Abstract
Studies in tissue culture cells have implicated p300 and CBP acetyltransferases in myogenic regulatory factor (MRF) mediated transcription and terminal differentiation of skeletal muscle cells. However, in vivo data placing p300 and CBP on myogenic differentiation pathways are not yet available. In this report we provide genetic evidence that p300 but not CBP acetyltransferase (AT) activity is required for myogenesis in the mouse and in embryonic stem (ES) cells. A fraction of embryos carrying a single p300 AT- deficient allele exhibit impaired MRF expression, delayed terminal differentiation and a reduced muscle mass. In mouse embryos lacking p300 protein, Myf-5 induction is severely attenuated. Similarly, ES cells homozygous for a p300 AT or a p300 null mutation fail to activate Myf5 and MyoD transcription efficiently, while Pax3, acting genetically upstream of these MRFs, is expressed. In contrast, ES cells lacking CBP AT activity express MyoD and Myf5 and undergo myogenic differentiation. These data reveal a specific requirement for p300 and its AT activity in the induction of MRF gene expression and myogenic cell fate determination in vivo.
Keywords: cell fate determination/differentiation/ES cells/HAT/myogenin
Introduction
Most sequence-specific transcription factors regulate gene expression by interacting with transcriptional co-regulators, which influence chromatin structure. Acetyltransferases (ATs) and chromatin remodelling complexes facilitate chromatin opening, while deacetylases (HDACs) and lysine 9-specific methyltransferases frequently contribute to gene silencing (Jenuwein and Allis, 2001; Narlikar et al., 2002). The transcriptional co-regulators CBP and p300 possess histone acetyltransferase activity (Bannister and Kouzarides, 1996; Ogryzko et al., 1996) and are capable of interacting with a large variety of transcription factors playing central roles in a wide range of cellular processes including proliferation, differentiation and apoptosis (for reviews see Shikama et al., 1997; Goodman and Smolik, 2000; Chan and La Thangue, 2001). Moreover, CBP and p300 exhibit tumour-suppressing activity, especially in hematopoietic cells (Kung et al., 2000; Rebel et al., 2002). A growing body of evidence indicates that p300 and CBP are also able to acetylate proteins other than histones, such as transcription factors, components of the transcription machinery and tumour suppressors (Gu and Roeder, 1997; Imhof et al., 1997; Boyes et al., 1998; Hung et al., 1999; Soutoglou et al., 2000), adding yet another level of control to their transcriptional regulatory potential.
The p300/CBP family is specific to multicellullar organisms and evolutionarily conserved from plants to humans (Bordoli et al., 2001b), suggesting that p300 and CBP are likely to play important roles in organ development and morphogenesis. Drosophila melanogaster and Caenorhabditis elegans only possess the cbp ortholog (Akimaru et al., 1997; Shi and Mello, 1998), while both p300 and CBP are present in humans and mice. The two proteins are highly related (63% identity) and their expression pattern during mouse development is almost identical (Partanen et al., 1999), suggesting that their respective functions overlap to some degree. However, there is evidence that they are not interchangeable, in particular from the analysis of p300 and cbp knockout mice which exhibit in part distinct phenotypes (Yao et al., 1998; Tanaka et al., 2000). For example, an increased incidence of haematological malignancies and abnormal skeletal patterning was reported in cbp but not in p300 heterozygous null mice (Tanaka et al., 1997; Kung et al., 2000). At the cellular level, it was shown that ribozyme-mediated ablation of p300, but not CBP, inhibits retinoic-acid-induced differentiation (Kawasaki et al., 1998). Similar studies in p300–/– fibroblasts implied that p300, but not CBP, is an essential retinoic acid receptor cofactor (Yao et al., 1998).
Previous in vitro studies have indicated that several ATs, including p300/CBP and PCAF, as well as HDACs are involved in the control of muscle differentiation (for review see McKinsey et al., 2001; Puri et al., 2001). During myogenesis, the primary myogenic regulatory factors (MRFs) MyoD and Myf5 are thought to cooperate with p300 and CBP (Eckner et al., 1996; Yuan et al., 1996; Puri et al., 1997a; Sartorelli et al., 1997) and with MEF2 transcription factors (Eckner et al., 1996; Sartorelli et al., 1997) to mediate activation of the secondary MRFs, myogenin and Mrf4. The primary MRFs specify myogenic identity of uncommitted somitic mesoderm cells. The secondary MRFs, in conjunction with MEF2 proteins, allow myoblasts to exit the cell cycle and to differentiate into myocytes and mature myofibers (for reviews see Sabourin and Rudnicki, 2000; Buckingham, 2001). This process requires both repression of genes associated with proliferation and activation of muscle-specific genes.
CBP and the GCN5-related acetyltransferase PCAF were shown to acetylate MyoD, thereby facilitating heterodimer formation with ubiquitously expressed E-box proteins encoded by the E2A gene (Puri et al., 1997b; Sartorelli et al., 1999; Polesskaya et al., 2000). However, disruption of Pcaf in the mouse is not lethal and does not ostensibly impair myogenesis (Xu et al., 2000; Yamauchi et al., 2000), and chemical inhibition of CBP AT activity still allows MyoD to activate myogenin in C2C12 cells (Polesskaya et al., 2001). Therefore the significance of PCAF- and/or CBP-mediated MyoD acetylation in vivo remains unclear.
In this report, we provide the first in vivo evidence that p300 indeed plays a role in skeletal myogenesis. A fraction of mouse embryos bearing a monoallelic mutation inactivating p300 AT activity exhibit impaired myogenesis and MRF expression. In contrast, embryos with an equivalent mutation in cbp do not manifest overt muscle defects. Moreover, embryonic stem cells lacking p300 or its AT activity are strongly impaired in their ability to activate MyoD and Myf5 during skeletal myogenesis, while ES cells deficient in CBP or its AT activity are still capable of forming myotubes. These results suggest that the AT activity of p300 is distinct from that of CBP and reveal an essential function of p300 AT activity in myogenic cell fate specification.
Results
Generation of heterozygous mice and homozygous ES cells deficient in p300 or CBP AT activity
We have used a knock-in strategy in ES cells to introduce the previously described AT-inactivating point mutation in p300 [residues WY(1466–1467)AS] or CBP [residues WY(1503–1504)AS] (Bordoli et al., 2001b). Integration of the mutation by homologous recombination of the p300 and cbp targeting vectors (Figure 1A and Supplementary figure S1A available at The EMBO Journal Online, respectively) was detected by Southern blot (Figure 1B; Supplementary figure S1B). Heterozygous ES cells were injected into blastocysts that gave rise to chimeric mice transmitting the AT-mutant allele to their progeny.
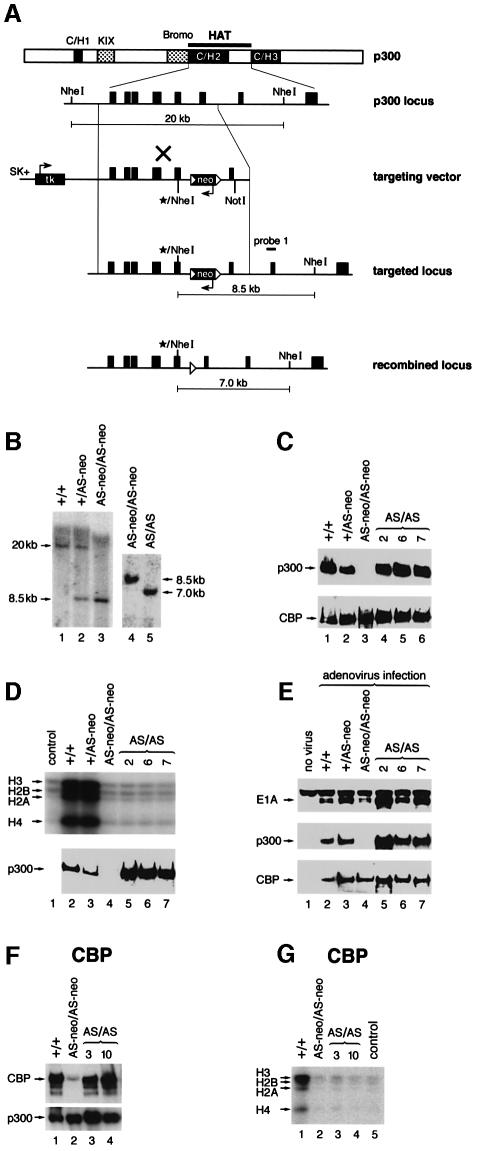
Fig. 1. Targeting strategy to generate p300 AT mutant ES cells and characterization of p300 or CBP AT-mutant ES cells. (A) Overall organization of the p300 protein, the wild-type p300 genomic locus, the targeting vector and the recombined loci is shown. The asterisks indicate the site of the mutation, which harbors a de novo NheI restriction site. (B) Southern blot with probe 1 analysing genomic DNA isolated from wild-type, heterozygous (p300+/AS-neo) and homozygous p300 KO (p300AS-neo/AS-neo) and p300 AT– (p300AS/AS) ES cells. The probes and the size of the genomic DNA fragments following NheI restriction digest are indicated in (A). (C and F) Western blot analysis showing p300 and CBP protein levels in (C) p300 KO and p300 AT– cells and (F) CBP KO (cbpAS-neo/AS-neo) and CBP AT– (cbpAS/AS) cells. (E) E1A binding assay: western blot analysis following E1A immunoprecipitation from ES cells infected with adenovirus using, from top to bottom, E1A, p300 or CBP antibodies. (D and G) HAT assay with (D) p300 KO and three independent p300 AT– cell lines and (G) CBP KO and two independent CBP AT– cell lines.
