Abstract
Gene transcription is required for long-term memory (LTM) formation. LTM formation is impaired in a male-specific manner in mice lacking either of the two Ca2+/calmodulin-dependent kinase kinase (Camkk) genes. Since altered transcription was suggested to cause these impairments in LTM formation, we used microarrays to screen for CaMKKβ-dependent gene expression changes. Here we show that the hippocampal mRNA expression of two splicing factors, splicing factor arginine/serine-rich 3 (Sfrs3/Srp20) and polypyrimidine tract-binding protein-associated splicing factor (Psf), is altered in CaMKKβ-deficient males. In wild-type (WT) mice, the basal expression level in the hippocampus is higher in males than in females, and the sex difference in Srp20 expression is detectable before puberty. Training in two hippocampus-dependent learning tasks, the spatial version of the Morris water maze (MWM) and background contextual fear conditioning, increases the hippocampal mRNA expression of both splicing factors in WT males. However, the increase in Srp20 mRNA expression occurs only in males and not in females, whereas the up-regulation of Psf expression occurs in both sexes. Importantly, control experiments demonstrate that the up-regulation of both splicing factors is specific for the learned associations after contextual fear conditioning. In summary, we provide the first evidence for a regulation of splicing factors during LTM formation and we suggest that alternative splicing contributes to sex differences in LTM formation.
The hippocampus is required for several types of memory, including spatial and contextual fear memory (for reviews, see Mizuno and Giese 2005; Morris 2006). Formation of long-term memory (LTM) requires gene transcription, which can be initiated by Ca2+ signals that are transduced by the Ca2+/calmodulin (CaM) kinase cascade (Silva and Giese 1994; for review, see Corcoran and Means 2001). This kinase cascade consists of CaM kinase kinase α (CaMKKα), CaM kinase kinase β (CaMKKβ), CaM kinase I (CaMKI), and CaM kinase IV (CaMKIV). The two CaMKKs phosphorylate CaMKI and CaMKIV to enhance the activity of these effector kinases (Takemoto-Kimura et al. 2003; Chow et al. 2005; Blaeser et al. 2006). The great majority of transcriptional studies on hippocampal LTM formation in rodents focus exclusively on males or disregard sex effects by analyzing male and female data together. While many behaviorally induced transcriptional events do not differ between the sexes, as is the case of expression of nerve growth factor-inducible gene B (Ngfi-b) (Mizuno et al. 2006), increasing lines of evidence suggest sex differences in transcription underlying hippocampus-dependent LTM formation (Kudo et al. 2004; Hebda-Bauer et al. 2005; Mizuno et al. 2006, 2007).
Our previous work demonstrated an unexpected male-specific role of the CaMKKs in hippocampal LTM formation (Peters et al. 2003; Mizuno et al. 2006, 2007). In male mice, CaMKKα is required for contextual fear LTM formation as well as the conditioning-induced activation of the transcription factor cAMP-responsive element binding protein (CREB) and transcription of the brain-derived neurotrophic factor (Bdnf) (Blaeser et al. 2006; Mizuno et al. 2006). In contrast, in female mice CaMKKα is not needed for contextual fear LTM formation and the regulation of Bdnf mRNA expression (Mizuno et al. 2006). Furthermore, in male, but not female mice, CaMKKβ is involved in spatial memory formation, CREB activation by spatial training, and the transcription-dependent phase of long-term potentiation (Peters et al. 2003; Mizuno et al. 2007). Using microarrays, we compared hippocampal gene expression between CaMKKβ-deficient and wild-type (WT) male mice after hippocampus-dependent spatial training (Mizuno et al. 2007). In this study we demonstrated that the gene encoding the glycosyl phosphatidyl-inositol anchor attachment protein 1 (Gaa1) requires CaMKKβ for up-regulation by spatial training in males (Mizuno et al. 2007). Here, we show that two splicing factors, the splicing factor arginine/serine-rich 3 (Sfrs3/Srp20) (Jumaa and Nielsen 1997; Yu et al. 2004) and polypyrimidine tract-binding (PTB) protein-associated splicing factor (Psf) (Chanas-Sacre et al. 1999) are differentially expressed in the naïve male hippocampus of CaMKKβ-deficient mice. Since increasing evidence suggests that alternative splicing is an important, but not well understood mechanism of brain plasticity (Bottai et al. 2002; Lee and Irizarry 2003; Nijholt et al. 2004; O'Connor et al. 2004; Rattiner et al. 2004; Beffert et al. 2005), we studied the expression of Srp20 and Psf in more detail. We demonstrate that Srp20 and Psf are expressed at higher levels in male than in female adult WT hippocampus. Training in two hippocampus-dependent learning tasks, the Morris water maze and contextual fear conditioning, up-regulate the hippocampal mRNA expression of both splicing factors in male mice. Moreover, Psf mRNA expression was up-regulated after behavioral training in both sexes, while Srp20 mRNA expression was only regulated in males. Control experiments in the contextual fear-conditioning task in male animals determined that Srp20 and Psf mRNA expressions are regulated by a learned context/tone-shock association and not by the context or shock alone. Thus, we provide the first evidence for the regulation of splicing factor expression during LTM formation, and we suggest that alternative splicing contributes to sex differences in LTM formation.
Results
Lack of CaMKKβ alters the levels of hippocampal Srp20 and Psf mRNA expression in naïve males, but not in females
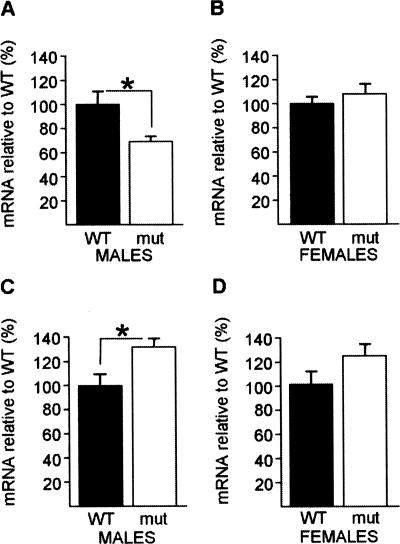
Affymetrix microarray analysis suggested that the hippocampal expressions of several genes are altered in male CaMKKβ-deficient mice in comparison with male WT mice after training in the hidden-platform version of the Morris water maze (Mizuno et al. 2007). The expression of 17 candidate genes was studied with quantitative real-time PCR (qPCR), and after the initial confirmation of a dys-regulation of Gaa1 (Mizuno et al. 2007) we investigated a putative change in the expression of Srp20 and Psf. Here, we tested whether the hippocampal mRNA expression of these two genes are affected by CaMKKβ deficiency per se by comparing mRNA levels in CaMKKβ-deficient mice and WT naïve mice of both sexes (Fig. 1). In males, hippocampal Srp20 mRNA expression was reduced in CaMKKβ-deficient mutants in comparison with WT mice (F(1,8) = 7.37, P < 0.05) (Fig. 1A). In contrast, hippocampal Psf mRNA expression was up-regulated in male mutants (F(1,11) = 5.95, P < 0.05) (Fig. 1C). Thus, in males, absent CaMKKβ signaling differentially affects hippocampal Srp20 and Psf mRNA expression.
Figure 1.
Lack of CaMKKβ results in altered Srp20 and Psf mRNA expression in the naïve hippocampus in males. (A) Hippocampal Srp20 mRNA expression was significantly reduced in naïve CaMKKβ-deficient males (n = 5) compared with naïve WT males (n = 5). (B) Hippocampal Srp20 mRNA expression levels did not differ between naïve mutant females (n = 6) and naïve WT females (n = 8). (C) Hippocampal Psf mRNA expression was significantly higher in naïve mutants males (n = 7) compared with naïve WT males (n = 6). (D) Hippocampal PSF mRNA levels did not differ significantly between naïve WT (n = 9) and mutant females (n = 6), although increasing the number of animals per group might have revealed a small difference between the female genotypes. Data are means ± SEM; (*) P < 0.05.
Based on the previous findings of a CaMKKβ male-specific role in inducing CREB phosphorylation by spatial training and in the transcription-dependent phase of LTP (Peters et al. 2003; Mizuno et al. 2007), we investigated whether the lack of CaMKKβ would also affect Psf and Srp20 mRNA expression in the female naïve hippocampus (Fig. 1B,D). In contrast to males, female mutants expressed Srp20 mRNA in the hippocampus at comparable levels to female WT mice (F(1,12) = 0.75, P = 0.40) (Fig. 1B). Furthermore, we observed that Psf mRNA levels were, on average, higher in mutant females when compared with WT; however, this difference did not reach statistical significance (F(1,13) = 2.83, P = 0.11). Increasing the number of animals per group might have revealed a small difference between the female genotypes (Fig. 1D).
Thus, in females, absent CaMKKβ signaling does not affect hippocampal Srp20 mRNA expression, while it might dys-regulate hippocampal Psf mRNA expression.
