Abstract
Species-specific antibody epitopes within several major immunoreactive protein orthologs of Ehrlichia species have recently been identified and molecularly characterized. In this study, dominant B-cell epitopes within the acidic (pI 5.35) ankyrin repeat-containing 200-kDa major immunoreactive protein (gp200) of Ehrlichia canis were defined. The E. canis gp200 gene (4,263 bp; 1,421 amino acids) was cloned and expressed as four (N-terminal, 1,107 bp; N-internal, 910 bp; C-internal, 1,000 bp; and C-terminal, 1,280 bp) overlapping recombinant proteins. The N-terminal, C-internal, and C-terminal polypeptides (369, 332, and 426 amino acids, respectively) were strongly recognized by antibody, and the major epitope(s) in these polypeptides was mapped to four polypeptide regions (40 to 70 amino acids). Smaller overlapping recombinant polypeptides (14 to 15 amino acids) spanning these regions identified five strongly immunoreactive species-specific epitopes that exhibited conformational dependence. The majority of the epitopes (four) were located in two strongly acidic (pI 4 to 4.9) domains in the distal N- and C-terminal regions of the protein flanking the centralized ankyrin domain-containing region. The amino acid content of the epitope-containing domains included a high proportion of strongly acidic amino acids (glutamate and aspartate), and these domains appear to have important biophysical properties that influence the antibody response to gp200.
Patients and dogs infected with Ehrlichia chaffeensis and Ehrlichia canis develop antibodies to a relatively well defined group of proteins that constitute the major immunoreactive proteins of Ehrlichia (4, 7, 17, 28). Many of these immunoreactive proteins and their corresponding orthologs have been identified and molecularly characterized in E. chaffeensis and E. canis (5, 15, 19, 21, 24, 25, 35, 36). The majority of the molecularly characterized immunoreactive proteins are secreted, serine/threonine rich, and strongly acidic and exhibit electrophoretic masses that are substantially larger than those predicted by amino acid sequences (5, 15, 19, 34, 35). Furthermore, the major immunodeterminants have been mapped to acidic serine-rich tandem repeats in many of these proteins (5, 19, 33, 35).
Recently, the largest major immunoreactive ehrlichial protein ortholog (gp200) of E. canis and E. chaffeensis has been identified and molecularly characterized (15). The recombinant E. canis gp200 N-terminal domain (P43) reacts strongly with antibodies in serum from dogs naturally and experimentally infected with E. canis (16, 17). The native and recombinant E. chaffeensis and E. canis gp200 orthologs exhibit molecular masses larger than those predicted by their amino acid sequences but lack serine-rich tandem repeats present in other ehrlichial proteins (15). However, the gp200s have ankyrin domains containing numerous ankyrin repeats (at least 21) that may mediate protein-protein interactions. The function of gp200 is unknown, but the protein is translocated to the nucleus of infected monocytes (23). gp200 exhibits homology with Anaplasma phagocytophilum AnkA (3), which is a type IV secretion substrate and is phosphorylated by host Abl-1 and Src tyrosine kinases (8, 13). AnkA also is translocated to the nucleus of infected neutrophils, where it binds DNA and may be involved in modulation of host cell gene transcription (26).
Elimination of Ehrlichia infection requires both cellular and humoral immune mechanisms. Although cell-mediated immune mechanisms are critically important in protection from intracellular pathogens, a number of studies have demonstrated an important role for humoral immunity in host defenses against ehrlichial pathogens (7, 30-32). Immunocompetent mice lacking B cells cannot clear a sublethal infection with Ixodes ovatus-carried ehrlichiae (32), and adoptive transfer of polyclonal immune serum protects severe combined immunodeficiency (SCID) mice from E. chaffeensis challenge (32). Specifically, protection has been demonstrated with antibodies directed against p28 of E. chaffeensis (11, 12, 25, 30), and studies with E. canis demonstrated that opsonization with antibodies resulted in the intracellular killing of the organism in vitro (10). SCID mice are protected from lethal infection by passive transfer of anti-Ehrlichia muris polyclonal antibody, but Fab antibody fragments are not protective (7).
The objective of this study was to define the epitopes involved in antibody recognition of gp200, a well-characterized immunoreactive ehrlichial protein. In this study, we determined that gp200 contains at least five major immunoreactive epitopes, the majority of which were localized to terminal domains dominated by strongly acidic amino acids. These domains appear to have important biophysical properties that influence the antibody response to gp200.
MATERIALS AND METHODS
Preparation of E. canis genomic DNA.
Genomic DNA was purified from E. canis (Jake strain) as previously described (18).
Anti-E. canis serum.
Convalescent anti-E. canis serum was collected from a dog (no. 2995) experimentally infected with E. canis.
PCR amplification of E. canis gp200 fragments.
Oligonucleotide primers were designed to amplify overlapping regions (28 fragments) containing potential E. canis gp200 epitopes (Table 1). Amplicons were generated from genomic E. canis DNA (HotMasterMix; Eppendorf, Westbury, NY) using the following thermal cycling profile: 94°C for 5 min; 30 cycles of 94°C for 30 s, annealing temperature (5°C less than the lowest primer melting temperature) for 30 s, and 72°C for the appropriate extension time (30 s/500 product base pairs); and 72°C for 7 min.
TABLE 1.
