Abstract
The presence of diadenosine oligophosphates (ApnA) in eukaryotic pathogens has been difficult technically to assess and thus is often overlooked. ApnA are a family of intercellular and intracellular signaling molecules and their biological activities differ relative to the number of phosphate moieties. The application of mass spectrometry to differentiate nucleotide phosphates has been limited by the high salt content in tissue extracts, enzymatic reactions or high performance liquid chromatography (HPLC) buffers, as well as the potential for sample loss when processing and desalting small biological samples. To address this problem a simple reverse phase HPLC (RP-HPLC) method using volatile organic buffers at low pH was developed to create elution profiles of adenosine and diadenosine phosphates. To test this method on a eukaryotic pathogen, small intravascular human filarial parasites (Brugia malayi) were extracted in phosphate buffered saline and a nucleotide phosphate profile was visualized by RP-HPLC. A major peak eluting at 10.4 min was analyzed directly by mass spectrometry and this confirmed the presence of significant quantities of diadenosine triphosphate, Ap3A. Application of this simplified RP-HPLC method will facilitate research on the normal and pathophysiological effects of ApnA particularly in situations when analysis of small biological samples is required.
Keywords: Diadenosine triphosphate, MALDI mass spectrometry, Filaria, Brugia malayi
1. Introduction
The presence and biological role of diadenosine oligophosphates (ApnA) in many eukaryotic pathogenic microbes has been difficult technically to assess and thus is often overlooked. ApnA are a family of potent intercellular and intracellular signaling molecules that affect a wide variety of cellular processes including vascular endothelial cell dependent function via stimulation of cell surface P2Y purine receptors [1–5]. Antagonists of ApnAreceptors are being studied as novel pharmacological tools that modulate vascular endothelial cell function and pathology [6]. The biological activity of a particular ApnA is dependent on both the number and configuration of the phosphates moieties. The general formula for diadenosine polyphosphates is “ApnA,” where “n” represents the number of phosphates bridging two adenosine moieties.
In mammalian cells, the ratio of Ap3A/Ap4A has been suggested as a means by which cell cycles are regulated. In humans, the tumor suppressor, FHIT, (fragile histidine triad) functions as an Ap3A hydrolase [7]. Increased Ap3A levels prevent apopotosis and thus sustain tumorigenesis [8]. In invertebrates, FHIT exists as a fusion protein with Nit, a member of the nitrilase superfamily [9]. In humans, ApnA also have been proposed as both an alternate energy storage form and intercellular messenger because of their relatively long half-life in blood, relative to monoadenosine phosphates [10]. Ap3A has been shown to mediate vasodilation via endothelial cell purine P2Y receptors, promotes thrombin-mediated platelet aggregation and can act as a neurotransmitter. Ap4A also promotes vasodilation, but has been shown to inhibit platelet aggregation, activate nitric oxide and induce apoptosis. Ap5A can be mitogenic, induce vasoconstriction and inhibit adenosine kinase [11–13]. The relative potencies of ApnA measured in a stably transfected human astrocytoma cell line bearing the purine receptor P2Y subset were measured as ADP > Ap3A > Ap6A > Ap2A > Ap5A > Ap4A [12]. Diadenosine polyphosphates also occur in human tears and facilitate tear secretion [14]. Ap3A and Ap4A activate different P2Y receptors in corneal epithelial cells and can either accelerate (Ap3A) or delay (Ap4A) the rate of cellular healing [15]. Recently it was shown that the asparaginyl-tRNA synthetase (AsnRS) of an important human intravascular parasite, Brugia malayi, is very highly expressed and has the capacity to synthesize Ap3A in vitro [16]. Aminoacyl-tRNA synthetases (AARS) are a family of multifunctional enzymes whose primary function is to charge cognate tRNAs with the correct amino acid; but the first established alternative functions for an AARS was the biosynthesis of Ap3A and Ap4A [17].
