Abstract
A cytopathogenic virus (designated the Aichi/2004 strain) was isolated in a human rectal adenocarcinoma cell line (HRT-18) from the ileum contents of a calf with diarrhea. Oval and elongated particles, approximately 100 to 170 nm in diameter, with club-shaped projections were seen in the infected culture supernatant, and torovirus-like (tubular and torus nucleocapsid) structures were seen in the infected cells by electron microscopy. An antiserum against bovine torovirus (BToV) reacted with the infected cells by immunofluorescence and neutralized the isolate. However, antisera against bovine coronavirus (BCV) failed to react with the infected cells by immunofluorescence or did not neutralize the isolate. Further, the isolate was positive for BToV by reverse transcription-PCR (RT-PCR) targeting fragments of the nucleocapsid (N), membrane (M), and spike (S) genes. Comparison of the nucleotide sequences of the PCR products with those of the published N, M, and S genes (476 to 497, 672, and 687 to 690 nucleotides, respectively) of toroviruses showed high sequence identities (up to 99.4%, 98.7%, and 94.9% for the N, M, and S genes, respectively) between the isolate and BToVs. In contrast, the isolate was negative for BCV by RT-PCR. In a serological survey of serum samples from 355 calves at 33 farms, 92% of calves were positive for neutralizing antibodies to the isolate. These results indicate that the isolate in this study was BToV and that BToV infection might be common in cattle in Japan. To our knowledge, this is the first isolation of BToV in tissue culture.
Toroviruses are members of the family Coronaviridae and are enveloped, positive-stranded RNA viruses that cause enteric diseases in animals and humans (11, 12, 17). The prototype torovirus is equine torovirus strain Berne (BEV), which was isolated from a horse with diarrhea in Berne, Switzerland, in 1972. BEV could be propagated in embryonic mule skin cells and is the only cell culture-adapted torovirus (29). Although culture-adapted BEV has not been demonstrated to be pathogenic experimentally, most adult horses possess neutralizing antibodies to BEV. In addition, neutralizing antibodies to BEV have been found in sera from cattle, goats, sheep, pigs, rabbits, and cats (30). Antigenic cross-reactivity between human and animal toroviruses has been demonstrated, for example, by hemagglutination inhibition and immunoelectron microscopy (1, 6).
Bovine torovirus (BToV), formerly called Breda virus, was originally isolated from diarrheic calves in Breda, IA, in 1979. BToVs cause diarrhea both in experimentally infected gnotobiotic calves and under field conditions (5, 7-9, 12, 14, 19, 32, 33). They infect villous and crypt enterocytes of the mid-jejunum, ileum, colon, and cecum, inducing villous atrophy and necrosis of the crypts in calves (12, 19, 31, 32). Respiratory infections with BToVs have also been reported (7). To date, BToVs have not been propagated in bovine tracheal organ culture, cell culture (e.g., primary calf kidney cells, primary bovine thyroid cells, HRT-18 cells, and MDBK cells), or embryonated eggs (32); rather, the only way to propagate the virus is via inoculation of gnotobiotic or colostrum-deprived calves. Immunological reagents for detecting BToV antigens and antibodies are therefore available in only a few laboratories, and consequently, details of the antigenic relationship between toroviruses and the epidemiological features of BToV infection remain unknown.
Here we describe the isolation in HRT-18 cells of a cytopathogenic BToV from the ileum of a dead calf with diarrhea, and we clarify the seroprevalence of the isolate in calves in Japan.
MATERIALS AND METHODS
Clinical specimens.
Intestinal contents (from the ileum and rectum) were collected from a dead calf (3 months old) with sporadic diarrhea at a beef farm in Aichi Prefecture, Japan, in 2004. The intestinal contents were diluted 1:10 in 0.01 M phosphate-buffered saline (pH 7.4), clarified by low-speed centrifugation at 3,000 × g for 10 min, and used for virus isolation and reverse transcription-PCR (RT-PCR). These samples were also tested for group A rotavirus (GAR) with an antigen detection kit (Dipstic-Rota; Eiken Chemical, Tokyo, Japan), for Salmonella species by a standard technique, and for Coccidium species and Cryptosporidium species by a sucrose flotation method. Serum samples from 355 calves aged 1 to 12 months at 33 dairy farms in Japan were collected and used for a serological survey against the isolate.
Virus isolation.
