Abstract
Mammals are aerobes that harbor an intestinal ecosystem dominated by large numbers of anaerobic microorganisms. However, the role of oxygen in the intestinal ecosystem is largely unexplored. We used systematic mutational analysis to determine the role of respiratory metabolism in the streptomycin-treated mouse model of intestinal colonization. Here we provide evidence that aerobic respiration is required for commensal and pathogenic Escherichia coli to colonize mice. Our results showed that mutants lacking ATP synthase, which is required for all respiratory energy-conserving metabolism, were eliminated by competition with respiratory-competent wild-type strains. Mutants lacking the high-affinity cytochrome bd oxidase, which is used when oxygen tensions are low, also failed to colonize. However, the low-affinity cytochrome bo3 oxidase, which is used when oxygen tension is high, was found not to be necessary for colonization. Mutants lacking either nitrate reductase or fumarate reductase also had major colonization defects. The results showed that the entire E. coli population was dependent on both microaerobic and anaerobic respiration, consistent with the hypothesis that the E. coli niche is alternately microaerobic and anaerobic, rather than static. The results indicate that success of the facultative anaerobes in the intestine depends on their respiratory flexibility. Despite competition for relatively scarce carbon sources, the energy efficiency provided by respiration may contribute to the widespread distribution (i.e., success) of E. coli strains as commensal inhabitants of the mammalian intestine.
The intestinal microflora is dominated by diverse anaerobes, providing both a health benefit to the host (15) and a barrier to infection (16, 21). Despite being present in substantially lower numbers, facultative anaerobes, primarily Escherichia coli and Enterococcus faecalis, are ubiquitous in mammalian intestines (40). While the intestine is commonly thought to be anaerobic (4), the tissues surrounding the lumen are oxygen rich and oxygen diffuses into the intestine at appreciable levels (20). Furthermore, oxygen from swallowed air is present in flatus (29). Oxygen in the intestine apparently has minimal impact on persistence of anaerobes, and it was recently shown that at least one predominant anaerobe, Bacteroides fragilis, respires oxygen at low concentrations (5). In contrast to obligate anaerobes, facultative anaerobes (e.g., E. coli) grow most rapidly when respiring oxygen and switch to anaerobic respiration in the absence of oxygen or to fermentation in the absence of alternative electron acceptors (17). However, the extent to which facultative anaerobes utilize oxygen to maximize their growth rate in the intestine is not known.
Nutrients consumed for growth of the microflora are thought primarily to be fermentable carbohydrates, the bulk of which are in the form of polysaccharides (37). E. coli colonizes the mouse intestine by growing within the polysaccharide-rich mucus layer covering the epithelium but is unable to degrade polysaccharides. Apparently, E. coli consumes the mono- and disaccharides released during degradation of mucosal polysaccharides and dietary fiber (8) by polysaccharide hydrolase enzymes secreted by members of the anaerobic microflora (11) and, perhaps, host colonic epithelial cells (6). Recent studies from our laboratory demonstrate that seven mucus-derived sugars contribute to E. coli colonization of the mouse intestine, suggesting that biochemical flexibility is key to its competitiveness in vivo (8). E. coli is nearly equally flexible in its respiratory metabolism (17), but nothing is known about the role of bacterial respiration for coupling ATP generation to carbohydrate oxidation in vivo. Thus, it is important to test the hypothesis that respiration confers a competitive advantage to E. coli in the intestine.
Enterohemorrhagic E. coli (EHEC), has an infectious dose for humans as low as 10 microorganisms and, following ingestion, grows rapidly to a population approaching a billion bacteria per gram of feces (24). Since colonization is the first step in the infection process, it is crucial to understand how EHEC colonizes the intestine because low numbers can survive transport to consumers in foodstuffs such as leafy vegetables, which have caused recent outbreaks in the United States (36). It is not known how EHEC acquires nutrients and generates energy for growth in vivo. While respiration is not a virulence factor per se, our experiments seek to establish the fundamental importance of housekeeping functions, such as energy metabolism, for pathogenesis. Since most mucosal pathogens are facultative anaerobes, these studies of E. coli may be extended to include many diseases.
Here we report the results of a systematic mutational analysis designed to identify which respiratory pathways contribute to the ability of commensal and pathogenic E. coli to colonize the streptomycin-treated mouse intestine. Our findings lead us to conclude that respiration provides an enormous competitive advantage to E. coli in vivo. The results challenge the traditional view that the intestine is strictly anaerobic (4). Instead, we obtained evidence that E. coli colonization of the mouse intestine is maximized by the ability to respire oxygen.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
The bacterial strains used in this study were derived from E. coli MG1655 Strr (streptomycin resistant), a K-12 strain (33), and E. coli EDL933 Strr, the prototypical O157:H7 strain (31). Cultures were grown at 37°C in Luria-Bertani (LB) medium with gyratory shaking at 250 rpm. Null alleles were constructed by using the allelic replacement method of Datsenko and Wanner (14), as described previously (8), such that target genes were deleted and replaced with kanamycin or chloramphenicol resistance cassettes (used as selectable markers in mouse colonization assays, as described below). The null allele strains are identified in the text by the genes that were deleted; single gene deletions began with the start codon and ended with the stop codon, and multiple gene deletions began with the start codon of the first gene deleted and ended with the stop codon of the last gene deleted. Strains containing multiple mutations were constructed by sequential allelic replacement; the first inserted cassette was removed with FLP recombinase (14), followed by subsequent allelic replacement(s) and removal of the insertion as necessary, leaving the selected marker in the last mutation made. Mutant strains were verified by phenotype analysis and DNA sequencing.
Phenotypic analysis.
MOPS [3-(N-morpholino) propanesulfonic acid] defined medium was used to grow cultures for growth curves, as described previously (8). Anaerobic cultures were grown in culture tubes filled to the top with N2-sparged medium, sealed, and incubated in Balch tubes. To test for nitrate or fumarate reductase activity, mutant strains were grown anaerobically overnight in MOPS medium with glycerol (1.6%) as the carbon source and either 50 mM nitrate or fumarate as the electron acceptor. Cell growth was monitored spectrophotometrically by the optical density at 600 nm.
High-performance liquid chromatography analysis.
