Abstract
Staphylococcus lugdunensis is a pathogen of heightened virulence that causes infections resembling those caused by Staphylococcus aureus rather than those caused by its coagulase-negative staphylococcal counterparts. Many types of S. lugdunensis infection, including native valve endocarditis, prosthetic joint infection, and intravascular catheter-related infection, are associated with biofilm etiology. Poly-N-acetylglucosamine (PNAG), a polysaccharide synthesized by products of the icaADBC locus, is a common mechanism of intercellular adhesion in staphylococcal biofilms. Here we report the characterization of ica homologues and the in vitro biofilm formation properties of a collection of S. lugdunensis clinical isolates. Isolates formed biofilms in microtiter wells to various degrees. Biofilm formation by most isolates was enhanced with glucose but diminished by sodium chloride or ethanol. icaADBC homologues were found in all S. lugdunensis isolates tested, although the locus organization differed substantially from that of other staphylococcal ica loci. icaR was not detected in S. lugdunensis, but a novel open reading frame with putative glycosyl hydrolase function is located upstream of the ica locus. icaADBC sequence heterogeneity did not explain the variability in biofilm formation among isolates. PNAG was not detected in S. lugdunensis extracts by immunoblotting with an anti-deacetylated PNAG antibody or wheat germ agglutinin. Confocal microscopy with fluorescently labeled wheat germ agglutinin showed a paucity of PNAG in S. lugdunensis biofilms, but abundant extracellular protein was visualized with SYPRO Ruby staining. Biofilms were resistant to detachment by dispersin B and sodium metaperiodate but were susceptible to detachment by proteases. Despite the genetic presence of icaADBC homologues in S. lugdunensis isolates, PNAG is not a major component of the extracellular matrix of in vitro biofilms formed by this species. Our data suggest that the S. lugdunensis biofilm matrix contains proteinaceous factors.
Staphylococcus lugdunensis, first described in 1988 (24), has gained notoriety for being an unusually virulent coagulase-negative staphylococcal species. S. lugdunensis is a human skin commensal that was found to colonize ∼20% of a sampled population (73). It is also a pathogen capable of causing a wide spectrum of diseases that are both typical and unexpected of a coagulase-negative Staphylococcus species. S. lugdunensis infections include skin and wound infection (31, 70), urinary tract infection (27), prosthetic joint infection (63), intravascular catheter infection (31), and native valve endocarditis (2, 56). Cases of S. lugdunensis native valve endocarditis are particularly serious, as they often present with a highly destructive and acute pattern more typical of endocarditis caused by Staphylococcus aureus rather than by other coagulase-negative staphylococci (CNS) (2, 56). Similar to other staphylococci, colonization of and formation of biofilms on host tissues or indwelling medical devices appear to be an important contributor to the pathogenesis of many infections caused by this organism.
Biofilms are complex and dynamic surface-associated microbial communities that are surrounded by a self-produced extracellular polymeric matrix (16). Biofilm formation protects microorganisms from challenging environmental conditions, subsequently rendering traditional antimicrobial therapies and host immune defense mechanisms ineffective against biofilm-associated bacteria (67). CNS and S. aureus are the two most common bacterial pathogens recovered from patients with nosocomial bloodstream infections (20), underscoring the significant health care burden resulting from staphylococcal biofilm formation on intravascular catheters or implanted prosthetic devices. Considering the inherent difficulty associated with treating biofilm infections, a thorough understanding of the mechanism(s) of staphylococcal biofilm formation will be essential in the development of new diagnostic and treatment approaches for these infections.
Staphylococcal biofilms form through an ordered process that begins with primary attachment of cells to a surface, which is followed by cellular proliferation, accumulation, and intercellular adhesion (45). Much work has been done to understand the molecular basis of intercellular adhesion, and it is now known that both polysaccharide and protein mechanisms can mediate staphylococcal biofilm accumulation (45, 54). The polysaccharide matrix polymer poly-N-acetylglucosamine (PNAG) (47), also called polysaccharide intercellular adhesion (46), is the best-characterized mechanism of staphylococcal intercellular adhesion. β-1,6-linked PNAG is produced by the genes of the icaRADBC locus, an operon of four biosynthetic genes (icaADBC) and icaR, an upstream gene encoding a divergently transcribed negative regulatory protein (9, 10, 28). S. aureus, Staphylococcus epidermidis, and Staphylococcus caprae harbor icaRADBC loci in their genomes (1, 10, 28), although the locus is absent in up to 45% of S. epidermidis clinical isolates (21). Homologues have been identified in other CNS species (21, 51), as well as several gram-negative genera, including Actinobacillus (33, 37), Bordetella (55), Yersinia (12), and Escherichia (75), emphasizing its evolutionary importance in biofilm adhesion. More recently, the involvement of proteins and other polymeric substances in S. epidermidis and S. aureus biofilm formation has become apparent (45, 54). Cell wall-associated proteins, such as Bap (biofilm-associated protein) in S. aureus (11) and Aap (accumulation-associated protein) in S. epidermidis (60), enable icaADBC-independent biofilm formation. Extracellular teichoic acids are also important constituents of the S. epidermidis biofilm matrix (41, 62).
Although our current understanding of staphylococcal biofilm formation is quite expansive, it derives predominantly from characterization and mechanistic studies focused on S. aureus and S. epidermidis. In this study, we have expanded this knowledge to include S. lugdunensis in order to begin to elucidate the mechanism of biofilm formation used by this organism. We characterized the properties of in vitro biofilm formation exhibited by a collection of clinical S. lugdunensis isolates. In addition, we identified and sequenced S. lugdunensis icaADBC homologues and assessed the role of this locus in biofilm formation.
(This work was presented, in part, at the 105th General Meeting of the American Society for Microbiology, 5 to 9 June 2005, Atlanta, GA; the International Symposium on Virulence Mechanisms of Bacterial Pathogens, 6 to 8 September 2006, Ames, IA; the 46th Interscience Conference on Antimicrobial Agents and Chemotherapy, 27 to 30 September 2006, San Francisco, CA; and the 4th American Society for Microbiology Conference on Biofilms, 25 to 29 March 2007, Quebec City, Quebec, Canada.)
MATERIALS AND METHODS
Microorganisms and growth conditions.
The microorganisms used in this study are listed in Table 1. Bacteria stored at −70°C were freshly streaked on sheep's blood agar or Trypticase soy agar before each replicate experiment. For microtiter plate biofilm formation and detachment assays, immunodetection assays, and confocal microscopy assays (unless stated otherwise), isolated colonies from 16- to 24-h-old plates were grown in Trypticase soy broth (BD BBL, Franklin Lakes, New Jersey) supplemented with 1% (wt/vol) sterile-filtered glucose (TSBgluc1%) with shaking at 130 rpm for 22 to 24 h. Unless otherwise noted, incubation was in ambient air at 37°C for all experiments.
TABLE 1.
Microorganisms used in this study
| Species and strain | Description | Reference or source |
|---|---|---|
| Staphylococcus epidermidis | ||
| RP62A | ATCC 35984; well-characterized biofilm-forming strain; icaADBC and PNAG positive | ATCC |
| CSF41498 | Biofilm-forming cerebrospinal fluid isolate; icaADBC and PNAG positive | 9 |
| Staphylococcus aureus | ||
| SA113 | ATCC 35556; well-characterized biofilm-forming strain; icaADBC and PNAG positive | ATCC |
| SA113 ica::tet | S. aureus SA113 with inactivated icaADBC locus; deficient for biofilm formation and PNAG production | 10 |
| RN4220 | Biofilm-forming strain; icaADBC and PNAG positive (weak) | 42 |
| Staphylococcus carnosus | ||
| TM300 | Non-biofilm-forming strain; icaADBC and PNAG negative | 26 |
| TM300(pCN27) | S. carnosus TM300 carrying plasmid pCN27 containing icaADBC cloned from S. epidermidis RP62A; biofilm and PNAG positive | 28 |
| Staphylococcus lugdunensis | ||
| IDRL-856 | Endocarditis isolate | 23 |
| IDRL-2394 | Prosthetic joint infection isolate | 23 |
| IDRL-2414 | Endocarditis isolate | 23 |
| IDRL-2492 | Endocarditis isolate | 23 |
| IDRL-2526 | Prosthetic joint infection isolate | 23 |
| IDRL-2554 | Prosthetic joint infection isolate | 23 |
| IDRL-2588 | Infected hematoma isolate | 23 |
| IDRL-2622 | Prosthetic joint infection isolate | 23 |
| IDRL-2639 | Paronychia isolate | 23 |
| IDRL-2640 | Folliculitis isolate | 23 |
| IDRL-2664 | Prosthetic joint infection isolate | 23 |
| IDRL-5204 | Prosthetic joint infection isolate | 23 |
| IDRL-5254 | Intravascular catheter infection isolate | 23 |
| IDRL-5256 | Prosthetic joint infection isolate | 23 |
| IDRL-5258 | Prosthetic joint infection isolate | 23 |
Microtiter plate biofilm formation assay.
Biofilm formation was assayed using a microtiter plate assay (8), with modifications as previously described (22). Briefly, cultures were adjusted with TSBgluc1% to match the turbidity of a 1.0 McFarland standard (∼1 to 2 × 108 CFU/ml) and diluted 1:50 in TSBgluc1% or TSBgluc1% containing various concentrations of sodium chloride (1 to 5%, wt/vol) or ethanol (0.5 to 4%, vol/vol). For experiments examining the effect of glucose concentration, bacteria were grown overnight, adjusted to the turbidity of a 1.0 McFarland standard, and diluted in TSB containing the corresponding amount of glucose being tested (0.5 to 5%, wt/vol). Two-hundred-microliter aliquots of each diluted culture were placed into four wells of 96-well microtiter plates (Nuclon Delta; Nalge Nunc International, Rochester, NY) and incubated for 24 h. Cell growth was measured by reading the optical density at 600 nm (OD600) on a microplate reader (Multiskan; Thermo Electron, Waltham, MA). The culture medium was discarded, and the wells were washed twice by fully submerging plates in deionized water to remove nonadherent cells and then air dried overnight. Biofilms were stained with 0.1% safranin for 1 min, rinsed under running tap water to remove excess stain, and air dried overnight. In order to ensure homogeneity among stained material in the wells, stained biofilms were resuspended in 200 μl of 30% glacial acetic acid, and the OD492 was measured. Wells containing uninoculated medium served as sterility controls and spectrophotometric blanks. Each condition was assayed three times on separate days with similar results.
Silicone elastomer disk biofilms and scanning electron microscopy (SEM).
Ten-millimeter-diameter disks were cut from 0.020-in.-thick nonreinforced medical grade silicone elastomer sheeting (Bentec Medical, Woodland, CA) with a skin biopsy punch and sterilized by autoclaving. An overnight culture of S. lugdunensis IDRL-2640 was adjusted to match the turbidity of a 1.0 McFarland standard and diluted 1:50 in TSBgluc1%. One-milliliter aliquots of the diluted culture or sterile medium were added to wells of a 24-well plate (Falcon; BD Biosciences, Franklin Lakes, NJ). Disks were placed in the bottoms of wells with sterile forceps and incubated for 24 h at 37°C in 5% CO2 to allow for biofilm formation. Following incubation, disks were removed from wells, soaked for 5 min in 1 ml of sterile phosphate-buffered saline (PBS) to remove planktonic bacteria, and stained with 1 ml 0.1% safranin for 1 min. Excess stain was removed by repeatedly dipping each disk in sterile water.
