Abstract
DNA damage and its repair can cause both local and global rearrangements of chromatin structure. In each case, the epigenetic information contained within this structure must be maintained. Using the recently developed method for the localized UV irradiation of cells, we analysed responses that occur locally to damage sites and global events trigged by local damage recognition. We thus demonstrate that, within a single cell, the recruitment of chromatin assembly factor 1 (CAF-1) to UV-induced DNA damage is a strictly local phenomenon, restricted to damage sites. Concomitantly, proliferating cell nuclear antigen (PCNA) locates to the same sites. This localized recruitment suggests that CAF-1 participates directly in chromatin structural rearrangements that occur in the vicinity of the damage. Use of nucleotide excision repair (NER)-deficient cells shows that the NER pathway—specifically dual incision—is required for recruitment of CAF-1 and PCNA. This in vivo demonstration of the local role of CAF-1, depending directly on NER, supports the hypothesis that CAF-1 ensures the maintenance of epigenetic information by acting locally at repair sites.
Keywords: chromatin assembly/DNA repair/epigenetics/UV damage
Introduction
Chromatin structure contains non-genetically encoded, mitotically heritable epigenetic information, which can take the form of DNA methylation, defined nucleosome position, specific inclusion of variant histones, post-translational modifications of histones and association of non-histone proteins and RNA (Jenuwein and Allis, 2001; Turner, 2002). This is essential for the regulation of genome function to ensure specific gene expression profiles in different cell types. Therefore beyond genetic integrity, epigenetic integrity is important to fully preserve cellular identity. It follows that after any perturbations, the restoration of chromatin to its original state is crucial.
At the nucleosome level chromatin can be rearranged or altered in different ways (comprehensively reviewed in Wolffe, 1998), including: (i) post-translational modification of histones by a variety of enzymes; (ii) remodelling, the increase in DNA accessibility performed by multi-subunit complexes containing a SWI/SNF family ATPase subunit; and (iii) disruption of nucleosomal structure by either partial or total removal of histones. Furthermore, in the nuclear context, chromatin is organized into higher order structures which can also potentially be altered, by unfolding or modification of non-histone proteins, for example. Our understanding of the form of these higher order structures and therefore their dynamic nature is still very limited. Nevertheless, at the nucleosomal level it is clear that chromatin structures are altered during processes that require access to the DNA, such as replication and transcription, but also DNA repair (for reviews see Fyodorov and Kadonaga, 2001; Green and Almouzni, 2002). How the structures disrupted during replication and transcription can be restored with the maintenance of epigenetic marks has generated a lot of interest in recent years. In the context of DNA repair processes, our understanding of the extent of chromatin structural changes, as well as the enzymes that perform them and those that ensure the maintenance of epigenetic information after damage to DNA, is less advanced.
NER is a highly conserved DNA repair pathway that removes bulky lesions, such as those caused by UV light. Analysis by nuclease sensitivity of repaired DNA in UV-irradiated cells indicated that the chromatin structure is rearranged during NER (Smerdon and Lieberman, 1978). These rearrangements were interpreted as resulting from changes at the nucleosomal level to allow the repair machinery to gain access, and subsequent nucleosomal restoration. The elegant systems available to study NER in vitro (Biggerstaff and Wood, 1999; Shivji et al., 1999), initially used to study repair of lesions in naked DNA, were used to demonstrate the inhibitory effect of the nucleosomal organization of DNA on NER, which can partly be relieved by nucleosome remodelling activities (for reviews see Thoma, 1999; Green and Almouzni, 2002). Based on such studies, the ‘Access Repair Restore’ model proposes that chromatin is inhibitory for repair and hence, in vivo, alteration of this structure is required in the proximity of the damage, concomitantly with, or just prior to, repair (Green and Almouzni, 2002). In addition to the inhibitory effect of chromatin due to restricted activity of the repair machinery at target sites, it is also important to consider the reduced detection of lesions within chromatin. Nucleo somal structure severely inhibits the binding of XPC, a major damage recognition protein for global genomic repair (GGR), to damaged DNA (Hara et al., 2000). This raises the following puzzling paradox: during NER, chromatin rearrangements result from the access requirements of repair factors, yet lesions cannot be detected in chromatin without such rearrangements, therefore repair cannot be initiated. This is a particular problem for cyclobutane pyrimidine dimers (CPD) that form within nucleosome cores, and also for highly compacted regions of the genome such as heterochromatin, as in both cases the damage may be particularly difficult to detect. However, such a detection paradox does not apply to transcription-coupled repair (TCR), where lesions are detected by stalling of the transcription machinery (Svejstrup, 2002). It has therefore recently been proposed that the detection of lesions during TCR could trigger global relaxation of chromatin structure throughout the genome, and hence enhance access of repair factors for GGR (Rubbi and Milner, 2003). This chromatin relaxation after UV irradiation appears to be independent of the NER repair machinery but dependent on p53 function, perhaps operating via the histone acetylase p300. Indeed, p53 was previously shown to be required for efficient GGR, although this was thought to be due to the p53-dependent induction of proteins that participate directly in repair (Ford and Hanawalt, 1997; Hwang et al., 1999). In fact the p53-dependent chromatin relaxation could also be due in part to indirect effects, as p53 is required for the induction of Gadd45, a protein that has been suggested to act as an accessibilty factor, particularly on UV-damaged or acetylated chromatin (Carrier et al., 1999).
