Abstract
Cystic fibrosis (CF) is an autosomal recessive disorder, the most common lethal genetic disease in Caucasians. Respiratory disease is the major cause of morbidity and mortality. Indeed, 95% of CF patients die of respiratory failure. Pseudomonas aeruginosa, an opportunistic pathogen, chronically infects the lungs of over 85% of CF patients. It is ineradicable by antibiotics and responsible for airway mucus overproduction that contributes to airway obstruction and death. The molecular mechanisms underlying this pathology are unknown. Here we show that P. aeruginosa activates a c-Src-Ras-MEK1/2-MAPK-pp90rsk signaling pathway that leads to activation of nuclear factor NF-κB (p65/p50). Activated NF-κB binds to a κB site in the 5′-flanking region of the MUC2 gene and activates MUC2 mucin transcription. These studies bring new insight into bacterial–epithelial interactions and more specifically into the molecular pathogenesis of cystic fibrosis. Understanding these signaling and gene regulatory mechanisms opens up new therapeutic targets for cystic fibrosis.
Cystic fibrosis (CF) is the most common lethal genetic disease in the Caucasian population (1). The mutation responsible for the disease is in the gene encoding the cystic fibrosis transmembrane regulator (CFTR), a chloride channel. This mutation gives rise to a chain of events involving chronic bacterial lung infection and mucus overproduction. Although there have been significant clues regarding the link between the mutation and chronic infection (2–4), the nature of the link between the mutation and mucus overproduction is completely unknown.
We recently showed that bacterial exoproducts up-regulate epithelial MUC2 mucin transcription (5). This suggests that CF-associated mucin overproduction occurs secondary to lung infection, a sine qua non of advanced disease. The molecular mechanisms underlying mucin induction are unknown. In the present study, we performed experiments to examine these mechanisms. Results showed that Pseudomonas aeruginosa, a common CF pathogen, activates the MUC2 mucin gene transcription by activation of a Src-dependent Ras-MEK1/2-ERK1/2-pp90rsk-NF-κB pathway. These studies provide insight into bacterial–host epithelial interactions and open up new therapeutic targets for CF.
MATERIALS AND METHODS
Reagents.
P. aeruginosa lipopolysaccharide (LPS) from serotype 10 was purchased from Sigma. PP1, PD98059, and caffeic acid phenethyl ester (CAPE) were purchased from Calbiochem.
Bacterial Strains and Culture Conditions.
The P. aeruginosa strains used in these studies were grown in M9 medium with aeration at 37°C to late logarithmic phase. The broth cultures were then centrifuged at 10,000 rpm in a Sorvall RC5C for 50 min. The supernatants containing bacterial exoproducts were sterilized by passage through a 0.22-μm polymer filter (Corning) and were then kept at −80°C until use. Bacterial culture supernatants were added to epithelial cell culture medium at a 1:4 dilution ratio.
Cell Culture.
Two MUC2-expressing epithelial cell lines were used in these studies. NCIH292 cells (a MUC2-expressing airway epithelial cell line) were maintained in RPMI 1640 medium. HM3 cells (a MUC2-expressing colon epithelial cell line) were maintained in Dulbecco’s modified Eagle’s medium. All media received additions of 10% fetal bovine serum, 100 units/ml penicillin, and 0.1 mg/ml streptomycin.
RNase Protection Assay (RPA).
The experiments were carried out with an RNA probe containing MUC2-specific sequence as described previously (5).
Cloning and Sequencing of the 5′-Flanking Region of Human MUC2 Gene, Plasmid Construction, Transfection, and Luciferase Assay.
The 5′-flanking region of the human MUC2 gene was cloned by screening a human placental 1FIXII genomic library, using the 5′ region of human MUC2 cDNA as the probe as described (5–7). The 5′-flanking region was sequenced by dideoxynucleotide sequencing. Deletional mutants of the 5′-flanking region DNA were obtained by combining restriction digestion of the upstream region of the gene and PCR amplification. The restriction DNA fragments or PCR-amplified fragments were ligated into a luciferase reporter gene. All junctions and identifications of the DNA sequences in the chimeric constructs were confirmed by DNA sequencing. The expression plasmid of a dominant-negative mutant form (pp90rskΔC) of the 90-kDa ribosomal S6 kinase (pp90rsk) was a gift from Warner Greene (Univ. of California, San Francisco). v-Src was a gift from J. Michael Bishop (University of California, San Francisco). SrcRF was a gift from Joan Brugge (Harvard Medical School, Boston). RasN17 was a gift from G. Cooper (Harvard Medical School). HMEK1 and HMEK1(K97R) were gifts from Alan Saltiel (Parke-Davis Pharmaceutical Research Division, Ann Arbor, MI). JNKK(K116R) was a gift from Michael Karin (Univ. of California, San Francisco). Transfection was performed by a standard electroporation method as described (5). P. aeruginosa culture supernatant and LPS were added to the transfected cells 42 hr after transfection. After 6 hr, the cells were harvested for luciferase assay. All transfections were carried out in triplicate. Luciferase activity was normalized with respect to β-galactosidase activity.