Oligonucleotide primers used to PCR amplify regions of E. canis gp200 for epitope mapping
| Targeta | Forward primer
|
Reverse primer
|
Amplicon size (bp) | ||
|---|---|---|---|---|---|
| Name | Sequence | Name | Sequence | ||
| gp200 | |||||
| Nt1-369 | 1f | ATGTCAGATCCAAAACAAGGT | 1107r | TCCATCTACAAGTCCAAAATCTAA | 1,107 |
| Ni360-663 | 1081f | AATTTAGATTTTGGACTTGTA | 1992r | CCCACGCTCACTTGGTAGGT | 911 |
| Ci651-983 | 1951f | AATTTTGTTGGGGATTCGTTA | 2952r | ACCTGCGTTATTCTTTTGAGTAAG | 1,001 |
| Ct980-1406 | 2935f | CAAAAGAATAACGCAGGTGATACA | 4221r | ACTAGGAGATGCTGCTTGTTGTTG | 1,286 |
| N-terminal region | |||||
| Nt8-186 (1) | 22f | GATCCAGAACAAAATCAAACTA | 564r | ACCTAAAACTGCATTCCTAACATCTG | 542 |
| Nt8-66 (2) | 22f | GATCCAGAACAAAATCAAACTA | 183r | ATATAAATCTTCACTCTCAGGA | 161 |
| Nt66-100 (3) | 196f | ATGCCTAAGGGTAAAAGAACTGC | 300r | TCTTGGCGGTAATGTAGGAGGTAAATC | 104 |
| Nt29-72 (4) | 85f | ATGCAGGAACAAGATCAGCAGCAG | 216r | AGCAGCAGTTCTTTTACCCTTAGG | 131 |
| Nt8-35 (5) | 22f | GATCCAGAACAAAATCAAACTA | 105r | CTGCTGCTGATCTTGTTCCT | 83 |
| Nt86-186 (6) | 265f | GATGATGAAGATTTACCTCCTACA | 564r | ACCTAAAACTGCATTCCTAACATCTG | 299 |
| C-internal region | |||||
| Ci650-765 (7) | 1948f | TTAAATTTTGTTGGGGATTCGTT | 2295r | ATTCTCACAATTAACATCAACTCCAG | 347 |
| Ci757-879 (8) | 2269f | ACTGGAGTTGATGTTAATTGTGAGAA | 2637r | AGATATTGCAGCATAAACCATTGG | 368 |
| Ci876-1024 (9) | 2626f | GCTGCAATATCTGGTAATGAGCA | 3072r | GTACCCTTCTTTGTCTCTGGCTGTT | 446 |
| Ci757-817 (10) | 2269f | ACTGGAGTTGATGTTAATTGTGAGAA | 2451r | TCCCTTTTCTTTTCTACCTGGAACTATC | 182 |
| Ci808-878 (11) | 2422f | GCGATAGTTCCAGGTAGAAAAGAAAAG | 2634r | TATTGCAGCATAAACCATTGGACG | 212 |
| Ci862-937 (12) | 2584f | GTTGAAGTTAATCGAAATAGTGAAATACGTC | 2811r | AGATTTACCAACATCACATCCTTCAGAAA | 227 |
| Ci933-985 (13) | 2797f | GATGTTGGTAAATCTGGAAAAGATGGTA | 2955r | ATCACCTGCGTTATTCTTTTGAGTAAGA | 158 |
| Ci979-1024 (14) | 2935f | CAAAAGAATAACGCAGGTGATACACCTT | 3072r | GTACCCTTCTTTGTCTCTGGCTGTT | 137 |
| Ci876-917 (15) | 2626f | GCTGCAATATCTGGTAATGAGCA | 2751r | TGCTACTGCAACCATAATTAAAGGATTTC | 125 |
| Ci915-947 (16) | 2743f | GCAGTAGCAGATGGTAATGCAGGTCTTC | 2841r | AGCATAATGTAACGCTGTATTACCATCTT | 98 |
| Ci942-985 (17) | 2824f | AATACAGCGTTACATTATGCTGTTAGTC | 2955r | ATCACCTGCGTTATTCTTTTGAGTAAGA | 131 |
| C-terminal region | |||||
| Ct1222-1395 (18) | 3665f | AATACGCGAAATAACTCTGAC | 4188r | TACCTGGGTAACTTCTGGTAAAC | 523 |
| Ct1222-1274 (19) | 3665f | AATACGCGAAATAACTCTGAC | 3822r | ATCATCACGAATACACTCTG | 157 |
| Ct1292-1340 (20) | 3874f | ATGAAGAAACTTGAGGCACGAG | 4020r | GAACGACACAGCACCACTACTTCTTG | 146 |
| Ct1222-1340 (21) | 3665f | AATACGCGAAATAACTCTGAC | 4020r | GAACGACACAGCACCACTACTTCTTG | 355 |
| Ct1266-1298 (22) | 3796f | ATGTCTTTATCAGAGTGTATTCGTG | 3894r | AGCTCGTGCCTCAAGTTTCTT | 98 |
| Ct1332-1371 (23) | 3994f | ATGGCAAGAAGTAGTGGTGCTGTGTCGTTC | 4113r | ATCATTTGACCCAAGACTAGTATCAGA | 119 |
| Ct1363-1404 (24) | 4087f | ATGTCTGATACTAGTCTTGGGTCAAATG | 4212r | TCGTGCTTGTTGTTGACTTACAGC | 125 |
Subscripts represent the amino acids contained in the fragment. Numbers in parentheses correspond to those in Fig. 2.
Recombinant gp200 protein expression and purification.
The four largest gp200 amplicons (910 to 1,280 bp), spanning ∼99% of the E. canis gp200 open reading frame, were cloned into the pUni/V5-His-TOPO Echo donor vector (Invitrogen, Carlsbad, CA). The donor vector is designed to recombine the insert into an acceptor vector with appropriate transcription regulatory and fusion protein coding sequences. The cloned donor vector was transformed into PIR1 Escherichia coli (Invitrogen) and selected on LB agar containing kanamycin (50 μg/ml). The resulting transformants were screened by PCR for correctly oriented inserts, and plasmids from the positive transformants were isolated and sequenced to verify proper orientation and frame. Correct donor vectors were recombined by Cre recombinase with the pRSET-E Echo acceptor vector (Invitrogen), which contains a loxP recombination site. Recombined vectors were transformed into TOP10 E. coli (Invitrogen) for plasmid propagation, and transformants were selected by growth on LB agar with kanamycin (50 μg/ml). Correct orientation and frame of the transformant inserts were verified as described above. These clones and the control plasmid pRSET-E/Uni-CAT (Invitrogen) were transformed into E. coli BL21(DE3)pLysS (Invitrogen), and protein expression was induced by addition of 1 mM isopropyl-β-d-thiogalactopyranoside (IPTG) to a liquid culture in log growth phase incubated for 3.5 h at 37°C. Following expression, cultures were pelleted at 3,000 × g for 20 min and frozen at −80°C.
All smaller recombinant E. canis gp200 fragments used for epitope mapping were amplified by PCR, cloned into the pBAD/TOPO ThioFusion expression vector (Invitrogen), and characterized similarly. Expression of the recombinant proteins in TOP10 E. coli (Invitrogen) was induced by adding 0.02% arabinose to cultures in log-phase growth. The recombinant E. canis gp200 proteins expressed from the recombined pUni/pRSET-E Echo vector were purified under denaturing conditions as described previously (5, 16). The recombinant peptides expressed from the pBAD/TOPO ThioFusion vectors were purified under native conditions as previously described (6). All recombinant proteins were quantitated with the bicinchoninic acid protein assay kit (Pierce, Rockford, IL).
Cloning of peptide epitope oligonucleotides.
Complementary 42- or 45-bp oligonucleotides encoding all epitopes (14- or 15-mer peptides) (Table 2) were annealed and cloned into the pBAD 102/Directional TOPO ThioFusion expression vector (Invitrogen) as previously described (5).
TABLE 2.