A Dionex DX-500-Microbore system was used with an IonPac AS11 column for anion analysis of mucus. Standards and blanks were analyzed, and standard curves were developed for a 1-mg sample of cecal mucus. Mouse cecal mucus was isolated from the cecum of CD-1 male mice and lyophilized as described previously (50). Regression coefficients for standard curves were calculated and used to demonstrate the linearity of the peak area with respect to concentration.
Mouse colonization experiments.
The streptomycin-treated mouse model has been used extensively to study colonization of the mouse large intestine by E. coli and Salmonella enterica serovar Typhimurium (8, 12, 22, 50). Briefly, three CD-1 male mice, 6 weeks of age, were given drinking water containing streptomycin sulfate (5 g/liter) for 24 h to remove the existing resident facultative microflora and then starved for food and water for 18 to 24 h. The mice were then fed approximately 105 CFU of both the wild-type and mutant strains in 1 ml of 20% sucrose. The wild-type strains were E. coli MG1655 Strr Nalr (nalidixic acid resistance) (33) and E. coli EDL933 Strr Nalr (31); Nalr was used to distinguish the wild-type (reference strain) from the null allele mutants in fecal plate counts. After the bacterial suspension was ingested, food and streptomycin-water were restored and fecal plate counts were determined at 5 h, 24 h, and on every other day thereafter for 15 days. Fecal samples were homogenized and diluted in 1% tryptone broth and plated on MacConkey agar containing either streptomycin (100 μg/ml) and nalidixic acid (50 μg/ml) to count the wild type or streptomycin and kanamycin (40 μg/ml) or chloramphenicol (30 μg/ml) to count the null allele mutants. Each colonization experiment was repeated on separate occasions, and the plotted values (in figures) represent the average for six mice. The log10 mean number of CFU per gram of feces ± the standard error for each strain in the mice was calculated for each time point. In all experiments, a difference between two strains of ≥10 CFU/g feces was statistically significant (i.e., P < 0.005 in Student's t test [two tailed with unequal variance]). The limit of detection in fecal plate counts was 102 CFU/g feces. To determine the role of strains during the maintenance stage of colonization, mice were colonized for 10 days with a mutant strain of E. coli EDL933, starved for food and streptomycin-water overnight, and fed 1010 CFU of the wild-type strain E. coli EDL933 Strr Nalr, after which food and streptomycin-water were replaced.
RESULTS
Colonization assays.
The preferred animal model for measuring the relative fitness of two bacterial strains for intestinal colonization is the streptomycin-treated mouse. Streptomycin treatment selectively removes facultative anaerobes while leaving the anaerobic microflora essentially intact; this opens a previously unavailable niche, which can then be colonized by newly introduced microorganisms such as E. coli (12, 27, 50). In this model, competing wild-type and mutant strains are fed together to mice and their populations are monitored in fecal plate counts. Previously we showed that colonization involves an initiation stage (5 h to 3 days postfeeding), in which nutrients are not limiting and the population increases from low to high numbers, and a maintenance stage (7 days post-feeding and beyond), in which nutrients are limiting and the population persists at a level correlated with the mutant strain's relative fitness for colonization (8). For this reason, the colonization data shown in Table 1 are given for day 1 (initiation) and day 9 (maintenance) of the 15-day-long experiments. To compare the bioenergetics of a commensal strain to those of a pathogenic strain, each of the respiratory mutations described in this report was constructed in E. coli MG1655 (3), derived from the human isolate E. coli K-12, and E. coli EDL933 (34), the prototypical strain of E. coli O157:H7. Please note that the streptomycin-treated mouse serves as a colonization model for EHEC strains, which do not cause disease in CD-1 mice (49). We found that each of the mutations tested in the commensal and pathogenic strains had a nearly identical impact on colonization. For this reason, colonization curves are shown for E. coli EDL933 experiments only.
TABLE 1.
Competitive colonization between respiratory mutants and wild-type E. coli strainsa
| Respiratory enzyme | Mutant(s) |
E. coli colonization (log10 CFU/g) by strain:
|
|||
|---|---|---|---|---|---|
| EDL933
|
MG1655
|
||||
| Day 1 | Day 9 | Day 1 | Day 9 | ||
| ATP synthetase | Δ(atpA-atpG) | 2.7 ± 0.1 | 5.6 ± 0.1 | 5.3 ± 0.5 | 5.9 ± 0.1 |
| Regulator of aerobic genes | ΔarcA | 4.7 ± 0.1 | 5.6 ± 0.2 | 3.0 ± 0.2 | 6.5 ± 0.3 |
| Regulator of anaerobic genes | Δfnr | 1.4 ± 0.2 | 5.6 ± 0.2 | 1.3 ± 0.1 | 6.8 ± 0.1 |
| Cytochrome bo oxidase | Δ(cyoA-cyoB) | 0.1 ± 0.1 | 0.4 ± 0.3 | 0.4 ± 0.1 | 0.8 ± 0.1 |
| Cytochrome bd oxidase | Δ(cydA-cydB) | 2.5 ± 0.1 | 5.2 ± 0.1 | 3.8 ± 0.2 | 5.0 ± 0.1 |
| Cytochrome bd oxidase (assembly) | Δ(cydD-cydC) | 2.4 ± 0.1 | 5.0 ± 0.2 | 3.3 ± 0.1 | 6.0 ± 0.2 |
| Nitrate reductase | ΔnarG | 0.3 ± 0.1 | 2.0 ± 0.1 | 0.4 ± 0.2 | 2.2 ± 0.1 |
| Nitrate reductase | ΔnarZ | 0.6 ± 0.1 | 0.1 ± 0.1 | 0.3 ± 0.1 | 0.1 ± 0.2 |
| Periplasmic nitrate reductase | Δ(napD-napA) | 0.4 ± 0.1 | 0.1 ± 0.1 | 0.2 ± 0.1 | 0.6 ± 0.1 |
| Nitrate reductases | ΔnarG, ΔnarZ | 0.9 ± 0.1 | 2.3 ± 0.1 | 0.1 ± 0.1 | 2.9 ± 0.2 |
| Nitrate reductases | ΔnarG, ΔnarZ, Δ(napD-napA) | 1.3 ± 0.1 | 3.1 ± 0.1 | 0.9 ± 0.1 | 3.8 ± 0.1 |
| Fumarate reductase | ΔfrdA | 0.8 ± 0.1 | 3.1 ± 0.2 | 0.1 ± 0.1 | 2.8 ± 0.2 |
Mice were fed 105 CFU each of a mutant and its wild-type parent. Mice were transferred to fresh cages every day, and feces no older than 24 h were assayed every other day for 15 days. At each time point, the log10 CFU/gram of feces for the mutant was subtracted from the log10 CFU/gram of feces for the wild type. The average ± standard error of the mean of day 1 and day 9 data from 6 mice is shown. Differences of at least 1 order of magnitude (10-fold) are in boldface type; all values shown in bold are statistically significant (P < 0.005; Student's t test).
