Abstract
Productive cis folding by the chaperonin GroEL is triggered by the binding of ATP but not ADP, along with cochaperonin GroES, to the same ring as non-native polypeptide, ejecting polypeptide into an encapsulated hydrophilic chamber. We examined the specific contribution of the γ-phosphate of ATP to this activation process using complexes of ADP and aluminium or beryllium fluoride. These ATP analogues supported productive cis folding of the substrate protein, rhodanese, even when added to already-formed, folding-inactive cis ADP ternary complexes, essentially introducing the γ-phosphate of ATP in an independent step. Aluminium fluoride was observed to stabilize the association of GroES with GroEL, with a substantial release of free energy (−46 kcal/mol). To understand the basis of such activation and stabilization, a crystal structure of GroEL–GroES–ADP·AlF3 was determined at 2.8 Å. A trigonal AlF3 metal complex was observed in the γ-phosphate position of the nucleotide pocket of the cis ring. Surprisingly, when this structure was compared with that of the previously determined GroEL–GroES–ADP complex, no other differences were observed. We discuss the likely basis of the ability of γ-phosphate binding to convert preformed GroEL–GroES–ADP–polypeptide complexes into the folding-active state.
Keywords: aluminium fluoride, beryllium fluoride, chaperonin, transition-state analogue
Introduction
A variety of molecular machines in the cell employ the energy of nucleotide triphosphates to carry out vital processes (Alberts, 1998). Among these, the collective of large ring assemblies known as chaperonins have been shown to provide essential kinetic assistance to the folding of many proteins to their native form (Sigler et al., 1998; Thirumalai and Lorimer, 2001; Hartl and Hayer-Hartl, 2002). Both an early organelle study (Ostermann et al., 1989) and subsequent in vitro reconstitution experiments (Goloubinoff et al., 1989; Gao et al., 1992) revealed an absolute requirement for ATP in providing such action. Notably, both ADP and a variety of ATP analogues, including ATPγS and AMP-PNP, failed to produce the native state.
Studies of the bacterial chaperonin, GroEL, have provided the most detailed picture of the action of ATP. Current understanding of the GroEL–GroES reaction cycle indicates that a GroEL ring progresses through several states: an open ring that is binding-competent, exposing a hydrophobic surface that can multivalently capture a non-native polypeptide; an ATP/GroES-bound ring that is folding-active, in which rigid body movements attendant to ATP/GroES binding have removed the hydrophobic surface away from the polypeptide, releasing it into a central cavity whose walls have become hydrophilic in character and which is capped by the cochaperonin GroES; and an ADP/GroES-bound ring that is weakened in affinity for GroES and ‘primed’ for allosteric ejection of the ligands by ATP binding to the opposite ring. The formation and turnover of these states have been recognized to be governed by the binding and hydrolysis of ATP. For example, the critical transition from a polypeptide-accepting state to the folding-active GroES-encapsulated state is driven by the cooperative binding of ATP (Yifrach and Horovitz, 1995), followed by GroES, to a polypeptide-bound ring (Weissman et al., 1995, 1996; Rye et al., 1997). ATP binding promotes the initial elevation and twisting movements of the apical domains (Ranson et al., 2001), whose full extent is driven and stabilized by GroES association (Roseman et al., 1996; Xu et al., 1997; Rye et al., 1999; see also Ma et al., 2000). These large rigid body movements are associated with ejection of substrate polypeptide into the GroES-encapsulated, so-called cis ring, where folding commences (Mayhew et al., 1996; Weissman et al., 1996; Rye et al., 1997). It came as a surprise that ATP hydrolysis is not required for triggering productive folding. That is, folding can proceed essentially quantitatively to the native state inside a GroES-bound GroEL ring in the absence of ATP hydrolysis, e.g. in a hydrolysis-defective mutant of GroEL (D398A). By contrast, release of the ligands, including refolded polypeptide, cannot occur while the ring occupies an ATP/GroES-bound state, because such GroEL–GroES–ATP complexes prove to be very stable against dissociation (Rye et al., 1997). Thus, the requirement for ATP hydrolysis in the chaperonin cycle was found to lie not in triggering folding, but in weakening such folding-active complexes; ‘priming’ them for release that is allosterically directed by ATP (and further accelerated by non-native polypeptide) binding to the opposite (trans) GroEL ring (Rye et al., 1997, 1999).
While these basic actions of ATP in the GroEL–GroES system have been identified, the issue has remained as to exactly what structural and functional changes the γ-phosphate of ATP is promoting. For example, ADP can also promote formation of GroEL–GroES complexes that look nearly identical using cryoelectron microscopy (30 Å resolution) to those formed in ATP (Roseman et al., 1996; Rye et al., 1999), but if a GroEL–GroES-requiring polypeptide was initially bound at GroEL, the substrate fails to be released into the central cavity and folded by such cis ternary complexes (Weissman et al., 1996; Rye et al., 1997). Why then, if a GroES-bound complex can be also formed in ADP, does only ATP trigger productive folding? At the other end of a folding cycle, after 8–10 s of productive folding inside a cis ATP ternary complex, ADP complexes produced by hydrolysis of cis ATP continue to be folding-active. Yet they are ‘primed’ for release of the ligands, i.e. weakened in the association of GroES with GroEL (Rye et al., 1997). What is the basis for this?
To address these questions, we have carried out both functional and structural studies here using complexes of ADP with aluminium fluoride and beryllium fluoride, which have been used in a variety of systems to simulate different states of the γ-phosphate along the reaction coordinate of ATP hydrolysis (e.g. Bigay et al., 1987; Fisher et al., 1995; Menz et al., 2001). Notably, such metal complexes have already been employed in the study of chaperonins of the type II category, present in archaebacteria (thermosome) and the eukaryotic cytosol (CCT), which harbour a built-in lid structure that protrudes from their apical domains. For example, an ADP·AlF3-complexed crystal structure of the thermosome has provided stereochemical information on the mechanism of ATP hydrolysis at chaperonins (Ditzel et al., 1998). A study of ADP·BeFx-complexed CCT revealed that this state exhibits little or no affinity for non-native actin substrate (Melki and Cowan, 1994). More recently, further studies of CCT complexed with ADP·AlFx, using equilibrium sedimentation and electron microscopy (EM) in one study, and small angle X-ray scattering and substrate sensitivity to proteolysis in another, indicated that the built-in lid structure is closed in this state, although whether associated polypeptide reaches native form was not determined (Melki et al., 1997; Meyer et al., 2003). Here we have studied ADP·AlFx complexes of GroEL–GroES both biochemically and crystallographically, determining the efficacy of ADP·AlFx in supporting folding, the energetics of formation of the folding-active state, i.e. of ADP, GroES and γ-phosphate binding, and the structure of this complex. The results are discussed in the context of the established and here further-resolved GroEL–GroES nucleotide cycle.
