Abstract
Streptococcus pyogenes is a ubiquitous and versatile pathogen that causes a variety of infections with a wide range of severity. The versatility of this organism is due in part to its capacity to regulate virulence gene expression in response to the many environments that it encounters during an infection. We analyzed the expression of two potential virulence factors, sagA and siaA (also referred to as pel and htsA, respectively), in response to conditions of varying cell densities and iron concentrations. The sagA gene was up-regulated in conditioned medium from a wild-type strain but not from sagA-deficient mutants, and the gene was also up-regulated in the presence of streptolysin S (SLS), the gene product of sagA, thus indicating that this gene or its product is involved in density-dependent regulation of S. pyogenes. By comparison, siaA responded in a manner consistent with a role in iron acquisition since it was up-regulated under iron-restricted conditions. Although siaA expression was also up-regulated in the presence of SLS and in conditioned media from both wild-type and sagA-deficient mutants, this up-regulation was not growth phase dependent. We conclude that sagA encodes a quorum-sensing signaling molecule, likely SLS, and further support the notion that siaA is likely involved in iron acquisition.
Streptococcus pyogenes is responsible for a variety of human diseases occurring at different body sites, such as pharyngitis, cellulitis, impetigo, necrotizing fasciitis, and streptococcal toxic shock syndrome. Multiple virulence factors enable this organism to colonize, evade immune defenses, and spread within the human host (6). These processes must be precisely regulated in order for S. pyogenes to survive and replicate within the numerous environmental challenges it encounters during infection. Consequently, the regulation of its gene expression is complex and involves an array of interacting regulators. Two such environmental challenges encountered by S. pyogenes during infection include high cell density, which can occur during tissue infections (20), and a lack of freely available iron within the human host.
Iron is an essential nutrient for pathogenic bacteria and is not readily available within the human host because it is located intracellularly and sequestered by a variety of iron-binding host proteins, such as hemoglobin and transferrins (11, 32). One of the means by which S. pyogenes has been proposed to obtain essential iron for growth is via SiaA, also referred to as HtsA, which is part of an ABC transporter involved in iron acquisition that acts by binding host hemoproteins (2, 19, 23). Liu and Lei (23) proposed that Shp, a streptococcal cell surface protein which is encoded on the same operon as siaA, acquires heme by binding hemoglobin and subsequently transferring the heme to SiaA (23).
Streptolysin S (SLS) production has also been proposed as an important part of the mechanism used by S. pyogenes to acquire intracellular iron by lysing host red blood cells (2, 10). SLS, an oxygen-stable, nonimmunogenic hemolysin with a broad cytolytic spectrum, is encoded by a nine-gene sag operon that was shown to be essential and sufficient for the production of SLS (3, 7, 8, 27). In addition to its putative role in iron acquisition, SLS is thought to be important for the pathogenesis of S. pyogenes because it is involved in inflammation, tissue injury, and resistance to phagocytic killing (3, 7, 22).
The sagA gene, also referred to as pel, encodes a bacteriocin-like peptide which functions as the basic structural unit of SLS (27). Further, sagA mRNA has also been implicated as a regulatory molecule (21, 24, 27) proposed to have an effect on virulence factors, such as M proteins, Sic, and SpeB (21, 24). Since sagA was proposed as a regulatory, bacteriocin-like molecule and we have previously shown that its expression increases with increasing cell density (3), it could act as a quorum-sensing molecule.
Quorum sensing is a density-dependent process which involves chemical signaling molecules that reach a critical threshold concentration with increasing cell density, resulting in altered gene expression (26). In gram-positive bacteria, signal molecules are usually oligopeptides secreted extracellularly by an ABC transporter (26). The peptide is detected by a two-component signal transduction system consisting of a sensor kinase and a response regulator, which relay the signal intracellularly, whereby the sensor kinase phosphorylates the response regulator, thus activating it (26). The phosphorylated response regulator then binds to DNA and alters target gene expression. Interestingly, several bacteriocins, which are a class of antimicrobial peptides produced by bacteria, including nisin of Lactococcus lactis (15) and subtilin of Bacillus subtilis (1), were shown to be regulated by quorum sensing (5). In fact, the structural peptides of nisin and subtilin were shown to function as the signaling molecules that induced their own expression upon activation of the density-dependent autoinduction loop (18). We propose that the bacteriocin-like sagA functions in a similar manner, whereby the structural peptide SLS induces its own expression in a density-dependent manner.
