Abstract
The Brucella abortus two-component regulatory system BvrR/BvrS controls the expression of outer membrane proteins (Omp) Omp3a (Omp25) and Omp3b (Omp22). Disruption of bvrS or bvrR generates avirulent mutants with altered cell permeability, higher sensitivity to microbicidal peptides, and complement. Consequently, the role of Omp3a and Omp3b in virulence was examined. Similar to bvrS or bvrR mutants, omp3a and omp3b mutants displayed increased attachment to cells, indicating surface alterations. However, they showed unaltered permeability; normal expression of Omp10, Omp16, Omp19, Omp2b, and Omp1; native hapten polysaccharide; and lipopolysaccharide and were resistant to complement and polymyxin B at ranges similar to those of the wild-type (WT) counterpart. Likewise, omp3a and omp3b mutants were able to replicate in murine macrophages and in HeLa cells, were resistant to the killing action of human neutrophils, and persisted in mice, like the WT strain. Murine macrophages infected with the omp3a mutant generated slightly higher levels of tumor necrosis factor alpha than the WT, whereas the bvrS mutant induced lower levels of this cytokine. Since the absence of Omp3a or Omp3b does not result in attenuation, it can be concluded that BvrR/BvrS influences additional Brucella properties involved in virulence. Our results are discussed in the light of previous works suggesting that disruption of omp3a generates attenuated Brucella strains, and we speculate on the role of group 3 Omps.
Members of the genus Brucella are intracellular bacterial pathogens of mammals (33). The ability of Brucella to invade and replicate in cells has been linked to its outer membrane (OM) properties as well as to structures built within the cell envelope (31, 32, 34). Among these, the lipopolysaccharide (LPS), the β-1,2-cyclic glucans, the type IV secretion system VirB, and the flagellum-like system are the most studied (2, 6, 17, 27). The notion that the Brucella OM plays a key role in virulence has been reinforced by the identification of the two-component regulatory system BvrR/BvrS, which controls the expression of at least two OM proteins (Omps), Omp3a and Omp3b, as well as the structure of the LPS (22, 28). Although they do not demonstrate obvious growth defects in vitro, the bvrS and bvrR mutants are avirulent in mice, displaying reduced invasiveness and replication in professional and nonprofessional phagocytes (7, 42).
Omp3a and Omp3b, also known as Omp25 and Omp22, respectively, belong to group 3 of the Brucella Omps (22, 37, 44, 45), a highly conserved family of up to seven members that includes some of the most abundant and immunogenic Brucella proteins (10). The function of group 3 Omps is not completely understood. The strong association of some of the members with LPS suggests that they play an important structural role in the OM (19, 37). In Brucella abortus, the gene encoding Omp31A is absent and the gene encoding Omp25b is truncated (23, 46), suggesting that neither of these Omps plays a significant role in Brucella virulence, although they still may participate in host preferences. Omp31A is a hemin-binding protein, and its expression is induced by iron limitation (11). The virulence of B. melitensis Rev1 Omp31A mutants, however, does not differ from the parental Rev1 counterpart (8). These apparent inconsistencies may be accounted for in part by the redundancy of iron uptake systems in Brucella (11). There are several reports indicating that B. abortus Omp3a is involved in virulence (14-16) and that it acts as a negative regulator of tumor necrosis factor alpha (TNF-α) production in human macrophages (24). Since the levels of Omp3a and Omp3b expression are severely diminished in B. abortus bvrS and bvrR mutants and these mutants are avirulent (22, 42), we decided to construct omp3a and omp3b knockout strains and to explore their biological characteristics. Although omp3a and omp3b mutants displayed some surface properties that distinguished them from the parental strain, we found that they did not reproduce the defects of the bvrS or bvrR mutants and remained virulent in the systems tested.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
The relevant characteristics of the bacterial strains and plasmids used in this study are listed in Table 1. Escherichia coli and Brucella strains were grown on tryptic soy broth, tryptic soy agar, or blood agar base (BAB). When needed, nalidixic acid (Nal; 25 μg/ml), kanamycin (Km; 50 μg/ml), or ampicillin (Amp; 100 μg/ml) was added to the cultures. Growth ability was tested in tryptic soy broth, brain heart infusion broth, and Gerhardt's modified minimal medium using an automatic microbiology analyzer (Bioscreen C; Labsystems, Vantaa, Finland).
TABLE 1.
Bacterial strains and plasmid constructs used in this study
| Strain or plasmid | Relevant characteristics | Reference or source |
|---|---|---|
| Strains | ||
| B. abortus | ||
| 2308 Nalr | Virulent WT strain, biotype 1, LPS-S, spontaneous Nalr mutant | 39 |
| bvrS mutant strain 2.13 | 2308 NalrbvrS::Tn5 | 42 |
| bvrR mutant strain 65.21 | 2308 NalrbvrR::Tn5 | 42 |
| bvrR+ strain 65.21p | 65.21 carrying plasmid pBBR2.13 | 42 |
| omp3a mutant 2308 | Nalromp3a::Km | This work |
| omp3b mutant 2308 | Nalromp3b::Km | This work |
| E. coli | ||
| SM10 (λ pir) | thi-1 thr leu tonA lacY supE recA::RP4-2-Tet::Mu Kmr; λ pir | 40 |
| XL1-Blue | TetrsupE44 hsdR17 recA1 endA1 gyrA96 thi relA1 lac F′ [proAB+lacIqlacZΔM15 Tn10 (Tetr)] | 38 |
| TOP 10 F′ | F′ [lacIq Tn10 (Tetr)] mcrA Δ(mrr-hsdRMS-mcrBC) φ80lacZΔM15 ΔlacX74 recA1 araD139 Δ(ara-leu) 7697 galU galK rpsL (Strr) endA1 nupG | Invitrogen |
| Plasmids | ||
| pCR2.1 | High-copy-number cloning vector; Ampr Kmr | Invitrogen |
| pUC4K | Plasmid containing kanamycin resistance cassette | Stratagene |
| pSK-oriT | pBluescript II SK(-) oriT RK2 Ampr | 43 |
| pBBR1MCS-4 | Intermediate-copy-number cloning vector; Ampr | 25 |
| pAC2553 | pUC19 containing Δomp3a::Km | A. Cloeckaert |
| pSK3aKm | pSK-oriT containing Δomp3a::Km (1.8 kb) | This work |
| pTA3a | pCR2.1 containing an internal fragment of B. abortus 2308 omp3a amplified by PCR (0.55 kb) | This work |
| pTA3b | pCR2.1 containing complete B. abortus omp3b gene amplified by PCR (1.1 kb) | This work |
| pTA3b− | pCR2.1 containing deleted omp3b gene (0.6 kb) | This work |
| pSK3b-Km | pSK-oriT containing Δomp3b::Km (1.9 kb) | This work |
| pBBR3b | pBBR1MCS-4 containing B. abortus omp3b gene (1.1 kb) | This work |
DNA and RNA manipulations.