To visualize biofilms by electron microscopy, disks were incubated as described above, soaked in 1 ml of sterile water for 5 min, and placed into Trumps fixative (4% formaldehyde and 1% glutaraldehyde in phosphate buffer, pH 7.3). Following critical-point drying and gold-palladium sputter coating, disks were imaged by cold-field emission SEM using a Hitachi S-4700 instrument (Hitachi Ltd., Tokyo, Japan).
DNA extraction.
Genomic DNA was prepared for Southern blotting and restriction site PCR from 200-ml cultures grown overnight in TSB at 37°C with 5% CO2. Cells were pelleted, resuspended in 4 ml of 200-μg/ml lysostaphin (Sigma-Aldrich, Saint Louis, MO), and incubated at 37°C for 30 min. An equal volume of DNA Stat-60 reagent (Tel-test, Inc., Friendswood, TX) was added, and cells were mixed by inversion and incubated for 20 min at room temperature prior to DNA extraction and precipitation as recommended by the manufacturer.
Low-stringency icaA and icaR Southern blotting.
Genomic DNA (∼5 μg) was digested with restriction enzyme EcoRI (Roche Applied Science, Indianapolis, IN) or HaeIII (Invitrogen, Corp., Carlsbad, CA), separated by electrophoresis on a 1% agarose gel, and transferred to a nylon membrane by downward capillary transfer (Nytran SuperCharge Turboblotter Kit; Whatman, Inc., Florham Park, NJ). Southern blotting and washes were performed under low-stringency conditions in digoxigenin (DIG) EasyHyb hybridization solution (Roche Applied Science) as suggested by the manufacturer. The PCR DIG probe synthesis kit (Roche Applied Science) was used to synthesize labeled probes from ica-positive S. aureus and S. epidermidis strains with primer pairs KFicaAF/KFicaAR (Table 2) for icaA or SAicaRF/SAicaRR and SEicaRF/SEicaRR (Table 2) for icaR, respectively. Bound probes were visualized on X-ray film with the chemiluminescent substrate CSPD (Roche Applied Science) following immunological detection with an alkaline phosphatase-labeled anti-DIG antibody (Roche Applied Science).
TABLE 2.
Oligonucleotides used in this study
| Oligonucleotide | Sequence |
|---|---|
| icaAR | 5′-CCTCTGTCTGGGCTTGACC-3′ |
| KFicaAF | 5′-GATGGAAGTTCTGATAATAC-3′ |
| KFicaAR | 5′-GTGAAAACACCTGAAATAGTATTGA-3′ |
| SAicaRF | 5′-TTGAAGGATAAGATTATTGATAAC-3′ |
| SAicaRR | 5′-TAGTAGCGAATACACTTCATC-3′ |
| SEicaRF | 5′-TTGAAAGATAAGATTATTGATAAC-3′ |
| SEicaRR | 5′-CATTTAACAGTGAATATACTTG-3′ |
| mRS-BamHI | 5′-GGTACCTAATACGACTCACTATANNNNNNNNNNGGATCC-3′ |
| mRS-EcoRI | 5′-GGTACCTAATACGACTCACTATANNNNNNNNNNGAATTC-3′ |
| mRS-Sau3AI | 5′-GGTACCTAATACGACTCACTATANNNNNNNNNNGATC-3′ |
| mRS-TacI | 5′-GGTACCTAATACGACTCACTATANNNNNNNNNNTCGA-3′ |
| KLF3F | 5′-CAAAAAAACCAAGGGTAAAG-3′ |
| KLF4R | 5′-ACCTAAAATAGACTTCTTATTTC-3′ |
| KLF5R | 5′-CCCATCACTAGATCATATTGT-3′ |
| KLF6F | 5′-ATGTTAGAACATTTTATCGAT-3′ |
| KLF6R | 5′-ATCGATAAAATGTTCTAACAT-3′ |
| KLF7F | 5′-AATCCGAAATTGGCTGCGGTA-3′ |
| KLF8F | 5′-TTTAACGAGGAAGAGACGATT-3′ |
| KLF9R | 5′-GAATCAGAACTTCTTGCCCA-3′ |
| KLF11R | 5′-TATTTGGAAACTCTAACGATA-3′ |
| KLF12F | 5′-AGCCACGCGCATTATGTCGAA-3′ |
| KLF13F | 5′-TGCTTGTTCCTGAGACGATAC-3′ |
| KLF14R | 5′-TCGTCTCTTCCTCGTTAAAACA-3′ |
| KLF15R | 5′-ATTAAAAAGGAAATACCT-3′ |
| KLF16F | 5′-AACGTCTTCGATGGGCACAAGG-3′ |
| KLF17F | 5′-CAGCAAAGCACACATTCGTTAGC-3′ |
| KLF18R | 5′-GTCGCAAACGCTCCTTTTTTAC-3′ |
| KLF20F | 5′-GAAAAGAAACACATATTGAT-3′ |
| KLF21R | 5′-GCGGGTCTTTACGCATATTA-3′ |
| KLF22F | 5′-GGCTGTTAAATGCTTTGGTCG-3′ |
| KLF25F | 5′-TTACTCCATAAACATCATCC-3′ |
| KLF26R | 5′-GTTGAAAATCAAGCATTGT-3′ |
| KLF27R | 5′-CATAGCTAGAAACAACCTGTC-3′ |
| KLF28F | 5′-CAATTTGTTATTGCGCTATTC-3′ |
| KLF29R | 5′-TACCTTTGACGTTGAGCG-3′ |
| KLF31F | 5′-CACATACCATTTCTAGTGC-3′ |
| KLF32F | 5′-TGCCGTTTGTCTTGTACTTC-3′ |
| KLF33R | 5′-ATTGATTAACTGACGAGATAC-3′ |
| KLF34R | 5′-ATTTTCCATCTATATCTCAC-3′ |
| KLF35R | 5′-GTTTACATTTTTCAATATATAG-3′ |
| KLF37R | 5′-TCCTTTTTCTGTTAAAAAATG-3′ |
| KLF38F | 5′-GGTATTAATCATGGCAAAGTTT-3′ |
| KLF43F | 5′-TATCAATAGTTGAATCGTATA-3′ |
| KLF45F | 5′-AATGGCTTAAAGCACATGGGG-3′ |
| KLF46F | 5′-GTAAAGAAGCGTTTGAGGCTG-3′ |
| KLF47R | 5′-GTACTTTTATATTTTGATTGC-3′ |
| KLF48F | 5′-GAAGGGATTCGCTATGGC-3′ |
| KLF49R | 5′-AATATAGCACAATAAGGA-3′ |
| KLF50F | 5′-TGTCATGCTGTGTGTTATTAT-3′ |
| KLF51F | 5′-CAAGCACATCATTGTATTCCG-3′ |
| KLF52R | 5′-ACCTAATTTACGCGATTCACTG-3′ |
| KLF53F | 5′-CGTTTTAAATACATTATTTTG-3′ |
| KLF54F | 5′-TTATTTATGTGTCGGTTGTTTC-3′ |
| KLF56R | 5′-TTACATAGGAGGACCTCTAAG-3′ |
| KLF57F | 5′-GTGATTACATCTGTCATTGCG-3′ |
| KLF57R | 5′-CGCAATGACAGATGTAATCAC-3′ |
| KLF58R | 5′-TGTGCTTGTGATACAGCGTG-3′ |
| KLF59F | 5′-TATGCTTCATTACGCATCACC-3′ |
| KLF60F | 5′-TATTTGGAAAGCACGATTAC-3′ |
| KLF60R | 5′-GTAATCGTGCTTTCCAAATA-3′ |
| KLF62R | 5′-TGTCTAACGAAGATGCAGGAC-3′ |
| KLF64F | 5′-TTCATATTCTGCAATAGCCTG-3′ |
| KLF65R | 5′-GTTGCGCATGTGTCGATATC-3′ |
| KLF66F | 5′-GGTGGCTATATTGGTTATAAC-3′ |
| KLF67R | 5′-ATGTTCCTTTAAAATCAAT-3′ |
| KLF68F | 5′-TTGGCATTGGTATTATTTTAC-3′ |
| KLF69F | 5′-GAATTCTATATTTGCCGCT |
| KLF70F | 5′-CAACCTGCGATGCGTGTTTAAT |
| KLF70R | 5′-ATTAAACACGCATCGCAGGTTG |
| KLF71F | 5′-CTGTTGTTGGAACGCTAGGTA-3′ |
| KLF72F | 5′-TTAGGGGACAGCTTCAGGCCA-3′ |
| KLF73F | 5′-TTATTTTTATGTTTGACTTT-3′ |
| KLF73R | 5′-AAAGTCAAACATAAAAATAA-3′ |
| KLF75R | 5′-GTTATTGATGCACGTCTTGG-3′ |
| KLF76F | 5′-ACGAAAATAAACAGTGTCT-3′ |
| KLF76R | 5′-AAGACACTGTTTATTTTCGT-3′ |
| KLF77F | 5′-TCGGAATCATTAATTTGAGAT-3′ |
| KLF78F | 5′-TAACTTTATTAATATAGATGA-3′ |
| KLF79F | 5′-AGGTCAAACGTCTACCC-3′ |
| KLF80R | 5′-AAAAGTCAGTTGGCGATG-3′ |
| KLF82R | 5′-AATGATATTGAAATACAGCG-3′ |
S. lugdunensis ica operon sequence acquisition by restriction site PCR.
Primers KFicaAF and icaAR (Table 2) (also called ICAF and ICAR, respectively, in a previous publication [64]), which were designed from regions of high homology between the icaA genes of S. aureus and S. epidermidis and were reportedly used to amplify icaA from S. lugdunensis (64), were used in standard PCRs with S. lugdunensis genomic DNA, AmpliTaq Gold, and buffer I (Applied Biosystems, Foster City, CA) under low-stringency annealing conditions (42°C). The resulting PCR products were sequenced with the same primers. Restriction site PCR (65), a primer walking strategy that couples an outward-facing primer of known sequence with a universal primer that recognizes a specific restriction enzyme recognition site (mRS-BamHI, mRS-EcoRI, mRS-Sau3AI, or mRS-TacI [Table 2]), was used to acquire the sequence of the entire region encompassing the icaADBC locus from S. lugdunensis IDRL-2414 and IDRL-2664. Outward-facing primers were paired with one of the restriction site PCR primers in standard PCRs. One microliter of this reaction product was used as the template in a second PCR containing the restriction site PCR primer and a specific primer located internal to the first-round primer. The resulting products were analyzed by gel electrophoresis, sequenced with the second-round specific primer, and verified bidirectionally (Table 2). Sequencing was performed on an ABI Prism 377 DNA sequencer with an ABI Prism BigDye Terminator cycle sequencing ready reaction kit (Perkin-Elmer Applied Biosciences, Foster City, CA) at the Mayo Clinic DNA Sequencing core facility. Primers were synthesized by the Mayo Clinic DNA Synthesis core facility or Integrated DNA Technologies, Inc. (Coralville, IA).
S. lugdunensis ica locus PCR screen.
Primer pairs KLF64F/KLF82R and KLF32F/KLF67R (Table 2) were used to amplify 7.6-kb and 4.9-kb products, respectively, from the region spanning the S. lugdunensis icaADBC locus with PCR SuperMix High Fidelity (Invitrogen). Template DNA was prepared by alkaline washing as previously described (21). S. lugdunensis IDRL-2414, IDRL-2664, IDRL-5204, IDRL-5256, and IDRL-5258 PCR products were bidirectionally sequenced with primers listed in Table 2. Sequence alignments and comparisons were performed with Sequencher software (Gene Codes Corp., Ann Arbor, MI).
Immunoblot detection of PNAG and other polysaccharides in stationary-phase or biofilm S. lugdunensis cells.
