Abstract
Signaling by fibroblast growth factors (FGFs) and their receptors has been previously implicated in control of cell proliferation, differentiation and migration. Here we report a novel role for signaling by the EGL-15 FGFR of Caenorhabditis elegans in controlling protein degradation in differentiated muscle. Activation of EGL-15, by means of a reduction of function mutation (clr-1) affecting an inhibitory phosphatase, triggers protein degradation in adult muscle cells using a pre-existing proteolytic system. This activation is not suppressed by mutation in either of the known genes encoding FGF ligands (egl-17 or let-756) but is well suppressed when both are mutated, indicating that either ligand is sufficient and at least one is necessary for FGFR activation. Activity of the Ras pathway through mitogen-activated protein kinase (MAPK) is required to trigger protein degradation. This is the first report that degradation of intracellular protein can be triggered by a growth factor receptor using an identified signal transduction pathway. The data raise the possibility that FGF-triggered proteolysis may be relevant to muscle remodeling or dedifferentiation.
Keywords: oncogene/proteolysis/Ras/remodeling/signal transduction
Introduction
Muscle accounts for roughly half the protein stores of the human body (Rooyackers and Nair, 1997), representing a reserve that can be mobilized by proteolysis in times of metabolic or energetic stress. Starvation, for example, promotes the loss of muscle mass. While it is not always clear why muscle wasting is seen in patients with cancer, diabetes, sepsis, heart failure or AIDS, muscle wasting itself can be the proximal cause of death (Mitch and Goldberg, 1996; Llovera et al., 1998; Molkentin and Dorn, 2001; Tisdale, 2001). Extensive study of the molecular mechanisms of proteolysis in muscle has resulted in identification of a number of extracellular factors, such as insulin, tissue necrosis factor-α (TNF-α) and interleukin 6 (Tisdale, 2001), which can regulate proteolysis, and a number of proteolytic systems (calpains, proteasome) which function in muscle (Attaix and Taillander, 1998; Jagoe and Goldberg, 2001). However, relatively little is known about the intramuscular signal transduction that mediates between extracellular factors and the regulation of proteolysis.
Proteolysis in muscle is often measured by release of labeled amino acids from total protein or by loss of muscle mass rather than by focusing on the fate of a well-defined substrate (Attaix and Taillander, 1998). Although soluble proteins provide the metabolic drive and regulation for muscle contraction and regulate gene activities in muscle, many prior studies of protein degradation have focused primarily on the insoluble proteins incorporated into myofibrils. The evidence (Solomon and Goldberg, 1996; Eble et al., 1999) indicates that soluble muscle proteins are degraded more rapidly than myofibrillar proteins because the latter are resistant to proteolysis while incorporated in supramolecular complexes and are liable for degradation primarily when disassembly renders them soluble. This has two crucial implications. First, the measured rates of degradation of myofibrillar proteins in vivo may reflect rate-limiting disassembly steps. Secondly, the proteolytic processes affecting proteins that are normally soluble may also contribute to degradation of myofibrillar proteins. However, there are few validated exemplars of endogenous muscle-specific soluble cytosolic proteins, and few tools are available for studying them.
It has become frequent practice to use ‘reporter’ genes to study the transcriptional and translational regulation of gene expression, and we have now extended that strategy to the study of protein degradation. To follow the in vivo degradation of a single protein in innervated muscle, we have utilized transgenic strains of Caenorhabditis elegans containing an unc-54::lacZ transgene under the control of a muscle myosin heavy-chain (unc-54 gene) promoter and enhancer (Okkema et al., 1993; Fire and Waterston, 1989) so that a chimeric ‘reporter’ protein is expressed specifically in the 95 body-wall and eight vulval muscle cells. The 146 kDa reporter consists of the N-terminal 263 amino acids of UNC-54 myosin fused (with a short intervening fragment) to Escherichia coli β-galactosidase. This protein contains only a fraction of the myosin ATPase domain and does not assemble into myofibrils, but remains soluble in the muscle cytosol where it forms active β-galactosidase tetramers (Zdinak et al., 1997).
We have shown that the reporter protein is continually expressed throughout larval development into early adulthood and is not degraded in well-fed adult animals (Zdinak et al., 1997). The fact that the protein forms active β-galactosidase tetramers and is completely stable in well-fed adults indicates that it forms a stable structure and is not subject to default degradation by a ‘misfolded-protein’ degradation pathway (Patil and Walter, 2001; Shen et al., 2001) or by a hypothetical intracellular pathway that degrades ‘foreign proteins’. In contrast with its stability in fed animals, the reporter is degraded via a proteasome-mediated pathway during starvation or denervation (Zdinak et al., 1997; Szewczyk et al., 2000), two conditions which can trigger muscle wasting in mammals. Furthermore, the degradation of the LacZ reporter closely paralleled that of a muscle-specific soluble Green Fluorescent Protein (GFP) reporter and of two endogenous muscle marker enzymes, adenylate kinase and arginine kinase (Fostel et al., 2003).
The use of C.elegans has also enabled the application of genetics to discover and study intracellular signal transduction components that regulate muscle proteolysis in vivo. Our laboratory reported that mutational activation of the Ras oncogene homolog LET-60 or its downstream effector mitogen-activated protein kinase (MAPK) is sufficient to provoke degradation of the β-galactosidase reporter protein in muscle (Szewczyk et al., 2002). This degradation is at least partially distinct from that seen during starvation or denervation, and does not appear to be mediated by the proteasome. Although the proteolytic system that degrades reporter in response to Ras activation has not yet been identified, the discovery of Ras as a potential intracellular regulator of muscle proteolysis was provocative, insofar as Ras is known to function downstream of factors which can regulate muscle proteolysis in mammals (e.g. insulin, TNF-α).
As a first step to understanding the physiological significance of Ras-induced protein degradation, it is important to identify the extracellular factors and surface receptors that might control Ras activity to regulate protein degradation. Cellular specificity in signaling appears to be achieved, at least in part, by distinct cell- or tissue-specific pairings of cell-surface receptors with downstream effectors (Tan and Kim, 1999). A given cell-surface receptor may have its signals transduced by different effectors in different tissues, and a given intracellular signal transduction pathway may, in different cells, receive signals from different surface receptors and/or signal to different downstream targets. For example, the Ras-MAPK pathway in C.elegans was first recognized as regulating the differentiation of hypodermal precursor cells to form the vulva in response to signal from the LET-23 receptor, a homolog of mammalian epidermal growth factor receptor (EGFR) (Sternberg et al., 1995; Sternberg and Han, 1998), but LET-23 EGFR also signals in a Ras-independent fashion via inositol-1,4,5-triphosphate to control ovulation (Clandinin et al., 1998). Our laboratory reported that mutational activation of LET-23 did not induce proteolysis in muscle (Szewczyk et al., 2002), consistent with the fact that EGFR is not reported to be expressed in muscle (Chang et al., 1999).