Plasmid and chromosomal DNA were extracted with QIAprep spin miniprep (QIAGEN GmbH, Hilden, Germany) and Ultraclean microbial DNA isolation (MO BIO Laboratories, Inc.) kits, respectively. Primers were synthesized by Sigma-Genosys (Haverhill, United Kingdom). For RNA manipulation, bacterial cultures adjusted to 109 CFU/ml were disrupted with 0.5% Zwittergent 3-16 at 37°C for 1 h. Then, total RNA was extracted using an RNeasy minikit (QIAGEN) according to the manufacturer's instructions. RNA (0.2 μg) was used as a template for the synthesis of cDNA with SuperScript III reverse transcriptase (RT) (Invitrogen) using the random hexamers from the same kit.
Construction and characterization of omp3a and omp3b mutants.
The B. abortus 2308 omp3a::Km mutant was constructed by homologous recombination between the chromosomal omp3a gene and plasmid pSK3aKm carrying an omp3a::Km construct from pAC2553 (Table 1). This omp3a::Km construct was obtained after cleavage of the B. abortus 2308 omp3a gene with StyI and the insertion of a kanamycin resistance cassette. In order to facilitate plasmid mobilization by conjugation, omp3a::Km was subcloned from pAC2553 into the mobilizable plasmid pSK-oriT (43) as an XbaI-SacI 2.1-kb fragment, generating plasmid pSK3aKm. This new construct was confirmed by PCR with primers Omp25U1 (5′-TGCGCTGCTGCCGTTCTCTG-3′) and Omp25L1 (5′-GGATCCGGCCAGATCATAGTTCTTGT-3′), which amplify a specific 547-bp fragment of the omp3a gene, and by double digestion with EcoRI and HindIII. Plasmid pSK3aKm was introduced into B. abortus 2308 by conjugation with E. coli SM10 (λ pir) (39). The first recombination event (vector integration in the Brucella chromosome) was selected by Nal and Km resistance, and the second recombination event (excision of the mutator plasmid) was selected by Amp sensitivity. To confirm the mutation, different colonies were screened by PCR with primers Omp25U1 and Omp25L1. EcoRV-digested genomic DNA from selected clones was analyzed by Southern blotting using pSK-oriT, pSK3aKm, and pTA3a (Table 1) as probes and by RT-PCR as previously described (28) with primers Omp25U1 and Omp25L1. Failure to express Omp3a was confirmed by Western blotting using OM fragments (18) or Sarkosyl-resistant fractions with anti-Omp3a monoclonal antibodies (MAbs) A70/06B05/A07 and A76/02C12/C11 (9, 42). The B. abortus omp3b::Km mutant was constructed as follows. First, B. abortus 2308 omp3b was amplified by PCR with specific primers BAF-1 (5′-CCCGGCTGTTACATATGCTG-3′) and BAF-2 (5′-CGCGCTGATATCGACATGAC-3′) and cloned into vector pCR2.1 (Invitrogen). The resulting plasmid pTA3b was used as a template for inverse PCR mutagenesis to delete omp3b. This plasmid was first denatured with 1 M NaOH and 1 mM EDTA for 15 min at 37°C, neutralized with 3 M sodium acetate (pH 4.8), purified with ethanol, and finally resuspended in water (13). The sample was then inverse amplified with primers IM3B-1 (5′-ACGCGTCGACGCCGGCCTGAACTACAA-3′) and IM3B-2 (5′-ACGCGTCGACGCGGCGACAGGGTCGTTAT-3′) carrying a restriction site for SalI (underlined in the primer sequences). After 5 min at 95°C, amplification was carried out for 30 cycles of 1 min at 95°C, 45 s at 63°C, and 45 s at 68°C, and a final extension at 68°C for 10 min. The 4.5-kb amplified fragment was purified, digested with SalI, and religated to generate plasmid pTA3b−. To facilitate the counterselection of the mutant, a Km resistance cassette from plasmid pUC4K (Amersham Pharmacia Biotech, NJ) was cloned in the SalI site. The mutated omp3b gene was subcloned into pSK-oriT as an EcoRI 1.9-kb fragment generating plasmid pSK3bKm. This construct was verified by PCR with primers BAF-1 and BAF-2 and by digestion with EcoRI and SalI. Plasmid pSK3bKm was introduced into B. abortus 2308 by conjugation with E. coli SM10 (λ pir) and Nalr Kmr Amps transconjugants were selected. The resulting colonies were screened by PCR with primers BAF-1 and BAF-2 and primers C3BK-2 (5′-CCGCGCGGACACCAAGCCTA-3′) and C3BK-1 (5′-CGGCGGCGTGACGGATGAAG-3′), which amplify a 1.9-kb fragment of the omp3b gene. Mutation was confirmed by Southern blotting using plasmids pSK-oriT, pSK3bKm, and pTA3b as probes and EcoRI-digested genomic DNA, as well as by RT-PCR with specific primers 3bZ-1 (5′-GCGCGCAGGTTGGTGGTT-3′) and 3bZ-2 (5′-GGATCCGCCGGCCTTGATCGAATG-3′), which amplify a specific 473-bp fragment of the omp3b gene. The absence of Omp3b was corroborated by two-dimensional (2D) gel analysis of the OM fragments of the omp3b mutant and the wild-type (WT) strain as previously described (22).
To determine the stability of the omp3a and omp3b mutants in vitro, bacteria were grown on BAB for 24 h and serial dilutions were plated on BAB, BAB-Nal, BAB-Km, and BAB-Amp. In vivo stability was determined in the mouse model. Groups of five BALB/c mice (see below) were inoculated intraperitoneally with 0.1 ml of a suspension containing 105 CFU of each bacterial strain, and 2 weeks later they were sacrificed and the spleens were removed. Each spleen was homogenized, and decimal dilutions were plated in triplicate samples on BAB, BAB-Nal, BAB-Km, and BAB-Amp. Mutants were considered stable in vivo and in vitro when viable counts were the same in all media. LPS stability (smooth LPS versus rough-type LPS) and the presence of native hapten (NH) polysaccharide was also verified by crystal violet staining, immunodiffusion, immunofluorescence, and Western blotting (1, 28). Regular typing and sensitivity to colorants and phages were determined as described elsewhere (1).
