Abstract
We developed the LipidQA (Lipid Qualitative/Quantitative Analysis) software platform to identify and quantitate complex lipid molecular species in biological mixtures. LipidQA can process raw electronic data files from the TSQ-7000 triple stage quadrupole and LTQ linear ion trap mass spectrometers from Thermo-Finnigan and the Q-TOF hybrid quadrupole/time-of-flight instrument from Waters-Micromass and could readily be modified to accommodate data from others. The program processes multiple spectra in a few seconds and includes a deisotoping algorithm that increases the accuracy of structural identification and quantitation. Identification is achieved by comparing MS2 spectra obtained in a data-dependent manner to a library of reference spectra of complex lipids that we have acquired or constructed from established fragmentation rules. The current form of the algorithm can process data acquired in negative or positive ion mode for glycerophospholipid species of all major head-group classes.
Keywords: glycerophospholipid, electrospray ionization, computer algorithm, quantitation, identification, database searching
INTRODUCTION
Lipids comprise a major class of complex biomolecules with great structural diversity [1]. The development of ESI/MS in the 1990's permitted direct analyses of complex lipids from liquid solutions as intact molecules [2, 3], and tandem MS permitted facile structural determinations of components of lipid mixtures [4-6]. This greatly simplified lipid analyses and led to ESI/MS/MS-based “high-throughput” approaches to characterizing biological lipid mixtures [3, 7]. Several factors complicate efforts to make such approaches routine: 1) Unfractionated biological extracts to which high throughput approaches are applied contain many lipid molecular species. 2) This complexity increases the likelihood that abundant isotope peaks of one molecular species will exhibit m/z values identical to those of (pseudo)molecular ions (e.g., [M-H]− or [M+H]+) of distinct molecular species, e.g., one that differs by a single degree of unsaturation, and it is thus important to perform precise deisotoping to achieve accurate quantitation. 3) Sample complexity also increases the difficulty of identifying lipid species from MS spectra only, which necessitates MS/MS analyses, and interpreting MS/MS spectra by direct inspection is time-consuming and requires conversance with lipid fragmentation patterns. 4) High throughput methods quickly generate huge amounts of data that are difficult to process manually. These factors motivate developing computerized algorithms to process data from high-throughput ESI/MS/MS analyses of lipids.
Recently, the program Fatty Acid Analysis Tool (FAAT) was developed to analyze high-resolution mass spectra of lipids obtained with FT-ICR instruments [8]. Another program (“Lipid Profiler”) employs a multiple precursor ion tandem MS scanning approach to identify and quantitate glycerolipid molecular species in mixtures [9], and the program LipidInspector [10] achieves lipid profiling by multiple precursor ion and neutral loss scanning driven by data-dependent MS/MS acquisition. Both Lipid Profiler and LipidInspector are based on algorithms for data acquisition with Applied Biosystems hybrid quadrupole/time-of-flight instruments that can perform multiple precursor ion scans in a single experiment. Neither those programs nor FAAT are readily applicable to MS data acquired with triple quadrupole or ion trap instruments.
Here we describe an approach to process data from high throughput MS analyses performed with ion trap, triple quadrupole, or quadrupole time-of-flight instruments that automatically identifies and quantitates complex lipid molecular species in mixtures. Unfractionated lipid extracts are directly infused and analyzed by ESI/MS with data-dependent switching to MS/MS mode. MS/MS data are searched against a library of reference spectra for complex lipids constructed from studies of standard glycerophospholipids of all major head-group classes [11-28] that we used to establish fragmentation rules [29] that facilitate identification of glycerophospholipids in biological mixtures [30-31]. Structure assignment permits determination of an elemental formula, and a theoretical isotope profile is then calculated and used in a deisotoping algorithm. Deisotoped MS data are normalized to internal standard peak intensity, and the normalized value is compared to a calibration curve to quantitate the lipid species. This value can be normalized to lipid phosphorus content to permit comparisons among samples. This approach is fully automated and facilitates processing of lipidomics data from high-throughput MS analyses.
MATERIALS AND METHODS
Lipid standards, solvents, and other chemicals
Synthetic lipid standards were purchased from Sigma Chemical (St. Louis, MO) or Avanti Polar Lipids (Alabaster, AL). Solvents, salts, and other chemicals were obtained from Fisher Chemical (Pittsburgh, PA).
Sample preparation for ESI/MS analyses
Standard samples were prepared in CHCl3/CH3OH 1:2 (v/v). Described methods [30] were used to extract lipids from mouse peritoneal exudate cells (PEC). For positive ion analyses, LiOH (final concentration 5 μmol/100 μL) was added to sample solutions to permit formation of [M+Li]+ adducts, which simplifies ESI/MS spectra by minimizing contributions of other metal ion adducts and facilitates informative fragmentation [11, 19-20]. For negative ion analyses, NH4OH (final concentration 0.3%) was added to facilitate formation of [M-H]− ions. Lipid phosphorus content was determined as described [32].
Data-dependent analyses of lipid mixtures
Q-TOF Micro hybrid quadrupole/time-of-flight mass spectrometer (Waters Corporation, Milford, MA) survey ESI/MS scans were integrated over 2 sec, and MS/MS scans were integrated over the period required to acquire an ion count exceeding 104 or over 30 sec. Switching from survey scan to MS/MS mode was performed in a data-dependent manner based on ESI/MS TIC intensity. The maximum MS/MS to survey scan ratio was three. The collision energy was 30 eV. Multi-point calibration was performed using a mixture of lipid standards. MS data were processed with MassLynx 3.5 software to produce peak list files.
For analyses on the LTQ (Thermo Electron corporation, Waltham, MA) linear ion trap instrument with Xcalibur 2.0, the maximum acquisition time of ESI/MS survey scans (recorded in profile mode) was set as 100 msec, and the maximum acquisition time of MS/MS scans (centroid mode) was set as 400 msec or integrated over an interval sufficient to acquire an ion count exceeding 104. Switching from survey scan to MS/MS mode was performed in a data-dependent manner as above. The maximum MS/MS to survey scan ratio was 3. Normalized collision energy was 15%. Multi-point calibration was performed using ultra-mark reagent (Finnigan). MS/MS spectra were extracted by the program extract_msn.exe (Finnigan) and processed with a data pretreatment program that is a component of LipidQA software to produce peak list files for MS/MS and MS analyses. Instrumental conditions for lipid analyses on the Finnigan TSQ-7000 triple stage quadrupole were described elsewhere [11-18] and included a collision energy of about 30 eV.