Results
Folding triggered by GroEL–GroES–ADP·AlFx or GroEL–GroES–ADP·BeFx
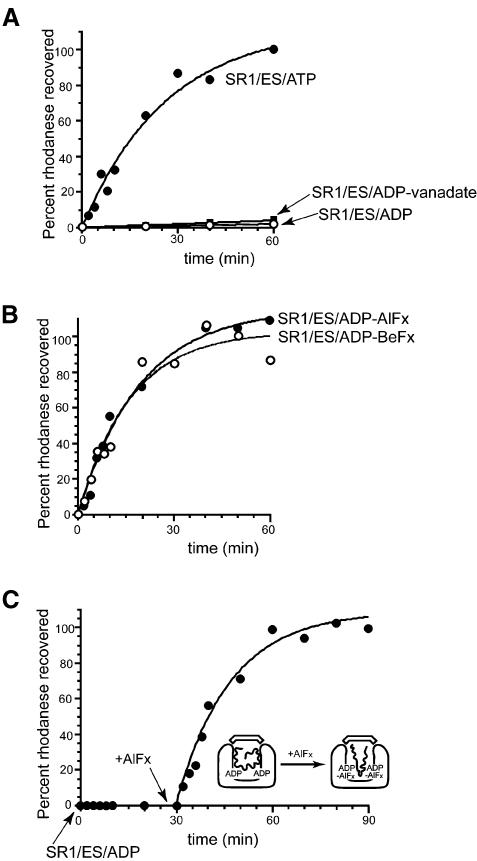
We tested whether a transition state analogue of ATP hydrolysis, ADP·AlFx, could trigger productive folding of the monomeric substrate protein, rhodanese, in a cis ternary complex. For this test, the single-ring chaperonin molecule, SR1, a minimal, cis-only version of GroEL, was utilized. This molecule binds non-native substrate protein in its central cavity and then, upon binding ATP and GroES, mediates productive folding. Non-native rhodanese was diluted from denaturant in the presence of SR1 to form a binary complex to which ADP, aluminium fluoride and GroES were added. The reaction mixture was assayed at varying times for acquisition of rhodanese activity, and we observed that the enzyme was reactivated with kinetics identical to that when ATP was used (compare Figure 1B with A). The ground state analogue, ADP·BeFx, was identically productive (Figure 1B), indicating that complexes simulating the γ-phosphate at several points along the reaction coordinate could function productively. By contrast, however, neither ADP nor the post-hydrolysis analogue ADP·vanadate (Goodno, 1982; Rayment, 1996) could trigger rhodanese refolding (Figure 1A). An additional substrate protein, malate dehydrogenase (MDH), was tested and exhibited the same behaviour (see Supplementary figure 1, available at The EMBO Journal Online). Similar incubations were carried out with binary complexes of rhodanese and wild-type, double-ring GroEL. Here also, ADP·AlFx and ADP·BeFx were productive, while ADP was not. The extent of recovery of rhodanese activity in these experiments was ∼50%, corresponding to the fraction of the ternary complexes predicted to be in the cis configuration (data not shown).
Fig. 1. AlFx and BeFx complexes support cis folding of the substrate protein rhodanese in the presence of ADP. (A and B) Time course of the recovery of rhodanese activity inside the complex formed between the single-ring GroEL mutant, SR1 and GroES. Binary complexes between urea-denatured rhodanese (1 µM) and SR1 (2 µM) were formed and mixed with GroES (4 µM) and (A) either 3 mM ATP, 5 mM ADP plus 10 mM vanadate, or 5 mM ADP, or (B) 5 mM ADP plus either AlFx [30 mM KF and 3 mM KAl(SO4)2] or BeFx (30 mM KF and 3 mM BeSO4). Enzymatic activity of the rhodanese monomer was assayed directly at the indicated times without disruption of the cis complex. Activity is expressed as a fraction of the final yield of the ATP-driven reaction at 60 min. (C) Ordered addition of the AlFx γ-phosphate analogue to a preformed, folding-inactive SR1–GroES–ADP–rhodanese complex also triggers rhodanese folding. A binary complex was formed between urea-denatured rhodanese (1 µM) and SR1 (2 µM), and mixed with GroES (4 µM) and 5 mM ADP. After 30 min, AlFx [30 mM KF and 3 mM KAl(SO4)2] was added to the mixture. Aliquots were taken for enzyme assay at the indicated times both before and after AlFx addition. In each panel, the results from a representative experiment are presented.
Folding triggered by addition of AlFx to a preformed GroEL–GroES–ADP–polypeptide complex
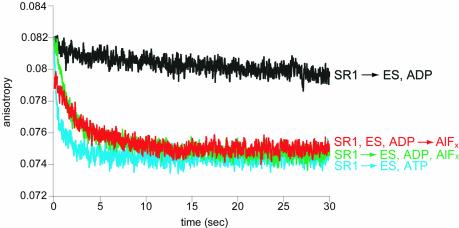
We asked whether adding aluminium fluoride separately to an already-formed folding-inactive (cis) SR1–GroES–ADP–polypeptide ternary complex could convert it to a folding-active state. That is, could one add, in effect, the γ-phosphate of ATP in a separate step? Remarkably, this order of addition triggered productive refolding of rhodanese, with kinetics identical to both the ATP and ADP·AlFx reactions (Figure 1C). Correspondingly, when the fluorescence anisotropy of a pyrene probe attached to rhodanese was examined, a rapid drop was observed when aluminium fluoride was added to an SR1–GroES–ADP–rhodanese complex (Figure 2), reproducing the previously observed behaviour upon GroES/ATP addition to SR1–rhodanese (Weissman et al., 1996). In contrast, addition of GroES/ADP to SR1–rhodanese failed to promote any change of anisotropy (Figure 2), reflecting that polypeptide failed to be displaced from the cavity wall by such an addition (see Supplementary figure 2). Thus, addition of an aluminium fluoride complex [or a beryllium fluoride complex (not shown)] could indeed simulate the addition of the γ-phosphate of ATP, sending the 470 kDa SR1–GroES–ADP assembly into its folding-productive state.
Fig. 2. Rhodanese is released into the cis cavity following addition of GroES and either ATP or ADP·AlFx to SR1–rhodanese. Binary complexes between urea-denatured, pyrene-labelled rhodanese and SR1 were mixed (1:1) in a stopped-flow apparatus with solutions containing 10 µM GroES and either 3 mM ATP (blue trace), 5 mM ADP (black trace) or 5 mM ADP and AlFx [3 mM KAl(SO4)2 and 30 mM KF] (green trace). The AlFx mixture alone was also mixed (1:1) in the stopped-flow with a solution of preformed SR1–rhodanese-GroES–ADP complex (red trace). The anisotropy of the pyrene label, reflecting the mobility of the polypeptide and its release from the cavity walls, was monitored as a function of time after mixing. Traces represent summations of 10−15 runs.
In a further experiment, we examined whether the activation of the folding process was a transient action, occurring only immediately following aluminium fluoride addition, or whether the activation of folding could occur at later times if polypeptide were effectively introduced into the central cavity of an already-formed GroEL–GroES–ADP·AlFx complex. To accomplish the latter, the substrate rhodanese was oxidatively cross-linked to the GroEL cavity wall of a cysteine-modified version of GroEL, effectively preventing its release and productive folding from occurring during the association of ADP, GroES and aluminium fluoride. After these additions had been made and the complex had been formed, the rhodanese was released into the central cavity by DTT reduction and its ability to fold was examined. Within the constraints of the inefficient binding of GroES under oxidizing conditions, production of native rhodanese was observed (Supplementary figure 3).