Immunological methods.
Cell envelope components were analyzed and immunodetected as described previously (22). Direct, indirect, and double immunofluorescence assays for the determination of extracellular and intracellular bacteria were performed as described elsewhere (21, 35). The levels of TNF-α in a supernatant medium of B. abortus-infected murine RAW 264.7 macrophages were measured by enzyme-linked immunosorbent assay (BD Biosciences, San Diego, CA) at different time intervals according to the manufacturer's instructions.
Sensitivity to polymyxin B, antibiotics, and nonimmune serum.
Sensitivity to polymyxin B was tested as described elsewhere (42). The sensitivity to several hydrophilic and hydrophobic antibiotics was tested as described previously (29). The sensitivity to the bactericidal action of human and bovine serum was estimated by the method described by Skurnik et al. (41).
Internalization, survival, and replication assays in cells and mice.
Ex vivo infection assays were performed with HeLa cells (ATCC CCL-2), murine RAW 264.7 macrophages (ATCC TIB-71), and human polymorphonuclear neutrophils (PMN). Cell cultures and gentamicin survival assays were performed as described previously (6, 7, 21, 35, 47). For double immunofluorescence analysis of Brucella-infected HeLa or RAW 264.7 cells, procedures described previously were followed (6, 35). Counts of intracellular and extracellular bacteria were performed for at least 50 infected cells and were expressed as the mean and standard deviation of the percentage of intracellular bacteria and the number of bacteria per infected cell. The percentage of cells with associated bacteria (intra- and extracellular) was expressed as the mean and standard deviation of numbers of cells with bound bacteria in five different ×400 magnification fields. Statistical analysis was performed using Student's t test. PMN were purified from defibrinated blood extracted from human donors with no history of brucellosis. One part of blood diluted with 1 part minimal essential medium containing 5% inactivated fetal calf serum, with 25 mM HEPES and 2 mM glutamine (Sigma-Aldrich, Co). Eight milliliters of diluted blood was layered on the top of a biphasic gradient of Ficoll-Histopaque (3 ml with a density of 1.077 ± 0.001 on the top of 3 ml with a density of 1.119 ± 0.001; Sigma) on a conic tube and centrifuged at 4°C for 30 min at 700 × g. Granulocytes were extracted from the interphase between the two Ficoll layers, washed in supplemented medium without antibiotics, and counted. Infections were performed with 2-ml plastic tubes by mixing 5 × 105 PMN with 5 × 106 Brucella cells in a total volume of 250 μl of medium without antibiotics. The tubes were incubated at 37°C for 1 h with mild agitation. Finally, the mixture was centrifuged, the supernatants were removed, and the cells were lysed with 100 μl 0.1% Triton X-100. Dilutions of cell lysates were plated on tryptic soy agar, and the bacterial CFUs were counted after 3 days of incubation. The number of internalized Brucella cells in the PMN was recorded by immunofluorescence, as described previously (35).
Female BALB/c mice were infected by the intraperitoneal route with 105 CFU of B. abortus omp3a, omp3b, or WT strains as described previously (42). For the bvrS or bvrR mutant, doses of 108 CFU were used. For each strain, 30 mice were inoculated and the numbers of CFU in spleens were determined at various times postinfection. Statistical comparisons were performed by the Fisher's protected least significant differences test.
RESULTS
B. abortus omp3a and omp3b mutants do not display significant phenotypic deviations from the WT.
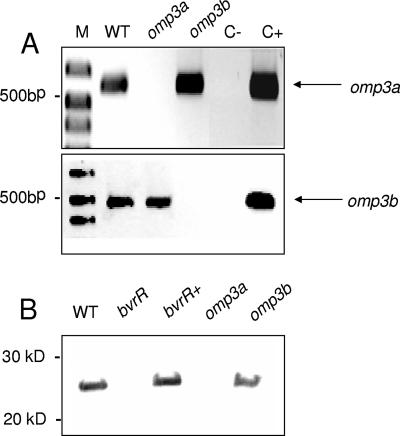
Independent disruption of omp3a and omp3b in B. abortus by the introduction of a kanamycin cassette was corroborated by RT-PCR and Western blotting. As expected, no transcription of specific mRNAs for omp3a or omp3b was detected by RT-PCR (Fig. 1A), and the absence of Omp3a in the omp3a mutant was demonstrated with MAbs against Omp3a (Fig. 1B). Antibodies against Omp3b are not currently available. However, 2D gel analysis of OM fragments of the omp3b mutant, the WT, and the omp3b mutant transformed with a plasmid encoding Omp3b demonstrated that protein spots similar to those previously shown to correspond to Omp3b isoforms (22) were absent in the omp3b mutant preparations and present in those of the WT and the omp3b mutant reconstituted strain (data not shown). In regular bacteriological media, both mutants and the WT showed similar growth patterns. These mutations did not affect the conventional phenotypic or metabolic properties described for B. abortus biotype 1 (1) or the ability of the mutants to grow in complex or defined media. The sensitivity to antibiotics such as doxycycline, gentamicin, streptomycin, chloramphenicol, penicillin, rifampin, and ciprofloxacin was similar to that of the WT but different from that of the bvrS or bvrR mutants, which displayed higher sensitivity (not shown). The LPS from the omp3a and omp3b mutants was smooth according to crystal violet staining, immunofluorescence, and sodium dodecyl sulfate-polyacrylamide gel electrophoresis analysis. Antibodies against Omp10, Omp16, Omp19, Omp2b, Omp1, and NH polysaccharide revealed quantities of these molecules in the omp3a and omp3b mutants similar to those in the WT strain (not shown).
FIG. 1.
Construction of B. abortus omp3a and omp3b mutants. (A) RT-PCR using specific primers for omp3a (top) and omp3b (bottom). Total RNA was extracted and retrotranscribed, and cDNA was amplified by PCR with primers Omp25U1/Omp25L1 and 3bZ-1/3bZ-2. Lanes: M, molecular size markers; WT, B. abortus 2308; omp3a, B. abortus omp3a mutant; omp3b, B. abortus omp3b mutant; C-, PCR-negative control with water; C+, PCR-positive control with Brucella genomic DNA. (B) Detection of Omp3a by Western blotting in cell envelope Sarkosyl-resistant fractions using anti-Omp3a MAbs. Lanes: WT, B. abortus 2308; bvrR, B. abortus bvrR::Tn5 mutant; bvrR+, reconstituted B. abortus bvrR+; omp3a, B. abortus omp3a deletion mutant; omp3b, B. abortus omp3b deletion mutant.
