Abstract
The multiple-stage ion-trap mass spectrometric approaches towards to the structural characterization of the monoacyl-PIM (triacylated PIM) and the diacyl-PIM (tetracylated PIM), namely, the PIM (diacylated PIM) consisting of one or two additional fatty acid substituents attached to the glycoside, respectively, were described. While the assignment and confirmation of the fatty acid substituents on the glycerol backbone can be easily achieved by the methods described in the previous article, the identity of the glycoside moiety and its acylation state can be determined by the observation of a prominent acylglycoside ion arising from cleavage of the diacylglycerol moiety ([M – H – diacylglycerol]−) in the MS2-spectra of monoacyl-PIM and diacyl-PIM. The distinction of the fatty acid substituents on the 2-O-mannoside (i.e., R3CO2H) from that on the inositol (i.e., R4CO2H) is based on the findings that the MS3-spectrum of [M – H – diacylglycerol]− contains a prominent ion arising from further loss of the fatty acid at the 2-O-mannoside (i.e., the [M – H – diacylglycerol – R3CO2H]− ion), while the ion arising from loss of the fatty acid substituent at the inositol (i.e., the [M – H – diacylglycerol – R4CO2H]− ion) is of low abundance. The fatty acyl moiety on the inositol can also be identified by the product-ion spectrum from MS4 of the [M – H – diacylglycerol – R3CO2H]− ion, which gives rise to a prominent ion corresponding to loss of R4CO2H. An [M – H – acylmannose]− ion was also observed in the MS2-spectra and, thus, the identity of the fatty acid substituent attached to 2-O-mannoside can be confirmed. The combined information obtained from the multiple-stage product-ion spectra from MS2, MS3, and MS4 permit the assignment of the complex structures of monoacyl-PIMs and diacyl-PIMs in a mixture isolated from M. bovis Bacillus Calmette Guérin.
Although ion-trap tandem mass spectrometry with ESI for structural characterization of complex structures of acyl-PIM2 was recently reported [1], structural detail revealed by multiple-stage ion-trap tandem mass spectrometry has not been described previously.
In the first part of this report, we described the IT multiple-stage mass spectrometric approaches for characterization of PI and PIM molecules from a lipid extract from M. bovis BCG [2]. In this subsequent report, we will describe the structural determination of the monoacyl- and diacyl PIMs in the extract, using the same mass spectrometric approaches. The assignment of the fatty acid substituents attached to the glycosides of monoacyl-PIM and of diacyl-PIM is based on the understanding that the fatty acyl moieties are connected to the O-6 position of the 2-O-linked Manp and to the O-3 position of the myo-inositol, respectively, based on the previous studies using NMR [3–8].
Materials and Methods
The isolation of PIMs from M. bovis Bacillus Calmette Guérin (BCG) and the IT multiple-stage mass spectrometry for structural characterization were described in the previous article [2]. The abbreviations and nomenclatures described earlier were also used [2].
Results and Discussion
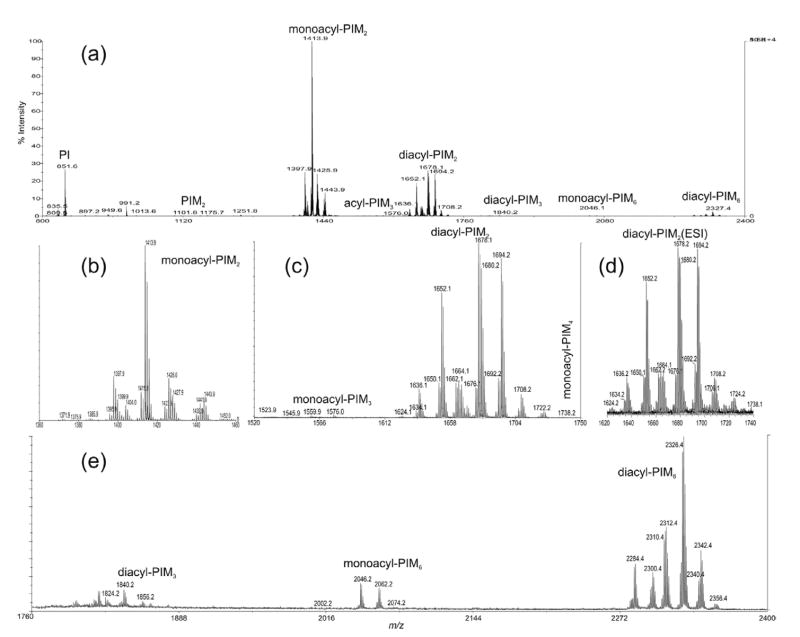
Figure 1a shows the negative-ion MALDI-TOF mass spectrum of the PIMs from M. bovis BCG obtained in the reflectron and delayed extraction mode. The homologous ions were seen for all the PIMs, including monoacyl-PIM2 (Figure 1b), diacyl-PIM2 (Figure 1c), diacyl-PIM3, monoacyl-PIM6, and diacyl-PIM6 (Figure 1e). The ESI mass spectrum of the [M – H]− ions of diacyl-PIM2 acquired by an IT instrument in a high-resolution scan mode (Figure 1d) is similar to the spectrum shown in Figure 1c. However, the sensitivity seen in the MALDI-TOF spectrum is at least 10 times greater, indicating that MALDI-TOF method may be suitable for profiling the molecular species of PIMs, while the multiple-stage IT mass spectrometry of the [M – H]− ions desorbed by ESI, as described below, is readily applicable for structural identification of the complex structures of PIMs (Table 1).
Figure 1.
The MALDI-TOF mass spectrum of the complex lipid extract from M. bovis BCG (a), and the separate mass spectra that represent monoacyl-PIM2s (b), monoacyl-PIM3s and diacyl-PIM2s (c), and diacyl-PIM3s, monoacyl-PIM6s and diacyl-PIM6s (e). The profile of the high-resolution ESI mass spectrum of diacyl-PIM2s as shown in (d) is identical to that shown in (c).
Table 1.
The composition of monoacyl- and diacyl-PIMs from M. bovis BCG
| [M – H]− | Structure | [M – H]− | Structure | [M – H]− | Structure |
|---|---|---|---|---|---|
| 1369.8 | 16:0-(16:0/16:1)-PIM2 | 1634.1 | (16:0,16:1)(18:1/16:0)-PIM2 | 1824.2 | (16:0,18:1)-(18:1/16:0)-PIM3 |
| 14:0-(16:0/18:1)-PIM2 | (16:0,16:0)(18:1/16:1)-PIM2 | 1840.2 | (16:0,18:1)-(19:0/16:0)-PIM3 | ||
| 18:0-(16:1/14:0)-PIM2 | 1636.1 | (16:0,16:0)(18:1/16:0)-PIM2 | (16:0,19:0)-(18:1/16:0)-PIM3 | ||
| 1371.9 | 16:0-(16:0/16:0)-PIM2 | 1650.1 | (16:0,16:1)(19:0/16:0)-PIM2 | 1856.2 | (16:0,19:0)-(19:0/16:0)-PIM3 |
| 1395.9 | 16:0-(18:1/16:1)-PIM2 | (16:0,16:0)(19:0/16:1)-PIM2 | |||
| 16:1-(18:1/16:0)-PIM2 | (16:0,16:0)(18:1/17:0)-PIM2 | 2284.3 | (16:0,16:0)-(18:1/16:0)-PIM6 | ||
| 16:0-(18:2/16:0)-PIM2 | (16:0,16:0)(19:1/16:0)-PIM2 | 2298.3 | (16:0,16:1)-(19:0/16:0)-PIM6 | ||
| 1397.9 | 16:0-(18:1/16:0)-PIM2 | 1652.1 | (16:0,16:0)(19:0/16:0)-PIM2 | 2300.3 | (16:0,16:0)-(19:0/16:0)-PIM6 |
| 18:0-(16:0/16:1)-PIM2 | (16:0,17:0)(18:0/16:0)-PIM2 | 2310.3 | (16:0,18:1)-(18:1/16:0)-PIM6 | ||
| 1399.9 | 16:0-(18:0/16:0)-PIM2 | 1662.1 | (16:0,18:1)(18:1/16:0)-PIM2 | 2312.3 | (16:0,18:0)-(18:1/16:0)-PIM6 |
| 1411.9 | 16:0-(19:0/16:1)-PIM2 | 1664.1 | (16:0,18:0)(18:1/16:0)-PIM2 | 2326.4 | (16:0,18:1)-(19:0/16:0)-PIM6 |
