Abstract
Dengue virus infections are a major cause of morbidity and mortality in tropical and subtropical areas in the world. Attempts to develop effective vaccines have been hampered by the lack of understanding of the pathogenesis of the disease and the absence of suitable experimental models for dengue viral infection. The magnitude of T-cell responses has been reported to correlate with dengue disease severity. Sixty Malaysian adults with dengue viral infections were investigated for their dengue virus-specific T-cell responses to 32 peptides antigens from the structural and nonstructural regions from a dengue virus isolate. Seventeen different peptides from the C, E, NS2B, NS3, NS4A, NS4B, and NS5 regions were found to evoke significant responses in a gamma interferon enzyme-linked immunospot (ELISPOT) assay of samples from 13 selected patients with dengue fever (DF) and dengue hemorrhagic fever (DHF). NS3 and predominantly NS3422-431 were found to be important T-cell targets. The highest peaks of T-cell responses observed were in responses to NS3422-431 and NS5563-571 in DHF patients. We also found almost a sevenfold increase in T-cell response in three DHF patients compared to three DF patient responses to peptide NS3422-431. A large number of patients' T cells also responded to the NS2B97-106 region. The ELISPOT analyses also revealed high frequencies of T cells that recognize both serotype-specific and cross-reactive dengue virus antigens in patients with DHF.
Dengue viruses belong to the genus Flavivirus, family Flaviviridae, and are subgrouped into four serotypes, Dengue virus serotype 1 (DV1) through DV4 (17, 23). Dengue virus infections are the most extensive vector-borne viral disease in humans. The virus is transmitted to humans via the bite of an infected Aedes aegypti mosquito (46). After an incubation period of 5 to 7 days, patients develop classical symptoms such as high fever, headache, rash, abdominal pain, myalgia, and arthralgia for 2 to 7 days. In the majority of patients, the disease causes a self-limiting febrile illness in which fever and symptoms normally abate. Reports have also revealed that almost 50% of infections are recognized to be clinically silent infections (12). Conversely, the disease can be severe and complicated, with thrombocytopenia, plasma leakage, bleeding, and hypovolemic shock, commonly referred to as dengue hemorrhagic fever (DHF), which occurs in 5 to 30% of cases and can be classified according to severity as grade I to IV (35, 54).
Epidemiological reports have revealed that dengue is endemic in 112 countries (44, 55). Approximately 2.5 to 3 billion people, living mainly in urban areas of tropical and subtropical regions, are potentially at risk of acquiring dengue viral infections (55). Estimates suggest that annually some 2.5 billion people are at risk of infection, and 100 million cases of dengue fever and half a million cases of DHF occur throughout the world, with a fatality rate in Asian countries of 0.5% to 3.5% (20). Children less than 15 years old make up 90% of the DHF subjects. According to epidemiologic studies in Malaysia, dengue fever (DF) has been endemic in Malaysia since 1902 and reached epidemic proportions in 1973. Reports propose that the age-specific morbidity rate was highest in the 10- to 19-year-old age group, followed by the 20- to 29-year-old group. The fatality rate of the disease was 5.4 cases per 100,000 in 1973 and reached 10.4 cases per 100,000 in 1987 (48). Currently, the fatality rate stands at 3 to 5% in the DHF group.
The major complication in dengue is that infection with one serotype during primary infection confers future protective immunity against that particular serotype but not against other serotypes during a secondary infection. This phenomenon was elucidated in the antibody-dependent enhancement theory (19). Halstead proposed that during secondary infection, the avidity or titer of pre-existing antibodies may not be sufficient to neutralize the second serotype and consequently increase the viral load through significant opsonization and replication of the virus in the macrophages. This mechanism, which leads to excessive immune activation with a storm of proinflammatory cytokine release, suggests a defect in vascular permeability, leading to the profound plasma leakage that is prominent in patients with severe disease (24, 29). However, this hypothesis alone could not explain the association of disease severity with viral load. Evidence clearly shows that at the defervescent stage, viral load falls abruptly as the fever abates, and at this point severe symptoms such as vascular leakage, hemorrhage, and shock appear. This observation suggested that it may be the host immune response to the virus rather than the virus itself that causes the pathology. It is believed that the disease severity might be caused by T-cell-mediated tissue damage and that the immune response may also contribute to vascular leakage by directly lysing infected endothelial cells. Others have reported that such virus-induced immunopathology can be distinguished in a number of other viral infections, such as respiratory syncytial virus and lymphocytic choriomeningitis virus infections, in mice (1, 6). Consequently, the T cells seem to play a vital role in immunopathogenesis of viral infections.
Identification of various peptides from all four serotypes will probably be necessary for the development of a universally immunogenic vaccine. Reports have shown that a dengue vaccine must provide solid and long-lasting protection against all four dengue virus serotypes. Others state that a safe vaccine should be polyvalent to avoid inducing a monotype-enhancing immune response that may lead to severe manifestation of the disease (9, 21, 43). Identification of peptides that could be protective or cause immunopathogenesis is therefore an important goal. In this study, T-cell responses in dengue infection were investigated in a selected cohort of naturally infected Malaysian patients. We selected 60 patients with primary and secondary dengue viral infections and measured the T-cell responses to a pool of dengue viral antigens in a selected cohort from which sufficient blood was available. Here, we report the frequencies of T-cell responses to dengue viral peptides of the structural and nonstructural regions.
MATERIALS AND METHODS
Study population.
The study was carried out from January 2005 to June 2006. Blood samples were obtained from clinically diagnosed dengue virus-infected adult patients who were admitted to the University Malaya Medical Centre (UMMC), Kuala Lumpur, Malaysia. Twenty to twenty-five milliliters of blood was collected from each patient. Blood samples were processed within 4 h, buffy coat extracted, and stored in liquid nitrogen till use. Patients with DF and DHF were classified according to World Health Organization criteria (54). Age, gender, race, and medical history were recorded for each patient. Clinical information, including observations regarding ascites, pleural effusion, and circulatory disturbance as a result of plasma leakage, was collected to enable disease classification. Ultrasonography or chest X ray was not performed routinely to detect potential low-level, clinically undetectable pleural effusion or ascites. Platelet counts and hematocrit values were recorded serially during hospitalization. Samples from healthy donors that were age, gender, and race matched with patients were obtained from the blood bank of UMMC as negative controls. Written informed consent was obtained from the patients. Ethical clearance was given by the Scientific and Ethical Committee at the UMMC.
Cell and serum isolation.
Peripheral blood mononuclear cells (PBMC) were isolated by the Ficoll-Hypaque (Lymphoprep; Axis-Shield, Oslo, Norway) density gradient centrifugation method (22). Cells were counted by the trypan blue exclusion method and resuspended at a concentration of 1 × 107/ml in freezing medium containing 90% fetal calf serum (JRH Biosciences Inc.) and 10% dimethyl sulfoxide and cryopreserved until use. Serum was obtained by centrifugation at 1,500 rpm for 5 min at 4°C and stored at −80 until use.