Complementary oligonucleotides encoding E. canis gp200 14- or 15-mer peptides
| Region | Oligonucleotide
|
|||
|---|---|---|---|---|
| Coding
|
Noncoding
|
|||
| Name | Sequence | Name | Sequence | |
| N-terminal region | pep1-cod | CACCCAGGAACAAGATCAGCAGCAGGGAGCAGTTGGTGGTGCTGTT | pep1-non | AACAGCACCACCAACTGCTCCCTGCTGCTGATCTTGTTCCTG |
| pep2-cod | CACCGGTGGTGCTGTTGGTAATAGTCCTATTGAAAGAGAGAGAGTA | pep2-non | TACTCTCTCTCTTTCAATAGGACTATTACCAACAGCACCACC | |
| pep3-cod | CACCAGAGAGAGAGTAGCTGCTCCTGAGAGTGAAGATTTATATACT | pep3-non | AGTATATAAATCTTCACTCTCAGGAGCAGCTACTCTCTCTCT | |
| pep4-cod | CACCGATTTATATACTGTGATTATACCTAAGGGTAAAAGAACTGCT | pep4-non | AGCAGTTCTTTTACCCTTAGGTATAATCACAGTATATAAATC | |
| pep5-cod | CACCAAAAGAACTGCTGCTCCAATTTTGGAAAGAAAGTCTCCTACTCCTGAA | pep5-non | TTCAGGAGTAGGAGACTTTCTTTCCAAAATTGGAGCAGCAGTTCTTTT | |
| C-internal region | pep6-cod | CACCAATACAGCGTTACATTATGCTGTTAGTCATTCAGATAAAGAG | pep6-non | CTCTTTATCTGAATGACTAACAGCATAATGTAACGCTGTATT |
| pep7-cod | CACCTCAGATAAAGAGTTTGGTAATAAAGCTATAAAGATATTAATT | pep7-non | AATTAATATCTTTATAGCTTTATTACCAAACTCTTTATCTGA | |
| pep8-cod | CACCAAGATATTAATTTCACGTAATAGTGTTGGGACTAATAGAGAT | pep8-non | ATCTCTATTAGTCCCAACACTATTACGTGAAATTAATATCTT | |
| pep9-cod | CACCACTAATAGAGATATTCTTACTCAAAAGAATAACGCAGGTGAT | pep9-non | ATCACCTGCGTTATTCTTTTGAGTAAGAATATCTCTATTAGT | |
| C-terminal region | pep10-cod | CACCCTAGTTAATACGCGAAATAACTCTGACGATACTGTTGCACAT | pep10-non | ATGTGCAACAGTATCGTCAGAGTTATTTCGCGTATTAACTAG |
| pep11-cod | CACCACTGTTGCACATTGTGCTCTTTTATCGGATATGAAATATGCT | pep11-non | AGCATATTTCATATCCGATAAAAGAGCACAATGTGCAACAGT | |
| pep12-cod | CACCATGAAATATGCTCAAAAGATACTTAAATCATGTAACCATGAT | pep12-non | ATCATGGTTACATGATTTAAGTATCTTTTGAGCATATTTCAT | |
| pep13-cod | CACCTGTAACCATGATACATTAGTGAGAGGAAATAGTAATAATCAA | pep13-non | TTGATTATTACTATTTCCTCTCACTAATGTATCATGGTTACA | |
| pep14-cod | CACCAGTAATAATCAATCTTTATCAGAGTGTATTCGTGATGATAGTAAA | pep14-non | TTTACTATCATCACGAATACACTCTGATAAAGATTGATTATTACT | |
| pep15-cod | CACCGATGATAGTAAATATAAAAAAGGTGGAATTTTTAGTAAGTCTTTA | pep15-non | TAAAGACTTACTAAAAATTCCACCTTTTTTATATTTACTATCATC | |
| pep16-cod | CACCAGAAGTAGTGGTGCTGTGTCGTTCAAACATGTGCAAGAAACAGGA | pep16-non | TCCTGTTTCTTGCACATGTTTGAACGACACAGCACCACTACTTCT | |
| pep17-cod | CACCCAAGAAACAGGAGTTGACACGTCTGGTCCTTCTGATATAGAAAGT | pep17-non | ACTTTCTATATCAGAAGGACCAGACGTGTCAACTCCTGTTTCTTG | |
| pep18-cod | CACCGATATAGAAAGTTTAGAGAGATTATCTGATACTAGTCTTGGGTCA | pep18-non | TGACCCAAGACTAGTATCAGATAATCTCTCTAAACTTTCTATATC | |
| pep19-cod | CACCAGTCTTGGGTCAAATGATTTTGATCAGCGAATGGCAGATTTAGAT | pep19-non | ATCTAAATCTGCCATTCGCTGATCAAAATCATTTGACCCAAGACT | |
| pep20-cod | CACCGCAGATTTAGATCAAGAAATAGCAAATATTGTTAGTGGTTTACCA | pep20-non | TGGTAAACCACTAACAATATTTGCTATTTCTTGATCTAAATCTGC | |
| pep21-cod | CACCACTCCTTTACCAGAAGTTACCCAGGTAGCTGTAAGTCAACAACAA | pep21-non | TTGTTGTTGACTTACAGCTACCTGGGTAACTTCTGGTAAACCACT | |
Gel electrophoresis and protein blotting.
Larger recombinant proteins and thioredoxin-fused recombinant proteins and peptides were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) on 4 to 12% NuPAGE gels (Invitrogen) with MOPS (morpholinepropanesulfonic acid) buffer as previously described (17). Smaller recombinant peptides were separated on 12% NuPAGE gels with MOPS running buffer. For some assays, total protein was visualized in the gel with Imperial Protein Stain (Pierce, Rockford, IL), according to the manufacturer's protocol. Recombinant proteins/peptides were transferred to a nitrocellulose membrane as described previously (17).
Western immunoblotting.
Immunoreactivities of recombinant gp200 fragments with anti-E. canis dog serum (no. 2995) were determined by Western immunoblotting as previously described (17) with primary antibodies diluted 1:500. Total membrane-bound protein was visualized by Ponceau S staining prior to membrane blocking. Recombinant peptides were detected with anti-V5, anti-Xpress, or anti-His (C-terminal) antibodies (Invitrogen) according to the manufacturer's protocol.
Synthetic peptides.
Custom peptides (Table 3) were synthesized and purified (Biosynthesis, Inc., Lewisville, TX) and used in an enzyme-linked immunosorbent assay (ELISA) to determine epitope reactivity with anti-E. canis serum.
TABLE 3.
Peptide sequences (14- and 15-mer) spanning the epitope-containing regions of E. canis gp200
| Epitope region (amino acid positions) | Peptide name | Sequence |
|---|---|---|
| N-terminal (29-84) | 1 | QEQDQQQGAVGGAV |
| 2 | GGAVGNSPIERERV | |
| 3 | RERVAAPESEDLYT | |
| 4 | DLYTVIIPKGKRTA | |
| 5 | KRTAAPILERKSPTP | |
| C-internal (942-985) | 6 | NTALHYAVSHSDKE |
| 7 | SDKEFGNKAIKILI | |
| 8 | KILISRNSVGTNRD | |
| 9 | TNRDILTQKNNAGD | |
| C-terminal (1222-1287) | 10 | LVNTRNNSDDTVAH |
| 11 | TVAHCALLSDMKYA | |
| 12 | MKYAQKILKSCNHD | |
| 13 | CNHDTLVRGNSNNQ | |
| 14 | SNNQSLSECIRDDSK | |
| 15 | DDSKYKKGGIFSKSL | |
| C-terminal (1333-1402) | 16 | RSSGAVSFKHVQETG |
| 17 | QETGVDTSGPSDIES | |
| 18 | DIESLERLSDTSLGS | |
| 19 | SLGSNDFDQRMADLD | |
| 20 | ADLDQEIANIVSGLP | |
| 21 | SGLPEVTQVAVSQQQ |
ELISA.