Electrophoretic Mobility-Shift Assay (EMSA).
Nuclear extracts from HM3 and NCIH292 cells were prepared according to ref. 8. Prior to extraction, cells were treated with P. aeruginosa culture supernatant for 6 hr. The protein concentration of the cell extract was determined using a bicinchoninic acid protein assay kit (Pharmacia LKB Biotechnology) using bovine albumin as standard. Various double-stranded oligonucleotide probes as indicated in each experiment were synthesized on the basis of the results of luciferase assay experiments described in Results. The oligonucleotide was labeled by [γ-32P]ATP and T4 polynucleotide kinase and purified on a nondenaturing 6% polyacrylamide gel. The probe was incubated at 37°C for 20 min with nuclear extract (5–10 μg of protein) in a solution containing 3 μg of poly(dI-dC) in 10 mM Hepes–KOH at pH 7.9, 210 mM NaCl, 0.75 mM MgCl2, 0.1 mM EDTA, 0.5 mM dithiothreitol, 0.5 mM PMSF, and 12.5% (vol/vol) glycerol. For antibody interaction, antibodies specific for CCATT-enhancer-binding protein (C/EBP), NF-κB p65, NF-κB p50, and Rel-C were added to the reaction during a 30-min preincubation on ice. Samples were applied to polyacrylamide gels under native conditions in high ionic strength buffer. The dried gel was exposed to x-ray film at −70°C for 2–12 hr with double intensifying screens.
RESULTS AND DISCUSSION
Activation of NF-κB Is Required for MUC2 Mucin Induction by P. aeruginosa.
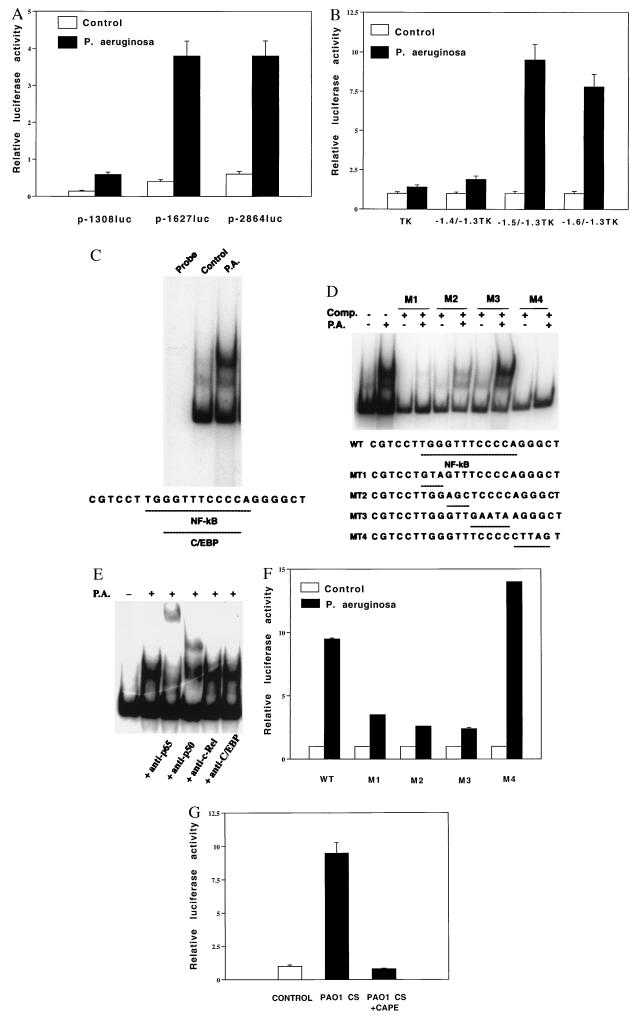
Although we previously showed that P. aeruginosa up-regulates MUC2 mucin gene transcription (5), the signal transduction pathways and transcriptional regulatory mechanisms are still unknown. Our first objective was to define P. aeruginosa-response elements in the 5′-flanking region of the MUC2 mucin gene and related transcription factors. Analysis of luciferase activity from a panel of MUC2-luciferase reporter gene deletion mutants revealed a response element between base pairs −1627 and −1308 (Fig. 1A). To more precisely define the P. aeruginosa-response element, we made heterologous constructs in which human MUC2 promoter base pair −1628/−1307, −1528/−1307, and −1430/−1307 regions were subcloned upstream of the thymidine kinase (TK) promoter. As shown in Fig. 1B, this suggested that the response element resides in the base pair −1528/−1430 region. To more accurately define the response element, we next performed EMSAs, using as probes 32P-labeled synthesized oligonucleotides corresponding to base pairs −1528/−1486 (A1) and −1485/−1430 (A2) and nuclear proteins from P. aeruginosa-treated and untreated HM3 and NCIH292 epithelial cells. A new protein complex was formed with probe A2, but not with A1 (data not shown). When we further performed EMSAs using oligonucleotides corresponding to base pairs −1485/−1459 (B1) and −1458/−1430 (B2), this protein complex was formed with probe B2, but not with B1 (Fig. 1C). Sequence analysis showed that this region contains overlapping C/EBP and NF-κB sites. Selective mutagenesis of these sites in nonradiolabeled probes disrupted competition for protein binding to radiolabeled wild-type probe, suggesting that NF-κB and/or C/EBP binding is involved in MUC2 mucin gene activation (Fig. 1D).