Sex differences in hippocampal Srp20 and Psf mRNA expression levels in naïve WT mice
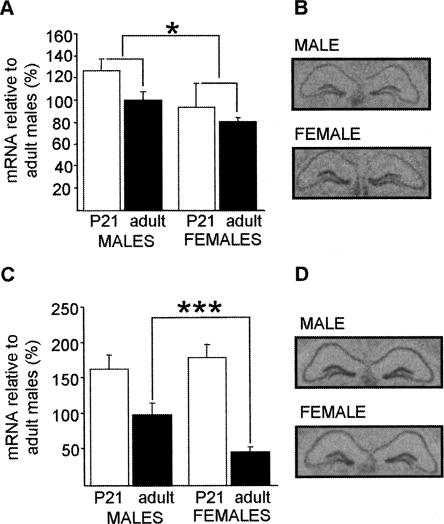
We found decreased expression of Srp20 in male CaMKKβ-deficient mice when compared with their WT counterparts, but no difference between genotypes in female animals. As this suggested different mechanisms of Srp20 regulation between the sexes, we sought to compare expression levels between male and female naïve WT mice (Fig. 2). As a number of studies have revealed that the onset of puberty influences gene expression (e.g., Koshibu and Levitt 2005), we analyzed age effects in the expression of Psf and Srp20 by comparing hippocampal mRNA levels in both sexes before puberty at postnatal day 21 (Omoto et al. 2005) and in adulthood, in both sexes. Two-way ANOVA of the hippocampal Srp20 mRNA expression levels showed that there was a significant effect of sex (F(1,20) = 7.40, P < 0.05), while an effect of age did not reach statistical significance (F(1,20) = 2.71, P = 0.12). We did not observe a sex x age interaction (F(1,20) = 0.65, P = 0.43) (Fig. 2A), indicating that sex differences in hippocampal Srp20 mRNA are detectable before the onset of puberty and remain in adulthood. Sex differences in Srp20 mRNA levels in adult mice were confirmed in Figures 5A and 6B, below.
Figure 2.
Sex differences in hippocampal Srp20 and Psf mRNA expression levels in WT mice. (A) Hippocampal Srp20 mRNA expression was significantly lower in females than in males independently of the age group considered (P21: five females, four males; adults: seven females, eight males). Sex differences in adult naïve animals were further confirmed (see Figs. 5A, 6B). (B) Srp20 is expressed in the three major hippocampal subfields in both male and female adult mice as determined by in situ hybridization. (C) Hippocampal Psf mRNA expression differed with age (significance stars not shown) and post hoc analysis suggested that the expression was significantly lower in adult females (n = 5) than in adult males (n = 8) and not in P21 mice (six females, four males). Higher hippocampal Psf mRNA expression in adult males than in adult females was also found in Figures 5C and 6D. (D) Psf is expressed in the three major hippocampal subfields in both male and female adult mice as determined by in situ hybridization. Data are means ± SEM; (*) P < 0.05; (***) P < 0.001.
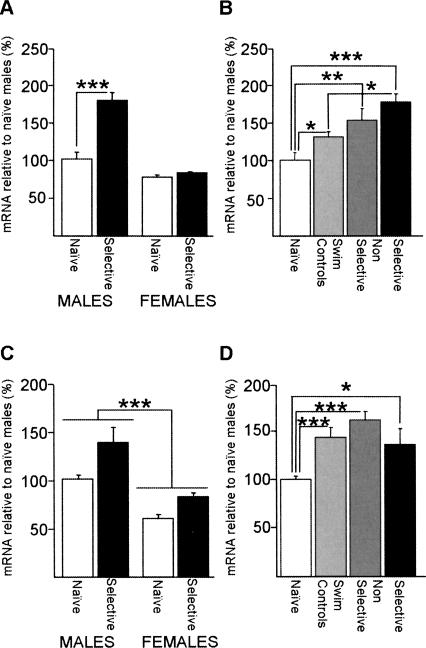
Figure 5.
Hippocampal Srp20 and Psf mRNA expressions are up-regulated after training in the MWM in WT mice, and the up-regulation of Srp20 expression occurs only in males. (A) Hippocampal Srp20 mRNA expression was significantly up-regulated in selective males, but not females. Post-hoc analysis also showed that naïve males expressed higher levels of Srp20 mRNA than naïve females (P < 0.05), which is not indicated for the ease of presentation. Males: seven naïve, five selective; females: seven naïve, six selective. (B) In males, Srp20 mRNA up-regulation occurred in the swim control and nonselective groups, but this was significantly smaller than in the selective group: seven naïves, seven swim controls, five nonselective, five selective. (C) Hippocampal Psf mRNA expression was significantly up-regulated by spatial training in both sexes as shown by two-way ANOVA. Additionally, males expressed significantly higher levels of hippocampal Psf mRNA than females. Males: eight naïve, five selective; females: five naïve, seven selective. (D) Psf mRNA up-regulation was not specific to MWM learning, as nonselective groups and swim controls express higher levels of Psf mRNA than the naïve group. Eight naïves, eight swim controls, six nonselective, five selective. Data are means ± SEM; (*) P < 0.05; (**) P < 0.01; (***) P < 0.001.
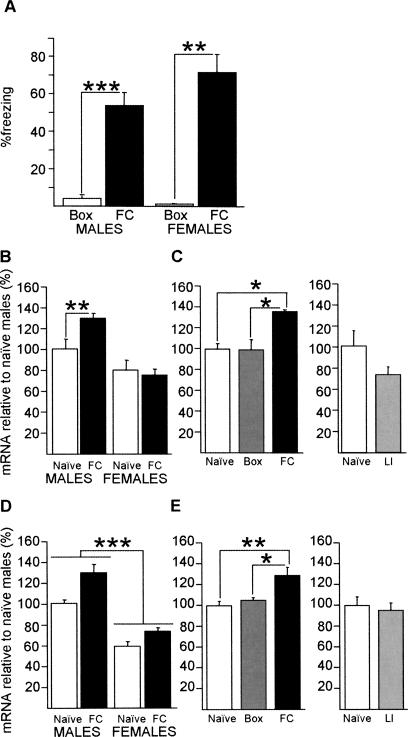
Figure 6.
Hippocampal Srp20 and Psf mRNA expressions are regulated by learned associations after contextual fear conditioning in WT mice; the up-regulation of Srp20 expression is male-specific. (A) Male (n = 7) and female mice (n = 3) showed contextual freezing 24 h after conditioning in comparison with a Box control group (eight males, four females). (B) Hippocampal Srp20 mRNA expression was significantly up-regulated by contextual fear conditioning in males, but not in females, as shown by post-hoc analysis. Females: six naïve, five contextually fear conditioned (FC); males: seven naïve, eight FC. (C) In males, hippocampal Srp20 mRNA expression levels did not differ between naïve mice (n = 7) and Box control mice (n = 6), while there was significantly increased expression in FC mice (n = 8). The LI shock control (n = 5) did not result in an up-regulation in comparison with naïve mice (n = 4). (D) Hippocampal Psf mRNA expression was significantly up-regulated by contextual fear conditioning in males (eight naïve, seven FC) and females (five naïve, seven FC) as shown by two-way ANOVA. Additionally, males expressed significantly higher levels of hippocampal Psf mRNA than females. (E) In males, hippocampal Psf mRNA expression levels did not differ between naïve mice (n = 8) and Box control mice (n = 6), while there was significantly increased expression in FC mice (n = 7). There was no difference in expression between naïve (n = 4) and LI control mice (n = 5). Data are means ± SEM; (*) P < 0.05; (**) P < 0.01; (***) P < 0.001.
Analysis of the hippocampal Psf mRNA expression demonstrated a significant effect of age (F(1,19) = 24.28, P < 0.001), reflected by the much higher values of Psf expression in P21 animals when compared with adults. We did not observe a significant effect of sex (F(1,19) = 0.84; P = 0.37), and no significant sex x age interaction (F(1,19) = 2.92, P = 0.10) (Fig. 2C). Post-hoc analysis suggested that males may express higher levels of hippocampal Psf mRNA than females in adulthood, but that there was no sex difference at postnatal day 21 (Fig. 2C). Higher hippocampal expression of Psf mRNA in adult males than in adult females was also found for the data presented in Figures 5C and 6D, below. Thus, in contrast to the sex difference of hippocampal Srp20 mRNA expression, the sex difference in hippocampal Psf mRNA expression only occurred after the onset of puberty. It is possible that the observed sex differences in Psf and Srp20 mRNA expression (Fig. 2A,C) are unique to the mouse strain considered; however, this appears unlikely, as preliminary analysis of microarray data for hippocampal gene expression in males and females of four mouse strains suggests sex-differences in mRNA levels of Srp20 and possibly Psf, regardless of strain (L.C. Schalkwyk, pers. comm.).
Our qPCR experiments demonstrated that the mRNA expression levels of Srp20 and Psf differ in the adult male and female hippocampus. To investigate whether this was due to localized differences in expression within the hippocampus, we performed in situ hybridizations in coronal sections of male and female adult brains. We found that in both sexes Srp20 (Fig. 2B) and Psf (Fig. 2D) are expressed throughout the whole hippocampus, suggesting that the observed sex differences in mRNA expression do not result from different patterns of expression across the three major hippocampal subfields.