| 16:1-(19:0/16:0)-PIM2 | (16:0,17:1)(19:0/16:0)-PIM2 | 2328.4 | (16:0,18:0)-(19:0/16:0)-PIM6 | ||
| 16:0-(19:1/16:0)-PIM2 | (16:0,16:0)(18:1/18:0)-PIM2 | 2340.4 | (17:0,18:1)-(19:0/16:0)-PIM6 | ||
| 17:0-(18:1/16:0)-PIM2 | 1676.1 | (16:0,18:1)(19:0/16:1)-PIM2 | 2342.4 | (16:0,19:0)-(19:0/16:0)-PIM6 | |
| 1413.9 | 16:0-(19:0/16:0)-PIM2 | (16:1,18:1)(19:0/16:0)-PIM2 | 2354.4 | (18:0,18:1)-(19:0/16:0)-PIM6 | |
| 1425.9 | 17:0-(19:0/16:1)-PIM2 | (16:0,19:0)(18:1/16:1)-PIM2 | 2356.4 | (18:0,18:0)-(19:0/16:0)-PIM6 | |
| 1427.9 | 17:0-(19:0/16:0)-PIM2 | (16:0,18:1)(19:1/16:0)-PIM2 | |||
| 16:0-(19:0/17:0)-PIM2 | (17:0,18:1)(18:1/16:1)-PIM2 | ||||
| 1441.9 | 18:0-(19:0/16:0)-PIM2 | (16:0,18:1)(18:1/17:0)-PIM2 | |||
| 16:0-(19:0/18:0)-PIM2 | 1678.1 | (16:0,18:1)(19:0/16:0)-PIM2 | |||
| 17:0-(19:0/17:0)-PIM2 | (16:0,19:0)(18:1/16:0)-PIM2 | ||||
| 1455.9 | 19:0-(19:0/16:0)-PIM2 | 1680.2 | (16:0,18:0)(19:0/16:0)-PIM2 | ||
| 18:0-(19:0/17:0)-PIM2 | (16:0,19:0)(18:0/16:0)-PIM2 | ||||
| 1692.2 | (17:0,18:1)(19:0/16:0)-PIM2 | ||||
| 1557.9 | 16:0-(18:1/16:1)-PIM3 | (16:0,19:1)(19:0/16:0)-PIM2 | |||
| 16:1-(18:1/16:0)-PIM3 | (16:1,19:0)(19:0/16:0)-PIM2 | ||||
| 1559.9 | 16:0-(18:1/16:0)-PIM3 | (16:0,19:0)(19:1/16:0)-PIM2 | |||
| 1573.9 | 16:0-(19:1/16:0)-PIM3 | 1694.2 | (16:0,19:0)(19:0/16:0)-PIM2 | ||
| 16:0-(19:0/16:1)-PIM3 | 1704.2 | (16:0,18:1)(19:0/18:1)-PIM2 | |||
| 16:0-(18:1/17:0)-PIM3 | (16:0,19:0)(18:1/18:1)-PIM2 | ||||
| 17:0-(18:1/16:0)-PIM3 | (18:0,18:1)(19:0/16:1)-PIM2 | ||||
| 16:1-(19:0/16:0)-PIM3 | (18:0,19:0)(18:1/16:1)-PIM2 | ||||
| 1575.9 | 16:0-(19:0/16:0)-PIM3 | (18:1,18:1)(19:0/16:0)-PIM2 | |||
| (16:0,20:1)(19:0/16:1)-PIM2 | |||||
| 1722 | 16:0-(18:1/16:0)-PIM4 | 1706.2 | (18:0,18:1)(19:0/16:0)-PIM2 | ||
| 1738 | 16:0-(19:0/16:0)-PIM4 | (18:0,19:0)(18:1/16:0)-PIM2 | |||
| (18:0,19:0)(18:0/16:1)-PIM2 | |||||
| 2020.1 | 16:0-(16:0/16:0)-PIM6 | (18:0,18:0)(18:1/17:0)-PIM2 | |||
| 2046.1 | 16:0-(18:1/16:0)-PIM6 | 1708.2 | (18:0,18:0)(19:0/16:0)-PIM2 | ||
| 2062.1 | 16:0-(19:0/16:0)-PIM6 | (17:0,18:0)(19:0/17:0)-PIM2 | |||
| 2074.1 | 18:0-(18:1/16:0)-PIM6 | (16:0,18:0)(19:0/18:0)-PIM2 | |||
| 2090.1 | 18:0-(19:0/16:0)-PIM6 | 1720.2 | (18:1,19:0)(19:0/16:0)-PIM2 | ||
| 2104.1 | 19:0-(19:0/16:0)-PIM6 | (16:0,19:0)(19:0/18:1)-PIM2 | |||
| (18:0,19:0)(19:0/16:1)-PIM2 | |||||
| (18:1,18:0)(19:0/17:0)-PIM2 | |||||
| 1722.2 | (18:0,19:0)(19:0/16:0)-PIM2 | ||||
| (16:0,19:0)(19:0/18:0)-PIM2 |
Characterization of Monoacyl-PIMs
The [M – H]− ions belong to the monoacyl-PIM2, monoacyl-PIM3, monoacyl-PIM4, and monoacyl-PIM6 in the monoacyl-PIM (triacylated PIM) family were observed (Table 1).
Monoacyl-PIM2
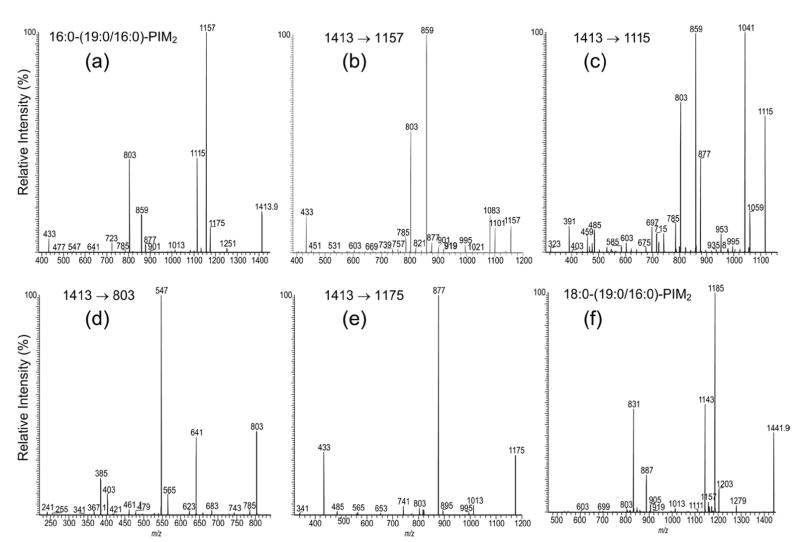
Among all of the PIMs isolated from M. bovis BCG, the subclass of monoacyl-PIM2 was the most abundant, while the ion at m/z 1413 was the most prominent in the family (Figure 1a, b). As shown in Figure 2a, the IT product-ion spectrum from MS2 of the m/z 1413 ion is dominated by the ions at m/z 1157 and 1115, arising from losses of the 16:0- and 19:0-fatty acid substituents, respectively. The ion at m/z 1157 is more abundant than the ion at m/z 1115, indicating that the 16:0- and 19:0-fatty acyl substituents are located at sn-2 and sn-1 of the glycerol backbone, respectively. The assignment is further examined by the MS3-spectra of the ions at m/z 1157 (1413 → 1157) (Figure 2b) and m/z 1115 (1413 → 1115) (Figure 2c). The former spectrum contains the ions at m/z 1101 (1157 – 56) and 1083 (1157 – 74), probably arising from the losses of the glycerol moieties involving a rearrangement process, similar to those observed for PI and PIM [2] and the two ions are nearly of equal abundance. The latter spectrum contains ions at m/z 1059 (1115 – 56) and m/z 1041 (1115 – 74) arising from the similar losses of the glycerol moieties. However, the ion at m/z 1058 is much more prominent than the ion at m/z 1041. The drastic differences in the profiles of the two spectra indicate that the MS3-spectra of the ions at m/z 1157 (1413 – 256) and 1059 (1413 – 298) can be used to distinguish the fatty acid substituent at sn-2 from that at sn-1. Therefore, the positions of the 16:0-fatty acyl substituent at sn-2 and of the 19:0-fatty acid moiety at sn-1 can be confirmed.
Figure 2.
The IT MS2 product-ion spectrum of the [M – H]− ion of 16:0-(19:0/16:0)-PIM2 at m/z 1413.9 (a), and its MS3 product-ion spectra of the ions at m/z 1157 (1413 → 1157) (b), at m/z 1115 (1413 → 1115) (c), at m/z 803 (1413 → 803) (d), and at m/z 1175 (1413 → 1175) (e); panel (f) illustrates the IT MS2 product-ion spectrum of the homologous ion at m/z 1441.9, arising from 18:0-(19:0/16:0)-PIM2.
The ion at m/z 859 (Figure 2a) arises from the combined losses of the 16:0- and 19:0-fatty acid moieties and the ion at m/z 877 arises from the combined losses of the fatty acyl moiety at sn-1 (or sn-2) either as an acid molecule and the fatty acyl substituent at sn-2 (or sn-1) or as a ketene molecule. The fragmentation pathways are consistent with the observation of the ions at m/z 859 and 877 in the MS3-spectra of the ions at m/z 1157 (1413 → 1157) (Figure 2b) and 1115 (1413 → 1115) (Figure 2c), which undergo further dissociation to eliminate the remaining fatty acid substituent at the glycerol backbone. The ion at m/z 877 in Figure 2c is more abundant than that in Figure 2b, suggesting that further loss of the 16:0-fatty acyl moiety at sn-2 as a ketene from the precursor [M – H – R1CO2H]− ion (i.e., m/z 1115) may be more favorable than further loss of the 19:0-fatty acyl ketene at sn-1 from the [M – H – R2CO2H]− ion (i.e., m/z 1157). These differences between the two spectra may also provide structural information for locating the fatty acyl substituents at the glycerol backbone [2].