Figure 1.
Characterization of NF-κB site and NF-κB in mucin MUC2 induction by P. aeruginosa. (A) Human MUC2 regulatory regions (base pairs −2864 to +14, −1627 to +14, and −1308 to +14) were subcloned upstream of a luciferase reporter gene in pGL2 basic vector and transfected into HM3 cells. (B) Human MUC2 regulatory regions (base pairs −1628 to −1307, −1528 to −1307, and −1430 to −1307) were subcloned upstream of a TK-32 promoter luciferase vector (named as −1.6/−1.3TK, −1.5/−1.3TK, and −1.4/−1.3TK, respectively) and transfected into HM3 cells. In both A and B, transfected cells were treated with either P. aeruginosa culture supernatants or vehicle for 6 hr prior to cell lysis. Luciferase activity was then assessed in P. aeruginosa-treated and nontreated cells. Nuclear proteins were incubated with a probe consisting of the 32P-labeled double-stranded oligonucleotides corresponding to the human MUC2 regulatory region base pairs −1458/−1430 in the absence (C) or presence of various mutant unradiolabeled oligonucleotides (M1 to M4) as indicated (D), or preincubated with antibodies to NF-κB p65, NF-κB p50, c-Rel, or C/EBP (E), and were subjected to EMSA. Probe incubation was with nuclear extracts (10 μg) from HM3 cells either treated or untreated with P. aeruginosa culture supernatant as indicated at the top. (F) Human MUC2 regulatory regions (base pairs −1528 to −1430) containing the wild type (WT) or various mutated sites within the region −1458/−1430 as indicated above were subcloned upstream of the TK-32 promoter in a luciferase vector (named as M1 to M4)) and transfected into HM3 cells. Transfected cells were treated with either P. aeruginosa culture supernatants or vehicle for 6 hr prior to cell lysis. Luciferase activity was then assessed in P. aeruginosa-treated and untreated cells. (G) Effect of NF-κB inhibitor CAPE. A human MUC2 construct p-2864luc was transfected into HM3 cells. Transfected cells were pretreated with or without CAPE (15 μg/ml) for 1 hr before P. aeruginosa treatment as indicated. In all the experiments shown above, transfections were carried out in triplicate. Values are the means ± SD; n = 3. Similar results were observed in another MUC2-expressing epithelial cell line, NCIH292.
Pursuing the possibility that NF-κB is indeed involved in MUC2 induction, we performed supershift assays using anti-NF-κB (p65, p50, and c-Rel) antibodies and an anti-C/EBPβ antibody reactive with α, β, δ, and CRP1. The NF-κB p65 and p50 antibodies supershifted the protein complex but the c-Rel and C/EBP antibodies did not (Fig. 1E), indicating that a NF-κB p65/p50 heterodimer accounts for the observed protein binding. That mutation of the NF-κB site (Fig. 1F) and that CAPE, an inhibitor of NF-κB-DNA binding (Fig. 1G) (9), abolished responsiveness of the wild-type MUC2 luciferase construct confirmed that NF-κB binding to the κB site in the regulatory region of MUC2 gene was critical for the up-regulation of the transcriptional activity of MUC2 induced by P. aeruginosa.
pp90rsk Mediates the Activation of NF-κB by P. aeruginosa.
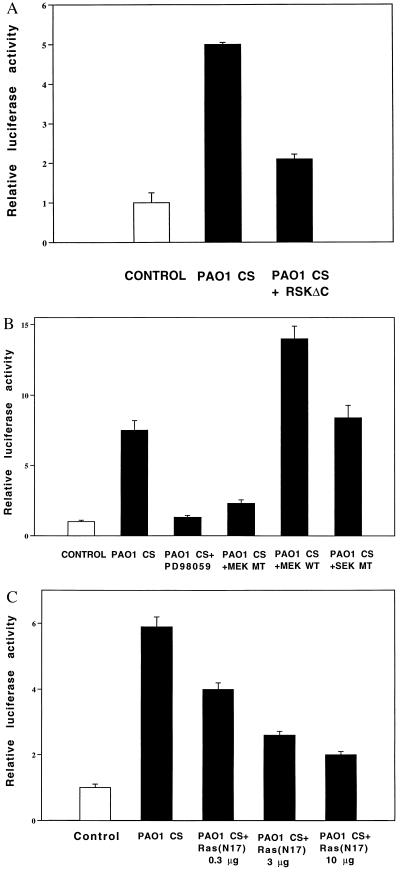
Despite extensive analysis of NF-κB by previous investigators, the signaling events upstream of its activation are poorly understood. We took advantage of the fact that the identity of some of the kinases responsible for phosphorylating the NF-κB-tethering protein IκB has recently been identified (10–15). Cotransfecting HM3 and NCIH292 cells with a dominant-negative mutant expression plasmid for the IκBα kinase pp90rsk (pp90rskΔC) prior to P. aeruginosa exposure reduced the response by approximately 70% (Fig. 2A). This finding indicates that P. aeruginosa up-regulation of MUC2 transcription is pp90rsk dependent.
