Abstract
In many vertebrates, extra-embryonic tissues are important signaling centers that induce and pattern the germ layers. In teleosts, the mechanism by which the extra-embryonic yolk syncytial layer (YSL) patterns the embryo is not understood. Although the Nodal-related protein Squint is expressed in the YSL, its role in this tissue is not known. We generated a series of stable transgenic lines with GFP under the control of squint genomic sequences. In all species, nodal-related genes induce their own expression through a positive feedback loop. We show that two tissue specific enhancers in the zebrafish squint gene mediate the response to Nodal signals. Expression in the blastomeres depends upon a conserved Nodal response element (NRE) in the squint first intron, while expression in the extra-embryonic enveloping layer (EVL) is mediated by an element upstream of the transcription start site. Targeted depletion experiments demonstrate that the zebrafish Nodal-related proteins Squint and Cyclops are required in the YSL for endoderm and head mesoderm formation. Thus, Nodal signals mediate interactions between embryonic and extra-embryonic tissues in zebrafish that maintain nodal-related gene expression in the margin. Our results demonstrate a high degree of functional conservation between the extra-embryonic tissues of mouse and zebrafish.
Keywords: Zebrafish, squint, cyclops, EVL, YSL, Nodal-related, mesoderm, endoderm, extra-embryonic, SB-505124
Introduction
In all multicellular organisms, cells differentiate according to their relative position in the embryo generating a highly reproducible pattern of cell fates. The body plan is established at early stages by specialized groups of cells called signaling centers. In many vertebrates, extra-embryonic tissues are the first signaling centers established, and act to induce the germ layers and form the major body axes (Beddington and Robertson, 1999; Schier and Talbot, 2005). In the mouse, for example, the first cell fate decision occurs during the cleavage stages and divides the embryo into embryonic and extra-embryonic lineages (Rossant and Tam, 2004). At later stages, signals from the extra-embryonic ectoderm and the extra-embryonic visceral endoderm are required to form the proximo-distal and anterio-posterior axes (Beddington and Robertson, 1999). In teleosts, the enveloping layer (EVL) is an extra-embryonic epithelial covering that forms during the cleavage stages and is sloughed off the embryo at later stages (Bouvet, 1976; Kimmel et al., 1990). Another extra-embryonic tissue is the yolk syncytial layer (YSL), which forms at the onset of zygotic expression (mid-blastula transition; MBT) when the blastomeres juxtaposed to the yolk fuse with each other and release their contents (Kimmel et al., 1995). While a potential signaling role for the EVL has not been tested, the signaling properties of the YSL are well documented (Oppenheimer, 1934; Solnica-Krezel, 1999). In transplant experiments, signals from the yolk can induce ectopic mesoderm (Mizuno et al., 1996). Conversely, mesoderm and endoderm fail to form when signals from the yolk are depleted by RNase injection (Chen and Kimelman, 2000). The essential signals produced by the YSL, however, are not known.
Nodal-related proteins form a conserved subclass of the TGF-β superfamily that act in all vertebrates to induce the mesoderm and endoderm, pattern all three germ layers and establish the left-right body axis (Schier, 2003). Consistent with these multiple functions, Nodal-related proteins are dynamically expressed throughout development. In the mouse, for example, nodal is expressed across the entire epiblast prior to gastrulation, but rapidly becomes restricted to the primitive streak and the visceral endoderm (Conlon et al., 1994; Zhou et al., 1993). At later stages, nodal is expressed in the node and left lateral plate mesoderm (LLPM) (Collignon et al., 1996). Genetic analysis indicates that Nodal signals have different roles in each domain. Conditional mutants showed that nodal is required in the node to establish left-right asymmetry (Brennan et al., 2002). By contrast, the primitive streak does not form in null nodal mutants, and the resulting embryos lack all mesodermal derivatives (Conlon et al., 1994; Zhou et al., 1993). Analysis of nodal mutant chimeras demonstrated that Nodal signals are required in the visceral endoderm for formation of the prechordal plate and anterior neural tissue (Varlet et al., 1997). Other genetic experiments indicate that Nodal signals in the epiblast act to pattern the extra-embryonic tissues (Brennan et al., 2001). Thus, in mammalian embryos, Nodal signals mediate reciprocal interactions between the embryonic and extra-embryonic tissues that are essential for embryonic development.
There are three nodal-related genes in zebrafish, but only two, squint (ndr1/sqt) and cyclops (ndr2/cyc), are required for mesoderm and endoderm formation (Feldman et al., 1998). The third nodal-related gene, southpaw (spaw/ndr3), is only expressed after gastrulation and is required to establish left-right asymmetry (Long et al., 2003). In the absence of sqt function, the zebrafish organizer, known as the embryonic shield, does not form (Feldman et al., 1998). These embryos subsequently recover, however, because Cyc signals induce mesoderm and endoderm during gastrulation (Dougan et al., 2003; Hagos and Dougan, 2007). At 24hpf, most sqt mutants are indistinguishable from wild type, but a variable minority have reduced prechordal plates and display mild cyclopia (Dougan et al., 2003; Heisenberg and Nusslein-Volhard, 1997). In contrast, all cyc mutants have reduced prechordal plate, resulting in cyclopia, and lack the floorplate (Hatta et al., 1991; Rebagliati et al., 1998b; Sampath et al., 1998). The defects in sqt; cyc double mutants are much more severe than either single mutant. These embryos lack all derivatives of the mesoderm and endoderm in the head and trunk, including the notochord, prechordal plate, trunk somites, pronephros, heart, blood and gut (Feldman et al., 1998). Thus, sqt and cyc have partially overlapping functions in germ layer formation.
Nodal signaling is mediated by a bipartite receptor complex containing the TGF-β Type I receptor, ALK4 and the Type II receptor, ActR-IIB (Reissmann et al., 2001). In order to bind and activate the ALK4/ActR-IIB receptor complex, Nodal-related proteins require the function of the Cripto/One-Eyed-pinhead (Oep) co-receptor (Cheng et al., 2003; Gritsman et al., 1999; Yeo and Whitman, 2001). ALK4 is a Ser/Thr kinase that phosphorylates cytoplasmic Smad2 and Smad3. PSmad2 or PSmad3 then dimerizes with Smad4 and translocates to the nucleus, where they activate transcription of target genes (Massague and Chen, 2000). The Smad heterodimers associate with any of several nuclear co-factors to stimulate gene expression, the most prominent of which are the winged-helix transcription factor FoxH1 and the paired-like homeodomain protein, Mixer (Kunwar et al., 2003). A few direct transcriptional targets of this pathway have been identified, including the nodal-related genes themselves (Meno et al., 1999). Conserved elements in the introns of Xenopus xnr1 and mouse nodal mediate the autoregulatory response (Brennan et al., 2001; Hyde and Old, 2000; Osada et al., 2000). In both species, this element drives expression in the LLPM after gastrulation (Hyde and Old, 2000; Osada et al., 2000; Saijoh et al., 2000). At earlier stages, transcription factors acting on this element boost expression levels in the margin in frog embryos, and mediate expression in the epiblast of mouse embryos (Brennan et al., 2001; Hyde and Old, 2000; Osada et al., 2000).
sqt is initially expressed during oogenesis, but its function during these stages is controversial (Gore et al., 2005; Gore and Sampath, 2002; Hagos et al., in press; Schier, 2005). In the zygote, sqt and cyc are expressed in three independent phases (Rebagliati et al., 1998a). sqt expression initiates in dorsal blastomeres soon after MBT (3hpf), under control of the dorsal determinant β-catenin (Bellipanni et al., 2006; Dougan et al., 2003). After initiation, sqt expression extends into the YSL and the EVL (Erter et al., 1998; Feldman et al., 1998). Although overexpression experiments demonstrated that Sqt signals in the YSL could induce overlying blastomeres to become dorsal mesoderm, it is not known if sqt is required in the YSL (Erter et al., 1998; Feldman et al., 1998). During the late blastula stages, sqt and cyc are co-expressed in all marginal blastomeres. Two lines of evidence indicate that expression in the marginal ring is independent of the earlier expression of sqt in the dorsal blastomeres. First, overexpressing β-catenin induces ectopic expression of sqt at 3.5hpf, but has no effect on expression at the margin (Dougan et al., 2003). Second, depletion of β-catenin eliminates the early dorsal expression of sqt, but does not effect sqt expression in the marginal ring (Bellipanni et al., 2006; Kelly et al., 2000). Although the T-box transcription factor VegT induces marginal expression of the nodal-related genes in Xenopus, the factors that induce this phase of nodal-related gene expression in zebrafish are not known (Stennard, 1998; White and Heasman, 2007). Expression of both sqt and cyc at this stage is maintained by an autoregulatory loop (Meno et al., 1999). In the third phase, sqt expression during gastrulation is maintained in a few blastomeres at the dorsal midline, called dorsal forerunners (Erter et al., 1998; Feldman et al., 1998; Rebagliati et al., 1998a). By contrast, cyc transcripts accumulate in the axial mesoderm (Rebagliati et al., 1998b; Sampath et al., 1998).
We have undertaken an analysis of sqt genomic sequences in order to understand the regulatory networks that control sqt expression in each of its domains. We generated a transgenic line containing sqt genomic sequences driving expression of a GFP reporter gene. This line faithfully reproduces the spatio-temporal expression pattern of endogenous sqt, and at the late blastula stage is expressed in the YSL as well as in the blastomeres. We show that expression in embryonic and extra-embryonic tissues is controlled by separable regulatory elements, including at least two elements that mediate the response to Nodal signals in different cell types. An element upstream of the transcription start site mediates the response to Nodal signaling specifically in the EVL cells. By contrast, a conserved Nodal response element (NRE) in the first intron is required for transgene expression in the blastomeres. We show that expression of the transgene in the blastomeres depends on Nodal signaling activity. Furthermore, expression of sqt and cyc in the blastomeres depend upon Nodal signals from the YSL. These experiments suggest that Nodal signals in the YSL act to stabilize nodal-related gene expression in the embryo margin by activating the Nodal autoregulatory pathway. Targeted depletion of Nodal signals from the YSL results in embryos lacking endoderm and head mesoderm, similar to the defects observed in mice lacking Nodal function in the visceral endoderm (Brennan et al., 2001; Varlet et al., 1997). Thus, our data provides strong genetic evidence for the functional conservation between the YSL and the visceral endoderm. This suggests a common evolutionary origin for teleost and mammalian extra-embryonic tissues, despite their profound morphological differences.
Materials and Methods
Zebrafish strain and morpholino injections
Wild-type embryos were obtained from natural crosses of Wik fish. Collected embryos were maintained at 28.5°C and staged according to morphological criteria (Kimmel et al., 1995). Translation blocking morpholinos against sqt and cyc have been described previously (Feldman and Stemple, 2001; Karlen and Rebagliati, 2001). The MO sequences are as follows: sqtMO: 5′-ATGTCAAATCAAGGTAATAATCCAC-3′; sqtMIS: ATcTgAAAATgAAGcTAATAATgCAC-3′; cycMO: 5′-GCGACTCCGAGCGTGTGCATGATG-3′; cycMIS: 5′-GCcACTgCGAGaGTGTGgATcATG-3′. Nucleotides in lower case represent mismatches. The sqtMOs were tagged with lissamine, while the cycMOs were tagged with fluorescein. Embryos were injected with each MO at 3hpf, soon after formation of the YSL. The distribution of the MO in the YSL was verified 2 hr after injection. Embryos with mislocalized MOs were removed from the experiments.
BAC recombineering
BAC 157J11 (CHORI-211 library) was identified as a 120 kb clone containing the entire sqt locus. We replaced the sqt first exon with the eGFP open reading frame by homologous recombination (Lee et al., 2001; Yu et al., 2000). We generated the pCS2EGFP-FRT-neo-FRT plasmid by inserting the sacII FRT-neo-FRT cassette from pIGCN21 into pCS2EGFP (Lee et al., 2001). We used the following primers to generate a recombination fragment: Forward targeting primer: 5′-CAGAGACTTTATTTCAATAACTGCGTGTGGATTATTACCTTGATTTGACATGGTGAGCAAGGGCGAGGA-3′; Reverse targeting primer: 5′-ACTTTTAGCGACGAGGCTCAAGACGGAGTCAGACTCGTAAAGAGTTGGATTCTATTCCAGAAGTAGTGAG-3′. Nucleotides in bold are homologous to sqt genomic sequences; the other nucleotides are homologous to the eGFP-FRT-neo-FRT cassette. This fragment was inserted into BAC 157J11 by homologous recombination, as described (Lee et al., 2001). To generate p9.9sqtGFPi, we used the gap repair method to excise GFP, the first intron, and 9.9kb upstream sequences from the modified BAC 157J11 (Liu et al., 2003). The gap-repair targeting vector was constructed in pCS2+, and consisted of a 490bp fragment homologous to a region 9.9kb upstream of the gfp open reading frame and a 390bp fragment homologous to sequences in the first intron. The upstream fragment was amplified from a preparation of BAC DNA using the following primers: Forward: 5′-TCATGGATCCGAAGATCAATTCAATCCCAT -3′; Reverse: 5′-TTACTCGAGCTAAATCAACGCTTAGACTT-3′. The forward primer contained a BamH1 site, and the reverse primer contained n XhoI site. To generate the downstream fragment, we used the following primers: Forward: 5′-TTGACCTCGCTAGACGCTGTAGCCTTGA-3′; Reverse: 5′-TCCATTCTAGAAGTGTTTAGGGCAGACAGGT-3′. The forward primer contained an Xho1 site and the reverse primer contained an XbaI site. The PCR products were cut with BamH1 and XhoI, or XhoI and XbaI, respectively, and were inserted into the BamH1 and XbaI sites of pCS2+ in a single, triple ligation reaction. The resulting plasmid, except for the XhoI site, was amplified by PCR using the following two primers: Clockwise Primer: 5′-CTAGACGCTGTAGCCTTGAT-3′; Counterclockwise primer: 5′-CTAAATCAACGCTTAGACTT. The PCR product was digested by Dpn in order to minimize false positives arising from vector DNA and transformed into bacteria containing the modified BAC157J11. The resulting plasmid, p9.9sqtGFPi, includes 9.9kb upstream of GFP and the first 603bp of the 631bp first intron.