Virus isolation was performed by using three cell lines: HRT-18, MDBK, and Vero cells. The HRT-18 cells were derived from a human rectal adenocarcinoma (24) kindly supplied by Hiroshi Kida, Hokkaido University. Confluent monolayers of these cells in 24-well plates were washed with Eagle minimal essential medium (EMEM) and inoculated with 0.1 ml of the intestinal-content suspensions. After adsorption for 60 min at 37°C, the cells were washed with EMEM and then received 0.5 ml of EMEM. The cells were incubated for 7 days at 37°C and examined for cytopathic effects (CPE). After incubation, the cells and supernatant were frozen and thawed once to harvest cell lysates, and subsequent passages were carried out in the same manner with 0.1 ml of cell lysates. After 2 passages, the isolates were cloned three times in HRT-18 cells by limiting dilution.
Physicochemical examination of the isolate.
The isolate was examined for stability to treatment with a lipid solvent (10% chloroform), acid (pH 3.0 at 22°C for 3 h), and heat (56°C for 30 min). The type of nucleic acid of the isolate was determined indirectly by examining the effect of 5-iodo-2′-deoxyuridine (IUdR) on viral replication as described previously (21). Bovine herpesvirus 1 was used as a control DNA virus. The isolate was also examined for size by filtration through membrane filters (pore sizes, 450, 200, 100, and 50 nm). The hemagglutination (HA) activity of the isolate was examined with 0.5% suspensions of mouse, chicken, and goose erythrocytes.
Immunofluorescence.
Confluent monolayers of HRT-18 cells in 6-well plates containing coverslips were inoculated with the isolate or bovine coronavirus (BCV) strain Mebus and incubated for 24 h at 37°C. Coverslips were fixed with acetone and incubated with a 1:100 dilution of gnotobiotic calf antiserum to BToV (kindly supplied by Linda Saif, The Ohio State University) (7) for 30 min at 37°C. After a wash in phosphate-buffered saline, coverslips were stained with fluorescein isothiocyanate (FITC)-conjugated anti-bovine immunoglobulin G (Kirkegaard & Perry Laboratories, Gaithersburg, MD). For examination of the BCV antigen, an anti-BCV polyclonal antiserum conjugated to FITC (Veterinary Medical Research & Development Inc., Pullman, WA) was applied at 37°C for 30 min.
Virus neutralization test.
Calf sera and antisera against BToV and BCV (26) were used for virus neutralization tests. Serial twofold dilutions of sera in 96-well tissue culture plates were incubated with an equal volume of a viral suspension containing 200 median tissue culture infective doses (TCID50)/0.05 ml of the isolate at 37°C for 1 h. An HRT-18 cell suspension containing 1.2 × 105 cells/ml was added with 0.1 ml of each serum-virus mixture, and the mixture was incubated at 37°C for 5 days. Virus-neutralizing antibody titers were expressed as the reciprocal of the highest dilution of serum that inhibited CPE completely.
RT-PCR.
Viral RNAs were extracted from the intestinal contents and infected cell supernatants using TRIzol LS reagent (Invitrogen Corp., Carlsbad, CA) according to the manufacturer's instructions. RT-PCR was carried out with an RNA PCR kit (AMV), version 3.0 (Takara Bio Inc., Shiga, Japan). Primer pairs targeting fragments of the nucleocapsid (N), membrane (M), and spike (S) genes of torovirus used for RT-PCR were those described by Smits et al. (22) and Hoet et al. (9). They were primers 1344 (5′-GAGAAAGAGCCAAGATGAATT-3′; positions 27761 to 27781) and 294 (5′-CTTACATGGAGACACTCAACCA-3′; positions 28403 to 28424) for the N genes, primers 1435 (5′-TCTTTGAAGATTGCCAAAA-3′; positions 25740 to 25758) and 1434 (5′-CATCTTCTAAAGATAAGTGG-3′; positions 26460 to 26479) for the M genes, and primers S5 (5′-GTGTTAAGTTTGTGCAAAAAT-3′; positions 20956 to 20976) and S3 (5′-TGCATGAACTCTATATGGTGT-3′; positions 21677 to 21697) for the S genes. These primer positions were based on the BToV strain Breda 1 genome (4). RT-PCR assays targeting the N gene of BCV described by Tsunemitsu et al. (25), the S gene of transmissible gastroenteritis virus (TGEV) described by Paton et al. (18), the M gene of porcine epidemic diarrhea virus (PEDV) described by Ishikawa et al. (10), and the 5′ noncoding region of bovine viral diarrhea virus described by Vilcek et al. (27) were carried out using the extracted RNAs.
