Abstract
Salmonella pathogenicity island 4 (SPI4) encodes a type I secretion system and the cognate substrate protein, SiiE. We have recently demonstrated that SiiE is a giant nonfimbrial adhesin involved in the adhesion of Salmonella enterica serovar Typhimurium to polarized epithelial cells. We also observed that under in vitro culture conditions, the synthesis and secretion of SiiE coincided with the activation of Salmonella invasion genes. These observations prompted us to investigate the regulation of SPI4 genes in detail. A novel approach for the generation of reporter gene fusions was employed to generate single-copy chromosomal fusions to various genes within SPI4, and the expression of these fusions was investigated. We analyzed the regulation of SPI4 genes and the roles of various regulatory systems for SPI4 expression. Our data show that the expression of SPI4 genes is coregulated with SPI1 invasion genes by the global regulator SirA. Expression of a SPI4 gene was also reduced in the absence of HilA, the central local regulator of SPI1 gene expression. Both SirA and HilA functions were required for the secretion of SiiE and the SPI4-mediated adhesion. Our data demonstrate that SPI4-mediated adhesion, as well as SPI1-mediated invasion, are tightly coregulated by the same regulatory circuits and induced under similar environmental conditions.
The pathogenesis of infections caused by Salmonella enterica involves the adaptation to and colonization of various environments within the infected host organism. S. enterica is a facultative intracellular pathogen that can colonize the intestinal lumen but also invades eukaryotic cells and subsequently proliferates within the “Salmonella-containing vacuole,” a membrane-bound intracellular niche. Successful pathogenesis of S. enterica requires rapid adaptation to these highly distinct environments and the coordinated regulation of basic metabolic functions, as well as sophisticated virulence determinants (reviewed in reference 34).
The transcriptional control of virulence gene expression in S. enterica is multifactorial and involves various levels of regulation (33). Virulence genes involved in invasion and intestinal pathogenesis, as well as virulence genes required for the intracellular lifestyle and systemic pathogenesis, are clustered in two complex regulons. Salmonella pathogenicity island 1 (SPI1) encodes a type III secretion system (T3SS) that mediates the translocation of effector proteins for the invasion of nonphagocytic cells by Salmonella (32). The expression of genes within SPI1 and of multiple loci outside of SPI1 that encode translocated effector proteins is under the control of the local regulator InvF (5). Within SPI1, the genes hilA, hilC, and hilD encode regulators, and HilA acts as a central regulator of the HilA regulon (4). SPI2 encodes a second T3SS, and effector proteins have a crucial role in the intracellular lifestyle and systemic pathogenesis of Salmonella (21). Similar to SPI1, there is a complex regulon consisting of genes within and further loci outside of SPI2 under the control of the SPI2-encoded two-component system SsrAB (16, 26, 36). Both, the HilA regulon and the SsrAB regulon are under the control of global regulatory systems that are also present in nonpathogenic enterobacteria. The HilA regulon is controlled by the two-component system BarA/SirA (1) and modulated by a large number of further regulatory systems (25). PhoPQ is a global regulator of more than 200 genes, and in S. enterica, this system is involved in adaptation to extra- and intracellular environments during infection (15).
In contrast to SPI1 and SPI2, the functions of other SPIs in Salmonella virulence is less well characterized. We have recently determined that SPI4 encodes a type I secretion system (T1SS) and SiiE, a large repetitive protein. SiiE is the substrate of the SPI4 T1SS and functions as a nonfimbrial adhesin in binding to epithelial cell surfaces (14, 28). Previous studies identified SPI4 as a factor for the intestinal colonization of calves (29), as well as for establishing persistent infections in the mouse model (22). During the analyses of secretion of SiiE and the functional characterization of the SPI4 T1SS, we observed that this secretion system is activated during culture growth in rich media at the late exponential growth phase. These growth conditions also activate the SPI1-dependent invasion functions. Interestingly, genes with proposed regulatory functions have not been identified within the SPI4 locus, which contains only six open reading frames (ORFs). SiiC, SiiD, and SiiF, i.e., the TolC-like outer membrane protein, the membrane fusion protein, and the transport ATPase, respectively, are the classical components of the T1SS. SiiA and SiiB are two additional SPI4-encoded proteins with unknown functions, but neither protein is required for the secretion of SiiE by the SiiCDF secretion system. Furthermore, we did not observe an effect of mutations in SiiA or SiiB on the expression of SiiE, suggesting that both proteins are not required for expression of other SPI4 genes (14).
A possible link between the regulation of SPI1 and SPI4 has been reported in a study by Ahmer et al. (1), who identified reporter fusions within SPI4 whose expression was dependent on the function of HilA, as well as SirA.
Based on extended knowledge of the genetic organization of SPI4 genes and the identification of SPI4 as an adhesion factor, we performed a comprehensive analysis of the regulation of SPI4 genes. In this study, we also made use of the recently developed Red reporter technique that allowed the generation of single-copy reporter fusions at precisely defined positions in the Salmonella chromosome (13). Our data indicate that SPI1 and SPI4 are coregulated, suggesting a synergistic function of both virulence systems during intestinal colonization and interaction with the intestinal mucosa.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
Salmonella enterica serovar Typhimurium NCTC 12023 was used as the wild-type (WT) strain, and all other strains used were isogenic derivatives of this strain. The strains used in this study are listed in Table 1. Bacterial strains were routinely grown in LB broth (1% yeast extract [BD], 0.5% Bacto-Tryptone [BD], 0.5% NaCl) or on LB plates containing antibiotics if required for the selection of plasmids. The minimal media PCN and PCN−P have been described previously (24) and were adjusted to pH 7.4 or pH 5.8 as indicated. For the analyses of secretion and reporter gene activities, bacterial strains were cultured overnight in glass test tubes with agitation at 37°C. If not otherwise stated, 100-ml baffled flasks containing 20 ml of prewarmed LB broth were inoculated 1:50 with the overnight cultures. The flasks were incubated on a rotary shaker at 150 rounds per minute at 37°C, and culture samples were collected at various time points after inoculation.
TABLE 1.
S. enterica serovar Typhimurium strains and plasmids used in this study
| Designation | Relevant characteristic(s) | Source or reference |
|---|---|---|
| NCTC 12028 | WT | Laboratory stock |
| MvP589 | ΔSPI4 FRT | 14 |
| MvP812 | ΔsiiF FRT | 14 |
| GVB311 | rpoE | 19 |
| SF1005 | rpoS | 12 |
| CS224 | sirA::Tn10 | 1 |
| CS015 | phoP::Tn10d-cam | 27 |
| CS022 | pho-24 | 27 |
| P4H2 | hilA::mTn5 | 8 |
| MvP531 | ΔssrB::aph | This study |
| MvP532 | ΔssrB FRT; aph deleted from MvP531 | This study |
| MvP568 | ΔslyA::aph | This study |
| MvP575 | ΔslyA FRT, aph deleted from MvP568 | This study |
| MvP586 | ΔompR/envZ FRT | This study |
| MvP608 | siiA::luc aph | This work |
| MvP609 | siiE::luc aph | This work |
| MvP610 | siiF::luc aph | This work |
| MvP618 | siiA::luc FRT, aph deleted from MvP608 | This work |
| MvP623 | siiA::luc FRT sirA::Tn10 | This work |
| MvP624 | siiA::luc aph ΔompR/envZ FRT | This work |
| MvP625 | siiA::luc aph ΔssrB FRT | This work |
| MvP626 | siiA::luc rpoE | This work |
| MvP627 | siiA::luc aph ΔslyA FRT | This work |
| MvP633 | siiA::luc rpoS | This work |
| MvP634 | siiA::luc FRT hilA::mTn5 | This work |
| MvP666 | sopE2::luc aph | This study |
| MvP690 | sopE2::luc aph ΔSPI4 FRT | This study |
| MvP746 | ΔinvF::aph | This study |
| MvP749 | sopE2::luc FRT, aph deleted from MvP666 | This study |
| MvP753 | sopE2::luc FRT sirA::Tn10 | This study |
| MvP754 | sopE2::luc FRT sirA::Tn10 | This study |
| MvP759 | ΔrfaH::aph | This study |
| MvP760 | ΔrfaH FRT, aph deleted from MvP759 | This study |
| MvP761 | ΔinvF FRT, aph deleted from MvP746 | This study |
| MvP848 | ΔhilA::aph | This study |
| MvP850 | ΔhilA FRT, aph deleted from MvP848 | This study |
| MvP889 | sipA::luc aph | This study |
| MvP971 | sipA::luc aph sirA::Tn10 | This study |
| MvP972 | sipA::luc aph ΔhilA FRT | This study |
| MvP974 | rtsAB::aph | This study |
| MvP976 | siiA::luc aph pho-24 | This study |
| MvP984 | siiA::luc aph phoP::Tn10d-cam ΔslyA FRT | This study |
| MvP988 | rtsAB::FRT, aph deleted from MvP974 | This study |
| MvP1030 | siiA::luc aph ΔrtsAB FRT | This study |
| Plasmids | ||
| pVV214 | hilA cloned in pACYC177, Ampr | 4 |
| pM2011 | sirA cloned in pBAD24, Ampr | W. D. Hardt, ETH Zürich, Switzerland |
Generation of reporter constructs and analysis of expression.