Dengue virus PCR and serology.
Dengue virus RNA was extracted from plasma samples with a QIAamp viral RNA minikit (QIAGEN). RNA was reverse transcribed and a one-step real-time reverse transcription-PCR assay for dengue virus was performed by employing TaqMan technology (56). Dengue virus infection was further confirmed for all samples serologically with an in-house capture immunoglobulin M (IgM) enzyme-linked immunosorbent assay (31). Primary and secondary dengue infection were defined based on IgG antibody titers determined by a hemagglutination inhibition test in paired acute- and convalescent-phase sera (11).
DNA extraction and HLA typing.
Human DNA was extracted with an AccuPrep genomic DNA extraction kit (Bioneer) following the manufacturer's instructions. A commercial HLA kit (Olerup SSP typing kit) was used to determine the HLA types.
Peptide prediction and synthesis.
Two predictive algorithms—SYFPEITHI (http://www.syfpeithi.de) and the RANKPEP (http://bio.dfci.harvard.edu/Tools/rankpep.html) program—were used to predict peptide antigens spanning the entire genome of structural antigens (capsid, pre-M/M, and envelope) and nonstructural viral antigens (NS1, NS2a, NS2b, NS3, NS4a, NS4b, and NS5) of DV2 isolates against major HLA types among the patient cohort. The complete sequences of dengue virus genome were obtained from GenBank (http://www.ncbi.nlm.nih.gov), and accession numbers are shown in Table 1. A total of 252 peptides were predicted to bind to HLA-A (HLA-A*02, HLA-A*24, HLA-A*03, and HLA-A*11) and HLA-B (HLA-B*08 and HLA-B*27). A total of 32 top-scoring peptides (based on binding affinities, as determined by the two programs, of ≥27) of 8 to 11 amino acids that associate with the predominant HLA-A and HLA-B types were selected and synthesized (Table 2). Peptides were synthesized by and purchased from JPT and were more than 90% pure according to high-pressure liquid chromatography analysis. The peptides were grouped into six mixtures of five or six peptides each for investigational convenience (pools A to F) (Table 2).
TABLE 1.
Viruses and GenBank accession numbers of the selected peptide sequences
TABLE 2.
Peptides sequences in each peptide pool analyzed by ELISPOT IFN- γ assay
| Peptide pool | Dengue virus region | Peptide sequence | Virus specificity | HLA type | Score |
|---|---|---|---|---|---|
| A | AnchC | ALVAFLRFL49-57 | DV2 | A*02/24 | 27 |
| AnchC | GRMLNILNR89-97 | DV2 | B*27 | 30 | |
| CP | RVSTVQQLTK22-31 | DV2 | A*03/11 | 30 | |
| CP | ILKRWGTIK65-73 | DV2 | A*03/11 | 32 | |
| PrM | QEKGKSLL17-24 | DV2 | A*02/24 | 30 | |
| B | E | MAKNKPTL34-41 | DV1, DV2, DV3 | B*08 | 34 |
| E | FLDLPLPWL213-221 | DV2 | A*02/24 | 27 | |
| E | SLSVSLVLV474-482 | DV2 | A*02/24 | 28 | |
| E | SLVLVGVVTL478-487 | DV2 | A*02/24 | 30 | |
| NS1 | SLRTTTASGK297-305 | DV2, DV4 | A*03/11 | 32 | |
| C | NS1 | ILSENEVKL78-86 | DV2 | A*02/24 | 28 |
| NS1 | PLKEKEENL337-345 | DV2 | B*08 | 38 | |
| NS2A | MLRTRVGTK22-30 | DV2 | A*03/11 | 30 | |
| NS2A | ALALGMMVLK126-135 | DV2 | A*03/11 | 29 | |
| NS2A | AVILQNAWK158-166 | DV2 | A*03/11 | 30 | |
| D | NS2B | ELERAADVK52-60 | DV2 | A*03/11 | 31 |
| NS2B | ILIRTGLLVI97-106 | DV2 | A*02/24 | 27 | |
| NS3 | RIKQKGIL25-32 | DV2 | B*08 | 34 | |
| NS3 | RIEPSWADVK64-74 | DV2 | A*03/11 | 30 | |
| NS3 | AIKRGLRTL112-120 | DV2 | A*02/24 | 27 | |
| NS3 | RVIDPRRCMK422-431 | DV2 | A*03/11 | 34 | |
| E | NS3 | DKKGKVVGL142-150 | DV2 | B*08 | 32 |
| NS4A | ALSELPETL44-52 | DV2 | A*02/24 | 27 | |
| NS4A | LLLLTLLATV55-64 | DV2 | A*02/24 | 31 | |
| NS4A | LLLTLLATV56-64 | DV2 | A*02/24 | 33 | |
| NS4B | LEKTKKDL6-13 | DV2 | B*08 | 30 | |
| NS4B | AIIGPGLQAK119-128 | DV1, DV2, DV3, DV4 | A*03/11 | 27 | |
| F | NS5 | NVREVKGLTK96-105 | DV2 | A*03/11 | 31 |
| NS5 | VLNPYMPSV182-190 | DV1, 2 | A*02/24 | 28 | |
| NS5 | KITAEWLWK375-383 | DV2, 3 | A*03/11 | 27 | |
| NS5 | KLAEAIFKL563-571 | DV2 | A*02/24 | 31 | |
| NS5 | AISGDDCVVK659-668 | DV2 | A*03/11 | 30 |
ELISPOT assay.
PBMC samples obtained 4 to 9 days after onset of fever from 60 patients experiencing a presumptive primary or secondary dengue infection were tested using a gamma interferon (IFN-γ)-based enzyme-linked immunospot (ELISPOT) assay kit (Mabtech, Stockholm, Sweden). Briefly, 96-well filtration plates (MultiScreenHTS, MPIPS4W10; Millipore) were precoated with 12 μg/ml of the anti-IFN-γ monoclonal antibody 1-DIK (Mabtech) and kept at 4°C till use. A total of 2 × 105 PBMC were then added in 100 μl of culture medium (RPMI-1640, 5% fetal calf serum [FCS], 1% l-glutamine) per well, and 10 μl of a peptide mixture or individual peptides at a final concentration of 18 μg/ml was subsequently placed in triplicate in each well. In every case, peptides recognized as being antigenic were retested as individual peptides. Consequently, each positive response was confirmed twice. Cells were cultured for 20 h at 37°C in a humidified 5% CO2 atmosphere. Plates were then washed with phosphate-buffered saline (PBS), and 100 μl of PBS containing 1 μg/ml of biotinylated the anti-IFN-γ monoclonal antibody 7-B6-1-biotin (Mabtech) was added to each well. After 2 h, plates were washed, and horseradish peroxidase streptavidin (BD Biosciences Pharmingen) was added at a dilution of 1:1,000 in PBS for 1 h. Spots were revealed by incubation with substrate for 20 min. The spots were then visualized and counted with a Zeiss ELISPOT reader. The average number of spots per well was used to express each experimental value as spot-forming cells (SFCs) per 106 PBMC. The number of positive SFCs was calculated after negative control values were subtracted. Responses were considered significant when a minimum of five SFCs were present per well, representing at least twice the number of SFCs in negative control well. All assays included positive (phytohemagglutinin) and negative (PBMC alone) controls. PBMC from healthy donors were also studied correspondingly with the pool peptides and individual peptides.