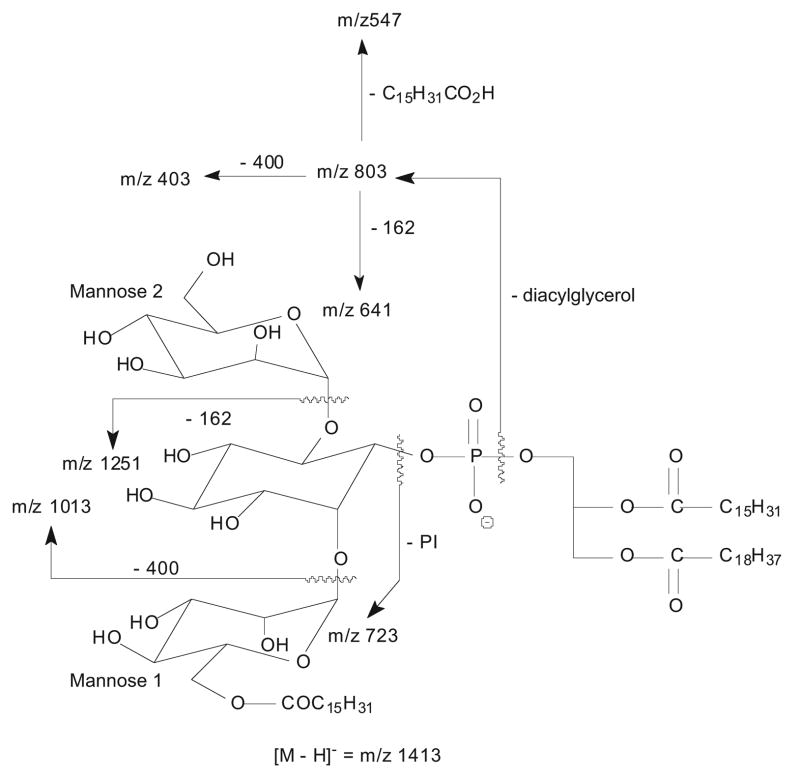
A 16:0-fatty acyl substituent attached to the glycoside is recognized by the presence of the ion at m/z 803 in Figure 2a, b, and c. This ion arises from loss of the diacylglycerol and is 238 Da higher than that of the analogous ion at m/z 565 observed for 19:0/16:0-PIM2 [2]. The presence of 16:0-fatty acyl substituent at the glycoside is also consistent with the presence of the ions at m/z 1251 (1413 – 162) and 1013 (1413 – 400), arising from losses of a mannosyl (probably mannose 2) and a palmitoylmannosyl (i.e., mannose 1 with an O-6 16:0-fatty acyl group) residues, respectively, along with the ion at m/z 723 arising from loss of 19:0/16:0-PI (Scheme 1) [2]. The presence of the ion at m/z1013 suggests that the 16:0-fatty acyl substituent is located on the mannosyl residue rather than on the inositol moiety.
Scheme 1.
The fragmentation pathways proposed for 16:0-(19:0/16:0)-PIM2.
Further dissociation of the ion at m/z 803 (1413 → 803) gives the ions at m/z 547 and 565, arising from losses of the 16:0-fatty acyl substituent as an acid and as a ketene molecules, respectively (Figure 2d). The spectrum also contains the ions at m/z 641 (803 – 162) and 623 (803 – 180), arising from loss of a mannosyl moiety, along with the ion at m/z 403 (803 – 400), corresponding to loss of a palmitoylmannosyl residue as described earlier. The MS4 product-ion spectra of the ions at m/z 803 originated from m/z 1115 (i.e., 1413 → 1115 → 803) and from m/z 1157 (i.e., 1413 → 1157 → 803) (data not shown) are also identical to that shown in Figure 2d. The presence of the m/z 803 ion in the MS3-spectra of m/z 1157 (Figure 2b) and m/z 1115 (Figure 2c) is consistent with the fact that a 16:0-fatty acyl moiety is attached to mannose 1.
The IT MS3-spectrum of m/z 1157 (Figure 2b) is dominated by the ion at m/z 859 arising from loss of the 19:0-fatty acyl substituent at sn-1, while the ion at m/z 901 corresponding to loss of the 16:0-fatty acyl substituent attached to mannose 1 is of low abundance. The results indicate that loss of the fatty acyl moiety at the mannosyl residue (assigned as mannose 1) is less favorable than that at sn-1 (or sn-2) of the glycerol backbone. This finding is also supported by the IT MS3-spectrum of the m/z 1175 ion (1413 → 1175) (Figure 2e), which is equivalent to a [M – H]− ion of 16:0-(19:0/0)-PIM2, arising from primary loss of the 16:0-fatty acyl substituent at sn-2 as a ketene. The spectrum is dominated by the ion at m/z 877 corresponding to loss of the 19:0-fatty acid at sn-1 and the ion expected at m/z 919 corresponding to loss of the 16:0-fatty acid substituent at mannose 1 (i.e., loss of R3CO2H) is not present [2]. This preferential loss of the fatty acid substituents at the glycerol backbone over that at the glycosyl moiety readily provides information to distinguish the fatty acyl substituents at glycerol backbone from that at mannose 1 and from that at the inositol (discuss later). The spectrum is also readily distinguishable from that arising from (19:0/16:0)-PIM2 [2], demonstrating the utility of IT tandem mass spectrometry in the differentiation of positional isomers such as (19:0/16:0)-PIM2 and 16:0-(19:0/0)-PIM2.
The IT MS2-spectrum of the ion at m/z 1441 (Figure 2f) is similar to that observed for the ion at m/z 1413 (Figure 2a) and the analogous ion arising from elimination of the diacylglycerol is seen at m/z 831, indicating that the molecule consists of a 18:0-fatty acyl substituent attached to mannose 1. The ions at m/z 1185 and 1143, arising from losses of the 16:0- and 19:0-fatty acyl substituents at sn-2 and sn-1, respectively, are prominent. However, the ion at m/z 1157 corresponding to loss of the 18:0-fatty acyl moiety is of low abundance, consistent with the notion that loss of the fatty acid on the glycosyl residue is less favorable than those attached to the glycerol backbone.
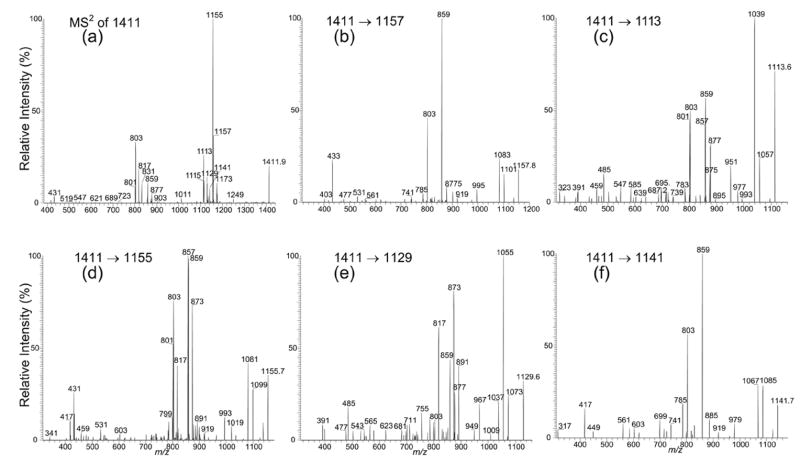
Nearly all of the acyl-PIM2 analyzed consists of more than one isomeric structure (Table 1). The complexity of the structures of the ions is shown by the ion at m/z 1411, which gives an MS2-spectrum (Figure 3a) that contains ions at m/z 1157, 1155, 1141, 1129, 1115, and 1113, reflecting the losses of the 16:1-, 16:0-, 17:0-, 18:1-, 19:1-, and 19:0-fatty acid moieties, respectively. The ions arising from losses of the various diacylglycerol moieties were observed at m/z 817, 803, and 801, indicating the presence of the molecular species consisting of a 17:0-, 16:0-, and a 16:1-fatty acyl substituents attached to mannose 1. The structures of the molecules are further unveiled by IT MS3 spectrometry.
Figure 3.
The IT MS2 product-ion spectrum of the ion at m/z 1411.9 (a), and its MS3 product-ion spectra of the ions at m/z 1157 (1411 → 1157) (b), at m/z 1113 (1411 → 1113) (c), at m/z 1155 (1411 → 1155) (d), at m/z 1129 (1411 → 1129) (e), and at m/z 1141 (1411 → 1141) (f). The product-ion spectra from MS2 and MS3 confirm that the ion at m/z 1411 consists of a major 16:0-(19:0/16:1)-PIM2 structure and several monoacyl-PIM2 isomers.
Further dissociation of the m/z 1157 ion (1411 → 1157) (Figure 3b), which originates from a primary loss of a 16:1-fatty acyl substituent, gives rise to the ions at m/z 1101 (1157 – 56) and 1083 (1157 – 74), by various losses of the glycerol residues as described earlier. The spectrum is nearly identical to that shown in Figure 2b, indicating that the m/z 1157 (1411 – 254) ion arises from m/z 1411 by loss of the 16:1-fatty acid moiety at sn-2, and thus the 19:0- and the 16:1-fatty acyl substituents can be assigned at sn-1 and sn-2, respectively, while the assignment of the 16:0-fatty acid moiety at mannose 1 is confirmed by the MS3-spectrum of the ion at m/z 803 (not shown), which is identical to that shown in Figure 2d.