Reporter strains harboring firefly luciferase in various positions in SPI4 were constructed by a modification of the Red-mediated recombination technique (6) that was described recently (13). Briefly, template plasmid p3121 harbors a cassette consisting of the promoterless firefly luciferase gene and the aph resistance gene flanked by FLP recombination target (FRT) sites. p3121 was used as a template to amplify by PCR a linear targeting construct that was used for transformation of S. enterica serovar Typhimurium harboring pKD46. A cassette consisting of luc and aph was inserted directly at the start codons of various genes. Proper insertion was confirmed by PCR and DNA sequencing, and if required, the aph cassette was removed by FLP-mediated recombination as described previously (6). This approach resulted in the generation of single-copy reporter fusions at various positions in SPI4, as well as to sopE2, sipA, and gyrA.
Reporter fusions were also assayed in the background of various strains deficient in specific regulatory systems. Mutant strains with ssrB, slyA, ompR/envZ, and hilA deleted were constructed by Red-mediated deletion as previously described. For further strain construction, the mutant alleles or reporter fusions were moved into the appropriate background by P22 transduction. The resulting strains are listed in Table 1.
Luciferase assay.
For quantification of expression, reporter strains were grown under various culture conditions, and aliquots of cultures were processed for assay of luc activity as previously described (7). All assays were done in triplicate and repeated at least three times on independent occasions.
Reverse transcription-PCR.
Bacterial cultures were grown in LB broth for 3.5 or 16 h, and cells were harvested by centrifugation for 5 min at maximal speed in a microcentrifuge at 4°C. The cell pellet was resuspended in RNA Protection Reagent (QIAGEN, Hilden, Germany) and incubated for 5 min at room temperature (RT). After centrifugation for 10 min at 5,000 × g, the supernatant was removed and the pellet was resuspended in 100 ml Tris-EDTA buffer containing 400 μg/ml lysozyme. The subsequent isolation of RNA with the RNeasy Kit (QIAGEN) was performed basically as described by the manufacturer. RNA recovery was quantified by determination of the optical density at 260 nm (OD260)/OD280. Aliquots containing 10 μg RNA were treated with RNase-free DNase (MBI Fermentas) to remove contaminating DNA. Subsequently, reverse transcription was performed using the cDNA synthesis kit (MBI Fermentas) with random hexamer primers as described by the manufacturer. Control reactions were performed without the addition of reverse transcriptase. Subsequently, PCR was performed using oligonucleotides listed in Table 2 and a program consisting of incubation at 95°C for 4 min 30 s and 33 cycles of 95°C for 30 s, 60°C for 30 s, and 72°C for 30 s. The reaction mixtures were analyzed by electrophoresis on 2% agarose gels. For image acquisition, a Kodak EDAS290 system was used with subsequent processing using Adobe Photoshop CS2.
TABLE 2.
Oligonucleotides used in this study
| Designation | Sequence |
|---|---|
| Red-mediated deletions | |
| SsrB-Red-Del-For | 5′-TATTATCTTAATTTTCGCGAGGGCAGCAAAATGAAAAATGGTGTAGGCTGGAGCTGCTTC-3′ |
| SsrB-Red-Del-Rev | 5′-CTCATCAAAATATGACCAATGCTTAATACCATCGGACGCCCCTGGCATATGAATATCCTCCTTA-3′ |
| SlyA-Red-Del-For | 5′-ACTGAAGCTACAGGTGCCAAGTGCGCAGCAAGCTAATTATGTGTAGGCTGGAGCTGCTTC-3′ |
| SlyA-Red-Del-Rev | 5′-TAAACCAGGCTTTACGTGTGGTCACATGGCCACACGTATGCATATGAATATCCTCCTTAG-3′ |
| OmpR-Red-Del-For | 5′-TACAATTTGTTGCGAACCTTTGGGAGTACAGACAATGCAAGTGTAGGCTGGAGCTGCTTC-3′ |
| EnvZ-Red-Del-Rev | 5′-GAGAAGAAAGGGAGGGTAATACCTCCCTTTCTTATGCCTCCATATGAATATCCTCCTTAG-3′ |
| HilA-Red-Del-For | 5′-AAGAGAATACACTATTATCATGCCACATTTTAATCCTGTTATTCCGGGGATCCGTCGACC-3′ |
| HilA-Red-Del-Rev | 5′-GGGGATCCTGTTTCCATCTTTTGAACCAAATATTGTCTTCTGTAGGCTGGAGCTGCTTCG-3′ |
| RtsAB-Red-Del-For | 5′-ATTTTTCAGAATAATTTTGATTTACACGCACATTTAATAAATTCCGGGGATCCGTCGACC-3′ |
| RtsAB-Red-Del-Rev | 5′-GCCTTGCCTACCACTCTACCAACATTTTAGGAAAAATTACTGTAGGCTGGAGCTGCTTCG-3′ |
| InvF-Red-Del-For | 5′-TGATTTTTAACTGGTGCTGACAACTATGCTAAATACGCAGGTGTAGGCTGGAGCTGCTTC-5′ |
| InvF-Red-Del-Rev | 5′-GTCTTCATTTGTCTGCCAATTGAATAATATTTGATAATTTACTATGAATATCCTCCTTAG-3′ |
| RfaH-Red-Del-For | 5′-AACTCTGGCAACCAACGCTAATCCAGATACGGCTTAAAGGGTGTAGGCTGGAGCTGCTTC-3′ |
| RfaH-Red-Del-For | 5′-GCCAGAACCTTATTTGAGGTTGTATTCTGAACGATCGCTACATATGAATATCCTCCTTAG-3′ |
| Red-mediated reporter fusions | |
| SiiA-Red-Luc-For | 5′-ACAAAAACATTTTATTCACAATGTAATATCAGGAGACAACATGGAAGACGCCAAAAACATAA-3′ |
| SiiA-Red-Rep-Rev | 5′-AACAGACAAACAAATAGCGGTAATGATTTATATATTTCACGTGTAGGCTGGAGCTGCTTC-3′ |
| SiiE-Red-Luc-For | 5′-ACTAAACGATTAGATACACCTTGAGAGTGAATATAATATTATGGAAGACGCCAAAAACATAA-3′ |
| SiiE-Red-Rep-Rev | 5′-GTAAACCCCCTCACCCAAAGGTGAGGGGCCTCCGTTATTAGTGTAGGCTGGAGCTGCTTC-3′ |
| SiiE-Red-Luc-For | 5′-ATAACGGAGGCCCCTCACCTTTGGGTGAGGGGGTTTACTTATGGAAGACGCCAAAAACATAA-3′ |
| SiiE-Red-Rep-Rev | 5′-CAGTACCACCTGATAACAGCGACAAGCGCTGCTTATTTTAGTGTAGGCTGGAGCTGCTTC-3′ |
| SopE2-Red-Luc-For | 5′-TATAGAAAATATTGCGAATAAGTATCTTCAGAATGCCTCCGAAGACGCCAAAAACATAAGAA-3′ |
| SopE2-Red-Rep-Rev | 5′-GCAAACCAGCGCCAATGCAGGTGTCGTTACCGTGGCGACCCGTGTAGGCTGGAGCTGCTTC-3′ |
| SipA-SDo-Luc-For | 5′-GGTTATTACTACCGTTGATGGCTTGCACATGCAGCGTTAACTAGGAGGATCAGCTATGGAAGACGCCAAAAACATA-3′ |
| SipA-Red-Rep-Rev | 5′-TCACTTTTTTGACTCTTGCTTCAATATCCATATTCATCGCCGTGTAGGCTGGAGCTGCTTC-3′ |
| RT PCR | |
| SiiA-Op-for | 5′-ATTGGTAGCAGGAAGCCAAG-3′ |
| SiiA-Op-rev | 5′-GCTCCGCTATTTTGAGCAGT-3′ |
| SiiAB-Op-For | 5′-TCCGCAGCATCAAGTACAAC-3′ |
| SiiAB-Op-Rev | 5′-TTTCTACTGGCGTCCTGAGC-3′ |
| SiiBC-Op-For | 5′-TGGGCTGAGAAAAGAAATGG-3′ |
| SiiBC-Op-Rev | 5′-AACAGCAACAGAGGGCTGAT-3′ |
| SiiCD-Op-For | 5′-GCGGAACGTTCACATACAAA-3′ |
| SiiCD-Op-Rev | 5′-CCTGACCATGAACCACTGAA-3′ |
| SiiDE-Op-For | 5′-TACCGGAGGAACAATTCAGC-3′ |
| SiiDE-Op-Rev | 5′-GAGAAACTTTTGCGCCTTTG-3′ |
| SiiEF-Op-For | 5′-ACTCCTCGGCGGATATTGT-3′ |
| SiiEF-Op-Rev | 5′-GCCGTTTCCGCACTTAAATA-3′ |
| SiiF-Op-for | 5′-AGTATTTTGACCGGGTCCTG-3′ |
| SiiF-Op-rev | 5′-GAAGCCGTAATGGTCGAAAC-3′ |
| Northern blot probes | |
| SiiA-probe-for | 5′-CAAAATAGCGGAGCCAATGA-3′ |
| SiiA-probe-rev | 5′-GGGAGTATGGCAAGGAAAAA-3′ |
| SiiF-probe-for | 5′-ACTCCTCGGCGGATATTGT-3′ |
| SiiF-probe-rev | 5′-CCACTCACACACACCATCCA-3′ |
Northern blotting.