Figure 2.
Involvement of Ras, MEK1/2, and pp90rsk in MUC2 mucin induction by P. aeruginosa. (A) Effect of coexpressing a dominant-negative mutant form of pp90rsk (pp90rskΔC) on P. aeruginosa-induced MUC2 up-regulation. p-2864luc (15 μg) was cotransfected with a dominant-negative mutant form of pp90rsk into HM3 cells. Forty hours later, HM3 cells were exposed to PAO1 culture supernatant (CS) 6 hr before harvesting. Luciferase activity was measured as described in the text. (B) Effect of PD98059 coexpressing a dominant-negative mutant form and wild-type form of MEK1/2 and a dominant-negative mutant form of SEK [JNKK9(K116R)] on P. aeruginosa-induced MUC2 up-regulation. HM3 cells were cotransfected with p-2864luc (15 μg) and a dominant-negative mutant form (3 μg) or a wild-type form (3 μg) of MEK1/2. Forty hours after transfection, HM3 cells transfected with only p-2864luc were pretreated with PD98059 (37 μM) (Calbiochem) for 30 min. All the cells were then exposed to PAO1 culture supernatant (CS) 6 hr before harvesting. (C) Effect of coexpressing a dominant-negative mutant form of Ras (RasN17) on P. aeruginosa-induced MUC2 up-regulation. HM3 cells were cotransfected with p-2864luc (15 μg) and a dominant-negative mutant form of Ras. The cells were then treated with PAO1 culture supernatant and lysed for luciferase assay. Similar results were observed in another MUC2-expressing epithelial cell line, NCIH292. In the experiments described above, all transfections were carried out in triplicate. Values are the means ± SD; n = 3.
The MEK1/2-ERK1/2 Pathway, but Not the SEK-JNK Pathway, Is Involved in MUC2 Mucin Induction by P. aeruginosa.
The signaling molecules upstream of the IκB kinases, predicted to vary with the nature of the activating stimulus, are still unknown. pp90rsk had previously been recognized as a mitogen-activated, serine/threonine ribosomal S6 kinase involved in the transduction of signals induced by stimuli that activate the Ras-Raf1-MEK1/2-ERK1/2 cascade (16, 17). Consistent with this, our earlier results had shown response sensitivity to the ERK1/2 inhibitor tyrphostin AG126 (5). Together, the data strongly suggested involvement of one or more MAP kinase pathways and led us to test the effect of inhibitors of MEK1/2, the kinase immediately upstream of ERK1/2, and SEK/JNKK, the kinase immediately upstream of c-Jun N-terminal kinase (JNK). As shown in Fig. 2B, cotransfection of HM3 and NCIH292 cells with a dominant-negative mutant expression plasmid of MEK1/2 [HMEK1 (K97R)] but not a dominant-negative mutant form of SEK [JNKK(K116R)] (18) reduced the response by approximately 60%. Consistent with this, preincubation of the cells with PD98059 (19), a specific chemical inhibitor of MEK1/2, abolished the response, and overexpression of wild-type MEK1 greatly enhanced the response. Together these results indicate that P. aeruginosa up-regulates MUC2 transcription by activation of the classical MEK1/2-ERK1/2 pathway in MUC2-expressing epithelial cells.
The Src-Dependent Ras Pathway Upstream of MEK1/2-ERK1/2-NF-κB Is Involved in P. aeruginosa-Induced MUC2 Up-Regulation.
