Abstract
Intrakingdom cell-to-cell communication and interkingdom cell-to-cell communication play essential roles in the virulence of enterohemorrhagic Escherichia coli (EHEC). Four signals, autoinducer 2 (AI-2), AI-3, and the human hormones epinephrine and norepinephrine, are important in this communication. The effect of these signaling compounds on the transcriptome of EHEC was examined in this study. We demonstrated that the luxS mutation affects primarily central metabolic genes in both pathogenic and nonpathogenic strains of E. coli and that addition of exogenous AI-2 does not fully restore the expression profile in a luxS-deficient strain lacking the ability to synthesize AI-2. Addition of AI-3 or epinephrine increased expression of the locus of enterocyte effacement regulon, which is known to play a pivotal role in EHEC virulence. Moreover, when epinephrine was added to the culture medium, the greatest number of gene alterations was observed. These alterations included a greater proportion of alterations in EHEC genes than in MG1655 genes, suggesting that epinephrine may be a global virulence signal. Detailed examination with real-time reverse transcriptase PCR (RT-PCR) confirmed the increases in virulence gene expression with addition of AI-3 and epinephrine. Additional studies with real-time RT-PCR examining the EHEC secreted effectors and putative fimbrial gene expression showed a variable expression profile, indicating that there is differential regulation of the secreted molecules. This study began to examine the global signaling networks in EHEC and revealed expression profiles that are signal and pathogen specific.
The human pathogen enterohemorrhagic Escherichia coli O157:H7 (EHEC) colonizes the human colon, resulting in the development of hemorrhagic colitis and hemolytic-uremic syndrome that may be fatal (36). Upon colonization of the colon, EHEC forms attaching and effacing (AE) lesions on the epithelial cells and produces Shiga toxin. Most of the genes involved in the formation of the AE lesions are in a chromosomal pathogenicity island called the locus of enterocyte effacement (LEE) (41). The LEE encodes a type III secretion system (TTSS) and effector proteins that are translocated into epithelial cells and cause extensive cytoskeletal rearrangements resulting in the formation of AE lesions (33, 34, 37). In addition to the LEE, EHEC's arsenal of virulence factors includes non-LEE-encoded effector proteins that are secreted through the LEE-encoded TTSS (8, 14, 21, 22, 25, 63) and may also include fimbriae that increase adherence or mediate colonization of epithelial cells (64, 65).
Regulation of the LEE genes is extremely complex and includes involvement of the global regulators H-NS (7, 26, 43, 50, 56, 66) and integration host factor (19) and the environment-dependent regulator Hha (52), which act to repress LEE transcription. Other regulators include the LysR transcriptional regulator QseA that positively regulates LEE by binding to ler (53, 55) and the ClpXP protease (31) that increases transcription of LEE by inhibiting GrlR repression and also by interacting with the stationary-phase sigma factor RpoS. RpoS also positively regulates transcription of LEE3 (31, 57), and the signaling molecule ppGpp can also increase transcription of the LEE (46). Many of the regulators mentioned above are common to both pathogenic and nonpathogenic strains of E. coli; however, a number of regulators are unique to EHEC. Encoded in the LEE, Ler (LEE-encoded regulator) is able to overcome H-NS-mediated repression and activate expression of the LEE2, LEE3, and LEE5 operons (26, 50, 56), and GrlR and GrlA repress and activate, respectively, transcription of ler (3, 14). The pch genes that are homologous to perC in enteropathogenic E. coli increase expression of the LEE genes (32). Finally, the transcriptional regulators that are encoded by eivF and etrA in a second, nonfunctional TTSS (ETT2) are negative regulators of the LEE (76).
EHEC also utilizes quorum sensing (QS) to regulate expression of its virulence and flagellar and motility genes (57-60). Initial investigations suggested that autoinducer 2 (AI-2) was the QS signal responsible for regulating expression of virulence genes in EHEC (57, 58); however, subsequent research using purified and in vitro-synthesized AI-2 demonstrated that the signaling molecule affecting the TTSS and motility was not AI-2 but was a distinct compound designated AI-3 (59). Differences in these molecules have been revealed by biochemical assays. The polar furanone AI-2 does not bind to C18 columns, whereas AI-3 binds to C18 columns and can be eluted only with methanol; and electrospray mass spectrometry revealed structural differences between AI-2 and AI-3 (9, 59). Moreover, the transcriptional assay for AI-2 is based on the production of bioluminescence in Vibrio harveyi, and AI-3 does not show any activity in this assay. Conversely, AI-3 activates transcription of the EHEC virulence genes, whereas AI-2 has no effect in this assay (59, 68).
AI-2 production is dependent upon the LuxS enzyme (4, 5, 61). LuxS plays a role in the metabolism of S-adenosylmethionine by converting S-ribosylhomocysteine into homocysteine and 4,5-dihydroxy-2,3-pentanedione (DPD). DPD is unstable and spontaneously cyclizes to form several different furanones, one of which is believed to be AI-2 (51). AI-3 is not dependent upon LuxS for synthesis; however, a luxS mutation leaves the cell with only one pathway to produce homocysteine, which may lead to diminished production of AI-3 (68). Additional regulation occurs through cross talk between EHEC and its host (59). EHEC senses the host hormones epinephrine and norepinephrine through the membrane protein QseC (11). QseC senses both AI-3 and epinephrine and thus functions in interkingdom cross-signaling (11). QseC is part of a two-component system, QseB/C, in which QseC is the sensor kinase and QseB is the response regulator. QseB/C activates transcription of the flagellar regulon responsible for swimming motility in EHEC (10). Furthermore, QseC plays an important role in EHEC pathogenesis, as the virulence of a qseC mutant was attenuated in a rabbit infection model (11).
A previous gene array analysis was performed in order to elucidate the role that QS plays in the regulation of EHEC virulence and physiology by comparing a luxS mutant strain of EHEC to wild-type (WT) EHEC (58). This analysis demonstrated that luxS regulation was pleiotropic and regulated numerous basic physiological functions, including cell division, motility, and genes involved in metabolism, as well as virulence (58). The fact that EHEC produces two AI molecules was not recognized at that time, nor was it known that EHEC responds to human hormones; thus, the specific role that each signaling molecule plays in gene regulation was not fully elucidated. The specific aim of this study was to determine more precisely how cell signaling by AI-2, AI-3, and epinephrine affect global gene expression in EHEC. Transcriptome analyses were performed to compare global gene expression in WT EHEC to gene expression in a luxS mutant grown without QS molecules added to the culture medium or grown with the signaling molecules AI-2, AI-3, and epinephrine added to the medium.
MATERIALS AND METHODS
Strains and culture and growth conditions.
WT EHEC strain 86-24 was used in this study. Strain 86-24 was isolated in 1986 from a patient in Seattle experiencing hemorrhagic colitis (24) and has been used extensively to study EHEC infection in animal models (13, 16, 36, 42, 54). The isogenic luxS mutant strain VS94 (58) was also used in this study. Dulbecco's modified Eagle's medium (DMEM) (Invitrogen) was used as the growth medium in all assays.
RNA extraction.
Cultures of strains 86-24 and VS94 were grown aerobically in LB medium at 37°C overnight and then were diluted 1:100 in DMEM and grown in a shaking incubator at 37°C. The tested compounds were added to the media at the following concentrations: 100 μM DPD (AI-2), 30 μM AI-3, and 50 μM epinephrine. RNA was extracted from three biological replicate cultures of each strain per condition at the late exponential growth phase (optical density at 600 nm, 1.0) using a RiboPure bacterial RNA isolation kit (Ambion).
Microarrays.
The GeneChip E. coli Genome 2.0 array system of the Affymetrix system was used to compare the gene expression in strain 86-24 to that in strain VS94 (luxS mutant) (with and without addition of signaling molecules to culture media). The GeneChip E. coli Genome 2.0 array includes approximately 10,000 probe sets for all 20,366 genes present in the following four strains of E. coli: K-12 lab strain MG1655, uropathogenic strain CFT073, O157:H7 enterohemorrhagic strain EDL933, and O157:H7 enterohemorrhagic strain Sakai (http://www.affymetrix.com/products/arrays/specific/ecoli2.affx). The RNA-processing, labeling, hybridization, and slide-scanning procedures were preformed as described in the Affymetrix Gene Expression Technical Manual (http://www.affymetrix.com/support/technical/manual/expression_manual.affx).