Sequence analysis.
PCR products were sequenced directly by cycle sequencing with an automatic sequencer (ABI PRISM 310; Applied Biosystems, Tokyo, Japan). Sequence data were analyzed by the Clustal W method (23) using Lasergene software (DNASTAR, Madison, WI). Phylogenetic trees were constructed using the neighbor-joining method (20) and were drawn with the MEGA (version 3.1) program (16).
Electron microscopy.
The infected cell culture supernatants were partially purified by ultracentrifugation through a 20% (wt/wt) sucrose cushion, negatively stained with 2% ammonium molybdate, and examined with an electron microscope (JEM-1011; JEOL Ltd., Tokyo, Japan). For ultrastructural observation, infected HRT-18 cells were fixed with 2% glutaraldehyde, postfixed with 1% osmium tetroxide, dehydrated in graded ethanol solutions, and embedded in an Epon mixture. Ultrathin sections were stained with a lead citrate-uranyl acetate solution and observed with an electron microscope.
Nucleotide sequence accession numbers.
The nucleotide sequences described in this paper have been submitted to the DDBJ nucleotide sequence database and are retrievable from GenBank. The accession numbers for fragments of the N, M, and S genes of strain Aichi/2004 are AB285125, AB285126, and AB285127, respectively.
RESULTS
Examination of intestinal contents.
Intestinal contents from the dead calf were negative for BCV, bovine viral diarrhea virus, TGEV, and PEDV by RT-PCR. These samples were also negative for Salmonella species, Coccidium species, and Cryptosporidium species. By contrast, both samples were positive for BoTV by RT-PCR targeting the N and S genes. The rectum contents were also positive for GAR by the antigen detection kit (the ileum contents were not tested).
Virus isolation in HRT-18 cells.
At the second passage, CPE appeared in HRT-18 cells inoculated with the ileum contents of the dead calf. CPE characterized by the enlargement of cells were observed 2 to 3 days after inoculation, and the cells were eventually detached from the plastic surface (Fig. 1). Infective titers of the isolates were a constant 105.8 to 106.8 TCID50/ml after passage 3. The experiments described below were conducted with the cloned isolate at passages 8 to 10. No cytopathogenic viruses were isolated from the rectum contents in HRT-18 cells. The samples were negative for virus isolation in other cell lines.
FIG. 1.
CPE in HRT-18 cells produced by BToV at passage 9. Cells are at day 5 after inoculation. Infected (A) and noninfected control (B) cells are shown.
Physicochemical properties of the isolate.
The isolate was inactivated by treatment with a lipid solvent and heating at 56°C for 30 min but was stable under acid treatment. IUdR inhibited the replication of bovine herpesvirus 1 (data not shown) but not that of the isolate, indicating that the isolate contains an RNA genome (Table 1). The isolate passed through the 450- and 200-nm-pore-size filters completely, and through the 100-nm-pore-size filter partially, but did not pass through the 50-nm-pore-size filter (Table 1). The isolate (105.3 TCID50/ml) hemagglutinated mouse erythrocytes (16 HA units) but not chicken and goose erythrocytes.
TABLE 1.
Physicochemical properties of BToV
| Treatment | Log TCID50/ml of BToV
|
|
|---|---|---|
| Treated | Not treated | |
| Chloroform (10%) | <1.8 | 5.6 |
| pH 3.0 for 3 h | 5.3 | 5.8 |
| 56°C for 30 min | 2.6 | 6.1 |
| IUdR (10−4.5 M) | 6.3 | 7.1 |
| Filtration | ||
| Pore size, 450 nm | 5.8 | 5.8 |
| Pore size, 200 nm | 5.8 | 5.8 |
| Pore size, 100 nm | 3.8 | 5.8 |
| Pore size, 50 nm | <1.8 | 5.8 |
Immunofluorescence.
BToV antigens were observed by indirect immunofluorescence as specific cytoplasmic fluorescence in HRT-18 cells that had been inoculated with the isolate and reacted with a gnotobiotic calf antiserum against BToV (Fig. 2A). This antiserum did not react with BCV antigens (Fig. 2C). By contrast, no fluorescent cells were observed when the infected cells were stained with an anti-BCV polyclonal antiserum conjugated to FITC that reacted with BCV antigens (Fig. 2B and D).