Statistical analysis.
A paired-sample t test analysis was used for comparisons across multiple groups of peptide pools. A P value of <0.05 was regarded as significant. The SPSS software package, version 14 (SPSS, Inc., Chicago, IL), was used for all analyses.
RESULTS
Characteristics of study population.
This study was carried out at the UMMC, Kuala Lumpur, Malaysia. We selected 60 adult patients with confirmed dengue virus infection and investigated their responses to dengue viral antigens. The patients' mean age was 30 years (range, 13 to 58 years), and they were selected from multiple racial backgrounds: Malay, Chinese, and Indian. The mean duration of illness was 5 days (range 4 to 9 days). Serology of plasma samples was carried out to determine the presence of IgM and IgG antibodies using standard assays. For the period of hospital admission, the average maximum hematocrit recorded was 48% (range, 42 to 54%) and the mean nadir of the platelet count was 35,129 × 106/ml (range, 5 × 106 to 82 × 106/ml). The characteristics of the patients with a positive SFC result in response to at least one peptide pool are shown in Table 3.
TABLE 3.
Dengue IgM assay, hemagglutination inhibition assay, and reverse transcription-PCR results for all subjects selected for peptide response study in the ELISPOT IFN-γ assay
| Patienta | Age (yr) | Sexb | Racec | Dengue type (grade)d | Day of onset | Result for IgMe | HIf | PCR resulte |
|---|---|---|---|---|---|---|---|---|
| Den0342* | 35 | F | M | DF | 6 | Pos | >10,240 | Neg |
| Den0427 | 20 | M | M | DF | 6 | Pos | 5,120 | Neg |
| Den0421* | 22 | M | M | DF | 6 | Pos | >10,240 | Neg |
| Den0392* | 23 | F | M | DF | 6 | Pos | >10,240 | Neg |
| Den0309 | 55 | M | C | DF | 7 | Pos | >10,240 | Neg |
| Den0301 | 25 | M | M | DF | 7 | Pos | >10,240 | Neg |
| Den0350 | 19 | M | M | DF | 7 | Pos | 640 | Neg |
| Den0360* | 25 | M | M | DF | 7 | Pos | >10,240 | Pos (DV1) |
| Den0228 | 55 | F | C | DF | 8 | Pos | >10,240 | Neg |
| Den0243* | 23 | M | C | DF | 8 | Pos | >10,240 | Neg |
| Den0277 | 16 | M | M | DF | 8 | Pos | 1280 | Neg |
| Den0347* | 37 | M | I | DF | 9 | Pos | >10,240 | Neg |
| Den0437 | 38 | M | M | DHF | 5 | Pos | 1280 | Neg |
| Den0235 | 38 | M | I | DHF | 5 | Pos | >10,240 | Neg |
| Den0381* | 40 | M | M | DHF | 5 | Pos | >10,240 | Pos (DV1) |
| Den0454 | 16 | M | I | DSS | 5 | Pos | 640 | Pos (DV3) |
| Den0416* | 17 | M | M | DHF | 6 | Pos | >10,240 | Pos (DV1) |
| Den0351* | 14 | M | I | DHF | 6 | Pos | 1280 | Pos (DV1) |
| Den0429 | 16 | M | M | DHF | 6 | Pos | >10,240 | Neg |
| Den0446 | 16 | F | I | DHF | 6 | Pos | >10,240 | Pos (DV1) |
| Den0367 | 20 | M | M | DHF | 6 | Pos | >10,240 | Neg |
| Den0426 | 25 | F | M | DSS | 6 | Pos | >10,240 | Neg |
| Den0344* | 19 | M | M | DHF | 7 | Pos | >10,240 | Pos (DV1) |
| Den0239 | 21 | M | M | DHF | 7 | Pos | 160 | Neg |
| Den0355* | 41 | M | M | DHF | 8 | Pos | >10,240 | Neg |
| Den0422 | 13 | M | I | DHF | 8 | Pos | >10,240 | Neg |
| Den0357 | 36 | M | I | DHF | 8 | Pos | >10,240 | Pos (DV1) |
| Den0356 | 24 | F | C | DHF | 8 | Pos | >10,240 | Neg |
| Den0380* | 20 | M | I | DHF | 8 | Pos | >10,240 | Pos (DV1) |
| Den0466 | 33 | F | C | DHF | 8 | Pos | >10,240 | Neg |
| Den0438* | 44 | M | C | DHF | 8 | Pos | >10,240 | Pos (DV1) |
| Den0386 | 14 | F | C | DHF | 9 | Pos | 1280 | Neg |
*, patient selected to study T-cell responses to individual peptides.
F, female; M, male.
C, Chinese; I, Indian; M, Malay.
DSS, dengue shock syndrome.
Pos, positive; Neg, negative.
HI, hemagglutination inhibition. Values are reciprocal antibody titers.
T-cell responses to peptide pools.
IFN-γ ELISPOT assays were performed to investigate T-cell responses of DF and DHF patients to multiple dengue viral antigens, derived from the structural (AnC, C, PrM, M, and E) and nonstructural (NS1, NS2A, NS2B, NS3, NS4A, NS4B, and NS5) regions of dengue viruses. Responses were considered positive when a minimum of 50 SFCs per 106 PBMC were present per well, representing at least twice the number of SFCs in negative control wells. This cutoff was determined based on the results of samples from healthy donors tested against all the peptide pools, which developed 0 to 45 SFC/106 PBMC (data not shown).
Overall, 12 DF and 20 DHF patients responded to at least to one peptide pool in the IFN-γ ELISPOT assay with values exceeding 50 SFC/106 PBMC (Table 4). All the DHF patients were documented to have clinically significant vascular permeability and/or plasma leakage. The leakage was manifested by a ≥20% increase in hematocrit values in eight DHF patients, and pleural effusion and/or ascites was detected by clinical examination in 12 DHF patients. Six patients with DF and 7 patients with DHF responded to all the peptide pools with >50 SFC/106 PBMC. Also, samples from only six (18.75%) patients (three with DF and three with DHF) responded to only one peptide pool. In addition, the magnitude of ELISPOT responses was higher in pools that contained peptides derived from nonstructural region of the dengue virus. More patients (10 DF and 15 DHF patients) recognized peptides from the NS3 region in peptide pool D than peptides from other peptide pools.
TABLE 4.