This study was undertaken to explore how S. pyogenes regulates the expression of sagA and siaA in response to various iron concentrations and cell densities. Furthermore, we hypothesized that sagA encodes a quorum-sensing signaling peptide based on its deduced peptide features, predicted cleavage, and posttranslational modifications (27) and on its response to cell density (3). We found that siaA was up-regulated in response to limiting iron conditions, thus adding strength to its role as a gene involved in iron acquisition. Moreover, sagA was shown to respond as a quorum-sensing signaling peptide since it was up-regulated both in a density-dependent manner and by SLS and was not induced in conditioned media from sagA-deficient mutants.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
S. pyogenes MGAS166 (M1 serotype) was used as the test strain for all in vitro experiments discussed in this study. Two strains, SBNH5(ΔSLS) (a nonhemolytic derivative of MGAS166 with a Tn916 insertion in the promoter region of the sag operon) (3), and NZ131:sagAΔcat (an M49 serotype group A Streptococcus with an in-frame allelic replacement of sagA by chloramphenicol) (7), were utilized to generate conditioned media. For all in vitro analyses of gene expression, overnight cultures of MGAS166 were grown in Todd-Hewitt (TH) broth (Difco Laboratories, MI) centrifuged at 39,410 × g at 4°C for 10 min, washed twice with either fresh TH broth or iron-restricted TH broth, and resuspended in the same. This overnight suspension was used at a dilution of 1/20 to inoculate the test media, incubated at 37°C in ambient air, and sampled for mRNA at various time points (2, 4, and 6 h postinoculation).
Preparation of test medium conditions.
In this study, TH broth was considered nonconditioned medium. Conditioned medium was prepared by growing MGAS166, SBNH5(ΔSLS), or NZ131:sagAΔcat in TH broth at 37°C in ambient air for 6 h (optical density at 600 nm [OD600] of 0.6 to 0.8). This culture was then centrifuged at 39,410 × g at 4°C for 15 min, and the cell-free supernatant was removed and filter sterilized using a Stericup filtration system (Millipore, Nepean). To prepare iron-restricted medium, TH broth was treated with the chelating resin Chelex-100 and supplemented with 0.55 mM of CaCl2, MgCl2, MnCl2, and ZnCl2 as described previously (31). This medium was supplemented with 1.0 μM or 1,000 μM ferric chloride or ferric citrate. Expression studies were also conducted in MGAS166 grown in TH broth containing 5.0 μg/ml of commercially available lyophilized SLS (Sigma-Aldrich, Oakville), which contains approximately 3% protein balanced by core RNA in addition to phosphate buffer salts and sodium chloride. Gene expression analysis under all the in vitro conditions tested was conducted with two independent cultures grown under identical conditions.
Growth rates.
The growth kinetics of MGAS166 in the presence of various concentrations of SLS and ferric chloride was analyzed using a Bioscreen microbiology reader (Bioscreen C; Labsystems, Helsinki, Finland). Overnight cultures of MGAS166 in TH broth were washed two times with either fresh TH broth or iron-restricted TH broth as mentioned above and subcultured in triplicate into microtiter plate wells containing 300 μl of test medium. Bioscreen parameters included growth at 37°C for 16 h with the OD600 recorded every 20 min. In addition, the OD600 was recorded for MGAS166 grown in conditioned medium from both sagA mutants [SBNH5(ΔSLS) and NZ131:sagAΔcat] and the wild-type strain (MGAS166) at 37°C for 0, 2, 4, and 6 h postinoculation. This experiment was performed with triplicate cultures.
Total RNA isolation.
S. pyogenes MGAS166 grown under the desired in vitro conditions was harvested by centrifugation at 39,410 × g at 4°C for 10 min, and the bacterial pellet was snap-frozen in liquid nitrogen and stored in −80°C until needed for further use. To isolate RNA, the bacterial pellet was resuspended in TRIzol reagent (Invitrogen, Ontario) and cells were lysed two times using an FP120 FastPrep machine (BIO 101, Mississauga, Ontario, Canada) at a speed of 6.0 for 20 s. RNA was then treated with DNase I and quantified by measuring absorbance at 260 nm, and its integrity was verified by agarose gel electrophoresis.
Real-time PCR analysis.