FIG. 2.
Immunofluorescence of HRT-18 cells infected with the isolate or BCV. (A and B) HRT-18 cells were infected with the isolate and reacted with an antiserum to either BToV (A) or BCV (B). (C and D) HRT-18 cells were infected with BCV and reacted with an antiserum to either BToV (C) or BCV (D).
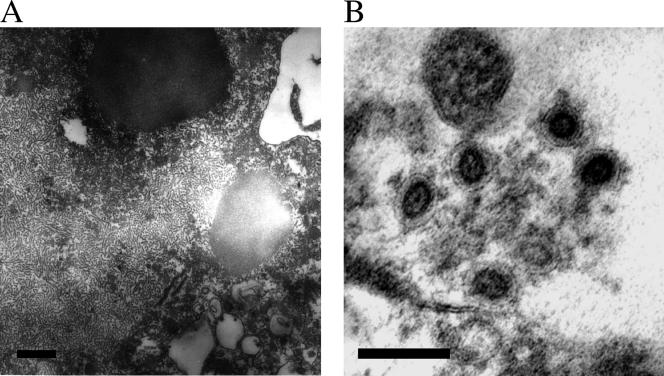
Electron microscopy.
Coronavirus-like particles were found in the infected HRT-18 cell culture supernatants. The particles were approximately 100 to 170 nm (average, 120 nm) in diameter and had club-shaped projections 15 to 20 nm long (Fig. 3). The infected HRT-18 cells showed cytoplasmic accumulations of tubular structures (Fig. 4A). A small number of concentric virions with a torus morphology were observed adjacent to the plasma membrane (Fig. 4B).
FIG. 3.
Electron micrograph of the Aichi/2004 isolate. The virus was partially purified from the supernatants of infected HRT-18 cells by ultracentrifugation through a 20% sucrose cushion and was negatively stained with 2% ammonium molybdate. Bar, 100 nm.
FIG. 4.
Electron micrographs of ultrathin sections of HRT-18 cells infected with BToV. (A) Accumulations of tubular structures are present in the cytoplasm. Bar, 500 nm. (B) Virions with electron-lucent centers were observed adjacent to the plasma membrane. Bar, 100 nm.
RT-PCR and sequence analysis of the isolate.
The isolate was positive for BToV by RT-PCR targeting the N, M, and S genes. Nucleotide sequences of the RT-PCR products from the isolate were determined and compared with those of the published N genes (476 to 497 nucleotides [nt] and 157 to 164 amino acids [aa]), M genes (672 nt and 233 aa), and S genes (687 to 690 nt and 228 to 229 aa) of toroviruses. For the N genes, a moderate to high degree of sequence identity (69.7 to 99.4% nucleotide identity and 67.5 to 98.8% amino acid identity) was observed between the isolate and BToVs, among which BToV strain B145 from The Netherlands showed the highest identity. For the M and S genes, sequence identities between the isolate and BToVs were very high (94.9 to 98.7% nucleotide identity and 97.3 to 100% amino acid identity for M genes and 94.3 to 94.9% nucleotide identity and 95.2% amino acid identity for S genes) (Table 2). The isolates were negative for BCV, TGEV, and PEDV by RT-PCR.
TABLE 2.