The immunoreactivities of recombinant and synthetic peptides were determined as previously described by ELISA (5). A synthetic 20-mer peptide (RNTTVGVFGLKQNWDGSAIS) previously determined to contain the hypervariable region 1 epitope of E. chaffeensis p28-19 (kindly provided by Xue-jie Yu, University of Texas Medical Branch) was used as a positive control for immunoreactivity (37). Primary antibody (canine anti-E. canis serum) was diluted 1:1,000, and secondary antibody (alkaline phosphatase-labeled goat anti-dog immunoglobulin G [heavy plus light chains]; Kirkegaard & Perry Laboratories, Gaithersburg, MD) was diluted 1:2,500. Substrate (Sure Blue; Kirkegaard & Perry) was added, and absorbance was measured with a tunable microplate reader (Molecular Devices, Sunnyvale, CA) at 650 nm. The absorbance of the sample in each well was expressed as the average optical density of three or four wells at 650 nm after subtraction of the thioredoxin-only blank wells.
Sequence analysis of mapped epitopes.
E. canis gp200 epitopes were examined for homology to other Ehrlichia sp. proteins (including gp200 orthologs) using the Basic Local Alignment Search Tool (BLAST) (http://www.ncbi.nlm.nih.gov/BLAST/).
RESULTS
gp200 amino acid composition and acidic domains.
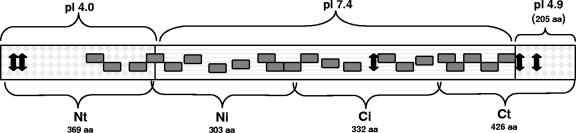
The overall gp200 (1,421 amino acids) composition was dominated by four hydrophobic (A, I, L, and V), four polar (S, T, N, and Q), two strongly acidic (D and E; 187 amino acids), and two strongly basic (K and R; 144 amino acids) amino acids. Interestingly, a large number (n = 118) of glycine (G) residues (high content in fibrous proteins) were also present. Three specific domains were identified according to amino acid composition and isoelectric point (Fig. 1). The distal terminal (Nt [amino acids 1 to 369] and Ct [last 205 amino acids]) polypeptides exhibited a substantially larger proportion (2:1) of strongly acidic amino acids (D and E) than did the internal region (848 amino acids; positions 370 to 1263) of the protein where ankyrin repeats were located, and the ratio of strongly basic and strongly acidic amino acids was equivalent. Consequently the isoelectric point of the internal ankyrin domain regions was slightly basic (pI 7.4). Although the acidic regions of the protein represented only ∼40% of the protein, the highly acidic domains greatly influenced the overall isoelectric point of gp200, resulting in a protein of acidic nature (pI 5.35).
FIG. 1.
Schematic of E. canis gp200 showing predicted isoelectric points (pIs) of acidic terminal domains and the slightly basic central ankyrin repeat (22 shaded boxes)-containing region. The cloned recombinant expressed regions (Nt, Ni, Ci, and Ct) are shown, and the approximate locations of mapped epitopes are designated by arrows. aa, amino acids.
Immunoreactivity of the major E. canis gp200 fragments.
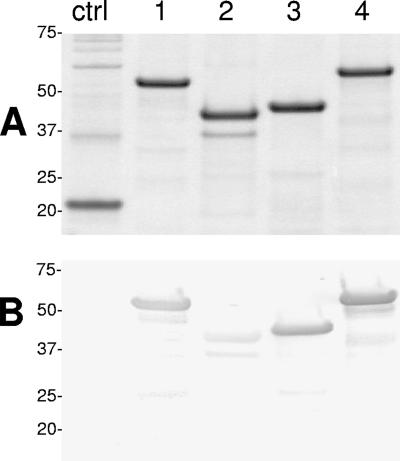
E. canis gp200 was cloned and expressed as four (∼1,000-bp) overlapping polypeptides: N-terminal (Nt; amino acids 1 to 369; pI 4.1), N-internal (Ni; amino acids 360 to 663; pI 6.0), C-internal (Ci; amino acids 651 to 983; pI 7.9), and C-terminal (Ct; amino acids 980 to 1406; pI 6.0). The Nt and Ct recombinant gp200 fragments (containing the acidic domains) exhibited substantially larger (∼6 kDa)-than-predicted molecular masses by SDS-PAGE (Fig. 2A). Three gp200 recombinant fragments (Nt, Ci, and Ct) reacted strongly with anti-E. canis antibody, but the Ni polypeptide exhibited a substantially weaker immunoreactivity (Fig. 2B). The recombinant proteins (Nt, Ni, Ci, and Ct) did not react with healthy dog sera (data not shown). Thus, the three strongly immunoreactive fragments (Nt, Ci, and Ct) were considered to have major B-cell epitopes and were investigated further.
FIG. 2.
(A) SDS-PAGE and total protein stain showing four E. coli-expressed overlapping E. canis gp200 recombinant proteins representing 99% of the open reading frame. Lane 1, Nt; lane 2, Ni; lane 3, Ci; lane 4, Ct. Molecular masses (kilodaltons) are shown on the left. (B) Corresponding Western immunoblot probed with polyclonal dog anti-E. canis serum (2995). Lanes and numbers at left are as defined for panel A. Negative controls (ctrl) are recombinant E. chaffeensis Dsb (22 kDa) (A) and recombinant chloramphenicol acetyltransferase (28 kDa) (B).
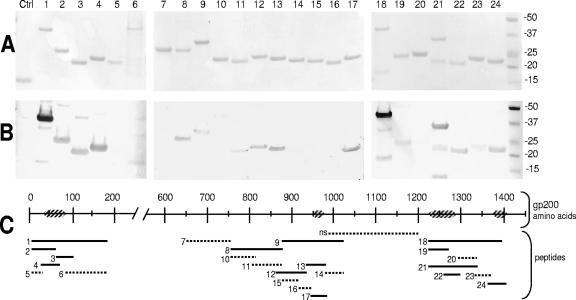
Major epitope-containing region in the Nt polypeptide.
