint sequences are given in Fig. 6. Two types of clones for the der(8) fusion gene were obtained; they differ by the presence (or absence) of a 6-bp stretch (TAACAG), encoding a TV doublet in the FGFR1 juxtamembrane domain (Fig. 6c), a feature previously described for wild-type FGFR1 cDNAs (38). The breakpoint is in intron 8, between FGFR1 exons 8 and 9 (Fig. 6 a and c), which encode the juxtamembrane domain of the receptor (22). Genomic DNA fragments containing portions of the FGFR1/FIM fusion gene, as well as the wild-type FGFR1 intron 8, were isolated. Breakpoint sequences were determined and compared with the nucleotide sequence of the junctions of introns and exons of the FGFR1 gene (20). FGFR1 intron 8 is 1.2 kb. It is interrupted 158 bp downstream of exon 8 and linked to an intronic FIM sequence that shares similarities with human Alu-Sp subfamily (not shown). This repetitive region may be the focus of the chromosome recombination (39).
Figure 6.
Schematic representation of the two partner genes and their fusion products. Vertical arrows indicate breakpoints in the FGFR1 and FIM proteins. (a) Nucleotide sequence of the junctions between exon 8 and intron 8, intron 8 and exon 9 (left and right ends, respectively), and of the breakpoint in intron 8 are indicated. (b) Schematic diagrams of the normal human FGFR1 and FIM proteins. FGFR1: three extracellular Ig-like domains (labeled I, II, and III), transmembrane domain (TM), intracellular region comprising juxtamembrane domain (JM), tyrosine kinase 1 and 2 subdomains (TK1, TK2) interrupted by a kinase insert (KI), and C-tail; FIM: N-terminal region with nine cysteine-rich motifs (▤), hydrophobic stretch rich in proline residues (▥), and a C-terminal putative bipartite nuclear localization signal (░⃞). (c) Schematic representation of the two resulting chimeric proteins. The nucleotide and deduced aa sequences near the breakpoints are shown below each protein. Portions of intronic nucleotide sequence corresponding to the FGFR1/FIM fusion gene are also shown. The boxed 6-bp sequence corresponds to the two FGFR1 variants. Upper- and lowercase letters correspond to exon and intron sequences, respectively. The chromosome position of genes encoding the four proteins is indicated on the right.
Constitutive Kinase Activity and Tyrosine Phosphorylation of FIM/FGFR1.
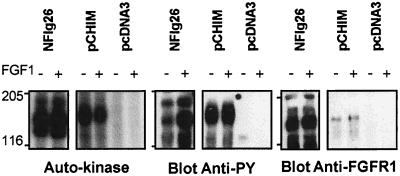
NIH 3T3 cells were transiently transfected with the FIM/FGFR1 cDNA to characterize the FIM/FGFR1 chimeric protein. Anti-C-FGFR1 immunoprecipitates were immunoblotted with anti-C-FGFR1 antibody to verify for proper expression of the chimeric construct and with anti-phosphotyrosine antibody to measure the tyrosine phosphorylation level of the kinase. Part of the immunoprecipitate was also assayed for autophosphorylation activity in the presence of [γ32P]ATP. The chimeric protein was detectable as a band of ≈150 kDa, slightly larger than FGFR1 (Fig. 7, Right, pCHIM and NFlg26 lanes, respectively). The chimeric protein had autophosphorylating activity, and was constitutively tyrosine phosphorylated regardless of FGF1 stimulation (Fig. 7 Left and Center, respectively). In contrast, tyrosine phosphorylation of FGFR1 was stimulated by FGF1 addition in NFlg26 cells (Fig. 7 Center). These results indicate that FIM/FGFR1 is a constitutively activated tyrosine kinase, and suggest that this activation may be mediated by dimerization of the N-terminal portion of the chimeric protein that contains putative zinc fingers.
Figure 7.
Autokinase activity and phosphorylation on tyrosine of FIM/FGFR1. The FGFR1 overexpressing cell line (NFlg26) and NIH 3T3 cells transiently transfected with either FIM/FGFR1 cDNA (pCHIM) or the empty vector (pcDNA3) were treated with (+) or without (−) FGF1. Immunoprecipitates using anti-C-FGFR1 antibody were analyzed for autokinase activity (Left) (autoradiography of 15 hr for NFlg26 and 45 hr for pCHIM and pcDNA3), phosphorylation on tyrosine after Western blot with an anti-phosphotyrosine antibody (4G10) and expression level by Western blot with anti-C-FGFR1 antibody (Right). The position of molecular mass standards (in kDa) is indicated.