While studying protein degradation in muscle induced by Ras activation (Szewczyk et al., 2002), we observed that temperature-sensitive activated-Ras mutants (Eisenmann and Kim, 1997) developed a Clear phenotype in which the gonads degenerate and the pseudocoelom fills with fluid. Both the Clear and protein-degrading phenotypes showed variable penetrance and variable expressivity, but were well correlated with each other in individual animals. The Clear phenotype had previously been associated with activation of the fibroblast growth factor receptor (FGFR) homolog EGL-15 (Kokel et al., 1998) by reduction of function mutation in clr-1, which encodes a receptor tyrosine phosphatase that negatively regulates the activity of EGL-15 FGFR (Kokel et al., 1998). In view of this correlation, it was suggested that EGL-15 FGFR signaling to cause the Clear phenotype is mediated by Ras (Schutzman et al., 2001). The abnormal fluid filling of Clear animals is unlikely to involve muscle directly (Borland et al., 2001), but altered EGL-15 activity has also been associated with defects in the post- embryonic migration of the sex myoblast cells that form the vulval muscles (DeVore et al., 1995). Although clr-1 is known to be expressed in body-wall muscle cells of C.elegans (Kokel et al., 1998), no phenotypes of clr-1 or egl-15 mutations have yet been associated with differentiated adult muscle.
Here we report that acute FGFR activation by a temperature-sensitive reduction-of-function mutation in clr-1 triggers protein degradation in muscle by a process that uses pre-existing signaling components and protease(s). This effect is not suppressed by reduction-of-function mutations in either of the known C.elegans FGF genes (egl-17 or let-756), but is well suppressed when both genes are mutated, implying that activation of EGL-15 FGFR requires at least one of the FGF ligands. Reporter degradation in clr-1 mutants requires the activities of GRB2, Ras, Raf, MEK and MAPK, demonstrating that protein degradation in response to FGFR activation requires signaling via the Ras-MAPK pathway. This is the first report that intracellular protein degradation can be triggered by a growth factor receptor using an identified signal transduction pathway.
Results
Activation of FGFR triggers protein degradation in muscle
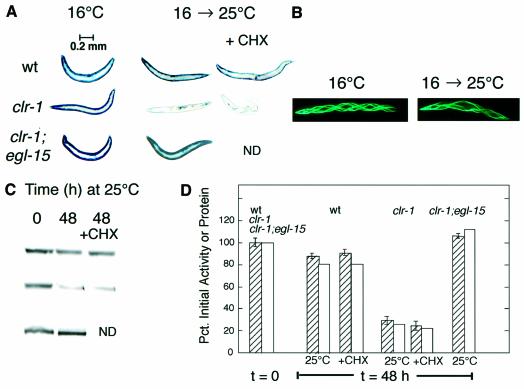
To test the possibility that FGFR activation might signal to promote proteolysis in muscle, we activated EGL-15 FGFR genetically. There are no existing gain-of-function mutations in egl-15, and the use of mutationally activated FGFR transgenes (Kokel et al., 1998) is potentially complicated by non-physiological levels of FGFR protein. Therefore we exploited a temperature-sensitive allele of clr-1(e1745ts), previously used to activate FGFR signaling (Chang et al., 2000; Schutzman et al., 2001). We constructed a strain carrying both the temperature- sensitive allele of clr-1 and an integrated unc-54::lacZ transgene whose protein product serves as a ‘reporter’ of proteolysis in body-wall muscle cells (Szewczyk et al., 2000; Fostel et al., 2003). Animals were grown to adulthood at permissive temperature (16°C) until full expression of the β-galactosidase reporter had occurred, and then were shifted to non-permissive temperature (25°C). The temperature upshift triggered a time- dependent degradation of pre-existing reporter protein (Figure 1). The decline in histochemical staining for β-galactosidase was confirmed by fluorimetric assay of β-galactosidase activity and by western blotting with monoclonal anti-β-galactosidase antibody (Figure 1). As controls, neither wild-type animals (at 16°C or 25°C) nor clr-1 mutants maintained at 16°C degraded the reporter protein (Figure 1). A muscle-specific GFP reporter, expressed in a soluble form in muscle cytosol, was also degraded in clr-1 mutants at 25°C (Figure 1), suggesting that the proteolytic system is not narrowly targeting some peculiar feature of the LacZ fusion protein. The temperature-dependent protein degradation in clr-1 mutant animals was strongly suppressed (Figure 1) by a reduction-of-function mutation in the kinase domain of egl-15 (DeVore et al., 1995), indicating that activation of EGL-15 FGFR can trigger protein degradation in muscle.
Fig. 1. FGFR activation induces proteolysis in muscle. Animals were grown to early to mid-adulthood at 16°C or for an additional 48 h at 25°C either without or with cycloheximide (CHX, 400 µg/ml) added 6 h before temperature upshift. (A) Histochemical staining for β-galactosidase activity (blue). Wild-type (top row), activated FGFR mutant clr-1(e1745ts) (middle row) or suppressed double mutants clr-1(e1745ts);egl-15(n1783) (bottom row). Blue objects inside the clr-1 animal at 25°C are embryos (cf. Figure 4A). (B) Soluble GFP in body-wall and vulval muscle is degraded (66% of 16°C control) in the clr-1(e1745ts) mutant after 48 h at 25°C. Helical twisting of the muscle bands is a phenotype associated with the rol-6 transformation marker. (C) Immunoblot (monoclonal anti-β-galactosidase antibody) of 146 kDa β-galactosidase reporter protein in 30-worm lysates. Each row in the blot corresponds to the row of stained worms in (A); each column corresponds to the columns of worms in (A). (D) Quantitation of 146 kDa LacZ fusion protein (open bars) by integrating bands at t = 48 h in (B) and of β-galactosidase activity (striped bars, means ± SD, N = 3) in 10-worm lysates. Values for each strain are given relative to the value for that strain at t = 0.
Since the animals had reached adulthood prior to temperature upshift, protein degradation in response to FGFR activation should not be the result of alterations in the muscles during development, but rather is triggered acutely upon FGFR activation. Furthermore, degradation appears to be the result of activation of pre-existing signaling components and protease(s), inasmuch as protein degradation was observed even in animals treated with the protein synthesis inhibitor cycloheximide either at the time of temperature upshift or 6 h prior to temperature upshift (Figure 1). Cycloheximide at this concentration completely inhibits protein synthesis, but does not itself affect the stability of the β-galactosidase reporter protein (Zdinak et al., 1997). The addition of cycloheximide also did not prevent the development of the Clear phenotype in clr-1(e1745ts) mutants at 25°C, implying that the Clear phenotype can develop independently of de novo protein synthesis. Degradation was not prevented by the proteasome inhibitors lactacystin (1 µM) or by Z-leu3-CHO (200 µM), consistent with observations for degradation induced by activated LET-60 Ras (Szewczyk et al., 2002).