Structure of a GroEL–GroES–ADP·AlFx complex resembles that of GroEL–GroES–ADP
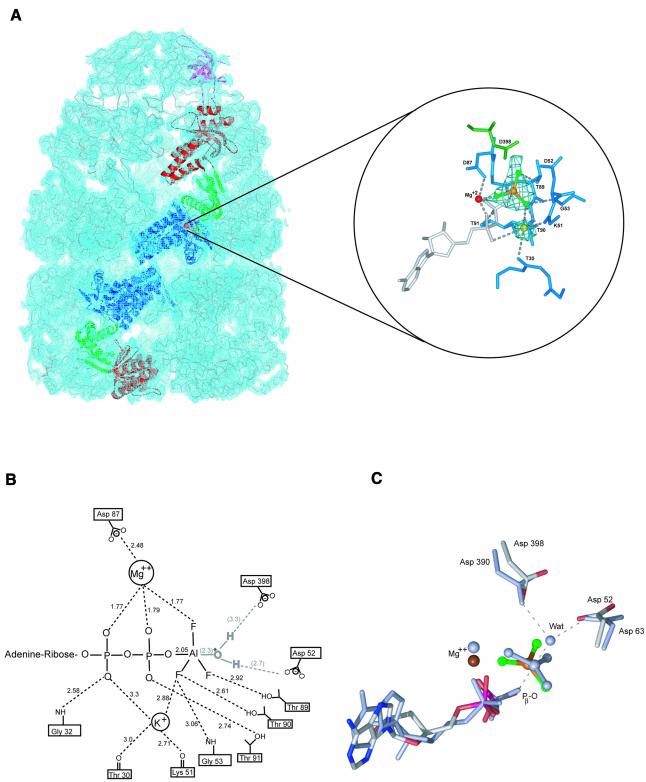
What structural changes are associated with binding ATP/GroES versus ADP/GroES to GroEL, or with binding aluminium fluoride to an already-formed GroEL–GroES–ADP complex? To address this, a GroEL–GroES–ADP·AlFx complex was crystallized and its structure solved by molecular replacement and refined against X-ray data to 2.8 Å resolution (R = 26.2%; Rfree = 27.8%; Table I). In the position of the equatorial nucleotide binding pocket of the cis ring, both ADP and a trigonal planar AlF3 moiety are clearly visible, the latter corresponding in position to the γ-phosphate of ATP, in σA-weighted difference and NCS-averaged maps (Figure 3A). The AlF3 directly contacts residues in the strictly conserved phosphate-binding loop (residues 87–91, DGTTT) through hydrogen bonds to the side-chain hydroxyls of Thr89 and Thr90 (Figure 3A and B; Table II). Asp398, a conserved residue in the intermediate domain whose mutation to Ala reduces ATP hydrolysis to 2% wild type (Rye et al., 1997) is positioned, along with Asp52, to interact with the AlF3, probably through an intermediate water molecule (Figure 3B and C). While this water was not well resolved in electron density, it was clearly observed in the complex of AlF3 with the thermosome (Ditzel et al., 1998), where virtually the same immediate contacts with an AlF3 ligand were observed (compare Asp63 and Asp390 with Asp52 and Asp398 in Figure 3C). By analogy with other AlFx-complexed NTPases, the AlF3 can be considered to mimic the trigonal bipyramidal geometry of the terminal phosphate undergoing nucleophilic attack by a water molecule (Scheffzek et al., 1997; Nassar et al., 1998). Here the negative charges on the aluminium fluoride are compensated by the backbone amide of Gly 53, as well as stabilizing interactions with the catalytic Mg+2 and what we interpret to be a second bound metal ion (Figure 3B). With respect to the latter, the strong electron density and the coordination geometry for this presumed metal ion suggests that it is K+, which was present at high levels in the crystallization mixture. This putative K+ ion stabilizes nucleotide binding by coordinating the α-phosphate, and it also stabilizes the γ-phosphate moiety through coordination with a fluoride ligand of AlF3, providing a direct mechanism for charge stabilization of the transition state. Notably, K+ is required for the ATPase activity of GroEL (Viitanen et al., 1990).
Table I. Data processing and refinement statistics for the GroEL–GroES–ADP·AlF3 complex.
| Spacegroup | P21212 |
| Cell dimensions (Å) | |
| a | 255.5 |
| b | 266.9 |
| c | 187.1 |
| Resolution (Å) | 50–2.8 |
| Unique reflections | 278 877 |
| Average redundancya | 4.0 (3.9) |
| Completeness (%)a,b | 90.3 (55.0) |
| I/σIa | 6.6 (1.3) |
| Rsym (%)a,c | 13.8 (60.1) |
| Refinement | |
| Reflections (|F|>0σ) | 278 591 |
| Number of protein atoms | 59 276 |
| Number of metal ions | 21 |
| Number of ADP molecules | 7 |
| R factor (%)d | 26.2 |
| Free R factor (%)e | 27.8 |
| R.m.s.d. bond lengths (Å) | 0.013 |
| R.m.s.d. deviation in bond angles (°) | 1.362 |
| Average B-factorf | 95.78 |
| Ramachandran statistics | |
| Most favorable (%) | 88.7 |
| Allowed (%) | 10.4 |
| Generously allowed (%) | 0.9 |
| Disallowed (%) | 0.0 |
aThe value for the highest resolution bin (2.87–2.8 Å) is given in parentheses.
bThe completeness is 91% to 3.0 Å. The completeness at 2.8 Å is low because these data were only recorded in the corners of the detector.
cRsym = Σ|Ih–<Ih>|/ΣIh, where <Ih> is the average over Friedel and symmetry equivalents.
dR factor = Σ||Fo|–|Fc||/Σ|Fo|, where Fc is the calculated structure factor.
eR free is as R factor but calculated for 2% of randomly chosen reflections that were omitted from the refinement.
fAverage isotropic B-factor derived from TLSANL (Howlin et al., 1993).
Fig. 3. Crystallographic model of the GroEL–GroES–ADP·AlF3 complex and illustration of the nucleotide-binding pocket. (A) Unbiased electron density map (light blue) calculated with coefficients Fobsexp(iαave) where the phases αave result from 7-fold NCS averaging, density modification, and phase extension starting from random phases. The map is contoured at 1σ and was calculated using all diffraction data between 50.0 Å and 2.8 Å resolution. A Cα trace is shown for the entire complex, with subunits from a single protomer highlighted as follows: the GroES subunit is shown in magenta; and the apical, intermediate and equatorial domains are shown in red, green and blue, respectively. (Inset) σA-weighted |Fo|−|Fc| electron density map contoured at 3.5 σ showing difference density for both AlF3, bound in the γ-phosphate position of the ATP binding pocket in the cis ring of GroEL−GroES, and a coordinating K+ metal ion. The protein is shown as a skeletal model, with relevant residues in the equatorial and intermediate domains coloured blue and green, respectively, ADP is white, the Mg2+ is a red sphere, K+ is a yellow sphere, and the trigonal AlF3 is shown as an orange (aluminium) and green (fluorine) ball-and-stick model. Interactions are indicated by grey dashed lines. (B) Schematic representation of the coordination of aluminium fluoride in the cis active site of GroEL–GroES. Interactions are shown as dotted lines. The catalytic water could not be unambiguously assigned, probably owing to the limited resolution of the diffraction data, and was not included in the final model, but is shown here to illustrate its likely interaction partners based on results from (C), a least-squares superposition of conserved interactions in the nucleotide binding site in GroEL–GroES and the thermosome complexed with ADP·AlF3 (Ditzel et al., 1998). The thermosome-derived side-chains ADP, AlF3, water (Wat) and Mg++ are shown in light blue.
Table II. Interactions in the nucleotide binding region for ADP- and ADP·AlF3-bound structures of GroEL–GroES.
| Interaction | GroEL–GroES–ADPa [length (Å)] | GroEL–GroES–ADP·AlF3 [length (Å)] |
|---|---|---|
| HOH (at γ-PO4)–ADP O3β | 2.16 | – |
| HOH (at γ-PO4)–Thr89 OG1 | 2.8 | – |
| Al3+–ADP O3β | – | 2.05 |
| F1–Mg2+ | – | 1.77 |
| F2–K+ | – | 2.88 |
| F2–Gly53 N | – | 3.06 |
| F2–Thr90 OG1 | – | 2.61 |
| F3–Thr89 OG1 | – | 2.92 |
| K+–Lys51 O | – | 2.71 |
| K+–Thr30 OG1 | – | 3.0 |
| K+–ADP O1α | – | 3.3 |
| HOH(*)–Al3+ | – | 2.3 |
| HOH(*)–Asp398 OD1 | – | 3.3 |
| HOH (*)–Asp52 OD1 | – | 2.7 |
aThe ADP structure (1AON) was re-refined (see Materials and methods, and Supplementary table I). A single-electron density peak was seen at the γ-phosphate position, and was modelled as a water molecule [denoted HOH (at γ-PO4)] based on coordination geometry and B-factor. HOH(*) is a putative water molecule, stabilized by Asp398 and Asp52, interacting with AIF3 (see text and Figure 3B and C).