Sex-dependent regulation of hippocampal Srp20, but not Psf, mRNA expression by spatial training in the Morris water maze (MWM) in WT mice
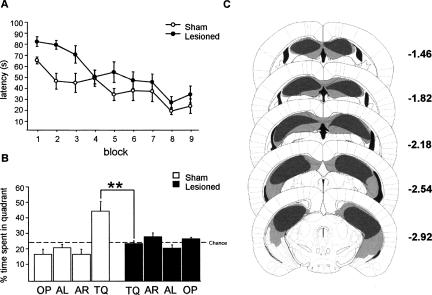
Dys-regulation of hippocampal Srp20 and Psf mRNA expression was shown to occur in adult naïve male CaMKKβ-deficient mice (Fig. 1A,C). Since male CaMKKβ-deficient mice display delayed spatial memory formation in the MWM (Peters et al. 2003), it was conceivable that hippocampus-dependent spatial training in the MWM could regulate hippocampal Srp20 and Psf mRNA expression in adulthood. We investigated this idea in our MWM set-up, in which hippocampal lesions impair spatial learning in both males (Angelo et al. 2003) and females (Fig. 3). We trained male (n = 11) and female (n = 12) WT mice in the MWM using a four trial/day training protocol for 6 d, and tested them in a probe trial at the end of the sixth day of training (Fig. 4). We subdivided the mice in two groups according to individual probe trial performance. These were designated selective and nonselective. Selective animals displayed a clear preference for the target quadrant (TQ) during the probe trial: searching more than 35% of the time in TQ and no similar percentage of time in any other quadrant. Nonselective animals did not display such preference.
Figure 3.
Hippocampal lesions in female 129B6F1 mice impair spatial memory formation in the Morris water maze. (A) Time to reach the hidden platform differed between the sham-lesioned and lesioned mice. Two-way ANOVA with repeated measures showed that there was a significant effect of treatment (F(1,13) = 5.9, P < 0.05) and training (F(8,104) = 14.9, P < 0.001), but no significant interaction (F(8,104) = 1.5, P = 0.18). (B) During the probe trial on day 3, the sham-lesioned mice (n = 8) searched selectively in the target quadrant (TQ) where the platform had been located (F(3,28) = 10.8, P < 0.0001), whereas the lesioned mice (n = 7) searched randomly (F(3,27) = 2.7, P = 0.07). Analysis of time spent in the target quadrant showed a significant effect of lesion (F(1,13) = 9.0, P < 0.01). (C) Minimum (black) and maximum (gray-shaded) of hippocampal lesions. Note that the maximal lesions are an overestimate, because neocortex overlying the hippocampus became detached during slice preparation. Data are means ± SEM; (**) P < 0.01. Adapted with permission from Elsevier © 2001, Paxinos and Franklin 2001.
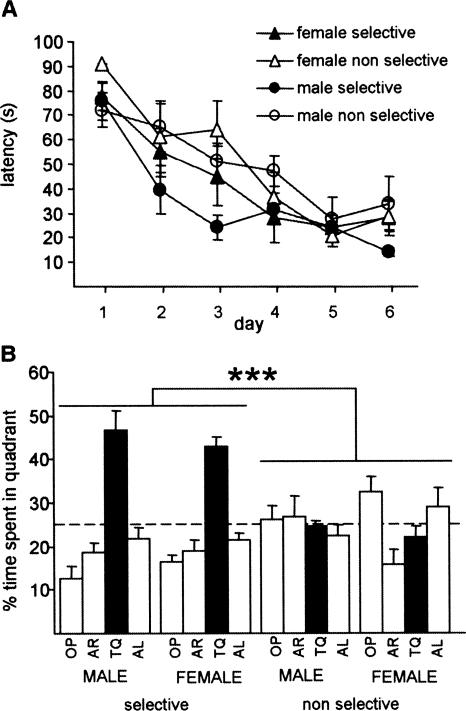
Figure 4.
Performance of male and female WT mice in the hidden-platform version of the Morris water maze (MWM). (A) Time to find the hidden platform decreased with training in all groups; planned comparisons suggest that selective males (n = 5) acquired the task faster than their nonselective counterparts (n = 6), while both female groups acquired the task at the same rate (seven selective, five nonselective). (B) In a probe trial given at the end of training, selective animals spent significantly more time swimming in the target quadrant than in any other quadrant (significance stars are omitted); while this was not observed in nonselective animals; no sex effects were observed. (TQ) Target quadrant; (AR) adjacent right quadrant; (AL) adjacent left quadrant; (OP) opposite quadrant. Data are means ± SEM; (***) P < 0.001.
A three-way ANOVA with repeated measures on latency to reach the platform revealed an overall improvement in acquisition across all groups (effect of training: P < 0.001) and no differences in acquisition between the sexes (F(1,19) = 0.70, P = 0.41). There were no significant two-way or three-way interactions between the factors considered. A larger sample size might have revealed improved acquisition in selective animals (F(1,19) = 4.09, P = 0.057), which was also suggested by a planned two-way ANOVA with repeated measures between selective and nonselective males, but not between the female groups (effect of group F(1,9) = 4.21, P = 0.07) (Fig. 4A).
A two-way ANOVA comparison of the search times during the probe trial with quadrant selectivity and sex as factors revealed no sex effects (F(1,19) = 1.54, P = 0.23), a significant difference between selective and nonselective groups (F(1,19) = 63.71, P < 0.001), and no sex x selectivity interaction (F(1,19) = 0.087, P = 0.77) (Fig. 4B). Planned one-way ANOVAs confirmed that, in both sexes, selective animals spent more time in TQ than their nonselective counterparts (males: F(1,9) = 29.87; P < 0.001; females: F(1,10) = 33.91; P < 0.001). Furthermore, there was a clear preference toward TQ in the selective groups (males: F(3,16) = 23.94, P < 0.001; post-hoc analysis: P < 0.001 for all TQ comparisons; females: F(3,24) = 37.47, P < 0.001; post-hoc analysis: P < 0.001 for all TQ comparisons), while in the nonselective groups, males searched randomly (males: F(3,20) = 0.37; P = 0.77) and females displayed a slight preference toward OP (F(3,16) = 4.42, P < 0.05; post-hoc analysis OP vs AR [P < 0.01] and OP vs TQ [P < 0.05]).
In order to investigate whether Srp20 and Psf mRNA levels were increased in animals that successfully learned the MWM task, we compared mRNA levels between naïve and selective animals of both sexes. Analysis of hippocampal Srp20 mRNA expression levels showed that there was an effect of training (F(1,21) = 31.7, P < 0.001), an effect of sex (F(1,21) = 66.0, P < 0.001), and a sex x training interaction (F(1,21) = 24.5, P < 0.001) (Fig. 5A). Post-hoc analysis revealed that naïve males expressed higher levels of Srp20 mRNA than naïve females (P < 0.05) and Srp20 mRNA was up-regulated in selective males (P < 0.001), but not in selective females (P = 0.63) (Fig. 5A). For Psf mRNA, a two-way ANOVA with training and sex as factors indicated effects of training (F(1,21) = 17.0, P < 0.001) and sex (F(1,21) = 44.2, P < 0.001), but no sex x training interaction (F(1,21) = 0.96, P = 0.34) (Fig. 5C). Thus, in contrast to the hippocampal Srp20 mRNA expression, the hippocampal Psf mRNA expression was up-regulated in both male and female WT mice after spatial training in the MWM.
Successful learning of the MWM task is generally assessed by probe trial performance. A number of studies have established direct correlations between levels of hippocampal transcription of certain genes and learning performance (Matsuyama et al. 1997; Balschun et al. 2003; Brightwell et al. 2004). For this reason we used the ‘’nonselective’’ group of animals (Fig. 4) as a control for spatial learning-induced transcriptional changes. Several factors intrinsic to the task such as novelty of the environment (Hall et al. 2000), physical activity (Vaynman et al. 2004), interaction with the human experimenter, and stress (Aguilar-Valles et al. 2005) are sufficient to trigger changes in gene expression. In a number of studies, “swim control’’ groups are used to account for the factors mentioned above. Different experimental designs have been used to define swim controls, including swimming in the absence of the platform (Shors et al. 2000; Cavallaro et al. 2002; Leil et al. 2003; Oh et al. 2003) or in a visible platform version of the task (Leil et al. 2002). Our swim control group consisted of mice that swam in the absence of a platform for the same number of trials and for the same amount of time per trial as trained animals, so that the amount of physical activity and the time of exposure to the room was matched to the groups trained in the hidden platform version of the task.
In order to investigate whether the regulation of these genes occurred specifically in selective animals, we compared Srp20 and Psf mRNA levels across four groups of male mice (naïve, swim control, nonselective, and selective). For Srp20 mRNA levels (Fig. 5B), one-way ANOVA pointed to significant differences across the four groups considered (F(3,20) = 9.47; P < 0.001). Post-hoc analysis confirmed the previously described up-regulation in selective animals when compared with naïves (P < 0.001), and also revealed significant up-regulation in nonselective animals (P < 0.01) and swim controls (P < 0.05). In addition, selective animals expressed higher levels of Srp20 mRNA than swim controls (P < 0.05), suggesting that novelty, physical activity, stress (or a combination of both) should only partly account for the up-regulation verified in selective animals.
Psf mRNA levels were significantly different between the four male groups considered (F(3,23) = 9.43, P < 0.001, one-way ANOVA). Training in the absence of a platform was sufficient to trigger Psf up-regulation, as levels of expression differed significantly between naïve and all of the remaining groups (post-hoc: naïve vs. nonselective and naïve vs. swim controls P < 0.001; naïve vs. selective P < 0.05), but not between swim controls and the selective and nonselective groups (P > 0.05) (Fig. 5D). Thus, as far as this task is concerned, Psf up-regulation does not seem to be specifically regulated by learning of the task. An up-regulation of Psf and Srp20 in selective and nonselective animals has been replicated in an independent cohort of animals (data not shown).