Possible roles of FGF-like ligands
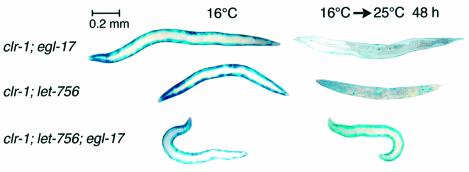
Having identified a role for EGL-15 FGFR in triggering muscle protein degradation, we next sought to identify the extracellular signal(s) to this receptor. EGL-15 FGFR has only two predicted FGF-like ligands in C.elegans: EGL-17 (Burdine et al., 1997) and LET-756 (Roubin et al., 1999). Reduction-of-function mutations in egl-17 affect the migration of the sex myoblasts during development (Stern and Horvitz, 1991), but do not suppress the Clear phenotype of clr-1(e1745ts) mutants (Borland et al., 2001). Strong loss-of-function mutations in let-756 are lethal (Roubin et al., 1999); non-lethal reduction-of-function alleles of let-756 cause the animals to be small and ‘starved’ in appearance, but do not affect sex myoblast migration (Roubin et al., 1999) and only weakly suppress the Clear phenotype (Borland et al., 2001). Even more puzzling, there appears to be no synthetic phenotype in egl-17;let-756 double mutants (Roubin et al., 1999). As shown in Figure 2, reduction-of-function mutation in either egl-17 or let-756 fails to prevent protein degradation in response to the clr-1 mutation, but there is significant suppression when both ligands are mutated. These observations suggest that either of the FGF-like ligands (EGL-17 or LET-756) is sufficient and at least one is necessary for the clr-1 mutation to activate EGL-15 in muscle to promote protein degradation. This is the first report that these two ligands have any effect on body-wall muscle and the first indication that they may serve partially or completely redundant functions. Furthermore, our data imply that there is some FGF ligand normally available to activate EGL-15 FGFR, although the present data cannot determine if this level is sufficient to result in actual FGFR activation in the genetic background of wild-type clr-1+. We will report elsewhere (N.J.Szewczyk, B.K.Peterson, L.P.Parkinson and L.A.Jacobson, in preparation) that such activation occurs in normal muscle, but does not result in protein degradation because the effector pathway is inhibited downstream.
Fig. 2. Reduction-of-function mutations in FGF ligands suppress FGFR-induced protein degradation. Activated FGFR animals [clr-1(e1745ts)] were grown to adulthood at 16°C (left) or for an additional 48 h at 25°C (right) prior to staining for β-galactosidase activity (blue). Single mutations in FGF ligand genes egl-17 or let-756 (top two rows) do not suppress FGFR-induced proteolysis (right column; cf. clr-1 mutant, Figure 1), but the two mutations in combination (bottom row) show suppression. At 16°C (left column) all mutants show normal β-galactosidase staining. Strains with let-756 also contain unc-32(e189).
Ras is necessary for FGFR-induced protein degradation
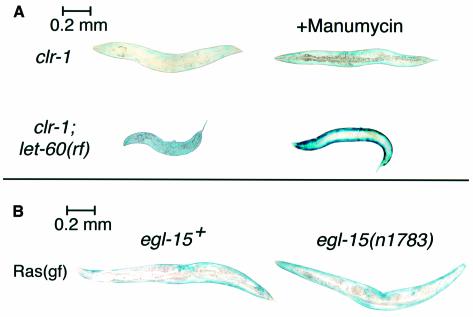
Ras activation is sufficient to promote reporter degradation in muscle (Szewczyk et al., 2002), and Ras acts downstream of EGL-15 FGFR signaling in generation of the Clear phenotype (Schutzman et al., 2001). Nevertheless, the phenotypes are separable; gap-1 mutants degrade muscle protein but do not become Clear (Szewczyk et al., 2002). Furthermore, there is no justification for assuming a priori that mature muscle cells contain all components required to couple EGL-15 FGFR to LET-60 Ras, or that FGFR might not have alternative downstream effectors in muscle. To test the possibility that Ras acts downstream of FGFR in promoting protein degradation, we determined whether protein degradation in response to FGFR activation was blocked when the ability of Ras to act as signal transducer was altered. Because strong loss-of-function mutations in let-60 (Ras) are lethal in C.elegans (Han and Sternberg, 1990), we tested instead a sublethal (Beitel et al., 1990) reduction-of-function (rf) mutation in let-60 (Ras) and found (Figure 3A) that this was not sufficient to block the protein degradation seen in clr-1 animals shifted to 25°C after adulthood. Taken at face value, this might have suggested (incorrectly) that Ras was not necessary for FGFR-induced protein degradation, but this is not a null allele of let-60. To decrease Ras activity further, but only from the time temperature upshift was imposed to activate EGL-15, clr-1(rf);let-60(rf) double mutants were treated with manumycin, a farnesyltransferase inhibitor that reduces Ras signaling in C.elegans (Hara and Han, 1995), presumably because Ras requires farnesylation to attain full activation (Schafer et al., 1989). Manumycin treatment of clr-1;let-60, but not clr-1 single mutants, blocked protein degradation at 25°C, confirming that Ras activity is necessary for FGFR-induced protein degradation (Figure 3A) and implying that a low level of Ras activity is sufficient. This suggests that EGL-15 FGFR signaling is enhanced by farnesylation of LET-60 Ras, as previously shown for LET-23 EGFR signaling to Ras (Hara and Han, 1995). Our observations are consistent with the report that this particular allele of let-60 does not prevent development of the Clear phenotype of clr-1 mutants, even though a null allele of let-60 does so in a genetic background that suppresses its lethality (Schutzman et al., 2001).
Fig. 3. Activated EGL-15 FGFR signals unidirectionally via Ras to induce protein degradation. (A) Activated FGFR [clr-1(e1745ts)] animals were grown to adulthood at 16°C and then for an additional 48 h at 25°C. Activated FGFR induction of protein degradation (upper left) is not blocked by reduction of Ras signal in let-60+ animals treated with 100 µM manumycin (top right) or by Ras reduction-of-function mutation let-60(n2021) (bottom left), but is blocked by the combination of manumycin and let-60(rf) (bottom right). (B) Animals were grown to early adulthood at 16°C, at which time they stain strongly for β-galactosidase (not shown), and were shifted to 25°C for 72 h before staining with X-gal. Protein degradation induced by temperature-sensitive activated-Ras mutation let-60(ga89ts) (left) at 25°C (Szewczyk et al., 2002) is not blocked by reduction-of-function mutation in the FGFR gene egl-15(n1783 (right).
To determine whether signaling from EGL-15 FGFR to Ras is unidirectional, we asked whether EGL-15 FGFR function affects Ras-induced protein degradation in muscle. Double mutants containing both a reduction-of-function allele of egl-15(n1783) and a temperature-dependent activated-Ras mutation let-60(ga89ts) were constructed. As shown in Figure 3B, activated-Ras mutants degraded reporter protein after shift to non-permissive temperature regardless of whether the genetic background contained wild-type egl-15+ or mutant FGFR. These results are consistent with the model that Ras acts downstream of FGFR in controlling muscle protein degradation.
As would be expected if FGFR signals positively via Ras, double mutants in which both FGFR and Ras are mutationally activated [clr-1(rf);let-60(gf)] have a synthetic phenotype that exaggerates the characteristics of each single mutant; in this case, the animals degraded the reporter protein even at a temperature (16°C) at which each single mutant appeared nearly wild type (unpublished data). This reflects the probability that the mutant gene products do not achieve fully normal function even at 16°C (Eisenmann and Kim, 1997) (cf. Figure 4D).
