Abstract
DNA adenine methyltransferase (Dam) not only regulates basic cellular functions but also interferes with the proper expression of virulence factors in various pathogens. We showed previously that for the human pathogen Yersinia enterocolitica, overproduction of Dam results in increased invasion of epithelial cells. Since invasion and motility are coordinately regulated in Y. enterocolitica, we analyzed the motility of a Dam-overproducing (DamOP) strain and found it to be highly motile. In DamOP strains, the operon encoding the master regulator of flagellum biosynthesis, flhDC, is upregulated. We show that the increased invasion is not due to enhanced expression of known and putative Y. enterocolitica invasion and adhesion factors, such as Inv, YadA, Ail, Myf fibrils, Pil, or Flp pili. However, overproduction of Dam no longer results in increased invasion for an inv mutant strain, indicating that Inv is necessary for increased invasion after overproduction of Dam. Since we show that overproduction of Dam results in an increased amount of rough lipopolysaccharide (LPS) molecules lacking O-antigen side chains, this implies that reduced steric hindrance by LPS might contribute to increased invasion by a Y. enterocolitica DamOP strain. Our data add an important new aspect to the various virulence-associated phenotypes influenced by DNA methylation in Y. enterocolitica and indicate that Dam targets regulatory processes modulating the composition and function of the bacterial surface.
The DNA adenine methyltransferase (Dam) of gammaproteobacteria catalyzes the methylation of adenine residues at the N6 position in GATC sequences. Methylation occurs directly after DNA replication with a delay, thereby leaving the newly synthesized daughter strand nonmethylated for a short period of time. Depending on their presence and their affinity, methylation-sensitive regulatory proteins either bind to the hemimethylated DNA during this time and prevent the subsequent methylation of GATC sequences or bind preferentially to fully methylated DNA. Therefore, the methylation status of GATC sequences has an impact on the binding of regulatory proteins and consequently is involved in the regulation of several basic processes of the bacterial cell, such as mismatch repair, chromosome replication, transposition, or transcription of genes (10, 56).
It is therefore not surprising that Dam influences the virulence properties of a variety of bacterial pathogens. Dam-overproducing (DamOP) and/or dam mutant strains of Salmonella enterica (dam), Vibrio cholerae (DamOP), Pasteurella multocida (DamOP), Aeromonas hydrophila (DamOP), Haemophilus influenzae (dam), Klebsiella pneumoniae (dam), Yersinia pestis (dam), or Yersinia pseudotuberculosis (DamOP, dam) show reduced virulence in animal models of infection (11, 20, 23, 25, 26, 30, 34, 42, 46, 48, 55). These in vivo outcomes are most likely the result of the effects of DNA adenine methylation on the regulation of diverse virulence functions (26). For example, it has been demonstrated that Dam influences the proper secretion and translocation of type III effector proteins in Y. pseudotuberculosis (DamOP), S. enterica (dam), or A. hydrophila (DamOP) (20, 23, 30, 31). Furthermore, host cell invasion and adhesion are affected in strains of S. enterica (dam) and H. influenzae (dam) with altered Dam methylation (23, 55). A variety of adhesive pili of S. enterica and Escherichia coli are phase-variably expressed, with adenine methylation stabilizing the ON or OFF phase (10). A changed methylation status also decreases the swimming motility of E. coli (dam), A. hydrophila (DamOP), and S. enterica (DamOP, dam) (4, 5, 20, 41).
The food-borne human pathogen Yersinia enterocolitica is able to cause different gastrointestinal syndromes, ranging from self-limiting enteritis to mesenteric lymphadenitis. In rare cases, Y. enterocolitica is able to disseminate to deeper tissues and cause systemic infections (9, 16). We used a DamOP strain as a tool to identify and analyze methylation-sensitive processes implicated in virulence of this pathogen. By using DamOP strains as well as dam mutant strains, it is possible to alter methylation patterns in regulatory regions of genes, whereby the affinity of transcription factors for these regions can be altered, thereby mimicking a situation that can also be found in vivo (26, 56). In previous studies, we could demonstrate that overproduction of the Dam enzyme in Y. enterocolitica leads to a relaxed Ca2+ regulation of the Yop/Ysc type III secretion system. This effect depends at least in part on a ClpP-mediated degradation of the regulatory protein LcrG (21). Furthermore, we found that the expression of several in vivo-expressed genes is not changed in DamOP strains. However, a DamOP strain cured of the pYV virulence plasmid shows an increased ability to invade cultured epithelial cells (22). Invasion processes are critical steps in Y. enterocolitica pathogenesis, since infection and colonization of the intestinal lymphoid tissue require transmigration of the bacteria from the intestinal lumen across the epithelial barrier (1, 24).
Therefore, we aimed at identifying the mechanisms leading to the hyperinvasive phenotype after overproduction of Dam in more detail. Furthermore, since the ability to invade epithelial cells is often coregulated with other virulence functions, we wondered if Dam influences these functions as well. In this study, we demonstrate that overproduction of Dam leads to an increased motility of Y. enterocolitica. Furthermore, we present data indicating that despite the hyperinvasive phenotype, the expression of a variety of known and putative surface proteins implicated in invasion and adhesion is reduced after overproduction of Dam. In relation to a new role for Dam in the regulation of bacterial pathogenesis, we present data that the lipopolysaccharide (LPS) O-antigen status is also changed, putatively increasing the accessibility of Inv and at least in part being responsible for the increased ability of the bacteria to invade host cells. Altogether, our data indicate that Dam influences diverse virulence-associated surface properties of Y. enterocolitica, thereby contributing to the multifaceted functions of DNA methylation in regulating virulence.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
Bacterial strains and plasmids used in this study are listed in Table 1. Unless otherwise indicated, all strains were grown in Luria-Bertani (LB) broth or on agar plates at 26°C for Y. enterocolitica or at 37°C for E. coli. Antibiotics were used as described previously (22). Overproduction of Dam was achieved by electroporating pTP166Kan or pTP166Kan-damΔ as a negative control into the corresponding Y. enterocolitica strain and inducing expression of dam from the Ptac promoter by the addition of 1 mM isopropyl-β-d-thiogalactopyranoside (IPTG) (also for cells used as negative control) 30 min after subculture for each assay. The construction of pTP166Kan and pTP166Kan-damΔ has been described in detail previously (22). Both constructs are derivatives of pTP166, which carries the E. coli dam gene under the control of the Ptac promoter (33). To allow propagation of the plasmids in Y. enterocolitica, the bla gene encoding ampicillin resistance was replaced by a kanamycin resistance cassette. In pTP166Kan-damΔ, the Ptac promoter and the 5′ part of the dam gene have been deleted.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Relevant characteristics | Source or reference |
|---|---|---|
| Y. enterocolitica strains | ||
| JB580v | ΔyenR (r− m+) Nalr, serogroup O:8 | 32 |
| GHY147 | JB580v, pTP166Kan-damΔ, Nalr Kanr | 22 |
| GHY150 | JB580v, pTP166Kan, Nalr Kanr | 22 |
| GHY174 | JB580v, flhD::lacZYA, Nalr Camr | This study |
| GHY175 | JB580v, flhD::lacZYA, pTP166Kan-damΔ, Nalr, Camr Kanr | This study |
| GHY176 | JB580v, flhD::lacZYA, pTP166Kan, Nalr Camr Kanr | This study |
| GHY287 | JB580v, inv::pEP-inv, Nalr Camr | This study |
| E. coli strains | ||
| DH5α | Φ80dΔ(lacZ)M15 Δ(argF-lac)U169 endA1 recA1 hsdR17(rK− mK+) deoR thi-1 supE44 gyrA96 relA1 | Gibco BRL |
| S17-1λpir | Tpr SmrrecA thi pro hsdR M+ RP4::2-Tc::Mu::Km Tn7λpir lysogen | 39 |
| Plasmids | ||
| pFUSE | Camr, mob+ (RP4), R6K ori (suicide vector) lacZYA | 6 |
| pEP185.2 | Camr, mob+ (RP4), R6K ori (suicide vector) | 32 |
| pTP166Kan | Kanamycin-resistant derivative (Amps) of pTP166 | 22 |
| pTP166Kan-damΔ | dam mutant derivative of pTP166Kan | 22 |
| pFUSE-flhD | flhD promoter fragment in pFUSE | This study |
| pEP-inv | Internal fragment of inv in pEP185.2 | This study |
Analysis of motility.