Nucleotide and amino acid sequence identities of the N, M, and S genes of BToV strain Aichi/2004 with those of toroviruses
| Torovirus straina (origin) | % Identityb of the following gene of BToV Aichi/2004 to that of the indicated virus:
|
|||||
|---|---|---|---|---|---|---|
| N
|
M
|
S
|
||||
| Nucleotide (476-497 nt) | Amino acid (157-164 aa) | Nucleotide (672 nt) | Amino acid (223 aa) | Nucleotide (687-690 bp) | Amino acid (228-229 aa) | |
| B145 (bovine) | 99.4 | 98.8 | 98.4 | 100.0 | 94.9 | 95.2 |
| B150 (bovine) | 98.1 | 96.9 | 98.7 | 100.0 | —c | — |
| B155 (bovine) | 98.6 | 96.9 | 98.7 | 100.0 | — | — |
| B156 (bovine) | 98.6 | 96.9 | — | — | — | — |
| B6 (bovine) | 98.6 | 98.1 | 98.2 | 100.0 | — | — |
| B1314 (bovine) | 96.9 | 95.0 | — | — | — | — |
| Breda 1 (bovine) | 69.7 | 67.5 | 94.9 | 97.3 | 94.3 | 95.2 |
| Berne (equine) | 68.3 | 66.2 | 84.4 | 92.8 | 76.1 | 76.9 |
| Markelo (porcine) | 93.0 | 93.1 | 79.3 | 90.6 | 66.1 | 58.7 |
| P4 (porcine) | 93.8 | 92.5 | 79.5 | 91.0 | — | — |
| P9 (porcine) | 93.4 | 92.5 | 79.6 | 90.1 | — | — |
| P10 (porcine) | 93.4 | 92.5 | 79.9 | 91.0 | — | — |
| P78 (porcine) | 95.3 | 94.4 | — | — | — | — |
The nucleotide sequences used were taken from the following accession numbers: for B145, AJ575388 (N gene), AJ575375 (M gene), and AJ575373 (S gene); for B155, AJ575386 (N gene) and AJ575377 (M gene); for B156, AJ575385 (N gene); for B1314, AJ575384 (N gene); for B6, AJ575389 (N gene) and AJ575374 (M gene); for Breda 1, AF076621 (N and M genes) and AY427798 (S gene); for Berne, D00563 (N gene), X52505 (M gene), and X52506 (S gene); for Markelo, J575358 (N gene), AJ575368 (M gene), and AJ575372 (S gene); for P10, AJ575361 (N gene) and AJ575371 (M gene); for P4, AJ575359 (N gene) and AJ575369 (M gene); for P9, AJ575360 (N gene) and AJ575370 (M gene); and for P78, AJ575362 (N gene).
Values for strains with the highest identity with Aichi/2004 are boldfaced.
—, no data available.
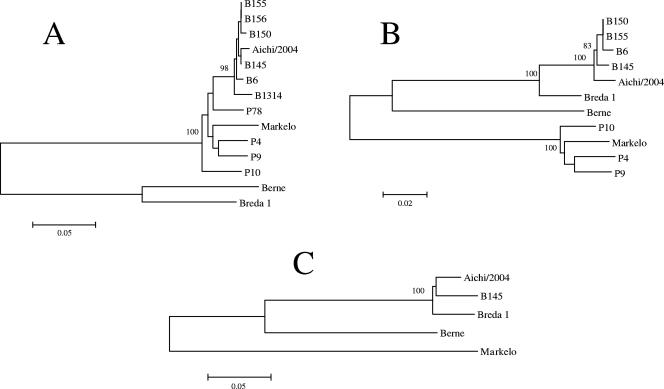
Phylogenetic analysis based on comparisons of the N, M, and S genes showed that the isolate belonged to the same cluster as BToV strains B145, B150, B155, B156, B6, and B1314 from Europe (Fig. 5). In general, the clusters of these genes were differentiated by host species (horse, cattle, and pig). However, the N gene of strain Breda 1 did not cluster with the N genes of other BToV strains (Fig. 5A).
FIG. 5.
Phylogenetic trees for the N genes (A), M genes (B), and S genes (C) of toroviruses and of the isolate, constructed using the neighbor-joining method (13) and drawn with the MEGA (version 3.1) program (9). Bootstrap values greater than 800 in 1,000 pseudoreplicates are shown as percentages. The accession numbers of the nucleotide sequences used are given in Table 2, footnote a.
Virus neutralization tests and serological survey.
An antiserum to BToV neutralized the isolate at a titer of 6,400. In contrast, an antiserum to BCV failed to neutralize the isolate (titer, <2).
In a serological survey, 325 of 355 calves (92%) at all 33 farms tested were positive for neutralizing antibodies to the isolate. In a comparison by age group, the antibody prevalence rate among young calves (up to the age of 3 months) was 100%, slightly higher than the rates for older calves. Geometric mean antibody titers were relatively high up to the age of 3 months and then stayed at low levels to the age of 9 months. Antibody titers rose at 10 to 12 months of age (Table 3).
TABLE 3.