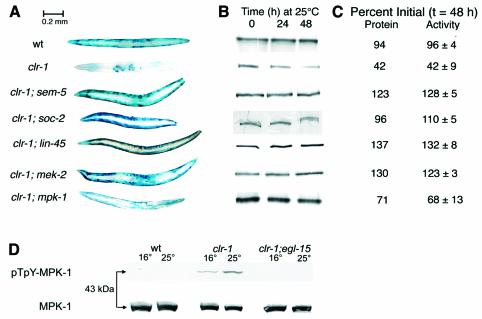
Fig. 4. Activated FGFR signals protein degradation via SEM-5 and the Raf–MAPK pathway. Animals were grown to adulthood at 16°C and then for an additional 48 h at 25°C. (A) Histochemical staining for β-galactosidase activity. Genotypes (all mutations are reduction-of-function alleles) are indicated on the left. (B) Immunoblot (monoclonal anti-β-galactosidase) of 146 kDa β-galactosidase fusion protein in 30-worm lysates. Each row, corresponding to the animals with genotypes in (A), shows the kinetics of loss of the 146 kDa band. (C) Quantitation of 146 kDa fusion protein by integrating the bands at t = 48 h in (B) and of β-galactosidase activity (means ± SD of triplicate samples) by fluorimetric assay of 10-worm lysates. Data are expressed as a percentage of the value for each genotype prior to temperature upshift. Rows correspond to the genotypes in (A). (D) Activation of MPK-1 in clr-1 mutants 4 h after shift to 25°C. Each lane contains lysate of 40 worms. Upper set blotted with monoclonal anti-pTpY-ERK; lower set blotted with polyclonal anti-ERK.
FGFR signals to Ras via SEM-5
In generation of the Clear phenotype, EGL-15 FGFR signals to LET-60 Ras via the GRB2 homolog SEM-5 (Clark et al., 1992) and the guanine nucleotide exchange factor (SOS1 homolog) LET-341 (Chang et al., 2000). SEM-5 likely directly interacts with LET-341, which in turn activates LET-60 (Ras). In support of a similar mechanism operating in muscle to couple FGFR signaling to Ras, clr-1 mutants carrying either of two reduction-of-function mutations in sem-5 failed to degrade the reporter protein after temperature shift (Figure 4). In contrast with LET-23 EGFR, which signals to Ras during hypodermal differentiation (Sternberg et al., 1995), EGL-15 (FGFR) does not appear to bind SEM-5 directly (Borland et al., 2001), suggesting that another unidentified protein, possibly of the FRS2 family (Kouhara et al., 1997), bridges the binding of EGL-15 to SEM-5 to allow for signal transduction.
FGFR signals protein degradation through the Raf–MEK–MAPK cascade
We have shown (Szewczyk et al., 2002) that activated-Ras signaling to trigger protein degradation in muscle is mediated by the protein kinases Raf (LIN-45), MEK (MEK-1) and MAPK (MPK-1). To test whether this pathway (as distinct from another Ras effector pathway) is necessary downstream of activated FGFR, we constructed double mutants containing the temperature-sensitive clr-1 mutation in combination with reduction-of-function mutations in lin-45 (Raf), mek-2 (MEK) or mpk-1 (MAPK). As shown in Figure 4, any one of these mutations suppressed the protein degradation triggered by activation of FGFR. The suppression by mpk-1 was only partial, but this is a non-null allele of mpk-1 that (unlike a null allele) does not fully suppress the Clear phenotype of clr-1 mutants (Schutzman et al., 2001). Western blots (Figure 4D) with antibody against double-phosphorylated (activated) pTpY-ERK confirmed that MPK-1 was activated when clr-1 mutants were shifted to 25°C (and more weakly at 16°C); no pTpY-MPK-1 was detected in wild-type or in suppressed clr-1;egl-15 double mutants. Taken together with the observation (Szewczyk et al., 2002) that MPK-1 activation is sufficient to induce protein degradation in muscle, these results indicate that FGFR signals protein degradation primarily via the Raf–MEK–MAPK cascade.
LET-60 (Ras) activation of LIN-45 (Raf) has been shown to be enhanced by the leucine-rich repeat protein SOC-2 (Li et al., 2000), distinct alleles of which were identified as clr-1 suppressors (Selfors et al., 1998) or as activated-Ras suppressors (Sieburth et al., 1998). Either of two soc-2 mutations (one from each category; neither is a null) blocked FGFR-induced protein degradation (Figure 4) and to a lesser extent Ras-induced protein degradation (unpublished data). It is not known why soc-2 blocks protein degradation signaled from FGFR better than from Ras. This may suggest the existence of parallel or bypass steps in these pathways, or it may be an indication of subtle differences in the interactions of mutant proteins with their multiple binding partners.
Discussion
FGFR activation triggers protein degradation in muscle
We have shown that FGFR activation, requiring at least one of two FGF ligands, acutely promotes protein degradation in muscle cells of C.elegans by signaling via SEM-5 (GRB2) to the Ras–Raf–MEK–MAPK cascade (Figure 5). This is the first report that intracellular protein degradation can be triggered by a growth factor receptor using an identified signal transduction pathway. FGFR activation provokes degradation of the soluble cytosolic β-galactosidase reporter protein, but phalloidin stains of animals with activated FGFR or activated Ras indicate that there is no obvious proteolysis of the myofibrils (unpublished data). The degradation of myofibrillar proteins and cytoplasmic proteins probably occurs by at least partially distinct mechanisms (Solomon and Goldberg, 1996), so there is no reason to think that degradation of the cytosolic β-galactosidase reporter protein in any way reports on degradation of normal myofibrillar UNC-54 myosin heavy chain. We believe that the LacZ and GFP proteins used in the work reported here are likely reporting on the activity of relatively non-selective systems that degrade soluble proteins in the muscle cytosol (Zdinak et al., 1997; Szewczyk et al., 2000; Szewczyk et al., 2002; Fostel et al., 2003). We also found that, upon Ras activation, a different LacZ reporter protein localized in muscle nuclei is degraded either much more slowly than cytosolic reporter protein or not at all (Szewczyk et al., 2002), suggesting that FGFR-induced protein degradation is at least partially selective with regard to subcellular location of the target proteins.
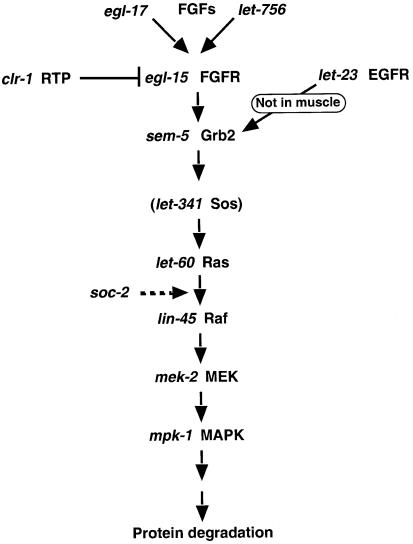
Fig. 5. Proposed pathway by which FGFR signals protein degradation in muscle. Signaling from LET-23 EGFR to Ras via Grb2 and Sos occurs in vulval precursor cells of the hypodermis (Clark et al., 1992; Chang et al., 2000) but not in muscle (Szewczyk et al., 2002).
Although physiologically or pathologically significant phenomena such as muscle wasting or muscle remodeling undoubtedly involve the net degradation of the supramolecular protein assemblies in myofibers, it seems rather unlikely that such changes are initially determined by proteolysis of the myofibrillar proteins. We believe that changes in myofibers are likely to be consequence rather than cause. The soluble proteins that power and regulate contraction, and that control gene expression in muscle, are more likely candidates to be the critical regulators. Even relatively small changes in the concentrations of such proteins might have profound functional consequences.