LipidQA software
The Lipid QA program is written in the language Visual C++. The fragment ion database and the lipid chemical formula database are constructed as ASCII code files with the appendices “.str” and “.fml”, respectively. LipidQA features include: 1) Different functions are used to select analytical objectives, e.g., quantitation only, identification, or simultaneous quantitation and identification. 2) The program can process peak list files generated by data systems for Finnigan LTQ and TSQ-7000 instruments and the Waters Q-TOF Micro instrument. 3) The program can accommodate multiple internal standards and calibration curves for complex lipid mixture analyses, and LipidQA automatically selects the appropriate internal standard and calibration curve based on its identification of the lipid molecular species without user input. 4) Users can choose to analyze a single lipid subclass or to analyze multiple subclasses simultaneously, e.g., glycerophosphatidic acid (GPA), glycerophospho-ethanolamine (GPE), -inositol (GPI), -serine (GPS), and –glycerol (GPG), in negative ion mode. Li+ adducts of glycerophosphocholine (GPC) species are analyzed in positive ion mode. Access to LipidQA software can be obtained through Haowei Song (hsong@im.wustl.edu), John Turk (jturk@wustl.edu), or the Washington University Biomedical Mass Spectrometry Resource website.
RESULTS AND DISCUSSION
Outline of LipidQA software and data-dependent lipid profiling
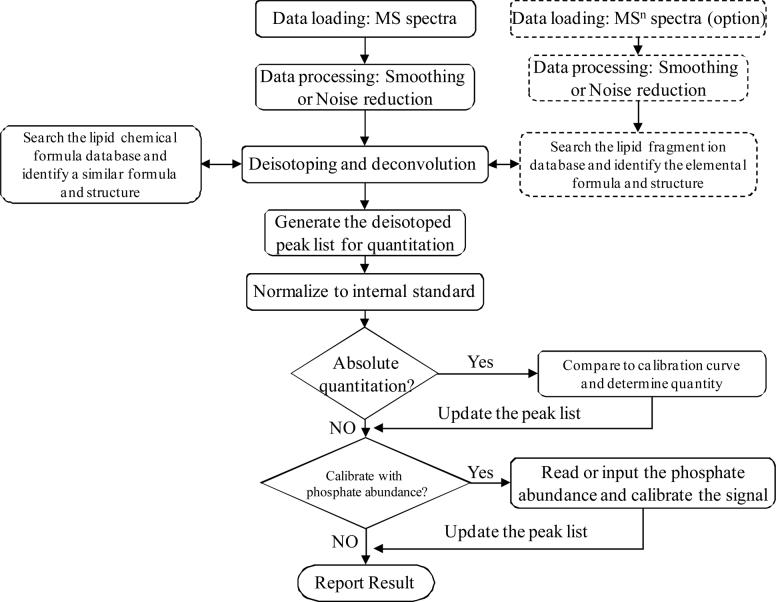
Figure 1 illustrates the working flow chart of the LipidQA program. First, peak list files of both MS and MS/MS spectra are loaded into memory and undergo data pretreatment that includes smoothing (5-point Triangular Smooth)/noise filtering. MS/MS data are searched against a fragment ion database from a library of reference spectra for complex lipids that we have acquired [11-28] or constructed from established fragmentation rules [29] to identify lipid molecular structures, and deduced chemical formulae are used to calculate theoretical isotope distributions subsequently used in the deisotoping algorithm. The m/z values of those ions in the MS spectra that do not correspond to a recorded MS/MS spectrum are searched against a lipid chemical formula database, and identified formulae are used to calculate theoretical isotope distributions for deisotoping, as described further below.
Figure 1.
Working flow chart of LipidQA program
Upon completing deisotoping and deconvolution steps, a peak list of monoisotopic masses of different lipid molecular species and intensities of deisotoped peaks is generated for quantitation. First, ion intensities are normalized to that of the internal standard ion. If the “Absolute Quantitation” feature is selected, relative intensity values are automatically compared to a calibration curve to quantitate the lipid species. If the “Lipid Phosphorus” normalization feature is selected and the lipid phosphorus content specified, the program automatically normalizes the quantity of each species to that value. Absolute or normalized quantitative results from different samples within a set are then combined into a data file that can be processed by the Excel application, e.g., to prepare data summaries or illustrations.
Lipid Identification: Fragment ion database searching
Identifying proteins by searching MSn spectra against databases [33, 34] is now routine, but identifying lipids still relies mainly on direct inspection of MS/MS spectra. Rules to explain fragmentation patterns of a wide variety of glycerolipids comprising hundreds of molecular species from different head-group classes have been established [11-29], and we have constructed a lipid fragment ion database that includes species with fatty acid side chain lengths of 10−30 carbon atoms and 0−6 double bonds. Searching the MS2 spectra of different lipid species against this database permits deduction of their structures and chemical formulae.
Table 1 lists the fragment ions (Column 1) and their relative intensities for tandem spectra of Li+ adducts of glycerophosphocholine (GPC) lipids [11, 19-20]. The ions [M+Li-59]+ (loss of trimethylamine), [M+Li-183]+ (loss of phosphocholine), and [M+Li-189]+ (loss of lithiated phosphocholine) identify the choline head-group, and constant neutral loss MS/MS scanning for these losses can be used to identify GPC lipids as a class. The ions [M+Li-R1CO2H]+, [M+Li-R2CO2H]+, [M+Li-59-R1CO2H]+, [M+Li-59-R2CO2H]+, [M+Li-R1CO2Li]+, [M+Li-R2CO2Li]+, [R1CO]+, and [R2CO]+ identify the fatty acid substituents. The relative intensities of ions arising from losses of the fatty acid substituents identify their positions on the glycerol backbone. Ions generated by losses of the sn-1 fatty acid substituent from Li+ adducts of GPC lipids are more abundant than those generated by losses of the sn-2 substituent [11].
Table 1.
Examples of the fragmentation rules and relative intensity rules used to construct the fragment ion database.
| Fragment Ions | Structural Feature Reflected* | Substituent Position | 16:0/18:1-GPC-Li+ | 18:1/16:0-GPC-Li+ | ||
|---|---|---|---|---|---|---|
| Fragment ion m/z | Observed Relative Intensity (%) | Fragment ion m/z | Observed Relative Intensity (%) | |||
| [M+Li]+ | NN | − | 766.58 | 9 | 766.58 | 22 |
| [M+Li-59]+ | HG | − | 707.51 | 7 | 707.51 | 10 |
| [M+Li-183]+ | HG | − | 583.51 | 47 | 583.51 | 39 |
| [M+Li-189]+ | HG | − | 577.51 | 100 | 577.51 | 100 |
| [M+Li-R1CO2H]+ | R1 | + | 510.34 | 8 | 484.32 | 10 |
| [M+Li-R2CO2H]+ | R2 | + | 484.32 | 5 | 510.34 | 5 |
| [M+Li-59-R1CO2H]+ | R1 | + | 451.27 | 17 | 425.25 | 14 |
| [M+Li-59-R2CO2H]+ | R2 | + | 425.25 | 7 | 451.27 | 7 |
| [M+Li-R1CO2Li]+ | R1 | + | 504.33 | 7 | 478.32 | 6 |
| [M+Li-R2CO2Li]+ | R2 | + | 478.32 | 6 | 504.33 | 5 |
| [R1CO]+ | R1 | − | 239.24 | 4 | 265.25 | 4 |
| [R2CO]+ | R2 | − | 265.25 | 5 | 239.24 | 3 |
| [C2H5PO4+Li]+ | HG | − | 131.01 | 6 | 131.01 | 3 |
| [C5H12N]+ | HG | − | 86.1 | 12 | 86.1 | 11 |
The indicated ions can contain information about the following structural features of the phospholipid molecule: NN (Number of Nitrogens); HG (head group); R1 (sn-1 substituent); R2 (sn-2 substituent); substituent position refers to sn-1 vs. sn-2. Positional assignments: [M+Li-R1CO2H]+ >[M+Li-R2CO2H]+, [M+Li-59-R1CO2H]+ >[M+Li-59-R2CO2H]+, [M+Li-R1CO2Li]+ >[M+Li-R2CO2Li]+
MS/MS spectra of 16:0/18:1-GPC-Li+ (Figure 2A) and of 18:1/16:0-GPC-Li+ (Figure 2B) contain the fragment ions listed in Table 1 and illustrate rules that establish identities and positions of fatty acid substituents. Positional assignments are based on the relative abundance of ions reflecting losses of fatty acid substituents rather than the relative intensities of the acylium ions [R1CO]+ and [R2CO]+ because the latter are variably susceptible to subsequent fatty acid-specific but not position-specific decomposition processes (Figure 3). Fragmentation patterns of other classes of glycerolipids are summarized in Supplemental Material.