Prevalence and titers of BToV (Aichi/2004) neutralizing antibody in calf sera grouped according to age, collected from 33 farms in Japan
| Age (mo) | No. tested | No. (%) positive | Antibody titer
|
|
|---|---|---|---|---|
| Geometric mean titera | Range | |||
| ≤2 | 24 | 24 (100) | 73.94 | 4-1,024 |
| 3 | 33 | 33 (100) | 68.16 | 2-≥2,048 |
| 4 | 36 | 34 (94) | 18.08 | <2-512 |
| 5 | 62 | 58 (93) | 22.36 | <2-256 |
| 6 | 55 | 51 (93) | 16.60 | <2-≥2,048 |
| 7 | 47 | 38 (81) | 19.02 | <2-≥2,048 |
| 8 | 48 | 45 (94) | 28.29 | <2-≥2,048 |
| 9 | 26 | 22 (85) | 29.11 | <2-≥2,048 |
| 10-12 | 24 | 20 (83) | 132.51 | <2-≥2,048 |
| Total | 355 | 325 (92) | 29.26 | <2-2,048 |
Antibody titer of antibody-positive sera.
DISCUSSION
In electron microscopy, toroviruses appear pleomorphic, with a spike-bearing envelope and a spherical, oval, elongated, or kidney-shaped morphology, measuring 100 to 140 nm in diameter (12, 28, 29, 32). A tubular structure considered to be the nucleocapsid is visible in the cytoplasm of cells infected with toroviruses. In cross sections, virions show the typical twin circular structures indicative of a torus morphology of the nucleocapsid (28, 29). In the present study, we isolated a cytopathogenic virus from a diarrheic calf by using HRT-18 cells, which possess high sensitivity to BCVs (2, 26). Negatively stained electron micrographs of the infected culture supernatants revealed the presence of pleomorphic particles with projections that were indistinguishable from BCVs. In contrast, ultrathin sections of the infected cells showed the cytoplasmic accumulation of tubular structures as well as concentric virions with a torus morphology, both of which are characteristics of toroviruses. Assessment of the physicochemical properties of the isolates showed the RNA genome as well as heat and lipid-solvent lability. The isolate stained with an antiserum to BToV, but not with an antiserum to BCV, by immunofluorescence. Further, an antiserum to BToV neutralized the isolate with a high titer, whereas an antiserum to BCV failed to neutralize it. Moreover, the nucleotide sequences of the isolate were markedly similar to those of published BToVs but not to those of BCVs. From these results, we concluded that the isolate in the present study was a BToV, and we designated it strain Aichi/2004. To our knowledge, this is the first report to describe the isolation of BToV in cell culture.
To date, equine torovirus BEV has been the only cell culture-adapted strain, suggesting that this strain is a mutant that has been modified in a way that enables it to replicate in cell culture (12). BToV did not grow in HRT-18 cells when it was first propagated in gnotobiotic calves in 1979 (32). The reason why strain Aichi/2004 could grow in HRT-18 cells in the present study remains unknown. Given that we employed no special methods for virus isolation, one possibility is that this isolate is a mutant virus that has been modified in a way that enables it to replicate in HRT-18 cells. However, an alternative possibility, which cannot be ruled out, is that the properties of the HRT-18 cells used in the present study may have changed to increase the torovirus tropism, despite the facts that no morphological change was observed at the microscopic level and the HRT-18 cells used here still possessed sensitivity to BCV. As possible evidence for this, the nucleotide sequences of the N, M, and S gene fragments were not unique to the isolate. In any case, further studies are needed to determine the full-length genome sequence of strain Aichi/2004 and to comprehensively compare the genetic properties of Aichi/2004 with those of BToV Breda 1, for which the full-length genome sequence has been determined (4).
Antigenic relatedness among BToVs, BEV, and human toroviruses has been identified (1, 6). In addition, porcine toroviruses have been found to be antigenically related to BToVs (15). In contrast, serological differences have been observed between BEV and BToVs (1). Further, BToVs have been subdivided into two serotypes (33). The cell culture-adapted Aichi/2004 strain is useful for clarifying the serotypic properties of toroviruses. In a genetic comparison, toroviruses were differentiated by host species into four genotypes (cattle, horses, humans, and pigs), whereas intertypic recombinations were observed (22). The present study indicates that BToV Aichi/2004 is genetically closer to the recently identified European BToVs than it is to the Breda 1 strain. Given that Breda 1 was detected more than 20 years ago, this genetic distance might reflect the time of detection (22).
BToVs have been reported to be enteric pathogens that cause mild to severe diarrhea in experimentally or naturally infected young calves. In the present study, the rectal contents were positive for GAR as well as BToV. Therefore, the cause of diarrhea in the calf was unknown. Experimental infection of calves is necessary to elucidate the pathogenicity of BToV strain Aichi/2004.