The major epitope(s) in the Nt region was identified by evaluating the immunoreactivities of six overlapping recombinant proteins (Fig. 3A). A smaller recombinant protein representing approximately one-half (amino acids 8 to 186) of the Nt region of gp200 was cloned and expressed, and Western blotting revealed that this smaller Nt polypeptide (designated Nt8-186) was strongly reactive with dog anti-E. canis serum (Fig. 3B). Nt8-186 was further divided into five overlapping polypeptides designated Nt8-66, Nt66-100, Nt29-72, Nt8-35, and Nt86-186. Two polypeptides, Nt8-35 and Nt86-186, were only weakly immunoreactive with anti-E. canis dog serum by Western blotting, while all other fragments were strongly immunoreactive (Fig. 3B). The region from amino acids 36 to 85 was identified in the strongly immunoreactive recombinant polypeptides but not in weakly reactive fragments; thus, this 51-amino-acid section of E. canis gp200 was identified as the major epitope-containing region and was located in a highly acidic domain (Fig. 1 and 3C). This polypeptide exhibited high serine/threonine/glutamate/aspartate (STED) content (∼31%) (Table 3).
FIG. 3.
Immunoreactivities of 24 overlapping recombinant polypeptides covering the major immunoreactive regions of the E. canis gp200. Lanes and lines: 1, Nt8-186; 2, Nt8-66; 3, Nt66-100; 4, Nt29-72; 5, Nt8-35; 6, Nt86-186; 7, Ci650-765; 8, Ci757-879; 9, Ci876-1024; 10, Ci757-817; 11, Ci808-878; 12, Ci862-937; 13, Ci933-985; 14, Ci979-1024; 15, Ci876-917; 16, Ci915-947; 17, Ci942-985; 18, Ct1222-1395; 19, Ct1222-1274; 20, Ct1292-1340; 21, Ct1222-1340; 22, Ct1266-1298; 23, Ct1332-1371; 24, Ct1363-1404. Nt, Ci, and Ct represent their locations in the large N-terminal, C-internal, and C-terminal regions of gp200, respectively (subscripts designate the amino acid numbers included in each fragment). (A) Membrane total protein staining. (B) Western immunoblots probed with polyclonal dog anti-E. canis serum. (C) Schematic of E. canis gp200 illustrating the locations of the 24 recombinant fragments and the epitope-containing regions. Solid lines, strongly immunoreactive recombinant gp200 fragments; dotted lines, weakly immunoreactive or nonimmunoreactive recombinant gp200 fragments; hatched boxes, epitope-containing regions; Ctrl, negative-control recombinant thioredoxin; ns, not shown. Molecular masses (kilodaltons) are shown on the right.
Major epitope-containing region in the Ci polypeptide.
The major epitope-containing region within the E. canis gp200 Ci polypeptide was identified by evaluating the immunoreactivity of 11 overlapping recombinant polypeptides spanning the Ci major fragment: Ci650-765, Ci757-879, Ci876-1024, Ci757-817, Ci808-878, Ci862-937, Ci933-985, Ci979-1024, Ci876-917, Ci915-947, and Ci942-985 (Fig. 3A). The five strongly immunoreactive polypeptides in this region, Ci757-879, Ci876-1024, Ci862-937, Ci933-985, and Ci942-985, spanned amino acids 756 to 1024. However, the major epitope-containing region was identified by excluding the amino acids contained within the remaining polypeptides which were only weakly immunoreactive or not immunoreactive (Fig. 3B). Therefore, the major epitope-containing region was located within amino acids 948 to 978 (Fig. 3C), which also exhibited a high STED content (∼29%) (Table 3).
Major epitope-containing region in the Ct polypeptide.
The epitope-containing regions in the Ct region of E. canis gp200 were identified by evaluating the immunoreactivity of smaller overlapping recombinant proteins (Fig. 3A). The large Ct region was divided roughly in half and expressed as two smaller polypeptides (amino acids 908 to 1203 and 1222 to 1395, respectively). The immunoreactivity of the distal half of the protein was strong, as determined by Western blotting with anti-E. canis dog serum (Fig. 3B); conversely, the other half was not immunoreactive (data not shown). The immunoreactive region (designated Ct1222-1395) was divided into six overlapping polypeptide fragments: Ct1222-1274, Ct1292-1340, Ct1222-1340, Ct1266-1298, Ct1332-1371, and Ct1363-1404. Four polypeptides, Ct1222-1274, Ct1222-1340, Ct1266-1298, and Ct1363-1404, were strongly immunoreactive with anti-E. canis antibodies, while two fragments (Ct1292-1340 and Ct1332-1371) were weakly immunoreactive (Fig. 3B). The epitope-containing regions within the Ct region of E. canis gp200 were determined by excluding the amino acids contained by the two weakly immunoreactive recombinant proteins, 1266 to 1340. Therefore, the regions including amino acids 1222 to 1290 and 1372 to 1404 were considered Ct epitope-containing regions and were located in highly acidic domains (Fig. 1 and 3C). Both epitope-containing regions had high STED content, 27% and 24%, respectively (Table 3).
Immunoreactivity of synthetic and recombinant polypeptides.
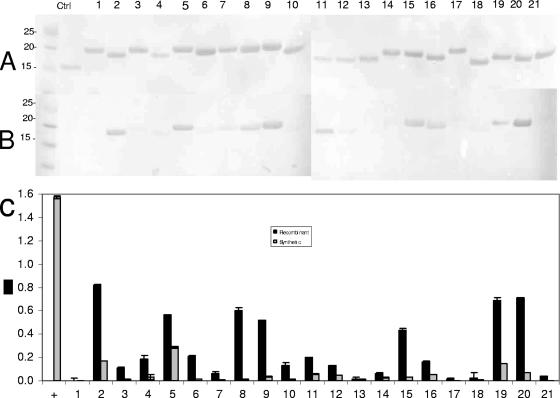
A total of 21 synthetic (14- and 15-mer) and corresponding recombinant overlapping polypeptides were generated to cover the sequences of the four major epitope-containing regions of E. canis gp200. Recombinant thioredoxin-fused peptides with amino acid sequences identical to the synthetic peptides were expressed in E. coli. Peptides 1 through 5 (14-mer) spanned the Nt epitope-containing region (amino acids 29 to 84), peptides 6 through 9 (14-mer) spanned the Ct epitope-containing region (amino acids 942 to 985), peptides 10 through 15 (14-mer) spanned one Ct epitope-containing region (amino acids 1222 to 1287), and peptides 16 through 21 (15-mer) spanned the distal Ct epitope-containing region (amino acids 1333 to 1402). All peptides overlapped by four amino acids (Table 3).
Western immunoblotting and ELISA were performed to determine the recombinant gp200 peptide immunoreactivity, and the immunoreactivities of the synthetic peptides were evaluated by ELISA. Recombinant peptides 2, 5, 9, 15, and 20 exhibited strong immunoreactivity, and peptides 8, 11, 16, and 19 were weakly immunoreactive by Western blotting (Fig. 4B). Seven recombinant peptides, 2, 5, 8, 9, 15, 19, and 20, were strongly immunoreactive by ELISA and were not identified by BLASTp as having significant homology with E. chaffeensis gp200 or any other ehrlichial protein (Fig. 4C). Most synthetic peptides exhibited reduced immunoreactivity compared to the corresponding recombinant expressed fusion polypeptides, indicating a conformational dependence of these epitopes (Fig. 4C). A 14-mer positive-control peptide derived from E. chaffeensis p28-19 was included to demonstrate peptide binding and immunoreactivity and was strongly reactive with convalescent dog anti-E. canis antibodies.