Hippocampal Srp20 and Psf mRNA levels are up-regulated by learned associations after contextual fear conditioning in male WT mice
Our expression studies after spatial training could not determine whether the observed up-regulation of Srp20 and Psf mRNA was specific for the formation of a spatial memory or elicited by factors intrinsic to the task. Classical conditioning tasks can be easily used to determine whether expression changes are specific for the learned conditioned-stimulus/unconditioned-stimulus association or whether they relate to one of the stimuli alone. Therefore, we investigated the regulation of Srp20 and Psf mRNA in a single-trial, hippocampus-dependent background contextual fear conditioning (Phillips and LeDoux 1994). We first established that in our set up a single training trial led to the formation of a contextual fear memory in both males and females (Fig. 6A). The regulation of hippocampal Srp20 and Psf mRNA expression after contextual fear conditioning was similar to that after spatial training. Srp20 mRNA expression levels differed between the sexes (F(1,22) = 24.1, P < 0.001), there was no effect of training (F(1,22) = 2.34, P = 0.14), but there was a significant sex x training interaction (F(1,22) = 4.74, P < 0.05) (Fig. 6B). Post-hoc analysis showed that Srp20 mRNA expression was up-regulated by training in males, but not in females (Fig. 6B). Thus, there was a male-specific up-regulation of hippocampal Srp20 mRNA expression after contextual fear conditioning in WT mice.
Analysis of Psf mRNA expression demonstrated an effect of sex (F(1,23) = 79.3, P < 0.001), an effect of training (F(1,23) = 16.6, P < 0.001), but no significant sex x training interaction (F(1,23) = 1.65, P = 0.21) (Fig. 6D). Thus, the hippocampal Psf mRNA expression was up-regulated after contextual fear conditioning for both sexes in WT mice.
We studied whether the conditioning-induced up-regulation of hippocampal Srp20 and Psf mRNA expression in males was specific for the learned associations using Box and latent inhibition (LI) groups. The Box group served as a control for the effects of the “conditioned stimulus’’ on gene expression, while the LI group controlled for the effects of shock alone, as the animals experienced a foot shock, but did not make a context-shock association (von Hertzen and Giese 2005; Lepicard et al. 2006). Srp20 mRNA expression was not up-regulated in LI controls (F(1,7) = 1.72, P = 0.23) and Box controls (F(2,18) = 4.75, P < 0.05; post-hoc analysis revealed no difference in expression between naïve males and Box controls) (Fig. 6C). Psf mRNA expression was also not up-regulated in LI controls (F(1,7) = 0.15, P = 0.71) and Box controls (F(2,18) = 7.91, P < 0.01, post-hoc analysis revealed no difference in expression between naïve males and Box controls) (Fig. 6E). Thus, the up-regulations of Srp20 and Psf mRNA expression in male WT mice after background contextual fear conditioning were specific for the context/tone-shock association.
Discussion
Our main finding is that the hippocampal mRNA expressions of two genes encoding splicing factors, Srp20 and Psf, are up-regulated during contextual fear memory formation as a specific result of the context/tone-shock association. This suggests that changes in splicing factor expression during LTM formation regulate alternative splicing. In addition, Srp20 up-regulation is sex-specific, occurring in males only, and implying that alternative splicing contributes to sex differences in LTM formation.
We ruled out the possibility that novelty and/or tone and shock alone contribute to the identified transcriptional changes by studying gene expression in a box control (exposure to the conditioning chamber without shock presentation) and in a latent inhibition control group (administration of shock after prolonged exposure to the context). In these control groups the levels of gene expression were similar to the naïve controls, thereby indicating that after contextual fear conditioning, the up-regulation of both Psf and Srp20 mRNA levels is specific for the learned context/tone-shock association. Thus, we conclude that the transcription of Srp20 and Psf is likely to be engaged during contextual fear memory formation.
At the time point considered (30 min after conditioning), only males showed up-regulated Srp20 mRNA levels, indicating that this transcriptional event is male specific. This was not due to technical issues with the female mRNA samples, because we could detect increased Psf mRNA expression after contextual fear conditioning in females. Our results are in agreement with the findings that the regulations of Bdnf, Gaa1, and Ngfi-b mRNA expression in the hippocampus after contextual fear conditioning, which are all specific for the context-shock association (Hall et al. 2000; von Hertzen and Giese 2005; Mizuno et al. 2007), can be sex-specific or common to both sexes (Mizuno et al. 2006, 2007). Additionally, contextual fear conditioning induces increased phosphorylation of CREB that is specific to male mice (Kudo et al. 2004) and male, but not female, CaMKKα mutant mice are impaired in contextual fear LTM formation (Blaeser et al. 2006; Mizuno et al. 2006). Taken together, these lines of evidence strengthen the hypothesis that molecular mechanisms elicited by training in hippocampus-dependent tasks can be sex-specific.
We have also studied hippocampal Srp20 and Psf mRNA expression after spatial training in the hidden-platform version of the MWM. The mRNA expression of both splicing factors was up-regulated after spatial training, with the increase in Srp20 mRNA unique to male mice, whereas the up-regulation of Psf mRNA expression was sex-independent. We investigated whether the transcriptional changes observed in males occurred specifically in animals that learned the task (selective), by assessing levels of gene expression in animals that failed to learn the task (nonselective) and in swim controls (mice exposed to the maze with no platform present for equal periods of time and experiencing similar levels of physical activity as the trained groups). Psf and Srp20 mRNA levels were increased in both control groups, and therefore, we conclude that these up-regulations are not specifically associated with successful learning of this spatial task, and can be related to other factors intrinsic to the task. However, this observation does not completely rule out a potential role for these genes in spatial learning. For example, the immediate-early gene activity-regulated cytoskeleton-associated protein (Arc/Arg3.1) is up-regulated to the same degree amongst animals trained in spatial and non-spatial versions of the MWM task (Guzowski et al. 2001), yet inhibition of ARC/ARG3.1 protein expression produces a significant impairment in spatial learning (Guzowski et al. 2000; Plath et al. 2006), thereby suggesting that transcriptional changes triggered by factors intrinsic to the task can be important in the formation of spatial memories. Taken together, our results suggest that, similarly to the contextual fear conditioning tasks, there are sex-dependent and sex-independent transcriptional events engaged during spatial memory formation.
With regard to Srp20 up-regulation in WT males after spatial training, it is interesting to note that naïve male, but not female, CaMKKβ-deficient mice have reduced Srp20 mRNA levels and impaired spatial memory formation (Peters et al. 2003; Mizuno et al. 2007). Therefore, it is conceivable that the reduced Srp20 expression contributes to the spatial memory deficits observed in male CaMKKβ-deficient mice. However, the reduced Srp20 expression in male CaMKKβ-deficient mice does not seem to account for deficits in contextual fear conditioning, as these mutants perform normally in this task (Peters et al. 2003; Mizuno et al. 2007). The finding of an up-regulation of Psf mRNA after training in the MWM in WT animals is harder to reconcile: basal levels of Psf expression are higher in CaMKKβ-deficient mutants than in WT mice and an up-regulation of Psf expression occurs in both sexes after spatial training and contextual fear conditioning. Therefore, it is unlikely that higher levels of Psf expression in CaMKKβ-deficient mice account for the spatial memory deficits. A possibility is that Psf up-regulation in naïve mutants occurs as a molecular compensation for lack of CaMKKβ, and this compensation might prevent deficits in contextual fear conditioning.
Sex differences in the hidden-platform version of the MWM have been debated (Jonasson 2005) and seem to be strongly dependent on the conditions of set-up and the amount of training (Frye 1995; Perrot-Sinal et al. 1996; Roof and Stein 1999). In our laboratory and training protocol, males and females displayed similar acquisition of the task and no overall differences in TQ preference in the probe trial. Furthermore, we have shown that both sexes require the hippocampus for spatial memory formation in our set up (Angelo et al. 2003; Fig. 3). The lack of sex differences at the behavioral level, both in the MWM and contextual fear conditioning, implies that the significance of the sex-specific transcriptional changes described may be related to differences in some, but not all, molecular mechanisms underlying memory formation. Furthermore, it has been suggested that, in rodents and in humans, the sexes use distinct spatial learning strategies, making use of either geometric or landmark cues (Sandstrom et al. 1998; Maguire et al. 1999; Roof and Stein 1999). Thus, it is conceivable that Srp20 is a marker gene for the male spatial learning strategy.
We have only investigated the transcriptional profile of our target genes 30 min after fear conditioning and 30 min after the probe trial in the MWM. A time-course study would be required to determine the temporal pattern of expression for these genes. Such an experiment would determine whether Srp20 expression is also regulated after behavioral training in females, but at a distinct time point from males. Should this be the case, this would imply sexual dimorphisms in the kinetics of transcriptional activation, which would in turn suggest the engagement of distinct signaling pathways. Differences between the hippocampal signaling pathways utilized by males and females is supported by our data; firstly, in contrast to males, in females, lack of CaMKKβ does not impact on Srp20 mRNA levels; and secondly, our studies with CaMKKβ and CaMKKα-deficient animals demonstrate that these two kinases are required for the formation of spatial and contextual memories, respectively, in males only (Mizuno et al. 2006, 2007).