ATP synthase is necessary for colonization.
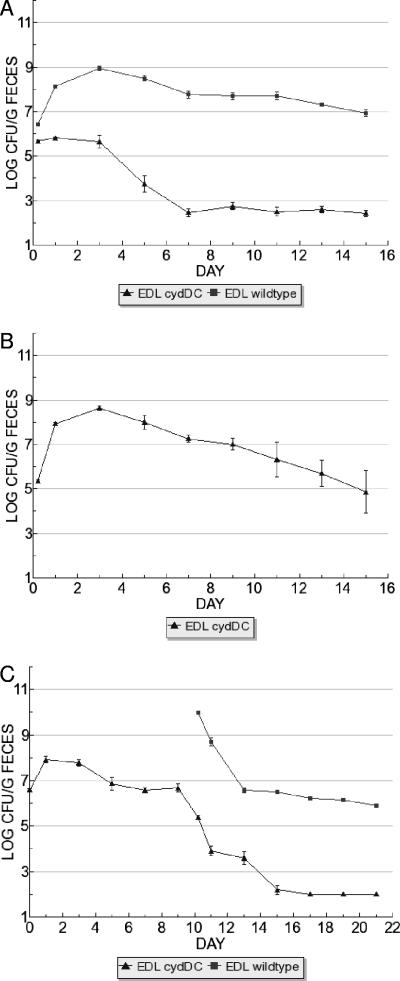
Since respiratory energy conservation, i.e., ATP generation, requires ATP synthase (32), we tested mutants that lack ATPase for their ability to compete with the wild type in the streptomycin-treated mouse colonization model. The ATPase mutant constructions deleted the atpA and atpG genes, which encode the F1 alpha and gamma subunits, respectively, resulting in strains capable only of fermentative energy metabolism (13). In competition with their respective wild-types, E. coli EDL933 Δ(atpA-atpG)::cat and E. coli MG1655 Δ(atpA-atpG)::cat mutants were eliminated from mice within 5 days (Table 1 and Fig. 1A). It is formally possible that mutations inadvertently introduced elsewhere on the genome caused the observed colonization phenotype. However, we would argue that this was not the case, for the following reasons. First, each of the allelic replacements described here and elsewhere (8, 28, 31, 33) was obtained with a frequency that varied less than 1 order of magnitude. Second, half of the mutants tested here and in similar studies (8) had no colonization defects. Third, the results were essentially identical in two different genetic backgrounds (EDL933 and MG1655). The failure of the Δ(atpA-atpG)::cat mutants to initiate colonization could be due to a general inability to grow in the intestine rather than an inability to compete with the wild type. To distinguish these possibilities, the mutants were fed alone to mice and found to colonize at wild-type levels (Fig. 1B) (data not shown). The ability of the respiratory-defective mutants to colonize alone indicates that fermentation is sufficient for growth of E. coli in the mouse intestine, but oxidative phosphorylation is essential for competition with respiratory-competent strains. This finding led us to consider whether one or more modes of respiration are crucial to occupation of intestinal niches formed by electron acceptor availability.
FIG. 1.
Respiratory mutants exhibited colonization defects in competitive colonization assays. E. coli EDL933 Δ(atpA-atpG) (A) and E. coli EDL933 Δ(cydA-cydB) (C) mutants were eliminated during competition with wild-type E. coli EDL933 but were able to colonize when fed alone to mice (B and D, respectively).
Respiration of oxygen is necessary for colonization.
Proof that aerobic respiration is essential for colonization by E. coli was obtained by competing mutants lacking the high-affinity cytochrome bd oxidase with their respective wild types. The Δ(cydA-cydB)::cat mutant constructions deleted genes encoding both subunits (I and II) of cytochrome bd oxidase, as shown previously (19). The E. coli EDL933 Δ(cydA-cydB)::cat and E. coli MG1655 Δ(cydA-cydB)::cat strains were eliminated by day 11 during competition with their wild-type parents (Table 1 and Fig. 1C). The fitness defect observed for the Δ(cydA-cydB)::cat strains was not merely a growth defect, since the mutants colonized when fed alone to mice (Fig. 1D) (data not shown). To confirm the requirement for high-affinity oxygen respiration, Δ(cydD-cydC)::cat mutants, which cannot assemble cytochrome bd oxidase in the membrane (39), were tested and found to have similar colonization defects (Table 1 and Fig. 2A). The Δ(cydD-cydC)::cat mutants were able to colonize when fed to mice alone (Fig. 2B) (data not shown). We note that in addition to CydDC being required for cytochrome bd oxidase assembly and activity (39), cydDC encodes a glutathione transport system (38), which if relevant might also be required for colonization. Since the Δ(cydA-cydB)::cat and Δ(cydD-cydC)::cat mutants failed to initiate colonization (i.e., 24 h), it was necessary to determine if they were also defective in the maintenance stage (beyond 7 days). Therefore, mice were precolonized (for 10 days) with E. coli EDL933 Δ(cydD-cydC)::cat and then challenged with the E. coli EDL933 wild type; the mutant was eliminated by the wild type in 7 days, confirming that functional cytochrome bd was important for maintenance of colonization (Fig. 2C).
FIG. 2.
Cytochrome bd oxidase assembly mutants of E. coli EDL933 exhibited colonization defects in competitive colonization assays. E. coli EDL933 Δ(cydD-cydC) was defective in competition with wild-type E. coli EDL933 (A) but colonized at wild-type levels when fed alone (B). The Δ(cydD-cydC) mutant exhibited a maintenance defect when mice were precolonized with E. coli EDL933 Δ(cydD-cydC) and challenged with wild-type E. coli EDL933 at day 10 (C).