Motility was analyzed by inoculating semisolid agar plates containing 0.3% agar with 3 μl of an overnight culture grown at 26°C. After incubation of the plates at 26°C or 37°C for 20 h, the diameters of the halos starting from the points of inoculation were compared. Experiments were performed at least in triplicate.
Quantification of gene expression.
The expression of flhDC was determined by reporter gene technology. For the construction of a Y. enterocolitica flhD promoter-lacZ fusion, a 750-bp fragment including the promoter region and the 5′ end of flhDC was amplified by PCR using the primer pair GH-flh1/GH-flh2 (Table 2) with genomic DNA of Y. enterocolitica JB580v as the template. The resulting fragment was digested with XbaI and ligated into the XbaI/SmaI-digested vector pFUSE, resulting in plasmid pFUSE-flhD. After conjugation of the plasmid to Y. enterocolitica JB580v from E. coli S17-1λpir, the merodiploid strain GHY174 (flhD-lacZYA flhD+) was generated (Table 1). Homologous integration into the chromosome was confirmed by Southern blot analysis (data not shown).
TABLE 2.
Oligonucleotides used in this study
| Oligonucleotide | Sequence (5′→3′)a |
|---|---|
| SF-ailRT3 | AGCTAGTTCTCTAATAGCCTG |
| SF-ailRT4 | TTTGGAAGTGGGTTGAATTGC |
| GH-flh1 | GCTCTAGAGGTTTACCTCTGCTGCCTTTTA |
| GH-flh2 | CGTACTCATTTTATACATCCC |
| SF-flp1RT1 | TTACGTTACAGCACAAGTTAATGCG |
| SF-flp1RT2 | ATGTTTGTCATGGATGTATCGACG |
| SF-invRT1 | CAGGCTAATATTATCGATCGG |
| SF-invRT2 | CCCGGTCATATCATTGTCATA |
| SF-myfART1 | CACCTGCCTTCCATCTGGTAATG |
| SF-myfART2 | AGAAAGTCGCTTCCACACGCTC |
| SF-pilLRT1 | CAAATGGTGGACATCACTATGCC |
| SF-pilLRT2 | TTCCCGATTCACATCATCAACC |
| SF-rnaYE1 | AATACCGCATAACGTCTTCG |
| SF-rnaYE2 | CTTCTTCTGCGAGTAACGTC |
| GH3067f | TCCGCTATTACAAGCCGAGT |
| GH3067r | CTGCGGATGCTTATCGGTAT |
| GH3071f | TCATAACCCAGGTTGCCATT |
| GH3071r | CGCGAGATCCATAACATGAA |
| GH3087f | GTGTGGAACCATGGAATGTG |
| GH3087r | CACTGACACCATCGCCATAC |
| GH-inv1 | CCATCGATCGCTGAACATAATGAGGCTTT |
| GH-inv2 | GCTCTAGATACGCTTTGACGTGAATGTCG |
GH-flh1 and GH-inv2 contain a restriction site for XbaI (underlined), and GH-inv1 contains a restriction site for ClaI (underlined).
To determine the expression of flhDC, overnight cultures of the corresponding Y. enterocolitica strains grown at 26°C were diluted in fresh LB medium and grown for 20 h at 26°C in the presence of 1 mM IPTG to induce overproduction of Dam from pTP166Kan. The bacterial cells were pelleted by centrifugation and washed with 0.85% (wt/vol) NaCl. Subsequently, β-galactosidase activities were quantified as described previously (36) and averaged from three independent experiments, each performed in triplicate.
The transcription of inv, ail, flp-1, pilL, myfA, ddhA, gne, and rosA was quantified by quantitative reverse transcription-PCR (qRT-PCR) as described recently (21). Briefly, overnight cultures of the DamOP strain of Y. enterocolitica and of the corresponding control strain grown at 26°C were diluted in fresh LB medium and grown at 26°C or 37°C for 4 h in the presence of 1 mM IPTG. After isolation of total RNA, DNase treatment, and randomly primed reverse transcription, relative gene expression was determined using the Light-Cycler system (Roche) and the QuantiTect SYBR Green PCR kit (QIAGEN). Experiments were performed at least in triplicate for each gene with the primer pairs SF-ailRT3 and SF-ailRT4, SF-flp1RT1 and SF-flp1RT2, SF-invRT1 and SF-invRT2, SF-myfART1 and SF-myfART2, SF-pilLRT1 and SF-pilLRT2, GH3067f and GH3067r (rosA), GH3071f and GH3071r (gne), and GH3087f and GH3087r (ddhA) (Table 2). The expression of the 16S rRNA was used as the housekeeping gene control using the primer pair SF-rnaYE1 and SF-rnaYE2 (Table 2).
Construction of an inv mutant strain.
To construct a Y. enterocolitica strain defective in Inv-mediated invasion of host cells, an internal fragment of the inv gene was amplified by PCR using the primers GH-inv1 and GH-inv2 (Table 2) and ligated into the suicide vector pEP185.2. The resulting plasmid, pEP-inv, was transferred to Y. enterocolitica from E. coli S17-1λpir by conjugation and integrated into the chromosome by homologous recombination with selection for chloramphenicol, directly resulting in an inv mutant strain. Homologous integration into the chromosome was confirmed by Southern blot analysis and Western blotting using Inv-specific antiserum (data not shown).
Determination of bacterial invasion and analysis of Inv levels.