Microarray data analysis.
The output from scanning a single replicate of the Affymetrix GeneChip E. coli Genome 2.0 array for each of the biological conditions was obtained using GCOS v 1.4 according to the manufacturer's instructions. Data were normalized using Robust Multiarray analysis (6, 30) at the RMAExpress website (http://rmaexpress.bmbolstad.com/). The resulting data were compared to determine features whose expression was increased or decreased in response to either the QS stimuli or inactivation of the luxS gene. Custom analysis scripts were written in Perl to complete multiple array analyses. The results of the array analyses were further confirmed using real-time reverse transcriptase PCR (RT-PCR) as described below. We note that the isolate used in these studies has not been sequenced and thus is not fully contained on the array and that differences in genome content are evident. Expression data can be accessed using accession number GSE7439 at the NCBI GEO database.
Real-time RT-PCR.
The primers used in the real-time assays were designed using Primer Express v1.5 (Applied Biosystems) (Table 1). The amplification efficiency and template specificity of each of the primer pairs were validated as described previously (69). The real-time RT-PCR was a one-step reaction performed with an ABI 7500 sequence detection system (Applied Biosystems), and the reaction mixtures were prepared as previously described (69).
TABLE 1.
Oligonucleotides used for real-time RT-PCR
| Gene | Forward primer (5′-3′) | Reverse primer (5′-3′) |
|---|---|---|
| escC | GCGTAAACTGGTCCGGTACGT | TGCGGGTAGAGCTTTAAAGGCAAT |
| escV | TCGCCCCGTCCATTGA | CGCTCCCGAGTGCAAAA |
| espA | TCAGAATCGCAGCCTGAAAA | CGAAGGATGAGGTGGTTAAGCT |
| eae | GCTGGCCCTTGGTTTGATCA | GCGGAGATGACTTCAGCACTT |
| espX3′ | AACCACGCAGTTCCCCATAA | GTTAGACAATTTAGAAAAACGATTGAGATG |
| espX3′ | GGGACAAATTTTAGCGGTTCTACA | CGTCCACTTTGTTGGTGTCTTAAT |
| espY5′ | CGTCGTACTAAAGCGCCATTT | ACTGAGGACAAAGTTAAGAGATTTGAGA |
| nleA | TGTTGAAGGCTGGAAGTTTGTTT | CCGCTACAGGGCGATATGTT |
| etrA | GCATTATTAGCATCCCAAAAGGA | AACGAACGAATGTCCAAGATCA |
| eivF | GGGAGTGTGGAAAGGGAACA | TGAATAGCACAACTTCTGATGCAA |
| Z3279 | ATGGCGCGGTTGGTGTA | CAACGAAAGTTTTACGCCATCA |
| Z4971 | CCTTAACCGCACTGGCGTTA | GGCTTTTTTCATCGTGGTGGTA |
| Z5223 | GCCCTTTTGAAATATTGACATTACC | GCCAAACGAGCGATTTTCC |
| stx2A | ACCCCACCGGGCAGTT | GGTCAAAACGCGCCTGATA |
| rpoA | GCGCTCATCTTCTTCCGAAT | CGCGGTCGTGGTTATGTG |
Real-time RT-PCR detection, quantification, and statistical analysis.
Data were collected using the ABI Sequence Detection 1.3 software (Applied Biosystems). All data were normalized to levels of rpoA and analyzed using the comparative critical threshold (CT) method (1). The expression levels of the target genes under the various culture conditions were compared using the relative quantification method (1). Real-time data are expressed below as the changes in expression levels compared to the WT levels. Statistical significance was determined by Student's t test, and a P value of ≤0.05 was considered significant.
RESULTS
In previous analyses of gene expression in WT EHEC and the luxS mutant, spotted amplicon-based arrays and hybridized EHEC cDNA were used with the E. coli K-12 array (58). We used the Affymetrix GeneChip technology as a starting point to examine the expression profiles of the entire E. coli genome with and without addition of QS signaling molecules.
Transcriptome comparison of WT strain 86-24 and luxS mutant VS94.
Inactivation of the luxS gene in E. coli 86-24 interrupts homocysteine synthesis from S-ribosylhomocysteine, preventing the production of AI-2 and diminishing the AI-3 QS signaling pathway in EHEC (58, 68). In the current study, a total of 710 genes were differentially expressed in the luxS mutant compared to the WT strain (Table 2), and the number of these genes that showed decreased expression (480 genes) was greater than the number that showed increased expression (230 genes).
TABLE 2.
Numbers of genes with altered expression as measured with the Affymetrix GeneChip array
| Strain used for comparison | Strain | No. of genes with:
|
||
|---|---|---|---|---|
| Increased expression | Decreased expression | No change in expression | ||
| 86-24 | VS94 | 230 | 480 | 9,497 |
| VS94 + DPD | 143 | 260 | 9,804 | |
| VS94 + epinephrine | 2,394 | 2,722 | 5,091 | |
| VS94 + AI-3 | 282 | 369 | 9,556 | |
| VS94 | VS94 + DPD | 261 | 142 | 9,804 |
| VS94 + epinephrine | 2,367 | 2,837 | 5,003 | |
| VS94 + AI-3 | 1,030 | 1,017 | 8,160 | |
The majority of the genes with an altered profile were derived from the E. coli MG1655 strain (∼39%). These genes represent a common E. coli backbone conserved among all E. coli pathovars, and many of the features encoded by the genes are associated with central metabolism and core biological processes. Not surprisingly, the proportions of altered features in EHEC isolates EDL933 (10.4%) and Sakai (8.3%) were higher than the proportion in E. coli CFT073 (3.7%), suggesting that the E. coli 86-24 strain is more similar to the other EHEC strains than to the uropathogenic isolate (40, 47, 72). The ratios of features with increased expression to features with decreased expression are similar in all of the pathovar subgroups (Table 3), with the exception of the intergenic regions, for which a significantly greater proportion of features showed decreased expression (7.7% with decreased expression versus 1% with increased expression). The reason for this altered profile is unclear, and this profile was not observed with any of the other stimuli. Perhaps the bias was a result of the probe selection process, since the intergenic regions are selected and the array represents an incomplete set. As a whole, these data suggest that the luxS mutation causes a metabolic deficiency that affects the central metabolism of most E. coli strains.
TABLE 3.