We cannot exclude the possibility that our observations correspond to the transcriptional readout of sexually dimorphic stress responses or stress-coping strategies (Westenbroek et al. 2003; Liu et al. 2006). However, as far as the contextual conditioning task is concerned, the contribution of stress alone is unlikely to have accounted for the transcriptional changes observed as handling, novelty, and exposure to aversive stimuli were not sufficient to trigger changes in mRNA levels. In addition, Psf and Srp20 mRNA levels were assessed in a microarray study aimed at studying stress effects on hippocampal gene expression in males and females from four different mouse strains. Preliminary results confirm sex differences in Srp20 and Psf mRNA levels independently of strain. Importantly, these results also indicate that Srp20 mRNA expression is not influenced by mild stress conditions, while suggesting the possibility that Psf mRNA expression might be regulated by stress (L.C. Schalkwyk, pers. comm.)
Interestingly, we have found that in adult naïve hippocampi both Srp20 and Psf mRNA expression levels are higher in males than in females, and that this does not appear to result from differences in the location of expression across the major hippocampal subfields. The sex difference in Srp20 mRNA expression level occurs before puberty, which is not the case for Psf mRNA. This suggests that the sexual dimorphisms in Srp20 and Psf mRNA in naïve animals are guided by different mechanisms. Consistent with this, lack of CaMKKβ signaling in naïve animals leads to distinct effects in hippocampal expression of these two genes; Srp20 mRNA expression is decreased in males only, while Psf mRNA expression appears to be increased in both sexes. The influence of estrogen in the regulation of genes and signaling pathways involved in memory formation is well documented (Sawai et al. 2002; Lee et al. 2004; Scharfman and MacLusky 2005). However, under our housing conditions, we failed to observe a robust estrous cycling pattern, and therefore it was not possible to correlate levels of gene expression with phase of estrous cycle. Further experiments, for instance, using ovarectomized mice, would be required to establish the influence of female sexual hormones in the expression of Psf and Srp20.
Recently, it has become apparent that alternative splicing is an important process to regulate gene expression for neuronal plasticity and memory formation (Beffert et al. 2005; Ule and Darnell 2006; Lee et al. 2007). Here, we provide evidence that mRNA levels of two splicing factors, Srp20 and Psf, are regulated in response to training in two hippocampus-dependent learning tasks, thus suggesting that these expression changes may contribute to the regulation of alternative splicing during memory formation. Interestingly, CaMKIV has been implicated in the regulation of alternative splicing in neurons (Xie et al. 2005). Here, we show that lack of the upstream kinase CaMKKβ leads to altered levels of expression of two splicing factors in the hippocampus. It is therefore possible that SRP20 and/or PSF contribute to changes in alternative splicing in response to CaMKIV signaling. Although the roles of PSF and SRP20 as splicing factors are well established (Patton et al. 1993; Jumaa and Nielsen 1997; Shav-Tal and Zipori 2002; Galiana-Arnoux et al. 2003; Sanford et al. 2005), not much is known about the function of SRP20 and PSF in neurons. SRP20 regulates splicing of exon 10 of the gene encoding the microtubule-associated protein tau (Yu et al. 2004), which might indicate a role for compartmentalized signaling during memory processes. The neuronal expression of Psf is established (Chanas-Sacre et al. 1999), but the transcripts it splices in these cells remain to be determined. Furthermore, SRP20 and PSF are multifunctional proteins, and in addition to splicing, they might regulate other nuclear processes. Additional functions of SRP20 include polyadenylation (Lou et al. 1998) and nucleocytoplasmic export of mRNA (Huang and Steitz 2001). PSF participates in the nuclear retention of misedited RNA (Zhang and Carmichael 2001), repression of transcription (Mathur et al. 2001; Urban and Bodenburg 2002), and DNA repair (for review, see Shav-Tal and Zipori 2002; Bladen et al. 2005). Further work will be necessary to determine the importance of the Srp20 and Psf mRNA expression changes on alternative splicing and memory formation. It remains to be established whether the mRNA up-regulations observed are reflected at the protein level and whether they impact on alternative splicing. Functional studies will be required to determine whether manipulation of Srp20 and Psf expression levels produce an impairment in a hippocampus-dependent memory task. Understanding of these aspects will be essential to clarify whether regulation of alternative splicing by PSF or SRP20 participates in memory encoding and whether this contributes to sex differences in memory formation.
Most mechanistic studies of hippocampus-dependent learning and memory use male rodents to avoid any bias due to the estrus cycle when using females. Here, we show that there are significant sex differences in hippocampal gene expression in rodents that are likely to impact on alternative splicing during memory formation. Thus, the results of our study emphasize the need to consider both sexes when analyzing mechanisms underlying memory formation.
Materials and Methods
Animals
Camkk2 null mutants (Peters et al. 2003) were obtained by intercrosses of nonsibling heterozygous mice in the (129/Sv x C57BL/6) F3–4 genetic background. The genotype was determined by PCR from tail biopsies with primers (Invitrogen): KKβ1: 5′-CAG CACTCAGCTCCAATCAA-3′; KKβ2: 5′-GCCACCTATTGCCTTGT TTG-3′; KKβ3: 5′-TAAGCACAAGCACTCATTCC-3′. WT mice used in behavioral and expression studies were obtained by intercrossing WT offspring of the F4 generation. All mice used for behavioral experiments were 8–16 wk old. We attempted to monitor the phase of the estrus cycle by observing vaginal color and opening over a period of 3 wk, but no robust cycling pattern was observed. For this reason, it was not possible to determine the phase of the estrus cycle in the females used for behavioral and transcriptional analyses. For transcriptional studies, a group of male and female littermates was sacrificed at the age of weaning, postnatal day 21. The experiments were conducted in accordance with the Animals (Scientific Procedures) Act 1986, UK.
Quantitative real-time PCR
From each mouse, the entire hippocampi were isolated, frozen, and homogenized in Trizol (Invitrogen) and total RNA was isolated with RNeasy mini-columns (QIAGEN). RNA (2.5 μg) from each sample was reverse transcribed using superscript II reverse transcriptase (Invitrogen). To rule out the possibility of contamination with genomic DNA, a control PCR discriminating between the hypoxanthine-guanine-phosphoribosyltransferase (Hprt) genomic DNA and cDNA was performed as previously described (von Hertzen and Giese 2005). The real-time PCR primers for Srp20, Psf, and Hprt were designed with Primer Express 2.0 software (Applied Biosystems): Srp20fwd: 5′-TGAGGATCCCCG AGATGCT-3′; Srp20rev :5′-CTTACACGGCAGCCACACAGT-3′; Psffwd: 5′-GGAGTTCCACCAGCAACCAT-3′; Psfrev: 5′-CTGCCC AAAGCGCTCAGT-3′; HPRTfwd: 5′-ATACAGGCCAGACTTTGTT GGATT-3′; HPRTrev: 5′-TCACTAATGACACAAACGTGATTCAA-3′. To measure mRNA levels, we used quantitative real-time PCR (qPCR). This relative quantification method is based on the normalization of target mRNA level to a housekeeping gene, Hprt (which acts as an RNA expression control). This allows direct comparison of levels of gene expression across samples from different age and sex groups, avoiding biases related to the amount of starting material (hippocampal tissue) and technical sources of variation in mRNA extraction and reverse transcription (for review, see Bustin 2000). Hprt is an X-linked gene that is subject to X chromosome inactivation, and Hprt mRNA levels do not differ between male and female cells (Gartler and Riggs 1983; Grant and Chapman 1988). QPCR was performed with SYBR Green as fluorescent marker on the ABI7000 PCR system (Applied Biosystems). Amplification reactions contained 1X SYBR Green Master Mix (Applied Biosystems), optimized primer concentrations (900 nM forward primer/900nM reverse primer), and 1 μL of cDNA (∼2.5 μg) in a final volume of 25 μL. Amplification reactions were performed in 96 well plates and amplification conditions were 95°C for 10 min, followed by 40 cycles of 95°C for 15 sec and 62°C for 60 sec. Dissociation curves were generated after amplification in order to monitor the purity of the product formed. Srp20 and Psf mRNA levels were normalized to Hprt mRNA levels by calculating differences in cycle threshold (Ct) between the target gene and Hprt (ΔCt). The average ΔCt from the calibrator group (adult naïve WT males [Fig. 1A,C], or adult naïve WT females [Fig. 1B,D]) was subtracted from each individual ΔCt, and the values logarithmically transformed to obtain a relative percentage of expression.
Samples with a ΔCt value deviating more than two standard deviations from the average of the group were excluded. QPCR experiments described in Figure 1 and Figure 6, C and E (naïve vs. latent inhibition), consisted of one QPCR plate with triplicates of the same sample; Ct values were averaged and samples were excluded if Ct values were not consistent across triplicates (samples excluded: SRp20, one WT, and two CaMKKβ-deficient males [Fig. 1A]; one WT female [Fig. 1B]; one naïve male [Fig. 6C]; PSF, one naïve male [Fig. 6E]). For the data presented in the remaining figures, each sample was run in one well per plate and ΔCt values averaged across plates and samples were excluded if ΔCt values were not consistent across plates (samples excluded: Srp20, one naïve male [Figs. 5A,B, 6B,C]; one selective female [Fig. 5A]; one swim control [Fig. 5B], one naïve female, one naïve female FC [Fig. 6B]; PSF, two naïve females [Figs. 2C, 5C, 6D], one FC male [Fig. 6D,E]).