FIG. 4.
Immunoreactivities of recombinant and synthetic overlapping 14- and 15-mer peptides (Table 3) spanning the epitope-containing regions of E. canis gp200. Positive control (Ctrl or +), E. chaffeensis p28-19 peptide epitope; lane and x-axis numbers correspond to the peptide numbers in Table 3. (A) Membrane total protein staining. (B) Western immunoblot probed with polyclonal dog anti-E. canis serum. (C) Immunoreactivities of the recombinant gp200 peptides and the corresponding synthetic gp200 peptides expressed as the average optical densities at 650 nm of quadruplet samples as determined by ELISA. The recombinant E. canis gp200 regions corresponding to the overlapping peptides are labeled under the x axis. Molecular masses (kilodaltons) are shown on the left.
Serine residues and antibody binding.
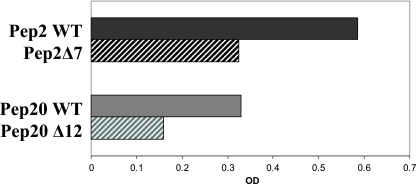
Serine is a frequently occurring amino acid in immunoreactive ehrlichial proteins and epitopes and was the second most common (n = 121) amino acid present in gp200. Recombinant peptides 2 and 20 were strongly reactive by ELISA, and each contained a single serine residue. Thus, these peptides were chosen for site-directed mutagenesis to examine the contribution of serine residues in antibody binding. Site-directed serine mutants (sequences shown in Table 3) of peptides 2 and 20 were substantially less immunoreactive by ELISA than were the corresponding wild-type peptides (Fig. 5), demonstrating that these serine residues are important determinants of these two epitopes.
FIG. 5.
Comparison of antibody reactivities as determined by ELISA between wild type (WT) and single serine-to-alanine mutant recombinant E. canis gp200 peptides 2 and 20 containing dominant epitopes. OD, optical density.
DISCUSSION
Antibodies are important for protection against Ehrlichia (7, 9, 10, 12, 31, 32), but the immunoprotective proteins and protective epitopes remain unknown. Many of the major immunoreactive proteins of E. canis and E. chaffeensis have been recently characterized, and most have serine-rich tandem repeats (5, 15, 16, 19, 34, 35). Furthermore, dominant antibody epitopes in these proteins have been mapped to acidic serine-threonine-rich regions and serine-rich tandem repeats in all of these proteins (5, 19, 33, 35). Thus, serine-rich acidic epitopes appear to be primary targets of the humoral immune responses against ehrlichiae and may be immunoprotective. In this study, five species-specific epitopes were identified primarily in terminal highly acidic domains of E. canis gp200.
The E. canis and E. chaffeensis gp200 orthologs have identical chromosomal locations and exhibit nucleic acid homology (∼50%) but have less amino acid identity (∼32%) (15). We previously reported that the N-terminal region (P43) (analogous to the Nt in this study) of gp200 was strongly recognized by antibody from E. canis-infected dogs and that antibodies directed at P43 were species specific (16). Ehrlichiae are known to have serologically cross-reactive antigens. The best-characterized examples are the major outer membrane proteins (p28/p30), map2, and other highly conserved immunoreactive proteins such as the chaperonin GroEL, disulfide oxidoreductase (Dsb), and ferric ion binding protein (Fbp) (1, 2, 6, 20, 24, 25, 29, 38). However, several of the epitopes characterized in the major immunoreactive proteins of E. canis and E. chaffeensis are molecularly distinct and elicit species-specific antibodies (5, 19). Consistent with our previous findings regarding the Nt epitopes (P43), the amino acid alignments of the mapped epitopes in gp200 (Nt, Ci, and Ct) identified no significant homology with E. chaffeensis gp200. Interestingly, all of the major immunoreactive protein orthologs (gp36/gp47, gp120/gp140, and gp19/VLPT) identified and characterized recently are antigenically distinct, further suggesting that cross-reactive antibodies generated between closely related Ehrlichia spp. are directed at more highly conserved proteins that are not necessarily considered to be major immunoreactive proteins.
The terminal epitope-containing domains exhibited high proportions of charged amino acids including glutamate and aspartate and the hydrophobic amino acid valine. The majority of the epitopes (except the Ci epitope [peptides 8 and 9]) were located within the first 100 amino acids or the last 200 amino acids of the protein, within these highly acidic terminal domains, flanking the ankyrin domain-containing region. The high proportion of acidic and polar amino acids observed in these terminal domains has been consistently observed in other characterized ehrlichial epitopes (5, 19). Furthermore, most of the characterized immunoreactive ehrlichial proteins (i.e., gp36, gp47, gp19, gp140, gp120, and gp200) are strongly acidic (pI ∼4 to 5.4). The role of the acidic, polar, and hydrophobic amino acids, such as glutamate, serine, and valine, is not currently understood, but the consistent presence of these amino acids in ehrlichial epitopes suggests that they constitute an important motif that interacts strongly with the host immune response. The predominance of antibody response to acidic ehrlichial proteins, particularly those containing tandem repeats, has not been described in relation to any other pathogen and suggests that an important and unique ehrlichia-host interaction occurs that directs the immune response towards these proteins.
The genome of E. canis has a revealed a serine/threonine bias in proteins associated with host-pathogen interactions (14). The polar amino acid serine was the second most frequently occurring amino acid in E. canis gp200 (n = 121), and the largest proportions of serine and threonine residues (73 residues combined; 17) were identified in the Ct region of E. canis gp200. They are also highly represented in other major immunoreactive ehrlichial proteins (5, 19, 34, 35). Many of these proteins are associated with host-pathogen interactions (14, 34). Serine was present in all (peptides 2, 5, 8, 15, 19, and 20) but one (peptide 9) of the peptides that exhibited the strong antibody recognition by E. canis-infected dog sera. Mutations of single serine residues present in peptides 2 and 20 did substantially reduce (50%) the immunoreactivity of the peptides, suggesting that serine residues are critical for the binding of antibodies to these epitopes. Notably, the high frequency of serine residues in immunoreactive ehrlichial proteins including gp200 has not been reported for other bacteria, but the high frequency of these residues suggests an important role in the pathobiology of ehrlichiae. Furthermore, there appears to be a direct relationship between the host immune response and acidic, serine-rich proteins.