IFN-γ responses to peptide pools A to F
| Dengue type and patient | No. of SFCs per 106 PBMC
|
|||||
|---|---|---|---|---|---|---|
| Pool A | Pool B | Pool C | Pool D | Pool E | Pool F | |
| DF | ||||||
| Den0342 | 170 | 85 | 165 | 170 | 380 | 160 |
| Den0427 | 240 | 145 | 205 | 170 | 180 | 235 |
| Den0421 | 105 | 135 | 105 | 150 | 160 | 100 |
| Den0392 | 45 | 0 | 110 | 170 | 0 | 20 |
| Den0309 | 10 | 5 | 35 | 15 | 60 | 10 |
| Den0301 | 65 | 20 | 0 | 75 | 45 | 205 |
| Den0350 | 10 | 10 | 35 | 50 | 20 | 30 |
| Den0360 | 165 | 265 | 145 | 225 | 210 | 185 |
| Den0228 | 55 | 20 | 0 | 25 | 0 | 0 |
| Den0243 | 260 | 245 | 145 | 430 | 75 | 135 |
| Den0277 | 45 | 30 | 25 | 50 | 40 | 60 |
| Den0347 | 130 | 190 | 230 | 435 | 210 | 100 |
| Range | 55-260 | 85-265 | 105-230 | 50-435 | 60-380 | 60-235 |
| Mean ± SEM | 148.75 ± 26.55 | 177.5 ± 28.16 | 157.86 ± 17.52 | 192.5 ± 43.97 | 182.14 ± 40.12 | 147.5 ± 21.07 |
| DHF | ||||||
| Den0437 | 65 | 5 | 0 | 35 | 15 | 50 |
| Den0235 | 55 | 120 | 230 | 70 | 85 | 130 |
| Den0381 | 120 | 145 | 40 | 565 | 100 | 160 |
| Den0454 | 15 | 20 | 10 | 45 | 80 | 40 |
| Den0416 | 0 | 0 | 0 | 120 | 155 | 280 |
| Den0351 | 255 | 220 | 220 | 215 | 230 | 100 |
| Den0429 | 80 | 55 | 5 | 0 | 0 | 20 |
| Den0446 | 85 | 140 | 150 | 140 | 155 | 110 |
| Den0367 | 64 | 97 | 105 | 92 | 102 | 51 |
| Den0426 | 0 | 0 | 10 | 75 | 40 | 0 |
| Den0344 | 170 | 135 | 150 | 235 | 150 | 100 |
| Den0239 | 0 | 0 | 10 | 350 | 75 | 0 |
| Den0355 | 0 | 50 | 0 | 75 | 0 | 0 |
| Den0422 | 55 | 20 | 25 | 10 | 20 | 10 |
| Den0357 | 75 | 80 | 35 | 25 | 110 | 45 |
| Den0356 | 85 | 130 | 150 | 155 | 150 | 60 |
| Den0380 | 150 | 150 | 225 | 780 | 165 | 130 |
| Den0466 | 100 | 45 | 45 | 85 | 50 | 0 |
| Den0438 | 10 | 0 | 5 | 425 | 300 | 75 |
| Den0386 | 50 | 45 | 65 | 55 | 30 | 45 |
| Range | 50-255 | 50-220 | 65-230 | 55-780 | 50-300 | 50-280 |
| Mean ± SEM | 100.64 ± 15.25 | 120.18 ± 14.62 | 161.88 ± 21.15 | 229.13 ± 55.10 | 136.21 ± 17.80 | 113.27 ± 19.79 |
Bold type indicates positive responses.
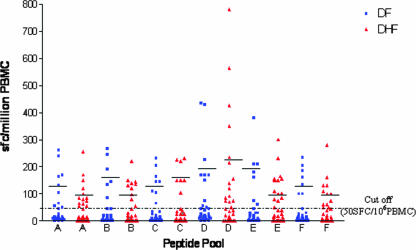
Spot numbers (>100 SFC/106) were higher for pools D and F in DHF patients, but the difference was not significant, as depicted in Fig. 1.
FIG. 1.
Levels of SFC in DF and DHF patients in response to peptide pools containing peptides from structural (AnC, C, PrM, and E) and nonstructural (NS1, NS2A, NS2B, NS3, NS4A, NS4B, and NS5) regions. Means after subtraction of control values (no peptides) are shown. The mean for each peptide pool is indicated by a bar. Responses were considered significant when a minimum of 50 SFCs per 106 PBMC were present per well, representing at least twice the number of SFCs in negative control wells.
T-cell responses to individual peptides.
The frequency of individual peptides from peptide pools was further analyzed in 13 patients from DF and DHF cases, who responded strongly to the peptide pools. Eight of the 13 selected patients had an IFN-γ response to one or more individual peptide from the C, NS2B, NS3, NS4A, NS4B, and NS5 regions. The highest values were obtained with peptides from the NS3 region in peptide pool D, with a range of 68 to 763 SFC/106 PBMC (Table 5). In this region, the highest responses were obtained with peptide NS3422-431 (RVIDPRRCMK); three patients (Den0380, Den0381, and Den0438) responded with 763, 755, and 726 SFC/106 PBMC (Table 5). Another peptide that evoked a strong response was NS4B119-128 (AIIGPGLQAK) in pool E, with 703 SFC/106 PBMC (Table 5). Also, patient Den0438 responded to 14 different types of peptides derived from the nonstructural region (NS2B, NS3, NS4A, and NS4B) in pools D and E, with values ranging from 356 to 726 SFC/106 PBMC (Table 5). Four (23.53%) of the 17 peptides recognized by PBMC contain sequences that were characterized previously as dengue virus T-cell epitopes (Table 6) (3, 36, 49).
TABLE 5.