For decades, physiological evidence has implicated unidentified parasite-derived substances that alter endothelial cell behavior during filarial disease. Filaria is an important intravascular nematode parasite of humans and domestic animals that cause a variety of severe morbidities [18]. These organisms are very small, non culturable in vitro and thus their acquisition in large amounts from infected hosts is problematic. Weller and Liu first reported the existence of a variety undefined low molecular weight bioactive compounds from filarial [19]. In the canine dog heartworm model (Dirofilaria immitis) endothelium-dependent pathways were shown to be responsible for altered pulmonary artery relaxation [20]. The animal filaria, Brugia pahangi, was reported to impair by an unknown mechanism bovine mesenteric lymphatic contractility and depress endothelium-dependent relaxation in the aorta of infected rats [21]. The serum of dogs infected with D. immitis exhibited similar effects on endothelium-dependent relaxation of rat aorta in vitro [22]. Many of these reported phenomena are consistent with the effects of a parasite derived ApnA.
The application of mass spectrometry to quantitatively identify ApnA in tissue extracts and thereby predict biological functions has been limited by problems attributable to the high salt buffers in enzymatic reactions or high performance liquid chromatography (HPLC) [23–25]. To address this problem, a reverse phase HPLC method was developed using volatile organic buffers to create profiles of adenosine and diadenosine phosphates at low pH. These methods were used to examine the human filarial parasite B. malayi, for the presence of ApnA.
2. Methods
ApnA standards were purchased from Sigma–Aldrich (St. Louis, Missouri, USA). Molecular mass values for each compound are: Ap3A (756.41), Ap4A (836.39), Ap5A (946.03), Ap6A (996.35), ATP (551.14), AMP (347.22), and ADP (427.20). Stock solutions (5–100 mM) of each standard were made in water and phosphate buffered saline pH 7.0 (Signma–Aldrich).
Separation of adenosine and diadenosine standards were compared using a Dionex Summit HPLC System with a High Pressure Binary Gradient Pump (Model P680A), Solvent rack with 4 analytical degasser channels (Model SOR-100A), UV/VIS Photodiode array detector (Model UVD 340U) at 225 nm, and the data system (Chromeleon™ software) with a 5µ reverse phase separation column (Acclaim 120, C18, 4.6 mm × 150 mm). The column was eluted with a linear gradient as presented in Table 1. The column was maintained at 25 °C and all experiments were conducted at room temperature.
Table 1.
Conditions for separation of adenosine- and diadenosine phosphates by reverse phase high performance liquid chromatography (RP-HPLC) at low pH using volatile organic buffers
| Time (min) | Flow (ml/min) | %Buffer Aa | %Buffer Bb | Curve |
|---|---|---|---|---|
| Initial | 0.80 | 100 | 0 | - |
| 3.00 | 0.80 | 100 | 0 | 5 |
| 43.00 | 0.80 | 80 | 20 | 5 |
| 53.00 | 0.80 | 30 | 70 | 5 |
| 55.00 | 0.80 | 30 | 70 | 5 |
| 60.00 | 0.80 | 100 | 0 | 5 |
| 90.00 | 0.80 | 100 | 0 | 9 |
Buffer A: 0.1% trifluoroacetic acid (TFA) in deionized water.
Buffer B: 90% acetonitrile and 0.098% TFA in deionized water.
Live adult B. malayi parasites were obtained from the NIH Filariasis Repository (Dr. John McCall, University of Georgia, Athens, GA). Fifty organisms (combined wet weight of 300 mg) were homogenized in 1ml of sterile phosphate buffered saline (PBS, Sigma–Aldrich) pH 7.0 and filtered through a 0.2 µ filter, and 20µl of this crude extract (~1 parasite = 6 mg) was analyzed by RP-HPLC.
The ApnA standard and sample fractions collected from the reverse phase-HPLC were examined by MALDI-MS and PSD-MALDI-MS. Data for each experiment was collected on a time-of-flight (TOF) Voyager-DE Pro-MALDI-TOF (Applied Biosystems, Framingham, Mass.) mass spectrometer. One microliter of saturated α-cyano-4-hydroxy-cinnamic acid (Sigma catalogue# C145505) matrix solution in 50% (v/v) acetonitrile/0.1% (v/v) trifluoroacetic acid was mixed with 1.0 µl standard. One microliter of the 1∶1 matrix∶sample solution was spotted onto a 100 well Voyager sample plate and allowed to air dry. This was repeated for the sample fraction. The molecular mass of the 10.4 min peak identified in adult B. malayi was compared to the Ap3A standard at 756.0 Da by matrix-assisted laser desorption/ionization mass spectrometry (MALDI-MS) and post source decay (PSD)-MALDI-MS. Data was collected in refletron mode combining 200 shots per spectra. Precursor ions from the sample and standard were selected for PSD-MALDI-MS. Laser intensity, mirror ratio, and guide wire percentages, were adjusted for maximum resolution. The fragmentation pattern of the sample was compared to the Ap3A standard.