Bacterial invasion in CHO-K1 cells and eukaryotic cell culture was performed as previously described using a standard gentamicin protection assay (22). To determine the amount of Inv after overproduction of Dam, bacteria were grown at 26°C or 37°C for 4 h and overproduction of Dam was induced as described above. Whole-cell lysates corresponding to 3 × 108 bacteria were separated by sodium dodecyl sulfate-12.5% polyacrylamide gel electrophoresis (PAGE) and transferred to a nitrocellulose membrane for immunoblot analysis. The nitrocellulose membrane was blocked for 45 min with 1% skim milk in phosphate-buffered saline (PBS) and incubated for 2 h at room temperature with a rabbit polyclonal antiserum specific for Inv (dilution, 1:2,500 in PBS-0.2% Tween 20), kindly provided by E. Bohn (Tübingen, Germany). After three washes with PBS-0.2% Tween 20, an alkaline phosphatase-conjugated secondary antibody (goat anti-rabbit; dilution, 1:7,500 in PBS; 45 min at room temperature) was used to detect Inv.
Analysis of LPS O-antigen status.
LPS was prepared as described previously by Bengoechea et al. (7). Briefly, overnight cultures of bacteria were diluted 1:10 in 5 ml of fresh LB medium and grown at 26°C for 3 h. Afterwards, cells were collected by centrifugation (12,000 × g, room temperature, 3 min) and resuspended in lysis buffer (2% deoxycholate, 4% 2-mercaptoethanol, 10% glycerol, and 0.002% bromophenol blue in 1 M Tris-HCl buffer, pH 6.8) in a volume adjusted according to the optical density at 600 nm of the cultures. Subsequently, the lysates were boiled for 10 min and subjected to 40 μg proteinase K for 3 h at 55°C. Samples were stored at −20°C until needed. LPS samples were analyzed by sodium deoxycholate-PAGE and silver staining as described previously (35, 53).
RESULTS
Overproduction of Dam alters motility of Y. enterocolitica by increasing expression of flhDC.
In a previous study, we showed that overproduction of the Dam enzyme in Y. enterocolitica leads to an increased ability to invade cultured epithelial cells independently of the pYV-encoded YadA protein (22). Invasion phenotypes are often related to functions of the flagellum and to motility, for example, in S. enterica or Listeria monocytogenes (17, 40). Interestingly, motility and the ability to invade eukaryotic cells are affected by Dam methylation in S. enterica (4, 5). Several studies with Y. enterocolitica indicate that expression of motility and the inv gene encoding Inv, the primary invasion factor in cell culture models, are coordinately regulated. Strains could be isolated which show a significant decrease in inv expression but are hypermotile when grown at 26°C. Furthermore, a clpB mutant of Y. enterocolitica is deficient in Inv production and in motility (2, 3).
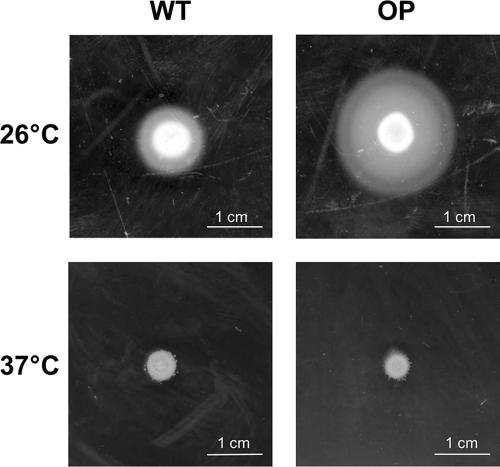
Due to these observations, we analyzed whether overproduction of Dam might also influence the motility of Y. enterocolitica. Therefore, we spotted a DamOP strain and a control strain on soft agar plates and incubated the plates at 26°C or 37°C. As shown in Fig. 1, the DamOP strain is indeed hypermotile at 26°C compared to the control strain (diameter of the halo, 2.3 ± 0.17 cm, compared to 1.4 ± 0.06 cm). Motility is temperature regulated in Y. enterocolitica, with bacteria being motile at 26°C but not at 37°C. Both the DamOP strain and the control strain are nonmotile at 37°C, indicating that overproduction of Dam does not induce the expression of motility under nonpermissive conditions but alters motility under permissive conditions.
FIG. 1.
Motility is increased after overproduction of Dam at 26°C. A Y. enterocolitica DamOP strain (OP; GHY150) and a control strain (WT; GHY147) were spotted on motility agar plates and incubated at 26°C or 37°C for 20 h. The agar plates shown are representative of at least three independent experiments.
Motility is regulated by a complex cascade, which is well characterized for Enterobacteriaceae, including Yersinia species (12, 47, 58). Interestingly, the promoter region of the flhDC operon, encoding the master regulator of flagellum biosynthesis, contains a nonmethylated GATC site in E. coli (54), indicating a putative role for Dam in the regulation of flhDC expression. Therefore, we speculated on whether overproduction of Dam might induce hypermotility in Y. enterocolitica via an upregulation of flhDC transcription. To analyze this, a lacZ transcriptional fusion to flhDC was constructed and the resulting β-galactosidase activity was monitored. After incubation for 20 h at 26°C, a mean flhDC transcription of 3,925 ± 430 Miller units was obtained for the DamOP strain, showing a 1.5-fold upregulation of flhDC transcription in comparison to results for the wild-type control strain (2,685 ± 55 Miller units; P = 0.0013). We and others have described similar modest effects on gene expression after overproduction of Dam for most Dam-regulated genes (21, 41). Furthermore, modest differences in flhDC expression can have a strong effect due to the multiplying effects of the downstream regulatory cascade. These results might indicate that the hypermotility after overproduction of Dam in Y. enterocolitica is mediated via an upregulation of the flhDC operon. We identified five GATC sequences in the 5′ untranslated region of the flhD gene, at 63 nucleotides (nt), 190 nt, 448 nt, 476 nt, and 693 nt (numbering is with respect to the flhD start codon), respectively, as potential targets of the Dam enzyme. However, we were not able to detect any differences in the methylation pattern of the flhDC upstream regulatory region by an adapted Southern blot analysis described previously (21). Our data indicate that all GATC sites present within this region are at least hemimethylated and that there is no switch from nonmethylated to fully methylated DNA after overproduction of Dam (data not shown). Nevertheless, a switch between hemimethylated and fully methylated DNA cannot be detected with the method used and remains a possibility; a similar mechanism was proposed for the Dam-dependent regulation of the flagellar gene fliC in S. enterica (5).
Overproduction of Dam reduces inv transcription and the steady-state level of Inv.