Pathovar-specific distribution of genesa
| Change in expression | No. (%) of genes in:
|
||||
|---|---|---|---|---|---|
| MG1655 (n = 4,070) | EDL933 (n = 1,787) | Sakai (n = 373) | CFT073 (n = 2,486) | Intergenic region (n = 1,297) | |
| 86-24 vs VS94 | |||||
| Decrease | 424 (10.42) | 266 (14.89) | 50 (13.40) | 124 (4.99) | 194 (14.96) |
| Marginal decrease | 15 (0.37) | 19 (1.06) | 4 (1.07) | 6 (0.24) | 16 (1.23) |
| Increase | 336 (8.26) | 189 (10.58) | 28 (7.51) | 54 (2.17) | 20 (1.54) |
| Marginal increase | 15 (0.37) | 8 (0.45) | 1 (0.27) | 3 (0.12) | 2 (0.15) |
| None | 3,280 (80.59) | 1,305 (73.03) | 290 (77.75) | 2,299 (92.48) | 1,065 (82.11) |
| Total | 4,070 | 1,787 | 373 | 2,486 | 1,297 |
| VS94 vs VS94 + DPD | |||||
| Decrease | 172 (4.23) | 82 (4.59) | 10 (2.68) | 75 (3.02) | 13 (1.00) |
| Marginal decrease | 11 (0.27) | 4 (0.22) | 2 (0.54) | 15 (0.60) | 1 (0.08) |
| Increase | 163 (4.00) | 121 (6.77) | 22 (5.90) | 46 (1.85) | 67 (5.17) |
| Marginal increase | 15 (0.37) | 15 (0.84) | 1 (0.27) | 6 (0.24) | 9 (0.69) |
| None | 3,709 (91.13) | 1,565 (87.58) | 338 (90.62) | 2,344 (94.29) | 1,207 (93.06) |
| Total | 4,070 | 1,787 | 373 | 2,486 | 1,297 |
| VS94 vs VS94 + epinephrine | |||||
| Decrease | 894 (21.97) | 257 (14.38) | 40 (10.72) | 94 (3.78) | 35 (2.70) |
| Marginal decrease | 17 (0.42) | 0 (0.00) | 2 (0.54) | 11 (0.44) | 2 (0.15) |
| Increase | 1,276 (31.35) | 1,057 (59.15) | 186 (49.87) | 454 (18.26) | 697 (53.74) |
| Marginal increase | 41 (1.01) | 21 (1.18) | 2 (0.54) | 14 (0.56) | 13 (1.00) |
| None | 1,842 (45.26) | 452 (25.29) | 143 (38.34) | 1,913 (76.95) | 550 (42.41) |
| Total | 4,070 | 1,787 | 373 | 2,486 | 1,297 |
| VS94 vs VS94 + AI-3 | |||||
| Decrease | 508 (12.48) | 212 (11.86) | 48 (12.87) | 68 (2.74) | 69 (5.32) |
| Marginal decrease | 17 (0.42) | 6 (0.34) | 0 (0.00) | 8 (0.32) | 4 (0.31) |
| Increase | 519 (12.75) | 228 (12.76) | 42 (11.26) | 107 (4.30) | 59 (4.55) |
| Marginal increase | 28 (0.69) | 25 (1.40) | 2 (0.54) | 4 (0.16) | 4 (0.31) |
| None | 2,998 (73.66) | 1,316 (73.64) | 281 (75.34) | 2,299 (92.48) | 1,161 (89.51) |
| Total | 4,070 | 1,787 | 373 | 2,486 | 1,297 |
The total number of genes assigned to the specific genomes included is 10,013. There are an additional 96 features that are used as controls and 99 features that are associated with phage and plasmids and thus not directly linked to a genome project. The total number of features on the array is 10,208.
Previously, Sperandio et al. (58) identified ∼400 MG1655 genes that were altered in the luxS mutant compared to the WT, representing ∼10% of the genes on the array using a conservative fivefold threshold for altered expression. If the analysis in this study was limited to the E. coli K-12 genes, a total of 280 genes had an altered profile. This is significantly less than the number of genes with an altered expression profile in the previous study (736 genes), in which a twofold threshold was utilized. Although the numbers of genes with an altered profile in the two studies are different, the array designs (amplicon versus 25-mer oligonucleotides) and analysis thresholds (absolute fold change versus normalization and algorithmic analysis) are also different.
Transcriptome modification with DPD.
The changes in gene expression caused by the luxS mutation in E. coli VS94 that are due to AI-2 signaling should be functionally complemented by addition of DPD to the growth medium (68). Indeed, the fewest differences in gene expression between 86-24 and VS94 occurred when DPD was added to the culture medium (Table 2). These data indicate that of the signaling molecules, AI-2 best complements the luxS mutation under the conditions examined. However, differences in gene expression between the WT strain and VS94 grown with DPD were evident; thus, addition of DPD to the growth medium does not completely compensate for the luxS mutation.
Comparisons between VS94 with DPD and VS94 resulted in the fewest differences in the transcriptional profile, with 403 altered genes (Table 2). Interestingly, when we compared the genes with altered expression profiles after the addition of DPD (i.e., 86-24 versus VS94 with DPD and VS94 versus VS94 with DPD), we observed 951 genes that were differentially regulated under these different conditions and only 18 genes that were similarly regulated by the addition of DPD. The genes that were regulated similarly in these conditions represent the minimal DPD-responsive set of genes. Further examination of the distribution of the pathovar-expressed genes after addition of DPD to a VS94 culture did not reveal any significant alterations in gene expression profiles.
Transcriptome modification with epinephrine.
The greatest transcriptome alteration was observed when epinephrine was added to the growth medium (expression of 5,204 genes was altered when VS94 was compared to VS94 with epinephrine [Table 2]). The activated genes included the LEE genes, stx2, the flagellar regulon genes (including flhDC), the genes encoding iron uptake systems, the gene encoding the Hfq protein (a chaperone involved in small regulatory RNA posttranscriptional regulation), and genes encoding several nucleoid proteins (H-NS, HU, FIS, and Hha, all reported to be involved in regulation of the LEE). Although initially overwhelming, the fact that epinephrine induces activation of several nucleoid proteins, mostly proteins involved in global repression of gene transcription, is consistent with this observation. The observed alterations in a large number of genes suggests that for assembly of energetically expensive structures such as the LEE-encoded TTSS and flagella (up-regulated by epinephrine), there is down-regulation of homeostatic genes.
Interestingly, a greater proportion of the EHEC-specific genes appeared to have an expression profile that was pathovar specific (Table 3). The expression of nearly 50 and 56% of the EHEC-specific genes from E. coli EDL933 and E. coli Sakai, respectively, was altered when epinephrine was added to the medium. Additionally, the CFT073 genes exhibited an altered expression profile with epinephrine treatment. These data contrast with the increased expression of ∼39% of the E. coli MG1655 genes and suggest that epinephrine preferentially activates virulence genes. This is consistent with previous studies that showed that E. coli senses and responds to this important hormone signal (11, 59, 68).
Additionally, in the intergenic regions there was a significant proportion of increased transcription; approximately 56% of the intergenic regions demonstrated increased expression. Most likely, there is increased regulation of upstream regions of the activated genes, as well as other features on the array, such as small RNAs (http://www.affymetrix.com/products/arrays/specific/ecoli2.affx). While there were a significant number of genes and features whose expression was increased with the epinephrine treatment, the proportion of genes whose expression was decreased in the E. coli MG1655 data set was also the greatest proportion observed in any culture condition (Table 3).
Transcriptome modification with AI-3.
Although the VS94 strain may produce AI-3, the concentration of synthesized AI-3 has been shown to be diminished compared to the concentration produced by the WT (68). When the expression profile was examined for the pathovar-specific distribution, a significant bias in terms of altered gene expression profiles was not apparent.
Effects on expression of LEE and Shiga toxin genes.
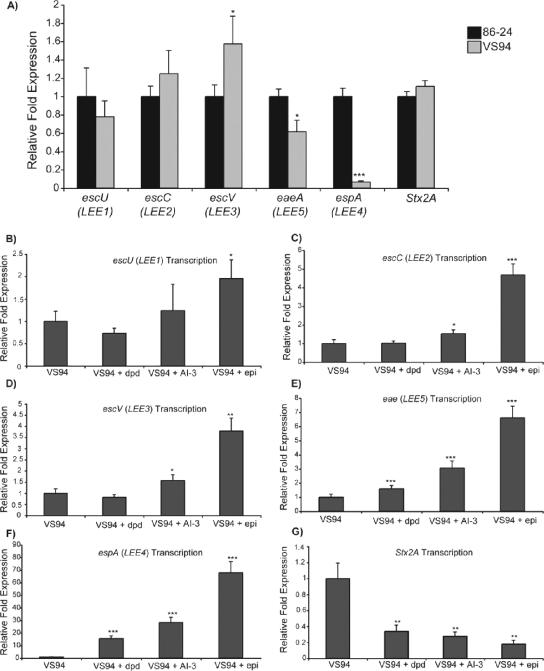
Real-time RT-PCR analyses provided interesting insights into how QS molecules contribute to regulation and expression of LEE genes (Fig. 1A to F). When VS94 was compared to WT EHEC, no significant differences in expression of LEE1 or LEE2 were apparent (Fig. 1A). However, expression of LEE3 was significantly increased, whereas expression of LEE4 and LEE5 was significantly decreased (Fig. 1A). Addition of DPD had variable effects on expression of LEE genes. When DPD was added to the medium, the expression of the LEE1 to LEE3 genes was similar to that in VS94 grown without any signaling molecules (Fig. 1B to D). The level of expression of LEE4 and LEE5 was significantly higher in the cultures grown with DPD; however, DPD did not enhance expression to the extent that was seen when either AI-3 or epinephrine was added to the growth medium.