PNAG production was assessed by immunoblotting essentially as previously described (10), with some modifications. For stationary-phase cells, bacteria were grown overnight in TSBgluc1% and equivalent amounts (1 to 2 ml) of cells, as determined by OD, were harvested. For biofilm cells, biofilms were established as described above in 48-well microtiter plates (Nuclon Delta; Nalge Nunc International) containing 500 μl of culture. Biofilms from two wells per organism were scraped with a pipette tip, resuspended in the culture medium, and pooled. Cell pellets were washed in sterile PBS, resuspended in 0.5 M EDTA, sonicated for 5 min at 40 kHz in a bath sonicator (Zenith Ultrasonics, Norwood, NJ), boiled for 5 min, and centrifuged. Supernatants were treated with 200 μg proteinase K for 30 min at 65°C and then for 15 min at 80°C to heat inactivate the enzyme. Extracts were spotted on nitrocellulose, and blots were blocked 1 h in 3% bovine serum albumin (BSA) in Tris-buffered saline (TBS). Blots were probed overnight at 4°C with a 1:5,000 dilution of goat anti-deacetylated PNAG antibody (48) in TBS-0.05% Tween 20 with 3% BSA. The anti-deacetylated PNAG antibody was kindly provided by Jerry Pier, Harvard Medical School. Blots were washed and probed with a 1:10,000 dilution of rabbit anti-goat horseradish peroxidase conjugate (Pierce, Rockford, IL) in TBS-0.05% Tween 20 with 3% skim milk for 1 to 4 h at room temperature. Alternatively, blots were probed with wheat germ agglutinin horseradish peroxidase conjugate (Sigma Aldrich), as previously described (34). Bound probes were visualized on X-ray film with the ECL chemiluminescence kit (Amersham Biosciences, Pittsburgh, PA).
The biotinylated lectin screening kit-I (Vector Laboratories, Burlingame, CA) was used to screen blots for the presence of other polysaccharides released from stationary-phase cells. Blots were blocked at room temperature for at least 1 h in TBS-0.1% Tween 20 with or without 5% BSA, depending on the optimized conditions for individual lectins, followed by 1 h of incubation at room temperature with 5 μg/ml lectin in TBS-0.1% Tween 20. Bound lectins were detected with the Vectastain Elite ABC kit (Vector Laboratories), as recommended by the manufacturer for Western blotting applications, and visualized on X-ray film with the ECL kit.
Scanned film images were adjusted with the brightness and contrast functions in Microsoft Office Picture Manager software.
Microtiter plate biofilm detachment assay.
Forty millimolar sodium metaperiodate (NaIO4) (Sigma-Aldrich) in water, 40 μg/ml purified recombinant dispersin B (kindly provided by Kane Biotech Inc., Winnipeg, Manitoba, Canada) in sodium phosphate buffer (50 mM sodium phosphate [pH 5.8], 100 mM NaCl), 100 μg/ml proteinase K (Roche Applied Science) in 10 mM Tris-HCl (pH 7.5), 10 U/ml trypsin (Promega Corp., Madison, WI) in 10 mM Tris-HCl (pH 7.5), 100 μg/ml chymotrypsin (Sigma-Aldrich) in 10 mM Tris-HCl (pH 7.5), and 100 μg/ml thermolysin (Sigma-Aldrich) in 10 mM Tris-HCl (pH 7.5) were tested for their ability to detach preformed S. lugdunensis biofilms from polystyrene microtiter plate wells. For certain experiments, proteinase K was inactivated by boiling for 40 min.
Biofilms grown in TSBgluc1% were formed in wells of microtiter plates and washed twice with deionized water, as described above for the microtiter plate biofilm formation assay. One hundred microliters of NaIO4, enzyme, or a suitable control was carefully added to minimize mechanical detachment of biofilms. Plates were incubated at 37°C for 2 h, and contents of wells were discarded and washed twice with deionized water. Plates were air dried overnight, stained with 0.1% safranin for 1 min, and processed as described above to quantify the amount of stained biofilm remaining after treatment, relative to that after treatment with the control reagent. Four wells were measured for each treatment condition. Assays were repeated two or three times on separate days with similar results.
CSLM.
Biofilms were grown for microscopy in four-well chambered coverglass (Lab-Tek II; Nalge Nunc International). Overnight cultures were adjusted and diluted 1:50 with TSBgluc1% as described above for the microtiter plate biofilm formation assay. One-milliliter aliquots were added to chamber wells and statically incubated for 20 to 24 h. Medium was removed from wells, and biofilms were rinsed with 1 ml PBS and stained for fluorescent confocal scanning laser microscopy (CSLM). To visualize PNAG among biofilm cells, biofilms were incubated in the dark for 15 min with 1 ml PBS containing 0.09 mg/ml wheat germ agglutinin-Oregon Green 488 conjugate (Molecular Probes, Eugene, OR) and 5 μg/ml FM 4-64 (Molecular Probes), a lipophilic styryl membrane dye that binds bacterial cell membranes (66). Stains were removed, and wells were rinsed with 2 ml PBS before imaging. Extracellular proteins among biofilm cells were visualized by incubation in the dark for 30 min with 1 ml undiluted SYPRO Ruby protein gel stain (Molecular Probes) containing 0.167 μM Syto-9 nucleic acid stain (Molecular Probes). Stains were removed before imaging.
Confocal images were acquired on an LSM510 equipped with an Axiovert 100 M inverted microscope using a Plan-Apochromat 100×/1.4 NA oil immersion objective (Carl Zeiss, Inc., Thornwood, NY). An argon laser was used to excite the fluorophores at wavelengths of 458 nm for SYPRO Ruby and 488 nm for Oregon Green (wheat germ agglutinin), FM 4-64, and Syto-9. Red fluorescence from SYPRO Ruby and FM 4-64 was detected with an LP 650 filter. Green fluorescence was detected from Oregon Green with a BP 505-550 filter and from Syto-9 with a BP 505-530 filter. Microscopy was performed on at least three different days. Images were prepared with the LSM510 software. Red/green fluorescence ratios to assess biofilm protein were calculated on SYPRO Ruby/Syto-9 images with KS 400 version 3.0 software (Carl Zeiss, Inc.). The fluorescence area (in square pixels) was averaged for images taken in two areas per biofilm from two independent biofilms.
Statistical analysis.
Data were analyzed with Student's t test using JMP 6.0.0 software (SAS Institute, Inc., Cary, NC).
Nucleotide sequence accession numbers.
Accession numbers for complete S. lugdunensis icaADBC operon sequences that have been deposited in GenBank, National Center for Biotechnology Information (http://www.ncbi.nlm.nih.gov) are as follows: S. lugdunensis isolate IDRL-2414, EF546620; S. lugdunensis isolate IDRL-2664, EF546621; S. lugdunensis isolate IDRL-5204, EF546622; S. lugdunensis isolate IDRL-5256, EF546623; and S. lugdunensis isolate IDRL-5258, EF546624.
RESULTS
S. lugdunensis clinical isolates form biofilm.
We recently described how antimicrobial agents affect biofilm formation by a collection of S. lugdunensis clinical isolates (23). A majority of the isolates were recovered from infections associated with biofilm etiology, including prosthetic joint infection, endocarditis, and intravascular catheter infection (Table 1). To further characterize S. lugdunensis biofilm formation, we evaluated this collection using in vitro biofilm formation assays under growth conditions known to support staphylococcal biofilm formation, namely, TSBgluc1% (13, 40).
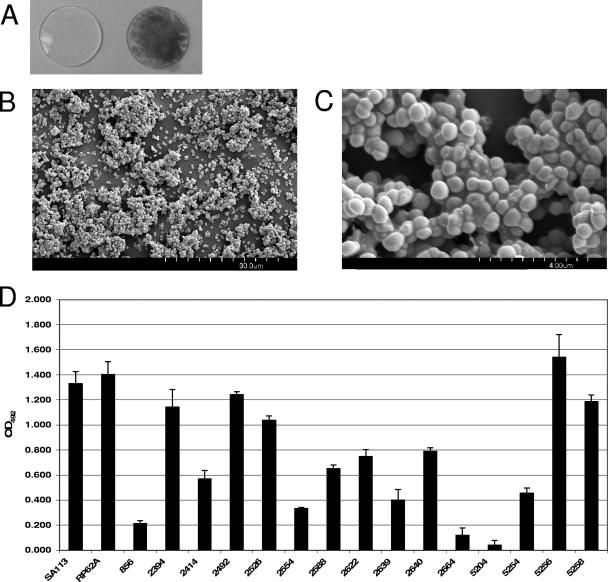
S. lugdunensis IDRL-2640, a folliculitis isolate, was incubated for 24 h in TSBgluc1% with a disk cut from silicone elastomer, a type of material used to manufacture intravascular catheters. Planktonic cells were removed by gentle washing and the disk was stained with safranin to visualize adherent bacteria. Figure 1A shows that the organism formed a strong confluent layer on the disk. SEM visualization of a duplicate disk revealed cells growing in clusters with the development of microcolonies across the disk surface (Fig. 1B). When viewed at higher magnification (Fig. 1C), cells appeared to be organized into a three-dimensional architecture held together by an extracellular polymeric substance. These properties are consistent with organisms growing as a biofilm.
FIG. 1.
Characterization of biofilm formation by S. lugdunensis clinical isolates. (A) S. lugdunensis biofilm formation on silicone elastomer. Disks cut from nonreinforced silicone elastomer sheeting were sterilized and incubated for 24 h in TSBgluc1% (left) or TSBgluc1% containing S. lugdunensis IDRL-2640 (right). Disks were rinsed and stained with safranin to visualize biofilms. (B) Scanning electron micrograph (magnification, ×1,800) of S. lugdunensis IDRL-2640 biofilm formed on a silicone elastomer disk after 24 h. (C) Higher-magnification (×13,000) SEM of biofilm shown in panel B. (D) Biofilm formation by S. aureus SA113, S. epidermidis RP62A, and 15 clinical S. lugdunensis isolates on polystyrene when grown in TSBgluc1% for 24 h. Biofilms were stained with safranin, resuspended in 30% glacial acetic acid, and quantified by OD492. Bars are the averages of measurements taken from four duplicate wells. Error bars represent the standard deviations. The assay was repeated three times, and representative data from a single replicate are shown.
The relative ability of each S. lugdunensis isolate to form biofilm was assessed using a microtiter plate biofilm formation assay. Figure 1D shows that all isolates were able to adhere to polystyrene. Compared to the strong biofilm-forming strains S. aureus SA113 and S. epidermidis RP62A, the biofilm formation phenotype varied widely among the S. lugdunensis isolates. In particular, isolates IDRL-2664 and IDRL-5204 were poor biofilm formers, whereas isolates IDRL-2394, IDRL-5256, and IDRL-5258 formed robust biofilms. This result is similar to previously observed diversities among clinically relevant S. aureus and CNS in their abilities to form biofilm in the microtiter plate assay (4, 14, 40). No correlation between the source of infection (Table 1) and the degree of biofilm formation existed for this set of staphylococci.