DISCUSSION
We report here the cloning and characterization of a gene, FIM, that maps to the 13q12 region, and is disrupted by a balanced t(8;13)(p12;q12) in a stem-cell MPD. The translocation results in the fusion of FGFR1, a tyrosine kinase receptor for members of the FGF family, to a protein, FIM. The two reciprocal fusion genes are transcribed in the malignant cells: FIM/FGFR1 from the der(13), and FGFR1/FIM from the der(8). FIM is widely expressed and could therefore drive the expression of the FIM/FGFR1 fusion transcript in the hematopoietic stem-cell compartment. We demonstrate that the chimeric receptor FIM/FGFR1 is constitutively phosphorylated in transfected cells. This fusion protein is likely to promote hematopoietic stem cell proliferation and leukemogenesis through constitutive activation of the FGFR1 kinase, either with the participation of normal FGFR1 substrates or by recruiting novel cytoplasmic proteins. The MPD syndrome associated with 8p12 rearrangement is characterized by T or B cell lymphoblastic lymphoma, eosinophilia, myeloid hyperplasia, and evolution toward acute myelogenous leukemia; it is likely to be due to transformation of a multilineage precursor (1, 3, 8). Our previous FISH studies showed that FGFR1 is involved in all cases with this malignant phenotype in which 8p12 is rearranged with 6q27, 9q32–34, or 13q12 (21). These findings strongly suggest that an altered FGFR1 protein could play the oncogenic role in the genesis of this myeloproliferative syndrome, leading to uncontrolled proliferation of stem cells.
The deduced sequence of the FIM protein contains several potentially important motifs: nine putative zinc fingers, a proline-rich stretch, and a putative bipartite nuclear localization signal. These structural features suggest that FIM may belong to a transcription regulator family, several members of which are involved in hematopoietic malignancies (40). FIM is structurally related to DXS6673E, a candidate gene for X-linked mental retardation in Xq13.1. DXS6673E is subject to X-inactivation and is disrupted in its 5′ untranslated region by a balanced t(X;13)(q13.1;q31) in a mentally retarded girl (34). These results show evidence for a family of molecules, the first two members of which, DXS6673E and the presently reported FIM molecule, are involved in translocations and rearranged in human diseases. The actual size of this gene family remains to be determined, a task that will be greatly facilitated by further characterization of the expressed sequence tag clones that we identified.
The four FGFRs (FGFR1–FGFR4) constitute a specific class of tyrosine kinase receptors. They play a crucial role in several processes, during development and adult life, through the binding of their numerous ligands, the fibroblast growth factors (reviewed in ref. 41). Activating mutations in human FGFR1, FGFR2, and FGFR3 were found in a variety of hereditary skeletal disorders (42, 43). FGFR1 is specifically involved in the Pfeiffer syndrome, an autosomal dominant craniosynostosis syndrome. We show here that FGFR1, which is disrupted by a translocation in a stem-cell MPD is also likely to have oncogenic potential. FGFR2 activation by a C terminus alteration through a chromosomal rearrangement was described in a rat osteosarcoma (44). Alterations of FGFR3, whose germline activating mutations are responsible for major forms of dwarfism (43, 45), were found in t(4;14)(p16;q32) translocations associated with multiple myeloma (46). Thus, distinct alterations of FGF receptors are associated with cancers—two of them, FGFR1 and FGFR3, are involved in two different types of hematopoietic malignancies—and in other human diseases. A pluriphenotypic behavior is observed with another tyrosine kinase receptor, RET, whose alterations lead to the developmental Hirschprung disease or to endocrine gland neoplasia (reviewed in ref. 47).
In human tumors, all the fusion events involving a tyrosine kinase receptor described to date produce chimeric proteins in which various portions of the N terminus of the normal receptor are replaced by other sequences (48–50). The resultant chimeric proteins contain structural and functional domains that differ from the wild-type proteins. The FIM/FGFR1 protein lacks the ligand-binding and transmembrane domains of FGFR1 that are replaced by the putative zing finger motifs of the FIM molecule while retaining the tyrosine kinase domain of FGFR1. The constitutive kinase activation of the chimeric protein is likely to be due to dimerization through the zinc finger like motifs of FIM, associated with the possible relocation of the mutant receptor to a new compartment of the cell. The elucidation of specific pathogenetic contributions of each fusion gene should give insight in the development and maintenance of the malignant phenotype. It will be interesting to determine whether t(8;13) cells are sensitive to the inhibitory effects of chemical compounds that interact with the catalytic domain of FGFR1 and induce conformational changes in the nucleotide-binding loop (51).