FIG. 1.
Transcriptional profiles of LEE and stx2A gene expression for WT EHEC and an isogenic luxS mutant (A) and for the luxS mutant grown with AI-2 (dpd), AI-3, or epinephrine (epi) (B to G), as measured by real-time RT-PCR and expressed as fold differences normalized to WT strain 86-24 (A) and the luxS mutant (B). The error bars indicate the standard deviations of the ΔΔCT values (1). Significance is indicated as follows: one asterisk, P ≤ 0.05; two asterisks, P ≤ 0.005; and three asterisks, P ≤ 0.0005.
Addition of either exogenous AI-3 or epinephrine increased expression of all of the LEE genes (Fig. 1B to F). AI-3 significantly increased expression of the LEE2 to LEE5 genes at the late-exponential phase of growth and also increased expression of LEE1 compared to the expression in VS94; however, the increased expression of LEE1 was not considered significant. A previous study showed that the most significant regulation of the LEE genes in the WT compared to a luxS mutant occurred at mid-exponential growth (69). This was most likely because a luxS mutant can still produce AI-3, albeit at a lower level than the WT, but by the time the strain reaches late-logarithmic phase, enough AI-3 has been produced to stimulate expression of the LEE (69). The current study showed that exogenous AI-3 still contributes to regulation of the LEE, even at the late-exponential growth phase, and further underscores the importance of AI-3 in EHEC pathogenesis.
Addition of epinephrine had the greatest effect on LEE gene expression. Expression of all five of the LEE operons was significantly increased when epinephrine was added to the culture medium. These data are congruous with the array data and reinforce epinephrine's role in EHEC pathogenesis.
The luxS mutation had no effect on expression of the stx2A gene (Fig. 1A); however, addition of any signaling molecule greatly decreased expression of Stx2A in the luxS mutant cultures (Fig. 1G). Although the expression was decreased, a similar trend was observed when expression of this gene was analyzed; expression of Stx2A was most similar in VS94 and VS94 with DPD, and the greatest differences in expression occurred between VS94 and VS94 with epinephrine.
Non-LEE-encoded effectors.
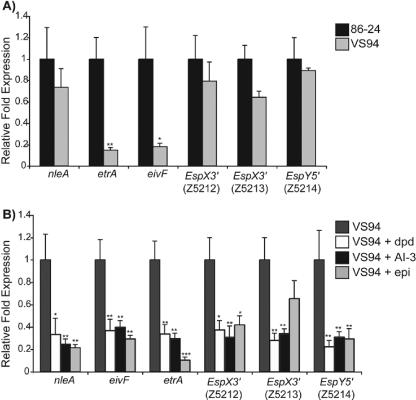
EHEC encodes many non-LEE effector proteins that are thought to be secreted (63) and that result in enhancement of virulence in the host. Several of these genes were selected for detailed analysis by real-time RT-PCR. NleA is encoded outside of the LEE, but it is secreted through the LEE-encoded TTSS. Once NleA enters the host, it localizes to the Golgi apparatus (25). Although its precise function is not understood, NleA appears to play a role in virulence in mouse model experiments as the virulence of an nleA mutant strain is attenuated (25, 45). Both the microarray and real-time RT-PCR data indicated that expression of nleA was not affected in the luxS mutant compared to the WT strain. However, addition of AI-2, AI-3, or epinephrine to luxS mutant cultures significantly decreased nleA expression (Fig. 2A and B). Again, the most significant differences were observed in cultures in which AI-3 or epinephrine was added to the medium; these differences were more significant than those observed when DPD was added.
FIG. 2.
Transcriptional profiles of nleA, etrA, eivF, and the secreted effectors EspX3′ (Z5212 and Z5213) and EspY5′ for WT EHEC and an isogenic luxS mutant (A) and for the luxS mutant grown with AI-2 (dpd), AI-3, or epinephrine (epi) (B), as measured by real-time RT-PCR and expressed as fold differences normalized to WT strain 86-24 (A) or the luxS mutant (B). The error bars indicate the standard deviations of the ΔΔCT values (1). Significance is indicated as follows: one asterisk, P ≤ 0.05; two asterisks, P ≤ 0.005; and three asterisks, P ≤ 0.0005.
Recently, several novel effectors proteins were identified in EHEC (63), and we performed a real-time RT-PCR analysis for the effectors EspX3′ (Z5212 and Z5213) and EspY5′ (Z5214). Similar to nleA, the luxS mutation did not alter expression of these genes (Fig. 2B), but addition of any of the signaling molecules (AI-2, AI-3, or epinephrine) reduced expression (Fig. 2B). Expression of Z5212 and Z5214 was significantly decreased in the cultures to which exogenous signaling molecules were added, and there were no significant differences that were dependent on the type of QS molecule added. Expression of Z5213 was significantly decreased when either DPD or AI-3 was added to the medium. Expression of this gene was decreased when epinephrine was added compared to the expression in VS94 grown in DMEM, but the difference was not considered significant. Addition of external signaling molecules had a decreased effect on the secreted effector genes.
ETT2-encoded regulators.
In addition to the LEE-encoded TTSS, EHEC contains a nonfunctional type III secretion system (ETT2) (27, 47). When expression of the genes was compared for WT and VS94, the expression was significantly decreased in the luxS mutant (Fig. 2A). Then, when we compared VS94 to VS94 grown with signaling molecules, addition of the signaling molecules further decreased expression of these genes (Fig. 2B). The transcriptional regulators encoded by eivF and etrA in the ETT2 have been shown to be negative regulators of the LEE (76). These data suggest that AI-3 and epinephrine may function not only by increasing expression of the LEE directly but also by inhibiting factors such as eivF and etrA that repress LEE (i.e., by repressing the repressors).
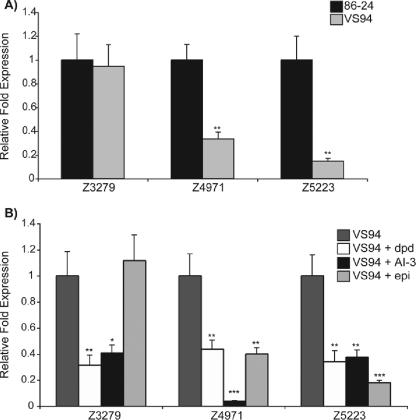
Fimbrial genes.
Attachment is the first step in colonization, and thus we wanted to examine the fimbrial genes on the array to determine if they were alternatively regulated in response to the quorum signals. Overall, epinephrine seemed to have the greatest effect of expression of known (fimZ, fimH, fimG, fimF, fimD, and fimC) and putative fimbrial genes. We performed real-time RT-PCR for three putative fimbrial genes. The putative major fimbrial subunit (Z4971) and a putative fimbrial chaperone (Z5223) were significantly down-regulated in the VS94 culture grown without signaling molecules compared to the expression in the WT (Fig. 3A). Addition of signaling molecules to cultures of VS94 caused further repression of these fimbrial genes. In contrast, there were no significant differences between the WT and the luxS mutant in the expression of the Z3279 gene encoding a putative fimbria-like protein (Fig. 3A). Addition of DPD or AI-3 caused further repression of this gene; however, expression was rescued to near-WT levels when epinephrine was added to the medium (Fig. 3B).
FIG. 3.
Transcriptional profiles of fimbrial gene expression for WT EHEC and an isogenic luxS mutant (A) and for the luxS mutant grown with AI-2 (dpd), AI-3, or epinephrine (epi) (B), as measured by real-time RT-PCR and expressed as fold differences normalized to WT strain 86-24 (A) or the luxS mutant (B). The error bars indicate the standard deviations of the ΔΔCT values (1). Significance is indicated as follows: one asterisk, P ≤ 0.05; two asterisks, P ≤ 0.005; and three asterisks, P ≤ 0.0005.