The heterozygous ES cells were made homozygous for the p300 or CBP AT mutation by cultivation in medium containing a high G418 concentration (Figure 1B; Supplementary figure S1B and C). These homozygous cells still harbour the neo gene and p300 protein levels were undetectable in p300AS-neo/AS-neo cells (Figure 1C, lane 3), while in cbpAS-neo/AS-neo cells the CBP protein levels were drastically diminished (reduced by more than 95%; Figure 1F, lane 2). Excision of the neo cassette (Figure 1B, lane 5, for p300 and Supplementary figure S1B, lanes 3 and 4, for CBP cells) restored p300 and CBP protein expression to levels comparable to those of wild-type cells, as illustrated in Figure 1C (lanes 4–6) for p300 and in Figure 1F (lanes 3 and 4) for CBP. Therefore excision of the neo cassette is required for full expression of AT-mutant p300 or CBP proteins. Furthermore, in agreement with previous in vitro studies (Bordoli et al., 2001a), CBP (Figure 1G) and p300 (Figure 1D) mutant proteins did not exhibit any AT activity. For simplicity, the AT-deficient cells will be referred to as p300 AT minus (AT–) and CBP AT–, and the p300AS-neo/AS-neo and the cbpAS-neo/AS-neo ES cells will be named p300 KO (knockout) and CBP KO, respectively.
In order to assess the conformational integrity of the mutant proteins, we have analysed their binding to the adenoviral E1A protein by immunoprecipitation. The AT-deficient proteins bound E1A as efficiently as wild-type p300 (Figure 1E, middle panel) or CBP (data not shown). These findings suggest that the AT mutation does not overtly perturb the overall structure of p300 and CBP and still allows binding of E1A to the C/H3 domain which is immediately adjacent to the AT domain (Figure 1A).
Full dose of p300 AT activity is required for normal muscle development in mice
The p300+/AS-neo and cbp+/AS-neo mice were viable and fertile. In vivo excision of the neo gene was performed by crossing these mouse lines with a CMV-Cre deleter strain whose Cre gene is active from the zygote state onwards (Schwenk et al., 1995). Genotyping of several E8.5–E11.5 embryos derived from such crosses revealed efficient and complete in vivo excision of the neo gene (data not shown).
p300+/AS embryos varied in size and did not survive embryogenesis. The majority of these embryos died between E12.5 and E16.5 due to heart failure or perinatally due to an inability to breathe (Shikama et al., 2003). In contrast, most of the cbp+/AS animals reached term, but died within the first 1–2 days following birth, probably owing to respiratory failure. Since most mice heterozygous for a p300 or cbp null allele are viable (Tanaka et al., 1997, 2000; Yao et al., 1998), the lethality observed in the two AT-mutant mice indicates that the AT-deficient alleles are dominant negative. Mouse strains harbouring these AT-mutant alleles can only be propagated as long as expression of the mutant locus is suppressed by the neomycin gene transcribed in opposite direction relative to the p300 or cbp gene. In this article, we have focused on skeletal muscle development in the two AT-mutant mouse lines and in ES cells.
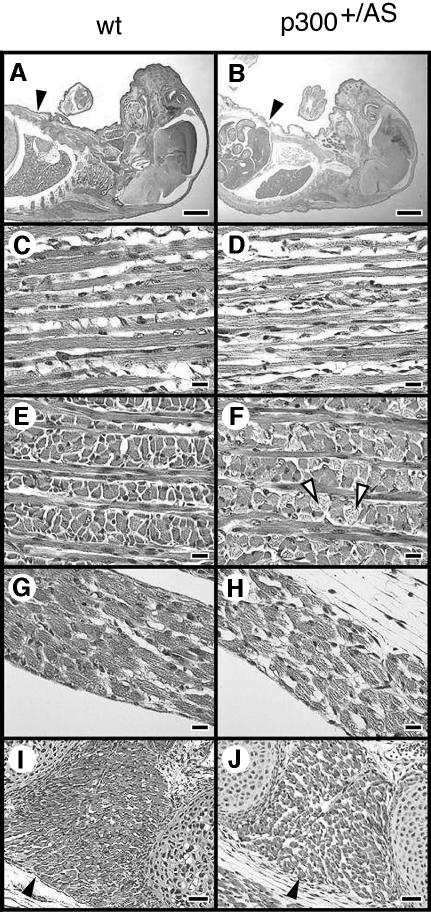
Histological analysis of E18.5 mouse embryos showed that in most of them the size of the muscle fibres was reduced and their interfibrillar space was increased, resulting in a loose and disorganized appearance (Figure 2, compare wt and p300+/AS panels). This was clearly visible for the trapezius muscle (Figure 2C and D), the tongue (Figure 2E and F), the diaphragm (Figure 2G and H) and the intercostal muscles (Figure 2I and J). In contrast, no such disorganized muscle phenotype was ever seen in cbp+/AS mice (data not shown). These observations indicate that hypaxial (tongue and limbs) as well as epaxial muscle formation is sensitive to a reduction in the dose of p300 but not CBP AT activity.
Fig. 2. Histological analysis of E18.5 wild-type and p300+/AS mouse embryos. HE staining of sagittal (A–D) or transverse (E and F) mouse sections. An overview of the embryos is shown in (A) and (B); black arrows point to the body wall. Enlarged views of the trapezius muscle (C and D), the tongue (E and F), the diaphragm (G and H) and the intercostal muscles (I and J). Scale bars: (A and B), 2 mm; (C–H), 100 µm; (I and J) 500 µm. White arrowheads mark muscle fibers in the trapezius (C and D) and loose muscle fibers in the tongue (F). Black arrowheads indicate the intercostal muscle located between the second and third sternebra (I and J).
A reduction in the size of muscle groups was visible in all p300+/AS embryos at E14.5 (Figure 3Ab and Ad), whereas cbp+/AS mice of this age had a largely intact muscle compartment (Figure 3Bb and Bd). In addition, sarcomeric actin protein expression levels were diminished in the back muscles in a fraction of E14.5 p300+/AS embryos (Figure 3Ac and Ad) but never in cbp+/AS embryos (Figure 3Bc and Bd). All E14.5 p300+/AS embryos also exhibited a peripheral edema (Figure 3Ab and Ad), which is in part due to cardiovascular malformations (Shikama et al., 2003) and may contribute to the impaired myogenesis by exerting pressure on muscle tissue. However, a reduction in sarcomeric actin expression was also observed in muscle groups that are not directly exposed to the edema, such as the limbs (data not shown) and the tongue (Figure 3Ae and Af). Moreover, a proportion of p300+/AS embryos exhibited a diminished or delayed expression of myosin heavy chain (MHC) at E10.5 (Figure 3C, right panels), a time point that is well before the appearance of the edema around E12.5. Based on these observations, the edema is unlikely to account fully for the compromised myogenesis in p300+/AS embryos. Rather, the presence of smaller muscle groups in all embryos and the partially penetrant reduction in terminal differentiation suggest that muscle formation is directly affected by p300 AT mutation.