S. lugdunensis biofilm formation is affected by environmental conditions.
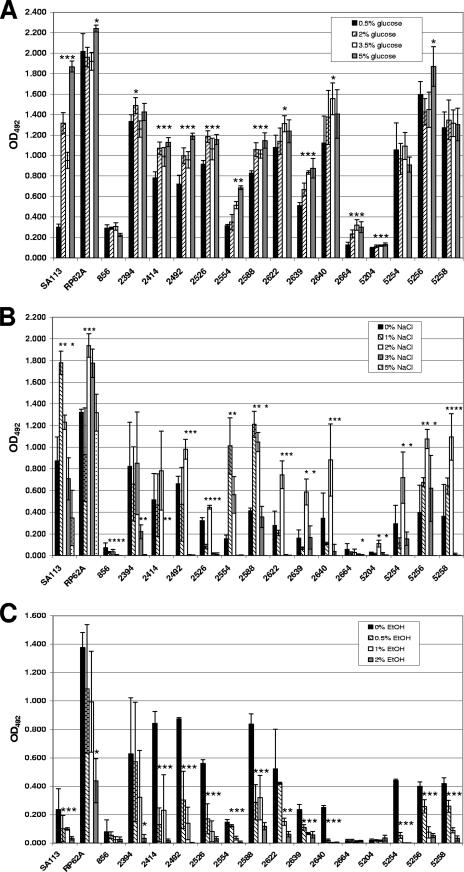
Changes in exogenous factors present in growth media that enrich or stress the growth environment, including glucose (7, 15, 44), increasing osmolarity (38, 44, 51), and alcohols (39, 40), influence S. aureus and S. epidermidis in vitro biofilm formation. We tested the effect of increasing concentrations of glucose, sodium chloride, and ethanol on the collection of S. lugdunensis isolates. Figure 2A shows that statistically measurable increases in biofilm formation in response to heightened glucose levels were observed for S. aureus SA113, S. epidermidis RP62A, and 80% (12/15) of the S. lugdunensis isolates. Three S. lugdunensis isolates, IDRL-856, IDRL-5254, and IDRL-5258, formed equivalent amounts of biofilm independent of the amount of glucose present in the environment.
FIG. 2.
Effect of environmental factors on S. lugdunensis biofilm formation. (A) Effect of increasing glucose concentrations on biofilm formation. S. aureus SA113, S. epidermidis RP62A, and S. lugdunensis clinical isolates were grown in wells of polystyrene microtiter plates in TSB supplemented with the indicated concentrations of glucose. (B) S. lugdunensis biofilm response to sodium chloride. Organisms were grown in microtiter plates in TSBgluc1% with the indicated concentrations of sodium chloride. (C) Biofilm formation in the presence of ethanol. Biofilm formation on polystyrene was assayed after growth in TSBgluc1% containing various concentrations of ethanol; 2% ethanol inhibited growth of S. lugdunensis isolates IDRL-2554, IDRL-2640, and IDRL-5254. In each graph, bars are the average biofilm formation in four wells. Error bars represent the standard deviations. Asterisks indicate statistically significant increases or decreases in biofilm formation compared to biofilm formation in the absence (B and C) or the lowest concentration (A) of each environmental factor tested (P < 0.05 by Student's t test). Data are representative of three replicate experiments with similar results.
Medium enrichment with sodium chloride at concentrations of up to 5% (wt/vol) has previously been shown to increase biofilm formation in S. aureus, S. epidermidis, and several other CNS (44, 51). We found that S. aureus SA113 and S. epidermidis RP62A biofilm levels increased, as expected, when grown with 1 to 2% or 1 to 3% sodium chloride, respectively (Fig. 2B). Likewise, 73% (11/15) of the S. lugdunensis isolates formed more biofilm in medium containing 1 or 2% sodium chloride. In contrast, higher concentrations of salt substantially reduced S. lugdunensis adherence to microtiter wells (Fig. 2B). Essentially no measurable quantity of biofilm could be detected when S. lugdunensis isolates were incubated with 5% sodium chloride. Compared to baseline biofilm formation, S. epidermidis RP62A biofilm was not affected in 5% sodium chloride, whereas the level of S. aureus SA113 biofilm was significantly reduced yet still discernible. Growth of all species tested was not affected by the increasing salt concentrations, as verified by measuring the OD of the biomass in each well prior to the removal of planktonic cells (data not shown). This suggests that high sodium chloride concentrations directly interfere with the process of S. lugdunensis biofilm formation.
Ethanol concentrations of up to 6% (vol/vol) are capable of stimulating biofilm formation by clinical S. epidermidis isolates (39). Interestingly, both of our positive biofilm-forming S. aureus and S. epidermidis reference strains exhibited significant reductions in biofilm production with increasing ethanol concentrations (Fig. 2C). Most ethanol concentrations also reduced the levels of biofilm production by all S. lugdunensis isolates that formed biofilm at a baseline OD492 of ≥0.150 (Fig. 2C). Not only did 2% ethanol prevent the adherence of all S. lugdunensis that formed measurable baseline biofilms, but 4% ethanol was bactericidal (data not shown) for all S. lugdunensis isolates. S. lugdunensis IDRL-2554, IDRL-2640, and IDRL-5254 were the only isolates killed after incubation in 2% ethanol. These results indicate that S. lugdunensis clinical isolates are more susceptible to ethanol than are S. epidermidis clinical isolates and that ethanol appears to be a negative regulator of S. lugdunensis biofilm formation.
Identification and sequencing of icaADBC homologues in S. lugdunensis.
There is overwhelming evidence demonstrating the importance of the icaADBC operon in staphylococcal biofilm formation (10, 28, 45, 54). Early reports suggested the presence of icaA in S. lugdunensis, as detected by PCR or Southern blotting under low-stringency hybridization conditions (10, 64), although no sequences were deposited in public databases. We were previously unable to detect icaA in S. lugdunensis prosthetic joint infection isolates with PCR primers designed to anneal to regions of high homology in the icaA gene sequences of three staphylococcal species (21). The genome of S. lugdunensis has not been sequenced, so the existence of ica gene homologues has remained ambiguous. We performed Southern blotting with S. aureus and S. epidermidis icaA probes under low-stringency conditions in order to determine whether the previously studied isolates truly lacked icaA. Hybridization signals were detected in all of the S. lugdunensis prosthetic joint infection isolates tested (data not shown), which is consistent with the work of Cramton et al. (10) and suggests that we were unable to detect icaA in S. lugdunensis by PCR due to mismatches between our primers and the S. lugdunensis icaA sequence. Using primer sequences reported by Sandoe and Longshaw (64) under low-stringency annealing conditions, we subsequently amplified and sequenced a short region of an icaA homologue from S. lugdunensis, which was extended in both the 5′ and 3′ directions from S. lugdunensis IDRL-2414 and IDRL-2664 using a primer walking technique called restriction site PCR (65).
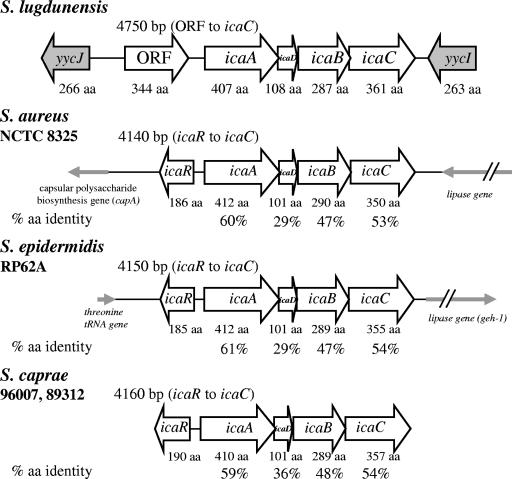
We sequenced and annotated a 7.6-kb region of the S. lugdunensis genome (Fig. 3) which included open reading frames (ORFs) with high degrees of similarity to icaADBC from S. aureus, S. epidermidis, and S. caprae. Surprisingly, an icaR homologue was not located upstream of icaA, as would be predicted from the conserved genomic organization of the locus in other staphylococcal species (1, 9, 10). Rather, a novel ORF lacking homology with known staphylococcal sequences (discussed below) was found directly upstream of, and in the same orientation as, icaA. The ORF-icaADBC genes span a 4.75-kb region that includes a 251-nucleotide intergenic region separating the ORF and icaA. ORFs with high degrees of similarity to yycJ and yycI from S. aureus and S. epidermidis flanked the ORF-icaADBC region on the 5′ and 3′ sides, respectively. The considerable distance separating yycJ and yycI is surprising, as these two genes otherwise occur as part of the YycFG two-component system operon that is highly conserved among gram-positive bacteria (18, 68, 69).
FIG. 3.
Genomic organization of the S. lugdunensis icaADBC locus. A 7.6-kb chromosomal region encompassing the S. lugdunensis icaADBC genes was sequenced from isolates IDRL-2414 and IDRL-2664. An ORF encoding a hypothetical glycosyl hydrolase was found in the position where icaR was expected. The ORF-icaADBC locus was found by PCR to be intact in all S. lugdunensis isolates tested in this work. icaADBC loci of S. epidermidis RP62A (ATCC 35984, GenBank accession number NC_002976), S. aureus NCTC 8325 (GenBank accession number NC_007795), and S. caprae strains 96007 (GenBank accession number AF246926) and 89318 (GenBank accession AF246927) are shown for comparison. Percent amino acid identities of S. lugdunensis genes with each homologue were determined with ClustalW. Slashes on the lipase genes indicate that they are not drawn to the same scale as the rest of the figure.
Inability to detect icaR in the S. lugdunensis genome.
The absence of icaR upstream of icaA led us to question whether it was located elsewhere in the genome of S. lugdunensis. No hybridization signals were detected upon low-stringency Southern blotting with probes generated from the full-length sequences of icaR from either S. aureus or S. epidermidis (data not shown). We also attempted low-annealing-temperature PCR with the primers used to generate the icaR Southern probes; however, this resulted in nonspecific annealing (data not shown). Although we cannot definitively conclude that icaR is absent in S. lugdunensis, these data strongly suggest that any icaR homologue would have very little similarity to known icaR sequences from other staphylococci.
A novel ORF with predicted glycosyl hydrolase activity.
The 1,035-nucleotide ORF located upstream of the icaA start codon is predicted to encode a 344-amino-acid protein that is not similar to any currently known staphylococcal sequences as determined by nucleotide and protein BLAST searches (http://www.ncbi.nlm.nih.gov/BLAST/). SignalP 3.0 (http://www.cbs.dtu.dk/services/SignalP/) (3) analysis of the sequence predicted that the hypothetical protein contains an N-terminal signal sequence between amino acid residues 25 and 26. The Conserved Domains Database (http://www.ncbi.nlm.nih.gov/Structure/cdd/cdd.shtml) identified a putative conserved domain within the predicted ORF amino acid sequence that closely resembles the glycosyl hydrolase family 20 catalytic domain (accession number pfam00728) and a group of N-acetyl-β-hexosaminidases (Chb) (accession number COG3525.2) that are involved in carbohydrate transport and metabolism (30). The substrate-binding pockets of family 20 glycosyl hydrolases are lined with several tryptophan residues that create a hydrophobic environment (71); such tryptophan residues are conserved in the predicted ORF amino acid sequence.
The translated protein sequences most closely related to the translated ORF sequence, as identified in a TBLASTX 2.2.16 search, were the dispersin B (DspB) homologues from Actinobacillus pleuropneumoniae and Actinobacillus actinomycetemcomitans (35, 37). Dispersin B is an N-acetyl-β-hexosaminidase that cleaves the β-1,6-linkages in polymers of N-acetylglucosamine (32, 35). A ClustalW (http://www.ebi.ac.uk/clustalw/) comparison showed the ORF amino acid sequence to be 26% identical to the dispersin B homologues from the two Actinobacillus species.
Contribution of ica locus sequence variability to biofilm formation ability.
We amplified the 7.6-kb genomic region spanning from yycJ through yycI (Fig. 3) in all S. lugdunensis isolates except IDRL-2492 and IDRL-2639 (data not shown). A 4.9-kb product encompassing the ORF-icaADBC region (Fig. 3) was amplified from the two remaining isolates, indicating that the ORF-icaADBC genes are intact in all isolates in our collection.