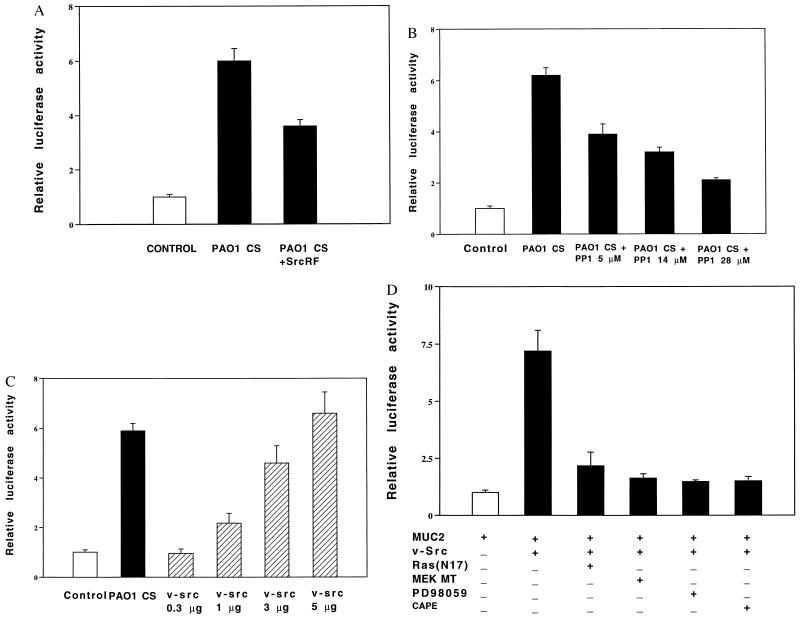
A key issue is how P. aeruginosa activates MEK1/2. On the basis of evidence that Ras is upstream of Raf-1-MEK1/2-ERK1/2, that c-Src is upstream of Ras (20–24), and that Src mediates bacterial-triggered signaling in macrophages, we investigated the involvement of Ras and Src in the P. aeruginosa response in epithelial cells. Cotransfection with a dominant-negative mutant form of Ras (RasN17) (25) reduced the response by 80% (Fig. 2C), and cotransfection with a dominant-negative mutant form of c-Src (SrcRF) inhibited the response by 45% (Fig. 3A) (26). Consistent with these results, the Src-specific chemical inhibitor PP1 reduced the response by 80% (Fig. 3B) (27), and cotransfection of HM3 and NCIH292 cells with v-Src greatly increased the response (Fig. 3C). We conclude from these data that the mucin induction by P. aeruginosa is dependent on both Ras and Src.
Figure 3.
Involvement of Src in MUC2 mucin induction by P. aeruginosa. (A and B) Inhibition of P. aeruginosa-induced MUC2 up-regulation by coexpressing a dominant-negative mutant form of c-Src (A) and Src inhibitor PP1 (B). p-2864luc was transfected into HM3 cells with (A) or without (B) a dominant-negative mutant form of c-Src. Forty hours later, HM3 cells were pretreated with (B) or without PP1 (A) (5, 14, and 28 μM) (Calbiochem) for 30 min and then exposed to PAO1 culture supernatant (CS) for 6 hr before harvesting. Luciferase activity was measured as described in the text. (C) Effect of coexpressing p-2864luc and v-Src. p-2864luc (15 μg) was cotransfected with v-Src (0.3, 1, 3, and 5 μg) into HM3 cells. Forty-eight hours after being transfected, HM3 cells were lysed for assessing luciferase activity. (D) Effect of PD98059, CAPE, and coexpressing a dominant-negative mutant form of Ras and MEK1/2 on v-Src-induced MUC2 up-regulation. p-2864luc (15 μg) was cotransfected with v-Src (3 μg) and a dominant-negative mutant form of either Ras or MEK1/2 into HM3 cells. Forty hours after being transfected, the cells transfected with only p-2864luc were pretreated with either PD98059 (37 μM, 30 min) or CAPE (15 μg/ml, 1 hr). All the cells were then exposed to P. aeruginosa for 6 hr. Forty hours after being transfected, HM3 cells were either exposed or not exposed to PAO1 culture supernatant (CS) 6 hr before harvesting for assessing luciferase activity. Similar results were observed in another MUC2-expressing epithelial cell line, NCIH292. In all the experiments described above, all transfections were carried out in triplicate. Values are the means ± SD; n = 3.
To assess the overall similarity between P. aeruginosa-triggered and v-Src-triggered signaling pathways, we asked whether signaling downstream of Src was sensitive to inhibitors already known to inhibit the P. aeruginosa-activated signal transduction pathway. We found that cotransfection of dominant-negative mutant forms of Ras (RasN17) and MEK1/2 [HMEK1(K97R)] or treatment with the MEK1/2 and NF-κB chemical inhibitors PD98059 and CAPE greatly reduced v-Src-activated responses (Fig. 3D). Consistent with this finding, there was no additive effect when P. aeruginosa supernatant was applied to HM3 and NCIH292 cells overexpressing v-Src (data not shown). These data strongly suggest overlap between the P. aeruginosa and v-Src signaling pathways.
The Endogenous MUC2 Gene and MUC2 Promoter-Driven Luciferase Reporter Gene Are Regulated Similarly.