High seroprevalence of BToV antibodies in the cattle population has been reported, with positive serum rates of 88.5%, 94%, and 55% in the United States, The Netherlands, and the United Kingdom, respectively (3, 13, 33). In the present serological survey, 92% of calves were positive for BToV neutralizing antibodies, suggesting that BToV infection in cattle is also common in Japan. The BToV antibodies at high titers in calves aged 3 months and younger might be maternally derived. An increase in antibody titers at the age of 10 to 12 months implies active infection at this time. Although BToV-associated diarrhea has not been reported in Japan, diagnosis of bovine diarrhea should include BToV examination. Further studies to reveal the epidemiological status of BToV infection in cattle are in progress.
Acknowledgments
We thank S. Fukuta, D. Nishi, T. Aita, M. Soledad, and N. Hattori for technical assistance.
Footnotes
Published ahead of print on 13 June 2007.
REFERENCES
- 1.Beards, G. M., D. W. Brown, J. Green, and T. H. Flewett. 1986. Preliminary characterisation of torovirus-like particles of humans: comparison with Berne virus of horses and Breda virus of calves. J. Med. Virol. 20:67-78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Benfield, D. A., and L. J. Saif. 1990. Cell culture propagation of a coronavirus isolated from cows with winter dysentery. J. Clin. Microbiol. 28:1454-1457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brown, D. W., G. M. Beards, and T. H. Flewett. 1987. Detection of Breda virus antigen and antibody in humans and animals by enzyme immunoassay. J. Clin. Microbiol. 25:637-640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Draker, R., R. L. Roper, M. Petric, and R. Tellier. 2006. The complete sequence of the bovine torovirus genome. Virus Res. 115:56-68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Duckmanton, L., S. Carman, E. Nagy, and M. Petric. 1998. Detection of bovine torovirus in fecal specimens of calves with diarrhea from Ontario farms. J. Clin. Microbiol. 36:1266-1270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Duckmanton, L., R. Tellier, C. Richardson, and M. Petric. 1999. The novel hemagglutinin-esterase genes of human torovirus and Breda virus. Virus Res. 64:137-149. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 7.Hoet, A. E., K. O. Cho, K. O. Chang, S. C. Loerch, T. E. Wittum, and L. J. Saif. 2002. Enteric and nasal shedding of bovine torovirus (Breda virus) in feedlot cattle. Am. J. Vet. Res. 63:342-348. [DOI] [PubMed] [Google Scholar]
- 8.Hoet, A. E., P. R. Nielsen, M. Hasoksuz, C. Thomas, T. E. Wittum, and L. J. Saif. 2003. Detection of bovine torovirus and other enteric pathogens in feces from diarrhea cases in cattle. J. Vet. Diagn. Investig. 15:205-212. [DOI] [PubMed] [Google Scholar]
- 9.Hoet, A. E., J. Smiley, C. Thomas, P. R. Nielsen, T. E. Wittum, and L. J. Saif. 2003. Association of enteric shedding of bovine torovirus (Breda virus) and other enteropathogens with diarrhea in veal calves. Am. J. Vet. Res. 64:485-490. [DOI] [PubMed] [Google Scholar]
- 10.Ishikawa, K., H. Sekiguchi, T. Ogino, and S. Suzuki. 1997. Direct and rapid detection of porcine epidemic diarrhea virus by RT-PCR. J. Virol. Methods 69:191-195. [DOI] [PubMed] [Google Scholar]
- 11.Jamieson, F. B., E. E. Wang, C. Bain, J. Good, L. Duckmanton, and M. Petric. 1998. Human torovirus: a new nosocomial gastrointestinal pathogen. J. Infect. Dis. 178:1263-1269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Koopmans, M., and M. C. Horzinek. 1994. Toroviruses of animals and humans. Adv. Virus Res. 43:233-273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Koopmans, M., U. van den Boom, G. Woode, and M. C. Horzinek. 1989. Seroepidemiology of Breda virus in cattle using ELISA. Vet. Microbiol. 19:233-243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Koopmans, M., L. van Wuijckhuise-Sjouke, Y. H. Schukken, H. Cremers, and M. C. Horzinek. 1991. Association of diarrhea in cattle with torovirus infections on farms. Am. J. Vet. Res. 52:1769-1773. [PubMed] [Google Scholar]