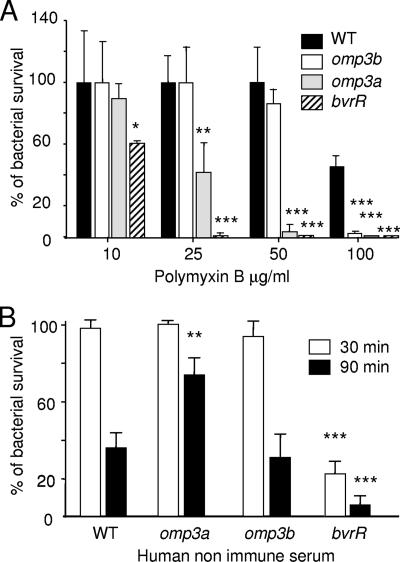
B. abortus omp3a and omp3b mutant resistance to polymixin B and complement.
The avirulent phenotype of the B. abortus bvrS and bvrR mutants correlates with their higher sensitivity to bactericidal cationic peptides and complement (28, 42). Although these features have been linked to structural alterations of the LPS molecule (28), the role of Omp3a and Omp3b, whose transcription is regulated by the BvrR/BvrS system, has not been explored. At 10 μg/ml of polymyxin B, the omp3a mutant displayed levels of polymixin B sensitivity between those of the WT and the bvrR mutant, whereas the omp3b mutant was as resistant as the WT (Fig. 2A). At 25 μg/ml of polymyxin B, the percent survival for the omp3a mutant was less than 50% and the percent survival for the omp3b mutant was similar to that of the WT. As expected, the percent survival of the bvrR mutant was close to zero. At 50 μg/ml, the omp3a mutant displayed very low levels of bacterial survival, but the level of survival of the omp3b mutant was similar to that of the WT strain. At concentrations as high as 100 μg/ml, both mutants showed practically no survival. Consistent with previous reports (28), the bvrR mutant was highly sensitive to the action of complement in normal serum after a 30-min incubation; however, both omp mutants were resistant (Fig. 2B). The bactericidal effect was more pronounced after 90 min of incubation in most of the strains. There were no significant differences between the omp3b mutant and the WT strain, but in contrast, the omp3a mutant displayed higher resistance than the WT strain. Similar results were obtained with bovine nonimmune serum (data not shown).
FIG. 2.
Sensitivity to polymixin B and human complement. (A) Bacterial survival in the presence of polymyxin B. The graph shows the percent survival after 1 h at 37°C with different peptide concentrations. (B) Bacterial survival after 30- or 90-min incubation with human nonimmune serum. Data represent the means ± standard deviations of percentages of viable bacteria in relation to a bacterial control without polymyxin B or a bacterial control with heat-inactivated human serum. Samples were compared using the Mann-Whitney U test. *, P < 0.05; **, P < 0.005; ***, P < 0.0005 (with respect to the WT strain).
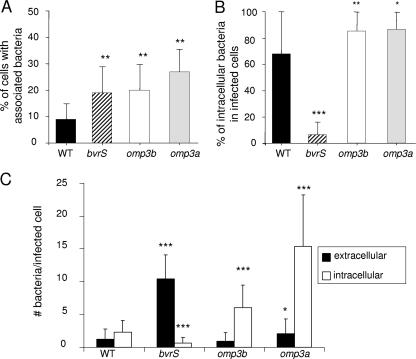
B. abortus omp3a and omp3b mutants invade, survive, and replicate within professional and nonprofessional phagocytes.
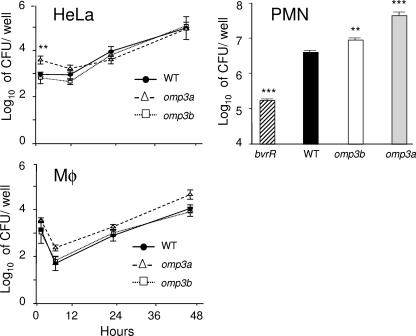
B. abortus bvrS and bvrR mutants are poor invaders and fail to survive and replicate within professional and nonprofessional phagocytes (42). To test whether their deficiency in Omp3a and Omp3b proteins could explain this phenotype, both omp mutants were evaluated with HeLa cells by using the gentamicin survival assay and double immunofluorescence microscopy to distinguish intracellular from extracellular bacteria (Fig. 3). Similar to the bvrS mutant, the omp3a and omp3b mutants attached to more cells than the WT (Fig. 3A). In contrast to the bvrS mutant, however, the percentage of intracellular bacteria was higher for these mutants than for the WT strain (Fig. 3B). Moreover, the absolute number of bacteria per cell was higher for all mutants than for the WT. As expected, the number of intracellular bacteria per cell was significantly higher for the omp3b and omp3a mutants and significantly lower for the bvrS mutant than for the WT (Fig. 3C). In order to analyze the sensitivity of the omp3a and omp3b mutants to the killing action of cells, HeLa cells, PMN, and RAW 264.7 cells were infected and the replication rates were compared with those of the WT strain and the avirulent bvrR mutant (Fig. 4). As reported previously (42), the bvrR mutant failed to replicate in nonprofessional phagocytic HeLa cells and macrophages (not shown). Although the omp3a mutant consistently displayed higher counts at initial times of infection, the replication levels of both omp mutants were not considerably different from that of the WT strain at later times (Fig. 4). Similar results were obtained with naïve bone marrow-derived murine macrophages (not shown). As expected, the bvrR mutant was readily killed by PMN, while both omp mutants displayed a resistance slightly higher than that of the WT strain (Fig. 4).
FIG. 3.
Gentamicin survival assay by double immunofluorescence 30 min postinfection in HeLa cells. (A) Proportion of cells with associated (intra- and extracellular) bacteria. (B) Proportion of intracellular bacteria with respect to the total of intra- and extracellular bacteria. (C) Absolute number of intra- and extracellular bacteria per infected cell. Data represent means ± standard deviations. Samples were compared using the Student t test. *, P < 0.05; **, P < 0.005; ***, P < 0.0005 (with respect to the WT strain).
FIG. 4.
Intracellular replication of B. abortus strains in epithelial HeLa cells, RAW 264.7 macrophages (Mφ), and PMN. Data represent means ± standard deviations of plate counts. Samples were compared using the Mann-Whitney U test. **, P < 0.005; ***, P < 0.0005 (with respect to the WT strain).