Bacterial cultures were grown in LB broth for 3.5 h, and cells were harvested by centrifugation for 5 min at maximal speed in a microcentrifuge at 4°C. The cell pellet was resuspended in RNA Protection Reagent (QIAGEN, Hilden, Germany) and incubated for 5 min at RT. After centrifugation for 10 min at 5,000 × g, the supernatant was removed and the pellet was resuspended in 100 μl Tris-EDTA buffer containing 400 μg/ml lysozyme. The subsequent isolation of RNA with the NucleoSpin RNA II kit (Macherey-Nagel, Düren, Germany) was performed basically as described by the manufacturer. The RNA was eluted with 92 μl RNase-free water and subsequently treated with TURBO DNase (Ambion, Applied Biosystems, Darmstadt, Germany). The RNA was further purified using the NucleoSpin RNA Clean Up kit (Macherey-Nagel) according to the manufacturer's instructions, eluted with 62 μl RNase-free water, and quantified by determination of the OD260/OD280. The RNA was ethanol precipitated, and the pellets were stored at −70°C. Northern blotting was carried out using the NorthernMax kit (Ambion). The blots were hybridized overnight with [32P]CTP-labeled DNA probes (Ready-To-Go labeling beads; GE Healthcare, Freiburg, Germany) derived from PCR products (the oligonucleotides are listed in Table 2). The blots were exposed to Hyperfilm (GE Healthcare) for at least 24 h.
Adhesion assays.
MDCK cells were seeded in 24-well plates (Greiner bio-one Cellstar) at a density of 1 × 105 cells per well. The cells were allowed to differentiate to polarized cell layers for 4 to 10 days in minimal essential medium (PAA, Cölbe, Germany) supplemented with 10% fetal calf serum (FCS), nonessential amino acids, and penicillin/streptomycin. The medium was changed to antibiotic-free medium 4 h before infection.
Bacteria were subcultured (1:30 from overnight cultures) in LB broth with appropriate antibiotics for 3.5 h and adjusted to an OD600 of 0.2 (adhesion assays) or an OD600 of 0.6 (immunofluorescence). For complementation of sirA with plasmid pM2011, expression was induced for 1 h with 0.01% arabinose or expression was repressed by the addition of 2% glucose for subcultivation. A master mixture of the inoculum was prepared in minimal essential medium (without penicillin/streptomycin), and the cells were infected for 25 min at a multiplicity of infection of 5 (adhesion assays) or 60 (immunofluorescence). The cells were washed with phosphate-buffered saline (PBS) five times and lysed with 500 μl prewarmed 0.1% Triton X-100 in PBS for 5 min at 37°C. Serial dilutions were made in prechilled PBS and plated on Mueller-Hinton plates (for viable-count experiments).
Quantification of SiiE by ELISA.
Antisera were raised in rabbits against the recombinant C-terminal moiety of SiiE (14). For detection of SiiE, culture supernatants were filter sterilized (0.45-μm Corning syringe filters), and aliquots of 50 μl were directly applied to 96-well polystyrene microtiter plates (Nunc MultiSorp) overnight at 4°C in a humid chamber. The plates were washed three times with 200 μl/well of PBS supplemented with 0.05% Tween 20 (PBS-Tween), and the rabbit anti-glutathione S-transferase-SiiE-C detection antibody diluted 1:1,000 in PBS plus 10% inactivated FCS (PBS-FCS) was applied for 2 h at RT. After five washes with PBS-Tween, 100 μl of the anti-rabbit horseradish peroxidase-coupled secondary antibody diluted 1:2,500 in PBS-FCS was added to each well for 30 min at RT. The wells were washed again seven times with PBS-Tween, and 50 μl of enzyme-linked immunosorbent assay (ELISA) horseradish peroxidase substrate (BD 555214) was added. After incubation in the dark at RT for 8 to 15 min, the reaction was stopped by the addition of 25 μl/well 1 M H3PO4. The OD450 was measured using a Dynatech MR5000 plate reader equipped with an appropriate filter set.
Immunofluorescence analyses.
The detection of SiiE on the surfaces of bacteria adhering to MDCK cells was performed basically as described before (14). Salmonella was detected using rabbit antiserum against lipopolysaccharide (LPS) O4 (BD, Heidelberg, Germany), and surface-associated SiiE was detected using a polyclonal antiserum raised against SiiE in mice. After permeabilization with 0.1% saponin, the actin cytoskeleton was labeled with a phalloidin-Texas Red conjugate (Molecular Probes, Invitrogen). For image acquisition, a Zeiss Axiovert 200 equipped with a Zeiss Axiocam MRm and filter sets for green fluorescent protein, Texas Red, and Cy5 was used. For image processing, the Zeiss Axiovision 4.5 package was used.
RESULTS
Transcriptional organization of SPI4 genes.
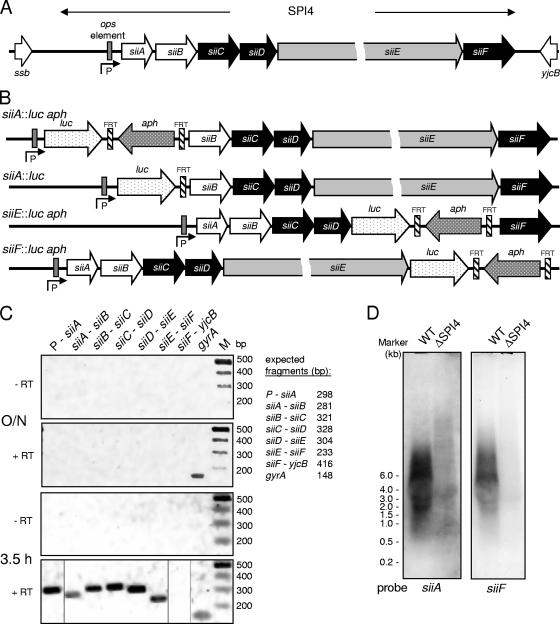
We previously defined the function of genes in SPI4 as encoding a T1SS for the secretion of the giant adhesin SiiE (14). SPI4 harbors six genes encoding the T1SS, SiiE, and proteins of unknown function (Fig. 1A). SiiE was secreted by Salmonella strains during growth in LB broth in the late exponential phase of culture. These culture conditions also induce the secretion of proteins involved in Salmonella invasion. We set out to investigate the regulation of SPI4 genes in more detail and constructed a set of strains harboring reporter gene fusions. For the strain construction, a novel approach was applied for “recombineering” of fusions to the reporter luciferase in single copies in the chromosome (13). This approach allowed the precise replacement of chromosomal genes by the reporter gene. All fusions were constructed in such a way that the second codon of the reporter was fused to the start codon of the gene under study. Thus, these reporter fusions measured the transcriptional activity (e.g., promoter strength) and the quality of ribosome binding sites. The positions of the reporter genes within SPI4 are indicated in Fig. 1B.