DNase-treated RNA samples were reverse transcribed by using a first-strand cDNA synthesis kit (MBI Fermentas, Ontario, Canada) in accordance with the recommendations of the supplier. Controls for cDNA synthesis included a no-RNA template sample and one without reverse transcriptase. The real-time PCR assays were performed in triplicate on each of the duplicate samples by using a SmartCycler system (Cepheid, Sunnyvale, CA) and a QuantiTect SYBR green PCR kit (QIAGEN, Ontario, Canada). Each 25-μl reaction mixture included 2 μl of cDNA (200 ng), 250 nmol of each primer (Table 1), and 2× SYBR green mix. The reactions were cycled in the SmartCycler by using the following parameters: 95°C for 15 min for the hot start, followed by 40 cycles of 94°C for 30 s, annealing at optimal temperature (Table 1) for 30 s, and primer extension at 72°C for 30 s. Gene expression analysis included the generation of standard curves for each gene and the utilization of the DNA gyrase A gene (gyrA) as an internal standard for normalizing gene expression, as described previously (30).
TABLE 1.
Primers and optimal annealing temperature used for the real-time PCR analysis of sagA, siaA, and gyrA
| Gene | Optimal annealing temp (°C) | Forward primer | Reverse primer |
|---|---|---|---|
| sagA | 50 | 5′-AGGAGGTAAACCTTATGTTA-3′ | 5′-TACCACCTTGAGAATTACCA-3′ |
| siaA | 65 | 5′-CAGCAGAGAATTGTAGCCACTTCG-3′ | 5′-CCCACACGCTTAACAGCATCATAG-3′ |
| gyrA | 65 | 5′-AGCGAGACAGATGTCATTGCTCAG-3′ | 5′-CCAGTCAAACGACGCAAACG-3′ |
Statistical analysis.
Statistical analysis was conducted using a single factor analysis of variance (ANOVA) such that relative expression levels were compared against 1.0, which indicates no change or equal levels of expression between experimental conditions being analyzed.
RESULTS
Growth kinetics of MGAS166.
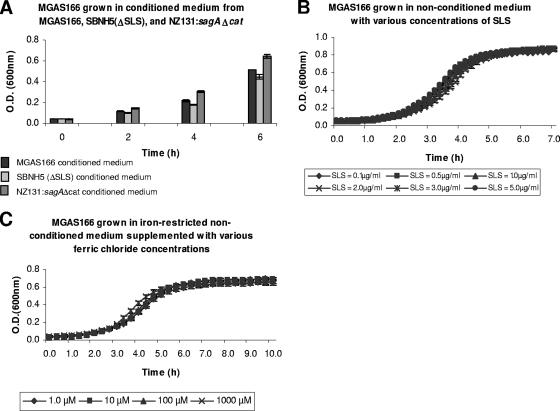
The conditions used to examine gene expression included nonconditioned medium and conditioned medium from sagA wild-type and mutant strains, varying iron concentrations, and the addition of SLS. Thus, to ensure that the growth conditions selected for gene expression analysis did not affect the growth of MGAS166, the growth kinetics of this organism were analyzed. MGAS166 was able to grow in the conditioned medium from MGAS166, SBNH5(ΔSLS), and NZ131:sagAΔcat with increasing cell densities from 0 to 6 h postinoculation (Fig. 1A). Similarly, since various concentrations of SLS, ranging from 0.1 to 5.0 μg/ml, did not inhibit the growth of MGAS166 (Fig. 1B), gene expression was analyzed in 5.0 μg/ml of SLS. A range of iron concentrations from 1.0 μM to 1,000 μM did not appear to alter the growth kinetics of MGAS166 (Fig. 1C). Since iron is necessary for growth and medium supplemented with essential divalent cations (such as CaCl2, MgCl2, MnCl2, and ZnCl2) results in the presence of trace iron, we chose to test gene expression at 1.0 and 1,000 μM of iron. Thus, relative expression of sagA and siaA was measured in low (1.0 μM) and high (1,000 μM) concentrations of iron.
FIG. 1.
Growth kinetics of MGAS166 in various conditioned media (A) and various concentrations of SLS (B) and ferric chloride (C). Each experiment represents an average of three independent cultures. Error bars indicate standard deviations.
Gene expression in nonconditioned medium.