We next asked whether the variability in biofilm formation among S. lugdunensis isolates (Fig. 1D) could be explained by differences in the ORF-icaADBC primary sequence. In addition to IDRL-2414 and IDRL-2664, we fully sequenced 7.6-kb PCR products from IDRL-5204, IDRL-5256, and IDRL-5258 (sequences deposited in GenBank). These isolates included a poor biofilm producer (IDRL-5204), intermediate biofilm producers (IDRL-2414 and IDRL-2664), and strong biofilm producers (IDRL-5256 and IDRL-5258) (Fig. 1D). The sequences of IDRL-2414, IDRL-2664, and IDRL-5204 were identical, but the sequences of the strongest biofilm formers were different at one or more locations. IDRL-5258 contained an R274Q change at amino acid 274 of IcaA. IDRL-5256 contained many single-nucleotide changes throughout the sequenced region. Eleven variations were found in the yycJ-ORF intergenic region, one variation occurred in the ORF-icaA intergenic region, and four variations were located between icaC-yycI. In addition, the following silent (noncoding) and coding mutations were found in each of the coding regions: yycJ, six silent; ORF, four silent, S26A, and E44D; icaA, eight silent; icaD, four silent; icaB, four silent and H23Q; icaC, three silent; and yycI, four silent. Despite the correlation of sequence variations found in the most proficient biofilm-forming isolates, it is not clear from these data whether the ORF-icaADBC locus primary sequence is a contributing factor in the relative ability of S. lugdunensis isolates to form biofilm.
Elucidation of the role of PNAG in S. lugdunensis biofilm formation.
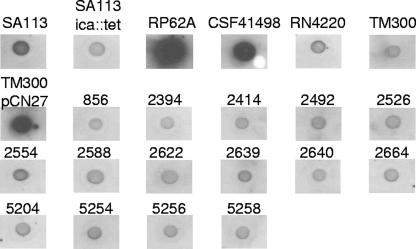
We tested the functional role of the S. lugdunensis icaADBC genes in biofilm formation by assaying for PNAG in biofilm or stationary-phase cells by immunoblotting with an anti-deacetylated PNAG antibody (Fig. 4 and data not shown). Figure 4 shows that PNAG was strongly detected in S. epidermidis strains RP62A and CSF41498 and in a PNAG-overexpressing strain of S. carnosus TM300 that carries plasmid pCN27 (28). PNAG production by S. aureus strains RN4220 and SA113 was less abundant, and PNAG was not detected in the negative control strain S. carnosus TM300. Unexpectedly, PNAG was not detected in any of the 15 S. lugdunensis isolates. PNAG was also not detected in extracts from stationary-phase cells harvested after overnight growth (data not shown).
FIG. 4.
Inability to detect PNAG in S. lugdunensis extracts by immunoblotting. Extracts from 24-h biofilm cells grown in TSBgluc1% were immunoblotted with an antibody raised against deacetylated PNAG. Cells were boiled in 0.5 M EDTA, and the supernatants were treated with proteinase K prior to blotting on nitrocellulose.
To confirm the lack of detection of PNAG from S. lugdunensis cells by an alternate method, stationary-phase cell extracts were probed with labeled wheat germ agglutinin. Signal levels from all S. lugdunensis strains were equivalent to those from the non-PNAG-producing S. aureus SA113 ica::tet and S. carnosus TM300 negative controls (data not shown).
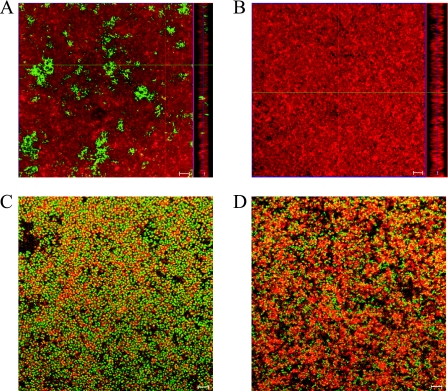
We used CSLM to visualize S. lugdunensis biofilms in comparison with biofilms formed by PNAG-producing S. epidermidis RP62A (Fig. 5A and B). Cells were stained with FM 4-64, a red lipophilic plasma membrane dye, and PNAG was stained with Oregon Green-conjugated wheat germ agglutinin. S. epidermidis RP62A formed a thick, multilayered biofilm interspersed with large and abundant structures of PNAG (Fig. 5A). In contrast, under identical microscopy settings, S. lugdunensis IDRL-2640 formed a thick and dense biofilm devoid of detectable PNAG (Fig. 5B). Similar results were observed with S. lugdunensis IDRL-5258 biofilms (data not shown). These images indicate that (i) S. lugdunensis isolates are able to form thick, multicellular biofilms and (ii) PNAG is not a major recognizable component of S. lugdunensis in vitro biofilms.
FIG. 5.
Assessment of the S. lugdunensis biofilm extracellular matrix with CSLM. Biofilms were grown in chambered coverglass wells, stained, and visualized by CSLM with a 100× oil immersion objection. (A and B) Microscopic visualization of the presence or absence of PNAG in the biofilm extracellular matrix of S. epidermidis RP62A (A) or S. lugdunensis IDRL-2640 (B). Large PNAG structures (green), stained with fluorescently labeled wheat germ agglutinin, were located among cells (red), stained with the lipophilic membrane dye FM 4-64, in S. epidermidis RP62A biofilms but not S. lugdunensis IDRL-2640 biofilms. Images show a single x-y slice from the center of the biofilm and the biofilm profile from the y-z axis (right). (C and D) Detection of extracellular proteins in biofilms of S. epidermidis RP62A (C) and S. lugdunensis IDRL-5258 (D). Proteins (red) were stained with SYPRO Ruby. Bacteria were stained with Syto-9, a vital nucleic acid stain. Yellow areas indicate protein and cellular colocalization. Bars, 5 μm.
Screen for other extracellular matrix polysaccharides.
We hypothesized that in the absence of PNAG, alternative polysaccharides may be present in the matrix of S. lugdunensis biofilms. Six lectins, i.e., concanavalin A (binds glucose and mannose), Dolichos biflorus agglutinin (binds N-acetylgalactosamine), soybean agglutinin (binds galactose and N-acetylgalactosamine), peanut agglutinin (binds galactose), Ricinus communis agglutinin I (binds galactose and N-acetylgalactosamine), and Ulex europaeus agglutinin I (binds fucose), were used to assay for other extracellular matrix polysaccharides in S. lugdunensis stationary-phase cell extracts by immunoblotting. Concanavalin A bound to all staphylococcal strains tested (data not shown), which was not unexpected based on its broad specificity for common sugars. Soybean agglutinin selectively bound to S. carnosus isolates, and Ricinus communis agglutinin I selectively bound to S. epidermidis and the S. carnosus PNAG-overexpressing strain (data not shown). However, none of the lectins bound to the S. lugdunensis extracts (data not shown), providing evidence that several other types of polysaccharides, in addition to PNAG, are not significant components of the S. lugdunensis biofilm matrix.
Detachment of S. lugdunensis biofilms with proteases but not carbohydrate-degrading reagents.
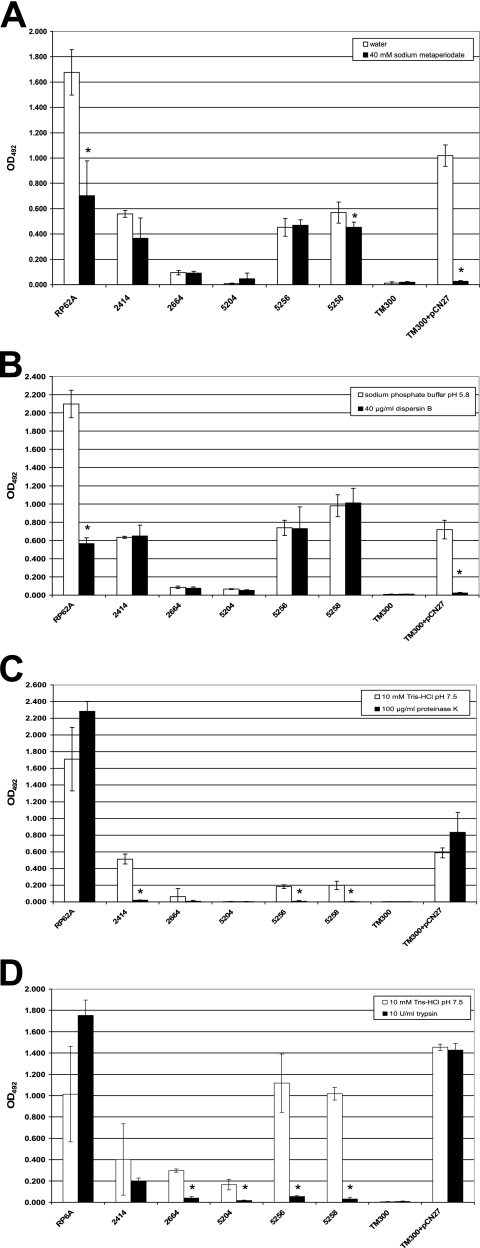
Several reports have recently demonstrated the existence of PNAG-independent, protein-mediated staphylococcal biofilm formation (5, 29, 59, 72). The paucity of PNAG in S. lugdunensis biofilms led us to examine the composition of the biofilm matrix with a chemical and enzymatic detachment approach. Sodium metaperiodate and dispersin B degrade PNAG, thereby releasing PNAG-containing biofilms from their associated surfaces (35, 36, 75). As expected, preformed biofilms of S. epidermidis RP62A and PNAG-overproducing S. carnosus TM300(pCN27) that were treated with sodium metaperiodate (Fig. 6A) and dispersin B (Fig. 6B) were susceptible to detachment by both reagents (P ≤ 0.001 by Student's t test). S. carnosus biofilms, which likely contain large amounts of PNAG without additional stabilizing factors, were essentially completely released from microtiter wells. In contrast, despite substantial detachment, much higher levels of S. epidermidis RP62A biofilms remained after treatment with either sodium metaperiodate or dispersin B. The biofilm matrix components of this strain are known to include extracellular teichoic acids and proteins, in addition to large amounts of PNAG (62), which may have assisted in protecting or stabilizing the biofilm from detachment. Confirmatory of our immunoblotting and microscopy data, biofilms of all S. lugdunensis isolates resisted detachment by dispersin B (Fig. 6B and data not shown). Sodium metaperiodate had moderate to little effect on the release of 53% (8/15) S. lugdunensis biofilms (Fig. 6A and data not shown). Seven S. lugdunensis biofilms (IDRL-2394, IDRL-2526, IDRL-2554, IDRL-2588, IDRL-2622, IDRL-2640, and IDRL-5258) demonstrated greater susceptibility to sodium metaperiodate (P ≤ 0.047 by Student's t test), suggesting that the biochemical constituents in the extracellular matrix of S. lugdunensis biofilms may vary among isolates.
FIG. 6.