It thus seems that in addition to local chromatin rearrangements that occur in response to DNA damage by UV, global chromatin relaxation must also be considered. In both of these cases some process will be required to restore the chromatin to its original state in order to ensure the stability of the epigenetic information.
In the case of UV damage, CAF-1 is currently the best candidate for a factor involved in the restoration of chromatin structure after repair. Initially identified by its ability to assemble nucleosomes onto nascent DNA strands in an SV40-based in vitro replication system (Smith and Stillman, 1989), CAF-1 was later shown to perform assembly of chromatin specifically onto plasmids that have been repaired by NER (Gaillard et al., 1996). Furthermore, in vitro, CAF-1 can be recruited to UV-damaged DNA in a PCNA-dependent manner (Moggs et al., 2000). However, although powerful, in vitro techniques are always limited for the analysis of chromatin as they never mimic the complexity of the in vivo situation. Furthermore, because such analyses tend to rely on analyses of repaired products, it has not been possible to use such systems to investigate direct links with the NER process. Supporting the in vitro connection between UV damage repair and CAF-1, Saccharomyces cerevisiae strains lacking the orthologues of CAF-1 are sensitive to UV (Kaufman et al., 1997; Game and Kaufman, 1999). In addition, in human cells, the chromatin-associated fraction of CAF-1 increases in response to UV irradiation (Martini et al., 1998). However, none of the studies to date had investigated whether CAF-1 function is required locally at damage sites, tightly linked to NER and its concomitent chromatin rearrangements, or alternatively whether CAF-1 participates in some global response to UV damage, perhaps linked to chromatin relaxation. To validate the importance of CAF-1 in an in vivo chromatin context and investigate a specific connection with NER in various mammalian cells, here we used a localized UV irradiation method to analyse repair sites in vivo. This allows direct comparison between irradiated and non-irradiated areas within a single nucleus (Moné et al., 2001), essential to determine whether CAF-1 acts locally to the damaged region, or across the whole genome at a distance from the damage. We demonstrate that CAF-1 and PCNA are concomitantly recruited specifically at sites of UV damage in vivo. This approach was then combined with the use of NER-defective cells lines (Volker et al., 2001) to show that CAF-1 recruitment is strictly dependent upon the incision stages of NER. This late action is consistent with CAF-1 being involved in the restoration of chromatin structures that have been locally disturbed during NER in vivo.