Acknowledgments
We are grateful to C. Mawas and D. Maraninchi for their encouragement; F. Coulier and P. Pontarotti for comments; M. Jaye (Rhône-Poulenc Rorer) for his kind gift of FGFR1 probes and NFlg26 cells; and to F. Birg for the critical reading of the manuscript. This work was supported by Institut National de la Santé et de la Recherche Médicale, Institut Paoli–Calmettes, and grants from Association pour la Recherche sur le Cancer, Comité des Bouches-du-Rhône de la Ligue Nationale contre le Cancer, Fondation de France, Fondation contre la Leucémie, and Fédération Nationale des Groupements des Entreprises Françaises dans la Lutte contre le Cancer. C.P. is the recipient of a Fellowship from the Société Française d’Hématologie.
Note Added in Proof
While this article was being processed, two other studies, by Xiao et al. (52) and Smedley et al. (53), described the molecular characterization of the t(8;13), and the 13q12 gene was named ZNF198 and RAMP, respectively.
Footnotes
This paper was submitted directly (Track II) to the Proceedings Office.
Abbreviations: FISH, fluorescent in situ hybridization; FGF, fibroblast growth factor; MPD, myeloproliferative disorder; SCID, severe combined immunodeficient; aa, amino acid.
Data deposition: The sequences reported in this paper have been deposited in the GenBank database (accession no. Y13472).
References
- 1.Rabbitts T. Nature (London) 1994;372:143–179. doi: 10.1038/372143a0. [DOI] [PubMed] [Google Scholar]
- 2.Soenen V, Chaffanet M, Preudhomme C, Dib A, Laï J-P, Fletcher J, Birnbaum D, Pébusque M-J. Genes Chromosomes Cancer. 1996;15:191–194. doi: 10.1002/(SICI)1098-2264(199603)15:3<191::AID-GCC9>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- 3.MacDonald D, Aguiar R, Mason P, Goldman J, Cross N. Leukemia. 1995;9:1628–1630. [PubMed] [Google Scholar]
- 4.Vannier J, Bizet M, Bastard C, Bernard A, Ducastelle T, Tron P. Leuk Res. 1984;8:647–657. doi: 10.1016/0145-2126(84)90013-4. [DOI] [PubMed] [Google Scholar]
- 5.Elsner S, Martin H, Rode C, Wassman B, Ganser A, Hoelzer D. Br J Haematol. 1994;87:124. [Google Scholar]
- 6.Friedhoff F, Rajendra B, Moody R, Alapatt T. Cancer Genet Cytogenet. 1983;9:391–394. doi: 10.1016/0165-4608(83)90088-2. [DOI] [PubMed] [Google Scholar]
- 7.Lewis J, Jenks H, Lazerson J. Am J Pediatr Hematol Oncol. 1983;5:265–269. [PubMed] [Google Scholar]
- 8.Oscier D, Mufti G, Gardiner A, Hamblin T. Med Genet. 1985;22:398–401. doi: 10.1136/jmg.22.5.398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jotterand Bellomo M, Muhlematter D, Wicht M, Delacretaz F, Schmidt P. Br J Haematol. 1992;81:307–309. doi: 10.1111/j.1365-2141.1992.tb08225.x. [DOI] [PubMed] [Google Scholar]
- 10.Nakayama H, Inamitsu T, Ohga S, Kai T, Suda M, Matsuzaki A, Ueda K. Br J Haematol. 1996;92:692–695. doi: 10.1046/j.1365-2141.1996.00386.x. [DOI] [PubMed] [Google Scholar]
- 11.van den Berg H, Kroes W, van der Schoot C, Dee R, Pals S, Bouts T, Slater R. Leukemia. 1996;10:1252–1253. [PubMed] [Google Scholar]
- 12.Fagan K, Hyde S, Harrison P. Cancer Genet Cytogenet. 1993;65:71–73. doi: 10.1016/0165-4608(93)90062-q. [DOI] [PubMed] [Google Scholar]