S. lugdunensis biofilms are susceptible to detachment by proteases but not carbohydrate-degrading reagents. Twenty-four-hour biofilms of S. epidermidis RP62A, S. carnosus TM300, S. carnosus TM300(pCN27), and S. lugdunensis clinical isolates formed in microtiter plates were washed and incubated with various chemical or enzymatic treatments for 2 h at 37°C to determine which agents could detach S. lugdunensis biofilms. Biofilm remaining in wells after treatment was stained with 0.1% safranin, resuspended in 30% glacial acetic acid, and quantitated by OD492. Results for five representative S. lugdunensis isolates that form biofilm to various degrees are shown here. (A) Sodium metaperiodate treatment. (B) Dispersin B treatment. (C) Proteinase K treatment. (D) Trypsin treatment. Bars are the average stained biofilm remaining in four wells after treatment. Error bars show the standard deviations. Asterisks indicate statistically significant decreases in biofilm detachment after treatment compared to biofilm detachment in the corresponding buffer (P < 0.05 by Student's t test). Data are representative of two or three replicate experiments with similar result
We next tested the ability of several proteases with various substrate specificities to detach biofilms of S. lugdunensis and the PNAG-containing control strains. Figure 6C shows that proteinase K had no effect on the PNAG-positive control strains, while biofilms of all S. lugdunensis isolates that formed substantial biofilms (Fig. 1D) were completely removed (P ≤ 0.005 by Student's t test). In order to demonstrate that the overwhelming detachment of S. lugdunensis biofilms was due to the enzymatic activity of proteinase K, we repeated the experiment with heat-inactivated proteinase K and found that the detachment effect was abolished (data not shown). Concordant results were obtained upon treatment of biofilms with trypsin (Fig. 6D and data not shown) (P ≤ 0.041 by Student's t test), except that two S. lugdunensis isolates (IDRL-2414 and IDRL-2526) were noticeably more resistant to release by trypsin. Two other proteases, thermolysin and chymotrypsin, were also found to selectively detach S. lugdunensis biofilms but not PNAG-containing biofilms formed by S. epidermidis RP62A or S. carnosus TM300(pCN27) (data not shown). These experiments provide evidence that proteins are important for S. lugdunensis in vitro biofilm formation on polystyrene when cells are grown in TSBgluc1%.
Visualization of extracellular proteins in S. lugdunensis biofilms by CSLM.
We stained biofilms of S. epidermidis RP62A and S. lugdunensis IDRL-5258 with the red fluorescent protein dye SYPRO Ruby in order to visualize extracellular proteins among biofilm cells, which were stained with the green nucleic acid stain Syto-9. Extracellular proteins were visible in biofilms of both S. epidermidis RP62A (Fig. 5C) and S. lugdunensis IDRL-5258 (Fig. 5D). S. lugdunensis biofilms appeared to contain more protein than S. epidermidis biofilms. To more accurately assess the relative abundance of extracellular protein per number of cells in biofilms formed by either organism, we calculated the ratio of protein to cell fluorescence. The average ratio of protein to cell fluorescence measured in S. lugdunensis IDRL-5258 biofilms was statistically higher than that in S. epidermidis RP62A biofilms (0.965 ± 0.285 [standard deviation] versus 0.385 ± 0.114, respectively; P < 0.01 by Student's t test), supporting the hypothesis that proteins are an important component of this organism's biofilm matrix.
DISCUSSION
The unusual disposition of S. lugdunensis to cause particularly virulent infections that are not typically associated with CNS attests to the unique behavior and nature of this organism. In this study, we examined in vitro biofilm formation characteristics and began to understand the mechanism of biofilm formation by a collection of S. lugdunensis clinical isolates from both biofilm- and non-biofilm-associated infections. Our work revealed that the process of biofilm formation in S. lugdunensis is substantially different from the mechanism of in vitro biofilm formation typically employed by S. epidermidis and S. aureus. All S. lugdunensis clinical isolates we studied contain icaADBC homologues, but not all isolates were proficient in forming biofilm on polystyrene. The phenotype of S. lugdunensis biofilms in response to the presence of sodium chloride and ethanol in the environment was significantly decreased, whereas biofilms of other staphylococci have been shown to increase under similar conditions (38-40, 44, 51). Moreover, detectable amounts of PNAG were lacking in S. lugdunensis biofilms. Rather, S. lugdunensis biofilms appear to be composed predominantly of proteins. Thus, the icaADBC locus, which is a common mechanism of biofilm formation in staphylococci (25), is present but not utilized in the formation of biofilms by clinical isolates of S. lugdunensis under in vitro conditions.
The clinical significance of S. lugdunensis as a notable human pathogen responsible for aggressive nosocomial and community-acquired infections has been well established in the 20 years since its initial description (74). However, our present comprehension of the virulence factors involved in S. lugdunensis pathogenesis is relatively limited (17, 43, 50, 52, 53, 57). In our experience working with S. lugdunensis (in accordance with Mitchell et al. [50]), we have been unable to introduce DNA and perform genetic analyses with any of the clinical isolates in our collection. This lack of a genetic system may partially explain the slow progress toward understanding S. lugdunensis virulence. Nevertheless, two virulence factors that facilitate primary attachment of bacteria to host tissues, von Willebrand factor-binding protein and fibrinogen-binding protein, have been characterized in S. lugdunensis (50, 52, 53). The binding of host proteins by S. lugdunensis is likely the first step of biofilm formation executed by this organism in vivo. Our results suggest that additional proteins, rather than the polysaccharide produced by the icaADBC locus, may be important in the subsequent steps of biofilm accumulation and intercellular adhesion.
Recent characterizations of icaADBC-negative S. epidermidis strains and icaADBC deletion mutants of S. aureus or S. epidermidis strains indicate that staphylococci are genetically equipped to form biofilm via multiple pathways (19, 29, 58, 72) and that the regulatory pathways overseeing ica-dependent and ica-independent biofilm formation are complex (54). Since it is not possible to generate icaADBC-null S. lugdunensis strains, we cannot definitively assert that the proteinaceous biofilms we describe in this work formed independently of the icaADBC locus. However, the decrease in S. lugdunensis biofilm formation in response to high concentrations of sodium chloride and ethanol (Fig. 2) is not consistent with ica-dependent biofilm formation responses that have been observed with other staphylococci under equivalent conditions (54). Recently, Hennig et al. showed that 4% sodium chloride inhibited biofilm formation by a PNAG-negative variant of S. epidermidis CSF41498 (29). These results are similar to the results we obtained with our icaADBC-positive, PNAG-negative S. lugdunensis isolates (Fig. 2B), suggesting that similar modes of regulation of protein-mediated biofilm formation may be present among these strains. Ethanol is known to enhance staphylococcal biofilm formation by down-regulating icaR transcription, thereby allowing continued transcription of icaADBC in the absence of IcaR-mediated repression of the locus (9). Although the apparent absence of icaR in the genomes of the isolates we studied provides a plausible explanation for the inability of ethanol to induce S. lugdunensis biofilm formation (Fig. 2C), as our data suggest that icaADBC does not substantially contribute to biofilm formation, it is unlikely that the ethanol effect observed here is mediated through the icaADBC locus. Obviously, regulation of S. lugdunensis biofilm formation proceeds by novel pathways.
Our data are in agreement with previous reports of biofilm formation by a limited number of S. lugdunensis isolates. Sandoe and Longshaw described an isolate of S. lugdunensis recovered from an infected ventriculoperitoneal shunt that adhered to polystyrene in a microtiter plate assay (64). They reported PCR amplification of icaA with >80% nucleotide sequence identity to that of S. epidermidis and S. aureus, although the sequence was not deposited in any publicly available sequence repository (64). We have confirmed the presence of icaA, as part of a predicted operon resembling those found in S. aureus, S. epidermidis, and S. caprae (Fig. 3), in our collection. However, our sequence analysis of icaA in five S. lugdunensis isolates revealed this gene to be only 59 to 61% identical at the amino acid level to its homologues in the other three species. The explanation for the discrepancy in sequence identity between our five isolates and the single isolate reported previously is unclear, although the low degree of icaA sequence identity between S. lugdunensis and other staphylococci provides an explanation for our failure to detect this gene by PCR in our previous work (21). More recently, 11 S. lugdunensis isolates from implanted medical devices were characterized by ica PCR, biofilm formation assays, and assessment of PNAG production (6). ica genes were detected in 6/11 strains, and only 3/11 formed biofilm in TSB with 0.25% glucose supplementation. In contrast, we found that all S. lugdunensis isolates had icaADBC, leading us to speculate that the authors’ primer design may have failed to amplify S. lugdunensis genes in the remaining isolates due to disparities in sequence between S. lugdunensis ica genes and the ica genes from which the primers were designed. Consistent with our findings, PNAG was not detected by immunoblotting in any of the reportedly ica-positive or ica-negative S. lugdunensis isolates (6). It is interesting to note that these authors grew isolates in brain heart infusion medium supplemented with 1% glucose for PNAG immunoblotting experiments, suggesting that our inability to detect in vitro PNAG production in S. lugdunensis isolates grown in TSBgluc1% was not related to the cultivation conditions.
Further characterization of the biofilm matrix components of two biofilm-forming S. lugdunensis strains that apparently lack icaA has been reported recently (5, 41, 61). It was found that biofilms formed by these two isolates were resistant to detachment by sodium metaperiodate and dispersin B but were susceptible to proteinase K and trypsin detachment (5, 41, 61). Chromatographic and nuclear magnetic resonance analysis of extracellular polymeric extracts of these two strains revealed low or no detectable levels of PNAG but did detect the strong presence of extracellular teichoic acids and protein components, suggesting that these isolates form protein and teichoic acid-mediated biofilms (41, 61). The results of the detachment assays with proteases and carbohydrate-degrading agents that we report here for our icaADBC-positive isolates (Fig. 6) are similar to those for the apparently icaADBC-negative S. lugdunensis isolates reported by Kogan et al. (41) and Sadovskaya et al. (61), indicating that protein-mediated biofilm formation is common among icaADBC-positive and (apparently) icaADBC-negative S. lugdunensis isolates. These authors did not study the extracellular matrix components of any ica-positive biofilm-forming strains in their collection (6). Based on our results, the presence of the ica locus in biofilm-forming S. lugdunensis isolates does not imply PNAG-dependent biofilm formation. Therefore, we advocate that these studies should be performed in order to determine whether teichoic acids, in addition to protein components, are found in the biofilms of the previously reported organisms (6).
Many unanswered questions emerge. Although our data do not reveal evidence for the function of the icaADBC homologues in S. lugdunensis biofilm formation, the fact that at least five independently isolated clinical isolates harbor intact ORFs for the entire locus suggests that there is an evolutionary advantage to maintain this region of the genome. It is possible that the icaADBC genes are expressed in vivo or under in vitro conditions different from those studied here. Aside from staphylococci, PNAG is found in the biofilm matrices of many organisms (12, 33, 37, 75), so it would be quite unexpected if the S. lugdunensis icaADBC locus is unimportant in all aspects of biofilm formation. Additionally, the genomic organization of the region containing the S. lugdunensis ica genes is extremely intriguing. The lack of icaR and the surprising finding that the S. lugdunensis yycJ and yycI homologues are not contained within a single operon raise the question of the evolutionary origin of this genomic region in this species. While we are not aware that yycFG homologues have been positively identified in S. lugdunensis, the yycFG two-component system is known to be essential in S. aureus (18, 49). Further exploration of the yyc homologues in S. lugdunensis will undoubtedly be interesting.
We are currently undertaking steps to determine the function and the role in biofilm formation, if any, of the novel ORF found upstream of S. lugdunensis icaA (Fig. 3). No sequences with similar homology can be identified in other staphylococci, but the similarity to the gene for the biofilm-degrading enzyme dispersin B, coupled with our observation that S. lugdunensis can form PNAG-independent biofilms, may indicate that S. lugdunensis possesses a unique mechanism to allow it to compete for biofilm niches with PNAG-dependent biofilm species.
In conclusion, we have shown that an icaADBC locus with a unique genomic organization is present in 15 clinical S. lugdunensis isolates, but PNAG was not a major component of the biofilms of these organisms. Instead, icaADBC-positive S. lugdunensis isolates formed PNAG-independent biofilms whose mechanism of adherence to polystyrene involved extracellular proteinaceous factors. Elucidation of the protein(s) that governs S. lugdunensis adhesion should yield information on additional biofilm genes present in this genetically intractable organism. In addition, further work is warranted to determine the conditions, if any, under which the S. lugdunensis ica locus is expressed. Once the involvement of icaADBC and other biofilm-associated genes is firmly established, we can begin to understand the novel regulatory pathways controlling S. lugdunensis biofilm formation.