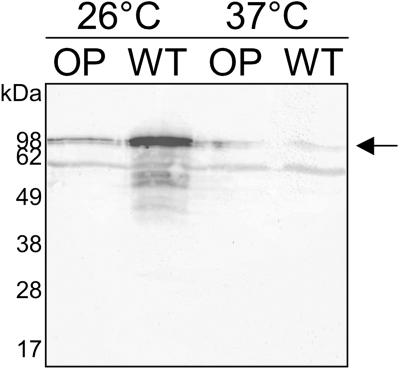
In a previous study, Young et al. showed that overexpression of flhDC from a plasmid in Y. enterocolitica resulted in an approximately twofold increase in inv transcription but in decreased amounts of Inv, suggesting transcriptional as well as posttranscriptional levels of regulation that are influenced directly or indirectly by FlhDC (57). Therefore, we assumed that the altered flhDC expression after overproduction of Dam might be responsible for the previously described increased invasion of epithelial cells by the DamOP strain by altering Inv levels (22). To address this, we incubated the DamOP strain and the control strain for 4 h at 26°C or 37°C and subsequently used qRT-PCR analysis to monitor inv transcription. Surprisingly, as shown in Table 3, we found that inv transcription is not upregulated but is downregulated 1.7-fold at 37°C and even 5.5-fold at 26°C after overproduction of Dam (P = 0.023 and 0.0008, respectively). To determine if the Dam-induced downregulation of inv transcription leads to a reduced protein level or if posttranscriptional levels of regulation, as proposed by Young et al. (57), might be involved in mediating the observed invasion phenotype, we analyzed whole-cell lysates of cells grown at 26°C or 37°C by immunoblotting with antibodies directed against the Inv protein. In accordance with the distinct downregulation of inv transcription at 26°C, the steady-state level of Inv was strongly reduced after overproduction of Dam at this temperature. After incubation at 37°C, the reduction of Inv amounts after overproduction of Dam was less obvious; however, the Inv level did not increase (Fig. 2). These data indicate that overproduction of Dam negatively affected expression of inv, further confirming the coordinated expression of motility and invasion properties in Y. enterocolitica. Nevertheless, despite the lowered expression of Inv, the DamOP bacteria had an altered ability to invade epithelial cells, suggesting that overproduction of Dam influences other functions associated with invasion in Y. enterocolitica, thereby compensating for reduced inv expression.
TABLE 3.
Relative gene expression after overproduction of Dam
| Gene, temp (°C) | Rel. expressiona | P valueb | Fold changec |
|---|---|---|---|
| inv, 26 | 0.181 ± 0.0194 | 0.0008* | ↓5.52 |
| inv, 37 | 0.573 ± 0.2061 | 0.023* | ↓1.75 |
| ail, 26 | 0.729 ± 0.0653 | 0.002* | ↓1.37 |
| ail, 37 | 0.875 ± 0.3599 | 0.51 | ↓1.14 |
| pilL, 26 | 0.363 ± 0.2505 | 0.014* | ↓2.75 |
| pilL, 37 | 0.902 ± 0.2339 | 0.519 | ↓1.11 |
| myfA, 26 | 0.902 ± 0.1761 | 0.393 | ↓1.11 |
| myfA, 37 | 0.922 ± 0.5869 | 0.83 | ↓1.08 |
| flp-1, 26 | 0.184 ± 0.1070 | 0.0001* | ↓5.43 |
| flp-1, 37 | 1.080 ± 0.8303 | 0.811 | ↑1.08 |
| ddhA, 26 | 0.680 ± 0.5963 | 0.32 | ↓1.47 |
| ddhA, 37 | 1.325 ± 0.2514 | 0.088 | ↑1.33 |
| gne, 26 | 0.599 ± 0.5557 | 0.2 | ↓1.67 |
| gne, 37 | 1.205 ± 0.3425 | 0.36 | ↑1.21 |
| rosA, 26 | 0.567 ± 0.5211 | 0.15 | ↓1.76 |
| rosA, 37 | 1.120 ± 0.2364 | 0.43 | ↑1.12 |
Relative expression. Relative mRNA amounts were determined by qRT-PCR and related to mRNA levels in control cells without overproduction of Dam, set as 1. Values are means ± standard deviations.
Significance of difference (P < 0.05) from level in control cells without overproduction of Dam was calculated by Student's t test and is indicated by an asterisk.
n-fold change (↓, downregulation; ↑, upregulation) compared to relative expression in control cells without overproduction of Dam.
FIG. 2.
Steady-state levels of Inv are decreased at 26°C after overproduction of Dam. Whole-cell lysates of a Y. enterocolitica DamOP strain (OP; GHY150) and a control strain (WT; GHY147) grown at 26°C or 37°C for 4 h were transferred to nitrocellulose and subjected to Western blot analysis using a polyclonal anti-Inv antiserum. An arrow indicates the expected size of full-length Inv.
Overproduction of Dam decreases the transcription of known and putative invasion factors.
The reduced steady-state levels of Inv after overproduction of Dam raised the question of which factor might be responsible for the observed hyperinvasive phenotype. Since invasion was analyzed with Y. enterocolitica strains cured of the pYV virulence plasmid, we could exclude a role for the plasmid-encoded invasion factor YadA (15, 22). A third Y. enterocolitica invasion factor is encoded by the ail gene (38). Although the ability of Ail to promote invasion is relatively modest, we could not exclude that Ail might play a more important role for the invasion phenotype under DamOP conditions. To address this question, we analyzed the putative influence of overproduction of Dam on ail transcription. To this end, we incubated the DamOP strain and a control strain at 26°C or 37°C and subsequently monitored ail transcription by qRT-PCR. As shown in Table 3, ail transcription is slightly downregulated at 26°C (1.37-fold; P = 0.002) but not at 37°C. Although this effect is moderate, the data indicate that the hyperinvasive phenotype caused by overproduction of Dam is at least not due to increased ail expression.
The downregulation of ail and inv transcription after overproduction of Dam suggested that further factors involved in invasion might be present in Y. enterocolitica. Pili and fimbriae often mediate binding to eukaryotic cells. Therefore, we analyzed if genes coding for Myf fibrils, a surface appendage homologous to the pH 6 antigen of Y. pseudotuberculosis (28, 29), are differentially transcribed after overproduction of Dam. Our qRT-PCR analyses show that overproduction of Dam does not influence myfA transcription, indicating that the Myf fibril is not involved in the hyperinvasive phenotype of DamOP Y. enterocolitica strains (Table 3).
Other operons encoding putative type IV pili have been identified in the Y. enterocolitica genome, which are encoded by the YAPIYE and YGI-1 pathogenicity islands (13, 14, 49, 50). To investigate the putative role of the YAPIYE (pil)-encoded and YGI-1 (flp/tad)-encoded pili in invasion/adherence of Y. enterocolitica after overproduction of Dam, we monitored the transcription of the pil and tad operons. To this end, we incubated the DamOP strain and the control strain of Y. enterocolitica as described above and performed qRT-PCR analyses with primers directed against the first genes of the operons, pilL and flp-1, respectively. Our results indicate that at 26°C the transcription of pilL and that of flp-1 are markedly downregulated after overproduction of Dam (pilL, 2.75-fold; P = 0.014; flp-1, 5.43-fold; P < 0.0001) (Table 3). At 37°C, pilL transcription is not significantly influenced by overproduction of Dam (P = 0.519). This also holds true for the mean transcription of flp-1 at 37°C (P = 0.811), although we obtained extremely variable results when comparing single experiments, which is mirrored in the considerable standard deviation (Table 3). Nevertheless, since the hyperinvasive phenotype of DamOP strains is highly reproducible and since we were not able to visualize pilus structures under these conditions on the bacterial surface by electron microscopy (data not shown), putative pili do not seem to be involved in the increased ability to invade tissue culture cells after overproduction of Dam.