FIG. 1.
Genetic and transcriptional organization of SPI4 genes. (A) SPI4 is a 23-kb locus consisting of six ORFs. siiCDF encode a T1SS for the secretion of the giant nonfimbrial adhesin SiiE. siiAB are not required for secretion and have unknown functions. P indicates the putative promoter of siiA to -F. An ops element has been identified in the promoter region (29). (B) Generation of reporter gene fusions in SPI4. The firefly luciferase (luc) reporter gene was introduced at various positions in SPI4 by a modification of the Red recombinase approach. If appropriate, the aph resistance cassette was removed by FLP-mediated recombination between FRT sites. (C) Transcriptional organization of SPI4 genes. WT Salmonella was grown in LB broth overnight (O/N) or for 3.5 h to late exponential phase, and total RNA was extracted. The RNA preparations were treated with DNase and subjected to reverse transcription (+ RT) if indicated. PCR was performed with various primer pairs, and the presence of amplification products was analyzed by agarose gel electrophoresis. (D) Detection of SPI4 transcripts by Northern blot analysis. WT Salmonella and the SPI4 deletion strain were subcultured for 3.5 h to late exponential phase. Total RNA was extracted, separated by agarose gel electrophoresis, and transferred to membranes. Hybridization was performed with radiolabeled probes complementary to siiA or siiF as indicated. The positions of RNA size markers are indicated.
Upstream of siiA, a large noncoding region is located. We have previously demonstrated that a DNA fragment comprising 720 bp of the region upstream of siiA has promoter activity (14). This promoter was used to activate the expression of individual sii genes for complementation of deletions in SPI4. Within the promoter region, an operon polarity suppressor (ops) element has been identified (29). ops elements are rare sequence motifs found in promoters of large operons (3). Further upstream, ssb is located, a gene that is conserved between S. enterica and Escherichia coli and thus is not part of the SPI4 locus. Sequence analyses indicated that the ORFs for siiB, siiC, and siiD overlap. siiA and siiB, siiD and siiE, and siiE and siiF are separated by noncoding regions of 107, 16, and 39 bp, respectively. This genetic organization would allow the presence of several transcriptional units within SPI4. For an experimental analysis of the transcriptional organization, reverse transcription-PCR analyses were performed (Fig. 1C). While the detection of transcripts was not possible if bacteria were used after growth overnight in LB broth, specific PCR products were obtained with RNA extracted from bacteria after culture for 3.5 h in LB broth. The transcripts for siiA to siiF were linked. No linkage between siiF and ORFs further downstream was detected.
We also performed Northern blot analyses for the detection of SPI4 transcripts (Fig. 1D). The use of probes specific for siiA and siiF resulted in hybridization signals in similar size ranges, suggesting the existence of one large transcript for siiABCDEF. However, precise determination of the sizes of the hybridizing bands was not possible due to the lack of suitable size markers. It should be noted that Northern blot analyses of very large bacterial mRNAs, such as the putative 23.5-kb transcript for SPI4, is rather uncommon.
Taken together, these data indicate that the six SPI4 genes form an operon and are likely to be cotranscribed as a large mRNA.
SPI4 genes are coexpressed with invasion genes of the HilA regulon.
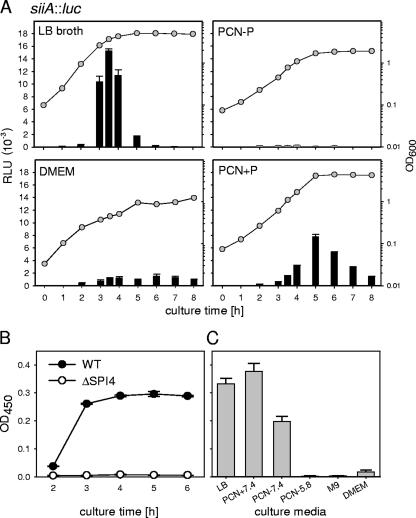
We next defined the in vitro conditions for the expression of SPI4 genes. A strain harboring a siiA::luc fusion was grown in various culture media previously reported to affect the expression of Salmonella virulence genes, and luciferase activities were quantified at various time points during culture (Fig. 2A). Only a very low level of reporter activity was observed if bacteria were cultured in PCN−P minimal medium at pH 5.8, conditions that induce the expression of SPI2 genes required for intracellular pathogenesis (24), as well as in M9 minimal medium (data not shown) or Dulbecco's modified Eagle medium for eukaryotic-cell culture. High levels of reporter activity were detected in PCN minimal medium adjusted to pH 7.4 and in LB broth. In both media, the highest activity was observed in the late exponential growth phase prior to entering the stationary phase. The reporter activity dropped rapidly after entrance into the stationary phase in LB broth, and the decrease was slower in PCN+P medium. As a complementary approach to assay the synthesis of a SPI4-encoded protein and the function of the SPI4-encoded T1SS, the secretion of SiiE was analyzed (Fig. 2B). An ELISA was developed for the quantification of SiiE secreted into the culture media. Secreted SiiE was detectable in the supernatants of WT Salmonella after 3 h of culture growth, and similar amounts of secreted SiiE were detected at later time points (Fig. 2B) and in the supernatants of overnight cultures (data not shown). No secreted SiiE was detected in the supernatant of the SPI4-deficient strain (Fig. 2B) or a siiF strain that is defective in the SPI4-encoded T1SS but expresses siiE (data not shown). We also analyzed the amounts of secreted SiiE after overnight culture in various media and observed that the largest amounts of SiiE were secreted during growth in LB broth or PCN+P medium at pH 7.4 (Fig. 2C).
FIG. 2.
Expression of siiA and secretion of SiiE during culture of S. enterica serovar Typhimurium in various growth media. (A) The reporter strain MvP618 harboring the siiA::luc fusion was grown overnight in LB broth. Various culture media, as indicated, were inoculated with 1/30 volume of the overnight culture, and bacteria were cultured in baffled glass flasks at 37°C with aeration. The culture growth was recorded by measurement of the OD600 (circles). Samples were taken at various time points during culture and processed for quantification of luciferase activity. Means for relative light units (RLU) and standard deviations are shown (bars). (B) WT S. enterica serovar Typhimurium or a SPI4 deletion strain was grown in LB broth, and samples were collected at various time points during culture. The bacterial cells were removed by centrifugation, and the culture supernatants were passed through 0.45-μm filters to remove residual bacteria. An ELISA was used to determine the relative amounts of secreted SiiE, and means and standard deviations of OD450 readings are given for triplicate samples. (C) The WT strain was grown for 14 h in various culture media as indicated (LB, Luria broth; PCN + 7.4, minimal medium with 25 mM Pi, pH 7.4; PCN-7.4, minimal medium with 0.4 mM Pi, pH 7.4; PCN-5.8, minimal medium with 0.4 mM Pi, pH 5.8; M9, minimal medium; DMEM, Dulbecco's modified Eagle medium, cell culture medium), and the secretion of the adhesin SiiE was analyzed by ELISA as described for panel B.
Based on these observations, we used growth in LB broth as the standard condition to study the expression of SPI4 genes. Since the expression of the siiA::luc fusion showed a rather sharp peak between 3 and 4 h of subculture and rapidly decreased at later time points, we decided to perform kinetic analyses of expression in all subsequent reporter assays.
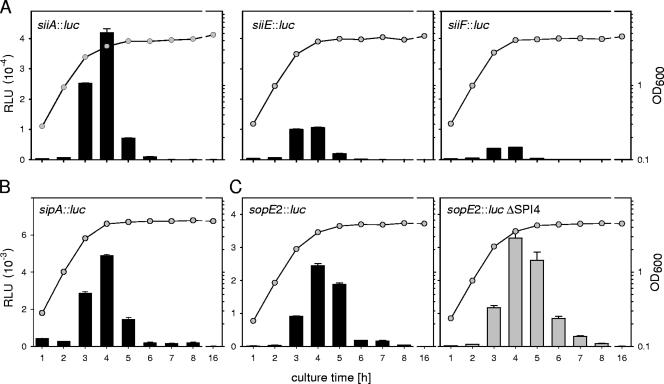
The expression levels of genes in various positions in SPI4 were compared. Strains harboring luciferase fusions to siiA, siiE, or siiF were subcultured in LB broth, and the reporter activities were quantified (Fig. 3A). The siiA fusion was expressed at the highest level compared to siiE, and siiF was expressed at the lowest level. The kinetics of expression were comparable for all fusions, with the highest activity levels between 3 and 4 h of subculture in LB broth. Since the fusions were generated in identical fashions, direct comparison of expression levels was possible and indicated that the maximal expression levels of siiE and siiF were 4.0-fold and 10.4-fold lower, respectively, than the expression of siiA.