- 13.Leslie J, Barker T, Glancy M, Jennings B, Pearson J. Br J Haematol. 1994;86:876–878. doi: 10.1111/j.1365-2141.1994.tb04847.x. [DOI] [PubMed] [Google Scholar]
- 14.Aguiar R, Macdonald D, Mason P, Cross N, Goldman J. Blood. 1995;86:834–835. [PubMed] [Google Scholar]
- 15.Behringer D, Schaefer H, Kunzmann R, Mertelsmann R, Dolken G. Leukemia. 1995;9:988–992. [PubMed] [Google Scholar]
- 16.Inhorn R, Aster J, Roach S, Slapak C, Soiffer R, Tantravahi R, Stone R. Blood. 1995;85:1881–1887. [PubMed] [Google Scholar]
- 17.Kempsi H, MacDonald D, Michalski A, Roberts T, Goldman J, Cross N, Cowell J. Genes Chromosomes Cancer. 1995;12:283–287. doi: 10.1002/gcc.2870120408. [DOI] [PubMed] [Google Scholar]
- 18.Naeem R, Singer S, Fletcher J. Genes Chromosomes Cancer. 1995;12:148–151. doi: 10.1002/gcc.2870120210. [DOI] [PubMed] [Google Scholar]
- 19.Michaux L, Mecucci C, Pereira Velloso E, Dierlamm J, Criel A, Louwagie A, van Orshoven A, van den Berghe H. Blood. 1996;87:1658–1659. [PubMed] [Google Scholar]
- 20.Johnson D, Lu J, Chen H, Werner S, Williams L. Mol Cell Biol. 1991;11:4627–4634. doi: 10.1128/mcb.11.9.4627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chaffanet M, Popovici C, Leroux D, Jacrot M, Adélaïde J, Dastugue N, Grégoire M-J, Hagemeijer A, Lafage-Potchitaloff M, Birnbaum D, et al. Oncogene. 1998;16:945–949. doi: 10.1038/sj.onc.1201601. [DOI] [PubMed] [Google Scholar]
- 22.Dionne C, Crumley G, Bellot F, Kaplow J, Searfoss G, Ruta M, Burgess W, Jaye M, Schlessinger J. EMBO J. 1990;9:2685–2692. doi: 10.1002/j.1460-2075.1990.tb07454.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lafage M, Pedeutour F, Marchetto S, Simonetti J, Prosperi M, Gaudray P, Birnbaum D. Genes Chromosomes Cancer. 1992;5:40–49. doi: 10.1002/gcc.2870050107. [DOI] [PubMed] [Google Scholar]
- 24.Chaffanet M, Imbert A, Adélaïde J, Le Paslier D, Wagner M, Wells D, Birnbaum D, Pébusque M-J. Cytogenet Cell Genet. 1996;72:63–68. doi: 10.1159/000134164. [DOI] [PubMed] [Google Scholar]
- 25.Sambrook J, Fritsch E, Maniatis T. Molecular Cloning: A Laboratory Manual. 2nd Ed. Plainview, NY: Cold Spring Harbor Lab. Press; 1989. [Google Scholar]
- 26.Wang J-K, Gao G, Goldfarb M. Mol Cell Biol. 1994;14:181–188. doi: 10.1128/mcb.14.1.181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bellot F, Crumley G, Kaplow J, Schlessinger J, Jaye M, Dionne C. EMBO J. 1990;10:2849–2854. doi: 10.1002/j.1460-2075.1991.tb07834.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang J-K, Goldfarb M. Oncogene. 1997;14:1767–1778. doi: 10.1038/sj.onc.1201021. [DOI] [PubMed] [Google Scholar]
- 29.Kozak M. Mamm Genome. 1996;7:563–574. doi: 10.1007/s003359900171. [DOI] [PubMed] [Google Scholar]
- 30.Kyte J, Doolittle R. J Mol Biol. 1982;157:105–132. doi: 10.1016/0022-2836(82)90515-0. [DOI] [PubMed] [Google Scholar]
- 31.Eisenberg D, Schwarz E, Komaromy M, Wall R. J Mol Biol. 1984;179:125–142. doi: 10.1016/0022-2836(84)90309-7. [DOI] [PubMed] [Google Scholar]
- 32.Klug A, Schwabe W. FASEB J. 1995;9:597–604. [PubMed] [Google Scholar]
- 33.Robbins J, Dilworth S, Laskey R, Dingwall C. Cell. 1991;64:615–623. doi: 10.1016/0092-8674(91)90245-t. [DOI] [PubMed] [Google Scholar]