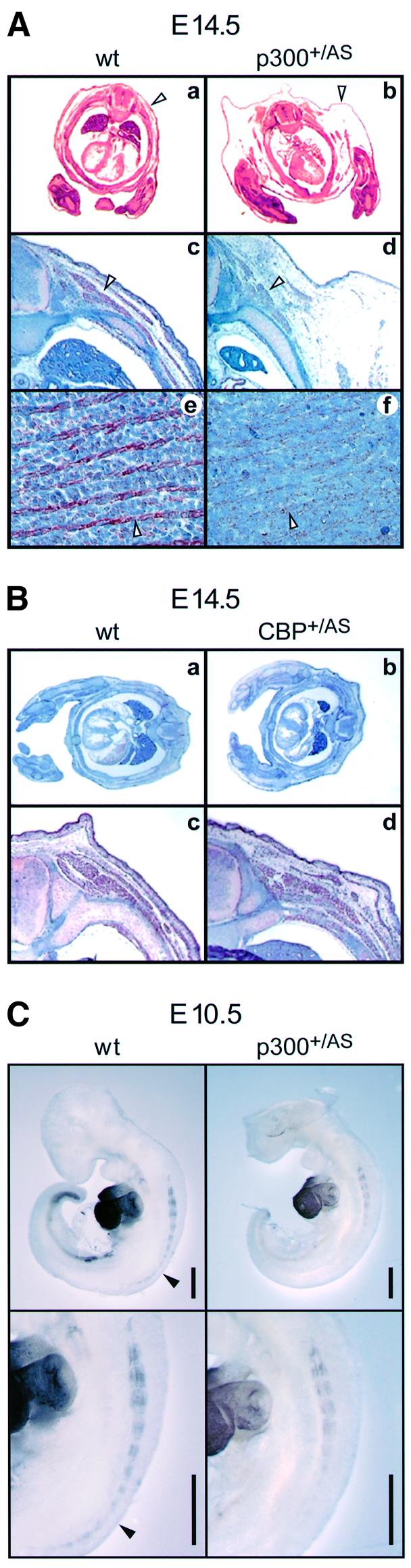
Fig. 3. Skeletal muscle structural protein expression in mouse embryos. (A) Transverse sections of wild-type (wt) and p300+/AS littermates aged E14.5 stained with HE (a and b) and with a sacromeric actin antibody (c–f). Enlarged view (20×) of the paravertebral area: arrowheads indicate the edema (a and b) and the back muscle mass (c and d). Enlarged view (40×) of the tongue (e and f); arrowheads highlight actin staining. (B) Transverse section of wild-type and cbp+/AS mice aged E14.5 stained with HE (a and b) and enlarged view (20×) of the paravertebral region following sarcomeric actin staining (c and d). (C) Myosin heavy-chain whole-mount immunostaining of E10.5 wild-type and p300+/AS littermates using MF20. Scale bar, 500 µm. Filled arrowheads indicate staining in the posterior region of the wild-type embryo which is absent in the p300+/AS littermate.
The reduction in muscle size was due to neither decreased proliferation nor increased apoptosis in p300+/AS embryos of the same size as wild-type littermates (Supplementary figure S2A and B, respectively). Migration of muscle precursor cells into the limb buds was also comparable between size-matched p300+/AS and wild-type littermates (Supplementary figure S2C). These findings suggest that the muscle defects of p300 AT mutant embryos are more likely to occur at the level of myogenic determination and differentiation, during maintenance of the differentiated state.
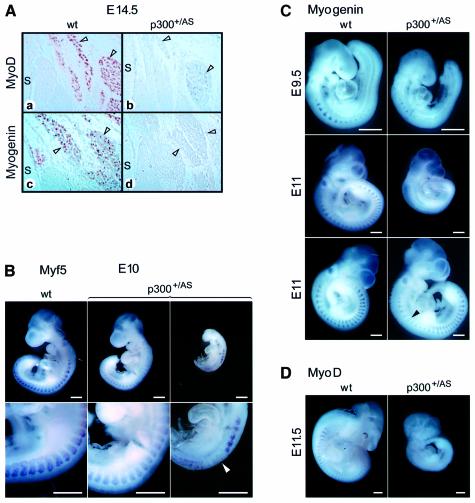
Since myoblast identity and terminal differentiation is determined by MRFs (Sabourin and Rudnicki, 2000; Buckingham, 2001), we investigated the expression levels of Myf-5, myogenin and MyoD in p300+/AS and wild-type embryos. Approximately 15% of E14.5 p300+/AS embryos exhibited a reduction in MyoD (Figure 4Aa and Ab) and myogenin (Figure 4Ac and Ad) protein expression. Similarly, RNA whole-mount in situ hybridization of embryos aged E9.5–E11.5 suggested that MRF expression is perturbed in a fraction of p300+/AS embryos relative to wild-type littermates. Thirty-seven embryos (16 wild type and 21 p300+/AS) were analysed. Eleven of the 21 p300+/AS embryos displayed an equivalent number of somites as wild-type littermates. Three of these mutant embryos showed diminished or delayed expression of Myf-5 or myogenin (Figure 4B, middle, and C, top and bottom). Among the 10 retarded p300+/AS embryos, two were of particular interest as they did not express myogenin or MyoD (Figure 4C, middle, and D) despite having reached a developmental stage (27 and 37 somites) where these transcripts are normally expressed (10–12 somite stage at E8.5 for myogenin and 35 somite stage at E10.5 for MyoD). In addition, another retarded p300+/AS embryo showed an irregular and disrupted Myf-5 expression pattern (Figure 4B, right). Taken together, these in vivo results show that monoallelic abrogation of p300 AT activity leads to a partial impairment of MRF expression, suggesting that the full complement of p300 AT activity is required to activate and/or sustain MRF gene transcription efficiently. Furthermore, the MRF activity itself is also likely to be affected by the p300 AT mutation since the impaired muscle formation was completely penetrant at E14.5 and E18.5 while the compromised MRF expression was not.
Fig. 4. MRF expression in mouse embryos. (A) MRF expression in the paravertebral region of E14.5 wild-type and p300+/AS embryos. MyoD (a and b) and myogenin (c and d) immunostaining. S, spinal chord; arrowheads indicate the muscle groups. (B–D) Whole-mount in situ hybridization of wild-type and p300+/AS littermate embryos aged between E9.5 and E11.5 showing (B) Myf5, (C) myogenin and (D) MyoD expression. The arrowhead in (B) highlights a disruption of the Myf5 expression pattern. Scale bars, 500 µm.
CBP and p300 AT activities are not required for embryoid body formation and proliferation
We have extended these in vivo analyses by investigating the myogenic differentiation potential of homozygous AT-deficient ES cells in which a requirement for AT activity should become more readily apparent than in the heterozygous mice. Pluripotent ES cells can form embryoid bodies (EBs) which are able to differentiate into a wide spectrum of cell types including neuronal, hematopoietic, skeletal muscle and bone cells (for a review see Desbaillets et al., 2000). In particular, skeletal myogenesis is well characterized in EBs and reflects the temporal and sequential MRF expression pattern seen during mouse development. (Braun and Arnold, 1994; Rohwedel et al., 1994; Weitzer et al., 1995).
All five cell lines (E14 wild type, p300 KO, p300 AT–, CBP KO and CBP AT–) efficiently formed EBs within 2 days (D2) (Figure 5A), suggesting that neither protein nor their intrinsic AT activity is required for this process. On D4 and D6, wild-type, p300 AT–, CBP KO and CBP AT– EBs showed an increase in diameter due to cell proliferation (Figure 5B). However, EBs derived from p300 KO cells remained small and showed reduced cell proliferation, a defect that was maintained throughout differentiation until confluency was reached. A similar finding was recently reported for p300 nullizigous EBs (Rebel et al., 2002). Since p300 AT deficient EBs did not show such a defect, these results indicate that a function of p300 other than the AT activity plays a role in proliferation of ES cells upon induction of differentiation.
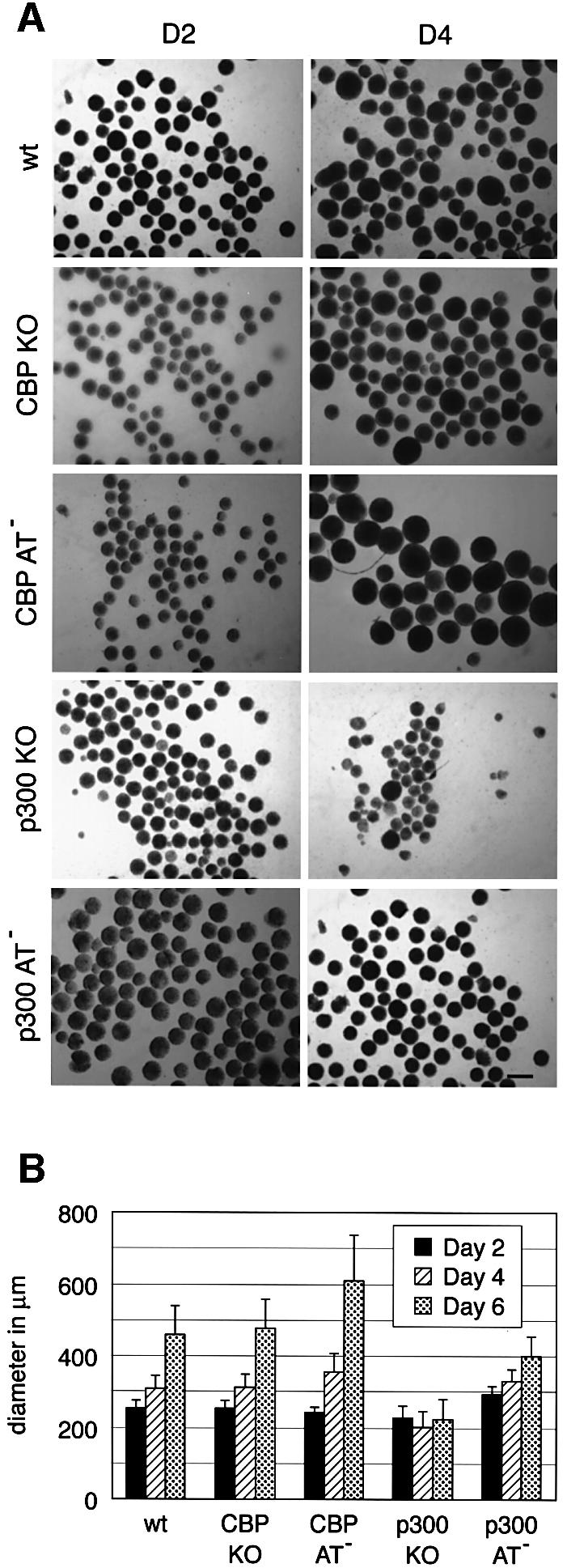
Fig. 5. Formation and proliferation of embryoid bodies. Wild-type E14, CBP KO, CBP AT–, p300 KO and p300 AT– ES cell lines were aggregated as hanging drops for 2 days to form EBs and transferred to bacterial dishes. (A) Microscope analysis of EBs at D2 and D4. The bar represents 400 µm. (B) Histogram representing the diameter of EBs derived from all five cell lines at D2, D4 and D6 of differentiation. The standard deviation represents the average of three independent experiments. Fifty EBs were measured per cell line at each time point.