3. Results
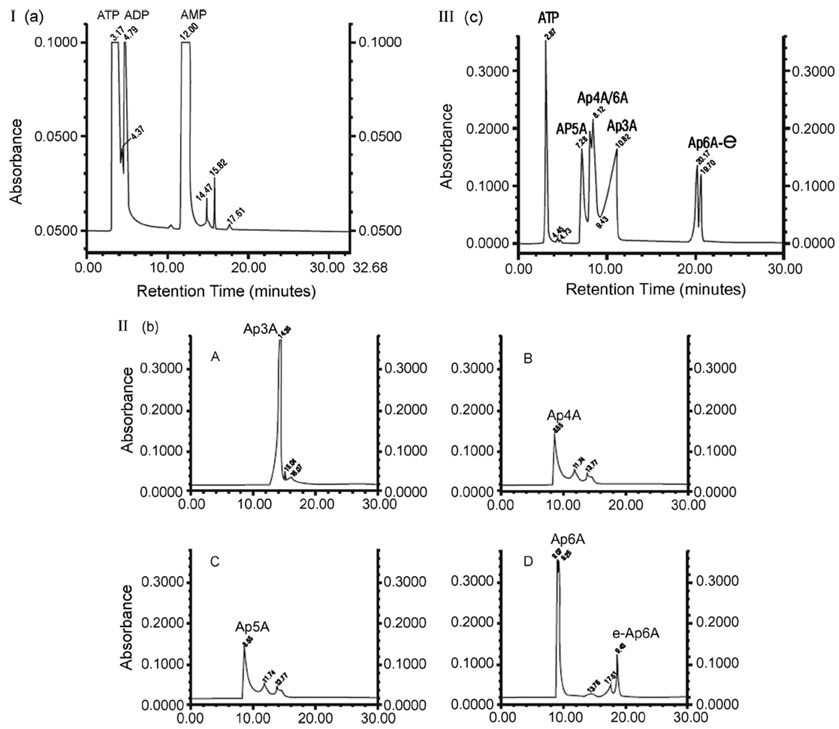
Reproducible chromatograms (n = 15) were obtained for ATP, ADP, AMP, Ap3A, Ap4A, Ap5A and Ap6A (Fig. 1). Commercially available sources of Ap6A often demonstrated two major peaks, consistent with Ap6A and Ap6A-epsilon. Mixtures of standards yielded reproducible separation of all diadenosine compounds from ATP and ADP, but did not distinguish AMP from Ap3A. Elution times for Ap6A were the longest (20.17±.5 min), followed by Ap3A (10.82±.49), Ap4A (8.12±.43 min) and lastly Ap5A (7.28±.27 min). ATP and ADP demonstrated short retention times of 3.12±0.49 and 4.71±0.51 min, respectively. MALDI mass spectrometry identified the correct mass of 756.41±0.5m/z of each Ap3A standard dissolved in PBS pH 7.0. Resolution at mass 756.40 calculated with 50% centroid is 5709 for this acquisition. The MALDI-MS was calibrated using an external calibration by creating a calibration file using the Ap3A standard. This file was then applied in the Instrument Control Panel when acquiring the data. Different lots of ApnA standards appeared to contain variable salt contamination, thus the observed mass of individual compounds could include one or more sodium atoms.
Fig. 1.
Panel I: Retention times for ATP, ADP and AMP reference standards. Panel II: Retention times for individual diadenosine phosphate reference standards: (A) Ap3A, (B) Ap4A, (C) Ap5A, (D) Ap6A. Panel III: Retention times for mixed reference standards, ATP and diadenosine phosphates. Chromatogram is optimized to visualize all compounds in a single figure.