We also examined the importance of the low-affinity cytochrome bo3 oxidase for colonization. The construction of Δ(cyoA-cyoB)::cat mutants deleted the genes encoding subunits I and II of the cytochrome bo3 oxidase, as shown previously (2). The results showed that E. coli EDL933 Δ(cyoA-cyoB)::cat and E. coli MG1655 Δ(cyoA-cyoB)::cat strains cocolonized with their respective wild-type parents, indicating that respiration of high oxygen levels was not necessary for colonization (Table 1). In summary, the colonization defects of the cytochrome bd oxidase mutants challenge the traditional view that the intestine is anaerobic (4). Instead, the results support the hypothesis that a microaerobic niche is critical for both establishing and maintaining E. coli in the intestine. Thus, a competitive advantage in vivo is conferred on strains that respire oxygen.
Aerobic respiratory control is necessary for colonization.
E. coli governs respiratory flexibility via the global regulators ArcA and Fnr. ArcA is a two-component regulator of several hundred genes (30) that responds to the oxidation state of the quinone pool, which is sensed by ArcB (18). Under high oxygen tension, E. coli expresses the low-affinity oxidase cytochrome bo3 (encoded by cyoABCDE) and the high-affinity oxidase cytochrome bd (encoded by cydAB) is repressed. Under microaerobic conditions, where oxygen scavenging may play an important role for growth and survival, ArcB phosphorylates ArcA, which represses the cyoABCDE operon and activates the cydAB and cydDC operons (1). Since the experiments described above indicated the importance of cytochrome bd, we reasoned that arcA mutants would have colonization defects. The construction of E. coli EDL933 ΔarcA::cat and E. coli MG1655 ΔarcA::kan deleted the gene encoding the response regulator of the ArcAB two-component system, as previously described (23). Although ΔarcA mutants colonized when fed alone (Fig. 3B) (data not shown), they could not compete with their respective wild types and were eliminated from mice within 3 days (Table 1 and Fig. 3A). While it is tempting to speculate that the colonization defect of the ΔarcA mutants resulted solely from failure to induce cydAB, the pleiotropic phenotype of the ΔarcA strain makes a number of alternative explanations possible. What is clear from this experiment is that appropriate regulation of aerobic respiratory genes is necessary for E. coli to be competitive in vivo.
FIG. 3.
Aerobic respiratory and anaerobic global regulatory mutants exhibited colonization defects in competitive colonization assays. E. coli EDL933 ΔarcA was eliminated during competition with wild-type E. coli EDL933 (A) but was able to colonize when fed alone (B). E. coli EDL933 Δfnr was eliminated during competition with wild-type E. coli EDL933 (C) but was able to colonize when fed alone (D).
Anaerobic control is necessary for colonization.
Induction of anaerobic processes in E. coli is controlled by Fnr, an oxygen-labile transcription factor, which activates transcription of hundreds of genes, including genes that encode respiratory pathways for nitrate and fumarate (25, 35). Since induction of anaerobic respiratory pathways requires Fnr, we constructed Δfnr::kan mutants to test the contribution of this global regulator during colonization. The E. coli EDL933 Δfnr::kan and E. coli MG1655 Δfnr::kan mutants were fed together with the respective wild types and found to initiate colonization (although not as well as the wild type), and then they were eliminated by day 5 (Table 1 and Fig. 3C). The defect was not an inability to grow in the intestine, since the Δfnr::kan mutants colonized when fed alone to mice (Fig. 3D) (data not shown). Since the Δfnr::kan mutants exhibit colonization defects in competition with the wild type, this implies that E. coli experiences conditions in the intestine that are required for Fnr function (7): i.e., anaerobic or nearly anaerobic conditions. Thus, we conclude that Fnr-dependent genes contribute to colonization success. While these results indicate that appropriate regulation of anaerobic respiration is important for colonization, the specific roles of the alternative pathways were unclear.
Nitrate reductase is necessary for colonization.
To determine which anaerobic respiratory pathways were used during colonization, we considered the in vivo role of nitrate reduction. Since E. coli has three systems for nitrate respiration, it was necessary to consider strains with mutations that eliminated each individually and in combination (17). Mutation of the primary nitrate reductase was accomplished by deleting narG (47) to create E. coli EDL933 ΔnarG::kan and E. coli MG1655 ΔnarG::kan. When ΔnarG::kan strains were fed together with their respective parent strains, they initiated colonization but then declined numerically (2.3 logs; P < 0.003) (Table 1 and Fig. 4A). To test the involvement in colonization of the secondary nitrate reductase, mutants were constructed in which narZ was deleted, the phenotype of which was described previously (46). Likewise, the involvement of the periplasmic nitrate reductase was tested with a construction that deleted napD and napA, which encode the assembly protein and large subunit of the reductase, respectively (46). In colonization assays, E. coli EDL933 ΔnarZ::cat, E. coli MG1655 ΔnarZ::cat, E. coli EDL933 Δ(napD-napA)::cat, and E. coli MG1655 Δ(napD-napA)::cat cocolonized with their wild-type parents, indicating that strains with these individual mutations had no phenotype in the intestine (Table 1).
FIG. 4.
Anaerobic respiration mutants exhibited colonization defects in competitive colonization assays. E. coli EDL933 ΔnarG (A), E. coli EDL933 ΔnarG ΔnarZ (B), E. coli EDL933 ΔnarG ΔnarZ Δ(napD-napA) (C), and E. coli EDL933 ΔfrdA (D) mutants were defective in competition with wild-type E. coli EDL933.