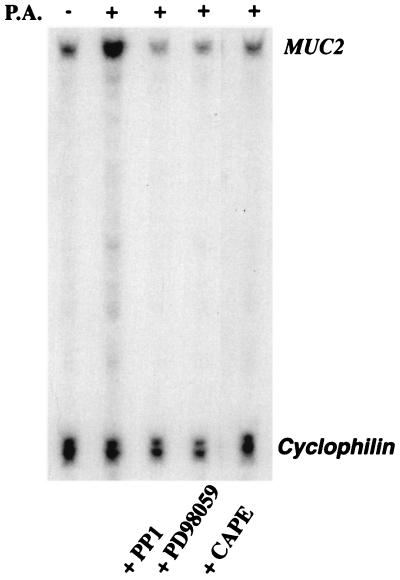
To confirm that the endogenous MUC2 gene and the MUC2 promoter-driven luciferase reporter gene are regulated similarly, we assessed the effects of the Src inhibitor PP1, the MEK1/2 inhibitor PD98059, and the NF-κB inhibitor CAPE on endogenous MUC2 mRNA induction by using RPAs. All these inhibitors strongly inhibited P. aeruginosa-induced MUC2 mRNA induction, confirming that the c-Src-activated signaling pathway revealed by luciferase reporter gene assays is indeed responsible for induction of the endogenous MUC2 gene (Fig. 4).
Figure 4.
Effects of inhibitors of Src, MEK1/2, and NF-κB on up-regulation of MUC2 mRNA induced by P. aeruginosa. HM3 cells were pretreated with PP1 (14 μM, 30 min), PD98059 (37 μM, 30 min), or CAPE (15 μg/ml, 60 min) and then exposed to PAO1 culture supernatant (CS) for 6 hr before harvesting for RNA extraction. RPA was then performed with the MUC2 gene-specific probe HAM1. A cyclophilin probe was included as a control to assess amount of RNA used in each hybridization reaction. Similar results were observed in another MUC2-expressing epithelial cell line, NCIH292.
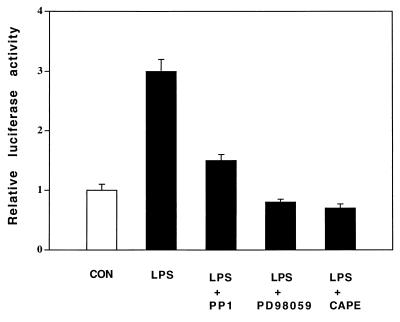
P. aeruginosa Culture Supernatant and LPS Up-Regulate MUC2 Transcription by Activation of the Same Src-Dependent MEK1/2-ERK1/2-NF-κB Pathway.
Despite the relatively detailed picture of the P. aeruginosa signaling pathway emerging from this study, the most upstream elements in the pathway have not yet been identified. With respect to the identity of the active factor in P. aeruginosa supernatant, our data indicate major if not exclusive involvement of LPS (Fig. 5). This raises additional questions, as details of LPS interaction with epithelial cell surfaces are not available. Unlike macrophages, most epithelial cells do not express CD14, the membrane-bound receptor that ligates LPS/LPS-binding protein (LBP) (28). Although many epithelial cells are known to ligate soluble (s)CD14 from serum, and epithelial responses to LPS are stronger in the presence than in the absence of serum, the P. aeruginosa response we observed was not inhibited by a neutralizing anti-CD14 antibody (data not shown), suggesting the presence of an alternate, as-yet-unidentified, receptor.
Figure 5.
Effects of inhibitors of Src, MEK1/2, and NF-κB on up-regulation of MUC2 transcription induced by P. aeruginosa LPS. HM3 cells were transfected with a MUC2 base pair −1,600/−1,300 TK promoter construct. Forty hours later, cells were pretreated with PP1 (14 μM, 30 min), PD98059 (37 μM, 30 min), or CAPE (15 μg/ml, 60 min) and then exposed to P. aeruginosa LPS (15 μg/ml) for 6 hr before harvesting for assessing luciferase activity. Similar results were observed in another MUC2-expressing epithelial cell line, NCIH292.
The chronology of pathogenic events leading to CF lung disease has been contested for decades. Although the data presented here cannot in themselves resolve the issue, they do provide an explanation for why CF lung disease is exacerbated by infection. In this respect they dovetail with observations of mice carrying mutations in the CFTR gene. The mouse data show that airway mucus transport velocity is reduced in mutant mice prior to infection (29). Davidson et al. (30) also showed that mutant mice had impaired mucociliary clearance from airways but no mucous cell metaplasia or mucus retention before infection. Once infection occurred, however, the mucus changes followed. Taken together, the in vivo mouse data and the in vitro findings reported here suggest a two-stage model of pathogenesis of CF lung disease.