In contrast, the profile of the ions at m/z 1039 (1113 – 74) and 1057 (1113 – 56) in the MS3-spectrum of the ion at m/z 1113 (1411 – C18H37CO2H) (1411 → 1113) (Figure 3c) is similar to that seen in Figure 2c, suggesting that the ion at m/z 1113 arises from the ion at m/z 1411 by loss of the 19:0-fatty acid at sn-1. The spectrum also contains ions at m/z 859 and 877, arising from losses of the 16:1-fatty acid moiety as acid and as ketene molecules at sn-2, respectively, along with the ion at m/z 803 that signifies that a 16:0-fatty acyl group is attached to mannose 1. The results are consistent with the assignment earlier that showed the ion at m/z 1411 is mainly a 16:0-(19:0/16:1)-PIM2 isomer.
In Figure 3c, ions at m/z 857 and 875, corresponding to losses of a 16:0-fatty acid moiety as an acid and as a ketene, respectively, along with the ion at m/z 801, corresponding to the phospholipoglycoside ion consisting of a 16:1-fatty acyl substituent at mannose 1 are also present. The results indicate that the m/z 1411 ion may also consist of a 16:1-(19:0/16:0)-PIM2 isomer. This structural assignment is supported by the MS3-spectrum of the ion at m/z 1155 (1411 – C15H31CO2H) (1411 → 1155) (Figure 3d), arising from primary loss of a 16:0-fatty acid moiety. The profile of the ions at m/z 1081 (1155 – 74) and 1099 (1155 – 56) resembles that of Panel b but differs from that of Panel c, suggesting that the 16:0-fatty acid is located at sn-2 in the structure of 16:1-(19:0/16:0)-PIM2. The spectrum also contains ions at m/z 857, arising from loss of the 19:0-fatty acid substituent at sn-1, along with the ion at m/z 801 reflecting that a 16:1-fatty acyl moiety is attached to mannose 1 and in agreement with the assigned 16:1-(19:0/16:0)-PIM2 structure.
In the same spectrum (Figure 3d), the ions at m/z 873 and 859 corresponding to losses of a 18:1- and a 19:1-fatty acid moieties, respectively, are also prominent. The m/z 859 ion is formed concurrently with the prominent ion at m/z 803, arising from a 16:0-(19:1/16:0)-PIM2 isomer, in agreement with the observation of the ion at m/z 1115 in Figure 3a, arising from loss of 19:1-fatty acid moiety, while the ions at m/z 873 and 817 may arise from a 17:0-18:1/16:0-PIM2 isomer. The assignment of this latter structure is further confirmed by the MS3-spectrum of the m/z 1129 (1411 – C17H33CO2H) (1411 → 1129) ion (Figure 3e), which contains the ions at m/z 1055 (1129 – 74) and 1073 (1129 – 56) with a pattern similar to that in Figure 3c, along with the ions at m/z 873 and 891, arising from losses of the 16:0-fatty acid as an acid and as a ketene at sn-2, respectively, while the ion observed at m/z 817 signifies that a 17:0-fatty acyl substituent is attached to mannose 1.
The presence of the ions at m/z 859 and 877 (Figure 3e) arising from loss of 17:0-fatty acyl substituent as an acid and as a ketene molecules, respectively, and of the ion at m/z 803 that signifies a 16:0-fatty acyl group attached to mannose 1, indicates that a 16:0-(18:1/17:0)-PIM2 structure may also exist. The MS3-spectrum of the m/z 1141 (1411 → 1141) ion (Figure 3f), formed from m/z 1411 by primary loss of a 17:0-fatty acid, shows that the profile of the ions at m/z 1067 (1141 – 74) and 1085 (1141 – 56) is similar to that in Figure 3b, supporting that indeed the 17:0-fatty acyl group is located at sn-2, while the prominent ion at m/z 859 arising from loss of 18:1-fatty acid identifies the fatty acid substituent at sn-1, and the ion at m/z 803 reveals the attachment of a 16:0-fatty acyl group to mannose 1 as described earlier.
Monoacyl-PIM3
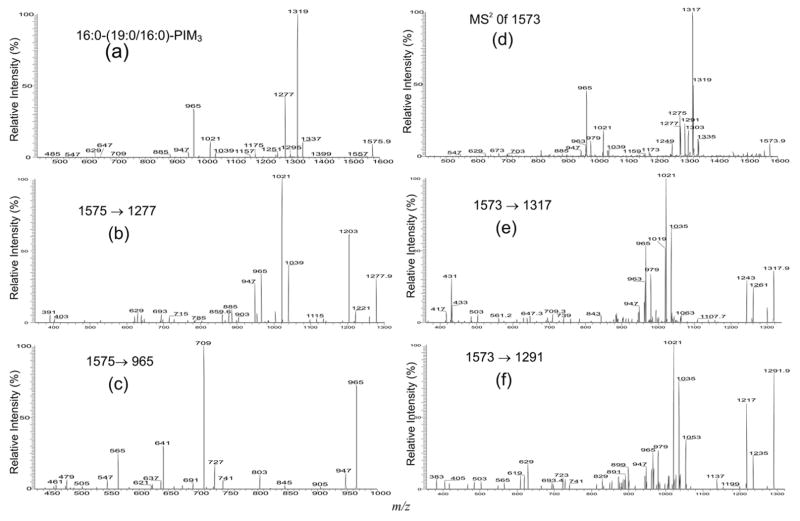
The major species belong to the monoacyl-PIM3 family was observed at m/z 1575 (Figure 1a, c), which is 162 Da heavier than the [M – H]− ion of 16:0-19:0/16:0-PIM2 at m/z 1413, indicating the presence of an additional mannose residue. The IT MS2-spectrum of the ion at m/z1575.9 (Figure 4a) is similar to that observed for 16:0-19:0/16:0-PIM2 (Figure 2a). The structurally informative ions are seen at m/z 1319 and 1337, which arise from losses of the 16:0-fatty acyl substituent at sn-2 as an acid and as a ketene, respectively, are more abundant than the ions at m/z 1277 and 1295, arising from the analogous losses of the 19:0-fatty acyl substituent at sn-1. The ions at m/z 965 and 885 arising from losses of a 19:0,16:0-diacylglycerol and a 19:0/16:0-PI moieties, respectively, are 162 Da higher than the analogous ion at m/z 803 and 723 observed in the spectrum of 16:0-19:0/16:0-PIM2 (Figure 2a), indicating that the compound may consist of a 16:0-19:0/16:0-PIM3 structure. The m/z 1251 ion, probably arising from cleavage of the α-(1→ 6) bond (between 6-manpα-1→ linked to O-6 of the myo-inositol) to eliminate of a dimannosyl residue, is similar to that observed for 16:0-19:0/16:0-PIM2 (Figure 2a), which eliminates a mannose by the same cleavage (Scheme 1) [2]. The ions arising from loss of the 16:0- (or 19:0) fatty acyl substituent, followed by loss of 19:0- (or 16:0) fatty acyl substituent as an acid or as a ketene molecule were observed at m/z 1021 (loss of two acids) and 1039 (loss of one acid and one ketene), respectively. The ions that are indicative of a PIM3 structure are observed at m/z 1175 (1575 – 238 – 162) and 1157 (1575 – 256 – 162), arising from the various losses of the palmitoylmannosyl moieties.
Figure 4.
The IT MS2 product-ion spectrum of the [M – H]− ion of 16:0-(19:0/16:0)-PIM3 at m/z 1575.9 (a), and its MS3 product-ion spectra of the ions at m/z 1277 (1575 → 1277) (b), and at m/z 803 (1575 → 965) (c); panel (d) illustrates the MS2 product-ion spectrum of the ion at m/z 1573.9, which is mainly a 16:0-19:1/16:0-PIM3 isomer. The MS3 product-ion spectra of the ions at m/z 1317 (1573 → 1317) (e), and at m/z 1291 (1573 → 1291) (f), revealed that isomers with 17:0-18:1/16:0-PIM3, 16:1-19:0/16:0-PIM3, 16:0-18:1/17:0-PIM3, and 17:0-18:1/16:0-PIM3 structures are also present. The assigned structures are further confirmed by the MS3-spectra of the ions at m/z 1303 (1573 → 1303), and at m/z 1275 (1573 → 1275) (shown in Supplementary Material).
The assignment of the 19:0- and 16:0-fatty acid substituents at sn-1 and sn-2 of the glycerol backbone is further confirmed by the IT MS3-spectrum of the ion at m/z 1319 (1575 → 1319) (shown in Supplementary Material which can be found in the electronic version of this article), which gives the ions at m/z 1263 (1319 – 56) and 1245 (1319 – 74) with a profile that indicates that the 16:0-fatty acid substituent is located at sn-2. The ions at m/z 1203 (1277 – 56) and 1185 (1277 – 74) seen in the IT MS3-spectrum of the ion at m/z 1277 (1575 → 1277) (Figure 4b) also confirm that the 19:0-fatty acid substituent is located at sn-1. Further dissociation of the ion at m/z965 (1575 → 965) (Figure 4c) gives rise to the ions at m/z 709 (965 – C15H31CO2H) and 727 (965 – C14H29CH = CO) by loss of the 16:0-fatty acid moiety attached to mannose 1, along with the ion at m/z 641 (965 – 162 × 2) by loss of a dimannosyl residue, and the ions at m/z 565 (965 – 238 – 162) and 547 (965 – 256 – 162) by losses of the palmitoylmannosyl moieties. The presence of the ions at m/z 565 and 547 is consistent with the observation of the ions at m/z 1175 and 1157 in Figure 4a, supporting the assignment that a 16:0-fatty acyl group (R3CO-) is attached to mannose 1, rather than to the inositol. The analogous ions were also observed for the ion at m/z 1559.9, which represents a 16:0-18:1/16:0-PIM3 structure (data not shown).