Taken together, by transcriptional analyses we were not able to identify an adherence/invasion factor that is responsible for the hyperinvasive phenotype of DamOP Y. enterocolitica strains. In contrast, overproduction of Dam in Y. enterocolitica results in downregulation of a relatively broad spectrum of genes encoding known and putative surface proteins conferring adhesion to and/or invasion of host cells.
Overproduction of Dam modulates LPS O-antigen status.
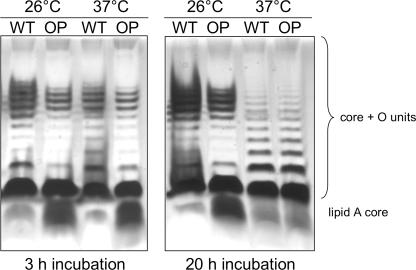
The observation that the transcription of invasion/adhesion factors is decreased or not changed and also that the amount of Inv protein is decreased in a DamOP strain but that at the same time the strain shows a hyperinvasive phenotype suggested that there might be a change in the surface structure of the DamOP strain increasing the accessibility of an invasion factor. Pierson showed that a mutation affecting the LPS profile of Y. enterocolitica increased the Ail-mediated entry into mammalian cells 20-fold, although the expression of ail was not changed (45). Furthermore, defects within the LPS O-antigen status lead to an increased motility and reduced transcription of the inv gene (7). Due to these observations, we wondered if the invasion and motility phenotype of a DamOP strain of Y. enterocolitica is caused or at least influenced by a modulation of the LPS structure. To analyze this in more detail, we incubated the DamOP strain and the control strain of Y. enterocolitica at 26°C or 37°C for 3 h and subsequently analyzed the LPS. As demonstrated in Fig. 3, overproduction of Dam indeed modulates the LPS O-antigen status. At 26°C and at 37°C, we detected increased amounts of lipid A core without O-antigen units after overproduction of Dam, whereas the proportion of core units with small O-antigen units was decreased. Interestingly, the amount of molecules with the preferred O-antigen chain length of about 7 to 10 repeats was not considerably changed after overproduction of Dam, putatively indicating that Dam methylation affects the stability of shorter LPS species or influences the addition of the first O-antigen units to the growing chain. However, in accordance with the study by Pierson (45), the increased occurrence of shorter LPS species after overproduction of Dam is a plausible explanation for an altered accessibility of an invasion factor on the bacterial surface; this might contribute to the hyperinvasive phenotype caused by overproduction of Dam despite the reduced expression of invasion and adhesion factors analyzed here.
FIG. 3.
The amount of lipid A core without conjugated O-antigen units is increased after overproduction of Dam. A DamOP strain (OP; GHY150) and a control strain (WT; GHY147) of Y. enterocolitica were grown at 26°C or 37°C for 3 h or 20 h. LPS was isolated by proteinase K treatment and analyzed by sodium deoxycholate-PAGE and silver staining.
Expression of LPS is temperature regulated in Y. enterocolitica; at 26°C, transcription of the O-antigen gene cluster is induced, while it is repressed at 37°C (8). Therefore, we repeated LPS isolation after 20 h of growth at 26°C and 37°C, since the effect of the growth temperature on the expression of LPS becomes most obvious in stationary-phase cultures. As can be seen in Fig. 3, the change in LPS structure after overproduction of Dam is still visible in overnight cultures grown at 26°C, but not at 37°C, when expression of the O-antigen cluster is repressed, indicating that overproduction of Dam does not interfere with the temperature regulation of LPS expression but modulates expression at the permissive temperature.
Overproduction of Dam does not influence transcription of LPS O-antigen genes.
Since overproduction of Dam modulates the LPS O-antigen status, we wondered if transcription of the corresponding genes might be directly or indirectly affected by DNA methylation. The O-antigen gene cluster consists of two transcriptional units controlled by the promoters Pwb1 and Pwb2, respectively, with transcription of the latter being indirectly modulated by the RosAB system (8). We performed qRT-PCR to analyze the transcription of ddhA, gne, and rosA, representing the first genes in the respective operons, in a strain overproducing Dam for 3 h and a control strain at 26°C and 37°C. As shown in Table 3, there was no significant change in the transcription of these genes in response to overproduction of Dam, indicating that the modulation of the LPS structure of DamOP Y. enterocolitica is mediated posttranscriptionally.
Inv is necessary for increased invasion after overproduction of Dam.
The qRT-PCR data revealed effects of Dam on the transcription of genes associated with adhesion and invasion, but the factor responsible for the observed hyperinvasive phenotype after overproduction of Dam remained elusive. We anticipated that if we mutated the factor necessary for increased invasion, the effect of overproduction of Dam would no longer be detectable. Since Inv is the major invasion factor of Y. enterocolitica, we constructed a mutant strain missing a functional Inv protein. Subsequently, CHO-K1 cells were infected with strain GHY287 (inv::pEP-inv) overproducing Dam from pTP166Kan or carrying the control plasmid pTP166Kan-damΔ. The DamOP strain and the control strain did not differ in their abilities to invade eukaryotic cells [GHY287 (pTP166Kan), 3.8% ± 1.99% invasion; GHY287 (pTP166Kan-damΔ), 4.01% ± 1.93% invasion]. These data show that an intact Inv protein is necessary for increased invasion after overproduction of Dam, indicating that Inv and not another adhesion/invasion factor is involved in the observed increased invasion phenotype after overproduction of Dam. These data further indicate that indeed the steric accessibility of Inv due to a changed LPS structure is involved in the increased invasion of a DamOP strain.
DISCUSSION
In previous studies, we were able to demonstrate that overproduction of the Dam enzyme in Y. enterocolitica affects the regulation of several virulence properties, such as type III secretion or invasion (21, 22). Since invasion processes are often coordinately regulated with other virulence functions, this study was conducted to analyze Dam's influence on further virulence-associated genes and to identify molecular mechanisms underlying the hyperinvasive phenotype caused by overproduction of Dam. Indeed, we could demonstrate that motility is increased after overproduction of Dam. This effect is mediated by an upregulation of flhDC encoding the master regulator of the flagellar biosynthesis cascade (58). However, we were not able to detect a direct switch from nonmethylated to methylated GATC sequences within the upstream regulatory region of flhDC after overproduction of Dam in Y. enterocolitica. In E. coli, a nonmethylated GATC sequence is present in the noncoding region of the flhDC operon (54). However, in a global screen for Dam-regulated genes of E. coli, flhDC could not be identified (41). Interestingly, in S. enterica the expression of fliC encoding a flagellar structural protein is influenced by DNA hemimethylation (5). Regulation by hemimethylation remains a possibility for the increased flhDC transcription after overproduction of Dam in Y. enterocolitica as well, since hemimethylation and methylation of both strands cannot be distinguished by the assay used. The hypothesis of direct regulation by Dam is further supported by the fact that the transcription of flhDC is regulated by H-NS and DNA structure, which is strongly affected by the methylation status (47, 56). In this context, it is interesting to note that Pérez-Gutiérrez and colleagues describe a relationship between the absence of LPS in rough mutants and H-NS expression, which underlies flhDC upregulation and altered secretion by the pYV-encoded Ysc type III secretion system (44). Since we previously described an effect of overproduction of Dam on type III secretion in Y. enterocolitica (21), the expression of LPS, motility, invasion, and type III secretion is obviously under the coordinated control of a complex regulatory network which can be modulated by Dam. This network might also include posttranscriptional effects via other factors. The ATP-dependent protease ClpP might be involved in this process, since the turnover of FlhDC is influenced by ClpXP in S. enterica and since clpP transcription is increased after overproduction of Dam in Y. enterocolitica, at least during incubation at 37°C (21, 51, 52). The detailed mechanism underlying the Dam-dependent increased motility in Y. enterocolitica remains to be elucidated.