Despite the presence of AlF3 and its formation of the bonds just described (Table II), when the structure was compared with the previously determined (Xu et al., 1997) and here further refined model of GroEL–GroES–ADP (see Materials and methods and Supplementary table I), we observed no significant difference in the tertiary or quaternary structure of the asymmetric GroEL–GroES complex [root mean square deviation (r.m.s.d.) ∼0.3 Å]. Local differences, at the level of apical domain secondary structure or in the GroES mobile loop, could also not be observed, although when a TLS (translation–libration–screw) refinement was carried out (Winn et al., 2001), the cis apical domains and GroES exhibited less local mobility in the ADP·AlF3 structure compared with the ADP structure (Supplementary figure 4). Concerns that lattice contacts in the ADP·AlF3 structure may have constrained potential domain movements (see Supplementary figure 5A) were addressed by crystallographic analysis of an SR1–GroES–ADP·AlFx complex. This weakly diffracting crystal lattice exhibited very sparse inter-molecular contacts (Supplementary figure 5B and table II). Although the diffraction was limited to 7.5 Å, the packing indicates that the apical and intermediate domains of the SR1 ring are not significantly shifted from their positions in the GroEL–GroES–ADP and GroEL–GroES–ADP·AlF3 complexes.
GroEL–GroES–ADP·AlF3 complexes are more stable to dissociation than GroEL–GroES–ADP complexes
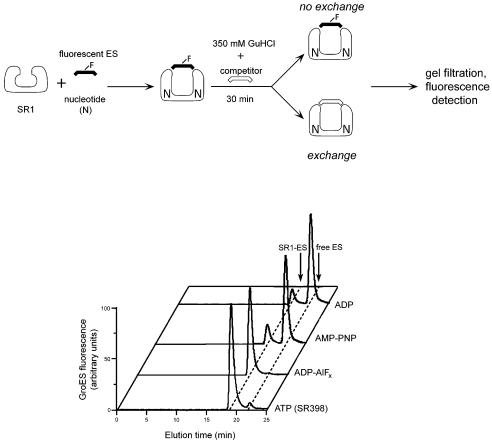
Despite the apparent similarity of GroEL–GroES–ADP·AlF3 and GroEL–GroES–ADP complexes as determined by X-ray crystallography at 3 Å, the former could trigger folding, whereas the latter could not. An additional effect of the presence of a ligand at the position of the γ-phosphate was also investigated. We had observed previously that a hydrolysis-defective mutant of GroEL, D398A, formed a very stable folding-active complex with GroES in the presence of ATP, resistant to exposure to 0.4 M GuHCl. By contrast, GroEL–GroES–ADP complexes readily dissociate; for example, in the absence of added ADP they dissociate during gel filtration (Rye et al., 1997). We examined the stability of the GroEL–GroES–ADP·AlFx complex by incubating fluorescently labelled GroES with SR1 in the presence of the various nucleotides, and assessing by in-line fluorescence detection whether GroES co-migrated with SR1 during gel filtration after exposure of the complexes to 0.35 M GuHCl for 30 min. In the case of ADP·AlFx, the complexes remained stable to such treatment, as had been observed for GroEL398–GroES–ATP complexes (Figure 4). By contrast, GroEL–GroES or SR1–GroES complexes formed with ADP (or with AMP-PNP or ATPγS) dissociated. Thus, the presence of the γ-phosphate or its addition as an AlFx analogue renders the GroEL–GroES complex very stable, effectively locking GroES onto the GroEL ring to which it is bound.
Fig. 4. Aluminium fluoride binding stabilizes the cis ADP complex, mimicking the effects of binding of intact ATP. The stability of the cis complex was probed by perturbation with a chaotrope (schematic). SR1 was incubated for 30 min at 25°C with fluorescently labelled GroES in the presence of either 5 mM ADP, 5 mM AMP-PNP or 3 mM ADP plus 3 mM KAl(SO4)2 and 30 mM KF; SR398 was similarly incubated with 3 mM ATP. Complexes were exposed to 0.35 M GuHCl for 30 min in the presence of excess unlabelled GroES competitor, then subjected to gel filtration with fluorescence detection. The results of a representative experiment are presented.
The energy landscape of cis complex formation: release of a large amount of free energy upon γ-phosphate binding
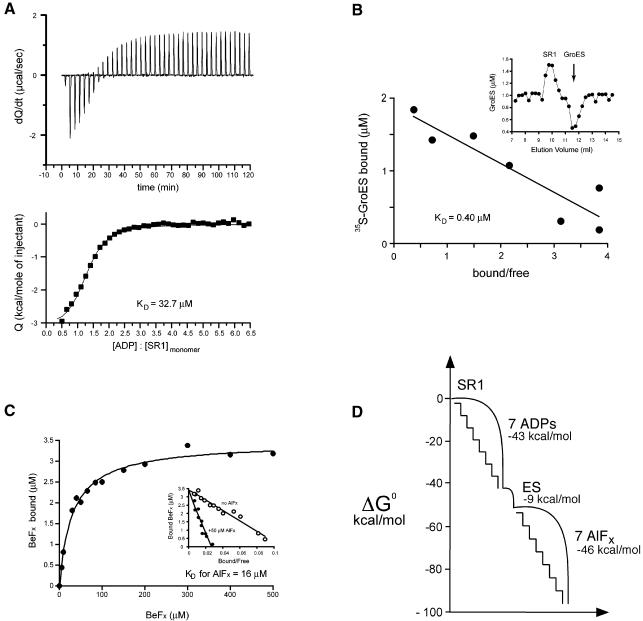
Because aluminium fluoride addition to a GroEL–GroES–ADP complex functionally mimicked the addition of the γ-phosphate, it was possible to assess the free energy change associated with γ-phosphate binding. In particular, the ability to separate productive cis complex formation into steps of ADP binding, GroES binding and aluminium fluoride binding enabled dissection of the energetics of complex formation. The free energy of ADP binding was determined by isothermal titration calorimetry (ITC), adding ADP to a solution of 12 µM SR1. An exothermic reaction was observed that showed saturation behaviour (Figure 5A). Fitting these data to a standard binding equation allowed an estimate of the number of sites (seven) and the KD for ADP (32.7 µM), from which the standard free energy of binding seven ADPs was calculated to be −43 kcal/mol. These results are comparable to those obtained by Inobe et al. (2001) in a similar ITC experiment (89 µM and −38.8 kcal/mol, respectively). The free energy of GroES binding was determined in a Hummel–Dreyer experiment by applying 0.5 nmol SR1 to a gel filtration column equilibrated with varying concentrations of radiolabelled GroES and 5 mM ADP, and measuring the amount of GroES eluting at the position of SR1 (as well as the corresponding deficiency at the position of GroES; see Figure 5B, inset). Plotting GroES bound versus bound/free (Figure 5B) allowed the determination of the KD for GroES (0.4 µM), enabling calculation of the free energy of GroES binding as −9 kcal/mol. Finally, to measure AlFx binding to SR1–GroES–ADP complexes, we employed a competitive binding assay between AlFx and [7Be]fluoride. Binding of [7Be]Fx by SR1–GroES–ADP was saturable and apparently non-cooperative (Figure 5C; see also Inobe et al., 2003). Aluminium fluoride (50 µM) exhibited competitive behaviour with respect to [7Be]Fx in binding to an SR1–GroES–ADP complex (Figure 5C, inset), allowing determination of the KD (16 µM) and calculation of the free energy of aluminium fluoride binding as −46 kcal/mol. The collective of free energy changes could therefore be plotted (Figure 5D), showing that there is a sizeable free energy change associated with aluminium fluoride binding, approximating that of binding ADP and GroES (particularly the nucleotide). This free energy change, much greater than the difference usually observed between native and unfolded conformations of a polypeptide, is associated with a strong stabilization of the GroEL–GroES complex and with immediate ejection of polypeptide off the cavity wall, into the cis chamber.