Severe impairment of skeletal myogenesis in ES cells lacking p300 AT activity
Having established EB formation of p300 and CBP AT-deficient cells, we assessed their myogenic differentiation potential by analysing MHC expression. After 44 days in culture (Figure 6A, left column), all cell lines were able to differentiate, albeit with strikingly different efficiencies. Wild-type and CBP mutant cell lines (KO and AT–) showed abundant striated myofibers, while both p300 mutant lines showed very few MHC-positive cells which exhibited little cell fusion and lacked striation as well as myofibrillar organization (Figure 6A). Quantification of the signal confirmed that wild-type and both CBP mutant lines displayed comparable MHC expression (Figure 6B). In contrast, p300 KO and AT– cells showed a 6- to 18-fold and a 12- to 38-fold reduction in MHC expression, respectively (Figure 6B). In keeping with the above results, skeletal actin and embryonic MHC mRNA levels were also reduced in p300 KO and AT– cell lines at D44 (Figure 6C). Differentiation of both p300 mutant cells was not merely delayed, since even at D66 these cells failed to form myofibers, which were abundant in wild-type and CBP mutant cells (Figure 6A, right column). Confluency of p300 KO cells was reached well before D66, suggesting that the differentiation defect is unlikely to be due to the limited proliferative capacity of these cells. Taken together, these data demonstrate that cells deficient in p300 but not CBP AT activity are largely unable to undergo terminal myogenic differentiation. However, the presence of weak MHC staining in p300 KO and AT– cells suggests that differentiation was not completely blocked, implying the existence of a myogenic differentiation pathway independent of p300.
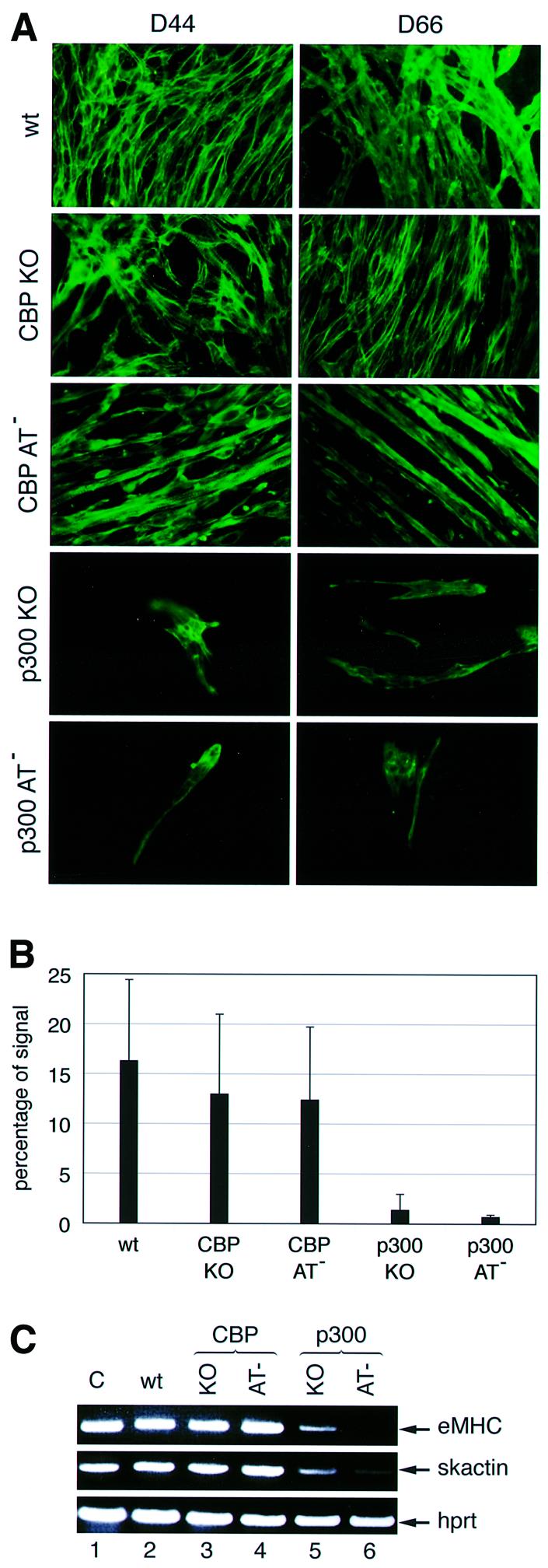
Fig. 6. Skeletal muscle formation during in vitro ES cell differentiation. The ES cell lines used were E14 wild type, CBP KO, CBP AT–, p300 KO and p300 AT–. (A) MHC expression at D44 or D66 of differentiation was determined by immunofluorescence. (B) The signal was quantified at D44 using the Adobe quantification toolpack™. The standard deviation is derived from the results of three independent experiments. (C) RT–PCR showing embryonic myosin heavy chain (eMHC) and skeletal actin (skactin) expression at D44. Hprt was used as an internal control. Lane 1, positive control with total RNA from an E11.5 embryo.
To exclude that p300 KO and AT– cells exhibit a general differentiation impediment, we subjected these cells to osteogenic differentiation (Phillips et al., 2001) and analysed the presence of mineralized bone nodules (Figure 7A). The percentage of EBs with calcium deposits was comparable for the five cell lines, ranging between 78% and 92% (Figure 7B). Similarly, the osteoblast-specific transcript osteocalcin was expressed at equivalent levels in all cell lines (Figure 7C). However, microscope examination revealed subtle differences in the fine structure of the bone nodules (J.-F.Roth et al., in preparation). Taken together, these results suggest that p300 KO and AT– cells are able to undergo osteogenesis as efficiently as wild-type and CBP cell lines, and thus do not manifest a general differentiation impairment.
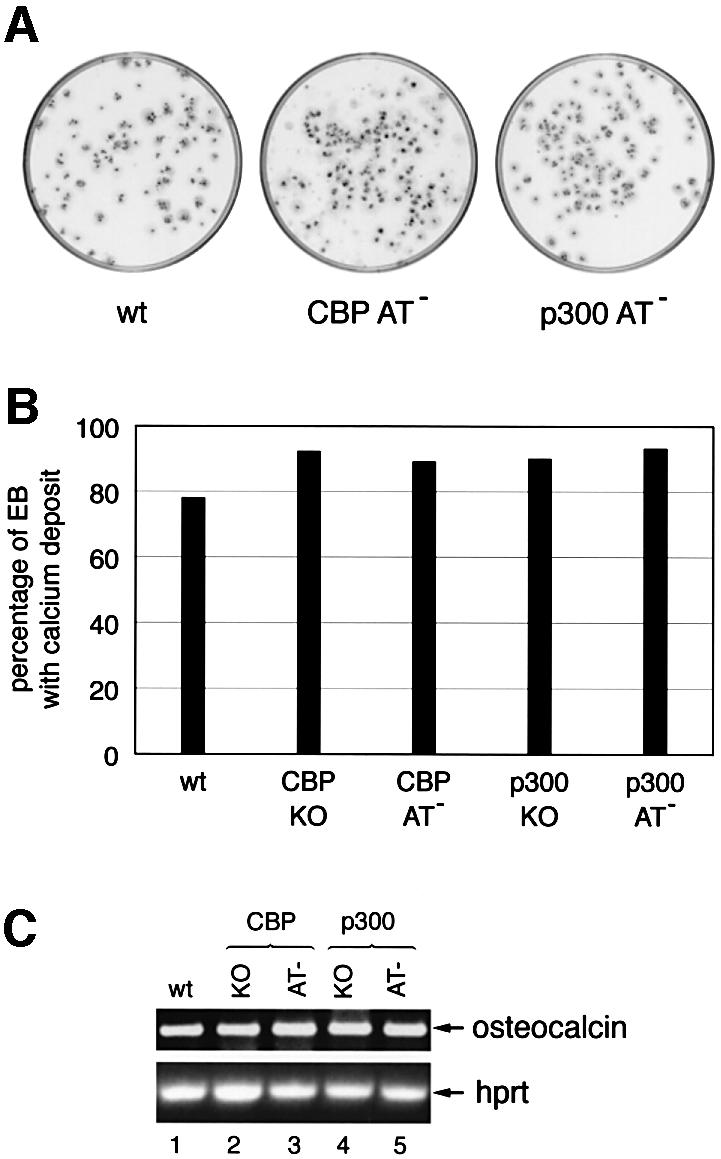
Fig. 7. In vitro osteogenesis. The ES cell lines used were E14, CBP KO, CBP AT–, p300 KO and p300 AT–. (A) Bone mineral deposition in EBs is indicative of osteogenesis and was visualized by alizarin red staining at D20. (B) Bar diagram illustrating the percentage of EBs containing calcium deposits. (C) Osteocalcin and Hprt expression analysed by RT–PCR of 150 EBs at D20 of osteogenesis.