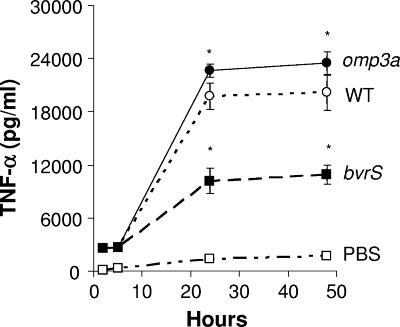
B. abortus omp3a and omp3b mutants induce higher levels of TNF-α in murine macrophages than the bvrS mutant.
It has been proposed that Omp3a from B. suis is involved in the inhibition of TNF-α production during infection of human macrophages (24) but not of murine macrophages (5, 12, 20). We measured the production of TNF-α in murine RAW 264.7 macrophages infected with omp3a or bvrS mutants (Fig. 5). Consistent with the rates of replication in macrophages, the levels of TNF-α induced by the omp3a mutant were in the same range as those of the WT. However, the levels of this cytokine induced by the attenuated bvrS mutant were significantly lower.
FIG. 5.
Induction of TNF-α in murine RAW 264.7 macrophages infected with different B. abortus strains. *, P < 0.05 (with respect to the WT strain).
B. abortus omp3a and omp3b mutants replicate in BALB/c mice.
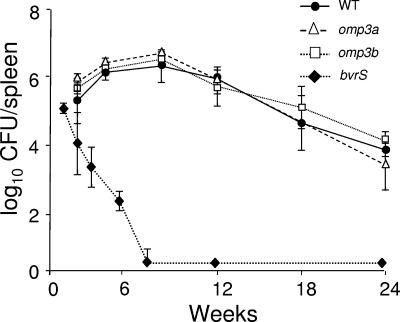
As described previously (42), the WT strain maintained high counts in the spleens of BALB/c mice 24 weeks after infection, while bvrS and bvrR mutants were eliminated within 3 weeks. Similar to the results obtained with cells, the replication of omp3a and omp3b mutants in mice did not significantly depart from that of the WT throughout the 24-week period of the experiment (Fig. 6).
FIG. 6.
Infection of the spleens of BALB/c mice with different B. abortus strains. Mice were infected intraperitoneally with 105 CFU/mouse, except for the bvrS mutant, for which the dose was 108 CFU/mouse. Values are means ± standard deviations (n = 5). The detection limit was 0.6 log CFU/spleen (3 to 4 CFU/spleen).
DISCUSSION
We have shown that the absence of either Omp3a or Omp3b does not lead to phenotypes resembling bvrS or bvrR mutants but rather to phenotypes similar to that of the WT strain. Indeed, both omp mutants replicated in macrophages and epithelial cells to the same extent as the WT, and more importantly, persisted in mice for up to 24 weeks, exhibiting profiles similar to those of the WT strain. There were, however, some discrete differences between the omp mutants and WT Brucella. For instance, both omp mutants bound more readily to epithelial cells (similar to bvrS and bvrR mutants) and were slightly more resistant to the killing action of human PMN. The omp3a mutant was more sensitive to polymixin B and more resistant to the bactericidal action of complement after 90 min than the WT strain and induced slightly larger amounts of TNF-α in murine macrophages than the WT (see below). These results and those obtained for the B. abortus omp3b mutant in the same set of experiments are slightly different from those recently reported for two Brucella ovis omp3a and omp3b mutants (4). The differences in experimental setups, as well as the fact that B. ovis is a rough bacterium, might explain these discrepancies. The reason why the B. abortus omp3a mutant is more resistant to the bactericidal action of complement than the WT remains elusive. It is not known why B. abortus is more resistant to complement than other bacteria. Although there is some evidence that indicates that this could be attributed to its LPS (30), we did not detect any differences between the mutants' LPS and the parent strain's LPS with the methods used. Nevertheless, the purpose of using these kinds of experiments is to reveal OM properties that might be altered in the mutants compared to the WT. In this sense, we could speculate that the absence of Omp3a but not of Omp3b or both in the mutant strains allows the exposure in the OM of other molecules that are able to activate complement in higher levels than the WT.
Besides the nonsignificant variations with respect to the WT at the LPS level, the B. abortus Omp mutants did not show important variations in the quantities of other cell envelope components, such as NH or β-cyclic glucans, Omp10, Omp16, Omp19, Omp2b, and Omp1, or display major differences in OM permeability or growth rates. However, this does not rule out the existence of additional changes in other surface molecules, given that group 3 Omps are highly abundant and they strongly associate with the LPS (19, 22, 36). A search of the B. abortus genome reveals more than 25 proteins and lipoproteins predicted to be located in the OM, a fact that is sustained by a recent proteomic analysis of B. abortus OM fragments (26). Therefore, it is possible that some of these proteins are also affected, as we have seen in 2D gels of OM fragments from these mutants (data not shown).
We observed that RAW 264.7 murine macrophages infected with the B. abortus omp3a mutant generated slightly larger amounts of TNF-α than the WT strain. These results are in agreement with those previously reported (5, 12, 20), showing that the production of TNF-α in murine macrophages is not related to Omp3a. However, the bvrS mutant, harboring very low quantities of Omp3a, Omp3b, and most likely other membrane defects (28), induced small amounts of TNF-α. This effect is probably due to phenotypic changes in the cell envelope influencing the biological behavior of the mutant strains.