In situ hybridization
Coronal sections (15 μm) of fresh frozen brains were mounted onto polylysine-coated superfrost slides (VWR), fixed in 4% paraformaldeheyde, and stored in 95% ethanol at 4°C. A total of 10 ng of probe oligonucleotides: Psf:5′-CTTTCTTCTCGTTGGC GTCTCATTTGTTCTTCCATCTCACGTTGGCGA-3′; Srp20:5′-GTGGTCCATAATAGCCAAAAGCCCGTTCTAATTCAGTCTTGTT TCCAT-3′, were labeled with [α-35S]dATP (Amersham Biosciences) using terminal deoxynucleotidil transferase (TdT; Promega) and purified through a Sephadex column. Labeled probes were diluted in hybridization buffer (50% formamide, 4X SSC, 25 mM sodium phosphate, 1 mM sodium pyrophosphate; 5X Denhardt’s solution) to a final concentration of 100–300 cpm/μL. Hybridization was performed in a humidified chamber at 42°C overnight. Slides were washed in 1X SSC for 10 min at room temperature (RT), two times in 1X SSC at 55°C, and briefly rinsed in 0.1X SSC, 70% ethanol, 95% ethanol at RT, and allowed to air dry. Slides were exposed to a 35S-sensitive autoradiographic film (Kodak) together with 14C microscale standards (Amersham Biosciences), for 1 wk.
Hippocampal lesions
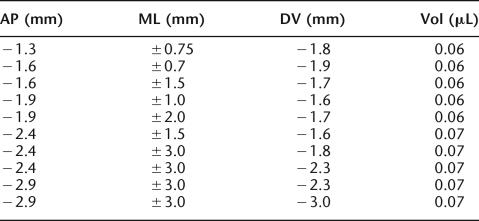
Female 129B6F1 mice were anesthetized with a mixture of isoflourane (Abbott) and O2, and then mounted in a Kopf stereotaxic frame. The scalp was shaved and surgically cleaned and then a midline incision was made that exposed the skull. The skull overlying the target area was removed and bilateral injections of ibotenic acid (Sigma) dissolved in PBS at a concentration of 10 mg/mL were made using a 5-μL Hamilton syringe with a 33-gauge needle at the following coordinates according to Paxinos and Franklin 2001:
After completion of the injections, the scalp was sutured and the mouse was returned to its home cage. Sham-lesioned mice were subjected to the same surgical procedure, but only PBS was injected into one of the dorsal hippocampal coordinates. All animals received a 0.1-mL subcutaneous injection of carprofen (Rimadyl; Pfizer Animal Health) after surgery was completed to provide pain relief. Behavioral testing began 7 d after surgery.
Histology
After completion of behavioral testing, all mice were given a lethal injection of sodium pentobarbitone (Euthatal; Animal Care Ltd.) and were perfused with physiological saline and 4% paraformaldehyde in PBS. The brains were removed and stored in 4% paraformaldehyde until they were coronally sectioned at 40 μm and stained with cresyl violet.
MWM experiments
Mice were trained in the hidden-platform version of MWM, which was shown to be hippocampus dependent in males (Angelo et al. 2003). Lesioned (n = 7) and sham-lesioned (n = 8) females were tested with 12 trials per day in blocks of four trials for 3 d. The maximal trial length was 90 sec, the intertrial interval was 60 sec, and the interblock interval was 60 min. A probe trial was given at the end of training on day 3 to assess for spatial memory. For the gene expression studies, we used a four trials/day training protocol for 6 d with 60-sec intertrial intervals and maximum trial duration of 90 sec. On the last day of training, animals were tested in a 90-sec probe trial. All trials were videotaped and images were analyzed using the HVS water program (HVS Image Ltd.). The mice were sacrificed 30 min after the probe trial to analyze hippocampal gene expression. Swim control groups were allowed to swim in the pool in the absence of the platform for the average duration of each training trial (depending on training day) for the same number of trials and days as the animals trained in the MWM. Naïve WT littermates were sacrificed on the same day.
Contextual fear conditioning
We used background contextual fear conditioning, which unlike foreground contextual fear conditioning, requires the dorsal hippocampus (Phillips and LeDoux 1994). This suggests a fuller engagement of the hippocampus in background versus foreground conditioning, which leads to an improved signal-to-noise ratio for detecting gene expression changes elicited by conditioning. The training protocol as well as control treatments for gene-expression studies were performed as described (von Hertzen and Giese 2005). Briefly, conditioning consisted of placing the mice in the training chamber (Campden Instruments); after 2 min, a tone (2.8 kHz, 80 dB) was played for 30 sec, the last 2 sec coinciding with a 0.75-mA foot shock, and 30 sec after the shock, the mice were removed from the chamber. A novelty control group-(Box) was trained under the same conditions, but did not receive a foot shock. A latent inhibition group was housed in the conditioning chamber overnight (12 h with food and water ad libitum), shocked in the morning, and removed from the chamber 30 sec after the shock. Under these conditions, the mice do not make a context-shock association (Lepicard et al. 2006). For hippocampal gene expression studies, WT mice were sacrificed 30 min after removal from the conditioning chamber and naïve WT littermates were sacrificed on the same day. The behavioral controls were tested for contextual fear memory 24 h after conditioning by measuring freezing upon re-exposure to the context.
Data analysis
Statisitical analyses were performed using SIGMASTAT (Systat Software Inc.). One- or two-way ANOVAs were used followed by Student-Newman-Keuls post-hoc analysis, when significant interactions were found. Two- and three-way repeated measures ANOVA, for analysis of MWM acquisition data (Fig. 4A), were performed using SPSS (SPSS, Inc.).
Acknowledgments
We thank Jeff Vernon and Dan Berwick for comments on an earlier version of this manuscript, Marco Peters for CaMKKβ-deficient mice for breeding, and Leo C. Schalkwyk, Institute of Psychiatry, King’s College London for correspondence of unpublished data. This study was supported by the Wellcome Trust, a Human Frontier Science Young Investigator Award, and a Medical Research Council Career Establishment grant for K.P.G. A.A.-M. is supported by a grant from Fundação para a Ciência e Tecnologia (SFRH BD 5804/2001) through the Graduate Programme in Areas of Basic and Applied Biology. The ABI7000 PCR system used in this study was funded by the Elton John AIDS Foundation.
Footnotes
Article is online at http://www.learnmem.org/cgi/doi/10.1101/lm.640307
References
- Aguilar-Valles A., Sanchez E., deGortari P., Balderas I., Ramirez-Amaya V., Bermudez-Rattoni F., Joseph-Bravo P. Analysis of the stress response in rats trained in the water-maze: Differential expression of corticotropin-releasing hormone, CRH-R1, glucocorticoid receptors and brain-derived neurotrophic factor in limbic regions. Neuroendocrinology. 2005;82:306–319. doi: 10.1159/000093129. [DOI] [PubMed] [Google Scholar]
- Angelo M., Plattner F., Irvine E.E., Giese K.P. Improved reversal learning and altered fear conditioning in transgenic mice with regionally restricted p25 expression. Eur. J. Neurosci. 2003;18:423–431. doi: 10.1046/j.1460-9568.2003.02746.x. [DOI] [PubMed] [Google Scholar]
- Balschun D., Wolfer D.P., Gass P., Mantamadiotis T., Welzl H., Schutz G., Frey J.U., Lipp H.P. Does cAMP response element-binding protein have a pivotal role in hippocampal synaptic plasticity and hippocampus-dependent memory? J. Neurosci. 2003;23:6304–6314. doi: 10.1523/JNEUROSCI.23-15-06304.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beffert U., Weeber E.J., Durudas A., Qiu S., Masiulis I., Sweatt J.D., Li W.P., Adelmann G., Frotscher M., Hammer R.E., et al. Modulation of synaptic plasticity and memory by Reelin involves differential splicing of the lipoprotein receptor Apoer2. Neuron. 2005;47:567–579. doi: 10.1016/j.neuron.2005.07.007. [DOI] [PubMed] [Google Scholar]
- Bladen C.L., Udayakumar D., Takeda Y., Dynan W.S. Identification of the polypyrimidine tract binding protein-associated splicing factor.p54(nrb) complex as a candidate DNA double-strand break rejoining factor. J. Biol. Chem. 2005;280:5205–5210. doi: 10.1074/jbc.M412758200. [DOI] [PubMed] [Google Scholar]