Figure 2. MS/MS spectra of 16:0/18:1-GPC-Li+ (A) and 18:1/16:0-GPC-Li+ (B).

Data acquired with a Finnigan TSQ-7000 via product ion scan mode.
Figure 3. MS/MS spectrum obtained from CAD of the ion of m/z 865.33 in a lipid extract from mouse peritoneal leukocytes.

A. Complete spectrum. B. Expanded spectrum from m/z 80 to 260. Data were acquired with a Micromass Q-TOF Micro mass spectrometer in negative ion mode.
In the database-searching algorithm, a similarity score (ID score) is generated for the comparison between the observed spectrum and that contained in the most closely matched library spectrum. The score is based on the percent matching fragment ion m/z values in the two spectra. After raw MS/MS data are smoothed and expressed in centroid mode, the 40 most intense peaks in the acquired MS/MS spectrum are selected to calculate the similarity score. Individual fragment ions of similar m/z value in acquired and reference spectra are compared pair by pair. If their m/z difference is less than the preset error tolerance (determined by instrument resolution), that fragment ion will be included in the similarity score calculation in equation 1.
| (1) |
Nmatch is the number of spectral lines in the acquired spectrum that match those in the reference spectrum. Nreference is the total number of spectral lines in the reference MS/MS spectrum. The relative intensities of the fragment ions arising from losses of fatty acid substituents are used to determine their positions on the glycerol backbone.
Rules that govern such positional assignments differ among phospholipid head-group classes [11-29]. In positive ion MS/MS spectra of [M+Li]+ ions of GPC lipids, ions that reflect loss of the sn-1 substituent are more abundant than those reflecting loss of the sn-2 substituent [11, 19-20], as illustrated in Table 1. In contrast, in negative ion MS/MS spectra of [M-H]− ions of GPE, GPI, GPA, GPG, and GPS lipids, ions that arise from loss of sn-2 substituent are more abundant than those arising from sn-1 substituent loss, as summarized in Supplemental Material.
MS and MS/MS spectra from 25 standard glycerophospholipid species of various head-group classes were used to optimize algorithms for data pretreatment, similarity score calculation, and quantitation. The performance of the algorithm was compared to results from direct inspection of re-analyses of the standards on each of the three mass spectrometers tested and upon analyses of biological extracts. Of the 25 standards used to optimize the algorithm, the lowest similarity score upon re-analyses on different instruments was 0.67 for 18:0/20:4-GPS analyzed on the Finnigan LTQ. Considering analyses of both standards and components of biological extracts, discrepancies between structural assignments achieved by direct inspection versus those by algorithm occurred in less than 5% of cases where the similarity score calculated by the algorithm exceeded 0.5, and that value is thus suggested as an appropriate threshold for confident structural assignments.
The performance of LipidQA was similar with each of the three instruments tested. Although tandem mass spectra of lipid standards differ among instruments, differences are primarily in relative abundances of ions within a spectrum rather than in the presence or absence of a given ion in spectra of the same compound obtained on these three instruments. The m/z values of the 40 most abundant ions in the tandem spectra of a given glycerolipid molecular species are virtually identical among these three instruments, although the relative abundances of certain ions in the spectra can be quite different.
For example, tandem spectra of [M-H]− ions of GPS and GPE lipids obtained on the TSQ are quite similar to those obtained on the Q-TOF, and carboxylate anions are the base ions in both spectra. In contrast, tandem spectra obtained on ion trap instruments, e.g., the LTQ, are dominated by ions reflecting losses of the fatty acid substituents. Nonetheless, the same sets of ions appear in spectra obtained on any of these instruments, and qualitative identification is thus unaffected. Moreover, although there are differences in relative abundances of ions in spectra obtained on different instruments within a spectrum, rules for preferential losses of sn-1 or sn-2 substituents are preserved, and positional assignments are also unaffected.
Correct identification of the structure and chemical formula of a lipid molecular species is necessary for accurate quantitation based on the deisotoped signal. Without MS/MS spectra, the theoretical formula used to calculate the isotope profile is obtained by searching the m/z value of the first isotope peak against a theoretical formula database. The matched formula might not be exactly correct if isobaric species of different lipid classes have been confused, and this can increase the error of quantitation. For example, 18:0/18:2-GPS and 18:2/22:6-GPE have similar [M-H]− m/z values of 786.6 and 786.51, respectively, but their isotope profiles differ. The elemental formula of 18:0/18:2-GPS is C42H77O10NP, and its isotope profile is 100, 48.26, 13.39 and 2.71 for relative abundances of monoisotopic [M-H]− and the first three [13C] isotopes. The elemental formula of 18:2/22:6-GPE (C45H73O8NP) yields an isotope profile for relative abundances of monoisotopic [M-H]− and the three [13C] isotopes is 100, 50.48, 14.07 and 2.82.
Using the isotope profile of 18:2/22:6-GPE rather than the correct profile for 18:0/18:2-GPS would increase the error in the deisotoping correction for the latter species, but much more serious errors would be introduced by causing quantitation to be based on a calibration curve for the wrong head-group class of lipids. Ion yield per mole of analyte is much more dependent on the nature of the head-group than on the identities of fatty acid substituents in phospholipid quantitation by ESI/MS [3, 4]. Identification based on MS/MS spectra searched against a fragment ion database is more likely to be accurate than are assignments based only on nominal m/z values of (pseudo) molecular ions, selected product ions, or ions reflecting specified neutral losses, as discussed further below.
Fragment ion database searching
When analyzing biological extracts without pretreatment or separation, extraneous peaks that do not represent bona fide components of biological mixtures are a major obstacle to accurate identification and quantitation of lipid species. In addition, some fatty acid substituents, especially polyunsaturates, yield fragment ions upon CAD that can be confused with distinct fatty acid substituents that are not contained in the molecular species being analyzed. Such factors complicate identification of lipid species by precursor ion or neutral loss scanning alone. The fragment ion database-searching algorithm of LipidQA achieves more reliable identification in such cases because Lipid QA considers data from the entire tandem mass spectrum and not just selected features.