The use of reporter proteins has allowed us to study the regulation of proteolysis but does not itself point to identification of normal targets of proteolysis, much as the expression of promoter fusions facilitates the study of where or when genes are expressed but not the study of the function of wild-type gene products. There are currently few tools available for studying endogenous muscle proteins in C.elegans. We are attempting to identify endogenous muscle proteins that are degraded in response to Ras or FGFR activation. The degradation of one or more such proteins in response to Ras or FGFR activation might be responsible for the deficits in locomotion that develop in these animals (DeVore et al., 1995; Szewczyk et al., 2002). The identification of endogenous substrates is complicated by the fact that some muscle-specific proteins do not exist exclusively in soluble form in the cytosol. Conversely, proteins that exist predominantly in myofibrils may also have soluble pools that are preferentially liable to proteolysis (Solomon and Goldberg, 1996). Thus one must be aware of the possibility that degradation of the cytosolic pool of a protein may be masked by the stability of other (possibly larger) pools of the same protein in supramolecular structures or even in other tissues. We have avoided all these difficulties by studying muscle-specific transgene-coded proteins localized exclusively to the cytosol, and the experimental system further allows us to study protein degradation without regard to any effects on protein synthesis. It is important to bear in mind that these reporter constructs provide no information about whether FGFR activation induces protein degradation in other cell types. There is no evidence, one way or another, to tell us if FGFR-induced (or Ras-induced) protein degradation is limited to muscle.
While both CLR-1 (Kokel et al., 1998) and LET-60 Ras (Dent and Han, 1998) have been shown to be expressed in muscle, EGL-15 has not. Thus it is plausible but unproven that FGFR acts directly in muscle to promote protein degradation. We will report elsewhere (N.J.Szewczyk, B.K.Peterson, L.P.Parkinson and L.A.Jacobson, in preparation) that intramuscular signaling from the DAF-2 insulin-receptor homolog opposes the effects of Ras or FGFR activation, implying that protein degradation in response to constitutive FGFR activation is indeed the result of intramuscular signaling.
FGFR effectors and signal strength
We have shown here that SEM-5 (GRB2), Ras, Raf, MEK and MAPK are necessary for FGFR-induced protein degradation, implying that the Ras–MAPK cascade is the primary route by which FGFR signals muscle protein degradation. MAPK activation alone was also sufficient to promote reporter degradation (Szewczyk et al., 2002), although the signaling steps that couple MAPK activation to proteolysis are not yet identified. We have identified a novel consequence of FGFR signaling in a novel cell type, but our data are completely consistent with the finding (Schutzman et al., 2001) that EGL-15 signals via Ras in non-muscle tissue (Borland et al., 2001) to generate the Clear phenotype.
The data of Figure 3 imply that only a relatively low level of Ras activity is required to trigger muscle protein degradation. It is also possible that multiple signal sources affecting the Ras pathway can be summed. One indication is that, unlike clr-1 or let-60(ga89ts) single-mutant animals, clr-1;let-60(ga89ts) double mutants degrade reporter protein at permissive temperature (16°C), suggesting some additive effect. A second prediction of a simple signal-summation model might be that reporter degradation at 25°C in a clr-1;let-60(ga89ts) double mutant should occur at a faster rate than in either single mutant. Seemingly contrary to this prediction, we find that in clr-1;let-60(ga89ts) animals at 25°C the rate of protein degradation (t1/2 ≈ 48 h) is faster than that in activated-Ras single mutants (t1/2 ≈ 72 h) but not demonstrably greater than that of activated-FGFR single mutants (t1/2 ≈ 48 h). One possibility is that the signal from activated-FGFR saturates the capacity of the protein-degradation system to respond, such that further increases in Ras signal in the double mutant do not elicit additional increases in degradation rate. There is little information about response saturation in this or any other in vivo signaling system.
A second class of possibilities is that activated-FGFR activates the Raf–MEK–MAPK segment of the pathway more fully than even maximal Ras activation alone. All members of the Raf–MEK–MAPK signaling cassette are potential targets for regulation by non-pathway members. For example, in other systems Raf may be regulated not only by Ras but also by Sprouty2 and Akt (Rommel et al., 1999; Yusoff et al., 2002). Finally, it remains possible that EGL-15 FGFR has unidentified Ras-independent effectors. Studies in other systems have identified a number of other potential downstream effectors of FGFR signaling (i.e. protein kinase C, phospholipase C) (Klint and Claesson-Welsh, 1999). We are currently investigating whether any of these effectors have a role in regulating proteolysis in C.elegans muscle, although it should be borne in mind that our data (Figures 3 and 4) show that Ras pathway activity is necessary downstream of EGL-15 to promote protein degradation.
Multiple receptors control protein degradation
While FGFR promotes protein degradation in muscle and EGFR does not (Szewczyk et al., 2002), FGFR is not the only surface receptor responsible for regulating muscle protein catabolism in C.elegans. We have shown (Szewczyk et al., 2000) that activation of nicotinic acetylcholine receptors (nAChR) opposes starvation-induced muscle protein degradation. However, nAChR stimulation does not block Ras-induced proteolysis, and reduction-of-function mutations in MEK or MAPK do not block denervation-induced proteolysis (Szewczyk et al., 2002), suggesting not only that multiple receptors regulate protein degradation, but that they may do so by distinct mechanisms. This inference is supported by the observation that denervation-induced protein degradation is prevented by proteasome inhibitors, but Ras-induced degradation is not (Szewczyk et al., 2002). Thus, not only can a signaling system (the Ras–MAPK system) receive external signals via different surface receptors in different cell types (EGFR in hypodermis versus FGFR in muscle), but a single cellular process (protein degradation) can also be controlled in a single cell type (muscle) by distinct surface receptors signaling via distinct mechanisms.
Implications for muscle
Whereas mammals have three distinct types of muscle (skeletal, smooth and cardiac), C.elegans has only striated and non-striated muscles. The unc-54::lacZ reporter gene is principally expressed in striated body-wall muscle and also in the non-striated vulval muscles. The protein degradation triggered by FGFR activation affects body-wall and vulval muscles (Figures 1, 2, 3 and 4), and thus affects both striated and non-striated muscle. C.elegans also contains pharyngeal, uterine, intestinal and anal depressor muscles, which are all non-striated, and which we have not studied for protein degradation because the unc-54::lacZ fusion is not significantly expressed in these cells.
Early experiments (Olwin and Hauschka, 1988; Moore et al., 1991) indicated that FGFRs were not expressed in mature skeletal muscle. Consequently, the loss of FGFR expression during myoblast maturation was proposed to be a key element in the terminal differentiation of muscle. More recently, however, there have been convincing demonstrations of the presence of FGFR protein and expression of FGFR mRNA in adult whole muscle (Sogos et al., 1998; Kastner et al., 2000). It has been suggested that differences in FGFR expression patterns vary with cell type (connective tissue, microsatellite, myofiber) (Sogos et al., 1998; Kastner et al., 2000), but no consensus has yet emerged on which receptors or receptor genes are expressed in which cell type(s), possibly due to differences in culture and detection methods. In contrast with the uncertain function of FGFR in mature mammalian skeletal muscle, there is an apparent function for at least FGFR1 in cardiac muscle since intracardiac administration of FGF -2 offers protection, via an FGFR1-mediated pathway, from cardiac dysfunction and damage in response to acute ischemia (Jiang et al., 2002). These results all suggest that FGFRs may have physiological functions in fully developed muscle, although these roles are likely to vary based upon cell and muscle type. C.elegans has only one FGFR while mammals have at least four, so our work offers little guidance about which FGFR(s) may be capable of stimulating protein degradation in mammalian muscle.