These data clearly demonstrate that, while dispensable for osteoblast differentiation, the AT activity of p300 is required for myogenic differentiation including efficient cell fusion leading to myofibre formation. CBP AT activity does not appear to be required for this process, pointing to a striking functional difference between the two AT activities.
Abrogation of p300 AT activity prevents Myf5 and MyoD gene expression
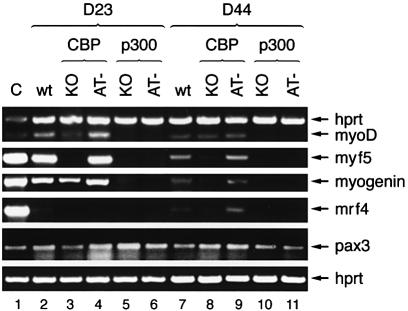
The expression pattern of the MRF genes during EB differentiation was analysed in order to determine at which stage skeletal muscle differentiation was blocked in both p300 mutant cells (Figure 8). At D23 and D44 (Figure 6A, right column), all four MRF transcripts were present in wild-type cell lines in the expected temporal pattern (Figure 8, lanes 2 and 7). In contrast, in p300 mutant cell lines, MRF genes failed to be efficiently induced (Figure 8, lanes 5, 6, 10 and 11). Since Myf5 and MyoD define myoblast identity, these results suggest that p300 is required for myoblast specification and acts genetically upstream of these two genes.
Fig. 8. Expression of myogenic regulatory factors in differentiated EBs. RNA expression levels of the indicated genes were determined by RT–PCR at D23 (lanes 2–6) and D44 (lanes 7–11). Lane 1, positive control using RNA from E10.5–E13.5 embryos. The ES cell lines used were E14 wild type (lanes 2 and 7), CBP KO (lanes 3 and 8), CBP AT– (lanes 4 and 9), p300 KO (lanes 5 and 10) and p300 AT– (lanes 6 and 11).
Unexpectedly, CBP KO and CBP AT– cells, which were both able to undergo muscle cell differentiation as assessed by MHC expression (Figure 6), differed in their MRF gene expression pattern. Unlike CBP AT– cells, which showed a pattern similar to wild-type cells (Figure 8, cf. lanes 4 and 2), Myf5 was not induced in CBP KO cells at D23. In addition, MyoD and myogenin induction was reduced (Figure 8, lane 3). At D44, CBP KO cells still exhibited lower levels of all MRF transcripts, except MyoD, when compared with wild-type and CBP AT– cells, (Figure 8, cf. lanes 7, 9 and 8, respectively). Interestingly, the depressed MRF levels correlated with a small but notable reduction in myotube formation observed in CBP KO relative to wild-type and AT– cells (Figure 6A). Given the previously suggested concept of two separate myogenic subpopulation defined by either MyoD or Myf5 expression (Braun and Arnold, 1996; for a review see Tajbakhsh and Buckingham, 2000), our data may reflect a deficiency in determination of the Myf5 subpopulation in cells lacking CBP. The fact that the CBP AT mutation does not interfere with Myf5 gene activation suggests that one or several domains of CBP, distinct from the AT domain, are required for this process. Importantly, this result also provides evidence that the mutation introduced in the AT domain leaves other CBP functions intact.
Finally, we have analysed Pax3 gene expression, since Pax3 and Myf5 have been identified as upstream regulators of MyoD (Maroto et al., 1997; Tajbakhsh et al., 1997). Pax3 has also been shown to be essential for migration of somitic muscle precursor cells to the limb buds (Bober et al., 1994). Pax3 mRNA levels were comparable in all cell lines at both D23 and D44 (Figure 8), suggesting that neither p300 nor CBP are essential for induction of Pax3 expression during ES cell differentiation. Nevertheless, since Pax3 is expressed in mesodermal and neuroectodermal tissues which are both generated upon in vitro differentiation, we cannot rule out the possibility that in this experiment a mesodermal defect in Pax3 expression could be partially masked by neuroectodermal Pax3 expression.
Impaired Myf 5 induction in p300 KO embryos
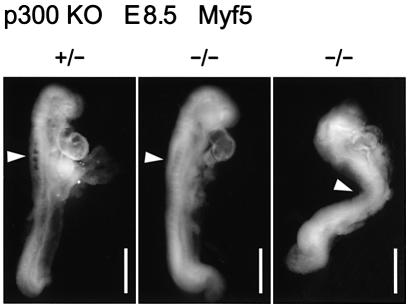
In order to complement the data obtained with the p300 KO ES cells, we have analysed Myf5 gene expression in p300–/– embryos. On a mixed 129SVJ × C57BL/6 background, these embryos die between E8.5 and E11 (Yao et al., 1998). However, on a pure C57BL/6 background, we did not encounter any p300–/– embryos developing and surviving beyond E8.5–E9.5. Therefore, Myf5 was the only MRF whose expression could be examined since it is induced around E7.75 in presomitic mesoderm (Ott et al., 1991). At E8.5, Myf5 expression was readily detectable in the first four somites of a p300+/– embryo, while it was much weaker in two p300–/– littermates (Figure 9). This result indicates that the absence of p300 leads to a compromised expression of Myf5 in vivo, thus confirming the findings with the p300 KO cells.
Fig. 9. Myf5 expression in E8.5 p300+/– and p300–/– littermates detected by whole-mount in situ hybridization. Arrowheads indicate the somites expressing Myf5. Scale bars, 500 µm.
Discussion
We have taken a genetic approach in mouse and ES cells to investigate the role of p300 and CBP AT activity during skeletal myogenesis. Unexpectedly, the results of both lines of enquiry indicate that p300 AT but not CBP AT activity is required for the induction and/or maintenance of MRF gene expression and muscle differentiation. Hence, p300 plays a critical role in myoblast cell fate determination by acting genetically upstream of Myf5 and MyoD genes. Moreover, while myogenesis is inhibited to a comparable degree in ES cells homozygous for either a p300 null or an AT mutation, this is not the case for equivalent CBP mutations. A CBP null mutation prevents efficient induction of Myf5, in contrast with the CBP AT mutation which has no effect. These observations suggest that during myogenesis, the AT activity plays a central role in the overall function of p300 whereas it appears to be largely dispensable in CBP.
All previous studies investigating the role of p300 and CBP in skeletal myogenesis were carried out in immortalized tissue culture cell lines such as C3H10T1/2 fibroblasts and C2C12 myoblasts, the latter already being committed to a myogenic fate and recapitulating aspects of terminal differentiation. Transient transfection and microinjection experiments with these cells have suggested that p300 and CBP cooperate with MyoD in the activation of the Cdk inhibitor p21 and of the myogenin gene, allowing terminal differentiation and myotube formation to take place (Eckner et al., 1996; Yuan et al., 1996; Puri et al., 1997a; Sartorelli et al., 1997). More recently, using the synthetic inhibitor LysCoA to block p300 and CBP AT activity, induction of the terminal differentiation markers MHC and muscle creatine kinase (MCK) was suggested to depend on p300/CBP AT activity (Polesskaya et al., 2001). This defect could in part be rescued by the overexpression of a wild type but not by a HAT mutant CBP protein lacking 138 amino acids. However, the above experiments could not address a possible role of p300 and CBP in the specification of the myogenic cell fate nor functionally distinguish between the two proteins.
Our analysis of skeletal myogenesis in AT-mutant and knockout mice provides the first evidence that p300 and its AT activity indeed play a role in this process in animals. The strength of our approach lies in the specificity of our point mutations and in our ability to assay myogenesis in its natural context without overexpression of genes. Taking advantage of ES cells recapitulating myogenesis more faithfully than myoblastic cell lines, we were able to distinguish p300 from CBP function and to show that p300 is required well before terminal differentiation, in particular at the level of myoblast specification. Moreover, we also demonstrate that CBP AT-deficient cells are capable of undergoing both myoblast specification and terminal differentiation.