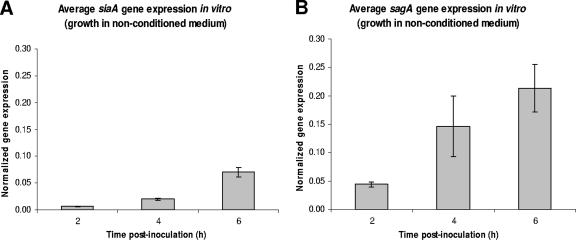
Both sagA and siaA gene expression in MGAS166 were analyzed in vitro for up to 6 h in nonconditioned medium. With increasing cell densities, an increase in the expression of both genes was observed (Fig. 2). The gyrA-normalized expression of siaA increased approximately 10-fold from 0.006 at 2 h to 0.070 at 6 h postinoculation (Fig. 2A). Similarly, the normalized sagA expression increased roughly fivefold from 0.044 at 2 h to 0.21 at 6 h postinoculation (Fig. 2B).
FIG. 2.
Average normalized expression of siaA (A) and sagA (B) in MGAS166 from duplicate cultures grown in nonconditioned medium at 2, 4, and 6 h postinoculation. Both siaA and sagA expression levels were normalized with gyrA. Error bars indicate standard deviations.
Gene expression in conditioned medium.
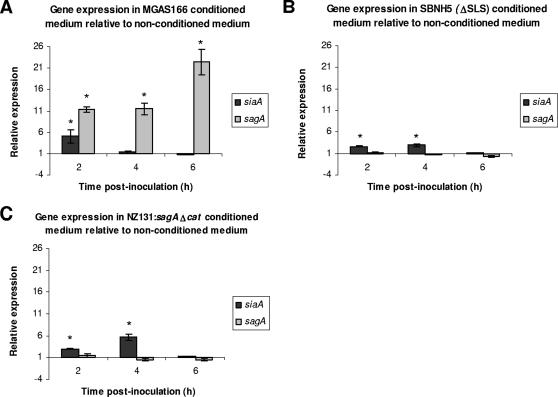
To determine whether sagA regulates gene expression in a density-dependent manner, gene expression was analyzed in the MGAS166 wild-type strain grown in conditioned medium from MGAS166 wild-type) and from two sagA-deficient mutants (SBNH5[ΔSLS] and NZ131:sagAΔcat). The relative levels of sagA and siaA gene expression were determined by a comparison of normalized gene expression in conditioned medium relative to that in nonconditioned medium. sagA was expressed at significantly higher levels in conditioned medium from MGAS166 relative to growth in nonconditioned medium throughout the time period tested (Fig. 3A). This dramatic up-regulation of sagA ranged from 11.3-fold at 2 h postinoculation to 22.4-fold at 6 h postinoculation. This relative up-regulation of sagA was not observed when MGAS166 was grown in the conditioned medium from the sagA mutants (Fig. 3B and C). In fact, there was no significant difference between the levels of sagA gene expression in these conditioned media relative to that in nonconditioned medium. By contrast, siaA expression was up-regulated in the conditioned media from MGAS166 as well as in media from the sagA-deficient mutants (Fig. 3). Interestingly, this up-regulation of siaA occurs earlier during the growth phase at 2 and 4 h postinoculation. Furthermore, there was no significant difference in the level of siaA expression at 6 h postinoculation in each of the conditioned media (MGAS166, SBNH5[ΔSLS], and NZ131:sagAΔcat) relative to that in the nonconditioned medium.
FIG. 3.
Expression of siaA and sagA during growth of MGAS166 in conditioned medium from MGAS166 (A), SBNH5(ΔSLS) (B), and NZ131:sagAΔcat (C) relative to nonconditioned medium as determined by real-time PCR analysis. Statistical significance (P < 0.05) as determined by a single factor ANOVA is indicated by the asterisk. Error bars indicate standard deviations.
Gene expression in 5.0 μg/ml of SLS.
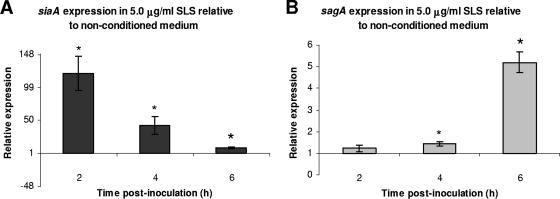
The expression of sagA and siaA was determined in MGAS166 grown in the presence of 5.0 μg/ml SLS, the gene product of sagA, to confirm its role as a signaling molecule. SLS up-regulated the expression of sagA at 2, 4, and 6 h postinoculation relative to its expression in nonconditioned medium (Fig. 4B). Although siaA was also up-regulated by SLS at 2, 4, and 6 h postinoculation, the highest level of siaA relative expression occurred at 2 h postinoculation and the lowest occurred at 6 h postinoculation, which was in contrast to sagA (Fig. 4A).