We have suggested (Szewczyk et al., 2002) that Ras activation first stimulates protein degradation and then stimulates protein synthesis (by stimulating transcription), and that this temporal sequence and/or the balance between these processes determine whether Ras signaling results in catabolic, anabolic or remodeling effects. Because FGFR signals via Ras, a similar argument can be made for FGFR signaling; a temporal separation of FGFR-induced protein degradation and FGFR-induced protein synthesis might facilitate a change in differentiated state or significant cellular or tissue remodeling. This should be considered in the context of recent speculation that FGFR activation in differentiated mammalian muscle might be a key feature of reversal of the differentiated state (Hughes, 2001), and the demonstration that expression of a downstream target of FGFR in muscle leads to both protein degradation and dedifferentiation (Odelberg et al., 2000).
FGFR-induced protein degradation may also be important for exercise-induced muscle remodeling. Exercise evokes increased FGF expression (Olfert et al., 2001), MAPK activation (Ryder et al., 2000) and degradation of both soluble and myofibrillar proteins in skeletal muscle (Dohm et al., 1987). Although these observations remain to be causally linked, our results hint that they may be linked and that FGFR activation in terminally differentiated myofibers or satellite cells may result in protein degradation needed as part of the remodeling process. In contrast with such an adaptive role, adventitious activation or overexpression of FGFR(s) might result in pathological proteolysis in muscle, as implied by the observation of FGFR4 overexpression in a patient with facioscapulohumeral muscular dystrophy (Saito et al., 2000). This suggests that FGFR activation in human muscle may affect myofibrillar proteins in a way that it apparently does not in C.elegans, although the time-scales are clearly very different.
EGL-15 FGFR has a known role in sex myoblast migration in C.elegans (DeVore et al., 1995), and migration is a classic case of cellular remodeling driven by dynamic changes in the actin cytoskeleton, which in turn may be regulated by proteolysis (Dourdin et al., 2001). This raises the interesting possibility that migration in response to an FGFR signal may involve intracellular protein degradation. Furthermore, not only myoblasts but also satellite cells in mammalian muscle respond to FGF (Johnson and Allen, 1995; Kastner et al., 2000; Suzuki et al., 2000), often migrate to the site of injury (Schultz and McCormick, 1994), and have a role in muscle regeneration (Floss et al., 1997). Thus it seems worth investigating whether protein degradation is involved in the function of myoblasts or satellite cells in wound healing or muscle remodeling.
Materials and methods
Nematode strains were maintained, grown and roughly age-synchronized as described (Zdinak et al., 1997), except that strains containing unsuppressed let-60(ga89ts) or clr-1(e1745ts) were routinely kept at 16°C. Genetic markers used in this study wereas follows: on LG I, him-1(e879), mek-2(ku114), unc-13(e51); on LG II, clr-1(e1745ts), unc-4(e120); on LG III, mpk-1(n2521), let-756(s2613), unc-32(e189); on LG IV, cha-1(p1182ts), dpy-13(e184), soc-2(n1774), soc-2(ku167), lin-45(sy96), unc-24(e138), him-8(e1489), let-60(ga89ts), let-60(n2021); on LG V, ccIs55(unc-54::lacZ); on LG X: sem-5(n1619), sem-5(n1779), egl-15(n1783), egl-17(n1377).
Strains containing unlinked mutations along with the reporter transgene ccIs55(unc-54::lacZ) or a muscle-specific GFP reporter (Wolkow et al., 2000; Fostel et al., 2003) were constructed by conventional genetic methods. Methods for gel electrophoresis, immunoblotting with monoclonal anti-β-galactosidase and fluorimetric assay of β-galactosidase activity were as described (Zdinak et al., 1997). Immunoblotting of MPK-1 after electrophoresis on 12% SDS–polyacrylamide gels used either polyclonal rabbit pan-ERK antibody (Santa Cruz sc-153) at 1:500 or monoclonal anti-pTpY-ERK (Sigma M-8159) at 1:2000 with detection by peroxidase-labeled donkey secondary antibodies and TMB substrate (KPL Laboratories). Animals were stained for β-galactosidase activity with 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-gal) as described (Zdinak et al., 1997) for 1–2 h at room temperature, governed by visual examination of stained control animals (wild type or mutants at permissive temperature) included with every experiment. Stained animals were photographed under bright-field illumination. The metered exposure is dominated by the bright-field background and depends little on the staining intensity of the animals being photographed, and thus is approximately constant from experiment to experiment. Live animals containing GFP were photographed using epifluorescence illumination with a Chroma 41001 filter set.
Acknowledgments
Acknowledgements
This work was supported by NSF grant MCB-0090734. We are grateful to A.Fire, D.Eisenmann, S.Kim, M.Han, G.Ruvkun and C.Wolkow for generously providing nematode strains, and to M.Stern and M.Sundaram for discussions and for sharing unpublished information. Many strains were from the Caenorhabditis Genetics Center, supported by the NIH National Center for Research Resources.