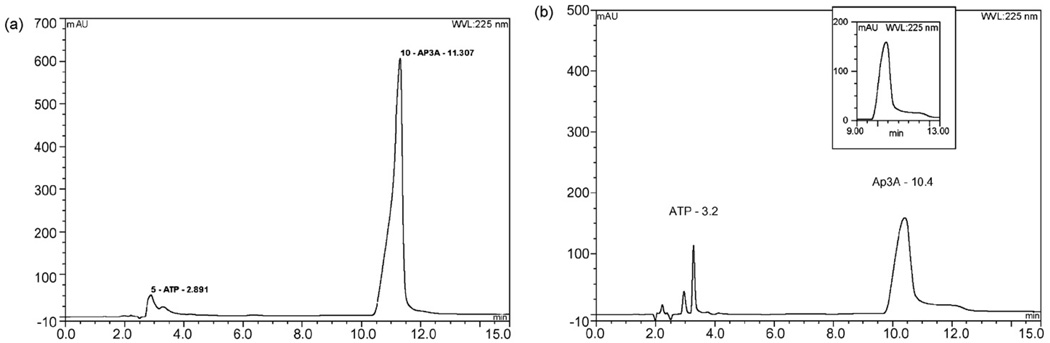
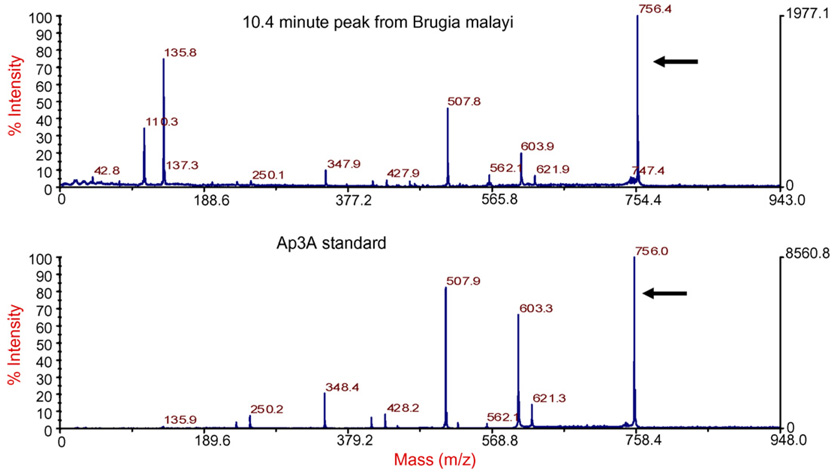
Chromatograms of B. malayi parasite extracts (n = 12) demonstrated a distinctive peak that eluted at 10.4±0.40 min that was similar to the Ap3A standard (Fig. 2, Panels A and B). MALDI mass measurement of the fraction and its fragmentation pattern were compared to published values for ApnA [24] and identified the compound as Ap3A (Fig. 3 and Table 2). Area-under-the-curve values for reference standard Ap3A (Fig. 2 panel A) and parasite extract (Fig. 2 panel B) indicated that the concentration of Ap3A in parasite extracts was 50 µM, or 1 µM per adult worm (6 mg wet weight per parasite). The amount of Ap3A in the parasites appeared as high as or greater than the level of compounds (e.g. ATP and ADP) with short elution times.
Fig. 2.
Panel A: Chromatogram of reference standard Ap3A shows retention time of 10.82±0.49 min. Injection volume was 25 µl of a 13 mM Ap3A stock solution. Panel B: Chromatogram from phosphate buffered saline extract of adult parasites. Fifty adults parasites (300 mg combined wet weight) were homogenized in 1 ml of phosphate buffered saline pH 7.0 (PBS) and undissolved particulates were removed with a 0.2 µ filter. 20 µl of the filtered PBS extract, the equivalent of one parasite (6 mg wet weight) was injected. Inset figure shows a characteristic peak at 10.4±0.4 min.
Fig. 3.
MALDI mass spectrometry analysis of 10.4 min peak from Brugia malayi demonstrated a fragmentation pattern (arrows) consistent with diadenosine triphosphate, Ap3A.
Table 2.