Since it has been reported that motility is required to initiate host cell invasion in Y. enterocolitica, it might be concluded that motility is the critical factor inducing the hyperinvasive phenotype after overproduction of Dam (57). Since this is true only for experiments in which contact between the bacteria and the host cells is not induced by centrifugation and since we determined the invasion frequencies of the DamOP strain and the control strain after inducing contact, increased motility after overproduction of Dam is not exclusively responsible for the hyperinvasive phenotype. Moreover, in our study we present evidence that increased flhDC transcription after overproduction of Dam goes along with a decreased inv transcription as well as a reduced (or at 37°C at least an unchanged) steady-state level of Inv, although it was reported that overexpression of flhDC from a plasmid results in increased inv transcription (57). A possible explanation for this discrepancy might be the fact that Dam has pleiotropic effects within a cell, representing a situation clearly different from overproduction of just one specific protein. Interestingly, a rough mutant of Y. enterocolitica is hypermotile and downregulates inv expression, further supporting a close regulatory connection of LPS expression, motility, and invasion (7). Interestingly, however, in contrast to the case with the DamOP strain, invasion is reduced in the rough mutant. This discrepancy might be explained by the difference between rough mutants without LPS on the one hand and a modified LPS structure after overproduction of Dam on the other. Alternatively, further effects of Dam on surface structures influencing invasion efficiency cannot be excluded. The invasion assay using an inv mutant strain overproducing Dam clearly indicates that Inv is necessary for the Dam-mediated hyperinvasive phenotype. This is surprising, since transcription of inv and also steady-state levels of Inv are decreased after overproduction of Dam. The data suggest an altered steric accessibility of the remaining Inv protein on the bacterial surface. This hypothesis is supported by the observation that mutations affecting LPS increased the Ail-mediated entry of Y. enterocolitica into host cells, although ail expression was not affected (45). Therefore, we postulated that a decreased amount of proteins promoting internalization might be accompanied by a changed LPS O-antigen status. Indeed, we could demonstrate that overproduction of Dam leads to an increase in the amount of rough LPS molecules lacking O-antigen side chains. Although the amount of core units with 7 to 10 repeats is not considerably changed, our data show that the population of LPS molecules is shifted to shorter species after overproduction of Dam. This indicates that overproduction of Dam results in an altered accessibility of Inv, with this being at least in part responsible for the Dam-induced hyperinvasive phenotype.
The amount of lipid A is strongly increased in DamOP Y. enterocolitica. Since lipid A is an important signal for the induction of the innate immune system in the host via Toll-like receptor 4, this might have implications for the course of an infection (37). However, since overproduction of Dam results in increased invasion of the wild-type strain but not the inv mutant strain, we can exclude the possibility that lipid A induces increased endocytosis in the invasion assay.
During infection of the host, Y. enterocolitica is facing various biotic surfaces composed of different cell types. For the various stages of infection, different factors might be predominant for promoting adhesion or internalization. This is supported by the fact that YadA, Inv, Ail, and MyfA are maximally expressed under conditions distinctly differing from each other in temperature, pH, or growth phase (18, 19, 28, 43). Furthermore, depending on the surface of the host cell, a competition between YadA and Inv for activation of different phagocytic responses was proposed for Y. pseudotuberculosis (27). Due to these observations, it can be speculated that overproduction of Dam does not directly influence the expression of an individual invasin/adhesin but influences a global program regulating the expression of diverse surface proteins with the outcome of adaptation to a specific environment during infection. Since it was reported that the LPS O-antigen status is involved in the regulation of the expression of virulence factors such as inv and flhDC (7), the LPS structure might well be a critical factor influenced by Dam and causing downstream effects, such as increased motility or reduced transcription of genes. Which additional factors might be involved in this putative global regulation network remains to be elucidated. An interesting player in this context might be ClpP, since ClpP modulates the expression of ail and type III secretion and since the transcription of clpP increases after overproduction of Dam (21, 43).
In conclusion, overproduction of Dam clearly leads to a new organization of the bacterial surface concerning proteins and appendages and of the LPS structure. DNA methylation by the Dam enzyme seems to contribute to an adaptation of the bacterial surface to specific stages during an infection, an idea that is also underlined by various reports of different pathogens with defects in virulence due to defects in DNA methylation (26).
Acknowledgments
We thank E. Bohn, Tübingen, Germany, for the gift of antibodies and L. Greune for electron microscopy.
This work was supported by Innovative Medical Research grants (IMF: HE120201 and HE110401) provided by the Medical School of the University of Münster and in part by grants from the Deutsche Forschungsgemeinschaft (SFB293/B5, HE3079/6-1, and Graduiertenkolleg 1409).
Editor: J. B. Bliska
Footnotes
Published ahead of print on 6 August 2007.