References
- Attaix D. and Taillander,D. (1998) The critical role of the ubiquitin-proteasome pathway in muscle wasting in comparison to lysosomal and Ca2+-dependent systems. In Rivett,A. (ed.), Advances in Molecular and Cellular Biology: Intracellular Protein Degradation, Vol. 27, JAI Press, Stamford, CT, pp. 235–266. [Google Scholar]
- Beitel G.J., Clark,S.G. and Horvitz,H.R. (1990) Caenorhabditis elegans ras gene let-60 acts as a switch in the pathway of vulval induction. Nature, 348, 503–509. [DOI] [PubMed] [Google Scholar]
- Borland C.Z., Schutzman,J.L. and Stern,M.J. (2001) Fibroblast growth factor signaling in Caenorhabditis elegans. BioEssays, 23, 1120–1130. [DOI] [PubMed] [Google Scholar]
- Burdine R.D., Chen,E.B., Kwok,S.F. and Stern,M.J. (1997) egl-17 encodes an invertebrate fibroblast growth factor family member required specifically for sex myoblast migration in Caenorhabditis elegans. Proc. Natl Acad. Sci. USA, 94, 2433–2437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang C., Newman,A.P. and Sternberg,P.W. (1999) Reciprocal EGF signaling back to the uterus from the induced C.elegans vulva coordinates morphogenesis of epithelia. Curr. Biol., 9, 237–246. [DOI] [PubMed] [Google Scholar]
- Chang C., Hopper,N.A. and Sternberg,P.W. (2000) Caenorhabditis elegans SOS-1 is necessary for multiple RAS-mediated developmental signals. EMBO J., 19, 3283–3294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clandinin T.R., DeModena,J.A. and Sternberg,P.W. (1998) Inositol trisphosphate mediates a RAS-independent response to LET-23 receptor tyrosine kinase activation in C.elegans. Cell, 92, 523–533. [DOI] [PubMed] [Google Scholar]
- Clark S.G., Stern,M.J. and Horvitz,H.R. (1992) C.elegans cell-signalling gene sem-5 encodes a protein with SH2 and SH3 domains. Nature, 356, 340–344. [DOI] [PubMed] [Google Scholar]
- Dent J.A. and Han,M. (1998) Post-embryonic expression pattern of C.elegans let-60 ras reporter constructs. Mech. Dev., 72, 179–182. [DOI] [PubMed] [Google Scholar]
- DeVore D., Horvitz,H. and Stern,M. (1995) An FGF receptor signaling pathway is required for the normal cell migrations of the sex myoblasts in C.elegans hermaphrodites. Cell, 83, 611–620. [DOI] [PubMed] [Google Scholar]
- Dohm G.L., Tapscott,E.B. and Kasperek,G.J. (1987) Protein degradation during endurance exercise and recovery. Med. Sci. Sports Exerc., 19, S166–S171. [PubMed] [Google Scholar]
- Dourdin N., Bhatt,A.K., Dutt,P., Greer,P.A., Arthur,J.S., Elce,J.S. and Huttenlocher,A. (2001) Reduced cell migration and disruption of the actin cytoskeleton in calpain-deficient embryonic fibroblasts. J. Biol. Chem., 276, 48382–48388. [DOI] [PubMed] [Google Scholar]
- Eble D.M., Spragia,M.L., Ferguson,A.G. and Samarel,A.M. (1999) Sarcomeric myosin heavy chain is degraded by the proteasome. Cell Tissue Res., 296, 541–548. [DOI] [PubMed] [Google Scholar]
- Eisenmann D.M. and Kim,S.K. (1997) Mechanism of activation of the Caenorhabditis elegans ras homologue let-60 by a novel, temperature-sensitive, gain-of-function mutation. Genetics, 146, 553–565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fire A. and Waterston,R.H. (1989) Proper expression of myosin genes in transgenic nematodes. EMBO J., 8, 3419–3428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Floss T., Arnold,H.H. and Braun,T. (1997) A role for FGF-6 in skeletal muscle regeneration. Genes Dev., 11, 2040–2051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fostel J.L., Coste,L.B. and Jacobson,L.A. (2003) Degradation of transgene-coded and endogenous proteins in the muscles of Caenorhabditis elegans. Biochem. Biophys. Res. Commun., in press. [DOI] [PubMed] [Google Scholar]
- Han M. and Sternberg,P.W. (1990) let-60, a gene that specifies cell fates during Caenorhabditis elegans vulval induction, encodes a ras protein. Cell, 63, 921–932. [DOI] [PubMed] [Google Scholar]
- Hara M. and Han,M. (1995) Ras farnesyltransferase inhibitors suppress the phenotype resulting from an activated ras mutation in Caenorhabditis elegans. Proc. Natl Acad. Sci. USA, 92, 3333–3337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes S.M. (2001) Muscle development: reversal of the differentiated state. Curr. Biol., 11, R237–R239. [DOI] [PubMed] [Google Scholar]
- Jagoe R.T. and Goldberg,A.L. (2001) What do we really know about the ubiquitin–proteasome pathway in muscle atrophy? Curr. Opin. Clin. Nutr. Metab. Care, 4, 183–190. [DOI] [PubMed] [Google Scholar]
- Jiang Z.S., Padua,R.R., Ju,H., Doble,B.W., Jin,Y., Hao,J., Cattini,P.A., Dixon,I.M. and Kardami,E. (2002) Acute protection of ischemic heart by FGF-2: involvement of FGF-2 receptors and protein kinase C. Am. J. Physiol. Heart Circ. Physiol., 282, H1071–H1080. [DOI] [PubMed] [Google Scholar]
- Johnson S.E. and Allen,R.E. (1995) Activation of skeletal muscle satellite cells and the role of fibroblast growth factor receptors. Exp. Cell Res., 219, 449–453. [DOI] [PubMed] [Google Scholar]
- Kastner S., Elias,M.C., Rivera,A.J. and Yablonka-Reuveni,Z. (2000) Gene expression patterns of the fibroblast growth factors and their receptors during myogenesis of rat satellite cells. J. Histochem. Cytochem., 48, 1079–1096. [DOI] [PubMed] [Google Scholar]
- Klint P. and Claesson-Welsh,L. (1999) Signal transduction by fibroblast growth factor receptors. Front. Biosci., 4, D165–D177. [DOI] [PubMed] [Google Scholar]
- Kokel M., Borland,C.Z., DeLong,L., Horvitz,H.R. and Stern,M.J. (1998) clr-1 encodes a receptor tyrosine phosphatase that negatively regulates an FGF receptor signaling pathway in Caenorhabditis elegans. Genes Dev., 12, 1425–1437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kouhara H., Hadari,Y.R., Spivak Kroizman,T., Schilling,J., Bar Sagi,D., Lax,I. and Schlessinger,J. (1997) A lipid-anchored Grb2-binding protein that links FGF-receptor activation to the Ras/MAPK signaling pathway. Cell, 89, 693–702. [DOI] [PubMed] [Google Scholar]
- Li W., Han,M. and Guan,K.L. (2000) The leucine-rich repeat protein SUR-8 enhances MAP kinase activation and forms a complex with Ras and Raf. Genes Dev., 14, 895–900. [PMC free article] [PubMed] [Google Scholar]
- Llovera M., Garcia-Martinez,C., Agell,N., Lopez-Soriano,F.J., Authier,F.J., Gherardi,R.K. and Argiles,J.M. (1998) Ubiquitin and proteasome gene expression is increased in skeletal muscle of slim AIDS patients. Int. J. Mol. Med., 2, 69–73. [PubMed] [Google Scholar]
- Mitch W.E. and Goldberg,A.L. (1996) Mechanisms of muscle wasting. The role of the ubiquitin–proteasome pathway. N. Engl. J. Med., 335, 1897–1905. [DOI] [PubMed] [Google Scholar]
- Molkentin J. and Dorn,I.G. (2001) Cytoplasmic signaling pathways that regulate cardiac hypertrophy. Annu. Rev. Physiol., 63, 391–426. [DOI] [PubMed] [Google Scholar]