- Blaeser F., Sanders M.J., Truong N., Ko S., Wu L.J., Wozniak D.F., Fanselow M.S., Zhuo M., Chatila T.A. Long-term memory deficits in Pavlovian fear conditioning in Ca2+/calmodulin kinase kinase α-deficient mice. Mol. Cell. Biol. 2006;26:9105–9115. doi: 10.1128/MCB.01452-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bottai D., Guzowski J.F., Schwarz M.K., Kang S.H., Xiao B., Lanahan A., Worley P.F., Seeburg P.H. Synaptic activity-induced conversion of intronic to exonic sequence in Homer 1 immediate early gene expression. J. Neurosci. 2002;22:167–175. doi: 10.1523/JNEUROSCI.22-01-00167.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brightwell J.J., Gallagher M., Colombo P.J. Hippocampal CREB1 but not CREB2 is decreased in aged rats with spatial memory impairments. Neurobiol. Learn. Mem. 2004;81:19–26. doi: 10.1016/j.nlm.2003.08.001. [DOI] [PubMed] [Google Scholar]
- Bustin S.A. Absolute quantification of mRNA using real-time reverse transcription polymerase chain reaction assays. J. Mol. Endocrinol. 2000;25:169–193. doi: 10.1677/jme.0.0250169. [DOI] [PubMed] [Google Scholar]
- Cavallaro S., D’Agata V., Manickam P., Dufour F., Alkon D.L. Memory-specific temporal profiles of gene expression in the hippocampus. Proc. Natl. Acad. Sci. 2002;99:16279–16284. doi: 10.1073/pnas.242597199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chanas-Sacre G., Mazy-Servais C., Wattiez R., Pirard S., Rogister B., Patton J.G., Belachew S., Malgrange B., Moonen G., Leprince P. Identification of PSF, the polypyrimidine tract-binding protein-associated splicing factor, as a developmentally regulated neuronal protein. J. Neurosci. Res. 1999;57:62–73. doi: 10.1002/(SICI)1097-4547(19990701)57:1<62::AID-JNR7>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- Chow F.A., Anderson K.A., Noeldner P.K., Means A.R. The autonomous activity of calcium/calmodulin-dependent protein kinase IV is required for its role in transcription. J. Biol. Chem. 2005;280:20530–20538. doi: 10.1074/jbc.M500067200. [DOI] [PubMed] [Google Scholar]
- Corcoran E.E., Means A.R. Defining Ca2+/calmodulin-dependent protein kinase cascades in transcriptional regulation. J. Biol. Chem. 2001;276:2975–2978. doi: 10.1074/jbc.R000027200. [DOI] [PubMed] [Google Scholar]
- Frye C.A. Estrus-associated decrements in a water maze task are limited to acquisition. Physiol. Behav. 1995;57:5–14. doi: 10.1016/0031-9384(94)00197-d. [DOI] [PubMed] [Google Scholar]
- Galiana-Arnoux D., Lejeune F., Gesnel M.C., Stevenin J., Breathnach R., Gatto-Konczak F. The CD44 alternative v9 exon contains a splicing enhancer responsive to the SR proteins 9G8, ASF/SF2, and SRp20. J. Biol. Chem. 2003;278:32943–32953. doi: 10.1074/jbc.M301090200. [DOI] [PubMed] [Google Scholar]
- Gartler S.M., Riggs A.D. Mammalian X-chromosome inactivation. Annu. Rev. Genet. 1983;17:155–190. doi: 10.1146/annurev.ge.17.120183.001103. [DOI] [PubMed] [Google Scholar]
- Grant S.G., Chapman V.M. Mechanisms of X-chromosome regulation. Annu. Rev. Genet. 1988;22:199–233. doi: 10.1146/annurev.ge.22.120188.001215. [DOI] [PubMed] [Google Scholar]
- Guzowski J.F., Lyford G.L., Stevenson G.D., Houston F.P., McGaugh J.L., Worley P.F., Barnes C.A. Inhibition of activity-dependent arc protein expression in the rat hippocampus impairs the maintenance of long-term potentiation and the consolidation of long-term memory. J. Neurosci. 2000;20:3993–4001. doi: 10.1523/JNEUROSCI.20-11-03993.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guzowski J.F., Setlow B., Wagner E.K., McGaugh J.L. Experience-dependent gene expression in the rat hippocampus after spatial learning: A comparison of the immediate-early genes Arc, c-fos, and zif268. J. Neurosci. 2001;21:5089–5098. doi: 10.1523/JNEUROSCI.21-14-05089.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall J., Thomas K.L., Everitt B.J. Rapid and selective induction of BDNF expression in the hippocampus during contextual learning. Nat. Neurosci. 2000;3:533–535. doi: 10.1038/75698. [DOI] [PubMed] [Google Scholar]
- Hebda-Bauer E.K., Watson S.J., Akil H. Cognitive performance is highly sensitive to prior experience in mice with a learning and memory deficit: Failure leads to more failure. Learn. Mem. 2005;12:461–471. doi: 10.1101/lm.94105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y., Steitz J.A. Splicing factors SRp20 and 9G8 promote the nucleocytoplasmic export of mRNA. Mol. Cell. 2001;7:899–905. doi: 10.1016/s1097-2765(01)00233-7. [DOI] [PubMed] [Google Scholar]
- Jonasson Z. Meta-analysis of sex differences in rodent models of learning and memory: A review of behavioral and biological data. Neurosci. Biobehav. Rev. 2005;28:811–825. doi: 10.1016/j.neubiorev.2004.10.006. [DOI] [PubMed] [Google Scholar]
- Jumaa H., Nielsen P.J. The splicing factor SRp20 modifies splicing of its own mRNA and ASF/SF2 antagonizes this regulation. EMBO J. 1997;16:5077–5085. doi: 10.1093/emboj/16.16.5077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koshibu K., Levitt P. Sex differences in expression of transforming growth factor-α and epidermal growth factor receptor mRNA in Waved-1 and C57Bl6 mice. Neuroscience. 2005;134:877–887. doi: 10.1016/j.neuroscience.2005.03.016. [DOI] [PubMed] [Google Scholar]
- Kudo K., Qiao C.X., Kanba S., Arita J. A selective increase in phosphorylation of cyclic AMP response element-binding protein in hippocampal CA1 region of male, but not female, rats following contextual fear and passive avoidance conditioning. Brain Res. 2004;1024:233–243. doi: 10.1016/j.brainres.2004.08.007. [DOI] [PubMed] [Google Scholar]
- Lee C.J., Irizarry K. Alternative splicing in the nervous system: An emerging source of diversity and regulation. Biol. Psychiatry. 2003;54:771–776. doi: 10.1016/s0006-3223(03)00375-5. [DOI] [PubMed] [Google Scholar]
- Lee S.J., Campomanes C.R., Sikat P.T., Greenfield A.T., Allen P.B., McEwen B.S. Estrogen induces phosphorylation of cyclic AMP response element binding (pCREB) in primary hippocampal cells in a time-dependent manner. Neuroscience. 2004;124:549–560. doi: 10.1016/j.neuroscience.2003.11.035. [DOI] [PubMed] [Google Scholar]
- Lee J.A., Xing Y., Nguyen D., Xie J., Lee C.J., Black D.L. Depolarization and CaM kinase IV modulate NMDA receptor splicing through two essential RNA elements. PLoS Biol. 2007;5:e40. doi: 10.1371/journal.pbio.0050040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leil T.A., Ossadtchi A., Cortes J.S., Leahy R.M., Smith D.J. Finding new candidate genes for learning and memory. J. Neurosci. Res. 2002;68:127–137. doi: 10.1002/jnr.10184. [DOI] [PubMed] [Google Scholar]
- Leil T.A., Ossadtchi A., Nichols T.E., Leahy R.M., Smith D.J. Genes regulated by learning in the hippocampus. J. Neurosci. Res. 2003;71:763–768. doi: 10.1002/jnr.10541. [DOI] [PubMed] [Google Scholar]
- Lepicard E.M., Mizuno K., Antunes-Martins A., von Hertzen L.S., Giese K.P. An endogenous inhibitor of calcium/calmodulin-dependent kinase II is up-regulated during consolidation of fear memory. Eur. J. Neurosci. 2006;23:3063–3070. doi: 10.1111/j.1460-9568.2006.04830.x. [DOI] [PubMed] [Google Scholar]
- Liu H.H., Payne H.R., Wang B., Brady S.T. Gender differences in response of hippocampus to chronic glucocorticoid stress: Role of glutamate receptors. J. Neurosci. Res. 2006;83:775–786. doi: 10.1002/jnr.20782. [DOI] [PubMed] [Google Scholar]
- Lou H., Neugebauer K.M., Gagel R.F., Berget S.M. Regulation of alternative polyadenylation by U1 snRNPs and SRp20. Mol. Cell. Biol. 1998;18:4977–4985. doi: 10.1128/mcb.18.9.4977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maguire E.A., Burgess N., O'Keefe J. Human spatial navigation: Cognitive maps, sexual dimorphism, and neural substrates. Curr. Opin. Neurobiol. 1999;9:171–177. doi: 10.1016/s0959-4388(99)80023-3. [DOI] [PubMed] [Google Scholar]
- Mathur M., Tucker P.W., Samuels H.H. PSF is a novel corepressor that mediates its effect through Sin3A and the DNA binding domain of nuclear hormone receptors. Mol. Cell. Biol. 2001;21:2298–2311. doi: 10.1128/MCB.21.7.2298-2311.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuyama S., Namgung U., Routtenberg A. Long-term potentiation persistence greater in C57BL/6 than DBA/2 mice: Predicted on basis of protein kinase C levels and learning performance. Brain Res. 1997;763:127–130. doi: 10.1016/s0006-8993(97)00444-7. [DOI] [PubMed] [Google Scholar]