REFERENCES
- 1.Autenrieth, I. B., and R. Firsching. 1996. Penetration of M cells and destruction of Peyer's patches by Yersinia enterocolitica: an ultrastructural and histological study. J. Med. Microbiol. 44:285-294. [DOI] [PubMed] [Google Scholar]
- 2.Badger, J. L., and V. L. Miller. 1998. Expression of invasin and motility are coordinately regulated in Yersinia enterocolitica. J. Bacteriol. 180:793-800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Badger, J. L., B. M. Young, A. J. Darwin, and V. L. Miller. 2000. Yersinia enterocolitica ClpB affects levels of invasin and motility. J. Bacteriol. 182:5563-5571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Badie, G., D. M. Heithoff, R. L. Sinsheimer, and M. J. Mahan. 2007. Altered levels of Salmonella DNA adenine methylase are associated with defects in gene expression, motility, flagellar synthesis, and bile resistance in the pathogenic strain 14028 but not in the laboratory strain LT2. J. Bacteriol. 189:1556-1564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Balbontin, R., G. Rowley, M. G. Pucciarelli, J. Lopez-Garrido, Y. Wormstone, S. Lucchini, F. Garcia-Del Portillo, J. C. Hinton, and J. Casadesus. 2006. DNA adenine methylation regulates virulence gene expression in Salmonella enterica serovar Typhimurium. J. Bacteriol. 188:8160-8168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bäumler, A. J., R. M. Tsolis, A. W. van der Velden, I. Stojiljkovic, S. Anic, and F. Heffron. 1996. Identification of a new iron regulated locus of Salmonella typhi. Gene. 183:207-213. [DOI] [PubMed] [Google Scholar]
- 7.Bengoechea, J. A., H. Najdenski, and M. Skurnik. 2004. Lipopolysaccharide O antigen status of Yersinia enterocolitica O:8 is essential for virulence and absence of O antigen affects the expression of other Yersinia virulence factors. Mol. Microbiol. 52:451-469. [DOI] [PubMed] [Google Scholar]
- 8.Bengoechea, J. A., L. Zhang, P. Toivanen, and M. Skurnik. 2002. Regulatory network of lipopolysaccharide O-antigen biosynthesis in Yersinia enterocolitica includes cell envelope-dependent signals. Mol. Microbiol. 44:1045-1062. [DOI] [PubMed] [Google Scholar]
- 9.Bottone, E. J. 1997. Yersinia enterocolitica: the charisma continues. Clin. Microbiol. Rev. 10:257-276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Casadesus, J., and D. Low. 2006. Epigenetic gene regulation in the bacterial world. Microbiol. Mol. Biol. Rev. 70:830-856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen, L., D. B. Paulsen, D. W. Scruggs, M. M. Banes, B. Y. Reeks, and M. L. Lawrence. 2003. Alteration of DNA adenine methylase (Dam) activity in Pasteurella multocida causes increased spontaneous mutation frequency and attenuation in mice. Microbiology 149:2283-2290. [DOI] [PubMed] [Google Scholar]
- 12.Chilcott, G. S., and K. T. Hughes. 2000. Coupling of flagellar gene expression to flagellar assembly in Salmonella enterica serovar Typhimurium and Escherichia coli. Microbiol. Mol. Biol. Rev. 64:694-708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Collyn, F., A. Billault, C. Mullet, M. Simonet, and M. Marceau. 2004. YAPI, a new Yersinia pseudotuberculosis pathogenicity island. Infect. Immun. 72:4784-4790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Collyn, F., M. A. Lety, S. Nair, V. Escuyer, A. Ben Younes, M. Simonet, and M. Marceau. 2002. Yersinia pseudotuberculosis harbors a type IV pilus gene cluster that contributes to pathogenicity. Infect. Immun. 70:6196-6205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cornelis, G. R., A. Boland, A. P. Boyd, C. Geuijen, M. Iriarte, C. Neyt, M. P. Sory, and I. Stainier. 1998. The virulence plasmid of Yersinia, an antihost genome. Microbiol. Mol. Biol. Rev. 62:1315-1352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cover, T. L., and R. C. Aber. 1989. Yersinia enterocolitica. N. Engl. J. Med. 6:16-24. [DOI] [PubMed] [Google Scholar]
- 17.Ellermeier, C. D., and J. M. Slauch. 2003. RtsA and RtsB coordinately regulate expression of the invasion and flagellar genes in Salmonella enterica serovar Typhimurium. J. Bacteriol. 185:5096-5108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ellison, D. W., M. B. Lawrenz, and V. L. Miller. 2004. Invasin and beyond: regulation of Yersinia virulence by RovA. Trends Microbiol. 12:296-300. [DOI] [PubMed] [Google Scholar]
- 19.El Tahir, Y., and M. Skurnik. 2001. YadA, the multifaceted Yersinia adhesin. Int. J. Med. Microbiol. 291:209-218. [DOI] [PubMed] [Google Scholar]
- 20.Erova, T. E., L. Pillai, A. A. Fadl, J. Sha, S. Wang, C. L. Galindo, and A. K. Chopra. 2006. DNA adenine methyltransferase influences the virulence of Aeromonas hydrophila. Infect. Immun. 74:410-424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fälker, S., M. A. Schmidt, and G. Heusipp. 2006. Altered Ca2+ regulation of Yop secretion in Yersinia enterocolitica after DNA adenine methyltransferase overproduction is mediated by Clp-dependent degradation of LcrG. J. Bacteriol. 188:7072-7081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fälker, S., M. A. Schmidt, and G. Heusipp. 2005. DNA methylation in Yersinia enterocolitica: role of the DNA adenine methyltransferase in mismatch repair and regulation of virulence factors. Microbiology 151:2291-2299. [DOI] [PubMed] [Google Scholar]
- 23.Garcia-Del Portillo, F., M. G. Pucciarelli, and J. Casadesus. 1999. DNA adenine methylase mutants of Salmonella typhimurium show defects in protein secretion, cell invasion, and M cell cytotoxicity. Proc. Natl. Acad. Sci. USA 96:11578-11583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Grutzkau, A., C. Hanski, H. Hahn, and E. O. Riecken. 1990. Involvement of M cells in the bacterial invasion of Peyer's patches: a common mechanism shared by Yersinia enterocolitica and other enteroinvasive bacteria. Gut 31:1011-1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Heithoff, D. M., R. L. Sinsheimer, D. A. Low, and M. J. Mahan. 1999. An essential role for DNA adenine methylation in bacterial virulence. Science 284:967-970. [DOI] [PubMed] [Google Scholar]
- 26.Heusipp, G., S. Fälker, and M. A. Schmidt. 2007. DNA adenine methylation and bacterial pathogenesis. Int. J. Med. Microbiol. 297:1-7. [DOI] [PubMed] [Google Scholar]
- 27.Hudson, K. J., J. B. Bliska, and A. H. Bouton. 2005. Distinct mechanisms of integrin binding by Yersinia pseudotuberculosis adhesins determine the phagocytic response of host macrophages. Cell. Microbiol. 7:1474-1489. [DOI] [PubMed] [Google Scholar]
- 28.Iriarte, M., and G. R. Cornelis. 1995. MyfF, an element of the network regulating the synthesis of fibrillae in Yersinia enterocolitica. J. Bacteriol. 177:738-774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Iriarte, M., J. C. Vanooteghem, I. Delor, R. Diaz, S. Knutton, and G. R. Cornelis. 1993. The Myf fibrillae of Yersinia enterocolitica. Mol. Microbiol. 9:507-520. [DOI] [PubMed] [Google Scholar]