This is illustrated by Figure 3A, which is the tandem spectrum from CAD of an ion of m/z 865 in negative ion ESI/MS analysis of a mouse peritoneal leukocyte lipid extract. This ion was determined by LipidQA to represent the [M-H]− ion of 22:6/22:6-GPG with high confidence (ID score = 0.77). The spectrum contains an intense ion at m/z 283 that could represent stearate (18:0) anion, and a precursor of m/z 283 scan might identify the parent at m/z 865 to be a stearate-containing lipid, such as 18:0/18:0-GPI ([M-H]− m/z = 865). That assignment would be consistent with the presence of an ion at m/z 241 in the spectrum (Figure 3B). An ion of that m/z value is observed in tandem spectra of GPI lipids and represents an inositol phosphate dehydration product. Direct inspection of the entire tandem spectrum, however, fails to reveal ions that reflect loss of 18:0, and such ions are expected in the tandem spectra of 18:0/18:0-GPI. In addition, other ions prominent in the reference spectrum [17] of 18:0/18:0-GPI in the LipidQA fragment ion database are absent, e.g., m/z 315, 297, 279, 259, and 223.
In fact, the ion at m/z 283 in the spectrum in Figure 3A does not represent stearate anion but arises from elimination of CO2 from docosahexaenoate anion [17, 29], and this was verified by obtaining the MS3 spectrum of m/z 327 on an ion trap instrument, which contained ions of m/z 283 and m/z 229 generated by sequential neutral losses of CO2. Erroneous assignment of the identity of the lipid molecular species at m/z 865 as 18:0/18:0-GPI that might have arisen from multiple precursor ion scanning for parents of both m/z 281 and 241 was thus avoided by LipidQA because it evaluated the entire tandem spectrum of the compound rather than selected features to achieve the correct identification of 22:6/22:6-GPG.
This identification is based on the observed tandem spectrum (Figure 3A) and fragmentation rules previously established from studies of standard GPG lipids [18] and subsequently applied to biological extracts containing GPG lipid species previously identified by other methods [28]. Features of the spectrum in Figure 3A that support identification of m/z 865.3 as the [M-H]− ion of 22:6/22:6-GPG include the base peak at m/z 327.2, which represents docosahexaenoate anion. Product ions m/z 537.3 and m/z 555.3 arise from loss from [M-H]− of 22:6 as a free fatty acid or as a ketene, respectively. The product ion m/z 463.256 arises from loss of both the glycerol head-group and 22:6 as a free fatty acid, and the ion m/z 209.052 arises from the phosphoglycerol head-group and is observed in spectra of all GPG lipid species. Thus, the structural assignment is unambiguous.
It should be noted that when an identification by LipidQA is said to be “correct” what is meant is that the algorithm makes the same assignment that the authors make when we directly inspect the spectrum, deduce identities of various ions in it, and then make a structural identification of the parent ion based on our deductions. The advantage of LipidQA is that it obviates laboriously slow direct inspection and deduction and permits high-throughput analyses of data that are interpreted as we would interpret them if we did inspect them directly.
Lipid quantitation: spectrum reduction and correction algorithms that include deisotoping, deconvolution, and correction for overlapping isotope envelopes produced by distinct lipid molecular species
Complex lipid mixtures from biological extracts contain many molecular species, and distinct species can exhibit overlapping isotope envelopes. Han and Gross [3] used two isotope correction factors to minimize this isotope effect on quantitative measurements. The type I correction factor converts the intensity of a monoisotopic peak to the total intensity of the entire isotopic cluster for the molecular species in question (“deisotoping”). The type II correction factor resolves the overlapping nature of the pseudomolecular ion peak of the species of interest with the M + 2 isotope peak from another species with a pseudomolecular ion of m/z value 2 Th lower than that for the first species (e.g., the second isotope of [M+Li]+ for 16:0/18:2-GPC has an m/z value identical to that of the monoisotopic [M+Li]+ ion for 16:0/18:1-GPC). Here we describe a strategy (derived from a deisotoping approach [35] developed by McLafferty) that simultaneously performs deisotoping and corrects isotopic overlap.
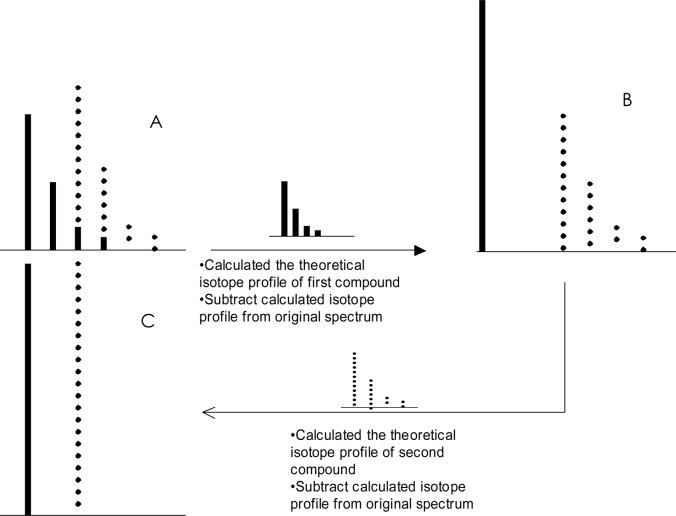
Figure 4 illustrates the deisotoping algorithm. ESI mass spectra can contain isotopic clusters of signals of different m/z values for the same molecular species with different charge states. Deconvoluting and deisotoping the spectrum reduces m/z values of all signals for a given species to a single monoisotopic value with z = 1 and simplifies interpretation. Raw data from an MS spectrum are processed by an adaptation of a method first developed for MALDI/TOF spectra [36], and the data are then further processed [37] by grouping peaks into clusters based on their charge state z (Figure 4A). The isotopic cluster peak spacing is 1/z. Deconvoluting spectra from high to low charge states avoids incorrect processing because intensities calculated for higher charge states fall between intensities of recorded signals [37].
Figure 4. Schematic illustration of deisotoping algorithms.
A, peaks are grouped into clusters based on their charge state z; B, theoretical isotope profile is calculated; and C, subtracted from the clustered spectrum to correct isotopic overlap
The deconvolution and deisotoping algorithm starts at the highest expected charge state zh for the processed spectrum and analyzes the spectrum from low to high m/z values in steps of one data point from the maximum expected charge Z = zh down to Z = 1 [37]. For each data point that contains signal, the corresponding mass is calculated as m/z × Z - (Z - 1). The theoretical isotope profile is calculated from the elemental formula identified from searching the fragment ion database or the theoretical formula database, and its contribution is subtracted from the clustered spectrum to correct isotopic overlap (Figure 4B). Monoisotopic singly charged signals are stored for each processed charge state in spectrum Sz. Combining calculated zh spectra Szh to S1 yields the final deconvoluted and deisotoped spectrum [37]. Deisotoped spectra from different charge states are combined, sorted, and used for quantitation (Figure 4C).