T-cell responses of DF and DHF patients to individual peptides
| Region | Sequence | Patient | No. of SFC/106 PBMC | HLA specificity | Reference |
|---|---|---|---|---|---|
| CP22-31 | RVSTVQQLTK | Den0243 | 103 | HLA-A*03/11 | 49 |
| E478-487 | SLVLVGVVTL | Den0355 | 57 | HLA-A*0201/24 | |
| NS2B52-60 | ELERAADVK | Den0438 | 375 | HLA-A*03/11 | |
| NS2B97-106 | ILIRTGLLVI | Den0342 | 148 | HLA-A*0201/*24 | |
| Den0344 | 168 | ||||
| Den0351 | 235 | ||||
| Den0360 | 350 | ||||
| Den0438 | 379 | ||||
| NS325-32 | RIKQKGIL | Den0342 | 240 | HLA-B*08 | |
| Den0360 | 235 | ||||
| Den0438 | 425 | ||||
| NS364-74 | RIEPSWADVK | Den0243 | 68 | HLA-A*03/11 | 49 |
| Den0347 | 203 | ||||
| Den0438 | 394 | ||||
| Den0360 | 213 | ||||
| NS3112-120 | AIKRGLRTL | Den0347 | 170 | HLA-A*0201/24 | |
| Den0360 | 173 | ||||
| Den0438 | 398 | ||||
| NS3142-150 | DKKGKVVGL | Den0421 | 215 | HLA-B*08 | 36 |
| Den0438 | 413 | ||||
| NS3422-431 | RVIDPRRCMK | Den0342 | 180 | HLA-A*03/*11 | |
| Den0380 | 763 | ||||
| Den0381 | 755 | ||||
| Den0344 | 83 | ||||
| Den0438 | 726 | ||||
| Den0392 | 131 | ||||
| Den0360 | 138 | ||||
| NS4A44-52 | ALSELPETL | Den0421 | 305 | HLA-A*0201/24 | |
| Den0347 | 90 | ||||
| Den0438 | 406 | ||||
| NS4A55-64 | LLLLTLLATV | Den0347 | 115 | HLA-A*0201/24 | |
| Den0438 | 344 | ||||
| NS4A56-64 | LLLTLLATV | Den0347 | 125 | HLA-A*0201/24 | 3 |
| Den0438 | 386 | ||||
| NS4B6-13 | LEKTKKDL | Den0438 | 356 | HLA-B*08 | |
| NS4B119-128 | AIIGPGLQAK | Den0421 | 205 | HLA-A*03/11 | |
| Den0347 | 70 | ||||
| Den0438 | 703 | ||||
| NS5182-190 | VLNPYMPSV | Den0438 | 194 | HLA-A*0201/24 | |
| NS5375-383 | KITAEWLWK | Den0438 | 68 | HLA-A*03/11 | |
| NS5563-571 | KLAEAIFKL | Den0416 | 722 | HLA-A*0201/24 |
TABLE 6.
Percentage of patients with responss to individual peptides from the NS3 region
| Region | Sequence | % of patients with response |
|---|---|---|
| NS325-32 | RIKQKGIL | 23.07 |
| NS364-74 | RIEPSWADVK | 30.76 |
| NS3112-120 | AIKRGLRTL | 23.07 |
| NS3142-150 | DKKGKVVGL | 15.38 |
| NS3422-431 | RVIDPRRCMK | 53.85 |
Frequency of T-cell responses to individual peptides from the NS3 region.
The IFN-γ ELISPOT assay revealed that the highest responses were to peptides derived from the NS3 region. The peptide NS3422-431 (RVIDPRRCMK) was among the most frequently recognized, with 53.85% of subjects responding to this peptide. Lower responses were noticed for the other four peptides in this region, with 30.76% of subjects responding to peptide NS364-74 (RIEPSWADVK), 23.07% to peptides NS3112-120 (AIKRGLRTL) and NS325-32 (RIKQKGIL), and 15.38% to peptide NS3142-150 (DKKGKVVGL) (Table 6). The region of NS3 with the strongest T-cell responses was at position 422 to 431. The responses ranged from 726 to 763 SFC/106 PBMC, which is almost sevenfold higher than responses to this peptide in other subjects (Table 5). A dengue virus serotype-specific PCR performed on plasma samples collected from these subjects revealed that they were infected with DV1 and were presumptively diagnosed as having secondary infections based on the high IgG titers. All three subjects triggered high T-cell response to peptides designed based on DV2. Therefore, the extent of sequence variation at the NS3 region between DV1 and DV2 was examined. We found that NS3422-431 (RVIDPRRCMK) in DV2 and NS3 (RVIDPRRCLK) in DV1 differed by only one amino acid (Table 7) . Three other peptides, NS325-32, NS364-74, and NS3112-120, also differed by only one amino acid between DV2 and DV1 (Table 7).
TABLE 7.
Amino acid variations in peptides derived from DV1 and DV2
| Peptide | Virus | Sequence |
|---|---|---|
| NS3422-431 | DV2 | RVIDPRRCMK |
| DV1 | RVIDPRRCLK | |
| NS364-74 | DV2 | RIEPSWADVK |
| DV1 | RLEPSWADVK | |
| NS3112-120 | DV2 | AIKRGLRTL |
| DV1 | AIKRKLRTL | |
| NS325-32 | DV2 | RIKQKGIL |
| DV1 | RILQRGLL |
DISCUSSION
Cytotoxic T lymphocytes (CTL) play an important role in the elimination of dengue virus-infected cells (28). Identification of antigenic peptides recognized by dengue virus-specific CTL may suggest new ways to suppress viral replication and prevent persistent infection. Multiple peptides from the conserved regions of the dengue virus would probably be essential in the development of a universally immunogenic vaccine (43). Several studies on dengue virus-specific T cells have been conducted in the context of T-cell clones generated against live attenuated dengue virus vaccines and, less frequently, from dengue patients (15, 26, 27). Over the last few years, studies of dengue patients have successfully identified several peptides that are specific for CD8+ and CD4+ CTL. In this study, T-cell epitopes restricted to HLA*02, HLA-A*11, and HLA*24, which are commonly expressed by the Malaysian population, were investigated (51).
In this study, the number of SFCs per 106 PBMC was higher in DHF patients than in DF patients. This is probably due to the higher frequency of activated CD8+ T cells circulating in patients with DHF than in those with DF (16, 41, 57). Others have reported that the circulating frequency of dengue virus peptide-specific CD8 T cells in DHF patients was 1:3,900 to 1:34,500 (34). This is comparable to frequencies seen in other acute viral diseases, such as influenza (1:6,000 to 1:111,000) (30), but lower than in chronic viral infection, such as that due to Epstein-Barr virus (1:100 to 1:2,500) (5). Seven patients were able to respond to more than one peptide from the same or different dengue viral regions. These results also are in agreement with those from other groups, showing that a single peptide can be recognized by more than one T-cell receptor (10, 25).
Peptides from the NS3 region were recognized by samples from almost half of the patients and also contained the largest number of antigenic peptides to which T cells preferentially appear to respond. This result is consistent with previous studies that have emphasized the importance of NS3 as a T-cell target (37). This is probably because the NS3 protein is highly conserved across all four dengue virus serotypes and contains motifs and charged residues that are essential for helicase activity and viral replication (39, 40). In the present study, patients with presumptive secondary dengue viral infection had high T-cell responses to the peptides studied. Hence, the antigenic peptides identified in this study are some of the targets of reactivated dengue virus-cross-reactive T cells that were possibly primed by a previous dengue virus infection (49).
Plasma leakage is a key feature often noticed in DHF. Reports have shown that the activation of T cells contributes to severe plasma leakage (14, 47). Virus entry into monocytes and macrophages results in the presentation of viral peptides on the cell surface, in association with HLA molecules. Interaction of these antigen-presenting cells with memory T cells induces proliferation and the production of proinflammatory cytokines such as IFN-γ and tumor necrosis factor alpha. These cytokines can directly affect vascular endothelial cells, resulting in plasma leakage (42, 47). The rapid induction of cross-reactive memory T cells (mainly NS3 specific) during secondary infection would be consistent with the increased incidence of DHF.