Fig. 5. Energetics of forming a folding-active cis complex in SR1, with discrete contributions of consecutive ADP, GroES and aluminium fluoride binding steps. (A) Isothermal calorimetric titration of SR1 with ADP at 25°C. (Upper panel) heat change produced at each injection of ADP; (bottom panel) the integrated data from the upper trace: the line was generated by the fit to the binding equation. A representative experiment is shown. ADP binding to SR1 is exothermic with a KD of 32.7 ± 1.4 µM (n = 3) and a stoichiometry of 7 ADP molecules/SR1 heptamer. (B) Affinity of SR1–ADP7 for GroES measured by a Hummel-Dreyer experiment. SR1 was chromatographed on a gel filtration column equilibrated with various amounts of [35S]GroES and 5 mM ADP; the inset shows a typical elution profile, here for 1 µM [35S]GroES. The area at the SR1 elution position was used to calculate the amount of bound GroES. Bound versus bound/free GroES was plotted (main figure), and the dissociation constant was determined from the slope of the line (−KD) to be 0.40 µM in two experiments. (C) Measurement of AlFx binding to SR1–ES–ADP by competition with [7Be]Fx. Main panel: the binding of [7Be]Fx to SR1–ES–ADP was measured by a spin-column assay (Materials and methods), and observed to be saturable and apparently non-cooperative (KD = 30 µM). (Inset) the effect of AlFx on [7Be]Fx binding is shown in an Eadie–Hofstee plot of the data from parallel binding experiments, one without and one with 50 µM AlFx. The maximal amount of [7Be]Fx bound (y-intercept) was unaltered in the presence of AlFx, demonstrating competitive binding between [7Be]Fx and AlFx. This allows the calculation of the KD for aluminium fluoride binding of 16 ± 1 µM (n = 3) (see Supplementary Materials and methods). (D) Estimated free energy transitions during activation of SR1. The standard free energy drops proceed in three stages [ADP binding (−42.7 kcal/mol), GroES binding (−8.7 kcal/mol) and aluminium fluoride binding (−45.8 kcal/mol)] and are computed at 25°C from the corresponding values of KD.
Discussion
Actions of the γ-phosphate of ATP at GroEL–GroES: ejecting polypeptide into the central cavity and stabilizing GroEL–GroES association
The foregoing functional studies, carried out with the addition of an aluminium fluoride complex to preformed, folding-inactive complexes of SR1–GroES–ADP or GroEL–GroES–ADP, show that binding of aluminium fluoride to such complexes is sufficient to trigger release of non-native polypeptide from the GroEL–GroES cavity wall into the central cavity, followed by folding to the native state in the cis cavity. Notably, the same productivity is observed for the ground state analogue beryllium fluoride, which is strictly tetra-coordinated and thus isomorphous to the γ-phosphate ground state (Bigay et al., 1987; Fisher et al., 1995), arguing that it is not the transition state of ATP hydrolysis, but the presence of the γ-phosphate, in what is presumably a continuum of early states generated along the hydrolysis reaction coordinate, which is sufficient to trigger ejection of substrate polypeptide into the central cavity and commencement of folding. Notably, a post-hydrolysis analogue, ADP·VO4, failed to trigger folding. Thus, it is the action of binding of the γ-phosphate moiety of ATP, forming a specific set of hydrogen bonds with both equatorial and intermediate domains of the GroEL–GroES machine, as observed here crystallographically with the aluminium fluoride analogue, that is sufficient to trigger initiation of protein folding. This agrees with earlier studies using the hydrolysis-defective D398A GroEL mutant, which showed that ATP/GroES binding, in the absence of ATP hydrolysis, was sufficient to trigger productive folding (Rye et al., 1997).
A further action of ATP/GroES binding, also observed earlier, was to stabilize the association of GroES with GroEL, effectively locking GroES onto GroEL and cementing together the cis chamber in which folding proceeds (Rye et al., 1997). Here also, addition of the aluminium fluoride complex to preformed GroEL–GroES–ADP complexes produced such stabilization, forming complexes that could not be dissociated even with 0.35 M GuHCl. Directly correlating with such stabilization of GroEL–GroES complexes was the release of a substantial amount of free energy upon binding of aluminium fluoride, i.e. ∼46 kcal/mol (Figure 5). Thus, the ATP state of a GroEL–GroES ring, the active and longest phase of the folding cycle, is also the most stable state.
Structural considerations: resemblance between GroEL–GroES–ADP·AlF3 and GroEL–GroES–ADP complexes
Surprisingly, the GroEL–GroES–ADP·AlF3 crystal structure determined here does not differ in the conformation of the intermediate and apical domains of its GroEL cis ring, nor in that of GroES, from those in the GroEL–GroES–ADP structure reported earlier (Xu et al., 1997) and re-refined here. The two structures, at ∼3 Å resolution, are superposable through these regions. Notably, an ongoing cryoEM study of D398A GroEL–GroES–ATP also fails to show any significant difference with either crystal structure, indicating that the similarity of the X-ray structures is not likely to be due to crystal lattice constraints (N.Ranson and H.Saibil, personal communication). Yet ATP or ADP·AlF3 can serve as a folding trigger and the GroEL–GroES complexes formed are extremely stable, whereas ADP cannot initiate a GroEL–GroES folding reaction and such a complex is much more readily dissociated. Can all of this be explained by local contacts between the γ-phosphate or aluminium fluoride complex and the equatorial and intermediate domains?
In the case of the stabilizing action of aluminium fluoride, it might be possible that contacts between the intermediate domain and the nucleotide pocket, for example between Asp398 and the γ-phosphate, could be serving effectively to stabilize an ‘arch’ structure that has GroES as its keystone. That is, formation of bonds that strengthen lateral support (Table II) at the level of the equatorial-intermediate contact, when rotationally multiplied 7-fold around the cis ring, may serve to strengthen the incorporation of the GroES ‘dome’ (see also the analysis of a thermosome–ADP·AlF3 complex in Ditzel et al., 1998). Additionally, the numerous contacts between the γ-phosphate and equatorial domain residues (Table II) would contribute to the overall stability of the folding-active structure.
Less clear from this structure, however, is how local bonding of aluminium fluoride in the equatorial domain could provide the impetus to eject polypeptide from the apical cavity walls of GroEL–GroES–ADP–polypeptide complexes. One possibility is that the structures of ADP complexes studied in the absence of non-native polypeptide do not reflect the structural properties of a cis ADP state formed in the presence of the substrate protein. Thus, the structure of a cis-ADP–polypeptide ring that serves as a starting point for rationalizing the effects of aluminium fluoride in triggering folding may be different from the GroEL–GroES–ADP structure, devoid of substrate polypeptide, determined previously. For example, biochemical studies, both earlier ones (Weissman et al., 1996; Rye et al., 1997) and those detailed here (Figure 2 and Supplementary figure 2), show that polypeptide remains associated with the cavity wall in de novo-formed GroEL–GroES–ADP complexes (where ADP/GroES has been added to GroEL-polypeptide binary complex). In such complexes, it may be that bound polypeptide acts as a load that pulls on the apical domains, resisting the ability of GroES to produce full elevation and twist of these domains. Shared occupancy of the apical domains could potentially involve either mutually exclusive interactions, with the GroES mobile loops interacting with some apical domains and polypeptide with others, or could possibly involve shared occupancy of different aspects of the same apical domains by both ligands. In either case, some or all of the apical domains of such GroEL–GroES–ADP–polypeptide complexes may be constrained in their degree of movement, remaining capable of undergoing significant further elevation and twist upon addition of aluminium fluoride, to reach a fully open position that is associated with release of substrate protein into the central cavity. Further structural analyses, particularly using cryoEM, may be able to resolve the state of de novo-formed cis ADP–polypeptide complexes in which both polypeptide and GroES are bound to the same GroEL ring.