Deletion analysis of p9.9sqtGFPi
p9.9sqtGFPi was digested with HindIII, which divided the upstream region into five fragments and one proximal fragment, which consisted of the GFP open reading frame (G), 923bp of genomic sequences upstream (p) and 603bp of the first intron (i) (Fig. 1). When injected into embryos, the proximal fragment, pGi, did not express GFP protein in the margin, except for variable expression in a few random cells. The sizes of the upstream HindIII fragments are as follows: fragment a= 431bp; b=746bp; c=3669bp; d=2629bp; e=1456bp. To determine which of the upstream fragments contained the necessary sequences to drive reporter gene expression in the margin, we ligated each HindIII fragment to pGi in pBluescript (Stratagene, La Jolla CA). We injected each of the five resulting constructs into embryos and assayed for GFP fluorescence during the blastula stages (5 hpf), and for expression in the Kupffer’s vesicle at 18hpf. pSqtapGFPi was the only construct to consistently display strong fluorescence in the entire margin and Kupffer’s vesicle. To determine if the ‘a fragment’ was capable of driving GFP expression on its own, we ligated it to the pax6 p0 promoter, which has been previously shown to work in zebrafish (Lakowski et al., 2007). We did not observe fluorescence when this construct was injected into embryos, indicating that the ‘a fragment’ is not sufficient to drive GFP expression. We used pSqtapGFPi as the starting point for further analysis.
FIG. 1. Construction and expression of transient sqtGFP lines.
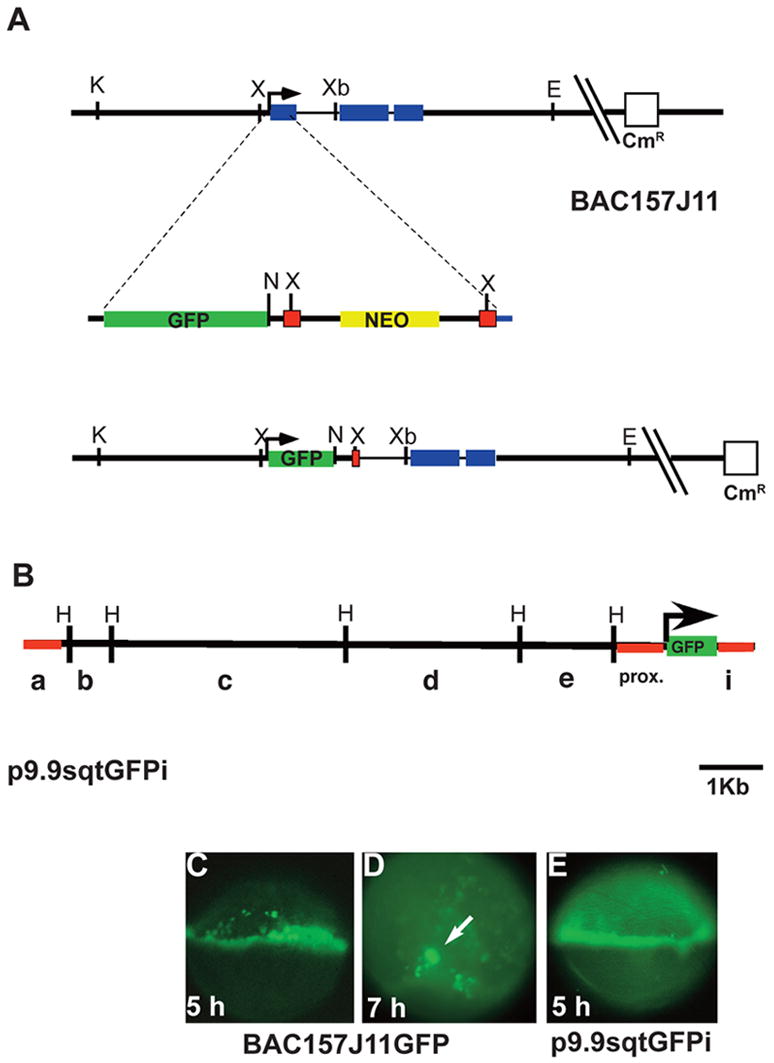
(A) Diagram of the sqt locus in BAC157J11. sqt has three exons (blue rectangles) separated by two introns, 600bp and 83bp long respectively. The start of transcription is indicated (arrow). Key restriction sites and the vector gene encoding chloramphenicol resistance are depicted. The GFP-FRT-neo-FRT cassette is diagrammed below the BAC, showing the site of integration in the first exon. The resulting engineered BAC clone is depicted below. After integration and excision of the gene encoding neomycin resistance, the gfp coding region replaces the sqt first exon. Green rectangle=GFP sequences; yellow rectangle=neomycin resistance gene; red squares=FRT recombination sites; blue rectangle=sqt coding sequences. (B) Diagram of p10sqtGFP following excision from the BAC by gap-repair. HindIII sites are indicated. Red lines indicate the sequences used to generate Tg-SqtapGFP and its derivatives. (C) Fluorescent image of a living, 5 hpf embryo injected with BAC157J11sqtGFP at the one-cell stage. Fluorescence appears throughout the margin, including the YSL, EVL and blastomeres. Perduring GFP expression is also observed in some cells farther from the margin. (D) Low magnification fluorescent image of a living, 7 hpf embryos injected with with BAC157J11sqtGFP. Fluorescence is observed in the dorsal forerunners (white arrows). Perdurant expression is also detected in blastomeres and EVL cells farther from the margin. (E) Fluorescent image of a living 5 hpf embryo injected with p9.9sqtGFPi at the one-cell stage. GFP fluorescence is detected around the entire margin, in the blastomeres, EVL and YSL. K=KpnI; X=XhoI; Xb=XbaI; E=EcoR1; N=Not1; H=HindIII.
To delete the 68 bp NRE from the sqt first intron, we ligated two PCR fragments including the sequences upstream or downstream of the NRE. To amplify the upstream fragment, we used the Forward primer 5′-AAATCCGCGGTTAGTGTGTGTGTATTA-3′, which has a SacII site and the Reverse primer 5′-AATTGAATTCTAAGCATAATACATGACT-3′, which has an EcoRI site. Nucleotides in bold are homologous to sqt genomic sequences. The downsteam fragment was amplified with the Forward primer: 5′-AATTGAATTCAGACAATAAGAATGTTCT-3′, which contains an EcoRI site. The Reverse primer, 5′-AATAGGGCCCTGTCTAAATGTGTATTGA-3′, has ApaI site. The fragments were digested with EcoR1 and inserted into pSqtapG to generate pSqtapGiΔNRE.
Injection of DNA and establishment of stable transgenic lines
For analysis of transiently expressed transgenes, we prepared DNA using the Qiagen BAC or plasmid purification Kit (Qiagen, Inc, Valencia CA). BAC DNA was diluted to 80ng/μl and 150pg was injected into the blastomeres of one-cell stage embryos. BAC DNA was stored at 4°C for up to four weeks. Transgene expression was observed in living embryos at the blastula stages (5 hpf). We also examined perdurant GFP expression in the Kupffer’s vesicle. The pattern, fluorescence intensity, and frequency of expression were all evaluated.
To generate stable transgenic lines, DNA fragments of interest were clone into the BamH1 and ApaI sites of a modified Tol2 vector (Kawakami et al., 2004; Urasaki et al., 2006). 150pg of transgene DNA was co-injected into the blastomere of the one-cell stage embryo with 100pg mRNA encoding the Tol2 transposase. F0 fish were raised to adulthood and intercrossed. F1 progeny were screened for GFP fluorescence in the margin at 30–50% epiboly, or in the Kupffer’s vesicle in the early somite stages.
Activation and inhibition of the Nodal pathway
To activate the Nodal pathway, we expressed TARAM-D, which is a mutated and constitutively activated form of the Nodal receptor (Renucci et al., 1996). Capped mRNA was synthesized using the Ambion mMessage mMachine™ Kit (Ambion, Inc., Austin TX). We injected 100pg mRNA into 1–2 cell stage embryos. To block the Nodal receptors, we used SB-505124, which is a specific inhibitor of the ALK 4,5,7 receptors and has been previously demonstrated to phenocopy embryos lacking nodal function (DaCosta Byfield et al., 2004; Hagos and Dougan, 2007).
in situ hybridization and sections
in situ hybridization was performed as previously described (Dougan et al., 2003). Embryos were processed to reveal expression of green fluorescent protein (gfp) (Cormack et al., 1996), goosecoid (gsc) (Stachel et al., 1993), floating head (flh) (Talbot et al., 1995), no-tail (ntl) (Schulte-Merker et al., 1994), myosin17 (my17/cardiac light chain myosin-2) (Yelon, 2001), mezzo (Poulain and Lepage, 2002), sox17 (Alexander and Stainier, 1999), sqt (Feldman et al., 1998) and cyc (Rebagliati et al., 1998b; Sampath et al., 1998). Following in situ hybridization, selected embryos were dehydrated in a series of 100% methanol, 100% ethanol, acetone and propylene oxide. Embryos were then incubated overnight in a 1:1 mixture of propylene oxide: Epon-Araldite, and for 1 hr in a 1:2 mixture of propylene oxide: Epon-Araldite. Following two washes in 100% Epon-Araldite, the resin was allowed to polymerize at 70°C for at least 18 hrs. Embryos were then cut into 3μm sections and mounted in Permount (Sigma-Aldrich, Inc., St. Louis MO).
Results
Isolation of sqt regulatory sequences
The strict spatio-temporal regulation of nodal-related gene expression is essential for germ layer formation (Schier, 2003). In order to understand the molecular mechanisms controlling nodal-related gene expression in zebrafish, we identified a 120kb BAC clone containing the entire sqt genomic locus. We used a BAC recombineering strategy to replace the first exon of sqt with the eGFP open reading frame (Fig. 1A) (Lee et al., 2001). After transient expression of the manipulated BAC clone, we observed GFP fluorescence at the margin in blastula stage embryos (5 hpf; Fig. 1C) and in a few dorsally located cells during gastrulation (Fig. 1D, arrow). This indicates that the BAC contains all the genomic sequences necessary to drive reporter gene expression in the marginal blastomeres and the dorsal forerunners.
To further narrow down the region necessary for sqt expression, we used the gap-repair method to generate a smaller clone from the BAC, consisting of 9.9 kb of genomic sequences upstream of gfp and 603bp of the first intron downstream of gfp (Fig. 1B; p9.9sqtGFPi) (Liu et al., 2003). After injection, we observed GFP fluorescence in the margin (Fig. 1E) and in the forerunner cells (data not shown). We conclude that this region contains enhancers responsible for driving sqt expression in the margin. To define a smaller region capable of driving GFP expression in the sqt pattern, we subdivided p9.9sqtGFPi into six fragments using convenient restriction sites (Fig. 1B). A fragment containing only the proximal 923bp of upstream genomic sequences and the 603bp intron did not express GFP, as indicated by the lack of fluorescence in living embryos at the blastula stage or later stages (Fig. 2A, construct SqtpGFPi; data not shown). Next, we wanted to determine which of the upstream fragments contained the sequences necessary for sqt expression. We observed GFP fluorescence when the distal-most ‘a fragment’ was inserted upstream of the proximal fragment (Fig. 2A, construct apGFPi, data not shown), but not when any of the other fragments were inserted at this position. This distal fragment is not capable of driving GFP expression when fused to a heterologous promoter (See Methods; data not shown). Thus, neither the ‘a fragment’ nor the ‘proximal fragment’ alone is sufficient to drive expression of a reporter gene in the margin of blastula stage embryos. This indicates that sequences in both fragments are necessary for expression in the margin. These results do not rule out the possibility that other important regulatory sequences are contained within the large BAC clone.
FIG. 2. Spatio-temporal expression of sqtGFP transgenes reflects endogenous sqt expression.