To illustrate how iterating deisotoping and deconvolution steps from high to low charge states resolves overlapping isotope clusters, consider a spectrum with two sets of singly charged ions that represent different molecular species: set A (m/z 650.5, 651.5, 652.5, 653.5) and set B (m/z 651.0, 652.0, 653,0, 654.0). The relative abundances of A and B in the mixture results in the peak series (expressed as m/z value/relative intensity): 650.5/100, 651.0/100, 651.5/36.9, 652.0/35.4, 652.5/8.6, 653.0/8.7, 653.5/1.5, 654.0/1.6. The ½ Th interval peak spacing suggests a doubly charged ion, and LipidQA first considers this possibility. The observed relative intensities of the ions in the series (650.5/100, 651.0/100, 651.5/36.9, 652.0/35.4) do not match the theoretical isotope profile, however, and LipidQA will not identify the ion series as a doubly charged but will proceed to the next lower charge state. The two ion series will thus be distinguished and deisotoped separately.
Table 2 illustrates the beneficial effects of deisotoping on the accuracy of quantitative measurements of standard samples by LipidQA. When amounts of two distinct compounds with overlapping isotope envelopes, e.g., 18:0/18:2-GPC ([M-H]− m/z 792) and 18:0/18:1-GPC ([M-H]− m/z 794), are similar, manual interpolation of raw intensity ratio data and LipidQA yield similar quantitative values. When amounts of two compounds differ greatly, the more abundant compound still yields a similar quantitative result both by manual analysis and LipidQA, but the amount of the less abundant compound is estimated less accurately by manual methods than by LipidQA, especially when the amount of the compound with the higher m/z value (e.g., 18:0/18:1-GPC, [M-H]− m/z 794) is lower than that of the compound with lower m/z value (e.g., 18:0/18:2-GPC, [M-H]− m/z 792). For example, with a sample that contained 16 μg of 18:0/18:2-GPC ([M-H]− m/z 792) and 3.2 μg of 18:0/18:1-GPC ([M-H]− m/z 794), the LipidQA quantitative estimate for 18:0/18:1-GPC was 3.5 ± 0.27 μg (relative error of 7.8% compared to the true value). Manual analysis yielded a quantitative estimate of 4.9 ± 2.7 μg for 18:0/18:1-GPC (relative error of 54.3%). The coefficient of variation of the manual method is thus 5.7-fold greater than that of the LipidQA program in this example.
Table 2.
Comparison of manual quantitation and LipidQA analysis. Samples were prepared with standard 18:0/18:2-GPC ([M+Li]+ m/z 792) and 18:0/18:1-GPC ([M+Li]+ m/z 794) in various relative abundances specified in the table. Data were acquired with a Finnigan LTQ mass spectrometer in positive ion mode.
| Sample NO. | 1 | 2 | 3 | 4 | 5 | 6 | 7 |
|---|---|---|---|---|---|---|---|
| Added 18:0/18:2-GPC (μg) | 1.6 | 3.2 | 8 | 16 | 16 | 16 | 16 |
| Manual Quantitation | 3.1(90.6) | 4.3(34.8) | 7.6(−4.9) | 15.4(−3.7) | 15.8(−1.1) | 15.5(−3.2) | 15.9(−0.8) |
| LipidQA | 3.0(88.8) | 4.3(34.8) | 8.0(−0.5) | 16.5(3.0) | 16.9(5.8) | 16.6(3.6) | 16.3(1.9) |
| Added 18:0/18:1-GPC (μg) | 16 | 16 | 16 | 16 | 8 | 3.2 | 1.6 |
| Manual Quantitation | 17.4(8.7) | 18.8(17.4) | 17.0(6.5) | 16.8(4.8) | 10.2(27.2) | 4.9(54.3) | 3.6(127.1) |
| LipidQA | 16.0(0.0) | 16.5(3.1) | 17.7(10.6) | 16.4(2.2) | 9.1(13.9) | 3.5(7.8) | 2.0(23.8) |
* Note: 2.8(80.4) means: calculated abundance (μg)/(Relative error (%))
Distinction between isomeric lipid species
The application of the deisotoping algorithm to the entire spectrum described above can determine the contribution of an isotope peak in the envelope of one compound to the intensity of an ion to which the pseudomolecular monoisotopic ion of a compound of higher molecular weight also contributes, as discussed above for 18:0/18:2-GPC and 18:0/18:1-GPC. In that case, the second [13C] isotope of the [M+Li]+ ion of 18:0/18:2-GPC has the same m/z value as the [M+Li]+ ion of 18:0/18:1-GPC (m/z 794).
Isomeric lipids are often encountered in biological extracts, and MS/MS data are required to distinguish them, as illustrated in spectra from a bacterial lipid extract (Figure 5). Peaks at m/z 849.5 and m/z 863.5 (Figure 5A) are identified by Lipid QA as [M-H]− ions of 18:0e/18:1-GPI (“e” denotes an sn-1 O-alkyl ether linkage to a fatty alcohol residue) and the diacyl species 18:0/18:1-GPI, respectively. For the small peak at m/z 861.42, two distinct, isomeric, diacyl species (18:0/18:2-GPI and 18:1/18:1-GPI) are identified from the MS/MS spectrum (Figure 5B) with similarly high ID scores (0.73 and 0.70, respectively, Table 3). Direct inspection of the MS/MS spectrum obtained from CAD of m/z 861.52 (Figure 5B) reveals product ions of m/z 279, 281 and 283 that represent carboxylate anions of linoleic (18:2), oleic (18:1), and stearic (18:0) acids, respectively. This reveals the presence of two isomers with different fatty acid substituents but the same number of total carbon atoms and double bonds, i.e., 18:0/18:2-GPI and 18:1/18:1-GPI. These assignments are supported by the presence of ions that reflect losses of 18:0, 18:1, and 18:2 as free fatty acids at m/z 577 ([M-H-R(18:0)CO2H]−), 579 ([M-H-R(18:1)CO2H]−), and 581([M-H-R(18:2)CO2H]−), respectively. LipidQA also computes the sum of intensities of ions (Table 3, third column) in the MS/MS spectrum attributable to each isomer to estimate their relative abundance.
Figure 5. Distinguishing different lipid molecular species with overlapping isotopic envelopes in a bacterial extract.

A, MS spectrum of a bacterial lipid extract in negative ion mode; B, MS/MS spectrum of peak at m/z 861.42. Data acquired with a Finnigan LTQ mass spectrometer.
Table 3.