Antibody epitopes mapped in other ehrlichial proteins appear to be single epitopes located in acidic serine-rich tandem repeats (5, 19, 33-35). However, we found numerous molecularly distinct epitopes in gp200 that were not located in tandem repeats. The number of epitopes identified in gp200 suggests that it is an important target of the host immune response against E. canis. Epitopes that have been mapped in other ehrlichial proteins are present in tandem repeat-containing proteins that are associated with host-pathogen interactions (27, 35). gp200 is translocated to the host cell nucleus (23), which has also been reported for AnkA, a similar ankyrin-containing protein found in A. phagocytophilum (26). Furthermore, AnkA has been proposed to be a type IV secreted virulence factor because of its potential ability to facilitate intracellular infection by activation of the host Abl-1 signaling pathway, recruitment of phosphatase SHP-1, and DNA binding (8, 13, 26). Although the role of antibodies in inhibiting the functional role of E. canis gp200 is unknown, inhibition of gp200 by the host immune response would likely result in an outcome beneficial to the host.
The immunoreactivity of the 21 synthetic peptides compared to the recombinant fusion peptides suggests that these epitopes have a conformational dependence, which is facilitated by fusion of the peptide to the thioredoxin fusion protein. Conformational dependence of antibody epitopes in Anaplasma and Ehrlichia spp. has been recognized in several proteins such as Fbp and map2 in E. canis and msp5 in Anaplasma marginale (2, 6, 13, 22). In this case, the strong antibody recognition of the synthetic E. chaffeensis p28-19 peptide demonstrated strong binding by synthetic peptides to the solid substrate used in this study. Therefore, the general lack of immunoreactivity of the synthetic peptides compared to the recombinant fusion gp200 peptides is most likely a result of improved solubility and appropriate epitope folding, resulting in increased antibody binding.
The E. canis and E. chaffeensis gp200s are the largest immunoreactive proteins (15, 17), and these unique ankyrin domain-containing proteins warrant further investigation. gp200-specific antibodies are elicited in dogs experimentally and naturally infected with E. canis (16, 17), and we have found that gp200 contains the largest number of distinct antibody epitopes reported in any ehrlichial immunoreactive protein. Additional studies are needed to determine the protection mediated by antibody against the E. canis gp200 epitopes to fully appreciate its potential as an immunoprotective antigen.
Acknowledgments
This work was supported by the National Institutes of Health (AI 071145-01), the Clayton Foundation for Research, and the UTMB Sealy Center for Vaccine Development.
Editor: V. J. DiRita
Footnotes
Published ahead of print on 6 August 2007.
REFERENCES
- 1.Alleman, A. R., A. F. Barbet, M. V. Bowie, H. L. Sorenson, S. J. Wong, and M. Belanger. 2000. Expression of a gene encoding the major antigenic protein 2 homolog of Ehrlichia chaffeensis and potential application for serodiagnosis. J. Clin. Microbiol. 38:3705-3709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alleman, A. R., L. J. McSherry, A. F. Barbet, E. B. Breitschwerdt, H. L. Sorenson, M. V. Bowie, and M. Belanger. 2001. Recombinant major antigenic protein 2 of Ehrlichia canis: a potential diagnostic tool. J. Clin. Microbiol. 39:2494-2499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Caturegli, P., K. M. Asanovich, J. J. Walls, J. S. Bakken, J. E. Madigan, V. L. Popov, and J. S. Dumler. 2000. ankA: an Ehrlichia phagocytophila group gene encoding a cytoplasmic protein antigen with ankyrin repeats. Infect. Immun. 68:5277-5283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chen, S. M., L. C. Cullman, and D. H. Walker. 1997. Western immunoblotting analysis of the antibody responses of patients with human monocytotropic ehrlichiosis to different strains of Ehrlichia chaffeensis and Ehrlichia canis. Clin. Diagn. Lab. Immunol. 4:731-735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Doyle, C. K., K. A. Nethery, V. L. Popov, and J. W. McBride. 2006. Differentially expressed and secreted major immunoreactive protein orthologs of Ehrlichia canis and E. chaffeensis elicit early antibody responses to epitopes on glycosylated tandem repeats. Infect. Immun. 74:711-720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Doyle, C. K., X. Zhang, V. L. Popov, and J. W. McBride. 2005. An immunoreactive 38-kilodalton protein of Ehrlichia canis shares structural homology and iron-binding capacity with the ferric ion-binding protein family. Infect. Immun. 73:62-69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Feng, H. M., and D. H. Walker. 2004. Mechanisms of immunity to Ehrlichia muris: a model of monocytotropic ehrlichiosis. Infect. Immun. 72:966-971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ijdo, J. W., A. C. Carlson, and E. L. Kennedy. 2007. Anaplasma phagocytophilum AnkA is tyrosine-phosphorylated at EPIYA motifs and recruits SHP-1 during early infection. Cell. Microbiol. 9:1284-1296. [DOI] [PubMed] [Google Scholar]
- 9.Lewis, G. E., Jr., S. L. Hill, and M. Ristic. 1978. Effect of canine immune serum on the growth of Ehrlichia canis within nonimmune canine macrophages. Am. J. Vet. Res. 39:71-76. [PubMed] [Google Scholar]
- 10.Lewis, G. E., Jr., and M. Ristic. 1978. Effect of canine immune macrophages and canine immune serum on the growth of Ehrlichia canis. Am. J. Vet. Res. 39:77-82. [PubMed] [Google Scholar]
- 11.Li, J. S., F. Chu, A. Reilly, and G. M. Winslow. 2002. Antibodies highly effective in SCID mice during infection by the intracellular bacterium Ehrlichia chaffeensis are of picomolar affinity and exhibit preferential epitope and isotype utilization. J. Immunol. 169:1419-1425. [DOI] [PubMed] [Google Scholar]
- 12.Li, J. S., E. Yager, M. Reilly, C. Freeman, G. R. Reddy, A. A. Reilly, F. K. Chu, and G. M. Winslow. 2001. Outer membrane protein-specific monoclonal antibodies protect SCID mice from fatal infection by the obligate intracellular bacterial pathogen Ehrlichia chaffeensis. J. Immunol. 166:1855-1862. [DOI] [PubMed] [Google Scholar]
- 13.Lin, M., A. den Dulk-Ras, P. J. Hooykaas, and Y. Rikihisa. 24 June 2007. Anaplasma phagocytophilum AnkA secreted by type IV secretion system is tyrosine phosphorylated by Abl-1 to facilitate infection. Cell. Microbiol. doi: 10.1111/j.1462-5822.2007.00985.x. [DOI] [PubMed]