Although the individual ΔnarZ::cat and Δ(napD-napA)::cat mutants did not exhibit colonization defects, there is reason to believe these gene systems should be expressed in the wild types under the conditions present in the intestine. First, the intestine contains regions that are microaerobic and others that are anaerobic (20), yet apparently is sufficiently anaerobic overall for Fnr control to be a factor in success of the entire E. coli population (Fig. 3). Second, we measured a concentration of 2.62 ± 0.21 mM nitrate in mouse cecal mucus (data not shown), a concentration which is physiologically relevant for controlling expression of the two nitrate-inducible systems (52). Third, E. coli cells isolated from the intestine show both logarithmic and stationary-phase characteristics (26). The narZYWV operon that encodes the secondary nitrate reductase is not regulated by anaerobiosis or nitrate but instead is RpoS dependent and induced in stationary phase (9). The napFDAGHBC operon that encodes the periplasmic nitrate reductase is maximally induced at 1 mM nitrate and is expressed at one-half of the maximal level at 2.5 mM (52). Thus, it is reasonable to expect that both the periplasmic and secondary nitrate reductases are expressed in the intestine. To investigate the phenotypes of these nitrate reductase mutations in the absence of the primary nitrate reductase, we constructed ΔnarG ΔnarZ::cat and ΔnarG ΔnarZ Δ(napD-napA)::cat mutants. In colonization assays, E. coli EDL933 ΔnarG ΔnarZ::cat and E. coli MG1655 ΔnarG ΔnarZ::cat mutants declined 3.2 log units (P < 0.000006) relative to the wild-type strain by day 15 (Table 1 and Fig. 4B). Also, the E. coli EDL933 ΔnarG ΔnarZ Δ(napD-napA)::cat and E. coli MG1655 ΔnarG ΔnarZ Δ(napD-napA)::cat mutants declined by 5 log units (P < 9 × 10−15) to populations just above the limit of detection (<103 CFU/g feces) by the conclusion of the experiments (Table 1 and Fig. 4D). The additive effect of the sequential nitrate reductase mutations in competition with the wild-type E. coli EDL933 in these experiments is statistically significant between the ΔnarG::kan and ΔnarG ΔnarZ::cat mutants (P < 0.003) and between the ΔnarG ΔnarZ::cat and ΔnarG ΔnarZ Δ(napD-napA)::cat mutants (P < 0.004). From these results, we conclude the primary nitrate reductase plays the larger role of the three systems in the mouse intestine. In contrast, the ΔnarZ and Δ(napD-napA) mutations affected colonization only in the ΔnarG background, suggesting a synergy, rather than mere redundancy, between the primary nitrate reductase and the other two systems. Apparently, conditions in the intestine signal induction of the three nitrate reductase gene systems, which together confer a competitive advantage to E. coli.
Fumarate reductase is necessary for colonization.
To test the importance of fumarate as an alternative electron acceptor in vivo, we constructed mutants with frdA, which are known to inactivate fumarate reductase (43). E. coli EDL933 ΔfrdA::kan and E. coli MG1655 ΔfrdA::kan mutants were fed together with their respective parent strains and found to initiate colonization but then declined by approximately 4 logs relative to the wild type (Table 1 and Fig. 4D). Thus, fumarate reductase mutants competed poorly, indicating that fumarate is used in the intestine as an alternative electron acceptor. Since fumarate was not detected in intestinal mucus (data not shown), it most likely was generated endogenously, giving rise to succinate as a fermentation product during anaerobiosis (41).
DISCUSSION
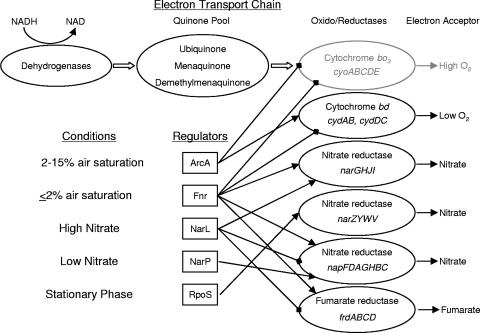
Figure 5 shows a model of the respiratory pathways that are critical for successful colonization by both EHEC and commensal E. coli. The results indicate that the gut is not strictly anaerobic, because the high-affinity cytochrome bd oxidase is required to successfully compete with the wild-type for colonization (Fig. 1 and 2). Also, anaerobic respiration of nitrate and fumarate is essential for E. coli in vivo (Fig. 4). We therefore conclude that success of E. coli in the gastrointestinal tract demands respiratory flexibility and use of the best available electron acceptor. The results allow us to deduce the intestinal environment as it is perceived by E. coli. Accordingly, the niches defined by mutational analysis of respiratory pathways should correspond with in vivo availability of exogenous electron acceptors (i.e., oxygen and nitrate) but not necessarily fumarate, which is generated endogenously by sugar degradation via central metabolism. These respiratory niches could be open to the entire population, wherein each bacterium simultaneously uses both oxygen and nitrate, or individual cells and microcolonies could each use different electron acceptors. If distinct microaerobic and anaerobic niches were to exist in the intestine, then mutants in the respective respiratory pathways would be maintained at populations corresponding to availability of the cogent electron acceptor when in competition with respiratory-competent wild types. However, since the loss of the high-affinity oxygen, nitrate, or fumarate respiratory pathways leads to the near or complete elimination of the entire population in competition with respiratory-competent strains (Fig. 1C and 4C and D), the data give us reason to think that both aerobic and anaerobic niches are equally crucial. Thus, the behavior of the E. coli respiratory mutants implies that the intestinal habitat is at one time microaerobic and at another time anaerobic. Indeed, oxygen tension in the intestine may fluctuate due to dynamic cycles of oxygen diffusion and respiratory consumption by facultative anaerobes.
FIG. 5.
Model of E. coli respiratory pathways. Respiratory oxidases and reductases used by E. coli in vivo are shown in black. The oxidase not affecting colonization is shown in gray. The environmental conditions affecting the key regulators of the genes encoding the oxido-reductases are shown. Activation is shown with arrowheads, and repression is shown with diamond heads.
Cole (10) postulated that the redundancy of respiratory systems and complexities of their regulation ready bacteria for changes in electron acceptor availability: when respiring nitrate, E. coli can respond rapidly to the “arrival” of oxygen or cope when nitrate is exhausted. Thus, the physiology, biochemistry, and genetic control of respiratory pathways provide flexibility that is thought to be essential for survival in a changing environment (10). It would appear this strategy is singularly important during animal colonization. We summarize these factors in Fig. 5. For E. coli to colonize the intestine, appropriate aerobic respiratory control (ArcA) and anaerobic control (Fnr) are required (Fig. 3). ArcA is most active under microaerobic conditions (i.e., oxygen tension of 2 to 15% of air saturation) (1), and Fnr is most active during transition to anaerobic conditions (i.e., oxygen tension of less than 2% of air saturation) (7). Whole-animal measurements indicated oxygen tensions in the 2 to 7% air saturation range for the mouse colon (20). The measured nitrate concentration in intestinal mucus (2.6 mM [see results]) is near that which results in maximal expression of both nitrate reductase systems (i.e., 1 mM for the periplasmic nitrate reductase and 7 mM for the primary nitrate reductase) (52). Regulation of the periplasmic nitrate reductase genes requires both NarL and NarP (45), while regulation of the primary reductase genes requires NarL only (44). Since NarP exerts its control at low nitrate levels (<4 mM) and NarL control dominates at higher nitrate levels (51), and since both the primary and periplasmic nitrate reductases are necessary for efficient colonization, this suggests that nitrate availability might fluctuate in the intestine. Thus, if the intestinal oxygen tension fluctuates in the anaerobic to microaerobic range and the nitrate concentration fluctuates in the 1 to 7 mM range, then cytochrome bd oxidase, the primary nitrate reductase, the periplasmic nitrate reductase, and fumarate reductase all will be expressed in vivo. Indeed, regulation of these gene systems is poised to be most responsive to changes in oxygen and nitrate availability in these concentration ranges (48).