Fragment ion database searching result for the MS/MS spectrum of m/z 861.42 from Figure 5A.
| Lipid | ID Score | MS/MS TIC |
|---|---|---|
| 14:0/22:2-GPI | 0.12 | 138 |
| 14:1/22:1-GPI | 0.12 | 138 |
| 14:2/22:0-GPI | 0.12 | 139 |
| 16:0/20:2-GPI | 0.19 | 138 |
| 16:1/20:1-GPI | 0.19 | 139 |
| 16:2/20:0-GPI | 0.19 | 138 |
| 18:0/18:2-GPI | 0.73 | 580 |
| 18:1/18:1-GPI | 0.70 | 533 |
| 20:0/20:0-GPG | 0.15 | 116 |
Profiling glycerophospholipid species by data-dependent transition from MS to MS/MS scanning and LipidQA analyses
Table 4 summarizes an experiment in which porcine brain lipid extracts were directly infused into the ESI source of a Q-TOF Micro mass spectrometer after adding internal standards and the quality control standard 17:0/17:0-GPS ([M-H]− m/z 762.5). Spectra were acquired in both negative and positive ion modes in a data-dependent manner, and data were processed to generate peak list files. LipidQA identified and quantitated a total of 36 distinct endogenous glycerophospholipid molecular species (Table 4), including 8 GPC, 10 GPE, 14 GPS, 2 GPI and 2 GPA lipid species with ID scores exceeding 0.5. Consistent with specifications provided by the supplier of the brain extract (Avanti, Alabaster, AL), GPE lipids accounted for 43% of the measured lipid phosphorus content; GPC lipids were the next most abundant components; and GPS lipid species accounted for 16% of total lipid phosphorus content and exhibited the greatest heterogeneity with 14 distinct molecular species (Table 4).
Table 4.
Glycerophospholipid profile of a lipid extract from porcine brain. ESI/MS(/MS) data for GPE, GPS, GPA, GPI were acquired with a Micromass Q-TOF Micro instrument in negative ion mode. GPC lipids were analyzed as Li+ adducts in positive ion mode. Mean values (± C.V.) are displayed (n=9). The designation "p" in p18:0/20:4-GPE denotes "plasmalogen," which is a glycerophospholipid with a 1-O-alk-1'-enyl (plasmenyl) linkage to a fatty aldehyde residue in the sn-1 position.
| Lipids | pmol/nmol Phosphate | Relative STDEV(%) | Lipid class/total |
|---|---|---|---|
| 18:0/18:1-GPA(701.5) | 2.58 | 12.40 | |
| 16:0/20:1-GPA(701.5) | 1.59 | 25.16 | GPA total 0.33% |
| 16:1/18:1-GPE(714.5) | 200.23 | 6.13 | |
| 18:0/18:1-GPE(744.5) | 65.48 | 2.73 | |
| 18:0/20:4-GPE(766.5) | 8.55 | 12.87 | |
| 18:1/20:1-GPE(770.5) | 40.51 | 6.37 | |
| 18:0/22:5-GPE(792.5) | 20.79 | 2.99 | |
| 18:0/22:4-GPE(794.5) | 24.86 | 10.05 | |
| p16:0/18:1-GPE(700.5) | 1.31 | 30.53 | |
| p16:0/20:4-GPE(722.5) | 18.32 | 15.61 | |
| p18:1/18:1-GPE(726.5) | 11.10 | 20.72 | |
| p18:0/20:4-GPE(750.5) | 26.80 | 7.46 | GPE total 43.54% |
| 18:0/18:1-GPI(863.5) | 2.56 | 32.06 | |
| 18:0/20:4-GPI(885.4) | 40.57 | 7.64 | GPI total 4.49% |
| 16:0/18:1-GPS(760.5) | 4.62 | 38.96 | |
| 18:0/18:1-GPS(788.5) | 33.93 | 9.14 | |
| 16:0/22:6-GPS(806.5) | 12.40 | 8.49 | |
| 18:0/20:4-GPS(810.5) | 8.47 | 10.50 | |
| 16:0/22:4-GPS(810.5) | 3.49 | 9.18 | |
| 18:1/20:1-GPS(814.5) | 3.22 | 13.04 | |
| 18:0/20:2-GPS(814.5) | 3.01 | 7.64 | |
| 18:0/20:1-GPS(816.5) | 5.16 | 18.80 | |
| 20:0/18:1-GPS(816.5) | 4.23 | 17.73 | |
| 18:0/20:0-GPS(818.5) | 2.11 | 30.33 | |
| 20:0/22:6-GPS(818.5) | 3.13 | 6.51 | |
| 18:0/22:6-GPS(834.4) | 48.21 | 9.96 | |
| 18:0/22:4-GPS(838.5) | 15.45 | 7.77 | |
| 22:6/22:6-GPS(878.4) | 9.45 | 8.40 | GPS total 16.32% |
| 16:0/18:1-GPC(766.5) | 105.87 | 5.12 | |
| 16:0/16:1-GPC(738.5) | 31.19 | 4.17 | |
| 16:0/16:0-GPC(740.5) | 66.01 | 7.27 | |
| 16:0/20:4-GPC(788.5) | 20.15 | 13.05 | |
| 18:0/18:2-GPC(792.5) | 19.77 | 17.45 | |
| 18:0/18:1-GPC(794.5) | 43.79 | 9.13 | |
| 16:0/22:6-GPC(812.5) | 10.10 | 13.86 | |
| 18:0/20:4-GPC(816.6) | 41.99 | 8.79 | GPC total 35.31% |
Statistical reproducibility of LipidQA measurements are reflected by the standard errors in Table 4, and coefficients of variation are generally less than 10% for components of mixtures that are at least 10% as abundant as the most abundant component. Greater variation is encountered for minor components. For example, the coefficient of variation is 38% for 16:0/18:1-GPS, which about 2% as abundant as 16:1/18:1-GPE in the mixture in Table 4. In additional evaluation of the ability of LipidQA to identify and provide reproducibly accurate quantitative measurements of lipid species, varied amounts of synthetic 17:0/17:0-GPS standard (2, 4, or 8 μg) were added to a bovine brain lipid extract and analyzed with endogenous glycerophospholipid species by LipidQA, which correctly identified the peak at m/z 762.5 as the [M-H]− ion of 17:0/17:0-GPS and accurately quantitated it with a relative error of only 3.8%, 1.1% and 3.1%, respectively, on triplicate measurements (not shown).
CONCLUSION AND PERSPECTIVE
We describe a computer software platform (LipidQA) to process electronic data automatically from ESI/MS(/MS) analyses of complex lipid mixtures, and it can identify and quantitate multiple molecular species in mixtures that contain several classes of lipids in less than 15 minutes. In contrast to other recently described programs for analyzing lipid MS data that are applicable only to specific mass spectrometers, LipidQA can process high throughput MS data acquired with ion trap, triple quadrupole, or quadrupole time-of-flight instruments. Quantitation directly from accumulated spectra obtained after direct infusion of lipid mixtures avoids variability introduced by multiple sample handling and chromatographic steps and also avoids differences in efficiency of formation of selected ions entailed in neutral loss or product ions scans.