- 14.Mavromatis, K., C. K. Doyle, A. Lykidis, N. Ivanova, M. P. Francino, P. Chain, M. Shin, S. Malfatti, F. Larimer, A. Copeland, J. C. Detter, M. Land, P. M. Richardson, X. J. Yu, D. H. Walker, J. W. McBride, and N. C. Kyrpides. 2006. The genome of the obligately intracellular bacterium Ehrlichia canis reveals themes of complex membrane structure and immune evasion strategies. J. Bacteriol. 188:4015-4023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.McBride, J. W., J. E. Comer, and D. H. Walker. 2003. Novel immunoreactive glycoprotein orthologs of Ehrlichia spp. Ann. N. Y. Acad. Sci. 990:678-684. [DOI] [PubMed] [Google Scholar]
- 16.McBride, J. W., R. E. Corstvet, E. B. Breitschwerdt, and D. H. Walker. 2001. Immunodiagnosis of Ehrlichia canis infection with recombinant proteins. J. Clin. Microbiol. 39:315-322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.McBride, J. W., R. E. Corstvet, S. D. Gaunt, C. Boudreaux, T. Guedry, and D. H. Walker. 2003. Kinetics of antibody response to Ehrlichia canis immunoreactive proteins. Infect. Immun. 71:2516-2524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.McBride, J. W., R. E. Corstvet, S. D. Gaunt, J. Chinsangaram, G. Y. Akita, and B. I. Osburn. 1996. PCR detection of acute Ehrlichia canis infection in dogs. J. Vet. Diagn. Investig. 8:441-447. [DOI] [PubMed] [Google Scholar]
- 19.McBride, J. W., C. K. Doyle, X. F. Zhang, A. M. Cardenas, V. L. Popov, K. A. Nethery, and M. E. Woods. 2007. Identification of a glycosylated Ehrlichia canis 19-kilodalton major immunoreactive protein with a species-specific serine-rich glycopeptide epitope. Infect. Immun. 75:74-82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.McBride, J. W., L. M. Ndip, V. L. Popov, and D. H. Walker. 2002. Identification and functional analysis of an immunoreactive DsbA-like thio-disulfide oxidoreductase of Ehrlichia spp. Infect. Immun. 70:2700-2703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.McBride, J. W., X. J. Yu, and D. H. Walker. 2000. A conserved, transcriptionally active p28 multigene locus of Ehrlichia canis. Gene 254:245-252. [DOI] [PubMed] [Google Scholar]
- 22.Munodzana, D., T. F. McElwain, D. P. Knowles, and G. H. Palmer. 1998. Conformational dependence of Anaplasma marginale major surface protein 5 surface-exposed B-cell epitopes. Infect. Immun. 66:2619-2624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nethery, K. A., C. K. Doyle, B. L. Elsom, N. K. Herzog, V. L. Popov, and J. W. McBride. 2005. Ankyrin repeat containing immunoreactive 200 kD glycoprotein (gp200) orthologs of Ehrlichia chaffeensis and Ehrlichia canis are translocated to the nuclei of infected monocytes, abstr. O-60. 4th Int. Conf. Rickettsiae Rickettsial Dis., Longrono, Spain.
- 24.Ohashi, N., A. Unver, N. Zhi, and Y. Rikihisa. 1998. Cloning and characterization of multigenes encoding the immunodominant 30-kilodalton major outer membrane proteins of Ehrlichia canis and application of the recombinant protein for serodiagnosis. J. Clin. Microbiol. 36:2671-2680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ohashi, N., N. Zhi, Y. Zhang, and Y. Rikihisa. 1998. Immunodominant major outer membrane proteins of Ehrlichia chaffeensis are encoded by a polymorphic multigene family. Infect. Immun. 66:132-139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Park, J., K. J. Kim, K. S. Choi, D. J. Grab, and J. S. Dumler. 2004. Anaplasma phagocytophilum AnkA binds to granulocyte DNA and nuclear proteins. Cell. Microbiol. 6:743-751. [DOI] [PubMed] [Google Scholar]
- 27.Popov, V. L., X. Yu, and D. H. Walker. 2000. The 120 kDa outer membrane protein of Ehrlichia chaffeensis: preferential expression on dense-core cells and gene expression in Escherichia coli associated with attachment and entry. Microb. Pathog. 28:71-80. [DOI] [PubMed] [Google Scholar]
- 28.Rikihisa, Y., S. A. Ewing, and J. C. Fox. 1994. Western immunoblot analysis of Ehrlichia chaffeensis, E. canis, or E. ewingii infections in dogs and humans. J. Clin. Microbiol. 32:2107-2112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sumner, J. W., K. G. Sims, D. C. Jones, and B. E. Anderson. 1993. Ehrlichia chaffeensis expresses an immunoreactive protein homologous to the Escherichia coli GroEL protein. Infect. Immun. 61:3536-3539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Winslow, G. M., E. Yager, and J. S. Li. 2003. Mechanisms of humoral immunity during Ehrlichia chaffeensis infection. Ann. N. Y. Acad. Sci. 990:435-443. [DOI] [PubMed] [Google Scholar]
- 31.Winslow, G. M., E. Yager, K. Shilo, E. Volk, A. Reilly, and F. K. Chu. 2000. Antibody-mediated elimination of the obligate intracellular bacterial pathogen Ehrlichia chaffeensis during active infection. Infect. Immun. 68:2187-2195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yager, E., C. Bitsaktsis, B. Nandi, J. W. McBride, and G. Winslow. 2005. Essential role for humoral immunity during Ehrlichia infection in immunocompetent mice. Infect. Immun. 73:8009-8016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yu, X. J., P. Crocquet-Valdes, L. C. Cullman, and D. H. Walker. 1996. The recombinant 120-kilodalton protein of Ehrlichia chaffeensis, a potential diagnostic tool. J. Clin. Microbiol. 34:2853-2855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yu, X. J., P. Crocquet-Valdes, and D. H. Walker. 1997. Cloning and sequencing of the gene for a 120-kDa immunodominant protein of Ehrlichia chaffeensis. Gene 184:149-154. [DOI] [PubMed] [Google Scholar]
- 35.Yu, X. J., J. W. McBride, C. M. Diaz, and D. H. Walker. 2000. Molecular cloning and characterization of the 120-kilodalton protein gene of Ehrlichia canis and application of the recombinant 120-kilodalton protein for serodiagnosis of canine ehrlichiosis. J. Clin. Microbiol. 38:369-374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yu, X. J., J. W. McBride, X. F. Zhang, and D. H. Walker. 2000. Characterization of the complete transcriptionally active Ehrlichia chaffeensis 28 kDa outer membrane protein multigene family. Gene 248:59-68. [DOI] [PubMed] [Google Scholar]
- 37.Zhang, J. Z., H. Guo, G. M. Winslow, and X. J. Yu. 2004. Expression of members of the 28-kilodalton major outer membrane protein family of Ehrlichia chaffeensis during persistent infection. Infect. Immun. 72:4336-4343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhang, Y., N. Ohashi, E. H. Lee, A. Tamura, and Y. Rikihisa. 1997. Ehrlichia sennetsu groE operon and antigenic properties of the GroEL homolog. FEMS Immunol. Med. Microbiol. 18:39-46. [DOI] [PubMed] [Google Scholar]