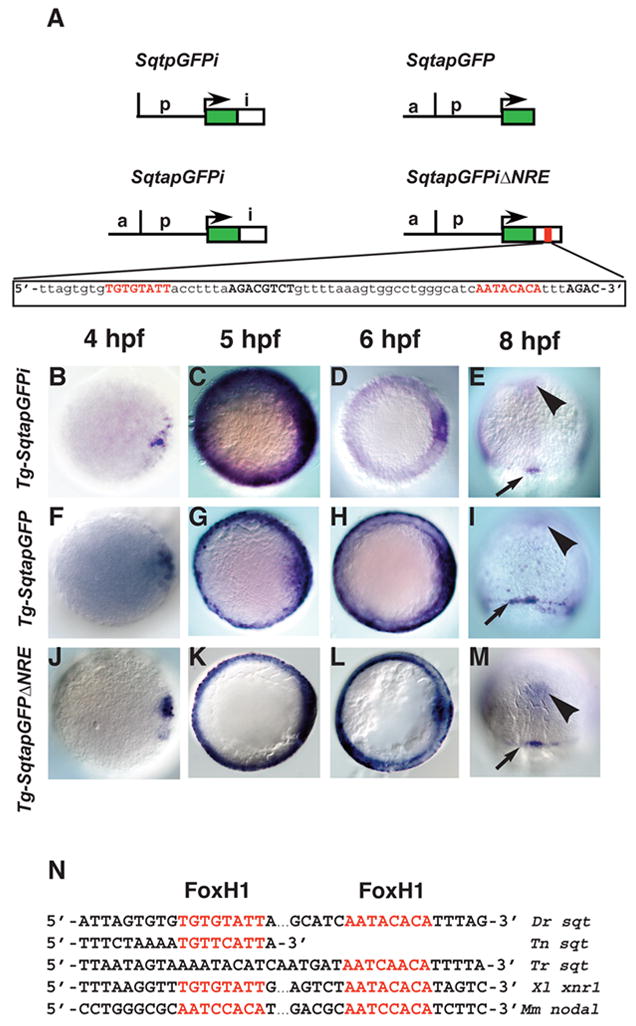
(A) Diagram of three constructs used to make stable transgenic lines with the Tol2 transposase. Each line consists of the GFP reporter gene and 1.4 kb of DNA from the sqt genomic region upstream of the transcription start site. Tg-SqtapGFPi contains 600bp of the sqt first intron inserted downstream of the GFP poly-Adenylation signal. Tg-SqtapGFP lacks the entire first intron. Tg-SqtapGFPiΔNRE is identical to Tg-SqtapGFPi, except that it lacks the 68 bp NRE sequence, shown below. The FoxH1 consensus sites are highlighted in red; Smad consensus binding sites are in black bold. Time-course of gfp expression in Tg-SqtapGFPi (B–E), Tg-SqtapGFP (F–I) and Tg-SqtapGFPiΔNRE (J–M). In all lines, gfp mRNA is induced in dorsal blastomeres soon after MBT (B, F, J). At 5 hpf, gfp is expressed in a ring around the entire margin in all lines (C, G, K). The ring of expression in Tg-SqtapGFP (G) and Tg-SqtapGFPiΔNRE (K) is thinner than the ring in Tg-SqtapGFPi. At 6 hpf, gfp expression in Tg-SqtapGFPi (D) and Tg-SqtapGFPiΔNRE (L) is absent from the margin, but expression persists in Tg-SqtapGFP (arrows) (H). At 8hpf, gfp is expressed in the dorsal forerunner cells in all three lines (red arrowheads) (E, I, M). In Tg-SqtapGFP, expression persists in marginal EVL cells throughout gastrulation (arrows) (I). In all three lines, ectopic gfp expression is detected at the midline (black arrowheads) (E, I, M). (N) The FoxH1 consensus binding sites in the sqt intron are conserved in Tetraodon nigroviridis (Tn), Takifugu rubripes (Tr), Xenopus laevis (Xl), and Mus musculus (Mm).
The temporal expression of sqt is controlled by sequences in the first intron
The inherent variability and mosaicism of transgenes following injection complicates analysis of their expression (Hsiao et al., 2001). To circumvent these problems, we used the Tol2 transposon to generate a stable transgenic line containing both the distal and proximal fragments upstream of gfp and 603bp of the first intron downstream (Fig. 2A; SqtapGFPi) (Kawakami et al., 2004; Urasaki et al., 2006). Despite relatively low fluorescence intensity of GFP protein at early stages, gfp mRNA was easily detected by in situ hybridization in all three phases of sqt expression (Fig. 2). Soon after MBT, gfp transcripts are detected in a few dorsal blastomeres (Fig. 2B). Like sqt, gfp is expressed in a marginal ring at 5 hpf and is down-regulated in the margin at 6 hpf (Fig. 2C, D). Transgene expression persists in the dorsal forerunner cells throughout gastrulation (Fig. 2E). Thus, Tg-SqtapGFPi recapitulates the major features of the endogenous sqt spatio-temporal expression pattern. We conclude that the factors that control sqt expression in each of its phases also act upon the transgene. Low levels of gfp expression in the axial mesoderm were observed in each of the three lines we generated, but do not reflect endogenous sqt expression (Fig. 2E, I, M, arrowheads). This raises the possibility that our construct lacks an element responsible for repressing sqt expression in the axial mesoderm.
Because important regulatory sequences have been identified in the first introns of nodal-related genes in Xenopus and mice, we asked whether the sqt intron contains essential regulatory elements (Norris et al., 2002; Osada et al., 2000). We generated a second line that lacks the intron sequences, but retains both the distal and proximal upstream fragments (Fig. 2A; SqtapGFP). In this line, gfp expression initiates soon after MBT (Fig. 2F), as does endogenous sqt (Erter et al., 1998; Feldman et al., 1998). At 5hpf, we observed a ring of gfp expression in the margin (Fig. 2G). Finally, at the onset of gastrulation, gfp was expressed in the dorsal forerunner cells and continues throughout gastrulation (Fig. 2I and data not shown). We conclude that the enhancer elements responsible for inducing sqt expression in the dorsal blastomeres, the embryo margin and in dorsal forerunner cells are included in the distal and proximal sqt genomic fragments upstream of GFP in the transgene. Consistent with this, we found three putative Tcf/Lef binding sites in the 923bp upstream of the sqt transcription start site, which are included in the transgene and could mediate activation of gene expression by β-catenin (data not shown) (Dorsky et al., 2002). By contrast, we found only one putative Tcf/Lef site in the region between −900bp to −1800bp upstream of the sqt transcription start site, suggesting that the clustering of sites close to the sqt transcription start site may be significant. The factors controlling sqt expression in the marginal ring have not been identified, but this domain of sqt expression is independent of β-catenin function (Bellipanni et al., 2006).
Distinct mechanisms control sqt expression in embryonic and extra-embryonic tissues
Reporter gene expression in Tg-SqtapGFP differed from gfp expression in Tg-SqtapGFPi and endogenous sqt expression in two ways. Firstly, the ring of gfp expression in Tg-SqtapGFP persists in marginal cells throughout gastrulation (Fig. 2H, I; compare with Fig. 2D, E). This indicates that sequences in the sqt first intron are required to repress gene expression at the margin during gastrulation. Secondly, gfp expression appeared narrower in Tg-SqtapGFP than in Tg-SqtapGFPi, and seemed confined to a more superficial layer of cells (Fig. 2C, G). To test this, we analyzed gfp expression in sections of 5hpf embryos from both lines (Fig. 3A, B). In Tg-SqtapGFPi, gfp is expressed in the YSL (Fig. 3A, under red line), blastomeres (Fig. 3A, above the red line) and EVL at roughly equivalent levels (Fig. 3A, arrow). This accurately reflects the expression pattern of sqt. In the absence of the intron, by contrast, gfp is strongly expressed in the YSL (Fig. 3B, under red line) and EVL (Fig. 3B, arrow), but is not detected in the blastomeres. The expression in the EVL appears as a narrow ring in whole mounts (Fig. 2G). These results indicate that genetically distinct pathways control sqt expression in the embryonic and extra-embryonic tissues. Rare blastomeres do express gfp, and these are invariably adjacent to the YSL or EVL (Fig. 3B, above red line). This staining could indicate that rare blastomeres in 5 hpf embryos share the regulatory program of the extra-embyronic tissues. We conclude that the intron contains a tissue specific enhancer responsible for driving expression in the blastomeres. This is consistent with results demonstrating that nodal-related genes in mice and frog contain conserved enhancers that drive tissue specific expression in the LLPM and boost nodal levels in the frog margin and mouse epiblast (Adachi et al., 1999; Brennan et al., 2001; Hyde and Old, 2000; Osada et al., 2000; Saijoh et al., 2003).
FIG. 3. Expression of gfp in the blastomeres depends on Nodal signaling.
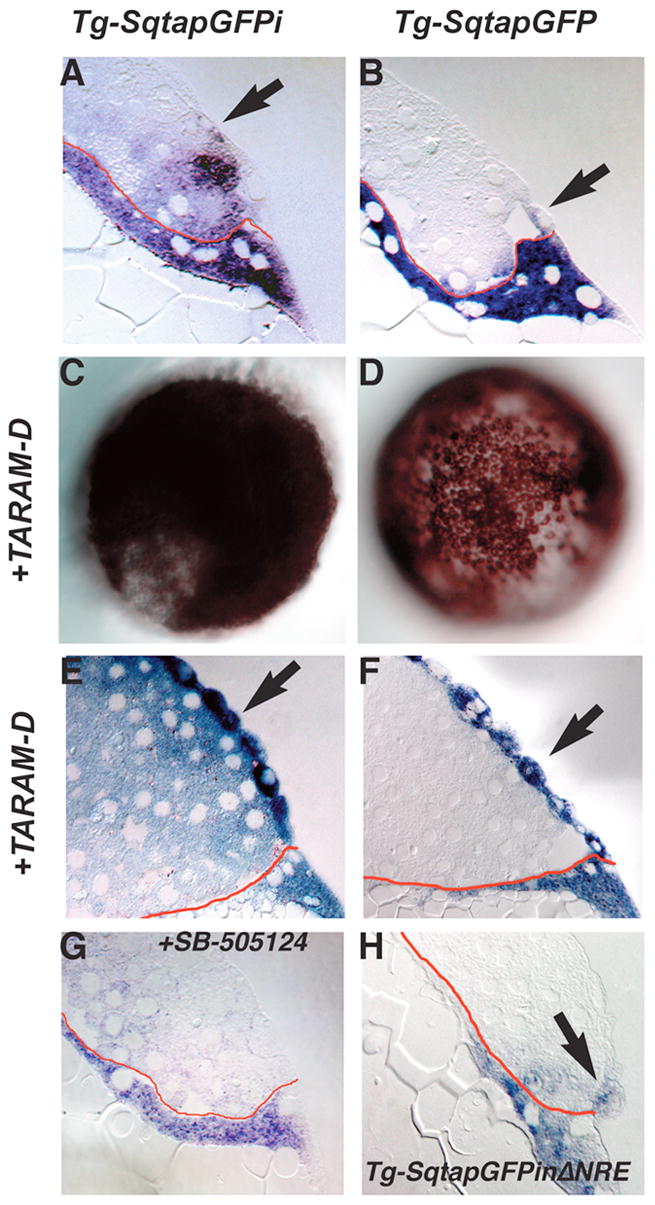
Distribution of gfp mRNA in sections of 5 hpf Tg-SqtapGFPi (A, E, G), Tg-SqtapGFP (B, F) and Tg-SqtapGFPiΔNRE (H) embryos stained to reveal gfp mRNA. Whole mounts of Tg-SqtapGFPi (C) and Tg-SqtapGFP (D) are also depicted. In all sections, the membrane that separates the YSL from the blastomeres is highlighted in red. (A) In Tg-SqtapGFPi, gfp transcripts are localized in the EVL (arrow), the blastomeres, and the YSL. (B) In Tg-SqtapGFP, gfp is expressed predominantly in the YSL and EVL (arrow). Expression is also observed in rare blastomeres adjacent to the YSL or EVL. (C, E) In response to activation of the Nodal pathway by ubiquitous expression of TARAM-D, gfp is ubiquitously expressed in Tg-SqtapGFPi embryos. In section, gfp transcripts are observed in all blastomeres, EVL cells (E, arrows), and the YSL. The EVL cells take on a more rounded appearance (E, F) In Tg-SqtapGFP embryos, TARAM-D induces gfp expression throughout the embryo (D), but only in the EVL cells (F). (G) When Nodal signaling is blocked by treatment with SB-505124 in Tg-SqtapGFPi embryos, gfp expression is reduced, but not eliminated in the YSL and EVL, and is eliminated in the blastomeres. (H) In Tg-SqtapGFPiΔNRE embryos, gfp expression is restricted to the EVL (arrow) and YSL.