The major limitations of the current version of LipidQA are low precision of quantitation and low sensitivity for identification of minor components of mixtures containing multiple components present is widely different amounts. Because switching from MS to MS/MS data acquisition is triggered by the signal to noise intensity of the MS ion current, some low abundance components fail to be detected. In addition, ion suppression effects with analyses of complex lipid mixtures by direct infusion precludes accurate quantitation of all components. Nonetheless, this approach is useful for high throughput lipid profiling of the major species in biological extracts when suppression of minor components is not considered a critical issue because it greatly accelerates data processing compared to laborious direct inspection.
Our current fragment ion database includes reference spectra for the major glycerophospholipid head-group classes, and it is planned to incorporate reference spectra for fragmentation of other lipid species, including sphingolipids, into future versions, which will extend its applicability.
ACKNOWLEDGMENTS
We thank Alan Bohrer for excellent technical assistance. This work was supported by United States Public Health Service Grants R37-DK34388, P41-RR00954, P60-DK20579, RO1-69455, and P30-DK56341.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Supplementary Material
REFERENCES
- 1.Fahy E, Subramaniam S, Brown HA, Glass CK, Merrill AH, Murphy RC, Jr., Raetz CRH, Russell DW, Seyama Y, Shaw W, Shimizu T, Spener F, van Meer G, VanNieuwenhze MS, White SH, Witztum JL, Dennis EA. A comprehensive classification system for lipids. J. Lipid Res. 2005;46:839–862. doi: 10.1194/jlr.E400004-JLR200. [DOI] [PubMed] [Google Scholar]
- 2.Murphy RC, Fiedler J, Hevko J. Analysis of nonvolatile lipids by mass spectrometry. Chem. Rev. 2001;101:479–526. doi: 10.1021/cr9900883. [DOI] [PubMed] [Google Scholar]
- 3.Han X, Gross RW. Shotgun lipidomics: Electrospray ionization mass spectrometric analysis and quantitation of cellular lipidomes directly from crude extracts of biological samples. Mass Spectrom. Rev. 2005;24:367–412. doi: 10.1002/mas.20023. [DOI] [PubMed] [Google Scholar]
- 4.Han X, Gubitosi-Klug RA, Collins BJ, Gross RW. Alterations in individual molecular species of human platelet phospholipids during thrombin stimulation: Electrospray ionization mass spectrometry-facilitated identification of the boundary conditions for the magnitude and selectivity of thrombin-induced platelet phospholipid hydrolysis. Biochemistry. 1996;35:5822–5832. doi: 10.1021/bi952927v. [DOI] [PubMed] [Google Scholar]
- 5.Pulfer M, Murphy RC. Electrospray mass spectrometry of phospholipids. Mass Spectrom. Rev. 2003;22:332–364. doi: 10.1002/mas.10061. [DOI] [PubMed] [Google Scholar]
- 6.Ekroos K, Chernushevich IV, Simons K, Shevchenko A. Quantitative profiling of phospholipids by multiple precursor ion scanning on a hybrid quadrupole time-of-flight mass spectrometer. Anal. Chem. 2002;74:941–949. doi: 10.1021/ac015655c. [DOI] [PubMed] [Google Scholar]
- 7.Schneiter R, Brugger B, Sandhoff R, Zellnig G, Leber A, Lampl M, Athenstaedt K, Hrastnik C, Eder S, Daum G, Paltauf F, Wieland FT, Kohlwein SD. Electrospray ionization tandem mass spectrometry (ESI-MS/MS) analysis of the lipid molecular species composition of yeast subcellular membranes reveals acyl chain-based sorting/remodeling of distinct molecular species en route to the plasma membrane. J. Cell Biol. 1999;146:741–754. doi: 10.1083/jcb.146.4.741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Leavell MD, Leary JA. Fatty Acid Analysis Tool (FAAT): An FT-ICR MS lipid analysis algorithm. Anal. Chem. 2006;78:5497–5503. doi: 10.1021/ac0604179. [DOI] [PubMed] [Google Scholar]
- 9.Ejsing CS, Duchoslav E, Sampaio J, Simons K, Bonner R, Thiele C, Ekroos K, Shevchenko A. Automated identification and quantification of glycerophospholipid molecular species by multiple precursor ion scanning. Anal. Chem. 2006;78:6202–6214. doi: 10.1021/ac060545x. [DOI] [PubMed] [Google Scholar]
- 10.Schwudke D, Oegema J, Burton L, Entchev E, Hannich JT, Ejsing CS, Kurzchalia T, Shevchenko A. Lipid profiling by multiple precursor and neutral loss scanning driven by the data-dependent acquisition. Anal. Chem. 2006;78:585–595. doi: 10.1021/ac051605m. [DOI] [PubMed] [Google Scholar]
- 11.Hsu FF, Bohrer A, Turk J. Formation of lithiated adducts of glycerophosphocholine lipids facilitates their identification by electrospray ionization tandem mass spectrometry. J. Am. Soc. Mass Spectrom. 1998;9:516–526. doi: 10.1016/S1044-0305(98)00012-9. [DOI] [PubMed] [Google Scholar]
- 12.Hsu FF, Turk J. Structural characterization of triacylglycerols as lithiated adducts by electrospray ionization mass spectrometry using low energy collisionally activated dissociation on a triple stage quadrupole instrument. J. Am. Soc. Mass Spectrom. 1999;10:587–600. doi: 10.1016/S1044-0305(99)00035-5. [DOI] [PubMed] [Google Scholar]
- 13.Hsu FF, Turk J. Distinction among isomeric unsaturated fatty acids as lithiated adducts by electrospray ionization mass spectrometry using low energy collisionally activated dissociation on a triple stage quadrupole instrument. J. Am. Soc. Mass Spectrom. 1999;10:600–613. doi: 10.1016/S1044-0305(99)00041-0. [DOI] [PubMed] [Google Scholar]
- 14.Hsu FF, Turk J. Characterization of phosphatidylethanolamine molecular species as lithiated adducts by triple quadrupole tandem mass spectrometry with electrospray ionization. J. Mass Spec. 2000;35:596–606. doi: 10.1002/(SICI)1096-9888(200005)35:5<595::AID-JMS965>3.0.CO;2-D. [DOI] [PubMed] [Google Scholar]
- 15.Hsu FF, Turk J. Charge-driven fragmentation processes in diacyl glycerophosphatidic acids upon low-energy collisional activation. A mechanistic proposal. J. Mass Spec. 2000;11:797–803. doi: 10.1016/S1044-0305(00)00151-3. [DOI] [PubMed] [Google Scholar]
- 16.Hsu FF, Turk J. Charge-remote and charge-driven fragmentation processes in diacyl glycerophosphoethanolamine upon low-energy collisional activation: A mechanistic proposal. J. Am. Soc. Mass Spectrom. 2000;11:892–899. doi: 10.1016/S1044-0305(00)00159-8. [DOI] [PubMed] [Google Scholar]