DISCUSSION
The data presented in this study provide a more complete picture of the transcriptional modifications that occur in E. coli due to bacterial signaling via AI-2 and AI-3 and interkingdom signaling with the host hormone epinephrine. A previous transcriptome analysis of E. coli 86-24 and the luxS mutant VS94 revealed that there was more-than-fivefold alteration of expression of ∼400 genes (235 up-regulated genes and 169 down-regulated genes) and alteration of expression of 736 genes when a less stringent twofold threshold was used (58). We observed 280 MG1655 genes with an altered profile in the luxS mutant in our arrays. It must be noted that the previous array utilized single E. coli K-12 (MG1655) amplicons for each feature, whereas the current Affymetrix GeneChip E. coli Genome 2.0 array contains the complete nonredundant gene complement of the laboratory-adapted isolate E. coli MG1655, two EHEC isolates, E. coli EDL933 and E. coli Sakai, and the uropathogenic E. coli isolate CFT073. Additionally, the Affymetrix GeneChip E. coli Genome 2.0 array contains 1,297 intergenic features, which the previous amplicon-based array did not. It was interesting that while the strain utilized in this study, E. coli 86-24, has not been sequenced and thus could not be completely contained on the array, we did find a significant number of E. coli CFT073 genes with an altered transcriptional profile. This suggests that E. coli 86-24 contains some regions that are shared with E. coli CFT073 and thus not with other sequenced EHEC strains. This is a further example of the mosaic nature of the E. coli genomes (72).
The function of AI-2 in bacterial signaling is an issue that is debated. Some studies have suggested that AI-2 is involved in biofilm formation and motility (15, 23, 28); however, it has also been suggested that AI-2 signaling is involved primarily in the regulation of metabolic processes (67, 71, 73, 74). In Salmonella, AI-2 regulation involves only genes that encode an ABC transporter termed Lsr (LuxS regulated) (62). This transporter has also been found in E. coli (75). In Salmonella and E. coli, the lsr genes share a high level of sequence homology, and functionally the proteins resemble sugar transporters. Similar to what occurs with other sugar transporters, import of AI-2 is strictly controlled (70, 75). AI-2 is synthesized and secreted during exponential growth and is imported in stationary phase when glucose becomes limiting (70, 75). In the presence of glucose, AI-2 is not imported because the lsr operon is not transcribed due to cyclic AMP-catabolite activator protein-mediated repression (70, 75). Indeed, gene expression profiles comparing E. coli MG1655 cultures grown in glucose-containing and glucose-free media showed that the lsr operon was induced only in the absence of glucose and that the luxS mutation in E. coli MG1655 affected mainly genes related to AI-2 production and transport (71). Moreover, a study using phenotype microarrays showed that the luxS mutation resulted in numerous metabolic changes, especially in the processes that involve nitrogen and carbon metabolism (68). Our data are congruous with studies that suggest a metabolic role for AI-2.
The AI-3 signaling molecule has been shown to activate the LEE and flagellar and motility genes in EHEC (11, 12, 59, 69). Addition of exogenous AI-3 significantly increased expression of the LEE in a luxS mutant; however, significant changes in global gene expression were not apparent under these conditions. This work demonstrates that even though the WT and the luxS mutant may make AI-3, there is not saturation of the receptor or the response mechanism as addition of exogenous AI-3 resulted in exacerbation of the virulent phenotype. More work is required to determine the level of AI-3 required for saturation of the EHEC system.
The greatest changes in gene expression occurred when epinephrine was added to the medium. The stress hormones epinephrine and norepinephrine are present in the gastrointestinal tract and modulate smooth muscle contraction, submucosal blood flow, and chloride and potassium secretion there (29). Norepinephrine is produced within adrenergic neurons present in the enteric nervous system (20), whereas epinephrine is synthesized in both the central nervous system and the adrenal medulla and is involved in systemic responses (48). The levels of norepinephrine and epinephrine in the intestine are in the micromolar range (17), similar to the levels that were used as signals in the present study. Moreover, during an EHEC infection, the integrity of the epithelial cell layer is compromised, causing bloody diarrhea and stressing the host; therefore, the concentrations of epinephrine and norepinephrine may be even higher.
Previous work has shown that AI-3 and epinephrine act synergistically (59, 69). Upon entry into the colonic lumen, EHEC responds to bacterial (commensal as well as pathogen)-produced AIs and activates motility. Then in close proximity to the host epithelium, the host hormones sustain and further alter the expression profile, allowing attachment though expression of the LEE genes, resulting in clinical disease presentation. This fine-tuning may allow the bacteria to respond favorably to various environmental situations.
The effect of the luxS mutation with or without added signals on the LEE operons was examined in detail. Comparisons of the luxS mutant and WT revealed significant differences only in the expression of LEE4 and LEE5, suggesting that additional mechanisms and/or signals are involved in the regulation of these operons. Addition of DPD had no effect on LEE1 to LEE3 but increased expression of LEE5 and LEE4, suggesting that the latter operons are controlled through AI-2 signaling in addition to the AI-3/epinephrine system. Although a luxS mutant still synthesizes a low level of AI-3, addition of exogenous AI-3 to the culture medium significantly increases expression of all the LEE genes at late-exponential growth phase (Fig. 1). Of the stimuli examined, epinephrine affected expression of the LEE genes to the greatest degree. Additionally, the identification of multiple stimuli that can activate the LEE regions suggests that there is a complex regulatory network for the virulence genes of EHEC.
Additional genes involved in EHEC virulence were also affected by addition of signaling molecules. Consistent with our observation that regulation of the stx2A/B genes occurs through several qse genes (unpublished data), signaling molecules directly affected expression of the Shiga toxin-producing genes. Other known regulatory proteins are produced in response to QS stimuli. For example, the level of QseA increased when epinephrine was added to the medium (59). QseA increases expression of the LEE (55) as well as QseE, a response regulator that controls transcription of the EspFu/TccP effector (8, 22, 49). Taken together, these data suggest that AI-3 and epinephrine are important regulators of the TTSS and virulence in EHEC.
The repertoire of EHEC's virulence factors extends beyond the LEE-encoded effectors, and similar to the LEE, these virulence factors are probably subjected to multiple complex levels of regulation. Interestingly, addition of the signaling molecules repressed expression of many non-LEE-encoded effectors. The luxS mutation did not have significant effects on gene expression of non-LEE effectors; however, addition of the signaling molecules significantly decreased expression.
While fimbriae are important virulence factors for enteropathogenic E. coli (7, 24, 43, 55) and for uropathogenic E. coli (2, 44), their role in EHEC adherence is not fully understood. In EHEC, fimbriae putatively increase adherence to epithelial cells (65) or aid in the formation of stable microcolonies (64). Other studies have proposed that fimbriae may be more important in effective colonization of the bovine gastrointestinal tract (38, 39). Mutation of ler in EHEC was associated with enhanced fimbrial expression (18). Our data show that regulation of known and putative fimbrial genes is different under different conditions.
The results reported in this study demonstrate that addition of exogenous AI-2 (in the form of DPD) cannot fully restore the metabolic defect caused by the luxS mutation. Epinephrine causes an EHEC-specific alteration of many genes, including those related to the pathogenesis of EHEC in humans (LEE genes and genes encoding toxins and fimbriae). Additional virulence traits seem to be activated by exogenous AI-3 even in the presence of endogenous AI-3, indicating that the system is not fully saturated by the production of endogenous AI-3. The gene sets altered by epinephrine and AI-3 do not entirely overlap, suggesting that regulation of the virulence traits in EHEC infection may follow a program based on the signal and level of signal sensed by the bacterium and may have a temporal component.
This study accurately mapped the transcriptome of EHEC in the presence of major QS signals and provided novel insight into the QS control of virulence in the presence of these stimuli.
Acknowledgments
We thank J. R. Falck from the University of Texas Southwestern Medical Center at Dallas for very generously providing the AI-3 used in this work.
This work was supported by the Burroughs Wellcome Fund, the Ellison Medical Foundation, and NIH grant AI053067.
Editor: A. Camilli
Footnotes
Published ahead of print on 16 July 2007.