It is important to point out that myogenesis is not completely inhibited in the absence of p300 or p300 AT activity since MRFs were expressed, albeit at very low levels, during skeletal muscle differentiation of p300 KO and AT– ES cells, and MHC could be detected by immunofluorescence. Similarly, p300–/– mice showed weak but detectable Myf5 gene expression at E8.5. Thus our results suggest the existence of p300 and p300 AT-independent pathways enabling myogenic specification and differentiation to occur. This observation points to a certain degree of redundancy among AT enzymes.
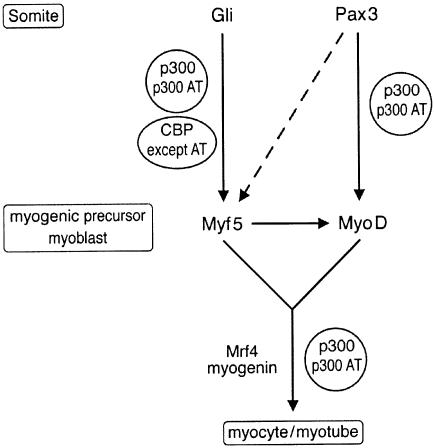
The defects observed in Myf5, myogenin and MyoD expression in a fraction of p300+/AS and in Myf5 expression in p300–/– embryos are indicative of a requirement of p300 and its AT activity at the level of MRF gene induction. This process is known to depend on the local balance between myogenesis-inducing (Wnt1, Wnt7 and Shh) and myogenesis-repressing (BMP/TGFβ family members) morphogens (for reviews see Bailey et al., 2001; Brent and Tabin, 2002). Accordingly, p300 and its AT activity may be required to convert these opposing morphogen signals into an appropriate transcriptional response leading to MRF activation. Transcription factors responding to Shh are the Gli family members which contribute to the activation of Myf5 (Tajbakhsh et al., 1998; Gustafsson et al., 2002). In turn, Myf5 induces myogenin (Kaul et al., 2000) and, together with Pax3, can contribute to MyoD activation (Maroto et al., 1997; Tajbakhsh et al., 1997). Thus we propose that p300 AT may cooperate with one or several of the transcription factors participating in Myf5 and MyoD gene induction (Figure 10). However, it is important to note that no Pax3 binding site is present in the MyoD core enhancer element, and thus its effect on MyoD activation may be indirect (for reviews see Borycki and Emerson, 1997; Arnold and Winter, 1998).
Fig. 10. Model placing p300, CBP and their respective AT activities on genetic pathways controlling skeletal myogenesis. Arrows represent an established link. Dashed arrows represent a suggested effect. Note that arrows do not necessarily reflect a direct effect.
In addition to playing a role at the level of MRF gene expression, our data are consistent with a requirement for p300 AT activity at the level of MRF transactivation potential. The defect in myofiber formation present in most E18.5 p300+/AS embryos does not fully correlate with impaired MRF expression that is only detectable in approximately 15% of the embryos. Because of this partial correlation, it is likely that the p300 AT mutation also affects the transcriptional activity of MRFs themselves, resulting in the generation of fewer myoblasts and less efficient myofiber formation. However, further analyses are required to address this issue in greater detail.
Previous in vitro studies have provided some evidence for a differential role of p300 and CBP during certain differentiation processes. Retinoic-acid-induced differentiation of F9 embryonal carcinoma cells was inhibited when p300, but not CBP, protein levels were reduced (Kawasaki et al., 1998). During hematopoiesis, p300 is required for differentiation of specific precursor cells (Kasper et al., 2002; Rebel et al., 2002), while CBP contributes to maintenance of the hematopoietic stem cell pool (Rebel et al., 2002). Enhancing the link between p300 and differentiation, our work demonstrates that it is mainly p300, but not CBP, which permits myogenic cell specification and differentiation in vivo. In addition, it is specifically the AT activity of p300 that is essential for this process, implying that the two ATs may modify distinct substrates. Indeed, a previous mutagenesis study has revealed several structural differences between p300 and CBP AT domains (Bordoli et al., 2001a). Since p300 and CBP modify histones with equal efficiency, at least in vitro, our observations raise the possibility that histones may not represent the most critical acetylation substrates of p300 during differentiation. Among the MRFs, MyoD has been demonstrated to be acetylated in vivo, and can be modified in vitro by PCAF, p300 and CBP on several lysine residues (Sartorelli et al., 1999; Polesskaya et al., 2000). Therefore it will be interesting to determine whether MyoD acetylation is diminished in p300 or CBP AT-deficient cells and whether altered acetylation of MyoD changes its transcriptional activity. Moreover, the availability of the AT-deficient ES cells will facilitate the identification of physiologically relevant acetylation substrates of p300 and CBP.
Materials and methods
Generation of homozygous p300 or CBP AT-deficient ES cells and mice
p300 and CBP AT mutations were introduced into E14, subclone KPA, cells using a knock-in strategy. The targeting vectors contained a floxed pgk-neo cassette in an intron flanking the mutated exon encoding a segment of the AT domain. The use of a weak neo allele allowed rapid subsequent generation of homozygous cells by raising G418 concentration to 5 mg/ml. The neo cassette was excised in vitro by electroporating ES cells with pPGK/CREbpA plasmid (kindly provided by K.Rajewsky), and clones sensitive to low G418 were isolated and analysed by Southern blot and PCR (primer sequences are available upon request).
To generate AT-mutant mice, p300+/AS-neo or cbp+/AS-neo ES cells were injected into blastocysts, which gave rise to chimeric mice transmitting the mutant allele to offspring. To remove the neo cassette, heterozygous mice were mated with CMV-Cre deleter mice (Schwenk et al., 1995). The efficiency of the neo cassette deletion was monitored by isolating DNA from the embryo proper and yolk sac. Neo cassette deletion was complete in most embryos, and the results from embryo and yolk sac DNA were matching. Subsequently, the genotype of the embryos was inferred from yolk sac DNA analysis.
Western blot analysis, immunoprecipitation and HAT assays
Total cell extracts were prepared in IPH buffer [50 mM Tris–HCl pH 8.0, 150 mM NaCl, 5 mM EDTA, 0.1 mM phenyl-methylsulfonyl fluoride (PMSF)]. Western blot and immunoprecipitation experiments were performed with the p300 antibody RW128 and the CBP antibody AC26 (Upstate Biotechnology). For E1A binding experiments, ES cells were infected with adenovirus Ad5 at a multiplicity of infection of 10. Following cell lysis in IPH buffer, the E1A protein was immunoprecipitated with M73 antibody and co-precipitated CBP and p300 proteins were detected by western blotting. HAT activity was determined by assaying the enzymatic activity of p300 or CBP proteins immunoprecipitated from 2 mg of total cell extract incubated with purified histones and acetyl-CoA (Sigma) as described previously (Bordoli et al., 2001b).
ES cell growth and differentiation
Wild-type E14 ES cells and their derivatives were maintained in Dulbecco′s modified Eagle′s medium (DMEM) supplemented with 15% defined fetal calf serum (FCS) (Hyclone), 2 mM l-glutamine, 100 IU penicillin, 100 µg/ml streptomycin, 20 mM HEPES buffer, 0.085 mM β-mercaptoethanol and 500 IU/ml leukaemia inhibitory factor (LIF) on dishes coated with 0.1% gelatine.
For skeletal muscle differentiation experiments, ES cells were maintained in Iscove′s modified Eagle′s medium (IMDM) supplemented with 15% FCS, 2 mM l-glutamine, 100 IU penicillin, 100 g/ml streptomycin, 450 µM monothioglycerol and 1× non-essential amino acids (Invitrogen). EBs were generated as described (Wobus et al., 2001). At day 0 (D0), 600 cells were allowed to aggregate into EB in 20 µl hanging drops. At D2, the EBs were transferred to bacterial dishes. At D5, the EBs were plated out. The medium was exchanged every 3 days.
Osteogenesis was induced as described previously (Gollner et al., 2001; Phillips et al., 2001).
Immunofluorescence
Differentiated EBs, attached to coverslips, were fixed in 3:1 methanol–acetic acid at –20°C for 20 min prior to staining with the MHC antibody MF 20 overnight at 37°C in a humidified chamber, following standard procedures. Quantification was performed using the Adobe quantification toolpack.
RT–PCR
RNA was extracted from 100–150 differentiated EBs using Trizol® reagent (Life Technologies) according to the manufacturer’s instructions. The RNA was treated with RNase-free DNase and 1 µg of total RNA was used per reaction. The reaction was performed using the Qiagen OneStep RT–PCR kit. The PCR primers are designed within different exons so that they amplify exon as well as intron sequences, allowing PCR products derived from mature or precursor transcripts to be distinguished. Primer sequences are available upon request.
Mouse histology
Haematoxylin–eosin (HE) staining was performed according to standard procedures. Immunohistochemistry on paraffin sections was performed using the histomouse Kit (Zymed), including a quenching step and using the Dako solution™ for unmasking. The antibodies used were anti α-sarcomeric actin (Sigma A2172), anti-MyoD (Santa Cruz Biotechnology M318) and anti-myogenin (Santa Cruz Biotechnology M225).