Consistent with the lack of important OM defects, the omp3a and omp3b mutants and the WT strain displayed almost identical replication rates in cells and similar numbers of colonies in mouse spleens. This is in sharp contrast with the fast clearing of the bvrS mutant. The omp3a and omp3b mutants bind to more cells and are internalized more by the cell population, and in absolute numbers, they show higher counts of total and internalized bacteria per cell than the WT strain. Therefore, and contrary to what is observed with the bvrS mutant, the internalization process in these bacteria is not impaired. This observation suggests that the two-component system is affecting the internalization process in a way not directly related to the presence of Omp3a or Omp3b. Although the omp3a and omp3b mutants are more efficiently internalized than the WT, they show replication and survival rates in cells and mice similar to those shown by the WT. The reason for this remains elusive. One possibility is that, in spite of their higher internalization rates, the actual absolute number of mutant bacteria reaching the final niche, i.e., the endoplasmic reticulum, is the same as the absolute number of WT bacteria. Overall, these results indicate that the absence of one of these two group 3 Omps does not generate attenuated B. abortus phenotypes. In previous reports, it has been described that a B. abortus 2308 omp3a deletion mutant opsonized with hyperimmune murine serum has delayed growth in late gestational bovine chorionic trophoblasts and cultured bovine macrophages. In fact, the percent survival of this mutant does not reach the levels of the WT after 48 h of infection (14). However, the data were normalized so that 100% survival represented the number of bacteria recovered after a 2-h incubation with macrophages. Our data show that both the omp3a and omp3b mutants were more efficiently internalized than the WT, and therefore their initial CFU counts were higher than those obtained for the WT. This difference is more obvious for HeLa cells than for murine macrophages. Conversely, during the following hours after infection but before replication occurs, the CFU counts decreased to the WT level. This shows that these mutants have an initially higher destruction rate than the WT strain, as suggested by Edmonds et al. (14). However, the CFU counts after 12 h of infection show that both mutants reached the same absolute numbers as the WT, indicating that the numbers of bacteria that replicate and consequently adapt to the intracellular niche are the same. Indeed, if our results are represented in the same way as they were by Edmonds et al. (14), the data are the same. Therefore, it is clear that increased efficiency of internalization compensates for augmented intracellular destruction.
Caro-Hernández et al. (4) showed that a B. ovis omp3b mutant is attenuated in a mouse model compared with the parent strain. However, and in contrast to what we observed with our B. abortus omp3b mutant, the B. ovis omp3b mutant has greater susceptibility to nonimmune serum and has growth defects, particularly when reaching the stationary phase. These differences might be explained from the OM physiology context; because B. ovis is a rough bacterium, it is more likely that the absence of Omp3b might result in drastic modifications of its biology. On the other hand, and consistent with our results, Caro-Hernández et al. also showed that a B. ovis omp3a mutant is virulent in the same mouse model (4). Conversely, Edmonds et al. (14, 15) reported that a B. abortus omp3a mutant was attenuated in cattle and in mice 18 to 20 weeks postinfection. In our mouse experiments, we did not detect significant variations in a 24-week follow-up period with respect to the WT. At the present time, we do not have an explanation for this discrepancy. However, there is more evidence consistent with our observations. Neither the kinetics of spleen infections nor the residual virulence of B. melitensis Rev1 in mice is modified by deletion of group 3 Omp31 (8). Although further research is needed, a hypothesis is that group 3 Omps are an interdependent and coordinated group of proteins that have become redundant to secure the presence of at least one member in the OM and to permit certain cell envelope plasticity in order to live in different environments. This would account for the role of BvrR/BvrS and for the somewhat surprising phenotype of the omp3a and omp3b mutants since, according to the hypothesis, the absence of one protein of the group will be balanced by one or more of the other members. In fact, it has been observed that knocking out omp25c, omp25d, or omp3b increases Omp25b production in B. suis, and on this basis, a compensatory regulation within group 3 Omps is suggested (37). Furthermore, a tight balance of the group 3 Omps seems to be essential for the integrity of the B. ovis membrane (4). Virulent WT B. abortus naturally lacks functional genes for group 3 Omp31 and Omp25b (23), showing that the absence of one or two members of this family is not decisive for virulence. All this indirect evidence has to be considered with caution because B. melitensis and B. abortus do not have the same profile of group 3 Omps and there are contradictory reports on the level of group 3 Omps in B. suis mutants with mutations in bvrR and/or bvrS (3, 37). Nevertheless, the compensatory hypothesis would explain why mutations in a single Omp do not cause marked phenotypic changes, while dysfunction in BvrR/BvrS has a profound influence, at least in B. abortus. In this regard, it can also be hypothesized that the BvrR/BvrS influence may extend directly or indirectly to other group 3 members, to other OM molecules, or beyond the OM structure. A proteomic analysis of the complete cell envelope of the B. abortus WT and avirulent bvrS and bvrR mutants indicates that this might be the case, since these mutants have small amounts of different group 3 Omps and altered quantities of other Omps and periplasmic components compared with the WT (26).
Acknowledgments
This work was partially supported by grant 8-N-2005 from NeTropica; grants from the MICIT/CONICIT, Costa Rica; a grant from Florida Ice & Farm; FIDA from the Universidad Nacional (UNA); and FS from the CONARE UNA/UCR agreement. Other grants were from the IFS (B/3456-1 and B/3456-2), the MEC of Spain (AGL2004-01162 and BIO2005-04985), and RTIC del FIS (objetivo 2, 2000-2006, G03/204). Fellowship support to L. Manterola from the Gobierno Vasco is gratefully acknowledged.
We thank C. Chacón and F. Mena for their assistance in the serologic analysis and A. Cloeckaert for providing some of the MAbs and plasmids used. We thank Samuel Wagner for the critical reading of the manuscript.
Editor: J. B. Bliska
Footnotes
Published ahead of print on 30 July 2007.
REFERENCES
- 1.Alton, G. G., L. M. Jones, R. D. Angus, and J. M. Verger. 1988. Techniques for the brucellosis laboratory. Institut National de la Recherche Agronomique, Paris, France.