To understand the pathway that controls sqt expression in the blastomeres, we asked if the intron mediates the response to Nodal signals. We injected embryos from both lines with mRNA encoding a mutated and constitutively activated version of the Nodal receptor, called TARAM-D (Renucci et al., 1996). This receptor acts in a cell autonomous manner to dorsalize embryos and induce expression of Nodal target genes (Aoki et al., 2002; Renucci et al., 1996). In response to excess Nodal signaling, the blastomeres adopt dorsal mesendodermal fates and the embryos fail to undergo epiboly (Hagos and Dougan, 2007; Shimizu et al., 2000). This accounts for the lack of doming and dramatically altered morphology of the YSL in TARAM-D injected embryos (Fig. 3E, F). In addition, EVL cells lose their normal squamous shape and become rounded (compare Fig. 3B, arrow and Figs. 3E, F, arrows). This indicates that EVL cells are capable of responding to activation of the Nodal pathway. If the intron is required to respond to Nodal signaling, then TARAM-D should induce ectopic gfp expression in Tg-SqtapGFPi, but not in the line lacking the intron. In contrast to this expectation, both lines exhibited a response to TARAM-D as indicated by a dramatic expansion of gfp expression apparent in whole mounts (Fig. 3C, D). The responses of the two lines are not equivalent, however, as revealed by sectioning embryos expressing TARAM-D. In Tg-SqtapGFPi embryos expressing TARAM-D, gfp is expressed in blastomeres throughout the entire embryo including all blastomeres and EVL cells and the YSL (Fig. 3E). gfp is expressed at roughly equivalent levels in the YSL and blastomeres. The higher level of expression in the EVL is likely due to the greater access of these cells to in situ reagents. Because there was no change in the pattern of gfp expression in the YSL, we could not determine if the YSL responds to TARAM-D. We conclude that Tg-SqtapGFPi contains all the sequences necessary for blastomeres and EVL cells to respond to Nodal signals. By contrast, in the line lacking the intron, gfp expression expands throughout the EVL in response to TARAM-D (Fig. 3F, arrows), but is not detected in the blastomeres (Fig. 3F). Thus an upstream enhancer mediates the response to Nodal signals specifically in the EVL. Thus, the Nodal signaling pathway acts by distinct mechanisms to induce transgene expression in the blastomeres and the EVL.
These results raised the possibility that gfp expression in the marginal blastomeres and EVL of Tg-SqtapGFPi embryos depends on Nodal signals. To test this, we utilized the chemical inhibitor of the Nodal receptors, SB-505124, to block Nodal signaling in Tg-SqtapGFPi embryos (DaCosta Byfield et al., 2004; Hagos and Dougan, 2007). 50μM of this compound is sufficient to phenocopy embryos lacking nodal-related gene function when added to embryos at 3hpf or earlier, whereas later treatments, or lower doses, generate milder phenotypes (Hagos and Dougan, 2007). When we treated Tg-SqtapGFPi embryos with 50μM SB-505124, overall levels of gfp expression are reduced, but they are reduced to a greater extent in the blastomeres and EVL than in the YSL (Fig. 3G). In some embryos, we do see expression in cells in the area of the EVL, but this probably represents expression in the dorsal forerunner cells (data not shown). In the YSL, gfp expression is consistently reduced, but not completely eliminated (Fig. 3G). This demonstrates that gfp expression in the blastomeres and EVL depends on activation of the Nodal-signaling pathway.
The sqt first intron contains an essential Nodal response element
We found two sequences in the sqt first intron that fit the FoxH1 consensus-binding site, TGT(T/G)(T/G)ATT, as defined by in vitro binding assays (Fig. 2A, SqtapGFPiΔNRE, red letters) (Zhou et al., 1998). This is similar to the “paired FAST” site defined in Xenopus (Fig. 2N) (Osada et al., 2000). These sites are located in close proximity to at least three core Smad binding sequences, AGAC (Fig. 2A, SqtapGFPiΔNRE, black bold letters) (Zawel et al., 1998). The introns of sqt orthologues in the pufferfish Tetraodon nigroviridis and Takifugu rubripes each contain a single putative FoxH1 binding site (Fig. 2N) (Aparicio et al., 2002; Jaillon et al., 2004). Since an estimated 200 Myr of evolution separates pufferfish from zebrafish, this shows a strong conservation of the intronic enhancer throughout the teleost lineage (Taylor et al., 2001).
To test whether this putative element is required for gfp expression in the blastomeres, we generated a transgenic line lacking both FoxH1 sites and associated Smad sites in the intron (Fig. 2A; Tg-SqtapGFPiΔNRE). In this line, gfp expression initiates in the dorsal blastomeres soon after MBT (Fig. 2J). In the late blastula stage (5 hpf), gfp is expressed in a narrow ring at the margin and strongly resembles reporter gene expression in Tg-SqtapGFP (Fig. 2K, compare with Fig. 2G). At the onset of gastrulation (6 hpf), gfp expression is induced in the dorsal forerunners (data not shown) and is maintained at the margin (Fig. 2L). In sections of embryos at this stage, gfp is expressed in the YSL, EVL and in a few marginal blastomeres close to the YSL and EVL (Fig. 3H). The expression in marginal blastomeres it is more frequent and stronger than in Tg-SqtapGFP, but much less than in Tg-SqtapGFPi (Fig. 3H, compare with Fig. 3A and 3B). This demonstrates that the deleted sequences are necessary for reporter gene expression in the blastomeres. Thus, we have defined a functional Nodal response element (NRE) within the sqt first intron, which is required for expression in the blastomeres. At mid-gastrulation (8 hpf), gfp expression is completely lost from the marginal blastomeres, EVL and YSL, but persists in the dorsal forerunners (Fig. 2M). This indicates that repression of gfp in the EVL occurs even in the absence of the NRE. Since gfp expression persists in the EVL in TgSqtapGFP (Fig. 2I), this means that sites outside of the NRE are also required for the proper regulation of sqt expression.
Expression of Nodal pathway components in extra-embryonic tissues
Our results from the transgene analysis raised the possibility that Nodal signals in the YSL are required to induce or maintain sqt expression in the blastomeres. If so, then nodal-related genes must be expressed in the YSL. Expression of sqt in the YSL has already been documented (Erter et al., 1998; Feldman et al., 1998). To determine if cyc is expressed in the YSL, we sectioned late blastula stage embryos (5 hpf) that were stained to reveal the localization of cyc transcripts. We found that cyc is expressed in marginal blastomeres, up to 6 tiers from the boundary of the YSL (Fig. 4A). Transcripts are concentrated in the perinuclear space as expected for a secreted protein, giving the stain a punctate appearance. We also detect cyc mRNA around the nuclei of the YSL (Fig. 4A, arrows). This demonstrates that the YSL is a source of both Sqt and Cyc signals that could act upon the overlying blastomeres.
FIG. 4. Expression of components of the Nodal-signaling pathway.
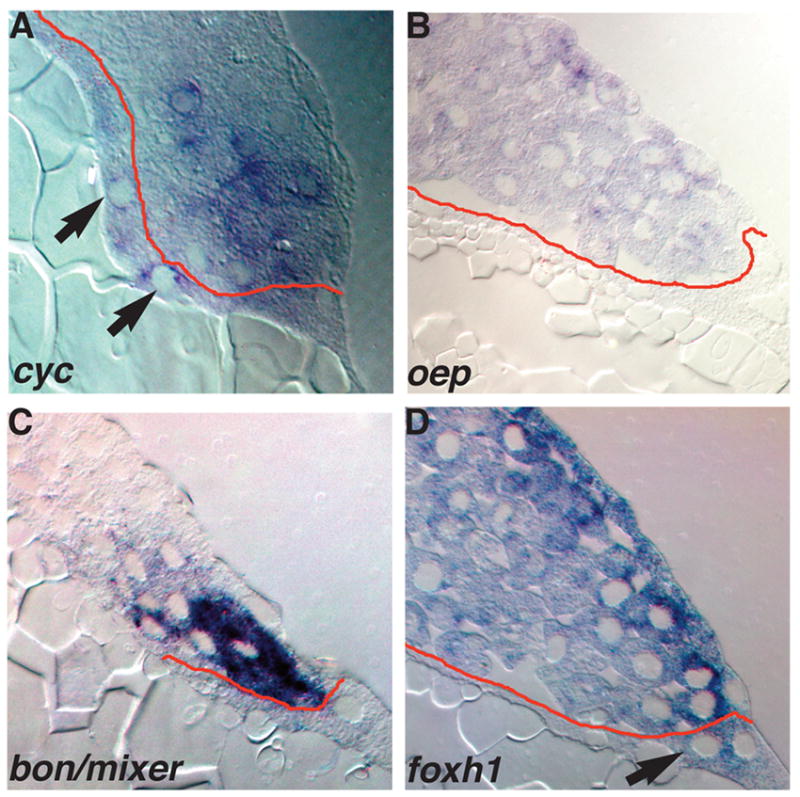
Sections of wild type embryos at 5 hpf (A) or 4.3 hpf (B–D) embryos stained for cyc (A), oep (B), bon/mixer (C) or foxh1 mRNA (D). (A) In 5 hpf embryos, cyc transcripts are distributed in a punctate pattern in marginal blastomeres within 5 rows of the YSL, and are also detected in the YSL (arrows). (B) oep is expressed in all blastomeres and EVL cells, but transcripts are excluded from the YSL. (C) bon/mixer is expressed exclusively in the blastomeres within 3–4 rows of the YSL. bon/mixer mRNA is not detected in the YSL or EVL. (D) foxh1 transcripts are found throughout the embryo, including all blastomeres and the YSL. foxh1 levels are elevated in the EVL and the cells immediately underneath the EVL.
Our results also suggest the possibility that Nodal signals in the blastomeres signal to the EVL and, perhaps, to the YSL. If so, then components of the Nodal signal transduction pathway should be expressed in the YSL and EVL. Therefore, we examined expression of the Oep co-receptor, and the FoxH1 and Bon/Mixer transcription factors in sections of mid-blastula stage (4hpf to 4.3 hpf) embryos. Despite the fact that maternal and zygotic oep transcripts are ubiquitously distributed in early embryos, oep mRNA is excluded from the YSL by 4hpf (Fig. 4B) (Zhang et al., 1998). Interestingly, oep transcripts are found in all EVL cells (Fig. 4B). By contrast, bon/mixer transcripts are excluded from the EVL and YSL at 4.3 hpf (Fig. 4C). bon/mixer mRNA is found exclusively in the marginal blastomeres, within five tiers of the YSL (Fig. 4C). At the same stage, foxh1 is expressed in all blastomeres as well as the YSL (Fig. 4D, arrow). Levels of foxh1 transcripts are elevated in the EVL and the layer of blastomeres immediately beneath the EVL (Fig. 4D). foxh1 is also expressed at lower levels in the interior blastomeres (Fig. 4D). We do not know if this difference in expression levels has functional significance, or if it reflects the greater accessibility of the superficial cells to the reagents for in situ hybridization. Nonetheless, the expression of Oep and FoxH1 in the EVL indicates that these cells are competent to respond to Nodal signals. Although the lack of oep transcripts in the YSL suggests that this tissue is not competent to respond to Nodal signals, it remains possible that Oep protein is produced from transcripts localized at earlier stages.
Sqt and Cyc signals in the YSL are required to pattern the embryo
To test if sqt and cyc are required in the YSL, we specifically knocked down their function in this tissue by targeted injection of antisense morpholinos (MOs) directed against sqt and cyc transcripts. Translation-blocking MOs against sqt and cyc have been described previously (Feldman and Stemple, 2001; Karlen and Rebagliati, 2001). Experiments with fluorescently labeled MOs showed that they act exclusively in a cell autonomous manner, since they do not diffuse across cell membranes (Amack and Yost, 2004). To confirm this, we examined the distribution of a lissamine-tagged control MO in live embryos two hours and 24 hours after injection at 3hpf (Fig. 5A, B). After 2 hours, the MO is distributed in a ring underneath the blastomeres, indicating that the MO is restricted to the YSL at this stage (Fig. 5A). An identical distribution was observed when fluorescently-tagged Dextran injected into the YSL (Feldman et al., 1998). At 24hpf, the fluorescence is still restricted to the yolk, demonstrating that the MO does not diffuse across the membrane into the blastoderm (Fig. 5B). Next, we injected 8ng each of fluorescently-tagged MOs against sqt and cyc, or mismatched controls, directly into the YSL soon after it forms at 3hpf. Embryos injected with both control MOs have normal embryonic shields at 6hpf (100%, N=24) (Fig. 5C, arrow), and develop a normal body axis at 24 hpf, with a notochord, somites and heart (100%, N=18) (Fig. 5D). When cyc function alone is depleted from the YSL, all embryos have normal shields (100%, N=36) (Fig. 5E, arrow). At 24hpf, the cyc-depleted embryos are indistinguishable from embryos injected with control MOs (100%, N=22) (Fig. 5F). When sqt function is depleted from the YSL, by contrast, the majority of embryos lack the embryonic shield (54%, N=11) (Fig. 5G). Previous results demonstrated that the organizer does not form in embryos lacking zygotic sqt function (Dougan et al., 2003; Feldman et al., 1998). Our results extend these previous studies by demonstrating that sqt function is required in the YSL to induce the morphological shield.
FIG. 5. sqt and cyc have partially overlapping roles in the YSL.