This consequently predicts that DHF patients would have higher levels of T-cell activation and clones of cross-reactive T cells that would be preferentially expanded.
In this study, several patients also responded to NS2B, NS4A, and NS4B. These proteins exhibit conserved hydrophobicity profiles among flaviviruses, suggesting that they are membrane-associated proteins (8). The NS2B region of 130 to 132 amino acids is involved in the protease function of the NS2B-NS3 complex. A 40-amino-acid hydrophilic domain of NS2B is essential for protease activity (13). NS4A and NS4B may be involved in membrane localization of NS3 and NS5 replication complexes via protein-protein interaction, since the NS3-NS5 complex is weakly associated with the membrane in spite of its hydrophilic characteristics (7, 53). Responsiveness to the NS5 protein, which is the most conserved flavivirus protein, was also seen. The RNA-dependent RNA polymerase activity has been suggested to reside in this domain, which is also important for viral replication (2).
Only one patient's samples responded to the structural regions of the dengue virus. The capsid protein contains a C-terminal hydrophobic domain and is present in infected cells. This hydrophobic domain may serve to localize assembly of nucleocapsid at the membrane site and function as a signal sequence for prM (4).The E protein is thought to contain peptides that predominantly induce virus-neutralizing antibody responses. This protein is responsible for virus attachment to susceptible cells, which results in virus growth. This antigen also mediates virus-specific membrane fusion, which presumably allows the newly infecting virus to escape the endocytic vesicle and initiate its intracellular replication cycle (18).
One of the limitations of this study was that only 32 candidate peptides were included, and most of these peptides with high binding specificities were derived from DV2 sequence. Although most of the PCR-positive cases represented DV1 infection, the DHF patients infected with DV1 mounted ELISPOT responses to dengue virus-specific peptides from DV2. This can be explained as a response to previous infection, as all patients in this study were experiencing a secondary infection. Thus, a previously mounted immune response against the virus is being boosted, a phenomenon referred to as “original antigenic sin” in the antibody response. The early cross-reactive T-cell response remains dominant in late convalescence, when the memory T-cell pool has been established. This result is in agreement with a study done with Thai schoolchildren where dominance of cross-reactive T cells was observed among dengue-virus specific memory T cells 12 months after secondary dengue virus infection (38). It is thought that peptides of nonstructural proteins recognized by both serotype-specific and serotype cross-reactive CD8+ CTL will have important implications for the design of effective subunit vaccines (33).
Besides immune status, several other risk factors have been proposed for development of DHF. These include infecting dengue virus, age, sex, malnutrition, and genetic background of the host (32). Human genetic background factors in DHF have not been extensively studied. Few studies suggest HLA association with dengue disease severity (45, 52). In this study we noticed that several peptides specific to HLA-A*02 and HLA-A*24 evoked very high IFN-γ responses. These results are in agreement with a study of DHF patients in Vietnam which found that polymorphisms at the HLA class 1 loci were significantly associated with DHF disease susceptibility. Others have reported that children with HLA-A*24 are more likely to develop DHF (49), while in adults HLA-A*0207 was associated with DHF in patients having secondary DV1 or DV2 infections only. HLA-A*0203 seems to be associated with less severe dengue, regardless of the secondary infecting virus serotype (50). We are currently conducting subtyping of the HLA*02 region to determine if the responses are also polymorphic.
In conclusion, two peptides, NS3422-431 and NS2B 97-106, were frequently recognized by T cells from dengue patients by an IFN-γ-based ELISPOT assay using synthetic dengue peptides with HLA-A*02/*24 and HLA-A*03/*11 binding restrictions. Some epitopes had been reported previously, and novel epitopes were also noted. These findings indicate a need to identify as many dengue-specific T-cell peptides as possible in a larger number of dengue patients with multiple HLA backgrounds to better understand how the immune system responds to dengue virus and contributes to pathogenesis.
Acknowledgments
We thank the clinicians and the nurses at the University Malaya Medical Center (UMMC), Kuala Lumpur, Malaysia, who assisted in recruiting dengue patients and collecting samples, Cynthia Gladys Lopez, at the blood bank of UMMC, for collection of control blood samples, and the patients and their families for their participation in this study.
This study was funded by the Academy of Science Malaysia, grant SAGA 66-02-03-0041.
We have no financial conflict of interest.
Footnotes
Published ahead of print on 13 June 2007.
REFERENCES
- 1.Aichele, P., K. Brduscha-Riem, S. Oehen, B. Odermatt, R. M. Zinkernagel, H. Hengartner, and H. Pircher. 1997. Peptide antigen treatment of naïve and virus-immune mice: antigen-specific tolerance versus immunopathology. Immunity 6:519-529. [DOI] [PubMed] [Google Scholar]
- 2.Bartholomeusz, A. I., and P. J. Wright. 1993. Synthesis of dengue virus RNA in vitro: initiation and the involvement of proteins NS3 and NS5. Arch. Virol. 128:111-121. [DOI] [PubMed] [Google Scholar]
- 3.Bashyam, H. S., S. Green, and A. L. Rothman. 2006. Dengue virus-reactive CD8+ T cells display quantitative and qualitative differences in their response to variant epitopes of heterologous viral serotype. J. Immunol. 176:2817-2824. [DOI] [PubMed] [Google Scholar]
- 4.Bulich, R., and J. G. Aaskov. 1992. Nuclear localization of dengue 2 virus core protein detected with monoclonal antibodies. J. Gen. Virol. 73:2999-3003. [DOI] [PubMed] [Google Scholar]