- 2.Arellano-Reynoso, B., N. Lapaque, S. Salcedo, G. Briones, A. E. Ciocchini, R. Ugalde, E. Moreno, I. Moriyón, and J. P. Gorvel. 2005. Cyclic beta-1,2-glucan is a Brucella virulence factor required for intracellular survival. Nat. Immunol. 6:618-625. [DOI] [PubMed] [Google Scholar]
- 3.Boigegrain, R.-A., I. Salhi, M.-T. Alvarez-Martinez, J. Machold, Y. Fedon, M. Arpagaus, C. Weise, M. Rittig, and B. Rouot. 2004. Release of periplasmic proteins of Brucella suis upon acidic shock involves the outer membrane protein Omp25. Infect. Immun. 72:5693-5703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Caro-Hernández, P., L. Fernández-Lago, M.-J. de Miguel, A. I. Martín-Martín, A. Cloeckaert, M.-J. Grillo, and N. Vizcaíno. 2007. Role of the Omp25/Omp31 family in the outer membrane properties and virulence of Brucella ovis. Infect. Immun. 75:4050-4061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Caron, E., A. Gross, J. P. Liautard, and J. Dornand. 1996. Brucella species release a specific, protease-sensitive, inhibitor of TNF-alpha expression, active on human macrophage-like cells. J. Immunol. 156:2885-2893. [PubMed] [Google Scholar]
- 6.Celli, J., C. de Chastellier, D. M. Franchini, J. Pizarro-Cerda, E. Moreno, and J. P. Gorvel. 2003. Brucella evades macrophage killing via VirB-dependent sustained interactions with the endoplasmic reticulum. J. Exp. Med. 198:545-556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chaves-Olarte, E., C. Guzmán-Verri, S. Meresse, M. Desjardins, J. Pizarro-Cerda, J. Badilla, J. P. Gorvel, and E. Moreno. 2002. Activation of Rho and Rab GTPases dissociates Brucella abortus internalization from intracellular trafficking. Cell. Microbiol. 4:663-676. [DOI] [PubMed] [Google Scholar]
- 8.Cloeckaert, A., I. Jacques, M. J. Grillo, C. M. Marin, M. Grayon, J. M. Blasco, and J. M. Verger. 2004. Development and evaluation as vaccines in mice of Brucella melitensis Rev. 1 single and double deletion mutants of the bp26 and omp31 genes coding for antigens of diagnostic significance in ovine brucellosis. Vaccine 22:2827-2835. [DOI] [PubMed] [Google Scholar]
- 9.Cloeckaert, A., J.-M. Verger, M. Grayon, M. S. Zygmunt, and O. Grépinet. 1996. Nucleotide sequence and expression of the gene encoding the major 25-kilodalton outer membrane protein of Brucella ovis: evidence for antigenic shift, compared with other Brucella species, due to a deletion in the gene. Infect. Immun. 64:2047-2055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cloeckaert, A., N. Vizcaino, J. Y. Paquet, R. A. Bowden, and P. H. Elzer. 2002. Major outer membrane proteins of Brucella spp.: past, present and future. Vet. Microbiol. 90:229-247. [DOI] [PubMed] [Google Scholar]
- 11.Delpino, M. V., J. Cassataro, C. A. Fossati, F. A. Goldbaum, and P. C. Baldi. 2006. Brucella outer membrane protein Omp31 is a haemin-binding protein. Microbes Infect. 8:1203-1208. [DOI] [PubMed] [Google Scholar]
- 12.Dornand, J., A. Gross, V. Lafont, J. Liautard, J. Oliaro, and J. P. Liautard. 2002. The innate immune response against Brucella in humans. Vet. Microbiol. 90:383-394. [DOI] [PubMed] [Google Scholar]
- 13.Dorrell, N., V. G. Gyselman, S. Foynes, S. R. Li, and B. W. Wren. 1996. Improved efficiency of inverse PCR mutagenesis. BioTechniques 21:604-608. [DOI] [PubMed] [Google Scholar]
- 14.Edmonds, M. D., A. Cloeckaert, N. J. Booth, W. T. Fulton, S. D. Hagius, J. V. Walker, and P. H. Elzer. 2001. Attenuation of a Brucella abortus mutant lacking a major 25 kDa outer membrane protein in cattle. Am. J. Vet. Res. 62:1461-1466. [DOI] [PubMed] [Google Scholar]
- 15.Edmonds, M. D., A. Cloeckaert, and P. H. Elzer. 2002. Brucella species lacking the major outer membrane protein Omp25 are attenuated in mice and protect against Brucella melitensis and Brucella ovis. Vet. Microbiol. 88:205-221. [DOI] [PubMed] [Google Scholar]
- 16.Edmonds, M. D., A. Cloeckaert, S. D. Hagius, L. E. Samartino, W. T. Fulton, J. V. Walker, F. M. Enright, N. J. Booth, and P. H. Elzer. 2002. Pathogenicity and protective activity in pregnant goats of a Brucella melitensis Δomp25 deletion mutant. Res. Vet. Sci. 72:235-239. [DOI] [PubMed] [Google Scholar]
- 17.Fretin, D., A. Fauconnier, S. Kohler, S. Halling, S. Leonard, C. Nijskens, J. Ferooz, P. Lestrate, R. M. Delrue, I. Danese, J. Vandenhaute, A. Tibor, X. DeBolle, and J. J. Letesson. 2005. The sheathed flagellum of Brucella melitensis is involved in persistence in a murine model of infection. Cell. Microbiol. 7:687-698. [DOI] [PubMed] [Google Scholar]
- 18.Gamazo, C., and I. Moriyón. 1987. Release of outer membrane fragments by exponentially growing Brucella melitensis cells. Infect. Immun. 55:609-615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gamazo, C., A. I. Vitas, I. Moriyón, I. López-Goñi, and R. Diaz. 1993. Brucella group 3 outer membrane proteins contain a heat-modifiable protein. FEMS Microbiol. Lett. 112:141-146. [DOI] [PubMed] [Google Scholar]
- 20.Gross, A., S. Spiesser, A. Terraza, B. Rouot, E. Caron, and J. Dornand. 1998. Expression and bactericidal activity of nitric oxide synthase in Brucella suis-infected murine macrophages. Infect. Immun. 66:1309-1316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Guzmán-Verri, C., E. Chaves-Olarte, C. von Eichel-Streiber, I. López-Goñi, M. Thelestam, S. Arvidson, J. P. Gorvel, and E. Moreno. 2001. GTPases of the Rho subfamily are required for Brucella abortus internalization in nonprofessional phagocytes: direct activation of Cdc42. J. Biol. Chem. 276:44435-44443. [DOI] [PubMed] [Google Scholar]