FIG. 3.
Expression levels of siiA, siiE, and siiF. (A) Reporter strains harboring luciferase fusions to siiA, ssiE, or siiF were grown overnight in LB broth. Aliquots of the cultures were used to inoculate fresh LB broth, and the cultures were grown at 37°C with aeration. The bacterial growth was recorded by reading the OD600 (circles). At various time points, culture samples were taken and processed for quantification of luc activities (bars). RLU, relative light units. The error bars indicate standard deviations. (B) Strains harboring the sopE2::luc fusion in the background of WT S. enterica serovar Typhimurium or the SPI4 deletion strain were generated and assayed as described for panel A.
Previous studies showed that the expression of SPI1 genes required for the invasion of host cells is induced during growth in LB broth and that bacteria in the late exponential growth phase show the highest invasiveness of epithelial cells (23). These observations indicate similar expression patterns of SPI1 and SPI4 genes. Since most published data on the expression of SPI1 genes were generated with β-galactosidase reporter fusions and therefore are not directly comparable, we constructed single-copy luc fusions to sipA and sopE2. sipA and sopE2 are representative members of the HilA regulon located within SPI1 and on a separate locus, respectively. SipA and SopE2 are translocated effector proteins of the SPI1-encoded T3SS. The kinetics of expression of the sipA::luc and sopE2::luc fusions were similar to those of the SPI4 fusions and showed a pronounced peak at 4 h of culture and a rapid decrease of reporter activity at later time points (Fig.3BC).
We also tested if the expression of sopE2 was affected by SPI4. In the background of a strain with the entire SPI4 locus deleted, the sopE2 fusion was expressed at levels comparable to and with kinetics similar to those observed in the WT background (Fig. 3C).
These data show that the HilA regulon and genes in SPI4 are activated by similar environmental conditions.
Roles of regulatory systems in SPI4 expression.
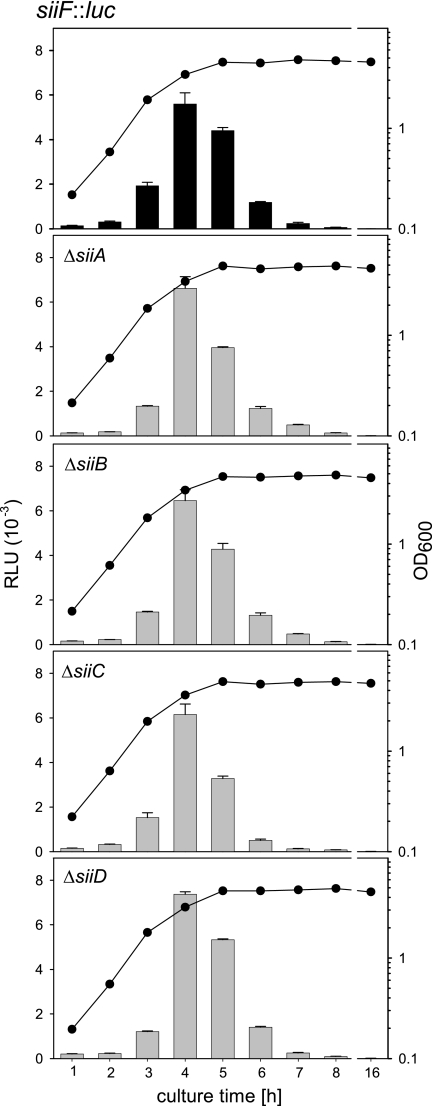
We next set out to characterize the regulatory cascade resulting in the coexpression of SPI1 and SPI4 genes. To investigate possible roles of SPI4-encoded proteins as regulators, we generated reporter fusions in the background of gene-specific deletions of SPI4 genes. As the roles of siiA and siiB in the function of SPI4 are not known, we analyzed the effects of the deletions of these genes, as well as of siiC and siiD, on the expression levels of the siiF::luc fusion, the most distal reporter in SPI4 (Fig. 4). The deletion of siiA, siiB, siiC, or siiD had no major effect on the maximal expression of the siiF::luc fusion and did not alter the kinetics of expression. Similarly, the deletion of siiF prevented secretion but did not significantly affect the levels of SiiE in bacterial cells (14). From these data, we conclude that SiiA and SiiB have no functions as regulators for the expression of SPI4 genes.
FIG. 4.
Roles of SPI4 genes in the expression of a reporter fusion to siiF. Strains harboring the siiF::luc fusion in the background of the WT strain or mutant strains harboring gene-specific deletions in siiA, siiB, siiC, or siiD were generated, and reporter activities were assayed as described in the legend to Fig. 3.
Taken together, our data indicate that siiA to siiF are transcribed as a large transcript under the control of a promoter upstream of siiA and that genes within SPI4 are not likely to affect the expression of siiA to -F. These data also suggest that the expression of SPI4 is most likely controlled by regulators encoded outside of SPI4. A previous study reported that SirA acts as a global regulator of Salmonella enteropathogenesis, controlling expression of SPI1, as well as SPI4, genes (1). BarA/SirA is a two-component regulatory system conserved between enterobacteriaceae with functions in the regulation of adaptation of bacteria to changing environments, as well as the regulation of virulence genes (35). PhoPQ and OmpR/EnvZ are further ubiquitous two-component systems with extended regulatory functions in pathogenic bacteria (reviewed in reference 34). In contrast, SsrAB is a SPI2-encoded two-component system required for the intracellular pathogenesis of Salmonella and controls the expression of the SPI2-encoded T3SS and cognate effectors (18). HilA is the SPI1-encoded central regulator of the HilA regulon, and InvF is a downstream regulator required for the expression of genes encoding substrate proteins of the SPI1 T3SS (see reference 11 for a recent review). RtsAB are further regulators affecting expression of the HilA regulon (10). The alternative sigma factors RpoE and RpoS are transcription factors with global effects on gene expression (17). SlyA is another regulator with effects on the expression of various virulence genes (31), while rfaH encodes an antitermination factor involved in the stabilization of very long transcripts (3).
We performed an analysis of the roles of various regulatory systems that have been implicated in the control of Salmonella virulence gene expression. To control the effects of the mutations in the various regulators on the stability of the reporter luciferase, we followed the luc activities in a strain with a luc fusion to the constitutively expressed gyrA gene (see reference 13 for details). With the exception of rpoE, no changes of more than twofold reduction or increase in reporter activities were observed in strains with defects in the various regulatory systems (data not shown). In the rpoE background, about fivefold-increased reporter activities were observed in the early exponential growth phase.
Using P22 transduction, strains were constructed that harbored the siiA::luc fusion in the background of mutations in genes encoding regulators. The effects of the mutations on the expression of the siiA::luc fusion were analyzed (Fig. 5A). Inactivation of ssrB, ompR/envZ, slyA, or phoP caused a two- to threefold reduction of the maximal expression of siiA::luc. Expression levels were not affected by a mutation in sigma factor rpoS. Expression was initiated with entry into the late exponential growth phase but was maintained at a high level in the early stationary phase. The rpoE mutation caused an ∼2-fold reduction of the maximal expression level. However, the kinetics of expression was altered in strains with defective RpoE. As a similar effect was observed for the gyrA::luc control fusion, this observation may indicate a global change in the gene expression pattern and/or protein turnover in the rpoE strain. The most dramatic effects were observed for strains harboring the siiA::luc fusion in the background of the sirA mutation or the pho-24 allele that results in a PhoP(Con) phenotype. Expression levels were 97- and 123-fold reduced in the sirA and pho-24 backgrounds, respectively (relative to the WT strain). Expression of siiA::luc in the hilA background was reduced by a factor of 6.3 compared to the maximum expression of the WT. The effects of the hilA mutations of other SPI4 genes were comparable, and we found 8.6- and 7.3-fold-reduced expression of siiE::luc and siiF::luc fusions, respectively, in the hilA background (data not shown). In addition to strain P4H2, harboring an insertion of the transposon mini-Tn5 in hilA, we generated a strain with a deletion of hilA using Red recombination (6). In all assays described here, the two hilA strains had identical characteristics (data not shown).