REFERENCES
- 1.Anonymous. 1997. Applied Biosystems Prism 7700 sequence detection system: user bulletin no. 2. Perkin-Elmer Corp., Norwalk, CT.
- 2.Bahrani-Mougeot, F. K., E. L. Buckles, C. V. Lockatell, J. R. Hebel, D. E. Johnson, C. M. Tang, and M. S. Donnenberg. 2002. Type 1 fimbriae and extracellular polysaccharides are preeminent uropathogenic Escherichia coli virulence determinants in the murine urinary tract Mol. Microbiol. 45:1079-1093. [DOI] [PubMed] [Google Scholar]
- 3.Barba, J., V. H. Bustamante, M. A. Flores-Valdez, W. Deng, B. B. Finlay, and J. L. Puente. 2005. A positive regulatory loop controls expression of the locus of enterocyte effacement-encoded regulators Ler and GrlA. J. Bacteriol. 187:7918-7930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bassler, B. L. 1999. How bacteria talk to each other: regulation of gene expression by quorum sensing. Curr. Opin. Microbiol. 2:582-587. [DOI] [PubMed] [Google Scholar]
- 5.Bassler, B. L., E. P. Greenberg, and A. M. Stevens. 1997. Cross-species induction of luminescence in the quorum-sensing bacterium Vibrio harveyi. J. Bacteriol. 179:4043-4045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bolstad, B. M., R. A. Irizarry, M. Astrand, and T. P. Speed. 2003. A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics 19:185-193. [DOI] [PubMed] [Google Scholar]
- 7.Bustamante, V. H., F. J. Santana, E. Calva, and J. L. Puente. 2001. Transcriptional regulation of type III secretion genes in enteropathogenic Escherichia coli: Ler antagonizes H-NS-dependent repression. Mol. Microbiol. 39:664-678. [DOI] [PubMed] [Google Scholar]
- 8.Campellone, K. G., D. Robbins, and J. M. Leong. 2004. EspFU is a translocated EHEC effector that interacts with Tir and N-WASP and promotes Nck-independent actin assembly. Dev. Cell 7:217-228. [DOI] [PubMed] [Google Scholar]
- 9.Chen, X., S. Schauder, N. Potier, A. VanDorssealaer, I. Pelczer, B. L. Bassler, and F. M. Hughson. 2002. Structural identification of a bacterial quorum-sensing signal containing boron. Nature 415:545-549. [DOI] [PubMed] [Google Scholar]
- 10.Clarke, M., and V. Sperandio. 2005. Transcriptional regulation of flhDC by QseBC and sigma 28 (FliA) in enterohaemorrhagic Escherichia coli. Mol. Microbiol. 57:1734-1749. [DOI] [PubMed] [Google Scholar]
- 11.Clarke, M. B., D. T. Hughes, C. Zhu, E. C. Boedeker, and V. Sperandio. 2006. The QseC sensor kinase: a bacterial adrenergic receptor. Proc. Natl. Acad. Sci. USA 103:10420-10425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Clarke, M. B., and V. Sperandio. 2005. Transcriptional autoregulation by quorum sensing Escherichia coli regulators B and C (QseBC) in enterohaemorrhagic E. coli (EHEC). Mol. Microbiol. 58:441-455. [DOI] [PubMed] [Google Scholar]
- 13.Dean-Nystrom, E. A., B. T. Bosworth, H. W. Moon, and A. D. O'Brien. 1998. Escherichia coli O157:H7 requires intimin for enteropathogenicity in calves. Infect. Immun. 66:4560-4563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Deng, W., J. L. Puente, S. Gruenheid, Y. Li, B. A. Vallance, A. Vazquez, J. Barba, J. A. Ibarra, P. O'Donnell, P. Metalnikov, K. Ashman, S. Lee, D. Goode, T. Pawson, and B. B. Finlay. 2004. Dissecting virulence: systematic and functional analyses of a pathogenicity island. Proc. Natl. Acad. Sci. USA 101:3597-3602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Domka, J., J. Lee, and T. K. Wood. 2006. YliH (BssR) and YceP (BssS) regulate Escherichia coli K-12 biofilm formation by influencing cell signaling. Appl. Environ. Microbiol. 72:2449-2459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Donnenberg, M. S., C. O. Tacket, S. P. James, G. Losonsky, J. P. Nataro, S. S. Wasserman, J. B. Kaper, and M. M. Levine. 1993. Role of the eaeA gene in experimental enteropathogenic Escherichia coli infection. J. Clin. Investig. 92:1412-1417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Eldrup, E., and E. A. Richter. 2000. DOPA, dopamine, and DOPAC concentrations in the rat gastrointestinal tract decrease during fasting. Am. J. Physiol. Endocrinol. Metab. 279:E815-E822. [DOI] [PubMed] [Google Scholar]
- 18.Elliott, S. J., V. Sperandio, J. A. Girón, S. Shin, J. L. Mellies, L. Wainwright, S. W. Hutcheson, T. K. McDaniel, and J. B. Kaper. 2000. The locus of enterocyte effacement (LEE)-encoded regulator controls expression of both LEE- and non-LEE-encoded virulence factors in enteropathogenic and enterohemorrhagic Escherichia coli. Infect. Immun. 68:6115-6126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Friedberg, D., T. Umanski, Y. Fang, and I. Rosenshine. 1999. Hierarchy in the expression of the locus of enterocyte effacement genes of enteropathogenic Escherichia coli. Mol. Microbiol. 34:941-952. [DOI] [PubMed] [Google Scholar]
- 20.Furness, J. B. 2000. Types of neurons in the enteric nervous system. J. Auton. Nerv. Syst. 81:87-96. [DOI] [PubMed] [Google Scholar]
- 21.Garmendia, J., G. Frankel, and V. F. Crepin. 2005. Enteropathogenic and enterohemorrhagic Escherichia coli infections: translocation, translocation, translocation. Infect. Immun. 73:2573-2585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Garmendia, J., A. D. Phillips, M. F. Carlier, Y. Chong, S. Schüller, O. Marches, S. Dahan, E. Oswald, R. K. Shaw, S. Knutton, and G. Frankel. 2004. TccP is an enterohaemorrhagic Escherichia coli O157:H7 type III effector protein that couples Tir to the actin-cytoskeleton. Cell. Microbiol. 6:1167-1183. [DOI] [PubMed] [Google Scholar]
- 23.Gonzalez-Barrios, A. F., R. Zuo, Y. Hashimoto, L. Yang, W. E. Bentley, and T. K. Wood. 2006. Autoinducer 2 controls biofilm formation in Escherichia coli through a novel motility quorum-sensing regulator (MqsR, B3022). J. Bacteriol. 188:305-316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Griffin, P. M., S. M. Ostroff, R. V. Tauxe, K. D. Greene, J. G. Wells, J. H. Lewis, and P. A. Blake. 1988. Illnesses associated with Escherichia coli O157:H7. Ann. Intern. Med. 109:705-712. [DOI] [PubMed] [Google Scholar]
- 25.Gruenheid, S., I. Sekirov, N. A. Thomas, W. Deng, P. O'Donnell, D. Goode, Y. Li, E. A. Frey, N. F. Brown, P. Metalnikov, T. Pawson, K. Ashman, and B. B. Finlay. 2004. Identification and characterization of NleA, a non-LEE-encoded type III translocated virulence factor of enterohaemorrhagic Escherichia coli O157:H7. Mol. Microbiol. 51:1233-1249. [DOI] [PubMed] [Google Scholar]
- 26.Haack, K. R., C. L. Robinson, K. J. Miller, J. W. Fowlkes, and J. L. Mellies. 2003. Interaction of Ler at the LEE5 (tir) operon of enteropathogenic Escherichia coli. Infect. Immun. 71:384-392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hayashi, T., K. Makino, M. Ohnishi, K. Kurokawa, K. Ishii, K. Yokoyama, C.-G. Han, E. Ohtsubo, K. Nakayama, T. Murata, M. Tanaka, T. Tobe, T. Iida, H. Takami, T. Honda, C. Sasakawa, N. Ogasawara, T. Yasunaga, S. Kuhara, T. Shiba, M. Hattori, and H. Shinagawa. 2001. Complete genome sequence of enterohemorrhagic Eschelichia coli O157:H7 and genomic comparison with a laboratory strain K-12. DNA Res. 8:11-22. [DOI] [PubMed] [Google Scholar]
- 28.Herzberg, M., I. K. Kaye, W. Petri, and T. K. Wood. 2006. YdgG (TqsA) controls biofilm formation in Escherichia coli K-12 through autoinducer 2 transport. J. Bacteriol. 188:587-598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Horger, S., G. Schultheiss, and M. Diener. 1998. Segment-specific effects of epinephrine on ion transport in the colon of the rat. Am. J. Physiol. Gastrointest. Liver Physiol. 275:G1367-G1376. [DOI] [PubMed] [Google Scholar]
- 30.Irizarry, R. A., B. Hobbs, F. Collin, Y. D. Beazer-Barclay, K. J. Antonellis, U. Scherf, and T. P. Speed. 2003. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics 4:249-264. [DOI] [PubMed] [Google Scholar]
- 31.Iyoda, S., and H. Watanabe. 2005. ClpXP protease controls expression of the type III protein secretion system through regulation of RpoS and GrlR levels in enterohemorrhagic Escherichia coli. J. Bacteriol. 187:4086-4094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Iyoda, S., and H. Watanabe. 2004. Positive effects of multiple pch genes on expression of the locus of enterocyte effacement genes and adherence of enterohaemorrhagic Escherichia coli O157:H7 to HEp-2 cells. Microbiology 150:2357-2571. [DOI] [PubMed] [Google Scholar]
- 33.Jarvis, K. G., J. A. Girón, A. E. Jerse, T. K. McDaniel, M. S. Donnenberg, and J. B. Kaper. 1995. Enteropathogenic Escherichia coli contains a putative type III secretion system necessary for the export of proteins involved in attaching and effacing lesion formation. Proc. Natl. Acad. Sci. USA 92:7996-8000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jerse, A. E., J. Yu, B. D. Tall, and J. B. Kaper. 1990. A genetic locus of enteropathogenic Escherichia coli necessary for the production of attaching and effacing lesions on tissue culture cells. Proc. Natl. Acad. Sci. USA 87:7839-7843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Reference deleted.