- Mizuno K., Giese K.P. Hippocampus-dependent memory formation: Do memory type-specific mechanisms exist? J. Pharmacol. Sci. 2005;98:191–197. doi: 10.1254/jphs.crj05005x. [DOI] [PubMed] [Google Scholar]
- Mizuno K., Ris L., Sanchez-Capelo A., Godaux E., Giese K.P. Ca2+/calmodulin kinase kinase α is dispensable for brain development but is required for distinct memories in male, though not in female, mice. Mol. Cell. Biol. 2006;26:9094–9104. doi: 10.1128/MCB.01221-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizuno K., Antunes-Martins A., Ris L., Peters M., Godaux E., Giese K.P. Calcium/calmodulin kinase kinase β has a male-specific role in memory formation. Neuroscience. 2007;145:393–402. doi: 10.1016/j.neuroscience.2006.11.056. [DOI] [PubMed] [Google Scholar]
- Morris R.G. Elements of a neurobiological theory of hippocampal function: The role of synaptic plasticity, synaptic tagging and schemas. Eur. J. Neurosci. 2006;23:2829–2846. doi: 10.1111/j.1460-9568.2006.04888.x. [DOI] [PubMed] [Google Scholar]
- Nijholt I., Farchi N., Kye M., Sklan E.H., Shoham S., Verbeure B., Owen D., Hochner B., Spiess J., Soreq H., et al. Stress-induced alternative splicing of acetylcholinesterase results in enhanced fear memory and long-term potentiation. Mol. Psychiatry. 2004;9:174–183. doi: 10.1038/sj.mp.4001446. [DOI] [PubMed] [Google Scholar]
- O’Connor V., Genin A., Davis S., Karishma K.K., Doyere V., De Zeeuw C.I., Sanger G., Hunt S.P., Richter-Levin G., Mallet J., et al. Differential amplification of intron-containing transcripts reveals long term potentiation-associated up-regulation of specific Pde10A phosphodiesterase splice variants. J. Biol. Chem. 2004;279:15841–15849. doi: 10.1074/jbc.M312500200. [DOI] [PubMed] [Google Scholar]
- Oh M.M., Kuo A.G., Wu W.W., Sametsky E.A., Disterhoft J.F. Watermaze learning enhances excitability of CA1 pyramidal neurons. J. Neurophysiol. 2003;90:2171–2179. doi: 10.1152/jn.01177.2002. [DOI] [PubMed] [Google Scholar]
- Omoto Y., Lathe R., Warner M., Gustafsson J.A. Early onset of puberty and early ovarian failure in CYP7B1 knockout mice. Proc. Natl. Acad. Sci. 2005;102:2814–2819. doi: 10.1073/pnas.0500198102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patton J.G., Porro E.B., Galceran J., Tempst P., Nadal-Ginard B. Cloning and characterization of PSF, a novel pre-mRNA splicing factor. Genes & Dev. 1993;7:393–406. doi: 10.1101/gad.7.3.393. [DOI] [PubMed] [Google Scholar]
- Paxinos G., Franklin K.B.J. The mouse brain in stereotaxic coordinates. Academic Press; San Diego: 2001. [Google Scholar]
- Perrot-Sinal T.S., Kostenuik M.A., Ossenkopp K.P., Kavaliers M. Sex differences in performance in the Morris water maze and the effects of initial nonstationary hidden platform training. Behav. Neurosci. 1996;110:1309–1320. doi: 10.1037//0735-7044.110.6.1309. [DOI] [PubMed] [Google Scholar]
- Peters M., Mizuno K., Ris L., Angelo M., Godaux E., Giese K.P. Loss of Ca2+/calmodulin kinase kinase β affects the formation of some, but not all, types of hippocampus-dependent long-term memory. J. Neurosci. 2003;23:9752–9760. doi: 10.1523/JNEUROSCI.23-30-09752.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips R.G., LeDoux J.E. Lesions of the dorsal hippocampal formation interfere with background but not foreground contextual fear conditioning. Learn. Mem. 1994;1:34–44. [PubMed] [Google Scholar]
- Plath N., Ohana O., Dammermann B., Errington M.L., Schmitz D., Gross C., Mao X., Engelsberg A., Mahlke C., Welzl H., et al. Arc/Arg3.1 is essential for the consolidation of synaptic plasticity and memories. Neuron. 2006;52:437–444. doi: 10.1016/j.neuron.2006.08.024. [DOI] [PubMed] [Google Scholar]
- Rattiner L.M., Davis M., Ressler K.J. Differential regulation of brain-derived neurotrophic factor transcripts during the consolidation of fear learning. Learn. Mem. 2004;11:727–731. doi: 10.1101/lm.83304. [DOI] [PubMed] [Google Scholar]
- Roof R.L., Stein D.G. Gender differences in Morris water maze performance depend on task parameters. Physiol. Behav. 1999;68:81–86. doi: 10.1016/s0031-9384(99)00162-6. [DOI] [PubMed] [Google Scholar]
- Sandstrom N.J., Kaufman J., Huettel S.A. Males and females use different distal cues in a virtual environment navigation task. Brain Res. Cogn. Brain Res. 1998;6:351–360. doi: 10.1016/s0926-6410(98)00002-0. [DOI] [PubMed] [Google Scholar]
- Sanford J.R., Ellis J., Caceres J.F. Multiple roles of arginine/serine-rich splicing factors in RNA processing. Biochem. Soc. Trans. 2005;33:443–446. doi: 10.1042/BST0330443. [DOI] [PubMed] [Google Scholar]
- Sawai T., Bernier F., Fukushima T., Hashimoto T., Ogura H., Nishizawa Y. Estrogen induces a rapid increase of calcium-calmodulin-dependent protein kinase II activity in the hippocampus. Brain Res. 2002;950:308–311. doi: 10.1016/s0006-8993(02)03186-4. [DOI] [PubMed] [Google Scholar]
- Scharfman H.E., MacLusky N.J. Similarities between actions of estrogen and BDNF in the hippocampus: Coincidence or clue? Trends Neurosci. 2005;28:79–85. doi: 10.1016/j.tins.2004.12.005. [DOI] [PubMed] [Google Scholar]
- Shav-Tal Y., Zipori D. PSF and p54(nrb)/NonO-multi- functional nuclear proteins. FEBS Lett. 2002;531:109–114. doi: 10.1016/s0014-5793(02)03447-6. [DOI] [PubMed] [Google Scholar]
- Shors T.J., Beylin A.V., Wood G.E., Gould E. The modulation of Pavlovian memory. Behav. Brain Res. 2000;110:39–52. doi: 10.1016/s0166-4328(99)00183-7. [DOI] [PubMed] [Google Scholar]
- Silva A.J., Giese K.P. Plastic genes are in! Curr. Opin. Neurobiol. 1994;4:413–420. doi: 10.1016/0959-4388(94)90104-x. [DOI] [PubMed] [Google Scholar]
- Takemoto-Kimura S., Terai H., Takamoto M., Ohmae S., Kikumura S., Segi E., Arakawa Y., Furuyashiki T., Narumiya S., Bito H. Molecular cloning and characterization of CLICK-III/CaMKIγ, a novel membrane-anchored neuronal Ca2+/calmodulin-dependent protein kinase (CaMK) J. Biol. Chem. 2003;278:18597–18605. doi: 10.1074/jbc.M300578200. [DOI] [PubMed] [Google Scholar]
- Ule J., Darnell R.B. RNA binding proteins and the regulation of neuronal synaptic plasticity. Curr. Opin. Neurobiol. 2006;16:102–110. doi: 10.1016/j.conb.2006.01.003. [DOI] [PubMed] [Google Scholar]
- Urban R.J., Bodenburg Y. PTB-associated splicing factor regulates growth factor-stimulated gene expression in mammalian cells. Am. J. Physiol. Endocrinol. Metab. 2002;283:E794–E798. doi: 10.1152/ajpendo.00174.2002. [DOI] [PubMed] [Google Scholar]
- Vaynman S., Ying Z., Gomez-Pinilla F. Hippocampal BDNF mediates the efficacy of exercise on synaptic plasticity and cognition. Eur. J. Neurosci. 2004;20:2580–2590. doi: 10.1111/j.1460-9568.2004.03720.x. [DOI] [PubMed] [Google Scholar]
- von Hertzen L.S., Giese K.P. Memory reconsolidation engages only a subset of immediate-early genes induced during consolidation. J. Neurosci. 2005;25:1935–1942. doi: 10.1523/JNEUROSCI.4707-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westenbroek C., Den Boer J.A., Ter Horst G.J. Gender-specific effects of social housing on chronic stress-induced limbic Fos expression. Neuroscience. 2003;121:189–199. doi: 10.1016/s0306-4522(03)00367-1. [DOI] [PubMed] [Google Scholar]
- Xie J., Jan C., Stoilov P., Park J., Black D.L. A consensus CaMK IV-responsive RNA sequence mediates regulation of alternative exons in neurons. RNA. 2005;11:1825–1834. doi: 10.1261/rna.2171205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Q., Guo J., Zhou J. A minimal length between τ exon 10 and 11 is required for correct splicing of exon 10. J. Neurochem. 2004;90:164–172. doi: 10.1111/j.1471-4159.2004.02477.x. [DOI] [PubMed] [Google Scholar]
- Zhang Z., Carmichael G.G. The fate of dsRNA in the nucleus: A p54(nrb)-containing complex mediates the nuclear retention of promiscuously A-to-I edited RNAs. Cell. 2001;106:465–475. doi: 10.1016/s0092-8674(01)00466-4. [DOI] [PubMed] [Google Scholar]