While the structural basis of the difference in the ability of de novo-formed cis ATP and ADP complexes to trigger folding remains to be explored further, it should be pointed out that, in the context of the physiological nucleotide cycle, no structural difference between the ATP and ADP states would necessarily be expected. That is, under normal conditions, ATP is the physiological nucleotide for triggering GroES binding and cis complex formation, binding with an order of magnitude greater affinity than ADP (Jackson et al., 1993; Burston et al., 1995; Cliff et al., 1999). Such ATP cis complexes support folding for a period of ∼8–10 s, after which hydrolysis ensues and a short-lived cis ADP complex is formed (Todd et al., 1994; Ranson et al., 1997). This ADP complex remains folding-active, with no detectable phase in folding associated with its formation, as judged from recovery of both enzyme activity and native fluorescence properties of folding proteins during this phase. This has been most definitively observed for SR1–GroES-polypeptide complexes, where addition of GroES and ATP to a polypeptide-SR1 complex triggers folding that continues unabated inside the GroEL–GroES chamber for many minutes beyond the single turnover of ATP to ADP, which occurs in this setting after 8–10 s (Weissman et al., 1996; Rye et al., 1997). Thus, based on this seamless transition, it might not be surprising that a cis ADP complex would exhibit a very similar structure to a cis ATP one.
It should be noted that polypeptide does not appear to rebind to the cavity wall in SR1–GroES–ADP complexes where folding is ongoing after the initial ATP-driven release, even though a substantial portion of input polypeptide remains non-native at least during the first few minutes of refolding. This is likely a function of the behaviour of both polypeptide and machine. Regarding polypeptide, it is likely to rapidly undergo conformational changes upon GroES/ATP-triggered release that may bury some of the hydrophobic surfaces originally bound to the cavity wall of an open ring, making such surfaces less accessible for rebinding. However, on the part of the machine itself, it may not be capable of re-exposing any significant hydrophobic surface to the cavity following ATP hydrolysis, because the apical domains, once fully mobilized upon initial exposure to ATP and GroES, have placed all of their hydrophobic surfaces into stable contacts with either GroES or the interface that is formed between them. Such surfaces do not become re-exposed following cis hydrolysis, despite the weakened affinity of GroEL for GroES, and polypeptide thus cannot compete for the removed hydrophobic apical surface. Thus, in a sense, there is some hysteresis to the behaviour of polypeptide binding within GroEL–GroES–ADP complexes, with de novo-formed ones able to retain polypeptide on the cavity walls, but ones formed from cis hydrolysis unable to do so. Yet it seems that the former, de novo-formed complexes are likely to be structurally different, as commented on above. Notably, such de novo cis-ADP ternary complexes are not likely to be populated physiologically, and so such a structural state, with both polypeptide and GroES bound to the same ring, is not of physiological significance. On the other hand, these complexes have been very informative concerning the action of the γ-phosphate, as shown here, allowing the observation of the role its binding plays in driving the GroEL–GroES machine across its own energy landscape to a thermodynamic minimum that is the folding-active state.
We thus conclude that in the progression of a physiological reaction, cis GroEL–GroES complexes, formed first in ATP, undergo the full excursion of apical domain movement, producing polypeptide release and commencement of folding inside a very stable GroES-encapsulated cis cavity. These complexes then hydrolyse to a cis-ADP state that does not undergo any significant structural changes at the level of the cis apical and intermediate domains or in GroES. The latter complex is a weakened one, however, as the result of loss of lateral arch supports by loss of the γ-phosphate, and is thus ‘primed’ for dissociation, normally triggered by ATP binding to the opposite ring. The polypeptide ligand is released from the dissociating cis chamber in either a conformational state committed to reaching native form without further binding to GroEL, or a non-native form that requires rebinding and a further trial at folding.
Materials and methods
Proteins
GroEL, SR1, SR398 (a single-ring version of the ATP hydrolysis-defective mutant D398A) and GroES were purified as described previously (Rye et al., 1997). GroES98C was labelled with fluorescein-5-maleimide as described previously (Rye, 2001). Chaperonin concentrations were determined by amino acid analysis. Bovine rhodanese was labelled with pyrene maleimide as described previously (Weissman et al., 1995).
Rhodanese folding
Binary complexes were formed by unfolding rhodanese in 50 mM Tris–HCl pH 7.4, 7 M urea, 10 mM DTT, 1 mM EDTA, followed by a 100-fold dilution into buffer A (50 mM Tris–HCl pH 7.4, 10 mM MgCl2, 5 mM KCl, 5 mM DTT, 20 mM sodium thiosulfate) containing chaperonins, as indicated in Figure 1. Refolding was initiated by the addition of nucleotide, and rhodanese activity assayed as described previously (Weissman et al., 1994).
Stopped-flow fluorescence anisotropy
Fluorescence anisotropy changes were measured by stopped-flow as described previously (Rye et al., 1997). For these experiments, 400 µM urea-denatured, pyrene-labelled rhodanese was diluted 100-fold into buffer A containing 4 µM SR1. This binary complex was loaded into one syringe. A solution containing 10 µM GroES and nucleotide with or without AlFx, as indicated in Figure 2, was loaded into the second syringe. In one experiment, syringe 1 was loaded with preformed SR1–rhodanese-GroES–ADP complex, and syringe 2 was loaded with 3 mM KAl(SO4)2 and 30 mM KF. Reactions were initiated by mixing equal volumes from each syringe.
Structure determination of GroEL–GroES–ADP·AlF3
Complexes were prepared and isolated as described previously (Xu et al., 1997), except that buffers included 3 mM KAl(SO4)2 and 30 mM KF. Crystals were obtained by microseeding and frozen as described (Xu et al., 1997), with KCl replacing sodium glutamate and 1 mM YbCl3 added to improve diffraction quality of the crystals. Data to 2.8 Å were collected at the Advanced Photon Source using the ID-19 beamline and were processed with DENZO and SCALEPACK (HKL Research; Otwinowski and Minor, 1997). Crystals belonged to the space group P21212 with unit cell dimensions a = 255.5 Å, b = 266.9 Å, c = 187.1 Å. A re-refined GroEL–GroES–ADP structure (see below) was positioned in the unit cell by molecular replacement using CNS (Crystallography and NMR System version 1.1) (Brunger et al., 1998). Unbiased electron density maps were calculated after 7-fold symmetry averaging, density modification and phase extension, starting from random phases in CNS. Based on the strong density seen in the γ-phosphate position, trigonal planar AlF3 was added to the model. The model was further refined using non-crystallographic symmetry restraints, torsion angle simulated annealing, and conjugate gradient minimization. Subsequent TLS refinement was performed in REFMAC (version 5.1.24) (Murshudov et al., 1999; Winn et al., 2001) using 49 TLS groups (for each of the seven protomers, three domains for each GroEL subunit, and one for GroES). The resulting TLS tensors were analysed using TLSANL (Howlin et al., 1993). Final refinement cycles with REFMAC gave a model with Rfree = 0.278 and Rwork = 0.262 at 2.8 Å. Water molecules were not included at this resolution. Coordinates and structure factors are available from the Protein Data Bank (PDB; accession code 1PCQ).
Re-refinement of GroEL–GroES–ADP
To ensure a fair comparison between the GroEL–GroES–ADP and GroEL–GroES–ADP·AlF3 structures, the GroEL–GroES–ADP model was re-refined using the same protocols for CNS and TLS refinement outlined above. Starting coordinates and observed amplitudes were obtained from the PDB (accession code 1AON). The cis apical domains were also remodelled using the highest available resolution apical domain structure (PDB accession code 1KID). Coordinates and structure factors are available from the PDB (accession code 1PF9).