- 36.Kaper, J. B., J. P. Nataro, and H. L. T. Mobley.2004. Pathogenic Escherichia coli. Nat. Rev. Microbiol. 2:123-140. [DOI] [PubMed] [Google Scholar]
- 37.Kenny, B., R. DeVinney, M. Stein, D. J. Reinscheid, E. A. Frey, and B. B. Finlay. 1997. Enteropathogenic E. coli (EPEC) transfers its receptor for intimate adherence into mammalian cells. Cell 91:511-520. [DOI] [PubMed] [Google Scholar]
- 38.Low, A. S., F. Dziva, A. G. Torres, J. L. Martinez, T. Rosser, S. Naylor, K. J. Spears, N. Holden, A. Mahajan, J. Findlay, J. Sales, D. G. E. Smith, J. C. Low, M. P. Stevens, and D. L. Gally. 2006. Cloning, expression, and characterization of fimbrial operon F9 from enterohemorrhagic Escherichia coli O157:H7. Infect. Immun. 74:2233-2244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Low, A. S., N. Holden, T. Rosser, A. J. Roe, C. Constantinidou, J. L. Hobman, D. G. E. Smith, J. C. Low, and D. L. Gally. 2006. Analysis of fimbrial gene clusters and their expression in enterohaemorrhagic Escherichia coli o157:H7. Environ. Microbiol. 8:1033-1047. [DOI] [PubMed] [Google Scholar]
- 40.Makino, K., K. Yokoyama, Y. Kubota, C. H. Yutsudo, S. Kimura, K. Kurokawa, K. Ishii, M. Hattori, I. Tatsuno, H. Abe, T. Iida, K. Yamamoto, M. Onishi, T. Hayashi, T. Yasunaga, T. Honda, C. Sasakawa, and H. Shinagawa. 1999. Complete nucleotide sequence of the prophage VT2-Sakai carrying the verotoxin 2 genes of the enterohemorrhagic Escherichia coli O157:H7 derived from the Sakai outbreak. Genes Genet. Syst. 74:227-239. [DOI] [PubMed] [Google Scholar]
- 41.McDaniel, T. K., K. G. Jarvis, M. S. Donnenberg, and J. B. Kaper. 1995. A genetic locus of enterocyte effacement conserved among diverse enterobacterial pathogens. Proc. Natl. Acad. Sci. USA 92:1664-1668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.McKee, M. L., A. R. Melton-Celsa, R. A. Moxley, D. H. Francis, and A. D. O'Brien. 1995. Enterohemorrhagic Escherichia coli O157:H7 requires intimin to colonize the gnotobiotic pig intestine and to adhere to HEp-2 cells. Infect. Immun. 63:3739-3744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mellies, J. L., S. J. Elliott, V. Sperandio, M. S. Donnenberg, and J. B. Kaper. 1999. The Per regulon of enteropathogenic Escherichia coli: identification of a regulatory cascade and a novel transcriptional activator, the locus of enterocyte effacement (LEE)-encoded regulator (Ler) Mol. Microbiol. 33:296-306. [DOI] [PubMed] [Google Scholar]
- 44.Mulvey, M. A., Y. S. Lopez-Boado, C. L. Wilson, R. Roth, W. C. Parks, J. Heuser, and S. J. Hultgren. 1998. Induction and evasion of host defenses by type 1-piliated uropathogenic Escherichia coli. Science 282:1494-1497. [DOI] [PubMed] [Google Scholar]
- 45.Mundy, R., L. Petrovska, K. Smollett, N. Simpson, R. K. Wilson, J. Yu, X. Tu, I. Rosenshine, S. Clare, G. Dougan, and G. Frankel. 2004. Identification of a novel Citrobacter rodentium type III secreted protein, EspI, and roles of this and other secreted proteins in infection. Infect. Immun. 72:2288-2302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nakanishi, N., H. Abe, Y. Ogura, T. Hayashi, K. Tashiro, S. Kuhara, N. Sugimoto, and T. Tobe. 2006. ppGpp with DksA controls gene expression in the locus of enterocyte effacement (LEE) pathogenicity island of enterohaemorrhagic Escherichia coli through activation of two virulence regulatory genes. Mol. Microbiol. 61:194-205. [DOI] [PubMed] [Google Scholar]
- 47.Perna, N. T., G. Plunkett, V. Burland, B. Mau, J. D. Glasner, D. J. Rose, G. F. Mayhew, P. S. Evans, J. Gregor, H. A. Kirkpatrick, G. Posfai, J. Hackett, S. Klink, A. Boutin, Y. Shao, L. Miller, E. J. Grotbeck, N. W. Davis, A. Lim, E. T. Dimalanta, K. D. Potamousis, J. Apodaca, T. S. Anantharaman, J. Lin, G. Yen, D. C. Schwartz, R. A. Welch, and F. R. Blattner. 2001. Genome sequence of enterohaemorrhagic Escherichia coli O157:H7. Nature 409:529-533. [DOI] [PubMed] [Google Scholar]
- 48.Purves, D., D. Fitzpatrick, S. M. Williams, J. O. McNamara, G. J. Augustine, L. C. Katz, and A. S. LaMantia (ed.). 2001. Neuroscience, 2nd ed. Sinauer Associates, Sunderland, MA.