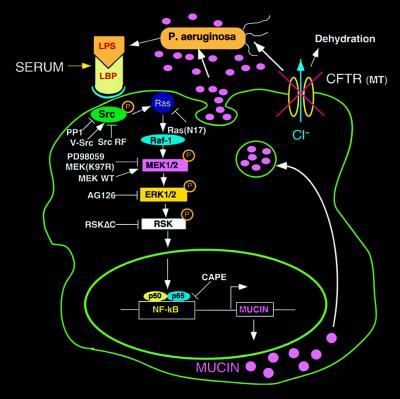
In stage 1, the CFTR mutation leads to P. aeruginosa retention and infection of the airways. In stage 2, the up-regulation of mucin transcription by P. aeruginosa via activation of the Src-Ras-MEK1/2-ERK1/2-pp90rsk-NF-κB pathway reported here causes airways to become plugged with mucin. The evolutionary forces responsible for the development of mucociliary clearance as a defense mechanism did not take into account that mutations in the CFTR gene of CF patients would transform a “mucociliary escalator” into a stagnant pool of mucus. Opportunistic pathogens such as P. aeruginosa find a niche in the stagnant mucus. Mucus, unable to execute its intended clearance function, paradoxically acts to nourish, support, and protect bacteria from antimicrobial agents (Fig. 6). It appears that this may be the means by which mucus, normally a substance protective to the host, interacts with bacteria in a vicious cycle leading to irreversible airway obstruction. A more extensive use of systems such as that described here should elucidate other facets of bacterial–epithelial interactions and open up new targets for therapeutic intervention.
Figure 6.
Schematic diagram showing steps in the signaling pathway by which P. aeruginosa up-regulates human MUC2 mucin gene transcription. As indicated, P. aeruginosa releases LPS, thereby activating a c-Src-Ras-Raf-1-MEK1/2-MAPK (ERK1/2)-pp90rsk pathway, which in turn leads to the activation of NF-κB and triggers MUC2 mucin transcription. LBP, LPS-binding protein. The overproduced mucin, in concert with abnormal airway lining fluid secondary to CFTR mutation, leads to airway mucus obstruction and lung failure.
Acknowledgments
We thank Drs. Warner Greene, Anthony DeFranco, J. Michael Bishop, G. Cooper, Alan Saltiel, Michael Karin, Joan Brugge, and John Hiscott for the various plasmids and antibodies that made this work possible. We thank Dr. Alice Prince for P. aeruginosa. We thank Drs. Zena Werb, Keith Yamamoto, Anthony DeFranco, and Synthia Mellon and Peilin Zhang for enlightening discussions, and we thank Dr. Zena Werb also for critically reading this manuscript. We also thank Mark Young for his help in photography and graphics. This work was supported by National Institutes of Health Grants HL 43762 and 24136 to C.B., an National Institute of Diabetes and Digestive and Kidney Diseases–University of California San Francisco Gene Therapy Core Center Grant, and a University of California San Francisco Graduate Student Research Award to J.-D.L.
ABBREVIATIONS
- CAPE
caffeic acid phenethyl ester
- CF
cystic fibrosis
- CFTR
CF transmembrane conductance regulator
- C/EBP
CCAAT-enhancer binding protein
- EMSA
electrophoretic mobility-shift assay
- ERK
extracellular signal-regulated kinase
- JNK
c-Jun amino-terminal kinase
- JNKK
c-Jun amino-terminal kinase kinase
- LPS
lipopolysaccharide
- MAPK
mitogen-activated protein kinase
- MEK
MAPK/ERK kinase
- NF-κB
nuclear factor κB
- pp90rsk
the 90-kDa ribosomal S6 kinase
- RPA
RNase-protection assay
- SEK
SAPK/ERK kinase
References
- 1.Collins F S, Wilson J M. Nature (London) 1992;358:708–709. doi: 10.1038/358708a0. [DOI] [PubMed] [Google Scholar]
- 2.Pier G B, Grout M, Zaidi T S, Oslen J C, Johnson L G, Yankasas J R, Goldberg J B. Science. 1996;271:64–67. doi: 10.1126/science.271.5245.64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Smith J J, Travis S M, Greenberg E P, Welsh M J. Cell. 1996;85:229–236. doi: 10.1016/s0092-8674(00)81099-5. [DOI] [PubMed] [Google Scholar]
- 4.Saiman L, Prince A. J Clin Invest. 1993;92:1875–1880. doi: 10.1172/JCI116779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Li J D, Dohrman A F, Gallup M G, Miyata S, Gum J R, Kim Y S, Nadel J A, Price A, Basbaum C B. Proc Natl Acad Sci USA. 1997;94:967–972. doi: 10.1073/pnas.94.3.967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gum J, Hicks J W, Kim Y S. Biochem J. 1997;325:259–267. doi: 10.1042/bj3250259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gum J, Hicks J W, Torbara N W, Siddiki B, Kim Y S. J Biol Chem. 1994;269:2440–2446. [PubMed] [Google Scholar]