SR1–GroES complex stability assay
Complexes were formed by mixing 10 µM SR1 with 7.5 µM ESf (fluorescein-labelled GroES98C) and nucleotide in buffer B (50 mM Tris–HCl pH 7.5, 10 mM MgCl2, 5 mM KCl, 1 mM DTT) with or without AlFx, as indicated in Figure 4. After 30 min at 25°C, an aliquot was diluted 10-fold into buffer B containing 350 mM GuHCl and 10 µM unlabelled GroES (final concentrations). After 30 min exposure to GuHCl, the sample was applied to a Tosohaas G4000SWxl HPLC gel filtration column equilibrated in buffer B; the elution of ESf was monitored using an in-line fluorescence detector. All chromatograms were normalized to total peak area.
Binding assays
The affinity of SR1 for ADP was determined by isothermal titration calorimetry. The calorimetric cell was filled with 12 µM SR1 in buffer C (50 mM Tris–HCl pH 7.4, 10 mM MgCl2, 10 mM KF, 1 mM DTT) and titrated with 10 mM ADP in buffer C. The heat due to binding of ADP to SR1 was obtained as the difference between the heat of reaction and the corresponding heat of dilution. The enthalpy, association constant and number of binding sites were then calculated from the total binding isotherm using the program Origin (OriginLab Corp.).
GroES binding to SR1–ADP complexes was measured by a Hummel–Dreyer assay as described previously (Weissman et al., 1996), except that 250 µl of 2 µM SR1 was applied to the column with 5 mM ADP and varying concentrations of 35S-labelled GroES in the running buffer.
The affinity of SR1–GroES–ADP for beryllium and aluminium fluoride complexes was determined using 7Be, obtained from Brookhaven National Laboratory, diluted with BeSO4 to an appropriate final specific activity. SR1–GroES–ADP complexes were formed by mixing 0.55 µM SR1, 2.2 µM GroES and 3.3 mM ADP in buffer C at 25°C. After 30 min, 90 µl of this complex was added to 10 µl of 7Be solution (containing ∼0.05–1.5 µCi of 7Be), and the sample was equilibrated for 30 min. Chaperonin-bound 7Be was removed from free 7Be using a 1 ml spin column (Bio-Gel P30; Bio-Rad) exchanged with buffer C. Non-specific binding of BeFx was corrected for by subtracting the amount of 7Be bound to SR1–GroES complexes formed in the presence of 5 mM AMP-PNP from the data for those generated with ADP. The dissociation constant for BeFx was determined by fitting the data to a standard binding isotherm. Affinity for aluminium fluoride was measured by its ability to compete with beryllium fluoride. In these experiments, 7Be stock solutions were supplemented with 500 µM KAl(SO4)2 to generate 50 µM AlFx after dilution with the SR1–GroES–ADP complex. The amount of 7Be bound was assayed as described above. The data show no difference in the maximum amount of bound 7Be in the presence or absence of AlFx (see Figure 5C, inset), indicating that AlFx acts as a competitive inhibitor and allowing a simple calculation of the dissociation constant for AlFx (see Supplementary material).
Supplementary data
Supplementary data are available at The EMBO Journal Online.
Acknowledgments
Acknowledgements
We thank members of the Sigler and Brunger laboratories for help with X-ray data collection and interpretation, especially Z.Xu, C.Schubert, Y.Korkhin, M.Breidenbach and S.Kaiser, and members of the Horwich laboratory for assistance with fluorimetry (B.Reid) and manuscript preparation (W.Fenton). We thank the staffs at APS SBC (Dr A.Joachimak) and the ALS. We also thank H.Saibil and N.Ranson for access to their preliminary cryoEM data on D398A GroEL complexes. C.C. was supported by an NSF predoctoral fellowship.
References
- Alberts B. (1998) The cell as a collection of protein machines: preparing the next generation of molecular biologists. Cell, 92, 291–294. [DOI] [PubMed] [Google Scholar]
- Bigay J., Deterre,P., Pfister,C. and Chabre,M. (1987) Fluoride complexes of aluminum or beryllium act on G-proteins as reversibly bound analogues of the γ-phosphate of GTP. EMBO J., 6, 2907–2913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunger A.T. et al. (1998) Crystallography & NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr. D, 54, 905–921. [DOI] [PubMed] [Google Scholar]
- Burston S.G., Ranson,N.A. and Clarke,A.R. (1995) The origins and consequences of asymmetry in the chaperonin reaction cycle. J. Mol. Biol., 249, 138–152. [DOI] [PubMed] [Google Scholar]
- Cliff M.J., Kad,N.M., Hay,N., Lund,P.A., Webb,M.R., Burston,S.G. and Clarke,A.R. (1999) A kinetic analysis of the nucleotide-induced allosteric transitions of GroEL. J. Mol. Biol., 293, 667–684. [DOI] [PubMed] [Google Scholar]
- Ditzel L., Lowe,J., Stock,D., Stetter,K.O., Huber,H., Huber,R. and Steinbacher,S. (1998) Crystal structure of the termosome, the archaeal chaperonin and homolog of CCT. Cell, 93, 125–138. [DOI] [PubMed] [Google Scholar]
- Fisher A.J., Smith,C.A., Thoden,J.B., Smith,R., Sutoh,K., Holden,H.M. and Rayment,I. (1995) X-ray structures of the myosin motor domain of Dictyostelium discoideum complexes with MgADP-BeFx and MgADP-AlF4. Biochemistry, 34, 8960–8972. [DOI] [PubMed] [Google Scholar]
- Gao Y., Thomas,J.O., Chow,R.L., Lee,G.-H. and Cowan,N.J. (1992) A cytoplasmic chaperonin that catalyzes β-actin folding. Cell, 69, 1043–1050. [DOI] [PubMed] [Google Scholar]
- Goloubinoff P., Christeller,J.T., Gatenby,A.A. and Lorimer,G.H. (1989) Reconstitution of active dimeric ribulose bisphosphate carboxylase from an unfolded state depends on two chaperonin proteins and Mg-ATP. Nature, 342, 884–889. [DOI] [PubMed] [Google Scholar]
- Goodno C.C. (1982) Myosin active-site trapping with vanadate ion. Methods Enzymol., 85, 116–123. [DOI] [PubMed] [Google Scholar]
- Hartl F.U. and Hayer-Hartl,M. (2002) Molecular chaperones in the cytosol: from nascent chain to folded protein. Science, 295, 1852–1858. [DOI] [PubMed] [Google Scholar]
- Howlin B., Butler,S.A., Moss,D.S., Harris,G.W. and Driessen,H.P.C. (1993) TLSANL: TLS parameter-analysis program for segmented anisotropic refinement of macromolecular structures. J. Appl. Crystallogr., 26, 622–624. [Google Scholar]
- Inobe T., Makio,T., Takasu-Ishikawa,E., Terada,T.P. and Kuwajima,K. (2001) Nucleotide binding to the chaperonin GroEL: non-cooperative binding of ATP analogs and ADP, and cooperative effect of ATP. Biochim. Biophys Acta, 1545, 160–173. [DOI] [PubMed] [Google Scholar]
- Inobe T., Kikushima,K., Makio,T., Arai,M. and Kuwajima,K. (2003) The allosteric transition of GroEL induced by metal fluoride-ADP complexes. J. Mol. Biol., 329, 121–134. [DOI] [PubMed] [Google Scholar]