- 5.Callan, M. F. C., L. Tan, A. Annels, et al. 1998. Direct visualisation of antigenic specific CD8+ T cells during the primary immune response to Epstein-Barr virus in vivo. J. Exp. Med. 187:1395-1402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cannon, M. J., P. J. Openshaw, and B. A. Askons. 1988. Cytotoxic T cells clear virus but augment lung pathology in mice infected with respiratory syncytial virus. J. Exp. Med. 168:1163-1168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chambers, T. J., C. Hahn, R. Galler, and C. M. Rice. 1990. Flavivirus genome organization, expression, and replication. Annu. Rev. Microbiol. 44:649-688. [DOI] [PubMed] [Google Scholar]
- 8.Chambers, T. J., R. Weir, A. Grakoui, D. W. McCourt, J. F. Bazan, R. J. Fletterick, and C. M. Rice. 1990. Evidence that the N-terminal domain of nonstructural protein NS3 from yellow fever virus is a serine protease responsible for site specific cleavage in the viral polyprotein. Proc. Natl. Acad. Sci. USA 87:8898-8902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chaturvedi, U. C., R. Shrivastava, and R. Nagar. 2005. Dengue vaccines: problems and prospects. Indian J. Med. Res. 121:639-652. [PubMed] [Google Scholar]
- 10.Chien, Y. H., and M. M. Davis. 1993. How αβ T cell receptors see peptide/MHC complexes. Immunol. Today 14:597. [DOI] [PubMed] [Google Scholar]
- 11.Clarke, D. H., and J. Cassals. 1958. Techniques for haemagglutination and haemagglutination-inhibition with arthropod-borne viruses. Am. J. Trop. Med. Hyg. 7:561. [DOI] [PubMed] [Google Scholar]
- 12.Endy, T. P., S. Chunsuttiwat, A. Nisalak, D. H. Libraty, S. Green, A. L. Rothman, D. W. Vaughn, and F. A. Ennis. 2002. Epidemiology of inapparent and symptomatic acute dengue virus infection: a prospective study of primary school children in Kamphaeng Phet, Thailand. Am. J. Epidemiol. 156:40-51. [DOI] [PubMed] [Google Scholar]
- 13.Falgout, B., R. H. Miller, and C. J. Lai. 1993. Deletion analysis of dengue virus type 4 nonstructural protein NS2B: identification of a domain required for NS2B-NS3 protease activity. J. Virol. 67:2034-2042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Green, S., D. W. Vaughn, S. Kalayanarooj, S. Nimmannitya, S. Suntayakorn, A. Nisalak, R. Lew, B. L. Innis, I. Kurane, A. L. Rothman, and F. A. Ennis. 1999. Early immune activation in acute dengue illness is related to development of plasma leakage and disease severity. J. Infect. Dis. 179:755-762. [DOI] [PubMed] [Google Scholar]
- 15.Green, S., I. Kurane, R. Edelman, C. O. Tacket, K. H. Eckels, D. W. Vaughn, C. H. Hoke, Jr., and F. A. Ennis. 1993. Dengue virus-specific human CD4+ T-lymphocyte responses in a recipient of an experimental live-attenuated dengue virus type 1 vaccine: bulk culture proliferation, clonal analysis, and precursor frequency determination. J. Virol. 67:5962-5967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Green, S., S. Pichyangkul, D. W. Vaughn, S. Kalayanarooj, S. Nimmannitya, A. Nisalak, I. Kurane, A. L. Rothman, and F. A. Ennis. 1999. Early CD69 expression on peripheral blood lymphocytes from children with dengue hemorrhagic fever. J. Infect. Dis. 180:1429-1435. [DOI] [PubMed] [Google Scholar]
- 17.Gubler, D. J. 1998. Dengue and dengue hemorrhagic fever. Clin. Microbiol. Rev. 11:480-496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gubler, D. J., and G. Kuno. 1999. Dengue and dengue hemorrhagic fever. CABI Publishing, Cambridge, MA.
- 19.Halstead, S. B. 1979. In vivo enhancement of dengue virus infection in rhesus monkeys by passively transferred antibody. J. Infect. Dis. 140:527-533. [DOI] [PubMed] [Google Scholar]
- 20.Halstead, S. B. 1999. Is there an inapparent dengue explosion? Lancet 353:1100-1101. [DOI] [PubMed] [Google Scholar]
- 21.Halstead, S. B., and J. Deen. 2002. The future of dengue vaccines. Lancet 360:1243-1245. [DOI] [PubMed] [Google Scholar]
- 22.Harris, R., and E. V. Ukayiofo. 1969. Rapid preparation for lymphocytes for tissue typing. Lancet 327:7615. [DOI] [PubMed] [Google Scholar]
- 23.Henchal, E. A., and R. Putnak. 1990. The dengue viruses. Clin. Microbiol. Rev. 3:376-396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Huang, Y. H., et al. 1997. Antibodies against dengue virus E protein peptide bind to human plasminogen and inhibit plasmin activity. J. Clin. Exp. Immunol. 110:35-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kalams, S. A., A. K. Johnson, A. K. Trocha, M. J. Dynan, H. S. Ngo, R. T. D'Aguila, J. T. Kurnick, and B. D. Walker. 1994. Longitudinal analysis of T cell receptor (TCR) gene usage by human immunodeficiency virus 1 envelope-specific cytotoxic T lymphocyte clones reveals a limited TCR repertoire. J. Exp. Med. 179:1261-1271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kurane, I., A. Meager, and F. A. Ennis. 1989. Dengue virus-specific human T cell clones: serotype crossreactive proliferation, interferon γ production, and cytotoxic activity. J. Exp. Med. 170:763-775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kurane, I., M. A. Brinton, A. L. Samson, and F. A. Ennis. 1991. Dengue virus-specific, human CD4+ CD8− cytotoxic T-cell clones: multiple patterns of virus cross-reactivity recognized by NS3-specific T-cell clones. J. Virol. 65:1823-1828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kurane, I., and F. A. Ennis. 1994. Cytotoxic T lymphocyte in dengue virus infection. Curr. Top. Microbiol. Immunol. 189:93-108. [DOI] [PubMed] [Google Scholar]
- 29.Kurane, I., and T. Takasaki. 2001. Dengue fever and dengue hemorrhagic fever: challenges of controlling an enemy still at large. Rev. Med. Virol. 11:301-311. [DOI] [PubMed] [Google Scholar]
- 30.Lalvani, A., R. Brookes, S. Hambleton, W. J. Britton, A. V. S. Hill, and A. J. McMichael. 1997. Rapid effector function in CD8+ memory T cells. J. Exp. Med. 186:859-865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lam, S. K., S. Devi, and T. Pang. 1987. Detection of specific IgM in dengue infections. Southeast. Asian J. Trop. Med. Public Health 18:532-538. [PubMed] [Google Scholar]
- 32.Leitmeyer, K. C., D. W. Vaughn, D. M. Watts, R. Salas, I. C. de Villalobos, C. Ramos, and R. Rico-Hesse. 1999. Dengue virus structural differences that correlate with pathogenesis. J. Virol. 73:4738-4747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Livingston, P. G., I. Kurane, L. C. Dai, Y. Okamato, C. J. Lai, R. Men, S. Karaki, M. Takiguchi, and F. A. Ennis. 1995. Dengue virus-specific, HLA-B35-restricted, human CD8+ cytotoxic T lymphocyte (CTL) clones: recognition of NS3 amino acids 500 to 508 by CTL clones of two different serotype specificities. J. Immunol. 154:1287-1295. [PubMed] [Google Scholar]