Several isomeric structures were observed for the ions at m/z 1573 and1557 (Table 1). The IT MS2-spectrum of the ion at m/z 1573.9 (Figure 4d) contains several sets of the ions at m/z 1317 and 1335, reflecting the losses of the 16:0-fatty acid substituent as an acid and as a ketene, respectively; at m/z 1303 and 1321, corresponding to the analogous losses of the 17:0-fatty acid substituent; at m/z 1291 and 1309, corresponding to the analogous losses of the 18:1-fatty acyl substituent; at m/z 1277 and 1295, corresponding to the analogous losses of the 19:1-fatty acyl substituent; at m/z 1275 and 1293, corresponding to the analogous losses of the 19:0-fatty acyl substituent. The presence of the 16:0-, 17:0-, and 16:1-fatty acyl substituents attached to the glycoside (i.e., mannose 1) is seen by the ions at m/z 965, 979, and 963. The presence of the 16:0-19:1/16:0-PIM3 structure is reflected by the set of the ions at m/z 1317, 1275, and 965, which signify the 16:0-, 19:1-, and 16:0-fatty acyl moieties at sn-2, sn-1 and at mannose 1, respectively. The assignment is consistent with the observation of the ion at m/z 1021, arising from the combined losses of the 16:0-fatty acid at sn-2 and the 19:1-fatty acid at sn-1, along with the ion at m/z 1249, arising from loss of a dimanosyl residue. The assignment of the 16:0-19:1/16:0-PIM3 structure is also supported by the MS3-spectrum of the m/z 1317 ion (1573 → 1317) (Figure 4e), which contains the ions at m/z 1261 (1317 – 56) and 1243 (1317 – 74) that show the pattern indicating that the ion at m/z 1317 arises from m/z 1573 due to loss of the 16:0-fatty acid substituent at sn-2. The spectrum is also dominated by the ion at m/z 1021 arising from further loss of the 19:1-fatty acid substituent at sn-1, along with the ion at m/z 965, consistent with the assigned structure.
In the same spectrum (Figure 4e), the ion pairs at m/z 1035 (1317 – 282) and 979, and at m/z 1019 (1317 – 298) and 963, probably arising from a 17:0-18:1/16:0-PIM3 and a 16:1-19:0/16:0-PIM3 structures, respectively, were also observed. The profile of the IT MS3-spectrum of the m/z 1277 ion (1573 → 1277) (not shown) is identical to that shown in Figure 4b, suggesting the presence of an isomer comprising a 19:1-fatty acid substituent at sn-1. Because the spectrum also contains a prominent ion at m/z 1021 reflecting loss of the 16:0-fatty acid substituent at sn-2, along with the ion at m/z 965 that reflect the attachment of a 16:0-fatty acid substituent to mannose 1, a 16:0-19:1/16:0-PIM3 structure can be confirmed.
Further dissociation of the ion at m/z 1291 (1573 – 282) (1573 → 1291, Figure 4f), gives rise to the ions at m/z 1217 (1291 – 74) and 1235 (1291 – 56) with a pattern similar to that in Figure 4b, supporting that the ion at m/z 1291 arises from loss of the 18:1-fatty acid substituent at sn-1 from m/z 1573. The ions at m/z 1035 (1291 – 256) and 1053 (1291 – 238), arising from further loss of a 16:0-fatty acid substituent as acid and as ketene, respectively, are formed together with the ion at m/z 979. These results support the earlier finding that the ion at m/z 1573 also represents a 17:0-18:1/16:0-PIM3 structure. The spectrum is also dominated by the ion at m/z 1021 (1291 – 270) along with 965, suggesting that a 16:0-18:1/17:0-PIM3 isomeric structure may also exist. The presence of this latter isomer is further supported by the IT MS3-spectrum of the ion at m/z 1303 (1573 → 1303) (shown in Supplementary Material), which shows that the m/z 1303 ([M – H – R2CO2H]−) ion also arises from m/z 1573 via loss of a 17:0-fatty acid at sn-2 and gives rise to the ion at m/z 1021 (1303 – 282) by a loss of 18:1-fatty acid substituent at sn-1, and to the ion at m/z 965 that signifies that a 16:0-fatty acid substituent is attached to mannose 1.
The set of the ions at m/z 1319, 1275, and 965 (Figure 4d), reflecting the 16:1-, 19:0-, and 16:0-fatty acyl moieties at sn-2, sn-1 and at mannose 1, respectively, probably arises from a 16:0-19:0/16:1-PIM3 structure. The assignment is further confirmed by the MS3-spectra of the ions at m/z 1275 (1573 → 1275) and at m/z 1319 (1573 → 1319) (shown in Supplementary Material). The spectra showed that the ions at m/z 1319 and 1275 are a [M – H – R2CO2H]− and a [M – H – R1CO2H]− ions, respectively. The IT MS3-spectrum of m/z 1275 is dominated by the ion at m/z 1021 (1275 – 254) arising from further loss of the 16:1-fatty acid at sn-2, along with the ion at m/z 965 that signifies the attachment of a 16:0-fatty acid moiety to mannose 1; the IT MS3-spectrum of m/z 1319 is dominated by the ion at m/z 1021 (1319 – 298), from loss of the 19:0-fatty acid substituent at sn-1, along with the ion at m/z 965, consistent with the attachment of a 16:0-fatty acid moiety to mannose 1. The combined information results in the assignment of the 16:0-19:0/16:1-PIM3 structure. Therefore, isomers with 16:0-19:1/16:0-PIM3, 17:0-18:1/16:0-PIM3, 16:1-19: 0/16:0-PIM3, 16:0-18:1/17:0-PIM3, and 16:0-19:0/16:1-PIM3 structures can be assigned for the ion at m/z 1573.
Monoacyl-PIM4 and monoacyl-PIM6
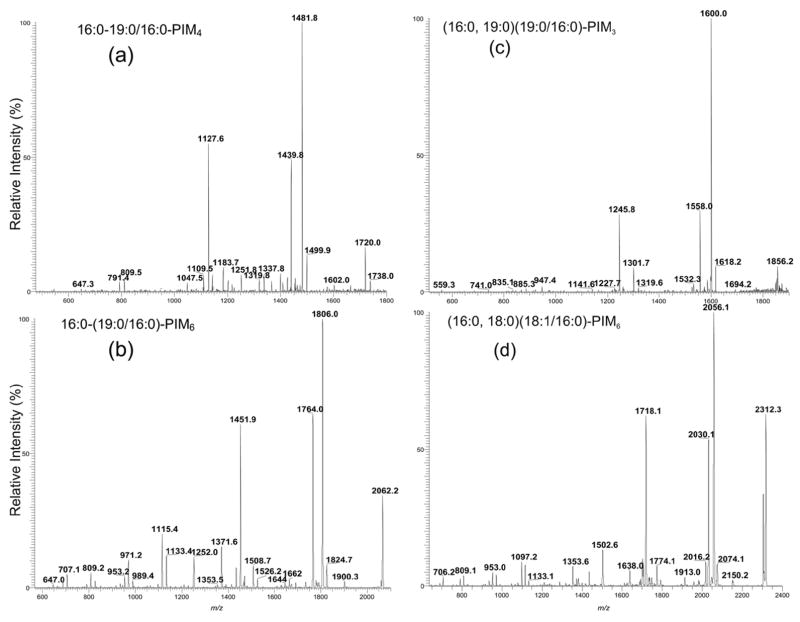
The two ions belong to the monoacyl-PIM4 family observed at m/z 1738 and 1722 were of low abundance and the ions corresponding to monoacyl-PIM5 were not observed (Figure 1c). The profile of the IT MS2-spectrum of the m/z 1738 ion (Figure 5a) is nearly identical to that observed for 16:0-19:0/16:0-PIM3 (Figure 4a) and the spectrum is dominated by the ions at m/z 1481 (1738.0 – 256.2) and 1439 (1738.0 – 298.3), indicating that the 16:0- and 19:0-fatty acyl substituents are located at sn-2 and sn-1, respectively. The ions arising from losses of the diacylglycerol and phosphatidylinositol moieties, respectively, are seen at m/z 1127, and 1047, while the ions arising from direct loss of the palmitoylmannosyl moiety are seen at m/z 1337 and 1319. These ions are 162 Da higher than the analogous ions observed in the MS2-spectrum of 16:0-19:0/16:0-PIM3, indicating that the ion at m/z 1738 is a 16:0-19:0/16:0-PIM4. The ions at m/z 809 and 791 may arise from further loss of the 16:0-fatty acyl substituent as a ketene and as an acid, respectively, from m/z 1047 (1738.0 – 690.4), which arises from loss of a 19:0/16:0-PA moiety from m/z 1738. In contrast, the analogous ions at m/z 647 and 629 observed for 16:0-19:0/16:0-PIM3 (Figure 4a) are of low abundance, consistent with the earlier findings that the abundances of the glycoside ions increase as the number of the hexose residue increases. The IT MS2-spectrum of the m/z 1722 ion (not shown) is also similar and represents a 16:0-18:1/16:0-PIM4 structure.
Figure 5.