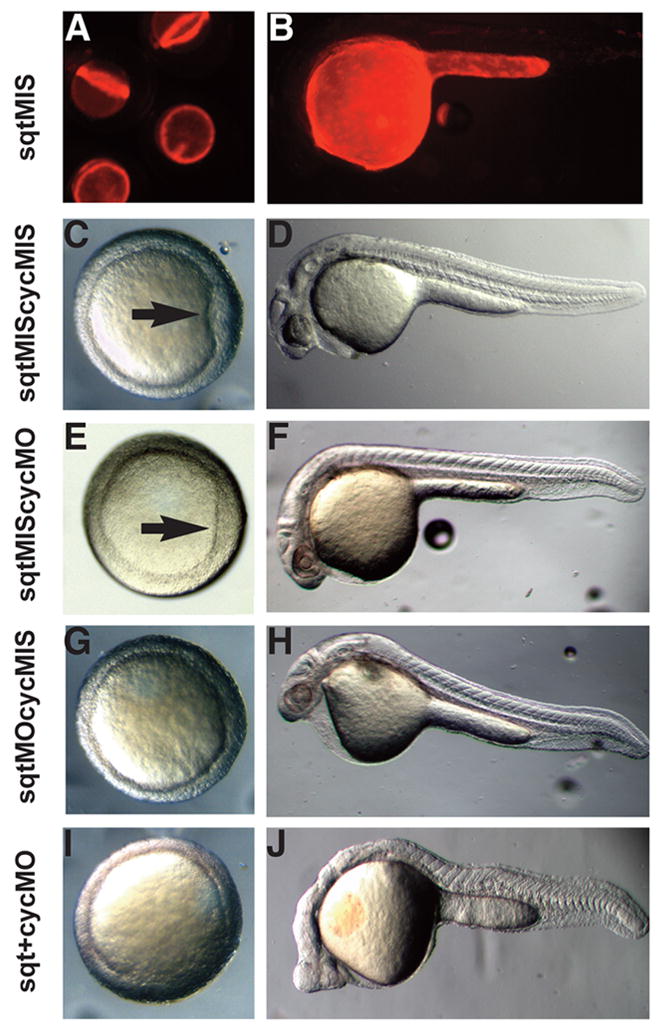
Images of live 6 hpf (A, C, E, G) or 24 hpf (B, D, F, H) embryos after targeted co-injection into the YSL of sqtMO and cycMO, or appropriate 5 bp mismatched control MOs. After co-injection of sqt and cyc mismatched MOs, the embryonic shield forms normally (A, arrow) and the body axis is indistinguishable from wild type (B). After co-injection of the sqtMIS MO and cycMO, the embryonic shield forms (C, arrow) and the body axis is normal (D). Embryonic shields do not form in many embryos following injection of the sqtMO and cycMIS MO (E) The majority of these embryos appear normal at 24 hpf (F), but many have reduced cardiac tissue. (G, H) After co-injection of sqtMO and cycMO, the vast majority of embryos lack embryonic shields (G, arrow). At 24 hpf, these embryos display severe cyclopia and have reduced notochord due to defects in axial mesoderm patterning (H). Paraxial mesoderm is less affected, as somites form in embryos co-injected with sqtMO and cycMO (H).
At 24 hpf, a distinct minority of embryos lacking sqt function in the YSL displayed mild cyclopia, indicating that a substantial fraction of the defective embryos at 6hpf have recovered (16%, N=54) (Fig. 5H). This is consistent with the observation that sqt mutants recover during gastrulation in a cyc-dependent manner (Dougan et al., 2003; Feldman et al., 1998; Hagos and Dougan, 2007). This raised the possibility that Cyc signals in the YSL could compensate for the loss of Sqt in the YSL. To test this, we simultaneously depleted both sqt and cyc function from the YSL. All of these embryos had reduced or missing shields at 6hpf (100%, N=23) (Fig. 5I) and the majority lack gsc expression at 5hpf, indicating dorsal mesoderm is not specified at this stage (Fig. 6B, 71%, N=28). These embryos also lack endoderm, as indicated by the absence mezzo and sox17 expression (Fig. 6E–H) (Alexander and Stainier, 1999; Poulain and Lepage, 2002). By 24hpf, the majority of these embryos display severe cyclopia and have narrow notochords (70%, N=20) (Fig. 5J). Consistent with this, flh expression is reduced during gastrulation (Fig. 6D) (Talbot et al., 1995). These results demonstrate that sqt and cyc have partially overlapping functions in the YSL and are required to induce the organizer and head mesoderm and endoderm.
FIG. 6. Sqt and Cyc in the YSL are required for endoderm and head mesoderm.
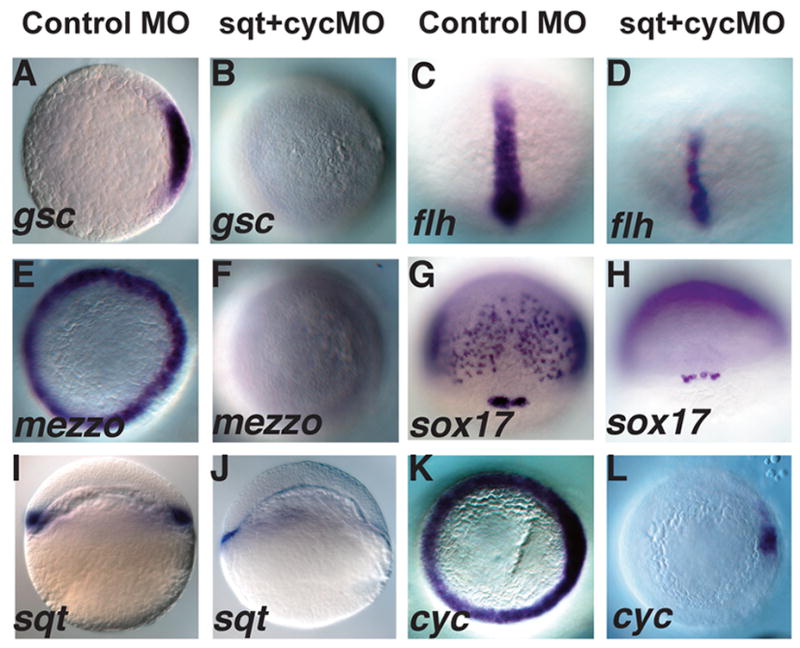
Embryos were injected with sqt and cyc mismatch MOs (A, C, E, G, I, K) or sqt and cyc MOs (B, D, F, H, J, L) and processed for in situ hybridization for mesoderm and endoderm marker genes. Control MOs do not affect expression of gsc (A), flh (C), mezzo (E), sox17 (G), sqt (I) or cyc (K). (B) By contrast, gsc expression is eliminated when sqt and cyc MOs are co-injected into the YSL. (D) flh expression is reduced, but not eliminated in these embryos. (F) These embryos also completely lack mezzo expression. (H) sox17 expression is eliminated from the endoderm progenitors, but not from the dorsal forerunners. (J) Extremely low levels of sqt expression are apparent when Sqt and Cyc signals are depleted from the YSL. (L) cyc expression is greatly reduced, except for a small patch of expression in the presumed dorsal blastomeres. In A–D; E–F, I–L, dorsal is to the right, when apparent.
Sqt and Cyc signals in the YSL are required to induce nodal-related gene expression
Our analysis of sqtGFP transgenes suggested that Nodal signals in the YSL induce or maintain sqt expression in the overlying blastomeres via the conserved NRE in the first intron. To test this, we asked if Nodal signals in the YSL are required for sqt and cyc expression in the embryo. When the two genes are simultaneously depleted from the YSL, expression of sqt (93%, N=55) (Fig. 6I, J) and cyc (63%, N=49) (Fig. 6K, L) are reduced in the majority of the embryos. Low levels of sqt expression are detected in these embryos, but these transcripts are located predominantly in the YSL (Fig. 6J), where the MO prevents translation of sqt mRNA. Since we did not detect sqt transcripts in the marginal blastomeres or in the EVL, this indicates that endogenous sqt, like our transgene, is controlled by a different regulatory program in the YSL than in the blastomeres. cyc is expressed in a few marginal blastomeres when Nodal signals are depleted from the YSL (Fig. 6L). Although the dorso-ventral axis is not apparent at this stage, we have previously demonstrated that dorsal expression of cyc is independent of earlier Nodal signals (Hagos et al., in press). Therefore in these experiments, it is likely that cyc is expressed in dorsal blastomeres. These domains of sqt and cyc expression account for the formation of trunk mesoderm in embryos lacking extra-embryonic Nodal signals. These results demonstrate that Nodal signals from the YSL are required for nodal-related gene expression in the overlying blastomeres.
Discussion
In this work, we present the first analysis of the regulatory elements that control nodal-related gene expression in zebrafish. Zygotic sqt is expressed in three independent phases. We identified a 1.9kb region of DNA that is sufficient to drive reporter gene expression in the endogenous sqt spatio-temporal expression pattern. This artificial promoter is comprised of three fragments from non-contiguous regions in the endogenous gene, including a 431bp distal element located 9.4kb upstream of the sqt transcription start site, the 923bp directly upstream of the sqt transcription start site and 603bp of the first intron. Since these sequences contain the cis-acting elements necessary to drive normal expression of sqt, we analyzed the roles of these sequences with the goal of better understanding the molecular mechanisms that control sqt expression in each of its phases. Although our transgenes accurately reflect many aspects of the endogenous sqt expression pattern, gfp expression behaved differently than endogenous sqt in some experiments. In the absence of Nodal signaling, for example, gfp expression persists in the YSL at a stage when endogenous sqt is not detected (Fig. 3G) (Meno et al., 1999). It is likely that this is due greater sensitivity of the gfp probe, compared to the sqt probe. Supporting this idea, in our transgenes we detect gfp expression in dorsal blastomeres as early as 2.5hpf, a half an hour before the earliest reported expression of sqt (3.0hpf) (data not shown) (Feldman et al., 1998). We cannot rule out the possibility, however, that our transgenes lack some elements necessary to fully recapitulate the endogenous pattern of sqt expression.
sqt is expressed in three independently controlled temporal phases
We found that the intron is not required for transgene expression in the dorsal blastomeres (Fig. 2F). This indicates that the sequences mediating the first phase of sqt expression are located in one of the two fragments upstream of the gfp coding region. Previous genetic analysis demonstrated that this expression depends on the dorsal determinant, β-catenin, but it is not known if sqt is a direct target (Bellipanni et al., 2006; Dougan et al., 2003). We found at least three potential binding sites for the β-catenin binding partner, Lef1/Tcf in the 923bp upstream of the transcription start site, and an additional site in the distal fragment (data not shown) (Dorsky et al., 2002). Although we did not perform deletion analysis of these elements, our data is consistent with the idea that sqt is a direct target of β-catenin. The element driving sqt expression in the dorsal forerunners during gastrulation must also be present in the sequences upstream of the gfp coding region, since all of our transgenes expressed gfp in the dorsal forerunners (Fig. 2E, I M). This enhancer remains to be identified.
Little is known about the factors that control expression of sqt at the margin during the blastula stages. This phase of sqt expression is particularly important, since studies conditionally inactivating the Nodal receptors show that Nodal signals are most active in patterning the germ layers during the mid-to-late blastula stages (3.5-5hpf) (Hagos and Dougan, 2007). Expression at this stage is independent of the earlier expression of sqt in the dorsal blastomeres, since depletion of β-catenin eliminates the early dorsal expression of sqt but does not effect sqt expression in the marginal ring (Bellipanni et al., 2006; Kelly et al., 2000). Our data indicates that proper expression of sqt at this stage involves the complex interaction of three different cell types.
sqt is expressed independently in three tissues in the margin
sqt is expressed in three distinct marginal tissues, including the marginal blastomeres and the extra-embryonic YSL and EVL (Erter et al., 1998; Feldman et al., 1998; Rebagliati et al., 1998a). Our data demonstrates that expression in each tissue is controlled by separable elements. Firstly, expression in the blastomeres is mediated by a conserved Nodal response element (NRE) within the first sqt intron (Fig. 3H). gfp expression in the blastomeres depends upon Nodal signaling (Fig. 3G), and the response to activation of the Nodal pathway in the blastomeres is mediated entirely by sequences in the first intron (Fig. 3F). Similarly, expression of the transgenes and endogenous sqt in the EVL depend upon Nodal signaling (Fig. 3G, 6J). Surprisingly, intron sequences are dispensable for expression in this tissue (Fig. 3B). This indicates that an element in one of the two fragments upstream of gfp is capable of mediating the response to Nodal signals in the EVL. Consistent with this idea, there are three FoxH1 consensus sites in the 923bp directly upstream of gfp in our transgenes (X. Fan and S. Dougan, unpublished observations). These sites could mediate the EVL response, but interestingly, they are not capable of mediating the response to TARAM-D in blastomeres (Fig. 3F). This indicates that there are significant functional differences between the FoxH1 sites, but it remains to be determined why the intron NRE mediates expression in the blastomeres and the upstream FoxH1 sites do not. The other known effector of the Nodal signaling pathway, Bon/Mixer, cannot mediate the response to Nodal signals in the EVL since it is not expressed in this tissue (Fig. 4C).
Finally, expression in the YSL is controlled by sequences upstream of gfp, but it is not clear if these sequences mediate a response to the Nodal-signaling pathway in the YSL like they do in the EVL (Fig. 3B, G). When Nodal signaling is reduced, expression of gfp in the blastomeres is reduced to a much greater extent than in the YSL (Fig. 3G). Similarly when embryos are depleted of extra-embryonic Nodal signals, endogenous sqt persists in the YSL after expression in the blastomeres and EVL is no longer detected (Fig. 6J). Both gfp and sqt are eventually lost from the YSL when Nodal signals are blocked, although gfp expression persists longer than sqt expression. The loss of gfp and sqt from the YSL could be due to decreased expression of an inducer of nodal-related gene expression, or to increased expression of an inhibitor.