Acknowledgments
We thank Robert Shanks, University of Pittsburgh Medical Center, Pittsburgh, PA, for providing protocols, suggesting experiments, and engaging in many valuable discussions. We acknowledge George O'Toole, Dartmouth Medical School, Hanover, NH, for providing S. aureus strains; Jerry Pier, Harvard Medical School, Boston, MA, for providing the anti-deacetylated PNAG antiserum; Paul Fey, University of Nebraska Medical Center, Omaha, NE, for S. carnosus TM300; Friedrich Götz, University of Tübingen, Tübingen, Germany, for S. carnosus carrying the pCN27 plasmid; James O'Gara, University College Dublin, Dublin, Ireland, for S. epidermidis CSF41498l; and Martin Strathmann and Hans-Curt Flemming, University of Duisburg-Essen, Duisburg, Germany, for providing microscopy protocols. We also thank Kane Biotech, Inc., for providing purified dispersin B. Scott Gamb, Mayo Clinic Electron Microscopy core facility, and Jim Tarara, Mayo Clinic Flow Cytometry/Optical Morphology core facility, provided excellent assistance with electron and confocal microscopy.
Funds for this work were provided, in part, by the Anderson Gift.
Editor: A. Camilli
Footnotes
Published ahead of print on 16 July 2007.
REFERENCES
- 1.Allignet, J., S. Aubert, K. G. Dyke, and N. El Solh. 2001. Staphylococcus caprae strains carry determinants known to be involved in pathogenicity: a gene encoding an autolysin-binding fibronectin and the ica operon involved in biofilm formation. Infect. Immun. 69:712-718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Anguera, I., A. Del Rio, J. M. Miro, X. Matinez-Lacasa, F. Marco, J. R. Guma, G. Quaglio, X. Claramonte, A. Moreno, C. A. Mestres, E. Mauri, M. Azqueta, N. Benito, C. Garcia-de la Maria, M. Almela, M. J. Jimenez-Exposito, O. Sued, E. De Lazzari, and J. M. Gatell. 2005. Staphylococcus lugdunensis infective endocarditis: description of 10 cases and analysis of native valve, prosthetic valve, and pacemaker lead endocarditis clinical profiles. Heart 91:e10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bendtsen, J. D., H. Nielsen, G. von Heijne, and S. Brunak. 2004. Improved prediction of signal peptides: SignalP 3.0. J. Mol. Biol. 340:783-795. [DOI] [PubMed] [Google Scholar]
- 4.Cafiso, V., T. Bertuccio, M. Santagati, F. Campanile, G. Amicosante, M. G. Perilli, L. Selan, M. Artini, G. Nicoletti, and S. Stefani. 2004. Presence of the ica operon in clinical isolates of Staphylococcus epidermidis and its role in biofilm production. Clin. Microbiol. Infect. 10:1081-1088. [DOI] [PubMed] [Google Scholar]
- 5.Chaignon, P., I. Sadovskaya, C. Ragunah, N. Ramasubbu, J. B. Kaplan, and S. Jabbouri. 2007. Susceptibility of staphylococcal biofilms to enzymatic treatments depends on their chemical composition. Appl. Microbiol. Biotechnol. 75:125-132. [DOI] [PubMed] [Google Scholar]
- 6.Chokr, A., D. Watier, H. Eleaume, B. Pangon, J. C. Ghnassia, D. Mack, and S. Jabbouri. 2006. Correlation between biofilm formation and production of polysaccharide intercellular adhesin in clinical isolates of coagulase-negative staphylococci. Int. J. Med. Microbiol. 296:381-388. [DOI] [PubMed] [Google Scholar]
- 7.Christensen, G. D., W. A. Simpson, A. L. Bisno, and E. H. Beachey. 1982. Adherence of slime-producing strains of Staphylococcus epidermidis to smooth surfaces. Infect. Immun. 37:318-326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Christensen, G. D., W. A. Simpson, J. J. Younger, L. M. Baddour, F. F. Barrett, D. M. Melton, and E. H. Beachey. 1985. Adherence of coagulase-negative staphylococci to plastic tissue culture plates: a quantitative model for the adherence of staphylococci to medical devices. J. Clin. Microbiol. 22:996-1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Conlon, K. M., H. Humphreys, and J. P. O'Gara. 2002. icaR encodes a transcriptional repressor involved in environmental regulation of ica operon expression and biofilm formation in Staphylococcus epidermidis. J. Bacteriol. 184:4400-4408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cramton, S. E., C. Gerke, N. F. Schnell, W. W. Nichols, and F. Gotz. 1999. The intercellular adhesion (ica) locus is present in Staphylococcus aureus and is required for biofilm formation. Infect. Immun. 67:5427-5433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cucarella, C., C. Solano, J. Valle, B. Amorena, I. Lasa, and J. R. Penades. 2001. Bap, a Staphylococcus aureus surface protein involved in biofilm formation. J. Bacteriol. 183:2888-2896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Darby, C., J. W. Hsu, N. Ghori, and S. Falkow. 2002. Caenorhabditis elegans: plague bacteria biofilm blocks food intake. Nature 417:243-244. [DOI] [PubMed] [Google Scholar]
- 13.Deighton, M. A., J. Capstick, E. Domalewski, and T. van Nguyen. 2001. Methods for studying biofilms produced by Staphylococcus epidermidis. Methods Enzymol. 336:177-195. [DOI] [PubMed] [Google Scholar]
- 14.de Silva, G. D., M. Kantzanou, A. Justice, R. C. Massey, A. R. Wilkinson, N. P. Day, and S. J. Peacock. 2002. The ica operon and biofilm production in coagulase-negative staphylococci associated with carriage and disease in a neonatal intensive care unit. J. Clin. Microbiol. 40:382-388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dobinsky, S., K. Kiel, H. Rohde, K. Bartscht, J. K. Knobloch, M. A. Horstkotte, and D. Mack. 2003. Glucose-related dissociation between icaADBC transcription and biofilm expression by Staphylococcus epidermidis: evidence for an additional factor required for polysaccharide intercellular adhesin synthesis. J. Bacteriol. 185:2879-2886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Donlan, R. M., and J. W. Costerton. 2002. Biofilms: survival mechanisms of clinically relevant microorganisms. Clin. Microbiol. Rev. 15:167-193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Donvito, B., J. Etienne, L. Denoroy, T. Greenland, Y. Benito, and F. Vandenesch. 1997. Synergistic hemolytic activity of Staphylococcus lugdunensis is mediated by three peptides encoded by a non-agr genetic locus. Infect. Immun. 65:95-100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dubrac, S., and T. Msadek. 2004. Identification of genes controlled by the essential YycG/YycF two-component system of Staphylococcus aureus. J. Bacteriol. 186:1175-1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fitzpatrick, F., H. Humphreys, and J. P. O'Gara. 2005. Evidence for icaADBC-independent biofilm development mechanism in methicillin-resistant Staphylococcus aureus clinical isolates. J. Clin. Microbiol. 43:1973-1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fluit, A. C., J. Verhoef, and F. J. Schmitz. 2001. Frequency of isolation and antimicrobial resistance of gram-negative and gram-positive bacteria from patients in intensive care units of 25 European university hospitals participating in the European arm of the SENTRY Antimicrobial Surveillance Program 1997-1998. Eur. J. Clin. Microbiol. Infect. Dis. 20:617-625. [DOI] [PubMed] [Google Scholar]
- 21.Frank, K. L., A. D. Hanssen, and R. Patel. 2004. icaA is not a useful diagnostic marker for prosthetic joint infection. J. Clin. Microbiol. 42:4846-4849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Frank, K. L., and R. Patel. 2007. Activity of sodium metabisulfite against planktonic and biofilm Staphylococcus species. Diagn. Microbiol. Infect. Dis. 57:355-359. [DOI] [PubMed] [Google Scholar]
- 23.Frank, K. L., E. J. Reichert, K. E. Piper, and R. Patel. 2007. In vitro effects of antimicrobial agents on planktonic and biofilm forms of Staphylococcus lugdunensis clinical isolates. Antimicrob. Agents Chemother. 51:888-895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Freney, J., Y. Brun, M. Bes, H. Meugnier, F. Grimont, P. A. D. Grimont, C. Nervi, and J. Fleurette. 1988. Staphylococcus lugdunensis sp. nov. and Staphylococcus schleiferi sp. nov., two species from human clinical specimens. Int. J. Syst. Bacteriol. 38:168-172. [Google Scholar]
- 25.Gotz, F. 2002. Staphylococcus and biofilms. Mol. Microbiol. 43:1367-1378. [DOI] [PubMed] [Google Scholar]
- 26.Gotz, F., B. Kreutz, and K. H. Schleifer. 1983. Protoplast transformation of Staphylococcus carnosus by plasmid DNA. Mol. Gen. Genet. 189:340-342. [Google Scholar]
- 27.Haile, D. T., J. Hughes, E. Vetter, P. Kohner, R. Snyder, R. Patel, and F. R. Cockerill III. 2002. Frequency of isolation of Staphylococcus lugdunensis in consecutive urine cultures and relationship to urinary tract infection. J. Clin. Microbiol. 40:654-656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Heilmann, C., O. Schweitzer, C. Gerke, N. Vanittanakom, D. Mack, and F. Gotz. 1996. Molecular basis of intercellular adhesion in the biofilm-forming Staphylococcus epidermidis. Mol. Microbiol. 20:1083-1091. [DOI] [PubMed] [Google Scholar]
- 29.Hennig, S., S. Nyunt Wai, and W. Ziebuhr. 2007. Spontaneous switch to PIA-independent biofilm formation in an ica-positive Staphylococcus epidermidis isolate. Int. J. Med. Microbiol. 297:117-122. [DOI] [PubMed] [Google Scholar]
- 30.Henrissat, B. 1991. A classification of glycosyl hydrolases based on amino acid sequence similarities. Biochem. J. 280:309-316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Herchline, T. E., and L. W. Ayers. 1991. Occurrence of Staphylococcus lugdunensis in consecutive clinical cultures and relationship of isolation to infection. J. Clin. Microbiol. 29:419-421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Itoh, Y., X. Wang, B. J. Hinnebusch, J. F. Preston III, and T. Romeo. 2005. Depolymerization of beta-1,6-N-acetyl-d-glucosamine disrupts the integrity of diverse bacterial biofilms. J. Bacteriol. 187:382-387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Izano, E. A., I. Sadovskaya, E. Vinogradov, M. H. Mulks, K. Velliyagounder, C. Ragunath, W. B. Kher, N. Ramasubbu, S. Jabbouri, M. B. Perry, and J. B. Kaplan. 2007. Poly-N-acetylglucosamine mediates biofilm formation and antibiotic resistance in Actinobacillus pleuropneumoniae. Microb. Pathog. 43:1-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jefferson, K. K., and N. Cerca. 2006. Bacterial-bacterial cell interactions in biofilms: detection of polysaccharide intercellular adhesins by blotting and confocal microscopy, Methods Mol. Biol. 341:119-126. [DOI] [PubMed] [Google Scholar]