- 30.Julio, S. M., D. M. Heithoff, D. Provenzano, K. E. Klose, R. L. Sinsheimer, D. A. Low, and M. J. Mahan. 2001. DNA adenine methylase is essential for viability and plays a role in the pathogenesis of Yersinia pseudotuberculosis and Vibrio cholerae. Infect. Immun. 69:7610-7615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Julio, S. M., D. M. Heithoff, R. L. Sinsheimer, D. A. Low, and M. J. Mahan. 2002. DNA adenine methylase overproduction in Yersinia pseudotuberculosis alters YopE expression and secretion and host immune responses to infection. Infect. Immun. 70:1006-1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kinder, S. A., J. L. Badger, G. O. Bryant, J. C. Pepe, and V. L. Miller. 1993. Cloning of the YenI restriction endonuclease and methyltransferase from Yersinia enterocolitica serotype O:8 and construction of a transformable R−M+ mutant. Gene 136:271-275. [DOI] [PubMed] [Google Scholar]
- 33.Marinus, M. G., A. Poteete, and J. A. Arraj. 1984. Correlation of DNA adenine methylase activity with spontaneous mutability in Escherichia coli K-12. Gene 28:123-125. [DOI] [PubMed] [Google Scholar]
- 34.Mehling, J. S., H. Lavender, and S. Clegg. 2007. A Dam methylation mutant of Klebsiella pneumoniae is partially attenuated. FEMS Microbiol. Lett. 268:187-193. [DOI] [PubMed] [Google Scholar]
- 35.Meissner, J., J. H. Krauss, U. J. Jürgens, and J. Weckesser. 1988. Absence of a characteristic cell wall lipopolysaccharide in the phototrophic bacterium Chloroflexus aurantiacus. J. Bacteriol. 170:3213-3216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Miller, J. H. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 37.Miller, S. I., R. K. Ernst, and M. W. Bader. 2005. LPS, TLR4 and infectious disease diversity. Nat. Rev. Microbiol. 3:36-46. [DOI] [PubMed] [Google Scholar]
- 38.Miller, V. L., and S. Falkow. 1988. Evidence for two genetic loci in Yersinia enterocolitica that can promote invasion of epithelial cells. Infect. Immun. 56:1242-1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Miller, V. L., and J. J. Mekalanos. 1988. A novel suicide vector and its use in construction of insertion mutations: osmoregulation of outer membrane proteins and virulence determinants in Vibrio cholerae requires toxR. J. Bacteriol. 170:2575-2583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.O'Neil, H. S., and H. Marquis. 2006. Listeria monocytogenes flagella are used for motility, not as adhesins, to increase host cell invasion. Infect. Immun. 74:6675-6681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Oshima, T., C. Wada, Y. Kawagoe, T. Ara, M. Maeda, Y. Masuda, S. Hiraga, and H. Mori. 2002. Genome-wide analysis of deoxyadenosine methyltransferase-mediated control of gene expression in Escherichia coli. Mol. Microbiol. 45:673-695. [DOI] [PubMed] [Google Scholar]
- 42.Oza, J. P., J. B. Yeh, and N. O. Reich. 2005. DNA methylation modulates Salmonella enterica serovar Typhimurium virulence in Caenorhabditis elegans. FEMS Microbiol. Lett. 245:53-59. [DOI] [PubMed] [Google Scholar]
- 43.Pederson, K. J., and D. E. Pierson. 1995. Ail expression in Yersinia enterocolitica is affected by oxygen tension. Infect. Immun. 63:4199-4201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pérez-Gutiérrez, C., C. M. Llompart, M. Skurnik, and J. A. Bengoechea. 2007. Expression of the Yersinia enterocolitica pYV-encoded type III secretion system is modulated by lipopolysaccharide O-antigen status. Infect. Immun. 75:1512-1516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pierson, D. E. 1994. Mutations affecting lipopolysaccharide enhance ail-mediated entry of Yersinia enterocolitica into mammalian cells. J. Bacteriol. 176:4043-4051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Robinson, V. L., P. C. Oyston, and R. W. Titball. 2005. A dam mutant of Yersinia pestis is attenuated and induces protection against plague. FEMS Microbiol. Lett. 252:251-256. [DOI] [PubMed] [Google Scholar]
- 47.Soutourina, O. A., and P. N. Bertin. 2003. Regulation cascade of flagellar expression in Gram-negative bacteria. FEMS Microbiol. Rev. 27:505-523. [DOI] [PubMed] [Google Scholar]
- 48.Taylor, V. L., R. W. Titball, and P. C. Oyston. 2005. Oral immunization with a dam mutant of Yersinia pseudotuberculosis protects against plague. Microbiology 151:1919-1926. [DOI] [PubMed] [Google Scholar]
- 49.Thomson, N. R., S. Howard, B. W. Wren, M. T. Holden, L. Crossman, G. L. Challis, C. Churcher, K. Mungall, K. Brooks, T. Chillingworth, T. Feltwell, Z. Abdellah, H. Hauser, K. Jagels, M. Maddison, S. Moule, M. Sanders, S. Whitehead, M. A. Quail, G. Dougan, J. Parkhill, and M. B. Prentice. 2006. The complete genome sequence and comparative genome analysis of the high pathogenicity Yersinia enterocolitica strain 8081. PLoS Genet. 2:e206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tomich, M., P. J. Planet, and D. H. Figurski. 2007. The tad locus: postcards from the widespread colonization island. Nat. Rev. Microbiol. 5:363-375. [DOI] [PubMed] [Google Scholar]
- 51.Tomoyasu, T., T. Ohkishi, Y. Ukyo, A. Tokumitsu, A. Takaya, M. Suzuki, K. Sekiya, H. Matsui, K. Kutsukake, and T. Yamamoto. 2002. The ClpXP ATP-dependent protease regulates flagellum synthesis in Salmonella enterica serovar Typhimurium. J. Bacteriol. 184:645-653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tomoyasu, T., A. Takaya, E. Isogai, and T. Yamamoto. 2003. Turnover of FlhD and FlhC, master regulator proteins for Salmonella flagellum biogenesis, by the ATP-dependent ClpXP protease. Mol. Microbiol. 48:443-452. [DOI] [PubMed] [Google Scholar]
- 53.Tsai, C. M., and C. E. Frasch. 1982. A sensitive silver stain for detecting lipopolysaccharides in polyacrylamide gels. Anal. Biochem. 119:115-119. [DOI] [PubMed] [Google Scholar]
- 54.Wang, M. X., and G. M. Church. 1992. A whole genome approach to in vivo DNA-protein interactions in E. coli. Nature 360:606-610. [DOI] [PubMed] [Google Scholar]
- 55.Watson, M. E., Jr., J. Jarisch, and A. L. Smith. 2004. Inactivation of deoxyadenosine methyltransferase (dam) attenuates Haemophilus influenzae virulence. Mol. Microbiol. 53:651-664. [DOI] [PubMed] [Google Scholar]
- 56.Wion, D., and J. Casadesus. 2006. N6-methyl-adenine: an epigenetic signal for DNA-protein interactions. Nat. Rev. Microbiol. 4:183-192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Young, G. M., J. L. Badger, and V. L. Miller. 2000. Motility is required to initiate host cell invasion by Yersinia enterocolitica. Infect. Immun. 68:4323-4326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Young, G. M., M. J. Smith, S. A. Minnich, and V. L. Miller. 1999. The Yersinia enterocolitica motility master regulatory operon, flhDC, is required for flagellin production, swimming motility, and swarming motility. J. Bacteriol. 181:2823-2833. [DOI] [PMC free article] [PubMed] [Google Scholar]