- 15.Kroneman, A., L. A. Cornelissen, M. C. Horzinek, R. J. de Groot, and H. F. Egberink. 1998. Identification and characterization of a porcine torovirus. J. Virol. 72:3507-3511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kumar, S., K. Tamura, and M. Nei. 2004. MEGA3: integrated software for molecular evolutionary genetics analysis and sequence alignment. Brief. Bioinform. 5:150-163. [DOI] [PubMed] [Google Scholar]
- 17.Lodha, A., N. de Silva, M. Petric, and A. M. Moore. 2005. Human torovirus: a new virus associated with neonatal necrotizing enterocolitis. Acta Paediatr. 94:1085-1088. [DOI] [PubMed] [Google Scholar]
- 18.Paton, D., G. Ibata, J. Sands, and A. McGoldrick. 1997. Detection of transmissible gastroenteritis virus by RT-PCR and differentiation from porcine respiratory coronavirus. J. Virol. Methods 66:303-309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pohlenz, J., N. Cheville, G. Woode, and A. Mokresh. 1984. Cellular lesions in intestinal mucosa of gnotobiotic calves experimentally infected with a new unclassified bovine virus (Breda virus). Vet. Pathol. 21:407-417. [DOI] [PubMed] [Google Scholar]
- 20.Saitou, N., and M. Nei. 1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4:406-425. [DOI] [PubMed] [Google Scholar]
- 21.Shimizu, M., J. Shirai, M. Narita, and T. Yamane. 1990. Cytopathic astrovirus isolated from porcine acute gastroenteritis in an established cell line derived from porcine embryonic kidney. J. Clin. Microbiol. 28:201-206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Smits, S. L., A. Lavazza, K. Matiz, C. Horzinek, M. P. Koopmans, and R. J. de Groot. 2003. Phylogenetic and evolutionary relationships among torovirus field variants: evidence for multiple intertypic recombination events. J. Virol. 77:9567-9577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tompkins, W. A., A. M. Watrach, J. D. Schmale, R. M. Schultz, and J. A. Harris. 1974. Cultural and antigenic properties of newly established cell strains derived from adenocarcinomas of the human colon and rectum. J. Natl. Cancer Inst. 52:1101-1110. [DOI] [PubMed] [Google Scholar]
- 25.Tsunemitsu, H., D. R. Smith, and L. J. Saif. 1999. Experimental inoculation of adult dairy cows with bovine coronavirus and detection of coronavirus in feces by RT-PCR. Arch. Virol. 144:167-175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tsunemitsu, H., H. Yonemichi, T. Hirai, T. Kudo, S. Onoe, K. Mori, and M. Shimizu. 1991. Isolation of bovine coronavirus from feces and nasal swabs of calves with diarrhea. J. Vet. Med. Sci. 53:433-437. [DOI] [PubMed] [Google Scholar]
- 27.Vilcek, S., A. J. Herring, J. A. Herring, P. F. Nettleton, J. P. Lowings, and D. J. Paton. 1994. Pestiviruses isolated from pigs, cattle and sheep can be allocated into at least three genogroups using polymerase chain reaction and restriction endonuclease analysis. Arch. Virol. 136:309-323. [DOI] [PubMed] [Google Scholar]
- 28.Weiss, M., and M. C. Horzinek. 1986. Morphogenesis of Berne virus (proposed family Toroviridae). J. Gen. Virol. 67:1305-1314. [DOI] [PubMed] [Google Scholar]
- 29.Weiss, M., F. Steck, and M. C. Horzinek. 1983. Purification and partial characterization of a new enveloped RNA virus (Berne virus). J. Gen. Virol. 64:1849-1858. [DOI] [PubMed] [Google Scholar]
- 30.Weiss, M., F. Steck, R. Kaderli, and M. C. Horzinek. 1984. Antibodies to Berne virus in horses and other animals. Vet. Microbiol. 9:523-531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Woode, G. N., J. F. Pohlenz, N. E. Gourley, and J. A. Fagerland. 1984. Astrovirus and Breda virus infections of dome cell epithelium of bovine ileum. J. Clin. Microbiol. 19:623-630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Woode, G. N., D. E. Reed, P. L. Runnels, M. A. Herrig, and H. T. Hill. 1982. Studies with an unclassified virus isolated from diarrheic calves. Vet. Microbiol. 7:221-240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Woode, G. N., L. J. Saif, M. Quesada, N. J. Winand, J. F. Pohlenz, and N. K. Gourley. 1985. Comparative studies on three isolates of Breda virus of calves. Am. J. Vet. Res. 46:1003-1010. [PubMed] [Google Scholar]