- 22.Guzmán-Verri, C., L. Manterola, A. Sola-Landa, A. Parra, A. Cloeckaert, J. Garin, J. P. Gorvel, I. Moriyón, E. Moreno, and I. López-Goñi. 2002. The two-component system BvrR/BvrS essential for Brucella abortus virulence regulates the expression of outer membrane proteins with counterparts in members of the Rhizobiaceae. Proc. Natl. Acad. Sci. USA 99:12375-12380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Halling, S. M., B. D. Peterson-Burch, B. J. Bricker, R. L. Zuerner, Z. Qing, L.-L. Li, V. Kapur, D. P. Alt, and S. C. Olsen. 2005. Completion of the genome sequence of Brucella abortus and comparison to the highly similar genomes of Brucella melitensis and Brucella suis. J. Bacteriol. 187:2715-2726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jubier-Maurin, V., R.-A. Boigegrain, A. Cloeckaert, A. Gross, M.-T. Alvarez-Martinez, A. Terraza, J. Liautard, S. Köhler, B. Rouot, J. Dornand, and J. P. Liautard. 2001. Major outer membrane protein Omp25 of Brucella suis is involved in inhibition of tumor necrosis factor alpha production during infection of human macrophages. Infect. Immun. 69:4823-4830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kovach, M. E., R. W. Phillips, P. H. Elzer, R. M. Roop II, and K. M. Peterson. 1994. pBBR1MCS: a broad-host-range cloning vector. BioTechniques 16:800-802. [PubMed] [Google Scholar]
- 26.Lamontagne, J., H. Butler, E. Chaves-Olarte, M. S. J. Hunter, M. T. C. Paquet, P. Kearney, L. Hamaidi, D. Chelsky, I. Moriyón, E. Moreno, and E. Paramithiotis. 2007. Extensive cell envelope modulation is associated with virulence in Brucella abortus. J. Proteome Res. 6:1519-1529. [DOI] [PubMed] [Google Scholar]
- 27.Lapaque, N., I. Moriyón, E. Moreno, and J. P. Gorvel. 2005. Brucella lipopolysaccharide acts as a virulence factor. Curr. Opin. Microbiol. 8:60-66. [DOI] [PubMed] [Google Scholar]
- 28.Manterola, L., I. Moriyón, E. Moreno, A. Sola-Landa, D. S. Weiss, M. H. Koch, J. Howe, K. Brandenburg, and I. López-Goñi. 2005. The lipopolysaccharide of Brucella abortus BvrS/BvrR mutants contains lipid A modifications and has higher affinity for bactericidal cationic peptides. J. Bacteriol. 187:5631-5639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Martínez de Tejada, G., and I. Moriyón. 1993. The outer membranes of Brucella spp. are not barriers to hydrophobic permeants. J. Bacteriol. 175:5273-5275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Moreno, E., D. T. Berman, and L. A. Boettcher. 1981. Biological activities of Brucella abortus lipopolysaccharides. Infect. Immun. 31:362-370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Moreno, E., and J.-P. Gorvel. 2004. Invasion, intracellular trafficking and replication of Brucella organisms in professional and non-professional phagocytes, p.287-312. In I. López-Goñi and I. Moriyón (ed.), Brucella: molecular and cellular biology. Horizon Bioscience, Norfolk, United Kingdom.
- 32.Moreno, E., and I. Moriyón. 2002. Brucella melitensis: a nasty bug with hidden credentials for virulence. Proc. Natl. Acad. Sci. USA 99:1-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Moreno, E., and I. Moriyón. 2006. The genus Brucella, p.315-456. In M. Dorkin, S. Falkow, E. Rosenberg, K.-H. Schleifer, and E. Stackebrandt (ed.), The prokaryotes,vol. 5. Springer-Verlag, New York, NY. [Google Scholar]
- 34.Moriyón, I., and I. López-Goñi. 1998. Structure and properties of the outer membranes of Brucella abortus and Brucella melitensis. Int. Microbiol. 1:19-26. [PubMed] [Google Scholar]
- 35.Pizarro-Cerdá, J., S. Méresse, R. G. Parton, G. van der Goot, A. Sola-Landa, I. Lopez-Goñi, E. Moreno, and J. P. Gorvel. 1998. Brucella abortus transits through the autophagic pathway and replicates in the endoplasmic reticulum of nonprofessional phagocytes. Infect. Immun. 66:5711-5724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Riezu-Boj, J. I., I. Moriyón, J. M. Blasco, C. Gamazo, and R. Díaz. 1990. Antibody response to Brucella ovis outer membrane proteins in ovine brucellosis. Infect. Immun. 58:489-494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Salhi, I., R.-A. Boigegrain, J. Machold, C. Weise, A. Cloeckaert, and B. Rouot. 2003. Characterization of new members of the group 3 outer membrane protein family of Brucella spp. Infect. Immun. 71:4326-4332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 39.Sangari, F., and J. Aguero. 1991. Mutagenesis of Brucella abortus: comparative efficiency of three transposon delivery systems. Microb. Pathog. 11:443-446. [DOI] [PubMed] [Google Scholar]
- 40.Simon, R., U. Priefer, and A. Puhler. 1983. A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in gram negative bacteria. Bio/Technology 1:784-791. [Google Scholar]
- 41.Skurnik, M., R. Venho, J. A. Bengoechea, and I. Moriyón. 1999. The lipopolysaccharide outer core of Yersinia enterocolitica serotype O:3 is required for virulence and plays a role in outer membrane integrity. Mol. Microbiol. 31:1443-1462. [DOI] [PubMed] [Google Scholar]
- 42.Sola-Landa, A., J. Pizarro-Cerda, M. J. Grillo, E. Moreno, I. Moriyón, J. M. Blasco, J. P. Gorvel, and I. López-Goñi. 1998. A two-component regulatory system playing a critical role in plant pathogens and endosymbionts is present in Brucella abortus and controls cell invasion and virulence. Mol. Microbiol. 29:125-138. [DOI] [PubMed] [Google Scholar]
- 43.Tibor, A., V. Wansard, V. Bielartz, R.-M. Delrue, I. Danese, P. Michel, K. Walravens, J. Godfroid, and J.-J. Letesson. 2002. Effect of omp10 or omp19 deletion on Brucella abortus outer membrane properties and virulence in mice. Infect. Immun. 70:5540-5546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Verstreate, D. R., M. T. Creasy, N. T. Caveney, C. L. Baldwin, M. W. Blab, and A. J. Winter. 1982. Outer membrane proteins of Brucella abortus: isolation and characterization. Infect. Immun. 35:979-989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Vizcaino, N., P. Caro-Hernandez, A. Cloeckaert, and L. Fernandez-Lago. 2004. DNA polymorphism in the omp25/omp31 family of Brucella spp.: identification of a 1.7-kb inversion in Brucella cetaceae and of a 15.1-kb genomic island, absent from Brucella ovis, related to the synthesis of smooth lipopolysaccharide. Microbes Infect. 6:821-834. [DOI] [PubMed] [Google Scholar]
- 46.Vizcaino, N., J. M. Verger, M. Grayon, M. S. Zygmunt, and A. Cloeckaert. 1997. DNA polymorphism at the omp-31 locus of Brucella spp.: evidence for a large deletion in Brucella abortus, and other species-specific markers. Microbiology 143:2913-2921. [DOI] [PubMed] [Google Scholar]
- 47.Weiss, D. S., K. Takeda, S. Akira, A. Zychlinsky, and E. Moreno. 2005. MyD88, but not Toll-like receptors 4 and 2, is required for efficient clearance of Brucella abortus. Infect. Immun. 73:5137-5143. [DOI] [PMC free article] [PubMed] [Google Scholar]