The IT MS2 product-ion spectra of the [M – H]− ions of 16:0-19:0/16:0-PIM4 at m/z 1737 *m/z 1738.0) (a), of 16:0-(19:0/16:0)-PIM6 at m/z 2061 (*m/z 2062.1) (b), of (16:0,19:0)(19:0/16:0)-PIM3 at m/z 1855 (*m/z 1856.2) (c), and of (16:0,18:0)(18:1/16:0)-PIM6 at m/z 2311 (*m/z 2312.3). Asterisk = with mass defect.
The subclass of monoacyl-PIM6 found in the extract is among the more prominent in the monoacyl-PIM family. The major ions were observed at m/z 2062 and m/z 2046 (Figure 1e), representing a 16:0-(19:0/16:0)-PIM6 and a 16:0-(18:1/16:0)-PIM6, respectively. The IT MS2-spectrum of the ion at m/z 2062 (Figure 5b) is dominated by the ions at m/z 1806 (2062.2 – 256.2) and 1764 (2062.2 – 298.3), arising from loss of the 16:0- and 19:0-fatty acid substituents at sn-2 and sn-1, respectively. The ions analogous to those observed for 16:0-19:0/16:0-PIM4 were observed at m/z 1451 and 1371, arising from losses of the 19:0/16:0-glycerol and the 19:0/16:0-PA moieties, respectively, as well as the ions at m/z 1115 (1371 – 256) and 1133 (1371 – 238) arising from m/z 1371 by further loss of the 16:0-fatty substituent as an acid and as a ketene, respectively. The above results indicate that the ion is a 16:0-(19:0/16:0)-PIM6. The prominence of the glycosyl ions at m/z 1115 and 1133 that possess an inositol and six mannose residues, again, is in agreement with the earlier notion that the glycosyl ions become more prominent as the number of the sugar residue increases. Similar results are also observed in the IT MS2-spectra of the ion at m/z 2046, 2020, 2074, 2090, and at 2104 which represent a 16:0-(18:1/16:0)-PIM6, 16:0-(16:0/16:0)-PIM6, a 18:0-18:1/16:0-PIM6, a 18:0-19:0/16:0-PIM6, and a 19:0-19:0/16:0-PIM6 isomer, respectively (data not shown). These ions may consist of more than one isomer. However, the structural detail is not available from the present data due to the limited resolution of our ITMS instrument that fails to achieve unit mass resolution for a mass over 2000 Da. Thus, a composite MSn spectrum arising from a range of various precursor ions (>5 Da) resulted.
Characterization of Diacyl-PIM Molecular Species
Diacyl-PIM2
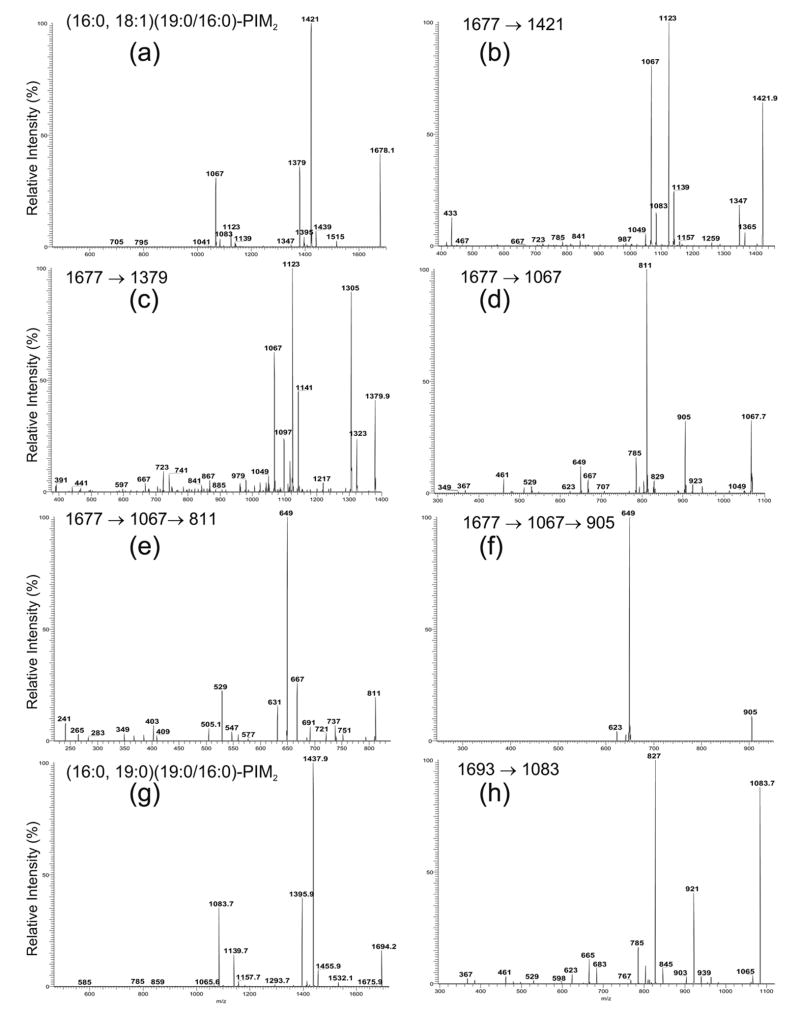
The negative-ion ESI/MS profile of the [M – H]− ions in the mass region corresponding to the diacyl-PIM2 as shown in Figure 1d is dominated by the ions at m/z 1678, 1694 and 1652. The MS2-spectrum of the m/z 1678.1 ion (Figure 6a) contains the major ions at m/z 1421 (1678.1 – 256.2) and 1439 (1678.1 – 238.2) arising from loss of the 16:0-fatty acyl substituent as an acid and as a ketene, respectively, and at m/z 1379 (1678.1 – 298.3) and 1397 (1678.1 – 280.3), arising from the analogous losses of the 19:0-fatty acyl substituent. The ions of the former set are respectively more abundant than the ions of the latter set, indicating that the 16:0- and 19:0-fatty acyl substituents are located at sn-2 and sn-1, respectively. The assignments are further confirmed by the MS3 product-ion spectra of the ions at m/z 1421 (Figure 6b) and 1379 (Figure 6c). The former spectrum contains ions at m/z 1365 (1421 – 56) and 1347 (1421 – 74) with a profile that shows that the ion at m/z 1421 is a [M– H – R2CO2H]− ion, while the latter spectrum contains ions at m/z 1323 (1379 – 56) and 1305 (1379 – 74) with a profile that is consistent with the findings that the ion at m/z 1379 is a [M – H – R1CO2H]− ion, arising from loss of the 16:0-fatty acid substituent at sn-1 from m/z 1678.
Figure 6.
The IT MS2 product-ion spectrum of the [M – H]− ions of (16:0, 18:1)(19:0/16:0)-PIM2 at 1677 (*m/z 1678.1) (a), and its MS3 product-ion spectra of the ions at m/z 1421 (1677 → 1421) (b), at m/z 1379 (1677 → 1379) (c), at m/z 1067 (1677 → 1061) (d), and its MS4 product-ion spectra at m/z 811 (1677 → 1067 → 811) (e), and at m/z 905 (1677 → 1067 → 905) (f). Panel (g) illustrates the MS2-spectrum of the (16:0, 19:0)(19:0/16:0)-PIM2 at 1693 (*m/z 1694.2) and its MS3-spectrum of the ion at m/z 1083 (1693 → 1083) (h). Asterisk = with mass defect.
The ion at m/z 1067 in Figure 6a arises from loss of 16:0/19:0-diacylglycerol and gives a M 3S -spectrum (1678 → 1067) (Figure 6d) that contains a prominent ion at m/z 811 arising from loss of a 16:0-fatty acid substituent, probably attached to mannose 1. This speculation is due to the fact that the profile of the spectrum is similar to that arising from m/z 803 (Figure 2d), an analogous ion that gives rise to a prominent ion at m/z 547 by loss of the 16:0-fatty acid substituent at mannose 1. In contrast, the ion at m/z 785 arising from loss of the 18:1-fatty acyl substituent, probably at the inositol is of low abundance. These results may indicate that loss of the fatty acid substituent at mannose 1 is more facile than loss of the fatty acid substituent at the inositol.
Further dissociation of the ion at m/z 811 (1678 → 1067 → 811) (Figure 6e) leads to the prominent ion at m/z649 by loss of a mannose residue, along with the ion at m/z 529 corresponding to loss of a 18:1-fatty acid substituent, probably at the inositol. The MS3-spectrum of the ion at m/z 905 (1678 → 1067 → 905) (Figure 6f) is dominated by the ion at m/z 649, arising from further loss of the 16:0-fatty acid substituent. However, the ion at m/z 623 arising from loss of an 18:1-fatty acid substituent is of low abundance. The results are consistent with the notion that loss of the 16:0-fatty acid substituent at mannose 1 is more facile than loss of the 18:1-fatty acid substituent at the inositol. The preferential loss of the fatty acyl substituent at mannose 1 over that at the inositol provides information to locate the position of fatty acid substituents in the glycoside.