FIG. 5.
Roles of global regulatory systems and specific regulators of virulence genes in SPI4 expression. (A) The activity of the siiA::luc reporter fusion was assayed in the background of the WT and various mutant strains as indicated. Assays were performed as described in the legend to Fig. 3. (B) The activity of the sopE2::luc reporter fusion was assayed in the background of the WT or a hilA or a sirA strain. (C) Effect of the PhoPQ system on SPI4 expression. The siiA::luc fusion was analyzed in the background of the WT or the phoP strain. Bacteria grown overnight in LB broth were used to inoculate LB broth (left) or PCN−P minimal medium adjusted to pH 5.8 (right) with 1/50 of the overnight culture. Reporter activities were assayed at various time points of culture as described above.
For comparison, we investigated the roles of SirA and HilA in the expression of sopE2 as a component of the HilA regulon (Fig. 5B). Here, the expression was reduced by factors of 102 and 175 in the backgrounds of hilA and sirA mutations, respectively.
The deletion of rfaH resulted in reduced expression of distal genes of the siiABCDEF operon, which is in line with the function of RfaH as an antiterminator during the transcription of large operons. The most severe effect was observed for the siiE::luc fusion, which showed an 8.1-fold-reduced reporter activity, whereas the maximal activities of the siiA and siiF fusions were less than 2-fold altered (data not shown).
The two-component regulatory system PhoPQ is considered a master regulator of Salmonella virulence traits and a central sensor for the transition from extracellular to intracellular life (reviewed in reference 15). As shown in Fig. 5A, phoP is not required for the expression of the siiA::luc fusion. However, it has been demonstrated that genes required for enteropathogenesis of Salmonella, such as SPI1 invasion genes, belong to the group of PhoPQ-repressed genes (prg). To investigate if SPI4 belongs to prg, we analyzed the expression of siiA::luc under PhoPQ-inducing conditions, i.e., during growth in PCN−P, minimal medium with limiting amounts of inorganic phosphate and acidic pH. These culture conditions result in activation of PhoPQ-activated genes and repression of prg. As shown in Fig. 5C, the expression levels of siiA::luc were comparable for WT and phoP strains during growth in LB. However, the expression in the phoP background remained at a high level in stationary phase. A more dramatic difference was observed in PCN−P medium, as the expression level for siiA::luc was about 10-fold higher for the phoP strain and remained at a high level during culture. Since the defect of the PhoPQ system abrogated the repression of siiA, we conclude that SPI4 gene expression is modulated by the PhoPQ system and that SPI4 genes belong to the prg.
In order to quantify the effects of the various regulators on the function of the SPI4-encoded T1SS, we quantified the amounts of SiiE secreted by S. enterica serovar Typhimurium and various mutant strains (Fig. 6). No SiiE was detected in the supernatants of siiE and siiF strains. The amounts of SiiE in the supernatants of ssrB, rpoE, and rpoS strains were reduced to about 70% of the WT level. The severe reduction in SiiE secreted by the sirA strains to 0.72% of the WT level was in line with the results of the reporter assays. A similar severe reduction in secreted SiiE was observed for the rfaH strain. The effect of the hilA defect was less severe, as secreted SiiE was reduced to 3.4% of the WT strain level.
FIG. 6.
Secretion of SiiE by WT Salmonella and various mutant strains. The WT and various mutant strains were subcultured in LB broth for 6 h. Cell culture supernatants were recovered, and the amount of secreted SiiE was quantified by ELISA as described in Materials and Methods. For normalization, the OD450 values determined by ELISA were divided by the cell density as determined by the OD600 of the bacterial cultures. The means and standard deviations for three assays are expressed as percentages of the secreted SiiE levels of the WT strain.
Requirement for HilA and SirA for Salmonella adhesion to polarized epithelial cells.
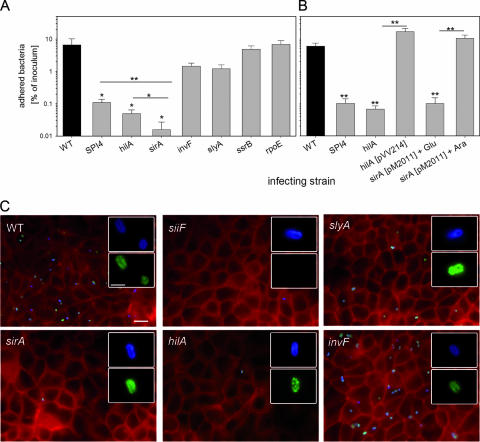
To correlate the effects of regulators on SPI4 expression with a virulence phenotype, adhesion experiments were performed with WT Salmonella, a SPI4 deletion strain, and strains with defects in the regulator HilA, SirA, InvF, SlyA, SsrB, or RpoE (Fig. 7). We previously described the role of SPI4 in the adhesion of S. enterica serovar Typhimurium to polarized epithelial cells and observed highly reduced adhesion of SPI4 mutant strains (14). In order to investigate the phenotypical consequences of SPI4 regulation by various regulatory systems, we quantified the effects of various defects in regulatory proteins on adhesion of S. enterica serovar Typhimurium to epithelial cells. Bacterial strains were subcultured in LB broth for 3.5 h prior to infection, and adhesion to polarized MDCK cells was analyzed by quantification of host cell-associated bacteria (Fig. 7A), as well as by qualitative inspection of immunostained cells (Fig. 7B). Under these assay conditions, 6.6% of the WT bacteria added to the apical side were adherent. Since the culture conditions used here induced SPI4-mediated adhesion, as well as SPI1-mediated invasion, the selectivity of the adhesion assay was controlled. We observed that the invasion-deficient invF strain was recovered in host cell lysates in 4.4-fold-lower numbers than the WT strain, while cell-associated bacterial counts for invasion-deficient hilA or sirA strains were highly reduced. Furthermore, the immunostaining procedure detected only extracellular, adherent bacteria, since host cells were not permeabilized prior to staining of the bacteria. Again, a dramatic difference in the number of cell-associated bacteria was observed for the invasion-deficient invF, hilA, and sirA strains. In accordance with our previous observations, the deletion of the entire SPI4 locus dramatically reduced adhesion (60.4-fold). The adhesion of mutant strains defective in HilA or SirA was reduced by factors of 165.8 and 414.4, respectively (Fig. 7A), and only very few adherent bacteria could be detected on the host cell surface (Fig. 7B). In contrast, defects in other regulators caused only minor (SlyA and InvF) or no significant (SsrB and RpoE) reduction in adhesion. SiiE is a secreted protein that is located on the bacterial cell surface only during contact with host cells, and we previously observed patches on adhering Salmonella positive for SiiE immunostaining (14). While the few host cell-associated bacteria of the siiF strain did not show any staining for SiiE, adhering bacteria of the other mutant strains showed patches of SiiE. Interestingly, SiiE staining was also observed on the few adhering sirA- or hilA-deficient bacteria. This may indicate that the defect in these regulators does not fully abolish expression of SPI4 but leads to highly reduced numbers of SPI4-expressing, and thus adherent, bacteria.
FIG. 7.
Roles of regulatory systems in the adhesion of S. enterica serovar Typhimurium to polarized epithelial cells. (A) Polarized monolayers of MDCK cells were infected with the S. enterica serovar Typhimurium WT strain and various strains defective in regulatory systems as indicated. At 25 min after the addition of the bacteria, the monolayers were washed to remove nonadherent bacteria. For the quantification of adherent bacteria, the host cells were lysed by the addition of PBS containing 0.1% Triton X-100, and serial dilutions of the lysates and the inoculum were plated onto agar plates for the quantification of CFU. The mean percentages of adherent bacteria are shown with standard deviations for experiments performed in triplicate. Statistical significance was calculated by Student's t test, and * and ** indicate P values of <0.05 and <0.01, respectively. (B) Adhesion assays were performed with various strains as described for panel A. Mutant strains defective in hilA or sirA were complemented with plasmids constitutively expressing hilA or expressing sirA under the control of the araC promoter, respectively. (C) Infection of MDCK cells grown on glass coverslips with various strains was performed as described for panel A. After 25 min of infection, the cells were fixed and subjected to immunostaining for SiiE (green) and Salmonella LPS antigen O4 (blue). Subsequently, the cells were permeabilized, and the host cell F actin was stained using Texas Red phalloidin (red). The micrographs show representative overviews of the apical sides of host cells, and the insets show individual adherent bacteria at higher magnification. The scale bar represents 10 μm and 2 μm for overviews and insets, respectively.