The inference that oxygen availability fluctuates because it is consumed by bacterial respiration suggests the interesting possibility that facultative anaerobes may make the intestinal environment more anaerobic. Indeed, this conclusion is supported by previous studies of the effect of streptomycin treatment on the mouse anaerobic microflora, which selectively removes facultative anaerobes (i.e., E. coli, enterococci, streptococci, and lactobacilli). Following administration of streptomycin, populations of strict anaerobes (e.g., bifidobacteria and clostridia) decreased, while populations of so-called “nanaerobes” were unchanged (22): e.g., Bacteroides fragilis, which respires oxygen when available in low concentrations (5). Thus, comparisons of the populations of anaerobes in mice with or without facultative anaerobes present support the hypothesis that oxygen-scavenging facultative anaerobes (e.g., E. coli) promote the stability of the predominantly anaerobic microflora, exemplifying how a minor member can have a large impact on an ecosystem.
Despite the apparent competitive advantage gained by oxygen respiration, the E. coli population is limited to between 108 and 109 CFU/g feces: i.e., E. coli represents between 1 in 1,000 and 1 in 10,000 bacteria in the intestine. The nutrient-niche hypothesis states that to be successful each species of the intestinal microflora must use at least one carbon source better than all other species (16). Corollary to this hypothesis, the population size of any member of the microflora is determined by the concentration of its preferred nutrient(s). The available concentrations of the seven sugars that contribute to colonization by E. coli MG1655 are quite low (12, 37). Since E. coli does not secrete polysaccharide-degrading enzymes, its preferred substrates are most likely provided by anaerobes, which degrade mucosal polysaccharides and dietary fiber and are thought to release the breakdown products for use by the host and other microbes (11). These facts lead to the conclusion that E. coli maximizes its growth yield by coupling oxidation of low nutrient concentrations to respiration in the intestine. This may be a general strategy of facultative anaerobes, which generally grow well on simple sugars but do not secrete polysaccharide-degrading enzymes. Thus, high-efficiency respiration may ensure the success of facultative anaerobes in the intestine, albeit always in lower numbers, by allowing them to maximize cell yield on scarce resources.
Since most mucosal pathogens are facultative anaerobes, our conclusions may extend to other mucosal pathogens. In support of this idea, Mycobacterium tuberculosis genes encoding cytochrome bd oxidase and the nitrate transporter were induced during mouse lung infection; a cytochrome bd oxidase mutant was attenuated during transition to chronic infection in mice (42). Likewise, Shigella flexneri cytochrome bd mutants showed decreased intracellular survival and attenuated virulence in mouse infections (53). These examples demonstrate the importance of respiration during infection by particular mucosal pathogens and support the idea that oxygen stimulates infectious disease by providing a competitive advantage for pathogens. Since there appears to be no distinction between enterohemorrhagic and commensal E. coli with respect to the respiratory pathways used in vivo, we suggest caution in targeting respiratory metabolism for combating EHEC infections because of potential collateral damage to commensal facultative anaerobes and the resulting instability of the intestinal microbiota (16).
In summary, we have shown that E. coli uses both aerobic and anaerobic respiratory pathways during colonization. The results presented in this study support the conclusion that the intestine is microaerobic and that aerobic bacterial respiration in the intestine is essential for competition and therefore successful colonization. Apparently, E. coli respires oxygen to optimize its reproduction in animals despite the low availability of its preferred carbon sources, which maximizes its colonization efficiency.
Acknowledgments
We thank Bruce Roe and David Laux for helpful comments.
This work was supported by grant AI48945 from the National Institutes of Health.
Editor: V. J. DiRita
Footnotes
Published ahead of print on 13 August 2007.
REFERENCES
- 1.Alexeeva, S., K. J. Hellingwerf, and M. J. Teixeira de Mattos. 2003. Requirement of ArcA for redox regulation in Escherichia coli under microaerobic but not anaerobic or aerobic conditions. J. Bacteriol. 185:204-209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Au, D. C.-T., R. M. Lorence, and R. B. Gennis. 1985. Isolation and characterization of an Escherichia coli mutant lacking the cytochrome o terminal oxidase. J. Bacteriol. 161:123-127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bachmann, B. J. 1996. Derivations and genotypes of some mutant derivatives of Escherichia coli K-12, p.2460-2488. In F. C. Neidhardt, R. Curtiss III, J. L. Ingraham, E. C. C. Lin, K. B. Low, B. Magasanik, W. S. Reznikoff, M. Riley, M. Schaechter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella: cellular and molecular biology, 2nd ed.,vol. 2. ASM Press, Washington, DC. [Google Scholar]
- 4.Backhed, F., R. E. Ley, J. L. Sonnenburg, D. A. Peterson, and J. I. Gordon. 2005. Host-bacterial mutualism in the human intestine. Science 307:1915-1920. [DOI] [PubMed] [Google Scholar]