- 17.Hsu FF, Turk J. Characterization of phosphatidylinositol, phosphatidylinositol-4-phosphate, and phosphatidylinositol-4,5-bisphosphate by electrospray ionization tandem mass spectrometry. A mechanistic study. J. Am. Soc. Mass Spectrom. 2000;11:986–999. doi: 10.1016/S1044-0305(00)00172-0. [DOI] [PubMed] [Google Scholar]
- 18.Hsu FF, Turk J. Studies on phosphatidylglycerol with triple quadrupole mass spectrometry with electrospray ionization. Fragmentation processes and structural characterization. J. Am. Soc. Mass Spectrom. 2001;12:1036–1043. [Google Scholar]
- 19.Hsu FF, Turk J. Electrospray ionization/tandem quadrupole mass spectrometric studies on phosphatidylcholines: The fragmentation processes. J Am. Soc. Mass Spectrom. 2003;14:352–363. doi: 10.1016/S1044-0305(03)00064-3. [DOI] [PubMed] [Google Scholar]
- 20.Hsu FF, Turk J, Thukkani AK, Messner MC, Wildsmith KR, Ford DA. Characterization of alkylacyl-, alk-1-enylacyl, and lyso subclasses of glycerophosphocholine lipids by tandem quadrupole mass spectrometry with electrospray ionization. J. Mass Spectrom. 2003;38:752–763. doi: 10.1002/jms.491. [DOI] [PubMed] [Google Scholar]
- 21.Hsu FF, Turk J, Shi Y, Groisman EA. Characterization of acylphosphatidylglycerols from Salmonella typhimurium by tandem mass spectrometry with electrospray ionization. J. Am. Soc. Mass Spectrom. 2004;15:1–11. doi: 10.1016/j.jasms.2003.08.006. [DOI] [PubMed] [Google Scholar]
- 22.Hsu FF, Turk J, Rhoades ER, Russell DG, Shi Y, Groisman EA. Structural characterization of cardiolipin by tandem quadrupole and multiple stage quadrupole ion-trap mass spectrometry with electrospray ionization. J. Am. Soc. Mass Spectrom. 2005;16:491–504. doi: 10.1016/j.jasms.2004.12.015. [DOI] [PubMed] [Google Scholar]
- 23.Hsu FF, Turk J. Structural studies on phosphatidylserine by tandem quadrupole and multiple stage quadrupole ion-trap mass spectrometry with electrospray ionization. J. Am. Soc. Mass Spectrom. 2005;16:1510–1522. doi: 10.1016/j.jasms.2005.04.018. [DOI] [PubMed] [Google Scholar]
- 24.Hsu FF, Turk J. Characterization of cardiolipin from Escherichia coli by electrospray ionization with multiple stage quadrupole ion-trap mass spectrometric analysis of [M-2H+Na]- ions. J. Am. Soc. Mass Spectrom. 2006;17:420–429. doi: 10.1016/j.jasms.2005.11.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hsu FF, Turk J. Characterization of cardiolipins as sodiated adducts by positive-ion electrospray ionization with multiple stage quadrupole ion-trap mass spectrometry. J. Am. Soc. Mass Spectrom. 2006;17:1146–57. doi: 10.1016/j.jasms.2006.04.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hsu FF, Turk J, Owens RM, Rhoades ER, Russell DG. Structural characterization of phosphatidyl-myo-inositol mannosides from Mycobacterium bovis Bacillus Calmette Guerin by multiple-stage quadrupole ion-trap mass spectrometry with electrospray ionization. I . PIMs and Lyso-PIMs. J. Am. Soc. Mass Spectrom. 2007;18:466–478. doi: 10.1016/j.jasms.2006.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hsu FF, Turk J, Owens RM, Rhoades ER, Russell DG. Structural characterization of phosphatidyl-myo-inositol mannosides from Mycobacterium bovis Bacillus Calmette Guerin by multiple-stage quadrupole ion-trap mass spectrometry with electrospray ionization. II . PIMs and Lyso-PIMs. J. Am. Soc. Mass Spectrom. 2007;18:479–492. doi: 10.1016/j.jasms.2006.10.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hsu FF, Turk J, Williams TD, Welti R. Characterization of phosphatidylglycerol in plant chloroplasts by multiple stage quadrupole ion-trap and tandem quadrupole mass spectrometry with electrospray ionization. J. Am. Soc. Mass Spectrom. 2007;18:783–790. doi: 10.1016/j.jasms.2006.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hsu FF, Turk J. Mechanisms for phospholipid fragmentation during electrospray ionization mass spectrometry with collision-induced dissociation. In: Byrdwell WC, editor. Modern Methods for Lipid Analysis by Liquid Chromatography-Mass Spectrometry and Related Techniques. American Oil Chemists Society Press; Champaign, IL: 2005. pp. 61–179. [Google Scholar]
- 30.Hsu FF, Ma Z, Wohltmann M, Bohrer A, Ramanadham S, Turk J. Electrospray ionization mass spectrometric analysis of human promonocytic U937 cell glycerolipids and evidence that differentiation is associated with membrane lipid composition changes that facilitate phospholipase A2 activation. J. Biol. Chem. 2000;275:16579–16589. doi: 10.1074/jbc.M908342199. [DOI] [PubMed] [Google Scholar]
- 31.Bao S, Bohrer A, Ramanadham S, Jin W, Zhang S, Turk J. Effects of stable suppression of Group VIA phospholipase A2 expression on phospholipid content and composition, insulin secretion, and proliferation of INS-1 insulinoma cells. J. Biol. Chem. 2006;281:187–198. doi: 10.1074/jbc.M509105200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hallberg A. Effects of starvation and different culture conditions on the phospholipid content of isolated pancreatic islets. Biochim. Biophys. Acta. 1984;796:328–335. doi: 10.1016/0005-2760(84)90134-6. [DOI] [PubMed] [Google Scholar]
- 33.MacCoss MJ, Wu CC, Yates JR. Probability-based validation of protein identifications using a modified SEQUEST algorithm. Anal. Chem. 2002;74:5593–5599. doi: 10.1021/ac025826t. [DOI] [PubMed] [Google Scholar]
- 34.Perkins DN, Pappin DJ, Creasy DM, Cottrell JS. Probability-based protein identification by searching sequence databases using mass spectroscopy data. Electrophoresis. 1999;20:3551–3567. doi: 10.1002/(SICI)1522-2683(19991201)20:18<3551::AID-ELPS3551>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 35.Horn DM, Zubarev RA, McLafferty FW. Automated reduction and interpretation of high resolution electrospray mass spectra of large molecules. J. Am. Soc. Mass Spectrom. 2000;11:320–332. doi: 10.1016/s1044-0305(99)00157-9. [DOI] [PubMed] [Google Scholar]
- 36.Samuelsson J, Dalevi D, Levander F, Rognvaldsson T. Modular scriptable and automated analysis tools for high-throughput peptide mass fingerprinting. Bioinformatics. 2004;20:3628–3635. doi: 10.1093/bioinformatics/bth460. [DOI] [PubMed] [Google Scholar]
- 37.Wehofsky M, Hoffman R. Automated deconvolution and deisotoping of electrospray mass spectra. J. Mass Spectrom. 2002;37:223–229. doi: 10.1002/jms.278. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.