Results
Recognition of DNA damage in vivo and initiation of repair
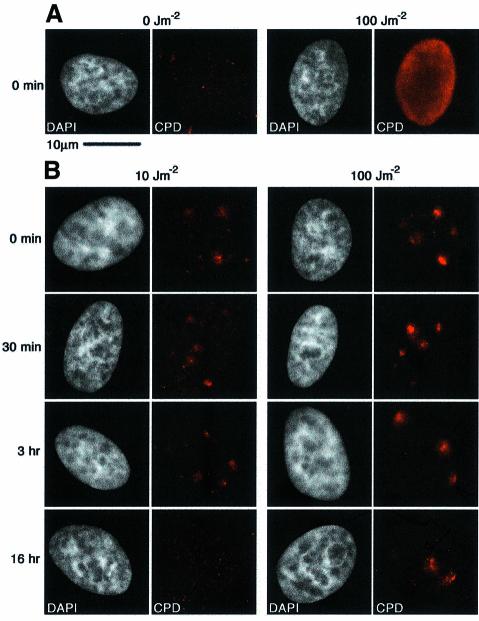
Damage caused in vivo by UV irradiation can be detected by indirect immunofluorescence using an antibody that recognizes CPD, specifically thymine dimers (Kamiya Biomedical) (Figure 1A). To create localized regions of damage within a single nucleus, HeLa cells grown on collagen/fibronectin-coated coverslips were covered with a millipore filter before irradiation (Moné et al., 2001; Volker et al., 2001). The filter used absorbs ∼98% of the applied dose, as measured with a dosimeter (Vilber Lourmat). Throughout our study we applied doses of between 10 and 200 J/m2 to the filter surface, therefore the average doses applied to the cells ranged from 0.2 to 4 J/m2, although the damage was locally concentrated with a maximum local dose of 200 J/m2. Such doses were low enough to permit cell survival after treatment. Localized damage was detectable immediately after irradiation, giving a signal that decreased in intensity after prolonged incubation in growth media at 37°C (Figure 1B). This decrease was dependent upon the initial dose applied; the signal was no longer detectable 16 h after a 10 J/m2 irradiation, but was still visible at this time after a 100 J/m2 dose, suggesting repair of CPD was not yet complete after this time (Figure 1B). Although the CPD signal was brighter in the cells that had been irradiated at 100 J/m2 than those irradiated at 10 J/m2, this difference was not striking, probably due to the high senstivity of the antibody that we use.
Fig. 1. In vivo formation and repair of localized DNA damage. (A) HeLa cells were UV-irradiated at 100 J/m2, or mock treated, and immediately fixed without detergent extraction. DNA damage was visualized by indirect immunofluorescence using a mouse monoclonal antibody against thymine dimers (CPD, red). (B) HeLa cells were locally irradiated through a polycarbonate UV-absorbing filter, at the doses indicated, followed by post- irradiation incubation for the times indicated on the left. DNA damage was detected by indirect immunofluorescence as in (A). (C) The recruitment of XPC protein to damage sites was visualized by indirect immunofluorescence following an irradiation dose of 100 J/m2 using a rabbit polyclonal antibody to XPC (green) and the mouse anti-thymine dimer monoclonal antibody (red). (D) The recruitment of stably expressed HA-tagged DDB2 protein to damage was visualized by indirect immunofluorescence following a dose of 100 J/m2 (or mock treatment) using a rat monoclonal antibody against the HA epitope (green) and the mouse anti-thymine dimer monoclonal antibody (red). (E) The local recruitment of PCNA, a protein involved in the repair synthesis stage of NER, to damage sites was visualized by indirect immunofluorescence after irradiation at 100 J/m2. Unless otherwise stated, all cells were treated with Triton prior to fixation to remove soluble nuclear proteins, and the DNA was visualized with DAPI (white). The scale bar in (A) represents 10 µm, the magnification used for all the images.
Because nucleosomal structure is inhibitory to repair in vitro, but repair can be enhanced by the addition of chromatin remodelling factors (Ura et al., 2001; Hara and Sancar, 2002) and increases in the amounts of acetylated histones have been detected after UV irradiation of human cells (Ramanathan and Smerdon, 1986; Brand et al., 2001), we asked whether changes in chromatin structure could be detected at damage sites. Specifically we looked for acetylation at specific residues of histone H3 and H4 and the recruitment of histone-modifying enzymes such as GCN5 and p300 or chromatin remodelling proteins such as ISWI. With the antibodies used we could not detect any major changes at sites of DNA damage after localized UV irradiation (data not shown; see Materials and methods for a list of antibodies). Furthermore, using the same antibodies we did not detect global changes after localized irradiation. Of course transient events or other modifications or factors could be involved for which we lack appropriate antibodies.