Molecular mass of fragments from the PSD-MALDI-MS spectra of the 10.8 min unknown peak (left column) obtained from Brugia malayi compared to published values for Ap3A (center column) [24]
| 10.4 min unknown peak | Published values for diadenosine triphosphate (Ap3A) | Fragment ion |
|---|---|---|
| 135.7933 | 136 | A’ |
| 231.9719 | 232 | A-2 H20 |
| 250.0788 | 250 | A-H20 |
| 347.9488 | 348 | Ap1 |
| 409.7073 | 409 | Ap2-H20 |
| 427.8537 | 428 | Ap2 |
| 507.7540 | 508 | Ap3 |
| 603.8803 | 605 | M-A’-H20 |
| 621.8699 | 622 | M-A’ |
| 756.3682 | 757 | M |
Formulae for various fragment ions are in column on the right. M indicates protonated parent ion; A’, adenine; A, adenosine; p, phosphate group (e.g., Ap3 is ATP).
4. Discussion
The method presented herein does not require extensive sample preparation prior to analysis by RP-HPLC or before analysis by MALDI mass spectrometry. In contrast, Hollah et al. used an ion pair reverse phase HPLC method to measure ApnA concentrations in human platelets; sample preparation required affinity chromatography with a dihydroxyboryl BioRex 70 resin, desalting by HPLC with a LiChrospher RP-18 column. Methods described by Jankowski et al. and Wright et al to identify and quantify diadenosine polyphosphate concentrations required either desalting or elution in high salt HPLC buffers to identify ApnA. Unnecessary sample processing or purification (i.e. desalting) prior to HPLC and mass spectrometry could conceivably result in underestimation of ApnA concentrations.
The combination of low pH and volatile organic buffers both prevents ApnA degradation by trace amounts of hydrolytic enzymes present in a filtered but potentially proteinacious biological sample extracted in aqueous buffers. While coelution of Ap3A and AMP might occur using the general conditions outlined herein, minor adjustment flow rates can improve separation of these two compounds if needed before mass spectrometry. Nonetheless, elution of sample fractions in acetonitrile-trifluoroacetic acid yields fractions that are directly compatible with matrices used in mass spectrometry. It is more likely then that initial sample handling (collection, freezing or other storage of tissues to be extracted) prior to analysis by RP-HPLC will be most responsible if quantitative sample loss occurs. The specific matrix reported herein for Ap3A identification, alpha-cyano-4-hydroxy-cinnamic acid is also compatible with general proteomic applications. Therefore, the RP-HPLC conditions described herein represent an improved method to analyze ApnA in complex biological samples. Furthermore, this method should be applicable to ion trap mass spectrometry, liquid chromatography electrospray ionization, (LCMS) systems with sample introduction via an HPLC. The observation that an intravascular eukaryotic parasite contains sizable levels of Ap3A has important implications for the study of disease pathogenesis in infectious diseases, and also provides a new method to evaluate pathophysiological mechanisms or pharmacological interventions involving diadenosine polyphosphates.
Acknowledgements
This work was supported in part by grants from the U.S. National Institutes of Health NIAID UO1 AI1053877 and R03TW1092-3 from Fogarty International Center. We thank Youli Milev and Rita Grantner for technical assistance.
References
- 1.Bochner B, Lee P, Wilson W, Cutler C, Ames B. Cell. 1984;37:225. doi: 10.1016/0092-8674(84)90318-0. [DOI] [PubMed] [Google Scholar]
- 2.Burnstock G, Williams M. J. Pharmacol. Exp. Ther. 2000;295:862. [PubMed] [Google Scholar]