In addition to the (16:0, 18:1)(19:0/16:0)-PIM2 isomer as assigned above, the m/z 1678 ion may also consist of a (16:0, 19:0)(18:1/16:0)-PIM2 isomer. This is based on the findings that the MS2-spectrum of m/z 1678 (Figure 6a) also contains the ions at m/z 1395 (1678.1 – 282.3), corresponding to loss of a 18:1-fatty acyl substituent, along with m/z 1083, which is identical to the phospholipoglycoside ion seen in the MS2-spectrum of (16:0, 19:0)(19:0/16:0)-PIM2 (Figure 6g, discuss later). The presence of the (16:0, 19:0)(18:1/16:0)-PIM2 structure is consistent with the fact that the MS 3-spectrum of the ion at m/z 1421 (Figure 6b) also contains the ion at m/z 1139, arising from further loss of the 18:1-fatty acid substituent at sn-1, and the phospholipoglycoside ion at m/z 1083, which contains both a 16:0- and a 19:0-fatty acyl substituents.
The MS2-spectrum of the m/z 1694.1 (Figure 6g) is also dominated by the ions at m/z 1437 (1694.1 – 256.2) and 1395 (1694.1 – 298.3), arising from losses of the 16:0-, 19:0-fatty acid substituents, respectively. The profile is similar to that shown in Figure 6a, indicating that the 16:0- and 19:0-fatty acyl moieties are located at sn-2, and sn-1, respectively. The ions at m/z 1531 and 1083 arise from loss of a mannosyl residue and loss of the diacylglycerol moiety, respectively, while the ion at m/z 1139 arises from the combined losses of the 16:0- and 19:0-fatty acid substituents. The identification of the fatty acid substituent attached to mannose 1 is recognized by the MS3-spectrum of the 1083 ion (1693 → 1083) (Figure 6h), which contains the prominent ion at m/z 827, arising from loss of 16:0-fatty acid substituent. In contrast, the ion at m/z 785, reflecting loss of the 19:0-fatty acyl substituent at the inositol is less abundant. The results suggest that the ion at m/z 1694 represents a (16:0, 19:0)(19:0/16:0)-PIM2 structure.
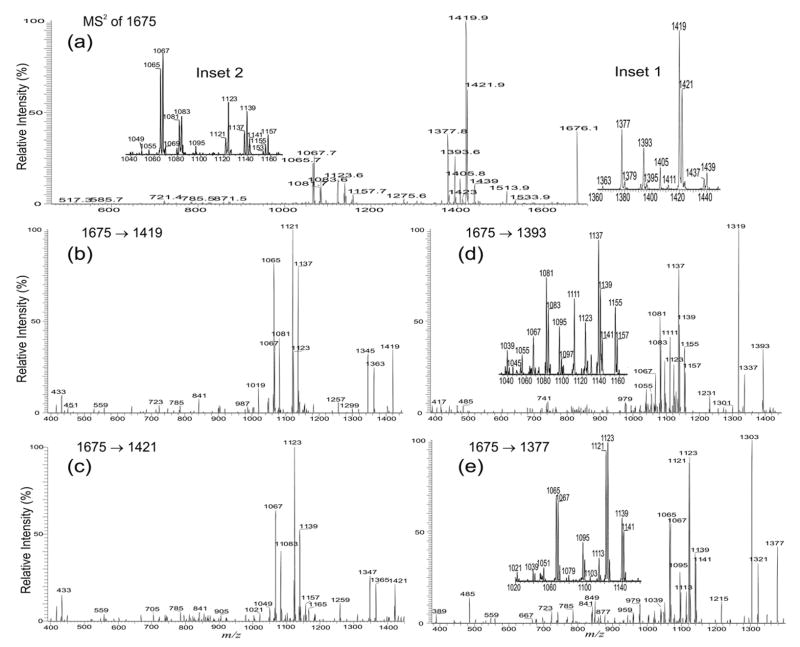
The complexities of the structures of the diacyl-PIM2 in the extract as revealed by multiple-stage IT mass spectrometry (Table 1) are demonstrated by the structural assignment of the ion at m/z 1676. The MS2 product-ion spectrum of the ion at m/z 1676.1 (Figure 7a) contains several sets of the fragment ions corresponding to losses of the 16:0-, 16:1-, 19:0-, 18:1-, 19:1-, and 17:0-fatty fatty acyl substituents as acids and as ketenes, respectively. These ions were seen in the sets of the ions at m/z 1419 and 1437; m/z 1421 and 1439; m/z 1377 and 1395; m/z 1393 and 1411; m/z 1379 and 1397; and at m/z 1405 and 1423, respectively (Figure 7a, inset 1). The ions reflecting the losses of the various diacylglycerol residues from the various isomers were observed at m/z 1067, 1065, 1083, and 1081, respectively, consistent with the presence of the ions at m/z 1121, 1123, 1137, and 1139, respectively (Figure 7a, inset 2), arising from the combined losses of the fatty acid substituents at sn-1 and at sn-2.
Figure 7.
The IT MS2 product-ion spectrum of the m/z 1675 (*m/z 1676.1) ion (a), and its MS3 product-ion spectra of the ions at m/z 1419 (1675 → 1419) (b), at m/z 1421 (1675 → 1421) (c), at m/z 1393 (1675 → 1393) (d), at m/z 1377 (1675 → 1377) (e). The combined information revealed that the ion is consisted of a major (16:0, 18:1)(19:0/16:1)-PIM2 structure along with five minor isomers (see text for detail). Asterisk = with mass defect.
The major (16:0, 18:1)(19:0/16:1)-PIM2 structure is revealed by the IT MS3-spectrum of the m/z 1419 ion (1675 → 1419) (Figure 7b), arising from primary loss of a 16:0-fatty acid. The profile of the fragment ions at m/z 1345 (1419 – 74) and 1363 (1419 – 56) confirms that the 16:0-fatty acid is located at sn-2, while the ions at m/z 1137, 1123, and 1121, arising from losses of the 18:1-, 19:1-, and 19:0-fatty acyl substituents at sn-2, respectively, indicate that the isomers with 18:1/16:0-, 19:1/16:0-, and 19:0/16:0-glycerol moieties, respectively, are also present. The ions at 1137, 1123, and 1121 are respectively formed together with the ions at m/z 1081, 1067, and 1065, which represent the phospholipoglycoside ions that possess a (16:0, 19:1), (16:0,18:1), and (16:1,18:1) fatty acyl substituents, residing at mannose 1 and inositol, respectively. The assignments of the fatty acyl substituents at mannose 1 or at the inositol are further confirmed by the MS3-spectra of m/z 1065, 1067, and 1081 (data not shown), which are similar to those observed earlier (Figure 6d and h) and are consistent with the presence of (16:1, 18:1)(19:0/16:0)-PIM2, (16:0, 18:1)(19:1/16:0)-PIM2, and (16:0, 19:1)(18:1/16:0)-PIM2 structures.
The profile of the IT MS3-spectrum of the ion at m/z 1421 (1675 → 1421) (Figure 7c) is similar to that arising from the ion at m/z 1419 (Figure 7b), indicating that the m/z 1421 ion arises from m/z 1675 by loss of the 16:1-fatty acyl substituent at sn-2. The ion at m/z 1123 arising from further losses of a 19:0 -fatty acyl substituents at sn-1, is formed together with the ion at m/z 1083, which represents a phospholipoglycoside anion that contains a 16:0-fatty acyl substituent at mannose 1 and a 19:0-fatty acyl substituent at inositol respectively, while the ion at m/z 1139, arising from loss of a 18:1-fatty acid substituent is formed together with the ion at m/z 1067, representing an analogous ion possessing a 16:0-fatty acyl substituent at mannose 1 and a 18:1-fatty acyl substituent at inositol. These results reveal the (16:0, 19:0)(18:1/16:1)-PIM2 and (16:0, 18:1) (19:0/16:1)-PIM2 structures.
In contrast, the IT MS3-spectrum of the m/z 1393 ion (1675 → 1393) (Figure 7d), arising from primary loss of a 18:1-fatty acid contains ions at m/z 1319 (1393 – 74) and 1337 (1393 – 56) with the feature that shows that the ion at m/z 1393 originates from loss of a 18:1-fatty acid at sn-1 from 1675. The sets of ions reflecting losses of the 16:0- (ions at m/z 1137 and 1155), 16:1- (ions at m/z 1139 and 1157), 18:1- (ions at m/z 1111 and 1129), and 17:0-fatty acid substituents (ions at m/z 1123 and 1141) as acids and ketenes, respectively, are also observed in the spectrum, indicating the presence of the isomers with (18:1/16:0)-, (18:1/16:1)-, (18:1/18:1)-, and (18:1/17:0)-glycerol residues. These ions are formed concurrently with the ions at m/z 1081, 1083, 1055, and 1067, respectively, which are the phospholipoglycoside anions containing (16:0, 19:1), (16:0,19:0), (16:0, 17:0), and (16:0, 18:1) fatty acyl substituents at mannose 1 and inositol, respectively. The results lead to the assignments of the two major (16:0, 19:1)(18:1/16:0)-PIM2 and (16:0, 19:0)(18:1/16:1)-PIM2 structures, as well as the two minor (16:0, 17:0)(18:1/18:1)-PIM2 and (16:0, 18:1)(18:1/17:0)-PIM2 species. The structures of (16:0, 19:1)(18:1/16:0)-PIM2 and (16:0, 19:0)(18:1/16:1)-PIM2 revealed here are consistent with those resulting from the MS3-spectra of m/z 1419 and 1421 as shown earlier.