FIG. 4.
Expression of siaA (A) and sagA (B) of MGAS166 in the presence of 5.0 μg/ml of SLS relative to nonconditioned medium as determined by real-time PCR analysis. Statistical significance (P < 0.05) as determined by a single factor ANOVA is indicated by the asterisk. Error bars indicate standard deviations.
Gene expression in iron-restricted medium.
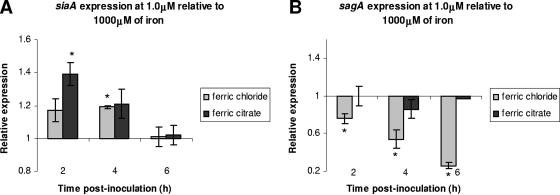
The effect of iron (ferric chloride and ferric citrate) on gene expression was analyzed in medium containing low (1.0 μM) iron concentration relative to medium containing high (1,000 μM) iron concentration. The siaA gene was significantly up-regulated in the presence of lower (1.0 μM) relative to higher (1,000 μM) concentrations of ferric chloride and ferric citrate at 2 and 4 h postinoculation, respectively (Fig. 5A). By 6 h postinoculation, there was no significant difference in the level of siaA expression in the presence of 1.0-μM and 1,000-μM concentrations of iron. Conversely, sagA was significantly down-regulated at 1.0 μM of ferric chloride relative to at 1,000 μM of ferric chloride (Fig. 5B). The down-regulation of sagA by low ferric citrate concentration relative to high ferric citrate concentration was evident at 4 h postinoculation (Fig. 5B).
FIG. 5.
Expression of siaA (A) and sagA (B) during growth of MGAS166 in low (1.0 μg/ml) relative to high (1,000 μg/ml) iron concentrations as determined by real-time PCR analysis. Statistical significance (P < 0.05) as determined by a single factor ANOVA is indicated by the asterisk. Error bars indicate standard deviations.
DISCUSSION
We explored the influence of iron and cell density on the expression of two virulence factors, sagA and siaA. The initial analysis of sagA and siaA expression in nonconditioned culture medium from 2 to 6 h indicated that the expression of each gene increased in a growth phase-dependent manner (Fig. 2), which is characteristic of quorum-sensing signaling molecules. However, when MGAS166 was grown in conditioned medium from the same strain, sagA but not siaA was expressed at significantly higher levels relative to growth in nonconditioned medium throughout the time period tested (Fig. 3A). Furthermore, sagA expression in conditioned medium also increased with increasing cell density from 2 to 6 h postinoculation. We confirmed that the cell density of the culture during growth was not inhibited by the conditioned medium (Fig. 1A). These results are consistent with the hypothesis that sagA encodes or influences activity of a signal molecule present in the conditioned medium and that the increased expression noted during laboratory growth is a result of increased cell density. In contrast, siaA was up-regulated only at 2 h postinoculation in the conditioned medium (Fig. 3A). Furthermore, by 4 and 6 h postinoculation, siaA expression returned to levels similar to that in nonconditioned medium. This pattern of up-regulation is not consistent with that of a quorum-sensing signaling molecule, which increases in expression in a growth-dependent manner.
The differences in gene expression between conditioned and nonconditioned medium suggested that the conditioned medium contained a signaling molecule; however, we did not conclusively identify the sagA gene product as being this molecule. In order to demonstrate sagA as a signaling molecule, MGAS166 was grown in conditioned media from sagA-deficient mutants of two serotypes (M1 and M49). These mutants differed slightly in their genetic backgrounds as the M1 mutant contained a transposon insertion in the sag operon promoter region that resulted in a mutation, which abrogated expression of the entire nine-gene operon (3). The M49 strain had a nonpolar in-frame deletion of the sagA gene, with the eight downstream genes being expressed (7). Most importantly both strains failed to produce an active SLS. We selected mutants of two different serotypes to determine whether varying the M serotype would alter the affect on sagA expression, particularly since the regulatory effect of sagA might be serotype dependent (3, 4, 7, 9, 21). The conditioned media from the mutants were characterized by the absence of a sagA transcript or SLS that is the product of this operon. In the absence of SLS, the conditioned medium did not up-regulate sagA expression relative to nonconditioned medium (Fig. 3B and C), indicating that it is indeed the sagA operon that is responsible for up-regulation. In contrast, there was little difference in the relative expression of siaA, regardless of whether the conditioned medium was from the MGAS166 parent strain or the sagA-deficient mutants (Fig. 3).