- 34.Loke, H., D. B. Bethell, C. X. T. Phuong, M. Dung, J. Schneider, N. J. White, N. P. Day, J. Farrar, and A. V. S. Hill. 2001. Strong HLA class I-restricted T cell responses in dengue hemorrhagic fever: a double-edged sword? J. Infect. Dis. 184:1369-1373. [DOI] [PubMed] [Google Scholar]
- 35.Malavige, G. N., S. Fernando, D. J. Fernando, and S. L. Seneviratne. 2004. Dengue viral infections. Postgrad. Med. J. 80:588-601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mangada, M. M., and A. L. Rothman. 2005. Altered cytokine responses of dengue specific CD4+ T cells to heterologous serotypes. J. Immunol. 175:2676-2683. [DOI] [PubMed] [Google Scholar]
- 37.Mathew, A., I. Kurane, A. L. Rothman, L. L. Zeng, M. A. Brinton, and F. A. Ennis. 1996. Dominant recognition by human CD8+ cytotoxic T-lymphocyte of dengue virus nonstructural proteins NS3 and NS1.2a. J. Clin. Investig. 98:1684-1691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mathew, A., I. Kurane, S. Green, H. A. Stephens, D. W. Vaughn, S. Kalayanarooj, S. Suntayakorn, D. Chandanayingyong, F. A. Ennis, and A. L. Rothman. 1998. Predominance of HLA-restricted cytotoxic T-lymphocyte responses to serotype-cross-reactive epitopes on nonstructural proteins following natural secondary dengue virus infection. J. Virol. 72:3999-4004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Matusan, A. E., M. J. Pryor, M. J., Davidson, A. D., and P. J. Wright. 2001. Mutagenesis of the dengue virus type 2 NS3 protein within and outside helicase motifs: effects on enzyme activity and virus replication. J. Virol. 75:9633-9643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Matusan, A. E., P. G. Kelley, M. J. Pryor, J. C. Whisstock, A. D. Davidson, and P. J. Wright. 2001. Mutagenesis of the dengue virus type 2 NS3 proteinase and the production of growth-restricted virus. J. Gen. Virol. 82:1647-1656. [DOI] [PubMed] [Google Scholar]
- 41.Mongkolsapaya, J., W. Dejnirattisai, X. N. Xu, S. Vasanawathana, N. Tangthawornchaikul, A. Chairunsri, S. Sawasdivorn, T. Duangchinda, T. Dong, S. Rowland-Jones, P. T. Yenchitsomanus, A. McMichael, P. Malasit, and G. Screaton. 2003. Original antigenic sin and apoptosis in the pathogenesis of dengue hemorrhagic fever. Nat. Med. 9:921-927. [DOI] [PubMed] [Google Scholar]
- 42.Mongkolsapaya, J., T. Duangchinda, W. Dejnirattisai, S. Vasanawathana, P. Avirutnan, A. Jairungsri, N. Khemnu, N. Tangthawornchaikul, P. Chotiyarnwong, K. Sae-Jang., M. Koch, Y. Jones, A. McMichael, X. Xu, P. Malasit, and G. Screaton. 2006. T cell responses in dengue hemorrhagic fever: are cross-reactive T cells suboptimal? J. Immunol. 176:3821-3829. [DOI] [PubMed] [Google Scholar]
- 43.Pervikov, Y. 2000. Development of dengue vaccine. Dengue Bull. 24:71-76. [Google Scholar]
- 44.Pinheiro, F. P., and S. J. Corber. 1997. Global situation of dengue and dengue hemorrhagic fever and its emergence in the Americas. World Health Stat. Q. 50:161-168. [PubMed] [Google Scholar]
- 45.Polizel, J. B., D. Bueno, J. E. L. Visentainer, A. M. Sell, S. D. Borelli, L. T. Tsuneto, M. M. O. Dalalio, M. T. M. Coimbra, and R. A. Moliterno. 2004. Association of human leukocyte antigen DQ1 and dengue fever in a white southern Brazilian population. Mem. Inst. Oswaldo Cruz Rio de Janeiro 99:559-562. [DOI] [PubMed] [Google Scholar]
- 46.Rothman, A. L. 2004. Dengue: defining protective versus pathologic immunity. J. Clin. Investig. 113:946-951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rothman, A. L., and F. A. Ennis. 1999. Immunopathogenesis of dengue haemorrhagic fever. Virology 257:1-6. [DOI] [PubMed] [Google Scholar]
- 48.Sekhar, K. C., and O. I. Huat. 1992. Epidemiology of dengue/dengue hemorrhagic fever in Malaysia—a retrospective epidemiological study 1973-1987. I. Dengue hemorrhagic fever. Asia. Pac. J. Public Health 6:15-25. [DOI] [PubMed] [Google Scholar]
- 49.Simmons, C. P., T. Dong, N. V. Chau, N. T. P. Dung, T. N. B. Chau, L. T. T. Thao, N. T. Dung, T. T. Hien, S. R. Jones, and J. Farrar. 2005. Early T-cell responses to dengue virus epitopes in Vietnamese adults with secondary dengue virus infections. J. Virol. 79:5665-5675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Stephens, H. A. F., R. Klaythong, M. Sirikong, D. W. Vaughn, S. Green, S. Kalayanarooj, T. P. Endy, D. H. Libraty, A. Nisalak, B. L. Innis, A. L. Rothman, F. A. Ennis, and D. Chandanayingyong. 2002. HLA-A and -B allele associations with secondary dengue virus infections correlate with disease severity and the infecting viral serotype in ethnic Thais. Tissue Antigens 60:309-318. [DOI] [PubMed] [Google Scholar]
- 51.Tee, Y. P., M. E. Phipps, and J. J. Bosco. 1999. Molecular HLA typing for bone marrow transplantation in Malaysia. Asia. Pac. J. Mol. Biol. Biotechnol. 7:29-37. [Google Scholar]
- 52.Wagenaar, J. F. P., A. T. A. Mairuhu, and E. C. M. van Gorp. 2004. Genetic influences on dengue virus infection. Dengue Bull. 28:126-134. [Google Scholar]
- 53.Wengler, G., T. Nowak, and E. Castle. 1990. Description of a procedure which allows isolation of viral nonstructural proteins from BHK vertebrate cells infected with the West Nile flavivirus in a state which allows their direct chemical characterization. Virology 177:795-801. [DOI] [PubMed] [Google Scholar]
- 54.World Health Organization. 2002. Dengue and dengue haemorrhagic fever. Fact sheet no. 117. World Health Organization, Geneva, Switzerland.
- 55.World Health Organization. 1999. Prevention and control of dengue and dengue hemorrhagic fever: comprehensive guidelines. WHO Regional Publication, SEARO, No. 29. World Health Organization, Geneva, Switzerland.
- 56.Yong, Y. K., H. T. Chong, C. T. Tan, and S. Devi. 2006. Rapid detection, serotyping and quantification of dengue viruses by TaqMan real-time one step RT-PCR. J. Virol. Methods 138:123-130. [DOI] [PubMed] [Google Scholar]
- 57.Zivna, I., S. Green, D. W. Vaughan, S. Kalayanarooj, H. A. Stephen, D. Chandanayingyong, A. Nisalak, A. Ennis, and A. L. Rothman. 2002. T Cell responses to an HLA-B*07-restricted epitope on a dengue NS3 protein correlate with disease severity. J. Immunol. 168:5959-5965. [DOI] [PubMed] [Google Scholar]