- 5.Baughn, A. D., and M. H. Malamy. 2004. The strict anaerobe Bacteroides fragilis grows in and benefits from nanomolar concentrations of oxygen. Nature 427:441-444. [DOI] [PubMed] [Google Scholar]
- 6.Beaulieu, J. F., and A. Quaroni. 1991. Clonal analysis of sucrase-isomaltase expression in the human colon adenocarcinoma Caco-2 cells. Biochem. J. 280:599-608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Becker, S., G. Holighaus, T. Gabrielczyk, and G. Unden. 1996. O2 as the regulatory signal for FNR-dependent gene regulation in Escherichia coli. J. Bacteriol. 178:4515-4521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chang, D. E., D. J. Smalley, D. L. Tucker, M. P. Leatham, W. E. Norris, S. J. Stevenson, A. B. Anderson, J. E. Grissom, D. C. Laux, P. S. Cohen, and T. Conway. 2004. Carbon nutrition of Escherichia coli in the mouse intestine. Proc. Natl. Acad. Sci. USA 101:7427-7432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chang, L., L. I. Wei, J. P. Audia, R. A. Morton, and H. E. Schellhorn. 1999. Expression of the Escherichia coli NRZ nitrate reductase is highly growth phase dependent and is controlled by RpoS, the alternative vegetative sigma factor. Mol. Microbiol. 34:756-766. [DOI] [PubMed] [Google Scholar]
- 10.Cole, J. 1996. Nitrate reduction to ammonia by enteric bacteria: redundancy, or a strategy for survival during oxygen starvation? FEMS Microbiol. Lett. 136:1-11. [DOI] [PubMed] [Google Scholar]
- 11.Comstock, L. E., and M. J. Coyne. 2003. Bacteroides thetaiotaomicron: a dynamic, niche-adapted human symbiont. Bioessays 25:926-929. [DOI] [PubMed] [Google Scholar]
- 12.Conway, T., K. A. Krogfelt, and P. S. Cohen. 29 December 2004, posting date. Chapter 8.3.1.2, The life of commensal Escherichia coli in the mammalian intestine. In A. Böck, R. Curtiss III, J. B. Kaper, F. C. Neidhardt, T. Nyström, K. E. Rudd, and C. L. Squires (ed.), EcoSal—Escherichia coli and Salmonella: cellular and molecular biology. ASM Press, Washington, DC. [DOI] [PubMed]
- 13.Cox, G. B., J. A. Downie, F. Gibson, and J. Radik. 1978. Genetic complementation between two mutant unc alleles (unc A401 and unc D409) affecting the Fl portion of the magnesium ion-stimulated adenosine triphosphatase of Escherichia coli K12. Biochem. J. 170:593-598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Datsenko, K. A., and B. L. Wanner. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. USA 97:6640-6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Eckburg, P. B., E. M. Bik, C. N. Bernstein, E. Purdom, L. Dethlefsen, M. Sargent, S. R. Gill, K. E. Nelson, and D. A. Relman. 2005. Diversity of the human intestinal microbial flora. Science 308:1635-1638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Freter, R. 1983. Mechanisms that control the microflora in the large intestine, p.33-54. In D. J. Hentges (ed.), Human intestinal microflora in health and disease. Academic Press, Inc., New York, NY.
- 17.Gennis, R. B., and V. Stewart. 1996. Respiration, p. 217-261. In F. C. Neidhardt, R. Curtiss III, J. L. Ingraham, E. C. C. Lin, K. B. Low, B. Magasanik, W. S. Reznikoff, M. Riley, M. Schaechter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella: cellular and molecular biology, 2nd ed., vol. 1. ASM Press, Washington, DC. [Google Scholar]
- 18.Georgellis, D., O. Kwon, and E. C. Lin. 2001. Quinones as the redox signal for the arc two-component system of bacteria. Science 292:2314-2316. [DOI] [PubMed] [Google Scholar]
- 19.Green, G. N., and R. B. Gennis. 1983. Isolation and characterization of an Escherichia coli mutant lacking cytochrome d terminal oxidase. J. Bacteriol. 154:1269-1275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.He, G., R. A. Shankar, M. Chzhan, A. Samouilov, P. Kuppusamy, and J. L. Zweier. 1999. Noninvasive measurement of anatomic structure and intraluminal oxygenation in the gastrointestinal tract of living mice with spatial and spectral EPR imaging. Proc. Natl. Acad. Sci. USA 96:4586-4591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hentges, D. J. 1983. Role of the intestinal microflora in host defense against infection, p.311-331. In D. J. Hentges (ed.), Human intestinal microflora in health and disease. Academic Press, New York, NY.
- 22.Hentges, D. J., J. U. Que, S. W. Casey, and A. J. Stein. 1984. The influence of streptomycin on colonization in mice. Microecol. Theor. 14:53-62. [Google Scholar]
- 23.Iuchi, S., and E. C. Lin. 1988. arcA (dye), a global regulatory gene in Escherichia coli mediating repression of enzymes in aerobic pathways. Proc. Natl. Acad. Sci. USA 85:1888-1892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kaper, J. B., J. P. Nataro, and H. L. Mobley. 2004. Pathogenic Escherichia coli. Nat. Rev. Microbiol. 2:123-140. [DOI] [PubMed] [Google Scholar]
- 25.Kiley, P. J., and H. Beinert. 1998. Oxygen sensing by the global regulator, FNR: the role of the iron-sulfur cluster. FEMS Microbiol. Rev. 22:341-352. [DOI] [PubMed] [Google Scholar]
- 26.Krogfelt, K. A., L. K. Poulsen, and S. Molin. 1993. Identification of coccoid Escherichia coli BJ4 cells in the large intestine of streptomycin-treated mice. Infect. Immun. 61:5029-5034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Laux, D. C., P. S. Cohen, and T. Conway. 2005. Role of the mucus layer in bacterial colonization of the intestine, p.199-212. In J. P. Nataro, P. S. Cohen, H. L. T. Mobley, and J. N. Weiser (ed.), Colonization of mucosal surfaces. ASM Press, Washington, DC.