Our data differs from those of Mangold et al. (24) who found that the addition of conditioned medium from a sagA-deficient mutant to an M1 serotype wild-type strain resulted in the up-regulation of sagA (24). However, by adding conditioned medium in a ratio of 1:1 to lag-phase cultures, these authors did not eliminate any inducing signals from the lag-phase cultures. We found that the addition of lag-phase cultures to conditioned medium also resulted in the up-regulation of sagA (data not shown). Furthermore, by using the conditioned medium immediately after preparation, we eliminated any potential effects that could result from freezing. Finally, real-time PCR analysis of mRNA expression is more sensitive than Northern analysis and avoids the problem of equalizing total cell numbers since the gyrA gene allows for the standardization of mRNA expression levels. Thus, we attribute this inconsistency between our results and those of Mangold et al. (24) to likely result from differences in experimental procedures.
Since SLS is the functional product of the sagA operon, its effect on gene expression was also evaluated. Various concentrations of exogenously added SLS were initially tested to ensure that the addition of SLS did not alter the cell density of the culture medium (Fig. 1B). The expression of sagA increased after the addition of SLS relative to nonconditioned medium, and this up-regulation followed a growth phase-dependent increase from approximately 1.5-fold at 2 h to roughly 5-fold at 6 h (Fig. 4B). Despite the presence of impurities in commercial SLS due to difficulties in purifying this toxin (8), its exogenous addition caused the induction of sagA similar to the pattern of induction observed with conditioned medium from the MGAS166 wild type. Interestingly, at 5.0 μg/ml, SLS did not induce sagA to levels as high as those in MGAS166-conditioned medium. The reasons for this result could be that (i) the concentration of SLS used for this analysis was either too high or too low or (ii) the presence of impurities in SLS could have altered its activity. Nevertheless, our data provide convincing evidence that the sag operon is indeed involved in signaling.
The addition of commercially available SLS also dramatically up-regulated siaA, and its effect was not growth phase dependent because the highest level of up-regulation (approximately 130-fold) was observed at 2 h rather than at 6 h (approximately 10-fold) (Fig. 4A). This result suggests that siaA could be a target gene for the SLS quorum-sensing system. Furthermore, the fact that siaA was not up-regulated in a growth phase-dependent manner indicated that it was not responding as a signaling molecule.
In addition to cell density and SLS, another environmental stimulus investigated in this study was iron, which has been studied poorly in relation to the virulence of S. pyogenes. Interestingly, one of the earliest associations between hemolysin production and iron acquisition by S. pyogenes was made by Griffiths and McClain (14). They utilized dialyzed brain heart infusion, which was chelated of all ions and then supplemented with essential cations and iron concentrations of up to 5.0 μg/ml, to determine the effect of iron concentration on hemolysin production. Griffiths and McClain showed that the hemolytic activity of S. pyogenes was affected by iron concentration, suggesting that the sag operon responded to iron as an extracellular signal. Furthermore, the hemolytic ability of S. pyogenes has been proposed as a means for this organism to acquire iron by lysing host cells (2, 10).
Iron additions at 1.0 and 1,000 μM were selected for relative gene expression analysis since neither appeared to significantly affect the growth kinetics of MGAS166 (Fig. 1C). Relative to higher iron concentrations, lower iron concentrations induced the expression of siaA but not of sagA (Fig. 5). These data are consistent with the finding that siaA is induced under limited iron conditions (2, 19). The sagA gene, however, was not up-regulated under the lower iron concentrations, though it was up-regulated at higher iron concentrations. There are two possible explanations for this result. First, once iron becomes accessible (low-iron concentrations) through uptake by the sia operon or another iron acquisition system of S. pyogenes, SLS need not be up-regulated to lyse red blood or other host cells in order to release intracellular iron. Second, one of the circumstances during which S. pyogenes could encounter conditions of high-iron concentrations during an infection is within macrophages following phagocytosis. Once phagocytosed, a bacterium is exposed to high levels of iron within the macrophages; this exposure stimulates the formation of damaging reactive oxygen species through the Fenton reaction (28). Although S. pyogenes possesses defense mechanisms against oxidative stress (12, 13, 16, 17, 29), it would also be advantageous for the organism to increase the production of its potent cytolysin SLS to allow it to escape from the macrophages. S. pyogenes has been shown to escape from the phagocytic vacuoles of polymorphonuclear leukocytes and escape into the cytoplasm where the bacteria not only remain viable but also are able to multiply (25). Although the mechanism by which this occurs is not known, it is possible that SLS plays a role in the process.