Whole-mount hybridization
Whole-mount in situ hybridization was performed as described (Wilkinson 1992). The antisense probes were labelled with digoxigenin and generated using the following plasmids: mouse MyoD from pEMC11s (a gift from A.Lassar), mouse Myf5 from pBS-Myf5 (kindly provided by T.Braun) and myogenin from pSP-myogenin, which contains a 1.4 kb EcoRI fragment of the rat myogenin gene. Whole-mount immunohistochemistry was performed according to standard procedures with MF20 antibody.
Supplementary data
Supplementary data are available at The EMBO Journal Online.
Acknowledgments
Acknowledgements
We thank Gisele Höpfl and Thomas Braun for technical advice, Anita Nussbaumer and Marianne Antony for maintenance of the mouse colony and Fritz Ochsenbein for helping with the preparation of graphics. This work was supported by a grant from the Forschungskommission of the University of Zürich (J.F.R and R.E), the Roche Research Foundation (N.S), and a grant and START fellowship from the Swiss National Science Foundation (R.E) and the Kanton of Zürich.
References
- Akimaru H., Hou,D.X. and Ishii,S. (1997) Drosophila CBP is required for dorsal-dependent twist gene expression. Nat. Genet., 17, 211–214. [DOI] [PubMed] [Google Scholar]
- Arnold H.H. and Winter,B. (1998) Muscle differentiation: more complexity to the network of myogenic regulators. Curr. Opin. Genet. Dev., 8, 539–544. [DOI] [PubMed] [Google Scholar]
- Bailey P., Holowacz,T. and Lassar,A.B. (2001) The origin of skeletal muscle stem cells in the embryo and the adult. Curr. Opin. Cell Biol., 13, 679–689. [DOI] [PubMed] [Google Scholar]
- Bannister A.J. and Kouzarides,T. (1996) The CBP co-activator is a histone acetyltransferase. Nature, 384, 641–643. [DOI] [PubMed] [Google Scholar]
- Bober E., Franz,T., Arnold,H.H., Gruss,P. and Tremblay,P. (1994) Pax-3 is required for the development of limb muscles: a possible role for the migration of dermomyotomal muscle progenitor cells. Development, 120, 603–612. [DOI] [PubMed] [Google Scholar]
- Bordoli L., Husser,S., Luthi,U., Netsch,M., Osmani,H. and Eckner,R. (2001a) Functional analysis of the p300 acetyltransferase domain: the PHD finger of p300 but not of CBP is dispensable for enzymatic activity. Nucleic Acids Res., 29, 4462–4471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bordoli L., Netsch,M., Luthi,U., Lutz,W. and Eckner,R. (2001b) Plant orthologs of p300/CBP: conservation of a core domain in metazoan p300/CBP acetyltransferase-related proteins. Nucleic Acids Res., 29, 589–597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borycki A.G. and Emerson,C.P. (1997) Muscle determination: another key player in myogenesis? Curr. Biol., 7, R620–R623. [DOI] [PubMed] [Google Scholar]
- Boyes J., Byfield,P., Nakatani,Y. and Ogryzko,V. (1998) Regulation of activity of the transcription factor GATA-1 by acetylation. Nature, 396, 594–598. [DOI] [PubMed] [Google Scholar]
- Braun T. and Arnold,H.H. (1994) ES-cells carrying two inactivated Myf-5 alleles form skeletal muscle cells: activation of an alternative Myf-5-independent differentiation pathway. Dev. Biol., 164, 24–36. [DOI] [PubMed] [Google Scholar]
- Braun T. and Arnold,H.H. (1996) Myf-5 and MyoD genes are activated in distinct mesenchymal stem cells and determine different skeletal muscle cell lineages. EMBO J., 15, 310–318. [PMC free article] [PubMed] [Google Scholar]
- Brent A. and Tabin,C. (2002) Developmental regulation of somite derivatives: muscle, cartilage and tendon. Curr. Opin. Genet. Dev., 12, 548. [DOI] [PubMed] [Google Scholar]
- Buckingham M. (2001) Skeletal muscle formation in vertebrates. Curr. Opin. Genet. Dev., 11, 440–448. [DOI] [PubMed] [Google Scholar]
- Chan H.M. and La Thangue,N.B. (2001) p300/CBP proteins: HATs for transcriptional bridges and scaffolds. J. Cell Sci., 114, 2363–2373. [DOI] [PubMed] [Google Scholar]
- Desbaillets I., Ziegler,U., Groscurth,P. and Gassmann,M. (2000) Embryoid bodies: an in vitro model of mouse embryogenesis. Exp. Physiol., 85, 645–651. [PubMed] [Google Scholar]
- Eckner R., Yao,T.P., Oldread,E. and Livingston,D.M. (1996) Interaction and functional collaboration of p300/CBP and bHLH proteins in muscle and B-cell differentiation. Genes Dev., 10, 2478–2490. [DOI] [PubMed] [Google Scholar]
- Gollner H., Dani,C., Phillips,B., Philipsen,S. and Suske,G. (2001) Impaired ossification in mice lacking the transcription factor Sp3. Mech. Dev., 106, 77–83. [DOI] [PubMed] [Google Scholar]
- Goodman R.H. and Smolik,S. (2000) CBP/p300 in cell growth, transformation and development. Genes Dev., 14, 1553–1577. [PubMed] [Google Scholar]
- Gu W. and Roeder,R.G. (1997) Activation of p53 sequence-specific DNA binding by acetylation of the p53 C-terminal domain. Cell, 90, 595–606. [DOI] [PubMed] [Google Scholar]
- Gustafsson M.K., Pan,H., Pinney,D.F., Liu,Y., Lewandowski,A., Epstein,D.J. and Emerson,C.P., Jr. (2002) Myf5 is a direct target of long-range Shh signalling and Gli regulation for muscle specification. Genes Dev., 16, 114–126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hung H.L., Lau,J., Kim,A.Y., Weiss,M.J. and Blobel,G.A. (1999) CREB-binding protein acetylates hematopoietic transcription factor GATA-1 at functionally important sites. Mol. Cell. Biol., 19, 3496–3505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imhof A., Yang,X.J., Ogryzko,V.V., Nakatani,Y., Wolffe,A.P. and Ge,H. (1997) Acetylation of general transcription factors by histone acetyltransferases. Curr. Biol., 7, 689–692. [DOI] [PubMed] [Google Scholar]
- Jenuwein T. and Allis,C.D. (2001) Translating the histone code. Science, 293, 1074–1080. [DOI] [PubMed] [Google Scholar]
- Kasper L.H., Boussouar,F., Ney,P.A., Jackson,C.W., Rehg,J., van Deursen,J.M. and Brindle,P.K. (2002) A transcription-factor-binding surface of coactivator p300 is required for haematopoiesis. Nature, 419, 738–743. [DOI] [PubMed] [Google Scholar]
- Kaul A., Koster,M., Neuhaus,H. and Braun,T. (2000) Myf-5 revisited: loss of early myotome formation does not lead to a rib phenotype in homozygous Myf-5 mutant mice. Cell, 102, 17–19. [DOI] [PubMed] [Google Scholar]
- Kawasaki H., Eckner,R., Yao,T.-P., Taira,K., Chiu,R., Livingston,D.M. and Yokoyama,K.K. (1998) Distinct roles of the co-activators p300 and CBP in retinoic-acid-induced F9-cell differentiation. Nature, 393, 284–289. [DOI] [PubMed] [Google Scholar]
- Kung A.L., Rebel,V.I., Bronson,R.T., Ch’ng,L.E., Sieff,C.A., Livingston,D.M. and Yao,T.P. (2000) Gene dose-dependent control of hematopoiesis and hematologic tumor suppression by CBP. Genes Dev., 14, 272–277. [PMC free article] [PubMed] [Google Scholar]
- Maroto M., Reshef,R., Munsterberg,A.E., Koester,S., Goulding,M. and Lassar,A.B. (1997) Ectopic Pax-3 activates MyoD and Myf-5 expression in embryonic mesoderm and neural tissue. Cell, 89, 139–148. [DOI] [PubMed] [Google Scholar]
- McKinsey T.A., Zhang,C.L. and Olson,E.N. (2001) Control of muscle development by dueling HATs and HDACs. Curr. Opin. Genet. Dev., 11, 497–504. [DOI] [PubMed] [Google Scholar]
- Narlikar G.J., Fan,H.Y. and Kingston,R.E. (2002) Cooperation between complexes that regulate chromatin structure and transcription. Cell, 108, 475–487. [DOI] [PubMed] [Google Scholar]
- Ogryzko V.V., Schiltz,R.L., Russanova,V., Howard,B.H. and Nakatani,Y. (1996) The transcriptional coactivators p300 and CBP are histone acetyltransferases. Cell, 87, 953–959. [DOI] [PubMed] [Google Scholar]