- 28.Leatham, M. P., S. J. Stevenson, E. J. Gauger, K. A. Krogfelt, J. J. Lins, T. L. Haddock, S. M. Autieri, T. Conway, and P. S. Cohen. 2005. Mouse intestine selects nonmotile flhDC mutants of Escherichia coli MG1655 with increased colonizing ability and better utilization of carbon sources. Infect. Immun. 73:8039-8049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Levitt, M. D., and J. H. Bond. 1980. Flatulence. Annu. Rev. Med. 31:127-137. [DOI] [PubMed] [Google Scholar]
- 30.Lynch, A. S., and E. C. C. Lin. 1996. Responses to molecular oxygen, p.1526-1538. In F. C. Neidhardt, R. Curtiss III, J. L. Ingraham, E. C. C. Lin, K. B. Low, B. Magasanik, W. S. Reznikoff, M. Riley, M. Schaechter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella: cellular and molecular biology, 2nd ed.,vol. 1. ASM Press, Washington, DC. [Google Scholar]
- 31.Miranda, R. L., T. Conway, M. P. Leatham, D. E. Chang, W. E. Norris, J. H. Allen, S. J. Stevenson, D. C. Laux, and P. S. Cohen. 2004. Glycolytic and gluconeogenic growth of Escherichia coli O157:H7 (EDL933) and E. coli K-12 (MG1655) in the mouse intestine. Infect. Immun. 72:1666-1676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mitchell, P. 1961. Coupling of phosphorylation to electron and hydrogen transfer by a chemi-osmotic type of mechanism. Nature 191:144-148. [DOI] [PubMed] [Google Scholar]
- 33.Møller, A. K., M. P. Leatham, T. Conway, P. J. M. Nuijten, L. A. M. de Haan, K. A. Krogfelt, and P. S. Cohen. 2003. An Escherichia coli MG1655 lipopolysaccharide deep-rough core mutant grows and survives in mouse cecal mucus but fails to colonize the mouse large intestine. Infect. Immun. 71:2142-2152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.O'Brien, A. D., A. R. Melton, C. K. Schmitt, M. L. McKee, M. L. Batts, and D. E. Griffin. 1993. Profile of Escherichia coli O157:H7 pathogen responsible for hamburger-borne outbreak of hemorrhagic colitis and hemolytic uremic syndrome in Washington. J. Clin. Microbiol. 31:2799-2801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Overton, T. W., L. Griffiths, M. D. Patel, J. L. Hobman, C. W. Penn, J. A. Cole, and C. Constantinidou. 2006. Microarray analysis of gene regulation by oxygen, nitrate, nitrite, FNR, NarL and NarP during anaerobic growth of Escherichia coli: new insights into microbial physiology. Biochem. Soc. Trans. 34:104-107. [DOI] [PubMed] [Google Scholar]
- 36.Pearson, H. 2007. The dark side of E. coli. Nature 445:8-9. [DOI] [PubMed] [Google Scholar]
- 37.Peekhaus, N., and T. Conway. 1998. What's for dinner? Entner-Doudoroff metabolism in Escherichia coli. J. Bacteriol. 180:3495-3502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pittman, M. S., H. C. Robinson, and R. K. Poole. 2005. A bacterial glutathione transporter (Escherichia coli CydDC) exports reductant to the periplasm. J. Biol. Chem. 280:32254-32261. [DOI] [PubMed] [Google Scholar]
- 39.Poole, R. K., F. Gibson, and G. Wu. 1994. The cydD gene product, component of a heterodimeric ABC transporter, is required for assembly of periplasmic cytochrome c and of cytochrome bd in Escherichia coli. FEMS Microbiol. Lett. 117:217-223. [DOI] [PubMed] [Google Scholar]
- 40.Savage, D. C. 1977. Microbial ecology of the gastrointestinal tract. Annu. Rev. Microbiol. 31:107-133. [DOI] [PubMed] [Google Scholar]
- 41.Sawers, R. G., and D. P. Clark. 27 July 2004, posting date. Chapter 3.5.3, Fermentative pyruvate and acetyl-coenzyme A metabolism. In A. Böck, R. Curtiss III, J. B. Kaper, F. C. Neidhardt, T. Nyström, K. E. Rudd, and C. L. Squires (ed.), EcoSal—Escherichia coli and Salmonella: cellular and molecular biology. ASM Press, Washington, DC.
- 42.Shi, L., C. D. Sohaskey, B. D. Kana, S. Dawes, R. J. North, V. Mizrahi, and M. L. Gennaro. 2005. Changes in energy metabolism of Mycobacterium tuberculosis in mouse lung and under in vitro conditions affecting aerobic respiration. Proc. Natl. Acad. Sci. USA 102:15629-15634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Spencer, M. E., and J. R. Guest. 1973. Isolation and properties of fumarate reductase mutants of Escherichia coli. J. Bacteriol. 114:563-570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Stewart, V. 1993. Nitrate regulation of anaerobic respiratory gene expression in Escherichia coli. Mol. Microbiol. 9:425-434. [DOI] [PubMed] [Google Scholar]
- 45.Stewart, V., P. J. Bledsoe, and S. B. Williams. 2003. Dual overlapping promoters control napF (periplasmic nitrate reductase) operon expression in Escherichia coli K-12. J. Bacteriol. 185:5862-5870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Stewart, V., Y. Lu, and A. J. Darwin. 2002. Periplasmic nitrate reductase (NapABC enzyme) supports anaerobic respiration by Escherichia coli K-12. J. Bacteriol. 184:1314-1323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Stewart, V., and C. H. MacGregor. 1982. Nitrate reductase in Escherichia coli K-12: involvement of chlC, chlE, and chlG loci. J. Bacteriol. 151:788-799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tseng, C.-P., J. Albrecht, and R. P. Gunsalus. 1996. Effect of microaerophilic cell growth conditions on expression of the aerobic (cyoABCDE and cydAB) and anaerobic (narGHJI, frdABCD, and dmsABC) respiratory pathway genes in Escherichia coli. J. Bacteriol. 178:1094-1098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wadolkowski, E. A., J. A. Burris, and A. D. O'Brien. 1990. Mouse model for colonization and disease caused by enterohemorrhagic Escherichia coli O157:H7. Infect. Immun. 58:2438-2445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wadolkowski, E. A., D. C. Laux, and P. S. Cohen. 1988. Colonization of the streptomycin-treated mouse large intestine by a human fecal Escherichia coli strain: role of growth in mucus. Infect. Immun. 56:1030-1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wang, H., and R. P. Gunsalus. 2003. Coordinate regulation of the Escherichia coli formate dehydrogenase fdnGHI and fdhF genes in response to nitrate, nitrite, and formate: roles for NarL and NarP. J. Bacteriol. 185:5076-5085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wang, H., C.-P. Tseng, and R. P. Gunsalus. 1999. The napF and narG nitrate reductase operons in Escherichia coli are differentially expressed in response to submicromolar concentrations of nitrate but not nitrite. J. Bacteriol. 181:5303-5308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Way, S. S., S. Sallustio, R. S. Magliozzo, and M. B. Goldberg. 1999. Impact of either elevated or decreased levels of cytochrome bd expression on Shigella flexneri virulence. J. Bacteriol. 181:1229-1237. [DOI] [PMC free article] [PubMed] [Google Scholar]