In conclusion, we demonstrated that sagA responds in a quorum-sensing manner, whereas siaA is stimulated under reduced iron conditions. Although SLS induced the aforementioned genes, only sagA expression responded in a growth-dependent manner. Future work analyzing the interplay between hemolysin production and iron acquisition will be useful for providing a comprehensive understanding of this pathogen.
Acknowledgments
This work was supported by an operating grant from Connaught Laboratories to Dennis G. Cvitkovitch and by infrastructure grants from the Canadian Foundation for Innovation and Ontario Innovative Trust. Additional support was provided by a CIHR Strategic Training Fellowship in Cell Signaling in Mucosal Inflammation and Pain (STP-53877).
Editor: V. J. DiRita
Footnotes
Published ahead of print on 16 July 2007.
REFERENCES
- 1.Banerjee, S., and J. N. Hansen. 1988. Structure and expression of a gene encoding the precursor of subtilin, a small protein antibiotic. J. Biol. Chem. 263:9508-9514. [PubMed] [Google Scholar]
- 2.Bates, C. S., G. E. Montanez, C. R. Woods, R. M. Vincent, and Z. Eichenbaum. 2003. Identification and characterization of a Streptococcus pyogenes operon involved in binding of hemoproteins and acquisition of iron. Infect. Immun. 71:1042-1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Betschel, S. D., S. M. Borgia, N. L. Barg, D. E. Low, and J. C. De Azavedo. 1998. Reduced virulence of group A streptococcal Tn916 mutants that do not produce streptolysin S. Infect. Immun. 66:1671-1679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Biswas, I., P. Germon, K. McDade, and J. R. Scott. 2001. Generation and surface localization of intact M protein in Streptococcus pyogenes are dependent on sagA. Infect. Immun. 69:7029-7038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cotter, P. D., C. Hill, and R. P. Ross. 2005. Bacteriocins: developing innate immunity for food. Nat. Rev. Microbiol. 3:777-788. [DOI] [PubMed] [Google Scholar]
- 6.Cunningham, M. W. 2000. Pathogenesis of group A streptococcal infections. Clin. Microbiol. Rev. 13:470-511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Datta, V., S. M. Myskowski, L. A. Kwinn, D. N. Chiem, N. Varki, R. G. Kansal, M. Kotb, and V. Nizet. 2005. Mutational analysis of the group A streptococcal operon encoding streptolysin S and its virulence role in invasive infection. Mol. Microbiol. 56:681-695. [DOI] [PubMed] [Google Scholar]
- 8.De Azavedo, J. C., K. Y. Salim, and D. J. Bast. 2006. SLS: one of the most potent and elusive of all bacterial toxins, p.728-736. In J. E. Alouf and M. R. Popoff (ed.), The comprehensive sourcebook of bacterial protein toxins. Academic Press, San Diego, CA.
- 9.Eberhard, T. H., D. D. Sledjeski, and M. D. Boyle. 2001. Mouse skin passage of a Streptococcus pyogenes Tn917 mutant of sagA/pel restores virulence, beta-hemolysis and sagA/pel expression without altering the position or sequence of the transposon. BMC Microbiol. 1:33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Eichenbaum, Z., E. Muller, S. A. Morse, and J. R. Scott. 1996. Acquisition of iron from host proteins by the group A streptococcus. Infect. Immun. 64:5428-5429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Genco, C. A., and D. W. Dixon. 2001. Emerging strategies in microbial haem capture. Mol. Microbiol. 39:1-11. [DOI] [PubMed] [Google Scholar]
- 12.Gibson, C. M., and M. G. Caparon. 1996. Insertional inactivation of Streptococcus pyogenes sod suggests that prtF is regulated in response to a superoxide signal. J. Bacteriol. 178:4688-4695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gibson, C. M., T. C. Mallett, A. Claiborne, and M. G. Caparon. 2000. Contribution of NADH oxidase to aerobic metabolism of Streptococcus pyogenes. J. Bacteriol. 182:448-455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Griffiths, B. B., and O. McClain. 1988. The role of iron in the growth and hemolysin (streptolysin S) production in Streptococcus pyogenes. J. Basic Microbiol. 28:427-436. [DOI] [PubMed] [Google Scholar]