Unlike most other nodal-related genes, sqt is not expressed in the left lateral plate mesoderm (LLPM) after gastrulation (Erter et al., 1998; Feldman et al., 1998; Rebagliati et al., 1998a). Despite this, the sqt NRE is similar to the intronic asymmetric enhancer (ASE) that drives expression in the LLPM of ascidians, frogs and mammals (Fig. 2N) (Hyde and Old, 2000; Osada et al., 2000). This suggests that the sqt NRE lacks critical sequences necessary for asymmetric expression. The ASE is comprised of two FoxH1 sites flanking a conserved core “TTG(G/C)CCA” motif (Osada et al., 2000). The sqt NRE contains two FoxH1 consensus sites, but lacks the conserved core (Fig. 2A). This raises the possibility that the TTG(G/C)CCA sequence binds the transcription factor complex that drives expression in the LLPM. Since we have not directly tested this idea, however, it remains possible that the sqt NRE lacks other sequences that drive asymmetric expression.
Nodal signals mediate interactions between embryonic and extra-embryonic tissues
Nodal signals are required during the mid-to-late blastula stages, a period of rapid cell division and cell intermixing (Hagos and Dougan, 2007). This raises the question of how a stable zone of Nodal signaling is maintained within such a dynamic cell population. We present several lines of evidence indicating that the YSL is a source of Nodal signals that are independent of the population of overlying blastomeres. First, expression of the transgene in the YSL is controlled by a different element than the one that controls expression in the blastomeres. Second, expression of a sqt transgene in the blastomeres is reduced when Nodal signaling is blocked with SB-505124, but expression in the YSL persists (Fig. 3G). Third, cyc is expressed in the YSL (Fig. 4A), along with sqt (Erter et al., 1998; Feldman et al., 1998). Finally, sqt and cyc expression in the blastomeres depends, in part, upon Nodal signaling in the YSL (Figs. 6I–L). Depletion of Sqt and Cyc signals in the YSL causes an overall reduction in Nodal signaling levels in the entire embryo. This in turn, results in a loss of endoderm and dorsal mesoderm (Figs. 5 and Fig. 6). Thus the YSL is a stable source of Nodal signals that act to impose a reproducible pattern upon a dynamic population of overlying blastomeres. The YSL must express other essential signals, however, since the defects resulting from degrading all mRNA in the YSL are much more severe than those from specific depletion of Sqt and Cyc (Chen and Kimelman, 2000). Nodal signals in the EVL may also act to stabilize expression in the blastomeres, but with the present technology it is not possible to deplete sqt and cyc function specifically in the EVL.
Our results suggest a model in which sqt expression in the margin is induced in the YSL by maternal transcription factors acting on an element upstream of the sqt transcription start site. Although we have not analyzed the cyc promoter, genetic data presented here and in previous studies indicates that cyc expression in the YSL must be induced independently of sqt function, otherwise Cyc signals could not compensate for the depletion of Sqt in the YSL (Fig. 5) (Dougan et al., 2003; Feldman et al., 1998). Sqt and Cyc signals from the YSL subsequently induce sqt expression in the blastomeres, acting through the NRE in the first intron (Fig. 7A). Sqt and Cyc signals also act through an upstream response element to induce sqt expression in the EVL (Fig 7A). We do not know if Nodal signals from the YSL can directly induce sqt expression in the EVL, or if Nodal signals from the blastomeres are required for sqt expression in the EVL. We expect that both tissues can act as sources, however, given the proximity of the EVL cells to the YSL and marginal blastomeres (Fig. 7A). Further experiments are necessary to determine if similar mechanisms control expression of cyc in these tissues.
FIG. 7. Conserved roles of extra-embryonic Nodal signaling in teleosts and mammals.
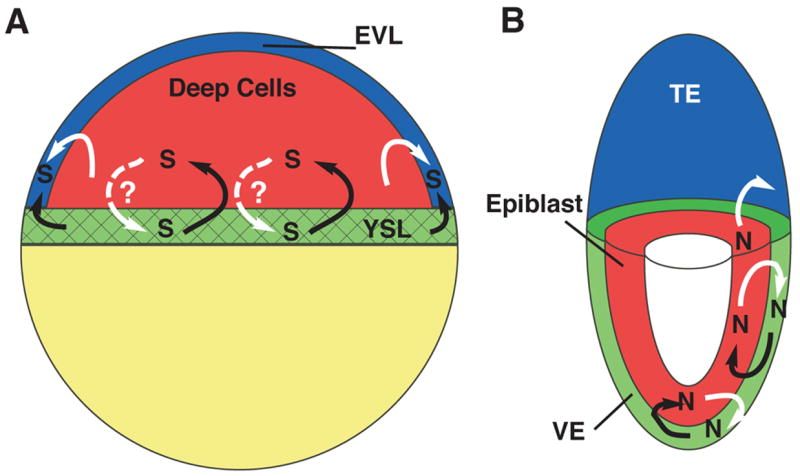
(A) Schematic of a blastula stage zebrafish embryo. Sqt signals (S) in the YSL (green, black arrows) induces sqt and cyc expression in the blastomeres (red). Sqt and Cyc in the YSL and/or the blastomeres induce expression in the EVL (blue). It is not clear if Nodal signals in the blastomeres induce or maintain expression in the YSL (dashed white arrows). (B) Schematic of a 6.0d mouse embryo. Nodal signals (N) in the visceral endoderm (green, black arrows) induce Nodal expression in the epiblast (red). Nodal signals in the epiblast (white arrows) maintain nodal expression in the visceral endoderm and pattern the extra-embryonic ectoderm, also known as the trophectoderm (blue). We have diagrammed these signaling interactions in the 6.0d mouse embryo for simplicity, but it is not clear when they occur during normal development. Black arrows indicate Nodal signaling from extra-embryonic tissues; White arrows indicate Nodal signaling from embryonic tissues. Solid lines depict interactions that have been demonstrated by experimental data; Dashed lines depict possible interactions. YSL=yolk syncytial layer; EVL=Enveloping Layer; VE=visceral endoderm; TE=trophectoderm.
The interactions between the YSL, EVL and marginal blastomeres serve to create a stable source of Nodal signaling at the margin despite the rapid cell divisions and intermixing that occurs during the blastula stages (Kimmel and Law, 1985; Kimmel and Warga, 1987). If a cell moves away from the margin beyond the range of Nodal signals, then it will stop expressing sqt or cyc. By contrast, sqt and cyc expression will be induced if a cell moves to a position within the range of Nodal signals at the margin. Since nodal-related gene expression is not simultaneously induced in the YSL and overlying blastomeres, there is a temporal gradient of sqt expression along the animal-vegetal axis. Cells that remain close to the YSL express sqt for a longer period than cells located farther from the margin. The length of time cells express sqt and cyc could have profound consequences on their eventual cell fate choice.
The conserved roles of teleost and mammalian extra-embryonic tissues
The interaction between embryonic and extra-embryonic tissues in zebrafish is remarkably similar to those previously described in other vertebrates. Previous groups have recognized that the mammalian visceral endoderm, teleost YSL and chick hypoblast each express orthologues of the Hex transcription factor (Ho et al., 1999; Martinez Barbera et al., 2000; Yatskievych et al., 1999). This has led to the suggestion that these tissues share a common evolutionary origin (Ho et al., 1999). We have extended this observation in several ways. First, we demonstrated that Nodal signals in the zebrafish YSL are required for head mesoderm and endoderm (Fig. 5, 6). Similarly, chimeric mutant mice lacking nodal function in the visceral endoderm have defects in the prechordal plate, which disrupts anterior neural development (Varlet et al., 1997). Thus, Nodal signals from the visceral endoderm pattern the epiblast (Fig. 7B). Secondly, in both species a conserved intronic NRE mediates the response to Nodal signals in the embryo (Fig. 2, 3) (Brennan et al., 2001). In the mouse, however, other factors can also induce expression in the epiblast, since expression of the transgene is reduced in the epiblast, but not completely eliminated, in the absence of the NRE (Brennan et al., 2001). Thus, Nodal signals in the YSL act by the same mechanism as those in the visceral endoderm to induce the prechordal plate. This strengthens the argument that the tissues share a common evolutionary origin. The fact that gfp expression in the YSL persists longer than it does in the blastomeres when Nodal signals are blocked suggests that expression of sqt in the YSL does not directly depend upon Nodal signaling (Fig. 3G). This idea is strengthened by our observation that sqt expression persists in the YSL, but not in blastomeres, when extra-embryonic Nodal signals are depleted (Fig. 6J).
One major difference between the species is that in the mouse, Nodal signals from the epiblast induce nodal expression in the visceral endoderm (Fig. 7B) (Brennan et al., 2001). By contrast, it is not clear if the teleost YSL responds to Nodal signals from the blastomeres. The Nodal effector, FoxH1 is expressed in the YSL, but oep mRNA is excluded from the YSL from an early stage (Fig. 4B, D). Since maternal oep is ubiquitously expressed, this suggests that maternal oep transcripts are rapidly cleared out of the YSL (Zhang et al., 1998). Injecting oep MOs into the YSL produced no phenotype (E. Hagos, B. Xu, R. Burdine, S. Dougan, unpublished observations). It remains possible, however, that Oep protein is still present, or that Nodal signals in the blastomeres induce other signals that act in an Oep-independent manner to maintain nodal-related gene expression in the YSL.
Our results also reveal striking parallels between the teleost EVL and the mammalian extra-embryonic ectoderm. In mice, the extra-embryonic ectoderm is derived from the trophectoderm, the outer layer of cells that form during compaction (Rossant, 2004). Similarly, the zebrafish EVL is an extra-embryonic tissue that forms during the cleavage stages and encases the cells that produce the embryo (Bouvet, 1976; Kimmel et al., 1990). In the mouse, Nodal signals in the epiblast pattern the extra-embryonic ectoderm by a Smad2-independent mechanism, although nodal itself does not appear to be expressed in this tissue (Fig. 7B) (Brennan et al., 2001). Several lines of evidence indicate that the zebrafish EVL is also patterned by Nodal signals. First, EVL cells are competent to respond to Nodal signals since they express the Nodal co-receptor, Oep, and the FoxH1 transcription factor (Fig. 4B, D). Second, both reporter gene expression and endogenous sqt expression are reduced in the EVL when Nodal signals are blocked by SB-505124 treatment or by loss of extra-embryonic Nodal signals (Fig. 3G; 6J). This indicates that the EVL cells require Nodal signals, like the mouse extra-embyronic ectoderm. Finally, the expansion of reporter gene expression in the EVL does not depend upon intron sequences (Fig. 3F). This demonstrates that the response to Nodal signals is mediated by a different mechanism in the EVL than in the blastomeres. We have not yet determined, however, if expression in the EVL is independent of Smad2 function. Nonetheless, our data indicate that in the zebrafish, as in the mouse, the Nodal pathway acts by different mechanisms to pattern different tissues. These parallels raise the possibility that the EVL and extra-embryonic ectoderm share a common evolutionary origin. Further gene expression analysis and functional tests are required to test if this is the case.
The evolutionary role of extra-embryonic tissues
The critical role ascribed to extra-embryonic tissues in patterning the embryo is unique to vertebrates. The tissues that respond to extra-embryonic Nodal signals are characterized by rapid proliferation, extensive cell movements and intermixing. In the zebrafish, the intermixing of the blastomeres during the blastula period has been well documented (Kimmel and Law, 1985; Kimmel and Warga, 1987). Cell movements are more extreme in the killifish blastula. In this species, the deep cells that form the embryo migrate in apparently random directions for hundreds of microns during the blastula period, when they respond to patterning signals from the YSL (Oppenheimer, 1934; Trinkaus, 1973). Similarly, the mouse epiblast is also a dynamic tissue (Tam et al., 1993; Varlet et al., 1997). In contrast to vertebrates, invertebrate chordates, such as amphioxus, completely lack extra-embryonic cells (Tung et al., 1960). The ascidian Ciona intestinalis does contain extra-embryonic cells, called test cells, but these have a protective function and are not thought to pattern early embryos (Sato and Morisawa, 1999). Although both ascidians and amphioxus contain migratory cell populations, cells in these embryos do not undergo the extensive intermixing that characterizes many vertebrate tissues (Holland et al., 1996; Jeffery et al., 2004). We suggest that the evolution of extra-embryonic sources of patterning signals freed cells to move and intermix to a greater extent than previously possible. This, in turn, may have increased the regulative properties of the embryo, permitting it to restore tissues following insult or injury. Supporting this idea, Xenopus laevis embryos do not have extra-embryonic tissues and do not undergo extensive intermixing during the blastula stages (Dale and Slack, 1987). Thus, to a first approximation, the ability of cells to move with respect to the source of Nodal signals correlates with the presence of extra-embryonic tissues.