- Jackson G.S., Staniforth,R.A., Halsall,D.J., Atkinson,T., Holbrook,J.J., Clarke,A.R. and Burston,S.G. (1993) Binding and hydrolysis of nucleotides in the chaperonin catalytic cycle: implications for the mechanism of assisted protein folding. Biochemistry, 32, 2554–2563. [DOI] [PubMed] [Google Scholar]
- Ma J., Sigler,P.B., Xu,Z. and Karplus,M. (2000) A dynamic model for the allosteric mechanism of GroEL. J. Mol. Biol., 302, 303–313. [DOI] [PubMed] [Google Scholar]
- Mayhew M., da Silva,A,C., Martin,J., Erdjument-Bromage,H., Tempst,P. and Hartl,F.U. (1996) Protein folding in the central cavity of the GroEL–GroES chaperonin complex. Nature, 379, 420–426. [DOI] [PubMed] [Google Scholar]
- Melki R. and Cowan,N.J. (1994) Facilitated folding of actins and tubulins occurs via a nucleotide-dependent interaction between cytoplasmic chaperonin and distinctive folding intermediates. Mol. Cell. Biol., 14, 2895–2904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melki R., Batelier,G., Soulié,S. and Williams,R.C. (1997) Cytoplasmic chaperonin containing TCP-1: structural and functional characterization. Biochemistry, 36, 5817–5826. [DOI] [PubMed] [Google Scholar]
- Menz R.I., Walker,J.E. and Leslie,A.G. (2001) Structure of bovine mitochondrial F(1)-ATPase with nucleotide bound to all three catalytic sites: implications for the mechanism of rotary catalysis. Cell, 106, 331–341. [DOI] [PubMed] [Google Scholar]
- Meyer A.S., Gillespie,J.R., Walther,D., Millet,I.S., Doniach,S. and Frydman,J. (2003) Closing the folding chamber of the eukaryotic cytosolic chapeornin requires the transition state of ATP hydrolysis. Cell, 113, 369–381. [DOI] [PubMed] [Google Scholar]
- Murshudov G.N., Vagin,A.A., Lebedev,A., Wilson,K.S. and Dodson,E.J. (1999) Efficient anisotropic refinement of macromolecular structures using FFT. Acta Crystallogr. D, 55, 247–255. [DOI] [PubMed] [Google Scholar]
- Nassar N., Hoffman,G.R., Manor,D., Clardy,J.C. and Cerione,R.A. (1998) Structures of Cdc42 bound to the active and catalytically compromised forms of Cdc42GAP. Nat. Struct. Biol., 5, 1047–1052. [DOI] [PubMed] [Google Scholar]
- Ostermann J., Horwich,A.L., Neupert,W. and Hartl,F.U. (1989) Protein folding in mitochondria requires complex formation with hsp60 and ATP hydrolysis. Nature, 341, 125–130. [DOI] [PubMed] [Google Scholar]
- Otwinowski Z. and Minor,W. (1997) Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol., 276, 307–326. [DOI] [PubMed] [Google Scholar]
- Ranson N.A., Burston,S.G. and Clarke,A.R. (1997) Binding, encapsulation and ejection: substrate dynamics during a chaperonin-assisted folding reaction. J. Mol. Biol., 266, 656–664. [DOI] [PubMed] [Google Scholar]
- Ranson N.A., Farr,G.W., Roseman,A.M., Gowen,B., Fenton,W.A., Horwich,A.L. and Saibil,H.R. (2001) ATP-bound states of GroEL captured by cryo-electron microscopy. Cell, 107, 869–879. [DOI] [PubMed] [Google Scholar]
- Rayment I. (1996) The structural basis of the myosin ATPase activity. J. Biol. Chem., 271, 15850–15853. [DOI] [PubMed] [Google Scholar]
- Roseman A.M., Chen,S., White,H., Braig,K. and Saibil,H.R. (1996) The chaperonin ATPase cycle: mechanism of allosteric switching and movements of substrate-binding domains in GroEL. Cell, 87, 241–251. [DOI] [PubMed] [Google Scholar]
- Rye H.S. (2001) Application of fluorescence resonance energy transfer to the GroEL–GroES chaperonin reaction. Methods, 24, 278–288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rye H.S., Burston,S.G., Fenton,W.A., Beechem,J.M., Xu,Z., Sigler,P.B. and Horwich,A.L. (1997) Distinct actions of cis and trans ATP within the double ring of the chaperonin GroEL. Nature, 388, 792–798. [DOI] [PubMed] [Google Scholar]
- Rye H.S., Roseman,A.M., Chen,S., Furtak,K., Fenton,W.A., Saibil,H.R. and Horwich,A.L. (1999) GroEL–GroES cycling: ATP and non-native polypeptide direct alternation of folding-active rings. Cell, 97, 325–338. [DOI] [PubMed] [Google Scholar]
- Scheffzek K., Ahmadian,M.R., Kabsch,W., Wiesmuller,L., Lautwein,A., Schmitz,F. and Wittinghofer,A. (1997) The Ras–RasGAP complex: structural basis for GTPase activation and its loss in oncogenic Ras mutants. Science, 277, 333–338. [DOI] [PubMed] [Google Scholar]
- Sigler P.B., Xu,Z., Rye,H.S., Burston,S.G., Fenton,W.A. and Horwich,A.L. (1998) Structure and function in GroEL-mediated protein folding. Annu. Rev. Biochem., 67, 581–608. [DOI] [PubMed] [Google Scholar]
- Thirumalai D. and Lorimer,G.H. (2001) Chaperonin-mediated protein folding. Annu. Rev. Biophys Biomol. Struct., 30, 245–269. [DOI] [PubMed] [Google Scholar]
- Todd M.J., Viitanen,P.V. and Lorimer,G.H. (1994) Dynamics of the chaperonin ATPase cycle: implications for facilitated protein folding. Science, 265, 659–666. [DOI] [PubMed] [Google Scholar]
- Viitanen P.V., Lubben,T.H., Reed,J., Goloubinoff,P., O’Keefe,D.P. and Lorimer,G.H. (1990) Chaperonin-facilitated refolding of ribulosebisphosphate carboxylase and ATP hydrolysis by chaperonin 60 (groEL) are K+ dependent. Biochemistry, 29, 5665–5671. [DOI] [PubMed] [Google Scholar]
- Weissman J.S., Kashi,Y., Fenton,W.A. and Horwich,A.L. (1994) GroEL-mediated protein folding proceeds by multiple rounds of binding and release of nonnative forms. Cell, 78, 693–702. [DOI] [PubMed] [Google Scholar]
- Weissman J.S., Hohl,C.M., Kovalenko,O., Kashi,Y., Chen,S., Braig,K., Saibil,H.R., Fenton,W.A. and Horwich,A.L. (1995) Mechanism of GroEL action: productive release of polypeptide form a sequestered position under GroES. Cell, 83, 577–587. [DOI] [PubMed] [Google Scholar]
- Weissman J.S., Rye,H.S., Fenton,W.A., Beechem,J.M. and Horwich,A.L. (1996) Characterization of the active intermediate of a GroEL–GroES-mediated protein folding reaction. Cell, 84, 481–490. [DOI] [PubMed] [Google Scholar]
- Winn M.D., Isupov,M.N. and Murshudo,G.N. (2001) Use of TLS parameters to model anisotropic displacements in macromolecular refinement. Acta Crystallogr. D. Biol. Crystallogr., 57, 122–133. [DOI] [PubMed] [Google Scholar]
- Xu Z., Horwich,A.L. and Sigler,P.B. (1997) The crystal structure of the asymmetric GroEL–GroES-(ADP)7 chaperonin complex. Nature, 388, 741–750. [DOI] [PubMed] [Google Scholar]
- Yifrach O. and Horovitz,A. (1995) Nested cooperativity in the ATPase activity of the oligomeric chaperonin GroEL. Biochemistry, 34, 5303–5308. [DOI] [PubMed] [Google Scholar]