These observations are in line with the effects of the regulators on SPI4 expression and stress the central role of the SirA pathway for activation of SPI4-mediated adhesion.
DISCUSSION
S. enterica serovar Typhimurium possesses gene clusters for as many as 13 different fimbrial adhesins and three nonfimbrial adhesins (MisL, ShdA, and BapA). Direct microscopic detection has been possible only for Fim and Agf fimbriae, and the conditions for expression of most of the other fimbriae have not been characterized so far. Humphries et al. (20) reported a systemic analysis of the expression of 11 of the 13 fimbrial adhesins of S. enterica serovar Typhimurium in vivo and in a bovine intestinal ligated-loop model. It is likely that most adhesins are expressed transiently in a temporally and spatially highly controlled fashion or that their functions are restricted to, and induced only in, specific host organisms.
SPI4 is a typical SPI with an enigmatic function in pathogenesis. Recently, a role of SPI4 in intestinal colonization by S. enterica in a bovine model was observed (29). We previously demonstrated the role of SPI4 genes in the synthesis of the giant nonfimbrial adhesin SiiE. In contrast to MisL and ShdA, but similar to BapA, SiiE is secreted by a cognate T1SS (14). SPI4 function is required only for adhesion to polarized epithelial cells. In this study, we demonstrate the coregulation of the SPI4-encoded secretion system and the adhesin SiiE with invasion genes of S. enterica serovar Typhimurium.
We utilized a novel approach for the precise generation of single-copy chromosomal reporter fusions in Salmonella (13). By comparing the reporter activities of various fusions in SPI4 under different growth conditions, it was evident that the expression of SPI4 genes was induced during growth in rich media, as well as in nonlimiting synthetic media, in the late exponential growth phase. There was a rapid induction at the end of the exponential phase, and expression rapidly dropped during the stationary growth phase. The comparison of the expression levels of sopE2 and sipA, encoding effector proteins of the SPI1 T3SS, indicated that the expression kinetics was similar to those of SPI4 genes.
The expression of SPI4 genes is tightly coregulated with genes of the HilA regulon required for host cell invasion. In accordance with an earlier report (1), we observed the pivotal role of SirA for the expression of SPI4. Other global regulators, such as OmpR/EnvZ, SlyA, PhoPQ, or the alternative sigma factors RpoE and RpoS, were not essential for the regulated expression of SPI4 genes. Also the local regulatory system SsrAB that is required for the expression of the SsrAB regulon, consisting of the SPI2-encoded T3SS and various effectors, and InvF, a local regulator of the expression of SPI1 genes, have no relevant contribution to the control of SPI4 gene expression. These data were corroborated by quantification of secreted SiiE and adhesion assays to test SPI4 function. The adhesion of bacteria grown under SPI4-inducing conditions was generally in line with the expression level of SPI4 in the background of various mutations. The SirA defect resulted in the most highly reduced adhesion, while adhesion was affected to only a minor extent by mutations in other regulators. However, the effect of a hilA defect on expression of SPI4 genes and adhesion was remarkable. A central role of HilA for the expression of SPI4 genes was suggested in previous reports (1, 10). Although SPI4-mediated adhesion was highly reduced in a hilA background, we observed that the maximal expression of reporter fusions in SPI4 was only 6.3- to 8.6-fold reduced in the hilA background compared to ∼100-fold-reduced expression observed in the sirA background. The amounts of SiiE secreted by the various strains as determined by ELISA were largely in accordance with the expression data (Fig. 6). In contrast to most HilA-regulated loci, no HilA box could be detected in the promoter region of SPI4. A previous study proposed the presence of HilA boxes in SPI4, but these sequences are located within the coding sequence of siiE and are unlikely to be the target of HilA (9). Previous reports described the regulation of reporter fusions in siiE generated by transposon mutagenesis (1, 10). Roles of SirA and HilA in the expression of siiE were observed (1), and more recently, Ellermeier and Slauch (10) demonstrated that the expression of a reporter fusion in siiE is dependent in HilA, RtsA, and RtsB but independent of InvF. The adhesion of a hilA strain was highly reduced, indicating that HilA may affect SPI4 expression in a posttranscriptional manner or that factors other than SPI4 contributing to adhesion are controlled by HilA. It should be considered that a certain proportion of the bacteria also invade host cells and cannot be distinguished from adherent bacteria in the quantitative assay used here. Therefore, the difference in adhesion between the SPI4 strain and the hilA or sirA strains might reflect the different invasiveness of the strains. At present, we cannot explain the highly reduced SPI4-mediated adhesion of the hilA strain despite the only mildly reduced SPI4 expression, and these discrepancies await further investigation. Our experimental analyses did not indicate the presence of regulators that are encoded by SPI4 genes. It is likely that yet-unknown transcriptional activators of SPI4 expression are present that are integrated in the regulatory cascade downstream of SirA. We are currently screening for these regulators.
A recent study reported the effect of RfaH on the expression of SPI4 genes (30). RfaH interacts with the ops element present in the promoter regions of large operons (2). RfaH acts as an antiterminator and stabilizes long transcripts, for example, those of genes for LPS O-antigen biosynthesis (3). The ops element has also been identified within the promoter region of the siiA to -F operon (29). We observed that in the background of an rfaH deletion the expression of siiE::luc, but not siiA::luc or siiF::luc, was reduced (data not shown). The rfaH deletion results in multiple alterations of the outer membrane, such as defective O-antigen biosynthesis. It is conceivable that the sensing of environmental signals leading to the expression of the SPI1-SPI4 operon is affected, as well as the assembly and function of the SPI4-encoded T1SS. We detected highly reduced amounts of SiiE in the rfaH background (Fig. 6).
Our study is the first observation of the direct coregulation of Salmonella invasion genes and an adhesin. Both the SPI1 T3SS and the SPI4 T1SS are induced in the late logarithmic growth phase, and expression is controlled by the global regulatory system SirA. The coregulation of SPI1 and SPI4 by the global regulator SirA, as well as the global regulator HilA, suggests that both virulence factors are required during the same phase of pathogenesis. Our previous analyses in a mouse model of intestinal inflammation indicate that SPI1, as well a SPI4, contribute to the inflammatory response of the mucosa (14). A further study by Morgan et al. (28) showed a role in colonization of the bovine intestine. On the cellular level, SPI4 mediates binding to the apical surfaces of polarized epithelial cells. Future work must reveal if this adhesion is required for the subsequent invasion of polarized epithelial cells by Salmonella.
Acknowledgments
We thank Alexander Sturm and Wolf-Dietrich Hardt for providing plasmid pM2011 and Cathy A. Lee for plasmid pVV214. We thank Britta Walter, Barbara Bodendorfer, and Andreas Burkovski for help with RNA experiments.
This work was supported by grants HE1964 of the Deutsche Forschungsgemeinschaft to M.H. M.H. thanks the Fonds der Chemischen Industrie for financial support.
Editor: J. B. Bliska
Footnotes
Published ahead of print on 16 July 2007.