Acknowledgments
We thank David Kimelman and Chris Bjornson helpful discussions and sharing reagents prior to publication; Will Talbot, Linda Holland and members of the Dougan and Lauderdale labs for helpful discussions and critical comments on the manuscript; Mary Ard for sectioning embryos; Chari Jefferson and Tabitha West for fish care. S. T. D. is a Georgia Cancer Coalition Distinguished Investigator. Work in the Dougan lab was supported by the Georgia Cancer Coalition and the American Cancer Society (RSG DDC-112979). R. D. B is the 44th Mallinckrodt Scholar of the Edward Mallinckrodt Junior Foundation. Work in the Burdine lab was assisted by grant #04-2405-CCR-E0 from the New Jersey State Commission on Cancer Research. Work in the Kawakami laboratory is supported by the NIH /NIGMS (R01 GM069382).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Adachi H, Saijoh Y, Mochida K, Ohishi S, Hashiguchi H, Hirao A, Hamada H. Determination of left/right asymmetric expression of nodal by a left side-specific enhancer with sequence similarity to a lefty-2 enhancer. Genes Dev. 1999;13:1589–600. doi: 10.1101/gad.13.12.1589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexander J, Stainier DY. A molecular pathway leading to endoderm formation in zebrafish. Curr Biol. 1999;9:1147–57. doi: 10.1016/S0960-9822(00)80016-0. [DOI] [PubMed] [Google Scholar]
- Amack JD, Yost HJ. The T box transcription factor no tail in ciliated cells controls zebrafish left-right asymmetry. Curr Biol. 2004;14:685–90. doi: 10.1016/j.cub.2004.04.002. [DOI] [PubMed] [Google Scholar]
- Aoki TO, Mathieu J, Saint-Etienne L, Rebagliati MR, Peyrieras N, Rosa FM. Regulation of nodal signalling and mesendoderm formation by TARAM-A, a TGFbeta-related type I receptor. Dev Biol. 2002;241:273–88. doi: 10.1006/dbio.2001.0510. [DOI] [PubMed] [Google Scholar]
- Aparicio S, Chapman J, Stupka E, Putnam N, Chia JM, Dehal P, Christoffels A, Rash S, Hoon S, Smit A, Gelpke MD, Roach J, Oh T, Ho IY, Wong M, Detter C, Verhoef F, Predki P, Tay A, Lucas S, Richardson P, Smith SF, Clark MS, Edwards YJ, Doggett N, Zharkikh A, Tavtigian SV, Pruss D, Barnstead M, Evans C, Baden H, Powell J, Glusman G, Rowen L, Hood L, Tan YH, Elgar G, Hawkins T, Venkatesh B, Rokhsar D, Brenner S. Whole-genome shotgun assembly and analysis of the genome of Fugu rubripes. Science. 2002;297:1301–10. doi: 10.1126/science.1072104. [DOI] [PubMed] [Google Scholar]
- Beddington RS, Robertson EJ. Axis development and early asymmetry in mammals. Cell. 1999;96:195–209. doi: 10.1016/s0092-8674(00)80560-7. [DOI] [PubMed] [Google Scholar]
- Bellipanni G, Varga M, Maegawa S, Imai Y, Kelly C, Myers AP, Chu F, Talbot WS, Weinberg ES. Essential and opposing roles of zebrafish beta-catenins in the formation of dorsal axial structures and neurectoderm. Development. 2006;133:1299–309. doi: 10.1242/dev.02295. [DOI] [PubMed] [Google Scholar]
- Bouvet J. Enveloping layer and periderm of the trout embryo (Salmo trutta fario L.)20U. Cell Tissue Res. 1976;170:367–82. doi: 10.1007/BF00219418. [DOI] [PubMed] [Google Scholar]
- Brennan J, Lu CC, Norris DP, Rodriguez TA, Beddington RS, Robertson EJ. Nodal signalling in the epiblast patterns the early mouse embryo. Nature. 2001;411:965–9. doi: 10.1038/35082103. [DOI] [PubMed] [Google Scholar]
- Brennan J, Norris DP, Robertson EJ. Nodal activity in the node governs left-right asymmetry. Genes Dev. 2002;16:2339–44. doi: 10.1101/gad.1016202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S, Kimelman D. The role of the yolk syncytial layer in germ layer patterning in zebrafish. Development. 2000;127:4681–9. doi: 10.1242/dev.127.21.4681. [DOI] [PubMed] [Google Scholar]
- Cheng SK, Olale F, Bennett JT, Brivanlou AH, Schier AF. EGF-CFC proteins are essential coreceptors for the TGF-beta signals Vg1 and GDF1. Genes Dev. 2003;17:31–6. doi: 10.1101/gad.1041203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collignon J, Varlet I, Robertson EJ. Relationship between asymmetric nodal expression and the direction of embryonic turning. Nature. 1996;381:155–8. doi: 10.1038/381155a0. [DOI] [PubMed] [Google Scholar]
- Conlon FL, Lyons KM, Takaesu N, Barth KS, Kispert A, Herrmann B, Robertson EJ. A primary requirement for nodal in the formation and maintenance of the primitive streak in the mouse. Development. 1994;120:1919–28. doi: 10.1242/dev.120.7.1919. [DOI] [PubMed] [Google Scholar]
- Cormack BP, Valdivia RH, Falkow S. FACS-optimized mutants of the green fluorescent protein (GFP) Gene. 1996;173:33–8. doi: 10.1016/0378-1119(95)00685-0. [DOI] [PubMed] [Google Scholar]
- DaCosta Byfield S, Major C, Laping NJ, Roberts AB. SB-505124 is a selective inhibitor of transforming growth factor-beta type I receptors ALK4, ALK5, and ALK7. Mol Pharmacol. 2004;65:744–52. doi: 10.1124/mol.65.3.744. [DOI] [PubMed] [Google Scholar]
- Dale L, Slack JM. Fate map for the 32-cell stage of Xenopus laevis. Development. 1987;99:527–51. doi: 10.1242/dev.99.4.527. [DOI] [PubMed] [Google Scholar]
- Dorsky RI, Sheldahl LC, Moon RT. A transgenic Lef1/beta-catenin-dependent reporter is expressed in spatially restricted domains throughout zebrafish development. Dev Biol. 2002;241:229–37. doi: 10.1006/dbio.2001.0515. [DOI] [PubMed] [Google Scholar]
- Dougan ST, Warga RM, Kane DA, Schier AF, Talbot WS. The role of the zebrafish nodal-related genes squint and cyclops in patterning of mesendoderm. Development. 2003;130:1837–1851. doi: 10.1242/dev.00400. [DOI] [PubMed] [Google Scholar]
- Erter CE, Solnica-Krezel L, Wright CV. Zebrafish nodal-related 2 encodes an early mesendodermal inducer signaling from the extra-embryonic yolk syncytial layer. Dev Biol. 1998;204:361–72. doi: 10.1006/dbio.1998.9097. [DOI] [PubMed] [Google Scholar]
- Feldman B, Gates MA, Egan ES, Dougan ST, Rennebeck G, Sirotkin HI, Schier AF, Talbot WS. Zebrafish organizer development and germ-layer formation require nodal- related signals. Nature. 1998;395:181–5. doi: 10.1038/26013. [DOI] [PubMed] [Google Scholar]
- Feldman B, Stemple DL. Morpholino phenocopies of sqt, oep, and ntl mutations. Genesis. 2001;30:175–7. doi: 10.1002/gene.1058. [DOI] [PubMed] [Google Scholar]
- Gore AV, Maegawa S, Cheong A, Gilligan PC, Weinberg ES, Sampath K. The zebrafish dorsal axis is apparent at the four-cell stage. Nature. 2005;438:1030–5. doi: 10.1038/nature04184. [DOI] [PubMed] [Google Scholar]
- Gore AV, Sampath K. Localization of transcripts of the Zebrafish morphogen Squint is dependent on egg activation and the microtubule cytoskeleton. Mech Dev. 2002;112:153–6. doi: 10.1016/s0925-4773(01)00622-0. [DOI] [PubMed] [Google Scholar]
- Gritsman K, Zhang J, Cheng S, Heckscher E, Talbot WS, Schier AF. The EGF-CFC protein one-eyed pinhead is essential for nodal signaling. Cell. 1999;97:121–32. doi: 10.1016/s0092-8674(00)80720-5. [DOI] [PubMed] [Google Scholar]
- Hagos EG, Dougan ST. Time-dependent patterning of the mesoderm and endoderm by Nodal signals in zebrafish. BMC Dev Biol. 2007;7:22. doi: 10.1186/1471-213X-7-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagos EG, Fan X, Dougan ST. The role of maternal Activin-like signals in zebrafish embryos. Dev Biol. doi: 10.1016/j.ydbio.2007.07.010. In Press. [DOI] [PubMed] [Google Scholar]
- Hatta K, Kimmel CB, Ho RK, Walker C. The cyclops mutation blocks specification of the floor plate of the zebrafish central nervous system. Nature. 1991;350:339–41. doi: 10.1038/350339a0. [DOI] [PubMed] [Google Scholar]
- Heisenberg CP, Nusslein-Volhard C. The function of silberblick in the positioning of the eye anlage in the zebrafish embryo. Dev Biol. 1997;184:85–94. doi: 10.1006/dbio.1997.8511. [DOI] [PubMed] [Google Scholar]
- Ho CY, Houart C, Wilson SW, Stainier DY. A role for the extra-embryonic yolk syncytial layer in patterning the zebrafish embryo suggested by properties of the hex gene. Curr Biol. 1999;9:1131–4. doi: 10.1016/s0960-9822(99)80485-0. [DOI] [PubMed] [Google Scholar]
- Holland ND, Panganiban G, Henyey EL, Holland LZ. Sequence and developmental expression of AmphiDll, an amphioxus Distal-less gene transcribed in the ectoderm, epidermis and nervous system: insights into evolution of craniate forebrain and neural crest. Development. 1996;122:2911–20. doi: 10.1242/dev.122.9.2911. [DOI] [PubMed] [Google Scholar]
- Hsiao CD, Hsieh FJ, Tsai HJ. Enhanced expression and stable transmission of transgenes flanked by inverted terminal repeats from adeno-associated virus in zebrafish. Dev Dyn. 2001;220:323–36. doi: 10.1002/dvdy.1113. [DOI] [PubMed] [Google Scholar]
- Hyde CE, Old RW. Regulation of the early expression of the Xenopus nodal-related 1 gene, Xnr1. Development. 2000;127:1221–9. doi: 10.1242/dev.127.6.1221. [DOI] [PubMed] [Google Scholar]
- Jaillon O, Aury JM, Brunet F, Petit JL, Stange-Thomann N, Mauceli E, Bouneau L, Fischer C, Ozouf-Costaz C, Bernot A, Nicaud S, Jaffe D, Fisher S, Lutfalla G, Dossat C, Segurens B, Dasilva C, Salanoubat M, Levy M, Boudet N, Castellano S, Anthouard V, Jubin C, Castelli V, Katinka M, Vacherie B, Biemont C, Skalli Z, Cattolico L, Poulain J, De Berardinis V, Cruaud C, Duprat S, Brottier P, Coutanceau JP, Gouzy J, Parra G, Lardier G, Chapple C, McKernan KJ, McEwan P, Bosak S, Kellis M, Volff JN, Guigo R, Zody MC, Mesirov J, Lindblad-Toh K, Birren B, Nusbaum C, Kahn D, Robinson-Rechavi M, Laudet V, Schachter V, Quetier F, Saurin W, Scarpelli C, Wincker P, Lander ES, Weissenbach J, Roest Crollius H. Genome duplication in the teleost fish Tetraodon nigroviridis reveals the early vertebrate proto-karyotype. Nature. 2004;431:946–57. doi: 10.1038/nature03025. [DOI] [PubMed] [Google Scholar]
- Jeffery WR, Strickler AG, Yamamoto Y. Migratory neural crest-like cells form body pigmentation in a urochordate embryo. Nature. 2004;431:696–9. doi: 10.1038/nature02975. [DOI] [PubMed] [Google Scholar]
- Karlen S, Rebagliati M. A morpholino phenocopy of the cyclops mutation. Genesis. 2001;30:126–8. doi: 10.1002/gene.1046. [DOI] [PubMed] [Google Scholar]
- Kawakami K, Takeda H, Kawakami N, Kobayashi M, Matsuda N, Mishina M. A transposon-mediated gene trap approach identifies developmentally regulated genes in zebrafish. Dev Cell. 2004;7:133–44. doi: 10.1016/j.devcel.2004.06.005. [DOI] [PubMed] [Google Scholar]
- Kelly C, Chin AJ, Leatherman JL, Kozlowski DJ, Weinberg ES. Maternally controlled (beta)-catenin-mediated signaling is required for organizer formation in the zebrafish. Development. 2000;127:3899–911. doi: 10.1242/dev.127.18.3899. [DOI] [PubMed] [Google Scholar]
- Kimmel CB, Ballard WW, Kimmel SR, Ullmann B, Schilling TF. Stages of embryonic development of the zebrafish. Dev Dyn. 1995;203:253–310. doi: 10.1002/aja.1002030302. [DOI] [PubMed] [Google Scholar]
- Kimmel CB, Law RD. Cell lineage of zebrafish blastomeres. III. Clonal analyses of the blastula and gastrula stages. Dev Biol. 1985;108:94–101. doi: 10.1016/0012-1606(85)90012-0. [DOI] [PubMed] [Google Scholar]
- Kimmel CB, Warga RM. Indeterminate cell lineage of the zebrafish embryo. Dev Biol. 1987;124:269–80. doi: 10.1016/0012-1606(87)90478-7. [DOI] [PubMed] [Google Scholar]