- 15.Gross, E., and J. L. Morell. 1971. The structure of nisin. J. Am. Chem. Soc. 93:4634-4635. [DOI] [PubMed] [Google Scholar]
- 16.Janulczyk, R., S. Ricci, and L. Bjorck. 2003. MtsABC is important for manganese and iron transport, oxidative stress resistance, and virulence of Streptococcus pyogenes. Infect. Immun. 71:2656-2664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.King, K. Y., J. A. Horenstein, and M. G. Caparon. 2000. Aerotolerance and peroxide resistance in peroxidase and PerR mutants of Streptococcus pyogenes. J. Bacteriol. 182:5290-5299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kleerebezem, M. 2004. Quorum sensing control of lantibiotic production; nisin and subtilin autoregulate their own biosynthesis. Peptides 25:1405-1414. [DOI] [PubMed] [Google Scholar]
- 19.Lei, B., M. Liu, J. M. Voyich, C. I. Prater, S. V. Kala, F. R. Deleo, and J. M. Musser. 2003. Identification and characterization of HtsA, a second heme-binding protein made by Streptococcus pyogenes. Infect. Immun. 71:5962-5969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Leonard, B. A., and A. Podbielski. 1999. Emerging density-dependent control systems in gram-positive cocci, p.315-331. In G. M. Dunny and S. C. Winans (ed.), Cell-cell signaling in bacteria. American Society of Microbiology, Washington, DC.
- 21.Li, Z., D. D. Sledjeski, B. Kreikemeyer, A. Podbielski, and M. D. Boyle. 1999. Identification of pel, a Streptococcus pyogenes locus that affects both surface and secreted proteins. J. Bacteriol. 181:6019-6027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Limbago, B., V. Penumalli, B. Weinrick, and J. R. Scott. 2000. Role of streptolysin O in a mouse model of invasive group A streptococcal disease. Infect. Immun. 68:6384-6390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liu, M., and B. Lei. 2005. Heme transfer from streptococcal cell surface protein Shp to HtsA of transporter HtsABC. Infect. Immun. 73:5086-5092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mangold, M., M. Siller, B. Roppenser, B. J. Vlaminckx, T. A. Penfound, R. Klein, R. Novak, R. P. Novick, and E. Charpentier. 2004. Synthesis of group A streptococcal virulence factors is controlled by a regulatory RNA molecule. Mol. Microbiol. 53:1515-1527. [DOI] [PubMed] [Google Scholar]
- 25.Medina, E., M. Rohde, and G. S. Chhatwal. 2003. Intracellular survival of Streptococcus pyogenes in polymorphonuclear cells results in increased bacterial virulence. Infect. Immun. 71:5376-5380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Miller, M. B., and B. L. Bassler. 2001. Quorum sensing in bacteria. Annu. Rev. Microbiol. 55:165-199. [DOI] [PubMed] [Google Scholar]
- 27.Nizet, V., B. Beall, D. J. Bast, V. Datta, L. Kilburn, D. E. Low, and J. C. De Azavedo. 2000. Genetic locus for streptolysin S production by group A Streptococcus. Infect. Immun. 68:4245-4254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ratledge, C., and L. G. Dover. 2000. Iron metabolism in pathogenic bacteria. Annu. Rev. Microbiol. 54:881-941. [DOI] [PubMed] [Google Scholar]
- 29.Ricci, S., R. Janulczyk, and L. Bjorck. 2002. The regulator PerR is involved in oxidative stress response and iron homeostasis and is necessary for full virulence of Streptococcus pyogenes. Infect. Immun. 70:4968-4976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Salim, K. Y., D. G. Cvitkovitch, P. Chang, D. J. Bast, M. Handfield, J. D. Hillman, and J. C. De Azavedo. 2005. Identification of group A Streptococcus antigenic determinants upregulated in vivo. Infect. Immun. 73:6026-6038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Spatafora, G., M. Moore, S. Landgren, E. Stonehouse, and S. Michalek. 2001. Expression of Streptococcus mutans fimA is iron-responsive and regulated by a DtxR homologue. Microbiology 147:1599-1610. [DOI] [PubMed] [Google Scholar]
- 32.Wandersman, C., and I. Stojiljkovic. 2000. Bacterial heme sources: the role of heme, hemoprotein receptors and hemophores. Curr. Opin. Microbiol. 3:215-220. [DOI] [PubMed] [Google Scholar]