- Moore J.W., Dionne,C., Jaye,M. and Swain,J.L. (1991) The mRNAs encoding acidic FGF, basic FGF and FGF receptor are coordinately downregulated during myogenic differentiation. Development, 111, 741–748. [DOI] [PubMed] [Google Scholar]
- Odelberg S.J., Kollhoff,A. and Keating,M.T. (2000) Dedifferentiation of mammalian myotubes induced by msx1. Cell, 103, 1099–1109. [DOI] [PubMed] [Google Scholar]
- Okkema P.G., Harrison,S.W., Plunger,V., Aryana,A. and Fire,A. (1993) Sequence requirements for myosin gene expression and regulation in Caenorhabditis elegans. Genetics, 135, 385–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olfert I.M., Breen,E.C., Mathieu-Costello,O. and Wagner,P.D. (2001) Skeletal muscle capillarity and angiogenic mRNA levels after exercise training in normoxia and chronic hypoxia. J. Appl. Physiol., 91, 1176–1184. [DOI] [PubMed] [Google Scholar]
- Olwin B.B. and Hauschka,S.D. (1988) Cell surface fibroblast growth factor and epidermal growth factor receptors are permanently lost during skeletal muscle terminal differentiation in culture. J. Cell Biol., 107, 761–769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patil C. and Walter,P. (2001) Intracellular signaling from the endoplasmic reticulum to the nucleus: the unfolded protein response in yeast and mammals. Curr. Opin. Cell Biol., 13, 349–355. [DOI] [PubMed] [Google Scholar]
- Rommel C., Clarke,B.A., Zimmermann,S., Nunez,L., Rossman,R., Reid,K., Moelling,K., Yancopoulos,G.D. and Glass,D.J. (1999) Differentiation stage-specific inhibition of the Raf–MEK–ERK pathway by Akt. Science, 286, 1738–1741. [DOI] [PubMed] [Google Scholar]
- Rooyackers O.E. and Nair,K.S. (1997) Hormonal regulation of human muscle protein metabolism. Annu. Rev. Nutr., 17, 457–485. [DOI] [PubMed] [Google Scholar]
- Roubin R., Naert,K., Popovici,C., Vatcher,G., Coulier,F., Thierry Mieg,J., Pontarotti,P., Birnbaum,D., Baillie,D. et al. (1999) let-756, a C.elegans fgf essential for worm development. Oncogene, 18, 6741–6747. [DOI] [PubMed] [Google Scholar]
- Ryder J.W., Fahlman,R., Wallberg Henriksson,H., Alessi,D.R., Krook,A. and Zierath,J.R. (2000) Effect of contraction on mitogen-activated protein kinase signal transduction in skeletal muscle. Involvement o the mitogen- and stress-activated protein kinase 1. J. Biol. Chem., 275, 1457–1462. [DOI] [PubMed] [Google Scholar]
- Saito A., Higuchi,I., Nakagawa,M., Saito,M., Uchida,Y., Inose,M., Kasai,T., Niiyama,T., Fukunaga,H. et al. (2000) An overexpression of fibroblast growth factor (FGF) and FGF receptor 4 in a severe clinical phenotype of facioscapulohumeral muscular dystrophy. Muscle Nerve, 23, 490–497. [DOI] [PubMed] [Google Scholar]
- Schafer W.R., Kim,R., Sterne,R., Thorner,J., Kim,S.H. and Rine,J. (1989) Genetic and pharmacological suppression of oncogenic mutations in ras genes of yeast and humans. Science, 245, 379–385. [DOI] [PubMed] [Google Scholar]
- Schultz E. and McCormick,K.M. (1994) Skeletal muscle satellite cells. Rev. Physiol. Biochem. Pharmacol., 123, 213–257. [DOI] [PubMed] [Google Scholar]
- Schutzman J.L., Borland,C.Z., Newman,J.C., Robinson,M.K., Kokel,M. and Stern,M.J. (2001) The Caenorhabditis elegans EGL-15 signaling pathway implicates a DOS-like multisubstrate adaptor protein in fibroblast growth factor signal transduction. Mol. Cell. Biol., 21, 8104–8116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selfors L.M., Schutzman,J.L., Borland,C.Z. and Stern,M.J. (1998) soc-2 encodes a leucine-rich repeat protein implicated in fibroblast growth factor receptor signaling. Proc. Natl Acad. Sci. USA, 95, 6903–6908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen X. et al. (2001) Complementary signaling pathways regulate the unfolded protein response and are required for C.elegans development. Cell, 107, 893–903. [DOI] [PubMed] [Google Scholar]
- Sieburth D.S., Sun,Q. and Han,M. (1998) SUR-8, a conserved Ras-binding protein with leucine-rich repeats, positively regulates Ras-mediated signaling in C.elegans. Cell, 94, 119–130. [DOI] [PubMed] [Google Scholar]
- Sogos V., Balaci,L., Ennas,M.G., Dell’era,P., Presta,M. and Gremo,F. (1998) Developmentally regulated expression and localization of fibroblast growth factor receptors in the human muscle. Dev. Dyn., 211, 362–373. [DOI] [PubMed] [Google Scholar]
- Solomon V. and Goldberg,A.L. (1996) Importance of the ATP–ubiquitin–proteasome pathway in the degradation of soluble and myofibrillar proteins in rabbit muscle extracts. J. Biol. Chem., 271, 26690–26697. [DOI] [PubMed] [Google Scholar]
- Stern M.J. and Horvitz,H.R. (1991) A normally attractive cell interaction is repulsive in two C.elegans mesodermal cell migration mutants. Development, 113, 797–803. [DOI] [PubMed] [Google Scholar]
- Sternberg P.W. and Han,M. (1998) Genetics of RAS signaling in C.elegans. Trends Genet., 14, 466–472. [DOI] [PubMed] [Google Scholar]
- Sternberg P.W., Lesa,G., Lee,J., Katz,W.S., Yoon,C., Clandinin,T.R., Huang,L.S., Chamberlin,H.M. and Jongeward,G. (1995) LET-23-mediated signal transduction during Caenorhabditis elegans development. Mol. Reprod. Dev., 42, 523–528. [DOI] [PubMed] [Google Scholar]
- Suzuki J., Yamazaki,Y., Guang,L., Kaziro,Y. and Koide,H. (2000) Involvement of Ras and Ral in chemotactic migration of skeletal myoblasts. Mol. Cell. Biol., 20, 4658–4665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szewczyk N.J., Hartman,J.J., Barmada,S.J. and Jacobson,L.A. (2000) Genetic defects in acetylcholine signalling promote protein degradation in muscle cells of Caenorhabditis elegans. J. Cell Sci., 113, 2003–2010. [DOI] [PubMed] [Google Scholar]
- Szewczyk N.J., Peterson,B.K. and Jacobson,L.A. (2002) Activation of Ras and the MAP kinase pathway promotes protein degradation in muscle cells of Caenorhabditis elegans. Mol. Cell. Biol., 22, 4181–4188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan P.B. and Kim,S.K. (1999) Signaling specificity: the RTK/RAS/MAP kinase pathway in metazoans. Trends Genet., 15, 145–149. [DOI] [PubMed] [Google Scholar]
- Tisdale M.J. (2001) Loss of skeletal muscle in cancer: biochemical mechanisms. Front. Biosci., 6, D164–D174. [DOI] [PubMed] [Google Scholar]
- Wolkow C.A., Kimura,K.D., Lee,M.S. and Ruvkun,G. (2000) Regulation of C.elegans life-span by insulinlike signaling in the nervous system. Science, 290, 147–150. [DOI] [PubMed] [Google Scholar]
- Yusoff P., Lao,D.H., Ong,S.H., Wong,E.S., Lim,J., Lo,T.L., Leong,H.F., Fong,C.W. and Guy,G.R. (2002) Sprouty2 inhibits the Ras/MAP kinase pathway by inhibiting the activation of Raf. J. Biol. Chem., 277, 3195–3201. [DOI] [PubMed] [Google Scholar]
- Zdinak L.A., Greenberg,I.B., Szewczyk,N.J., Barmada,S.J., Cardamone Rayner,M., Hartman,J.J. and Jacobson,L.A. (1997) Transgene-coded chimeric proteins as reporters of intracellular proteolysis: starvation-induced catabolism of a lacZ fusion protein in muscle cells of Caenorhabditis elegans. J. Cell. Biochem., 67, 143–153. [PubMed] [Google Scholar]