REFERENCES
- 1.Ahmer, B. M., J. van Reeuwijk, P. R. Watson, T. S. Wallis, and F. Heffron. 1999. Salmonella SirA is a global regulator of genes mediating enteropathogenesis. Mol. Microbiol. 31:971-982. [DOI] [PubMed] [Google Scholar]
- 2.Artsimovitch, I., and R. Landick. 2002. The transcriptional regulator RfaH stimulates RNA chain synthesis after recruitment to elongation complexes by the exposed nontemplate DNA strand. Cell 109:193-203. [DOI] [PubMed] [Google Scholar]
- 3.Bailey, M. J., C. Hughes, and V. Koronakis. 1997. RfaH and the ops element, components of a novel system controlling bacterial transcription elongation. Mol. Microbiol. 26:845-851. [DOI] [PubMed] [Google Scholar]
- 4.Bajaj, V., C. Hwang, and C. A. Lee. 1995. hilA is a novel ompR/toxR family member that activates the expression of Salmonella typhimurium invasion genes. Mol. Microbiol. 18:715-727. [DOI] [PubMed] [Google Scholar]
- 5.Darwin, K. H., and V. L. Miller. 1999. InvF is required for expression of genes encoding proteins secreted by the SPI1 type III secretion apparatus in Salmonella typhimurium. J. Bacteriol. 181:4949-4954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Datsenko, K. A., and B. L. Wanner. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. USA 97:6640-6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Deiwick, J., T. Nikolaus, S. Erdogan, and M. Hensel. 1999. Environmental regulation of Salmonella pathogenicity island 2 gene expression. Mol. Microbiol. 31:1759-1773. [DOI] [PubMed] [Google Scholar]
- 8.Deiwick, J., T. Nikolaus, J. E. Shea, C. Gleeson, D. W. Holden, and M. Hensel. 1998. Mutations in Salmonella pathogenicity island 2 (SPI2) genes affecting transcription of SPI1 genes and resistance to antimicrobial agents. J. Bacteriol. 180:4775-4780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.De Keersmaecker, S. C., K. Marchal, T. L. Verhoeven, K. Engelen, J. Vanderleyden, and C. S. Detweiler. 2005. Microarray analysis and motif detection reveal new targets of the Salmonella enterica serovar Typhimurium HilA regulatory protein, including hilA itself. J. Bacteriol. 187:4381-4391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ellermeier, C. D., and J. M. Slauch. 2003. RtsA and RtsB coordinately regulate expression of the invasion and flagellar genes in Salmonella enterica serovar Typhimurium. J. Bacteriol. 185:5096-5108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ellermeier, J. R., and J. M. Slauch. 2007. Adaptation to the host environment: regulation of the SPI1 type III secretion system in Salmonella enterica serovar Typhimurium. Curr. Opin. Microbiol. 10:24-29. [DOI] [PubMed] [Google Scholar]
- 12.Fang, F. C., S. J. Libby, N. A. Buchmeier, P. C. Loewen, J. Switala, J. Harwood, and D. G. Guiney. 1992. The alternative sigma factor katF (rpoS) regulates Salmonella virulence. Proc. Natl. Acad. Sci. USA 89:11978-11982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gerlach, R. G., S. Hölzer, D. Jäckel, and M. Hensel. 2007. Rapid engineering of bacterial reporter gene fusions using Red recombination. Appl. Environ. Microbiol. 73:4234-4242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gerlach, R. G., D. Jäckel, B. Stecher, C. Wagner, L. Lupas, W. D. Hardt, and M. Hensel. 2007. Salmonella pathogenicity island 4 encodes a giant non-fimbrial adhesin and the cognate type 1 secretion system. Cell. Microbiol. 9:1834-1850. [DOI] [PubMed] [Google Scholar]
- 15.Groisman, E. A. 2001. The pleiotropic two-component regulatory system PhoP-PhoQ. J. Bacteriol. 183:1835-1842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hansen-Wester, I., B. Stecher, and M. Hensel. 2002. Type III secretion of Salmonella enterica serovar Typhimurium translocated effectors and SseFG. Infect. Immun. 70:1403-1409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hengge-Aronis, R. 2002. Stationary phase gene regulation: what makes an Escherichia coli promoter σS-selective? Curr. Opin. Microbiol. 5:591-595. [DOI] [PubMed] [Google Scholar]
- 18.Hensel, M. 2000. Salmonella pathogenicity island 2. Mol. Microbiol. 36:1015-1023. [DOI] [PubMed] [Google Scholar]
- 19.Humphreys, S., A. Stevenson, A. Bacon, A. B. Weinhardt, and M. Roberts. 1999. The alternative sigma factor, σE, is critically important for the virulence of Salmonella typhimurium. Infect. Immun. 67:1560-1568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Humphries, A. D., M. Raffatellu, S. Winter, E. H. Weening, R. A. Kingsley, R. Droleskey, S. Zhang, J. Figueiredo, S. Khare, J. Nunes, L. G. Adams, R. M. Tsolis, and A. J. Bäumler. 2003. The use of flow cytometry to detect expression of subunits encoded by 11 Salmonella enterica serotype Typhimurium fimbrial operons. Mol. Microbiol. 48:1357-1376. [DOI] [PubMed] [Google Scholar]
- 21.Kuhle, V., and M. Hensel. 2004. Cellular microbiology of intracellular Salmonella enterica: functions of the type III secretion system encoded by Salmonella pathogenicity island 2. Cell Mol. Life Sci. 61:2812-2826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lawley, T. D., K. Chan, L. J. Thompson, C. C. Kim, G. R. Govoni, and D. M. Monack. 2006. Genome-wide screen for Salmonella genes required for long-term systemic infection of the mouse. PLoS Pathog. 2:e11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lee, C. A., and S. Falkow. 1990. The ability of Salmonella to enter mammalian cells is affected by bacterial growth state. Proc. Natl. Acad. Sci. USA 87:4304-4308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Löber, S., D. Jäckel, N. Kaiser, and M. Hensel. 2006. Regulation of Salmonella pathogenicity island 2 genes by independent environmental signals. Int. J. Med. Microbiol. 296:435-447. [DOI] [PubMed] [Google Scholar]
- 25.Lucas, R. L., C. P. Lostroh, C. C. DiRusso, M. P. Spector, B. L. Wanner, and C. A. Lee. 2000. Multiple factors independently regulate hilA and invasion gene expression in Salmonella enterica serovar typhimurium. J. Bacteriol. 182:1872-1882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Miao, E. A., and S. I. Miller. 2000. A conserved amino acid sequence directing intracellular type III secretion by Salmonella typhimurium. Proc. Natl. Acad. Sci. USA 97:7539-7544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Miller, S. I., A. M. Kukral, and J. J. Mekalanos. 1989. A two-component regulatory system (phoP phoQ) controls Salmonella typhimurium virulence. Proc. Natl. Acad. Sci. USA 86:5054-5058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Morgan, E., A. J. Bowen, S. C. Carnell, T. S. Wallis, and M. P. Stevens. 2007. SiiE is secreted by the Salmonella enterica serovar Typhimurium pathogenicity island 4-encoded secretion system and contributes to intestinal colonization in cattle. Infect. Immun. 75:1524-1533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Morgan, E., J. D. Campbell, S. C. Rowe, J. Bispham, M. P. Stevens, A. J. Bowen, P. A. Barrow, D. J. Maskell, and T. S. Wallis. 2004. Identification of host-specific colonization factors of Salmonella enterica serovar Typhimurium. Mol. Microbiol. 54:994-1010. [DOI] [PubMed] [Google Scholar]
- 30.Nagy, G., V. Danino, U. Dobrindt, M. Pallen, R. Chaudhuri, L. Emody, J. C. Hinton, and J. Hacker. 2006. Down-regulation of key virulence factors makes the Salmonella enterica serovar Typhimurium rfaH mutant a promising live-attenuated vaccine candidate. Infect. Immun. 74:5914-5925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Navarre, W. W., T. A. Halsey, D. Walthers, J. Frye, M. McClelland, J. L. Potter, L. J. Kenney, J. S. Gunn, F. C. Fang, and S. J. Libby. 2005. Co-regulation of Salmonella enterica genes required for virulence and resistance to antimicrobial peptides by SlyA and PhoP/PhoQ. Mol. Microbiol. 56:492-508. [DOI] [PubMed] [Google Scholar]
- 32.Patel, J. C., and J. E. Galan. 2005. Manipulation of the host actin cytoskeleton by Salmonella—all in the name of entry. Curr. Opin. Microbiol. 8:10-15. [DOI] [PubMed] [Google Scholar]
- 33.Rhen, M., and C. J. Dorman. 2005. Hierarchical gene regulators adapt Salmonella enterica to its host milieus. Int. J. Med. Microbiol. 294:487-502. [DOI] [PubMed] [Google Scholar]
- 34.Rychlik, I., and P. A. Barrow. 2005. Salmonella stress management and its relevance to behaviour during intestinal colonisation and infection. FEMS Microbiol. Rev. 29:1021-1040. [DOI] [PubMed] [Google Scholar]
- 35.Teplitski, M., R. I. Goodier, and B. M. Ahmer. 2003. Pathways leading from BarA/SirA to motility and virulence gene expression in Salmonella. J. Bacteriol. 185:7257-7265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Worley, M. J., K. H. Ching, and F. Heffron. 2000. Salmonella SsrB activates a global regulon of horizontally acquired genes. Mol. Microbiol. 36:749-761. [DOI] [PubMed] [Google Scholar]