The profile of the IT MS3-spectrum of the m/z 1377 ion (1675 → 1377) (Figure 7e) is similar to that observed for the m/z 1393 ion, indicating that the ion at m/z 1377 arises from m/z 1675 via further elimination of the 19:0-fatty acyl substituent at sn-1. Again, the ions formed from the loss of the 16:0-fatty acyl substituents at sn-2 as an acid and as a ketene are observed at m/z 1121 and 1139, respectively, while the ion pairs from the analogous losses of the 16:1- and 18:1-fatty acyl substituents are seen at m/z 1123 (1377 – 254) and 1141 (1377 – 236), and at m/z 1095 (1377 – 282) and 1113 (1377 – 264), respectively. These ions are formed together with the ions at m/z 1065, 1067 and 1039 that represent the phospholipoglycosides possessing a (16:1, 18:1), (16:0, 18:1), and a (16:0, 16:1)-fatty acyl substituents, respectively. Therefore, isomers with (16:1, 18:1)(19:0/16:0)-PIM2, (16:0, 18:1)(19:0/16:1)-PIM2, and (16:0, 16:1)(19:0/18:1)-PIM2 structures can be deduced. The assignments of the (16:1, 18:1)(19:0/16:0)-PIM2 and (16:0, 18:1)(19:0/16:1)-PIM2 structures are consistent with the results deduced from the MS3-spectra of m/z 1419 and 1421.
Diacyl-PIM3
Three minor ions at m/z 1824.2, 1840.2 and 1856.2 arising from diacyl-PIM3 were observed (Figure 1e). The IT MS2-spectrum of the m/z 1856.2 ion (Figure 5c) contains the analogous ions as seen for (16:0,19:0)(19:0/16:0)-PIM2 at m/z1694 (Figure 6g). The ions at m/z 1600 (1856.2 – 256.2) and 1558 (1856.2 – 298.3) arise from losses of the 16:0-fatty acyl substituent at sn-2 and the 19:0-fatty acyl substituent at sn-1, respectively. The ion at m/z 1245, arising from loss of the 19:0/16:0-diacylglycerol moiety is 162 Da higher than the analogous ion at m/z 1083 as shown in Figure 6g, indicating that the ion consists of a (16:0, 19:0)(19:0/16:0)-PIM3 structure. This is consistent with the presence of the ion at m/z 1532 (1856 – 324), arising from loss of a diman-nosyl residue. Similar MS2 product-ion spectra were also observed for the ions at m/z 1824 and 1840 (not shown), arising from a (16:0, 18:1)(18:1/16:0)-PIM3 and a (16:0, 18:1)(19:0/16:0)-PIM3 structures, respectively.
Diacyl-PIM6
The ions corresponding to diacyl-PIM4 and diacyl-PIM5 were not observed, but the species from diacyl-PIM6 are abundant (Figure 1e). The structural characterization is exemplified by the ion at m/z 2312.3, which gives the IT MS2-spectrum (Figure 5d) that is dominated by the ion at m/z 2056 (2312.3 – 256.2), arising from loss of the 16:0-fatty acid substituent at sn-2, along with the ions at m/z 2030 (2312.3 – 282.3), arising from loss of the 18:1-fatty acid substituent at sn-1. The results suggested that the compound probably consists of a (18:1/16:0)-glycerol backbone, consistent with the observation of the ion at m/z 1718, arising from m/z 2312 by loss of the (18:1/16:0)-diacylglycerol residue. The observation of the ion at m/z 1502 (2312 – 162 × 5) arising from loss of a pentama-nnosyl chain, indicates that the molecule is a diacyl-PIM6. The recognition of the 16:0-fatty acid situated at sn-2 is further confirmed by the MS3-spectrum of the ion at m/z 2056 (2312 → 2056) (data not shown), which contains the ions at m/z 1982 (2056 – 74) and 2000 (2056 – 56) with a profile that shows the m/z 2056 ion is indeed a product-ion of m/z 2312 from loss of the 16:0-fatty acid substituent at sn-2.
Further dissociation of the ion at m/z 1718 (shown in Supplementary Material) gives rise to the ion at m/z 1462, arising from loss of the 16:0-fatty acid substituent attached to mannose 1. The ions at m/z 1556 (1718 – 162), 1394 (1718 – 2 × 162), 1232 (1718 – 162 × 3), 1069 (1718 – 162 × 4), and 907 (1718 – 162 × 5), arise from losses of the various polymannosides linked to the 2-O-inositol, respectively. The results are consistent with the notion that the molecule is a diacyl-PIM6. The observation of the ion at m/z 1317 (1718 – 400.4) arising from loss of the palmitoylmannosyl residue attached to 6-O-inositol is also consistent with the fact that a 16:0-fatty acid substituent is attached to mannose 1. The MS4-spectrum of the ion at m/z 1317 (2312 → 1718 → 1317) (data not shown) gives a prominent ion at m/z 1033 (1317 – 284) by further loss of the 18:0-fatty acid substituent, indicating that a 18:0-fatty acid substituent is attached to the inositol. The above information suggests that the ion at m/z 2312 is mainly a (16:0,18:0)(18:1/16:0)-PIM6.
The characterization of the diacyl-PIM6s that consist of multiple isomeric structures is not conclusive, again, owing to the limitation of our current ion-trap instrument that is unable to specifically select the precursor ions that are greater than 2000 Da as described earlier.
Conclusions
The present method clearly demonstrates that multiple-stage ion-trap mass spectrometry with ESI is readily applicable for structural characterization of phosphatidylinositol-mannosides, including monoacyl- and diacyl-PIMs, as well as PI and PIMs that were described earlier [2]. Although this approach provides neither the information applicable for establishing the linkage of the fatty acid substituent to the glycoside in monoacyl-PIMs (i.e., to the O-6 position of the O-2 linked Manp), nor the information that establishes the linkage of the additional fatty acid substituent to the C3 position of the myo-inositol for diacyl-PIMs, the assignment of the fatty acid substituents attached to the 2-O-linked Manp, the myo-inositol, and to the glycerol backbone that are located at sn-1 or sn-2 can be easily achieved. More importantly, the structures of the molecular species that consist of various isomers can be revealed in detail, using the present approach. However, the implementation of an ion-trap instrument with sufficient resolution for selecting the high m/z precursor ions, such as those from diacyl-PIM6s, is necessary for their unambiguous structural assignment.
Acknowledgments
This research was supported by US Public Health Service Grants P41-RR-00,954, R37-DK-34,388, P60-DK-20,579, and P01-HL-57,278 and P30-DK56341.
References
- 1.Yague G, Segovia M, Valero-Guillen PL. Phospholipid Composition of Several Clinically Relevant Corynebacterium Species as Determined by Mass Spectrometry: An Unusual Fatty Acyl Moiety is Present in Inositol-Containing Phospholipids of Corynebacterium urealyticum. Microbiology. 2003;149:1675–1685. doi: 10.1099/mic.0.26206-0. [DOI] [PubMed] [Google Scholar]
- 2.Hsu FF, Turk J, Owens R, Rhoades ER, Russell DG. Structural Characterization of Phosphatidylinositol Mannosides from Mycobacterium bovis BCG by Multiple-Stage Quadrupole Ion-Trap Mass Spectrometry with Electrospray Ionization. I. PIMsLyso-PIMs. J Am Soc Mass Spectrom. 2007;18:466–478. doi: 10.1016/j.jasms.2006.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Khoo KH, Dell A, Morris HR, Brennan PJ, Chatterjee D. Structural Definition of Acylated Phosphatidylinositol Mannosides from Mycobacterium tuberculosis: Definition of a Common Anchor for Lipomannan Lipoarabinomannan. Glycobiology. 1995;5:117–127. doi: 10.1093/glycob/5.1.117. [DOI] [PubMed] [Google Scholar]
- 4.Gilleron M, Bala L, Brando T, Vercellone A, Puzo G. Mycobacterium tuberculosis H37Rv Parietal and Cellular Lipoarabinomannans: Characterization of the Acyl- and Glycoforms. J Biol Chem. 2000;275:677–684. doi: 10.1074/jbc.275.1.677. [DOI] [PubMed] [Google Scholar]
- 5.Gilleron M, Ronet C, Mempel M, Monsarrat B, Gachelin G, Puzo G. Acylation State of the Phosphatidylinositol Monnosides from Mycobacterium bovis Bacillus Calmette Guérin and Ability to Induce Granuloma and Recruit Natural Killer T Cells. J Biol Chem. 2001;276:34896–34904. doi: 10.1074/jbc.M103908200. [DOI] [PubMed] [Google Scholar]
- 6.Gilleron M, Quesniaux VFJ, Puzo G. Acylation State of the Phosphatidylinositol Hexammanosides from Mycobacterium bovis Bacillus Calmette Guérin and Mycobaterium tuberculosis H37Rv and Its Implication in Toll-Like Receptor Response. J Biol Chem. 2003;278:29880–29889. doi: 10.1074/jbc.M303446200. [DOI] [PubMed] [Google Scholar]
- 7.Nigou J, Gilleron M, Brando T, Puzo G. Structural Analysis of Mycobacterial Lipoglycans. Appl Biochem Biotechnol. 2004;118:253–268. doi: 10.1385/abab:118:1-3:253. [DOI] [PubMed] [Google Scholar]
- 8.Nigou J, Gilleron M, Puzo G. Lipoarabinomannans: Characterization of the Multiacylated Forms of the Phosphatidyl-Myo-Inositol Anchor by NMR Spectroscopy. Biochem J. 1999;337:453–460. [PMC free article] [PubMed] [Google Scholar]