- 8.Nogami H, Ohmori H, Li J D, Gallup M G, Gum J, Kim Y, Basbaum C. Gene. 1997;198:191–201. doi: 10.1016/s0378-1119(97)00314-4. [DOI] [PubMed] [Google Scholar]
- 9.Natarajan S, Singh T, Burke D, Grunberger B, Aggarwal B. Proc Natl Acad Sci USA. 1996;93:9090–9095. doi: 10.1073/pnas.93.17.9090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.DiDonato J, Hayakawa M, Rothwarf D, Zandi E, Karin M. Nature (London) 1997;388:548–554. doi: 10.1038/41493. [DOI] [PubMed] [Google Scholar]
- 11.Ghoda L, Lin X, Greene W. J Biol Chem. 1997;272:21281–21288. doi: 10.1074/jbc.272.34.21281. [DOI] [PubMed] [Google Scholar]
- 12.Schouten G I, Schouten G J, Vertegaal A C, Whiteside S T, Israel A, Toebes M, Dorsman J C, van der Eb A J, Zantema A. EMBO J. 1997;16:3133–3144. doi: 10.1093/emboj/16.11.3133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Baeuerle P A, Baltimore D. Cell. 1996;87:13–20. doi: 10.1016/s0092-8674(00)81318-5. [DOI] [PubMed] [Google Scholar]
- 14.Beauparlant P, Kwon H, Clarke M, Lin R, Sonenberg N, Wainberg M, Hiscott J. J Virol. 1996;70:5777–5785. doi: 10.1128/jvi.70.9.5777-5785.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Baldwin A S. Annu Rev Immunol. 1996;14:649–681. doi: 10.1146/annurev.immunol.14.1.649. [DOI] [PubMed] [Google Scholar]
- 16.Stokoe D, Campbell DG, Nakielny S, Hidaka H, Leevers S J, Marshall C, Cohen P. EMBO J. 1992;11:3985–3994. doi: 10.1002/j.1460-2075.1992.tb05492.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Treisman R. Curr Opin Cell Biol. 1996;8:205–215. doi: 10.1016/s0955-0674(96)80067-6. [DOI] [PubMed] [Google Scholar]
- 18.Minden A, Lin A, Claret F, Abo A, Karin M. Cell. 1995;81:1147–1157. doi: 10.1016/s0092-8674(05)80019-4. [DOI] [PubMed] [Google Scholar]
- 19.Aless D R, Cuenda A, Cohen P, Dudley D T, Saltiel A R. J Biol Chem. 1993;270:27489–27494. doi: 10.1074/jbc.270.46.27489. [DOI] [PubMed] [Google Scholar]
- 20.Howe L, Leevers S J, Gomez N, Nakielny S, Cohen P, Marshall C J. Cell. 1992;71:335–352. doi: 10.1016/0092-8674(92)90361-f. [DOI] [PubMed] [Google Scholar]
- 21.Moodie S, Willumsen B, Weber M, Wolfman A. Science. 1993;260:1658–1661. doi: 10.1126/science.8503013. [DOI] [PubMed] [Google Scholar]
- 22.Stokoe D, McCormick F. EMBO J. 1997;16(9):2384–2396. doi: 10.1093/emboj/16.9.2384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wood K W, Sarnecki C, Roberts T M, Blenis J. Cell. 1992;68:1041–1050. doi: 10.1016/0092-8674(92)90076-o. [DOI] [PubMed] [Google Scholar]
- 24.Katz M E, McCormick F. Curr Opin Genet Dev. 1997;7:75–79. doi: 10.1016/s0959-437x(97)80112-8. [DOI] [PubMed] [Google Scholar]
- 25.Feig L A, Cooper G M. Mol Cell Biol. 1988;8:2472–2478. doi: 10.1128/mcb.8.6.2472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mukhopadhyay D, Tsiokas L, Zhou X M, Foster D, Brugge J S, Sukhatme V P. Nature (London) 1995;375:577–581. doi: 10.1038/375577a0. [DOI] [PubMed] [Google Scholar]
- 27.Hanke J A, Gardner J P, Dow R L, Changelian P S, Brissette W H, Weringer E J, Pollok B A, Connelly P A. J Biol Chem. 1996;271:695–701. doi: 10.1074/jbc.271.2.695. [DOI] [PubMed] [Google Scholar]
- 28.Sweet M J, Hume D A. J Leukocyte Biol. 1996;60:8–26. doi: 10.1002/jlb.60.1.8. [DOI] [PubMed] [Google Scholar]
- 29.Zahm J M, Gaillard D, Dupuit F, Hinnrasky J, Porteous D, Dorin J R, Puchelle E. Am J Physiol. 1997;272:C853–C859. doi: 10.1152/ajpcell.1997.272.3.C853. [DOI] [PubMed] [Google Scholar]
- 30.Davidson D J, Dorin J R, McLachlan G, Ranaldi V, Lamb D, Doherty C, Govan J, Porteous D J. Nat Genet. 1995;9:351–357. doi: 10.1038/ng0495-351. [DOI] [PubMed] [Google Scholar]