- 34.van der Maarel S, Scholten I, Huber I, Philippe C, Suijkerbuijk R, Gilgenkrantz S, Kere J, Cremers F, Ropers H. Hum Mol Genet. 1996;5:887–897. doi: 10.1093/hmg/5.7.887. [DOI] [PubMed] [Google Scholar]
- 35.Gemmill R, Chumakov I, Scott P, Waggoner B, Rigault P, Cypser J, Chen Q, Weissenbach J, Gardiner K, Wang H, Pekarsky Y, Le Gall I, et al. Nature (London) 1995;377:299–319. doi: 10.1038/377299a0. [DOI] [PubMed] [Google Scholar]
- 36.Armstrong E, Vainikka S, Partanen J, Korhonen J, Alitalo R. Cancer Res. 1992;52:2004–2007. [PubMed] [Google Scholar]
- 37.Allouche M, Bayard F, Clamens S, Fillola G, Sie P, Amalric F. Leukemia. 1995;9:77–86. [PubMed] [Google Scholar]
- 38.Hou Z, Kan M, McKeehan K, McBride G, Adams P, McKeehan W. Science. 1991;251:665–668. doi: 10.1126/science.1846977. [DOI] [PubMed] [Google Scholar]
- 39.Chissoe S, Bodenteich A, Wang Y F, Wang Y P, Burian D, Clifton S, Crabtree J, Freeman A, Iyer K, Jian L, et al. Genomics. 1995;27:67–82. doi: 10.1006/geno.1995.1008. [DOI] [PubMed] [Google Scholar]
- 40.Strout M, Caligiuri M. Curr Opin Oncol. 1997;9:8–17. doi: 10.1097/00001622-199701000-00003. [DOI] [PubMed] [Google Scholar]
- 41.Goldfarb M. Cytokine Growth Factor Rev. 1996;7:311–325. doi: 10.1016/s1359-6101(96)00039-1. [DOI] [PubMed] [Google Scholar]
- 42.Muenke M, Schell U, Hehr A, Robin N, Losken H, Schinzel A, Pulleyn L, Rutland P, Reardon W, Malcolm S, Winter R. Nat Genet. 1994;8:269–274. doi: 10.1038/ng1194-269. [DOI] [PubMed] [Google Scholar]
- 43.Wilkie A, Morriss-Kay G, Jones E, Heath J. Curr Biol. 1995;5:500–507. doi: 10.1016/s0960-9822(95)00102-3. [DOI] [PubMed] [Google Scholar]
- 44.Lorenzi M, Horii Y, Yamanaka R, Sakaguchi K, Miki T. Proc Natl Acad Sci USA. 1996;93:8956–8961. doi: 10.1073/pnas.93.17.8956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Muenke M, Schell U. Trends Genet. 1995;11:308–313. doi: 10.1016/s0168-9525(00)89088-5. [DOI] [PubMed] [Google Scholar]
- 46.Chesi M, Nardini E, Brents L, Schröck E, Ried T, Kuehl M, Bergsagel L. Nat Genet. 1997;16:260–264. doi: 10.1038/ng0797-260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Edery P, Eng C, Munnich A, Lyonnet S. BioEssays. 1997;19:389–395. doi: 10.1002/bies.950190506. [DOI] [PubMed] [Google Scholar]
- 48.Sawyers C, Denny C. Cell. 1994;77:171–173. doi: 10.1016/0092-8674(94)90307-7. [DOI] [PubMed] [Google Scholar]
- 49.Golub T, Barker G, Lovett M, Gilliland D. Cell. 1994;77:307–316. doi: 10.1016/0092-8674(94)90322-0. [DOI] [PubMed] [Google Scholar]
- 50.Morris S, Kirstein M, Valentine M, Dittmer K, Shapiro D, Saltman D, Look A. Science. 1994;263:1281–1284. doi: 10.1126/science.8122112. [DOI] [PubMed] [Google Scholar]
- 51.Mohammadi M, McMahon G, Sun L, Tang C, Hirth P, Yeh B, Hubbard S, Schlessinger J. Science. 1997;276:955–960. doi: 10.1126/science.276.5314.955. [DOI] [PubMed] [Google Scholar]
- 52.Xiao S, Nalabolu S R, Aster J C, Ma J, Abruzzo L, Jaffe E S, Stone R, Weissman S M, Hudson T J, Fletcher J A. Nat Genet. 1998;18:84–87. doi: 10.1038/ng0198-84. [DOI] [PubMed] [Google Scholar]
- 53.Smedley D, Hamoudi R, Clark J, Warren W, Abdul-Rauf M, Somers G, Venter D, Fagan K, Cooper C, Shipley J. Hum Mol Genet. 1998;7:637–642. doi: 10.1093/hmg/7.4.637. [DOI] [PubMed] [Google Scholar]