- 3.Kisselev L, Jusesen J, Wolfson A, Frovola L. FEBS Lett. 1998;427:157. doi: 10.1016/s0014-5793(98)00420-7. [DOI] [PubMed] [Google Scholar]
- 4.Schluter H, Tepel M, Zidek W. J. Auton. Pharmacol. 1996;16:357. doi: 10.1111/j.1474-8673.1996.tb00053.x. [DOI] [PubMed] [Google Scholar]
- 5.Vartanian A, Alexandrov I, Prudowski I, Mclennan A, Kisselev L. FEBS Lett. 1999;256:175. doi: 10.1016/s0014-5793(99)00956-4. [DOI] [PubMed] [Google Scholar]
- 6.Williams M, Jarvis MF. Biochem. Pharmacol. 2000;59:1173. doi: 10.1016/s0006-2952(99)00341-x. [DOI] [PubMed] [Google Scholar]
- 7.Barnes L, Garrison P, Siprashvili Z, Guranowski A, Robinson AK, Ingram SW, Croce CM, Ohta M, Huebner K, et al. Biochemistry. 1996;35:11529. doi: 10.1021/bi961415t. [DOI] [PubMed] [Google Scholar]
- 8.Kowara R, Karaczyn A, Fivash M, Kaasprzak K. Chem. Res. Toxicol. 2002;15:319. doi: 10.1021/tx010112j. [DOI] [PubMed] [Google Scholar]
- 9.Pace H, Hodawkekar S, Draganescue A, Hugant J, Bieganowski P, Pekarsky Y, Croce C, Brenner C. Current Biol. 2000;10:907. doi: 10.1016/s0960-9822(00)00621-7. [DOI] [PubMed] [Google Scholar]
- 10.Luthje J, Ogilve A. Eur. J. Biochem. 1987;173:241. doi: 10.1111/j.1432-1033.1988.tb13990.x. [DOI] [PubMed] [Google Scholar]
- 11.Delaney S, Blackburn GM, Geiger JD. Eur. J. Pharmacol. 1997;332:35. doi: 10.1016/s0014-2999(97)01057-1. [DOI] [PubMed] [Google Scholar]
- 12.Patel K, Barnes A, Camacho J, Paterosco C, Boughtflower R, Cousens D, Marshall F. Euro. J. Pharmacol. 2001;430:203. doi: 10.1016/s0014-2999(01)01401-7. [DOI] [PubMed] [Google Scholar]
- 13.Stachon A, Stegemann H, Hohage H, Rahn K, Schlatter E. Cell Physiol. Biochem. 1998;8:175. doi: 10.1159/000016280. [DOI] [PubMed] [Google Scholar]
- 14.Pintor J, Carracedo G, Alsonso MC, Bautista A, Pereal P. Pflugers Arch. 2002;443:432. doi: 10.1007/s004240100696. [DOI] [PubMed] [Google Scholar]
- 15.Mediero A, Pral A, Pintor J. Invest. Ophthalmol. Vis. Sci. 2006;47:4500. doi: 10.1167/iovs.06-0209. [DOI] [PubMed] [Google Scholar]
- 16.Kron M, Petridis M, Milev Y, Leykam J, Hartlein M. Mol. Biochem. Parasitol. 2003;129:33. doi: 10.1016/s0166-6851(03)00080-x. [DOI] [PubMed] [Google Scholar]
- 17.Kron M, Haetlein M. The Aminoacyl-tRNA Synthetases. In: Ibba M, Francklyn C, Cusack S, editors. Landes Bioscience. Texas, USA: Georgetown; 2003. p. 397. [Google Scholar]
- 18.Ottesen E. In: Infectious Diseases. Cohen J, Powderly W, editors. London, England: Mosby; 2004. p. 1607. [Google Scholar]
- 19.Liu L, Weller PF. J. Clin. Invest. 1992;38:1113. doi: 10.1172/JCI115691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mupanomda M, Williams JF, Mackenzie CD, Kaiser L. J. Appl. Physiol. 1997;82:389. doi: 10.1152/jappl.1997.82.2.389. [DOI] [PubMed] [Google Scholar]
- 21.Kaiser L, Mupanomuda M, Williams JF. Am. J. Trop. Med. Hygiene. 1996;54:386. doi: 10.4269/ajtmh.1996.54.386. [DOI] [PubMed] [Google Scholar]
- 22.Lamb V, Williams JF, Kaiser L. Am. J. Vet. Res. 1993;54:2056. [PubMed] [Google Scholar]
- 23.Jankowski J, Jankowski V, Laufer U, Van der Giest M, Henning L, Tepel M, Azidek W, Schulter H. Arterioscler. Thromb. Vasc. Biol. 2003;23:1231. doi: 10.1161/01.ATV.0000075913.00428.FD. [DOI] [PubMed] [Google Scholar]
- 24.Wright M, Boonyalai N, Tanner JA, Hindley AD, Miller AD. FEBS J. 2006;273:3534. doi: 10.1111/j.1742-4658.2006.05361.x. [DOI] [PubMed] [Google Scholar]
- 25.Hollah P, Hausberg M, Kosch M, Barenbrock M, Letzel M, Schlatter E, Rahm K. J. Hypertens. 2001;19:237. doi: 10.1097/00004872-200102000-00010. [DOI] [PubMed] [Google Scholar]