- Ott M.O., Bober,E., Lyons,G., Arnold,H. and Buckingham,M. (1991) Early expression of the myogenic regulatory gene, Myf-5, in precursor cells of skeletal muscle in the mouse embryo. Dev. Suppl., 111, 1097–1107. [DOI] [PubMed] [Google Scholar]
- Partanen A., Motoyama,J. and Hui,C.C. (1999) Developmentally regulated expression of the transcriptional cofactors/histone acetyltransferases CBP and p300 during mouse embryogenesis. Int. J. Dev. Biol., 43, 487–494. [PubMed] [Google Scholar]
- Phillips B.W., Belmonte,N., Vernochet,C., Ailhaud,G. and Dani,C. (2001) Compactin enhances osteogenesis in murine embryonic stem cells. Biochem. Biophys. Res. Commun., 284, 478–484. [DOI] [PubMed] [Google Scholar]
- Polesskaya A., Duquet,A., Naguibneva,I., Weise,C., Vervisch,A., Bengal,E., Hucho,F., Robin,P. and Harel-Bellan,A. (2000) CREB-binding protein/p300 activates MyoD by acetylation. J. Biol. Chem., 275, 34359–34364. [DOI] [PubMed] [Google Scholar]
- Polesskaya A., Naguibneva,I., Fritsch,L., Duquet,A., Ait-Si-Ali,S., Robin,P., Vervisch,A., Pritchard,L.L., Cole,P. et al. (2001) CBP/p300 and muscle differentiation: no HAT, no muscle. EMBO J., 20, 6816–6825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puri P.L., Avantaggiati,M.L., Balsano,C., Sang,N., Graessmann,A., Giordano,A. and Levrero,M. (1997a) p300 is required for MyoD-dependent cell cycle arrest and muscle-specific gene transcription. EMBO J., 16, 369–383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puri P.L., Sartorelli,V., Yang,X.J., Hamamori,Y., Ogryzko,V.V., Howard,B.H., Kedes,L., Wang,J.Y., Graessmann,A.et al. (1997b) Differential roles of p300 and PCAF acetyltransferases in muscle differentiation. Mol. Cell, 1, 35–45. [DOI] [PubMed] [Google Scholar]
- Puri P.L., Iezzi,S., Stiegler,P., Chen,T.T., Schiltz,R.L., Muscat,G.E., Giordano,A., Kedes,L., Wang,J.Y. et al. (2001) Class I histone deacetylases sequentially interact with MyoD and pRb during skeletal myogenesis. Mol. Cell, 8, 885–897. [DOI] [PubMed] [Google Scholar]
- Rebel V.I., Kung,A.L., Tanner,E.A., Yang,H., Bronson,R.T. and Livingston,D.M. (2002) Distinct roles for CREB-binding protein and p300 in hematopoietic stem cell self-renewal. Proc. Natl Acad. Sci. USA, 99, 14789–14794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohwedel J., Maltsev,V., Bober,E., Arnold,H.H., Hescheler,J. and Wobus,A.M. (1994) Muscle cell differentiation of embryonic stem cells reflects myogenesis in vivo: developmentally regulated expression of myogenic determination genes and functional expression of ionic currents. Dev. Biol., 164, 87–101. [DOI] [PubMed] [Google Scholar]
- Sabourin L.A. and Rudnicki,M.A. (2000) The molecular regulation of myogenesis. Clin. Genet., 57, 16–25. [DOI] [PubMed] [Google Scholar]
- Sartorelli V., Huang,J., Hamamori,Y. and Kedes,L. (1997) Molecular mechanisms of myogenic coactivation by p300: direct interaction with the activation domain of MyoD and with the MADS box of MEF2C. Mol. Cell. Biol., 17, 1010–1026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sartorelli V., Puri,P.L., Hamamori,Y., Ogryzko,V., Chung,G., Nakatani,Y., Wang,J.Y. and Kedes,L. (1999) Acetylation of MyoD directed by PCAF is necessary for the execution of the muscle program. Mol. Cell, 4, 725–734. [DOI] [PubMed] [Google Scholar]
- Schwenk F., Baron,U. and Rajewsky,K. (1995) A cre-transgenic mouse strain for the ubiquitous deletion of loxP-flanked gene segments including deletion in germ cells. Nucleic Acids Res., 23, 5080–5081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Y. and Mello,C. (1998) A CBP/p300 homolog specifies multiple differentiation pathways in Caenorhabditis elegans. Genes Dev., 12, 943–955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shikama N., Lyon,J. and Thangue,N.B.L. (1997) The p300/CBP family: integrating signals with transcription factors and chromatin. Trends Cell Biol., 7, 230–236. [DOI] [PubMed] [Google Scholar]
- Shikama N., Lutz,W., Kretzschmar,R., Sauter,N., Roth,J.F., Marino,S., Wittwer,J., Scheidweiler,A. and Eckner,R. (2003) Essential function of p300 acetyltransferase activity in heart, lung and small intestine formation. EMBO J., 22, 5175–5185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soutoglou E., Katrakili,N. and Talianidis,I. (2000) Acetylation regulates transcription factor activity at multiple levels. Mol. Cell, 5, 745–751. [DOI] [PubMed] [Google Scholar]
- Tajbakhsh S. and Buckingham,M. (2000) The birth of muscle progenitor cells in the mouse: spatiotemporal considerations. Curr. Top. Dev. Biol., 48, 225–268. [DOI] [PubMed] [Google Scholar]
- Tajbakhsh S., Rocancourt,D., Cossu,G. and Buckingham,M. (1997) Redefining the genetic hierarchies controlling skeletal myogenesis: Pax-3 and Myf-5 act upstream of MyoD. Cell, 89, 127–138. [DOI] [PubMed] [Google Scholar]
- Tajbakhsh S., Borello,U., Vivarelli,E., Kelly,R., Papkoff,J., Duprez,D., Buckingham,M. and Cossu,G. (1998) Differential activation of Myf5 and MyoD by different Wnts in explants of mouse paraxial mesoderm and the later activation of myogenesis in the absence of Myf5. Dev. Suppl., 125, 4155–4162. [DOI] [PubMed] [Google Scholar]
- Tanaka Y., Naruse,I., Maekawa,T., Masuya,H., Shiroishi,T. and Ishii,S. (1997) Abnormal skeletal patterning in embryos lacking a single Cbp allele: a partial similarity with Rubinstein–Taybi syndrome. Proc. Natl Acad. Sci. USA, 94, 10215–10220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka Y., Naruse,I., Hongo,T., Xu,M., Nakahata,T., Maekawa,T. and Ishii,S. (2000) Extensive brain hemorrhage and embryonic lethality in a mouse null mutant of CREB-binding protein. Mech. Dev., 95, 133–145. [DOI] [PubMed] [Google Scholar]
- Weitzer G., Milner,D.J., Kim,J.U., Bradley,A. and Capetanaki,Y. (1995) Cytoskeletal control of myogenesis: a desmin null mutation blocks the myogenic pathway during embryonic stem cell differentiation. Dev. Biol., 172, 422–439. [DOI] [PubMed] [Google Scholar]
- Wilkinson D.G. (1992) Whole mount in situ hybridisation of vertebrate embryos. In Wilkinson,D.G. (ed.), In Situ Hybridization: A Practical Approach. IRL Press, Oxford, UK, pp. 75–82. [Google Scholar]
- Wobus A.M., Guan,K. and Pich,U. (2001) In vitro differentiation of embryonic stem cells and analysis of cellular phenotypes. Methods Mol. Biol., 158, 263–286. [DOI] [PubMed] [Google Scholar]
- Xu W., Edmondson,D.G., Evrard,Y.A., Wakamiya,M., Behringer,R.R. and Roth,S.Y. (2000) Loss of Gcn5l2 leads to increased apoptosis and mesodermal defects during mouse development. Nat. Genet., 26, 229–232. [DOI] [PubMed] [Google Scholar]
- Yamauchi T., Yamauchi,J., Kuwata,T., Tamura,T., Yamashita,T., Bae,N., Westphal,H., Ozato,K. and Nakatani,Y. (2000) Distinct but overlapping roles of histone acetylase PCAF and of the closely related PCAF-B/GCN5 in mouse embryogenesis. Proc. Natl Acad. Sci. USA, 97, 11303–11306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao T.-P., Oh,S.P., Fuchs,M., Zhou,N.-D., Ch’ng,L.-E., Newsome,D., Bronson,R.T., Livingston,D.M. and Eckner,R. (1998) Gene dosage-dependent embryonic development and proliferation defects in mice lacking the transcriptional integrator p300. Cell, 93, 361–372. [DOI] [PubMed] [Google Scholar]
- Yuan W., Condorelli,G., Caruso,M., Felsani,A. and Giordano,A. (1996) Human p300 protein is a coactivator for the transcription factor MyoD. J. Biol. Chem., 271, 9009–9013. [DOI] [PubMed] [Google Scholar]