- Kimmel CB, Warga RM, Schilling TF. Origin and organization of the zebrafish fate map. Development. 1990;108:581–94. doi: 10.1242/dev.108.4.581. [DOI] [PubMed] [Google Scholar]
- Kunwar PS, Zimmerman S, Bennett JT, Chen Y, Whitman M, Schier AF. Mixer/Bon and FoxH1/Sur have overlapping and divergent roles in Nodal signaling and mesendoderm induction. Development. 2003;130:5589–99. doi: 10.1242/dev.00803. [DOI] [PubMed] [Google Scholar]
- Lakowski J, Majumder A, Lauderdale JD. Mechanisms controlling Pax6 isoform expression in the retina have been conserved between teleosts and mammals. Dev Biol. 2007 doi: 10.1016/j.ydbio.2007.04.015. [DOI] [PubMed] [Google Scholar]
- Lee EC, Yu D, Martinez de Velasco J, Tessarollo L, Swing DA, Court DL, Jenkins NA, Copeland NG. A highly efficient Escherichia coli-based chromosome engineering system adapted for recombinogenic targeting and subcloning of BAC DNA. Genomics. 2001;73:56–65. doi: 10.1006/geno.2000.6451. [DOI] [PubMed] [Google Scholar]
- Liu P, Jenkins NA, Copeland NG. A highly efficient recombineering-based method for generating conditional knockout mutations. Genome Res. 2003;13:476–84. doi: 10.1101/gr.749203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long S, Ahmad N, Rebagliati M. The zebrafish nodal-related gene southpaw is required for visceral and diencephalic left-right asymmetry. Development. 2003;130:2303–16. doi: 10.1242/dev.00436. [DOI] [PubMed] [Google Scholar]
- Martinez Barbera JP, Clements M, Thomas P, Rodriguez T, Meloy D, Kioussis D, Beddington RS. The homeobox gene Hex is required in definitive endodermal tissues for normal forebrain, liver and thyroid formation. Development. 2000;127:2433–45. doi: 10.1242/dev.127.11.2433. [DOI] [PubMed] [Google Scholar]
- Massague J, Chen YG. Controlling TGF-beta signaling. Genes Dev. 2000;14:627–44. [PubMed] [Google Scholar]
- Meno C, Gritsman K, Ohishi S, Ohfuji Y, Heckscher E, Mochida K, Shimono A, Kondoh H, Talbot WS, Robertson EJ, Schier AF, Hamada H. Mouse Lefty2 and zebrafish antivin are feedback inhibitors of nodal signaling during vertebrate gastrulation. Mol Cell. 1999;4:287–98. doi: 10.1016/s1097-2765(00)80331-7. [DOI] [PubMed] [Google Scholar]
- Mizuno T, Yamaha E, Wakahara M, Kuroiwa A, Takeda H. Mesoderm induction in zebrafish. Nature. 1996;383:131–2. [Google Scholar]
- Norris DP, Brennan J, Bikoff EK, Robertson EJ. The Foxh1-dependent autoregulatory enhancer controls the level of Nodal signals in the mouse embryo. Development. 2002;129:3455–68. doi: 10.1242/dev.129.14.3455. [DOI] [PubMed] [Google Scholar]
- Oppenheimer JM. Experiments on Early Developing Stages of Fundulus. Proc Natl Acad Sci U S A. 1934;20:536–8. doi: 10.1073/pnas.20.9.536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osada SI, Saijoh Y, Frisch A, Yeo CY, Adachi H, Watanabe M, Whitman M, Hamada H, Wright CV. Activin/nodal responsiveness and asymmetric expression of a Xenopus nodal-related gene converge on a FAST-regulated module in intron 1. Development. 2000;127:2503–14. doi: 10.1242/dev.127.11.2503. [DOI] [PubMed] [Google Scholar]
- Poulain M, Lepage T. Mezzo, a paired-like homeobox protein is an immediate target of Nodal signalling and regulates endoderm specification in zebrafish. Development. 2002;129:4901–14. doi: 10.1242/dev.129.21.4901. [DOI] [PubMed] [Google Scholar]
- Rebagliati MR, Toyama R, Fricke C, Haffter P, Dawid IB. Zebrafish nodal-related genes are implicated in axial patterning and establishing left-right asymmetry. Dev Biol. 1998a;199:261–72. doi: 10.1006/dbio.1998.8935. [DOI] [PubMed] [Google Scholar]
- Rebagliati MR, Toyama R, Haffter P, Dawid IB. cyclops encodes a nodal-related factor involved in midline signaling. Proc Natl Acad Sci U S A. 1998b;95:9932–7. doi: 10.1073/pnas.95.17.9932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reissmann E, Jornvall H, Blokzijl A, Andersson O, Chang C, Minchiotti G, Persico MG, Ibanez CF, Brivanlou AH. The orphan receptor ALK7 and the Activin receptor ALK4 mediate signaling by Nodal proteins during vertebrate development. Genes Dev. 2001;15:2010–22. doi: 10.1101/gad.201801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renucci A, Lemarchandel V, Rosa F. An activated form of type I serine/threonine kinase receptor TARAM-A reveals a specific signalling pathway involved in fish head organiser formation. Development. 1996;122:3735–43. doi: 10.1242/dev.122.12.3735. [DOI] [PubMed] [Google Scholar]
- Rossant J. Lineage development and polar asymmetries in the peri-implantation mouse blastocyst. Semin Cell Dev Biol. 2004;15:573–81. doi: 10.1016/j.semcdb.2004.04.003. [DOI] [PubMed] [Google Scholar]
- Rossant J, Tam PP. Emerging asymmetry and embryonic patterning in early mouse development. Dev Cell. 2004;7:155–64. doi: 10.1016/j.devcel.2004.07.012. [DOI] [PubMed] [Google Scholar]
- Saijoh Y, Adachi H, Sakuma R, Yeo CY, Yashiro K, Watanabe M, Hashiguchi H, Mochida K, Ohishi S, Kawabata M, Miyazono K, Whitman M, Hamada H. Left-right asymmetric expression of lefty2 and nodal is induced by a signaling pathway that includes the transcription factor FAST2. Mol Cell. 2000;5:35–47. doi: 10.1016/s1097-2765(00)80401-3. [DOI] [PubMed] [Google Scholar]
- Saijoh Y, Oki S, Ohishi S, Hamada H. Left-right patterning of the mouse lateral plate requires nodal produced in the node. Dev Biol. 2003;256:161–73. doi: 10.1016/s0012-1606(02)00121-5. [DOI] [PubMed] [Google Scholar]
- Sampath K, Rubinstein AL, Cheng AM, Liang JO, Fekany K, Solnica-Krezel L, Korzh V, Halpern ME, Wright CV. Induction of the zebrafish ventral brain and floorplate requires cyclops/nodal signalling. Nature. 1998;395:185–9. doi: 10.1038/26020. [DOI] [PubMed] [Google Scholar]
- Sato Y, Morisawa M. Loss of test cells leads to the formation of new tunic surface cells and abnormal metamorphosis in larvae of Ciona intestinalis (Chordata, ascidiacea) Dev Genes Evol. 1999;209:592–600. doi: 10.1007/s004270050293. [DOI] [PubMed] [Google Scholar]
- Schier A. Nodal Signaling in Vertebrate Development. Annu Rev Cell Dev BIol. 2003;19:589–621. doi: 10.1146/annurev.cellbio.19.041603.094522. [DOI] [PubMed] [Google Scholar]
- Schier AF. Axis formation: squint comes into focus. Curr Biol. 2005;15:R1002–5. doi: 10.1016/j.cub.2005.11.051. [DOI] [PubMed] [Google Scholar]
- Schier AF, Talbot WS. Molecular Genetics of Axis Formation in Zebrafish. Annu Rev Genet. 2005 doi: 10.1146/annurev.genet.37.110801.143752. [DOI] [PubMed] [Google Scholar]
- Schulte-Merker S, van Eeden FJ, Halpern ME, Kimmel CB, Nusslein-Volhard C. no tail (ntl) is the zebrafish homologue of the mouse T (Brachyury) gene. Development. 1994;120:1009–15. doi: 10.1242/dev.120.4.1009. [DOI] [PubMed] [Google Scholar]
- Shimizu T, Yamanaka Y, Ryu SL, Hashimoto H, Yabe T, Hirata T, Bae YK, Hibi M, Hirano T. Cooperative roles of Bozozok/Dharma and Nodal-related proteins in the formation of the dorsal organizer in zebrafish. Mech Dev. 2000;91:293–303. doi: 10.1016/s0925-4773(99)00319-6. [DOI] [PubMed] [Google Scholar]
- Solnica-Krezel L. Pattern formation in zebrafish--fruitful liaisons between embryology and genetics. Curr Top Dev Biol. 1999;41:1–35. doi: 10.1016/s0070-2153(08)60268-9. [DOI] [PubMed] [Google Scholar]
- Stachel SE, Grunwald DJ, Myers PZ. Lithium perturbation and goosecoid expression identify a dorsal specification pathway in the pregastrula zebrafish. Development. 1993;117:1261–74. doi: 10.1242/dev.117.4.1261. [DOI] [PubMed] [Google Scholar]
- Stennard F. Xenopus differentiation: VegT gets specific. Curr Biol. 1998;8:R928–30. doi: 10.1016/s0960-9822(98)00012-8. [DOI] [PubMed] [Google Scholar]
- Talbot WS, Trevarrow B, Halpern ME, Melby AE, Farr G, Postlethwait JH, Jowett T, Kimmel CB, Kimelman D. A homeobox gene essential for zebrafish notochord development. Nature. 1995;378:150–7. doi: 10.1038/378150a0. [DOI] [PubMed] [Google Scholar]
- Tam PP, Williams EA, Chan WY. Gastrulation in the mouse embryo: ultrastructural and molecular aspects of germ layer morphogenesis. Microsc Res Tech. 1993;26:301–28. doi: 10.1002/jemt.1070260405. [DOI] [PubMed] [Google Scholar]
- Taylor JS, Van de Peer Y, Braasch I, Meyer A. Comparative genomics provides evidence for an ancient genome duplication event in fish. Philos Trans R Soc Lond B Biol Sci. 2001;356:1661–79. doi: 10.1098/rstb.2001.0975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trinkaus JP. Surface activity and locomotion of Fundulus deep cells during blastula and gastrula stages. Dev Biol. 1973;30:69–103. doi: 10.1016/0012-1606(73)90049-3. [DOI] [PubMed] [Google Scholar]
- Tung TC, Wu SC, Tung YY. The developmental potencies of the blastomere layers in Amphioxus egg at the 32-cell stage. Sci Sin. 1960;9:119–41. [PubMed] [Google Scholar]
- Urasaki A, Morvan G, Kawakami K. Functional dissection of the Tol2 transposable element identified the minimal cis-sequence and a highly repetitive sequence in the subterminal region essential for transposition. Genetics. 2006;174:639–49. doi: 10.1534/genetics.106.060244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varlet I, Collignon J, Robertson EJ. nodal expression in the primitive endoderm is required for specification of the anterior axis during mouse gastrulation. Development. 1997;124:1033–44. doi: 10.1242/dev.124.5.1033. [DOI] [PubMed] [Google Scholar]
- White JA, Heasman J. Maternal control of pattern formation in Xenopus laevis. J Exp Zoolog B Mol Dev Evol. 2007 doi: 10.1002/jez.b.21153. [DOI] [PubMed] [Google Scholar]
- Yatskievych TA, Pascoe S, Antin PB. Expression of the homebox gene Hex during early stages of chick embryo development. Mech Dev. 1999;80:107–9. doi: 10.1016/s0925-4773(98)00204-4. [DOI] [PubMed] [Google Scholar]
- Yelon D. Cardiac patterning and morphogenesis in zebrafish. Dev Dyn. 2001;222:552–63. doi: 10.1002/dvdy.1243. [DOI] [PubMed] [Google Scholar]
- Yeo C, Whitman M. Nodal signals to Smads through Cripto-dependent and Cripto-independent mechanisms. Mol Cell. 2001;7:949–57. doi: 10.1016/s1097-2765(01)00249-0. [DOI] [PubMed] [Google Scholar]
- Yu D, Ellis HM, Lee EC, Jenkins NA, Copeland NG, Court DL. An efficient recombination system for chromosome engineering in Escherichia coli. Proc Natl Acad Sci U S A. 2000;97:5978–83. doi: 10.1073/pnas.100127597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zawel L, Dai JL, Buckhaults P, Zhou S, Kinzler KW, Vogelstein B, Kern SE. Human Smad3 and Smad4 are sequence-specific transcription activators. Mol Cell. 1998;1:611–7. doi: 10.1016/s1097-2765(00)80061-1. [DOI] [PubMed] [Google Scholar]
- Zhang J, Talbot WS, Schier AF. Positional cloning identifies zebrafish one-eyed pinhead as a permissive EGF-related ligand required during gastrulation. Cell. 1998;92:241–51. doi: 10.1016/s0092-8674(00)80918-6. [DOI] [PubMed] [Google Scholar]
- Zhou S, Zawel L, Lengauer C, Kinzler KW, Vogelstein B. Characterization of human FAST-1, a TGF beta and activin signal transducer. Mol Cell. 1998;2:121–7. doi: 10.1016/s1097-2765(00)80120-3. [DOI] [PubMed] [Google Scholar]
- Zhou X, Sasaki H, Lowe L, Hogan BL, Kuehn MR. Nodal is a novel TGF-beta-like gene expressed in the mouse node during gastrulation. Nature. 1993;361:543–7. doi: 10.1038/361543a0. [DOI] [PubMed] [Google Scholar]