- 35.Kaplan, J. B., C. Ragunath, N. Ramasubbu, and D. H. Fine. 2003. Detachment of Actinobacillus actinomycetemcomitans biofilm cells by an endogenous beta-hexosaminidase activity. J. Bacteriol. 185:4693-4698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kaplan, J. B., C. Ragunath, K. Velliyagounder, D. H. Fine, and N. Ramasubbu. 2004. Enzymatic detachment of Staphylococcus epidermidis biofilms. Antimicrob. Agents Chemother. 48:2633-2636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kaplan, J. B., K. Velliyagounder, C. Ragunath, H. Rohde, D. Mack, J. K. Knobloch, and N. Ramasubbu. 2004. Genes involved in the synthesis and degradation of matrix polysaccharide in Actinobacillus actinomycetemcomitans and Actinobacillus pleuropneumoniae biofilms. J. Bacteriol. 186:8213-8220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Knobloch, J. K., K. Bartscht, A. Sabottke, H. Rohde, H. H. Feucht, and D. Mack. 2001. Biofilm formation by Staphylococcus epidermidis depends on functional RsbU, an activator of the sigB operon: differential activation mechanisms due to ethanol and salt stress. J. Bacteriol. 183:2624-2633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Knobloch, J. K., M. A. Horstkotte, H. Rohde, P. M. Kaulfers, and D. Mack. 2002. Alcoholic ingredients in skin disinfectants increase biofilm expression of Staphylococcus epidermidis. J. Antimicrob. Chemother. 49:683-687. [DOI] [PubMed] [Google Scholar]
- 40.Knobloch, J. K., M. A. Horstkotte, H. Rohde, and D. Mack. 2002. Evaluation of different detection methods of biofilm formation in Staphylococcus aureus. Med. Microbiol. Immunol. 191:101-106. [DOI] [PubMed] [Google Scholar]
- 41.Kogan, G., I. Sadovskaya, P. Chaignon, A. Chokr, and S. Jabbouri. 2006. Biofilms of clinical strains of Staphylococcus that do not contain polysaccharide intercellular adhesin. FEMS Microbiol. Lett. 255:11-16. [DOI] [PubMed] [Google Scholar]
- 42.Kreiswirth, B. N., S. Lofdahl, M. J. Betley, M. O'Reilly, P. M. Schlievert, M. S. Bergdoll, and R. P. Novick. 1983. The toxic shock syndrome exotoxin structural gene is not detectably transmitted by a prophage. Nature 305:709-712. [DOI] [PubMed] [Google Scholar]
- 43.Lambe, D. W., Jr., K. P. Ferguson, J. L. Keplinger, C. G. Gemmell, and J. H. Kalbfleisch. 1990. Pathogenicity of Staphylococcus lugdunensis, Staphylococcus schleiferi, and three other coagulase-negative staphylococci in a mouse model and possible virulence factors. Can. J. Microbiol. 36:455-463. [DOI] [PubMed] [Google Scholar]
- 44.Lim, Y., M. Jana, T. T. Luong, and C. Y. Lee. 2004. Control of glucose- and NaCl-induced biofilm formation by rbf in Staphylococcus aureus. J. Bacteriol. 186:722-729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mack, D., A. P. Davies, L. G. Harris, H. Rohde, M. A. Horstkotte, and J. K. Knobloch. 2007. Microbial interactions in Staphylococcus epidermidis biofilms. Anal. Bioanal. Chem. 387:399-408. [DOI] [PubMed] [Google Scholar]
- 46.Mack, D., W. Fischer, A. Krokotsch, K. Leopold, R. Hartmann, H. Egge, and R. Laufs. 1996. The intercellular adhesin involved in biofilm accumulation of Staphylococcus epidermidis is a linear beta-1,6-linked glucosaminoglycan: purification and structural analysis. J. Bacteriol. 178:175-183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Maira-Litran, T., A. Kropec, C. Abeygunawardana, J. Joyce, G. Mark III, D. A. Goldmann, and G. B. Pier. 2002. Immunochemical properties of the staphylococcal poly-N-acetylglucosamine surface polysaccharide. Infect. Immun. 70:4433-4440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Maira-Litran, T., A. Kropec, D. A. Goldmann, and G. B. Pier. 2005. Comparative opsonic and protective activities of Staphylococcus aureus conjugate vaccines containing native or deacetylated staphylococcal poly-N-acetyl- beta-(1-6)-glucosamine. Infect. Immun. 73:6752-6762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Martin, P. K., T. Li, D. Sun, D. P. Biek, and M. B. Schmid. 1999. Role in cell permeability of an essential two-component system in Staphylococcus aureus. J. Bacteriol. 181:3666-3673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mitchell, J., A. Tristan, and T. J. Foster. 2004. Characterization of the fibrinogen-binding surface protein Fbl of Staphylococcus lugdunensis. Microbiology 150:3831-3841. [DOI] [PubMed] [Google Scholar]
- 51.Moretro, T., L. Hermansen, A. L. Holck, M. S. Sidhu, K. Rudi, and S. Langsrud. 2003. Biofilm formation and the presence of the intercellular adhesion locus ica among staphylococci from food and food processing environments. Appl. Environ. Microbiol. 69:5648-5655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Nilsson, M., J. Bjerketorp, B. Guss, and L. Frykberg. 2004. A fibrinogen-binding protein of Staphylococcus lugdunensis. FEMS Microbiol. Lett. 241:87-93. [DOI] [PubMed] [Google Scholar]
- 53.Nilsson, M., J. Bjerketorp, A. Wiebensjo, A. Ljungh, L. Frykberg, and B. Guss. 2004. A von Willebrand factor-binding protein from Staphylococcus lugdunensis. FEMS Microbiol. Lett. 234:155-161. [DOI] [PubMed] [Google Scholar]
- 54.O'Gara, J. P. 2007. ica and beyond: biofilm mechanisms and regulation in Staphylococcus epidermidis and Staphylococcus aureus. FEMS Microbiol. Lett. 270:179-188. [DOI] [PubMed] [Google Scholar]
- 55.Parise, G., M. Mishra, Y. Itoh, T. Romeo, and R. Deora. 2007. Role of a putative polysaccharide locus in Bordetella biofilm development. J. Bacteriol. 189:750-760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Patel, R., K. E. Piper, M. S. Rouse, J. R. Uhl, F. R. Cockerill III, and J. M. Steckelberg. 2000. Frequency of isolation of Staphylococcus lugdunensis among staphylococcal isolates causing endocarditis: a 20-year experience. J. Clin. Microbiol. 38:4262-4263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Paulsson, M., A. C. Petersson, and A. Ljungh. 1993. Serum and tissue protein binding and cell surface properties of Staphylococcus lugdunensis. J. Med. Microbiol. 38:96-102. [DOI] [PubMed] [Google Scholar]
- 58.Qin, Z., X. Yang, L. Yang, J. Jiang, Y. Ou, S. Molin, and D. Qu. 2007. Formation and properties of in vitro biofilms of ica-negative Staphylococcus epidermidis clinical isolates. J. Med. Microbiol. 56:83-93. [DOI] [PubMed] [Google Scholar]
- 59.Rohde, H., E. C. Burandt, N. Siemssen, L. Frommelt, C. Burdelski, S. Wurster, S. Scherpe, A. P. Davies, L. G. Harris, M. A. Horstkotte, J. K. Knobloch, C. Ragunath, J. B. Kaplan, and D. Mack. 2007. Polysaccharide intercellular adhesin or protein factors in biofilm accumulation of Staphylococcus epidermidis and Staphylococcus aureus isolated from prosthetic hip and knee joint infections. Biomaterials 28:1711-1720. [DOI] [PubMed] [Google Scholar]
- 60.Rohde, H., C. Burdelski, K. Bartscht, M. Hussain, F. Buck, M. A. Horstkotte, J. K. Knobloch, C. Heilmann, M. Herrmann, and D. Mack. 2005. Induction of Staphylococcus epidermidis biofilm formation via proteolytic processing of the accumulation-associated protein by staphylococcal and host proteases. Mol. Microbiol. 55:1883-1895. [DOI] [PubMed] [Google Scholar]
- 61.Sadovskaya, I., P. Chaignon, G. Kogan, A. Chokr, E. Vinogradov, and S. Jabbouri. 2006. Carbohydrate-containing components of biofilms produced in vitro by some staphylococcal strains related to orthopaedic prosthesis infections. FEMS Immunol. Med. Microbiol. 47:75-82. [DOI] [PubMed] [Google Scholar]
- 62.Sadovskaya, I., E. Vinogradov, S. Flahaut, G. Kogan, and S. Jabbouri. 2005. Extracellular carbohydrate-containing polymers of a model biofilm-producing strain, Staphylococcus epidermidis RP62A. Infect. Immun. 73:3007-3017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Sampathkumar, P., D. R. Osmon, and F. R. Cockerill III. 2000. Prosthetic joint infection due to Staphylococcus lugdunensis. Mayo Clin. Proc. 75:511-512. [DOI] [PubMed] [Google Scholar]
- 64.Sandoe, J. A., and C. M. Longshaw. 2001. Ventriculoperitoneal shunt infection caused by Staphylococcus lugdunensis. Clin. Microbiol. Infect. 7:385-387. [DOI] [PubMed] [Google Scholar]
- 65.Sarkar, G., R. T. Turner, and M. E. Bolander. 1993. Restriction-site PCR: a direct method of unknown sequence retrieval adjacent to a known locus by using universal primers. PCR Methods Appl. 2:318-322. [DOI] [PubMed] [Google Scholar]
- 66.Sharp, M. D., and K. Pogliano. 1999. An in vivo membrane fusion assay implicates SpoIIIE in the final stages of engulfment during Bacillus subtilis sporulation. Proc. Natl. Acad. Sci. USA 96:14553-14558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Stewart, P. S., and J. W. Costerton. 2001. Antibiotic resistance of bacteria in biofilms. Lancet 358:135-138. [DOI] [PubMed] [Google Scholar]
- 68.Szurmant, H., M. A. Mohan, P. M. Imus, and J. A. Hoch. 2007. YycH and YycI interact to regulate the essential YycFG two-component system in Bacillus subtilis. J. Bacteriol. 189:3280-3289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Szurmant, H., K. Nelson, E. J. Kim, M. Perego, and J. A. Hoch. 2005. YycH regulates the activity of the essential YycFG two-component system in Bacillus subtilis. J. Bacteriol. 187:5419-5426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Tan, T. Y., S. Y. Ng, and W. X. Ng. 2006. Clinical significance of coagulase-negative staphylococci recovered from nonsterile sites. J. Clin. Microbiol. 44:3413-3414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Tews, I., A. Perrakis, A. Oppenheim, Z. Dauter, K. S. Wilson, and C. E. Vorgias. 1996. Bacterial chitobiase structure provides insight into catalytic mechanism and the basis of Tay-Sachs disease. Nat. Struct. Biol. 3:638-648. [DOI] [PubMed] [Google Scholar]
- 72.Toledo-Arana, A., N. Merino, M. Vergara-Irigaray, M. Debarbouille, J. R. Penades, and I. Lasa. 2005. Staphylococcus aureus develops an alternative, ica-independent biofilm in the absence of the arlRS two-component system. J. Bacteriol. 187:5318-5329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.van der Mee-Marquet, N., A. Achard, L. Mereghetti, A. Danton, M. Minier, and R. Quentin. 2003. Staphylococcus lugdunensis infections: high frequency of inguinal area carriage. J. Clin. Microbiol. 41:1404-1409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.von Eiff, C., G. Peters, and C. Heilmann. 2002. Pathogenesis of infections due to coagulase-negative staphylococci. Lancet Infect. Dis. 2:677-685. [DOI] [PubMed] [Google Scholar]
- 75.Wang, X., J. F. Preston III, and T. Romeo. 2004. The pgaABCD locus of Escherichia coli promotes the synthesis of a polysaccharide adhesin required for biofilm formation. J. Bacteriol. 186:2724-2734. [DOI] [PMC free article] [PubMed] [Google Scholar]