- 49.Reading, N. C., A. G. Torres, M. M. Kendall, D. T. Hughes, K. Yamamoto, and V. Sperandio. 2007. A novel two-component signaling system that activates transcription of an enterohemorrhagic Escherichia coli effector involved in remodeling of host actin. J. Bacteriol. 189:2468-2476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sánchez-SanMartín, C., V. H. Bustamante, E. Calva, and J. L. Puente. 2001. Transcriptional regulation of the orf19 gene and the tir-cesT-eae operon of enteropathogenic Escherichia coli. J. Bacteriol. 183:2823-2833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Schauder, S., K. Shokat, M. G. Surette, and B. L. Bassler. 2001. The LuxS family of bacterial autoinducers: biosynthesis of a novel quorum-sensing signal molecule. Mol. Microbiol. 41:463-476. [DOI] [PubMed] [Google Scholar]
- 52.Sharma, V. K., and R. L. Zuerner. 2004. Role of hha and ler in transcriptional regulation of the esp operon of enterohemorrhagic Escherichia coli O157:H7. J. Bacteriol. 186:7290-7301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sharp, F. C., and V. Sperandio. 2007. QseA directly activates transcription of LEE1 in enterohemorrhagic Escherichia coli. Infect. Immun. 75:2432-2440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sheoran, A. S., S. Chapman-Bonofiglio, B. R. Harvey, J. Mukherjee, G. Georgiou, A. Donohue-Rolfe, and S. Tzipori. 2005. Human antibody against Shiga toxin 2 administered to piglets after the onset of diarrhea due to Escherichia coli O157:H7 prevents fatal systemic complications. Infect. Immun. 73:4607-4613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sperandio, V., C. C. Li, and J. B. Kaper. 2002. Quorum-sensing Escherichia coli regulator A: a regulator of the LysR family involved in the regulation of the locus of enterocyte effacement pathogenicity island in enterohemorrhagic E. coli. Infect. Immun. 70:3085-3093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sperandio, V., J. L. Mellies, R. M. Delahay, G. Frankel, J. A. Crawford, W. Nguyen, and J. B. Kaper. 2000. Activation of enteropathogenic Escherichia coli (EPEC) LEE2 and LEE3 operons by Ler. Mol. Microbiol. 38:781-793. [DOI] [PubMed] [Google Scholar]
- 57.Sperandio, V., J. L. Mellies, W. Nguyen, S. Shin, and J. B. Kaper. 1999. Quorum sensing controls expression of the type III secretion gene transcription and protein secretion in enterohemorrhagic and enteropathogenic Escherichia coli. Proc. Natl. Acad. Sci. USA 96:15196-15201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sperandio, V., A. G. Torres, J. A. Girón, and J. B. Kaper. 2001. Quorum sensing is a global regulatory mechanism in enterohemorrhagic Escherichia coli O157:H7. J. Bacteriol. 183:5187-5197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sperandio, V., A. G. Torres, B. Jarvis, J. P. Nataro, and J. B. Kaper. 2003. Bacteria-host communication: the language of hormones. Proc. Natl. Acad. Sci. USA 100:8951-8956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sperandio, V., A. G. Torres, and J. B. Kaper. 2002. Quorum sensing Escherichia coli regulators B and C (QseBC): a novel two-component regulatory system involved in the regulation of flagella and motility by quorum sensing in E. coli. Mol. Microbiol. 43:809-821. [DOI] [PubMed] [Google Scholar]
- 61.Surette, M. G., M. B. Miller, and B. L. Bassler. 1999. Quorum sensing in Escherichia coli, Salmonella typhimurium, and Vibrio harveyi: a new family of genes responsible for autoinducer production. Proc. Natl. Acad. Sci. USA 96:1639-1644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Taga, M. E., J. L. Semmelhack, and B. L. Bassler. 2003. The LuxS-dependent autoinducer AI-2 controls the expression of an ABC transporter that functions in AI-2 uptake in Salmonellas typhimurium. Mol. Microbiol. 50:1411-1427. [DOI] [PubMed] [Google Scholar]
- 63.Tobe, T., S. A. Beatson, H. Taniguchi, H. Abe, C. M. Bailey, A. Fivian, R. Younis, S. Mattthews, O. Marches, G. Frankel, T. Hayashi, and M. J. Pallen. 2006. An extensive repertoire of type III secretion effectors in Escherichia coli 0157 and the role of lambdoid phages in their dissemination. Proc. Natl. Acad. Sci. USA 103:14941-14946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Torres, A. G., J. A. Giron, N. T. Perna, V. Burland, F. R. Blattner, F. Avelino-Flores, and J. B. Kaper. 2002. Identification and characterization of lpfABCC′DE, a fimbrial operon of enterohemorrhagic Escherichia coli O157:H7. Infect. Immun. 70:5416-5427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Torres, A. G., K. J. Kanack, C. B. Tutt, V. Popov, and J. B. Kaper. 2004. Characterization of the second long polar (LP) fimbriae of Escherichia coli O157:H7 and distribution of LP fimbriae in other pathogenic E. coli strains. FEMS Microbiol. Lett. 238:333-344. [DOI] [PubMed] [Google Scholar]
- 66.Umanski, T., I. Rosenshine, and D. Friedberg. 2002. Thermoregulated expression of virulence genes in enteropathogenic Escherichia coli. Microbiology 148:2735-2744. [DOI] [PubMed] [Google Scholar]
- 67.Vendeville, A., K. Winzer, K. Heurlier, C. M. Tang, and K. R. Hardie. 2005. Making ‘sense’ of metabolism: autoinducer-2, LuxS and pathogenic bacteria. Nat. Rev. Microbiol. 3:383-396. [DOI] [PubMed] [Google Scholar]
- 68.Walters, M., M. P. Sircili, and V. Sperandio. 2006. AI-3 synthesis is not dependent on luxS in Escherichia coli. J. Bacteriol. 188:5668-5681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Walters, M., and V. Sperandio. 2006. Autoinducer 3 and epinephrine signaling in the kinetics of locus of enterocyte effacement gene expression in enterohemorrhagic Escherichia coli. Infect. Immun. 74:544-5455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Wang, L., Y. Hashimoto, C.-Y. Tsao, J. J. Valdes, and W. E. Bentley. 2005. Cyclic AMP (cAMP) and cAMP receptor protein influence both synthesis and uptake of extracellular autoinducer 2 in Escherichia coli. J. Bacteriol. 187:2066-2076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Wang, L., J. Li, J. C. March, J. J. Valdes, and W. E. Bentley. 2005. luxS-dependent gene regulation in Escherichia coli K-12 revealed by genomic expression profiling. J. Bacteriol. 187:8350-8360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Welch, R. A., V. Burland, G. Plunkett, P. Redford, P. Roesch, D. Rasko, E. L. Buckles, S.-R. Liou, A. Boutin, J. Hackett, D. Stroud, G. F. Mayhew, D. J. Rose, S. Zhou, D. C. Schwartz, N. T. Perna, H. L. Mobley, M. S. Donnenberg, and F. R. Blattner. 2002. Extensive mosaic structure revealed by the complete genome sequence of uropathogenic Escherichia coli. Proc. Natl. Acad. Sci. USA 99:17020-17024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Winzer, K., K. R. Hardie, N. Burgess, N. Doherty, D. F. Kirke, M. T. Holden, R. Linforth, K. A. Cornell, A. J. Taylor, P. J. Hill, and P. Williams. 2002. LuxS: its role in central metabolism and the in vitro synthesis of 4-hydroxy-5-methyl-3(2H)-furanone. Microbiology 148:909-922. [DOI] [PubMed] [Google Scholar]
- 74.Winzer, K., K. R. Hardie, and P. Williams. 2002. Bacterial cell-to-cell communication: sorry, can't talk now—gone to lunch! Curr. Opin. Microbiol. 5:216-222. [DOI] [PubMed] [Google Scholar]
- 75.Xavier, K. B., and B. L. Bassler. 2005. Regulation of uptake and processing of the quorum-sensing autoinducer AI-2 in Escherichia coli. J. Bacteriol. 187:238-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Zhang, L., R. R. Chaudhuri, C. Constantinidou, J. L. Hobman, M. D. Patel, A. C. Jones, D. Sarti, A. J. Roe, I. Vlisidou, R. K. Shaw, F. Falciani, M. P. Stevens, D. L. Gally, S. Knutton, G. Frankel, C. W. Penn, and M. J. Pallen. 2004. Regulators encoded in the Escherichia coli type III secretion system 2 gene cluster influence expression of genes within the locus for enterocyte effacement in enterohemorrhagic E. coli O157:H7. Infect. Immun. 72:7282-7293. [DOI] [PMC free article] [PubMed] [Google Scholar]