We analysed both early (damage recognition) and late (repair synthesis) stages of NER in our experimental conditions. The XPC protein is the damage recognition protein for NER that operates on non-transcribed DNA (Batty and Wood, 2000; Volker et al., 2001). Triton-extracted cells fixed immediately after local irradiation at 100 J/m2 showed no specific enhancement of XPC signal at the damage sites (Figure 1C). However, 5 min after irradiation, the association of XPC with damage sites was clearly visible (Figure 1C). In these HeLa cells this colocalization was markedly reduced after 30 min post-irradiation incubation, suggesting that after initial binding the XPC protein is released from damage sites (Figure 1C). In vitro, XPC has a high affinity for 6-4 photoproducts (Hey et al., 2002), which are repaired more rapidly than CPD in vivo (Mitchell et al., 1985); this may account for the rapid loss of XPC association from damage sites. These data are consistent with recent publications that also show localized recruitment of XPC to damage sites (Volker et al., 2001; Wang et al., 2003) the slight kinetic differences may be due the different cell types used.
Even more rapid than the recruitment of XPC was the recruitment of another damaged DNA binding protein, DDB2 [the smaller (p48) subunit of damaged-DNA-binding protein 2, defective in XP-E (de Laat et al., 1999)]. Using HeLa cells carrying an integrated, epitope-tagged version of the DDB2 gene (Groisman et al., 2003), we detected recruitment of tagged-DDB2 protein to damaged sites immediately after localized irradiation at 100 J/m2 (Figure 1D). This recruitment was even observed if the cells were irradiated on ice and immediately fixed (not shown). The DDB2 protein was retained at damage sites for at least 1 h (Figure 1D). These data are consistent with a recent study, using a different epitope-tagged version of DDB2 (Wakasugi et al., 2002). It should be noted that in both cases the tagged DDB2 protein is not subject to the same transcriptional regulation as the endogenous DDB2 (Hwang et al., 1999). These data demonstrate that the early damage recognition events of NER can occur and are detectable very rapidly within chromatin in vivo.
Repair synthesis, which occurs after removal of the damaged oligonucleotide, is performed by polymerases delta and epsilon, which depend on the processivity factor PCNA (Wood and Shivji, 1997). HeLa cells were locally irradiated and Triton-extracted, and PCNA localization was analysed by indirect immunofluorescence. Cells that were in S-phase at the time of fixation display characteristic localization of PCNA at replication foci; these strongly staining cells are easy to identify (Martini et al., 1998) and were not analysed further. In cells outside S-phase, we detected PCNA localized to regions of DNA damage 30 min after 100 J/m2 (Figure 1E). This shows that later stages of NER operate and can be readily detected in vivo.
Recruitment of chromatin assembly activity to damage sites
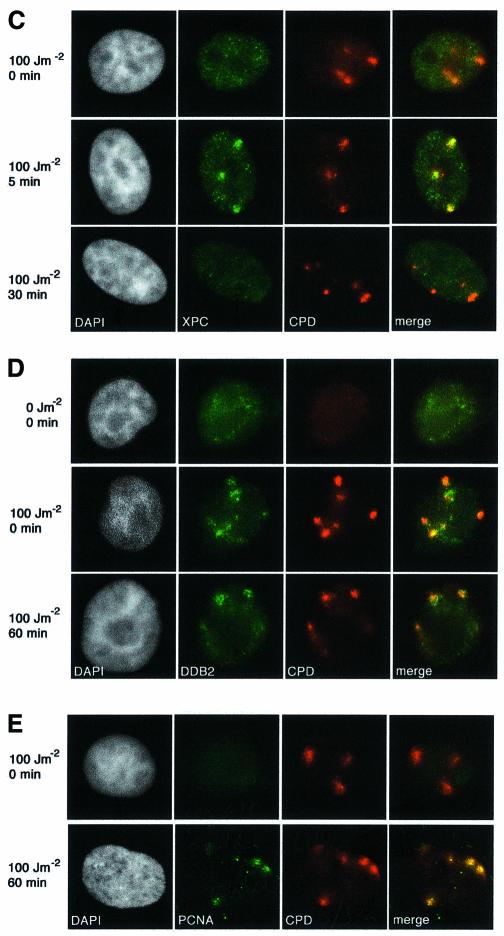
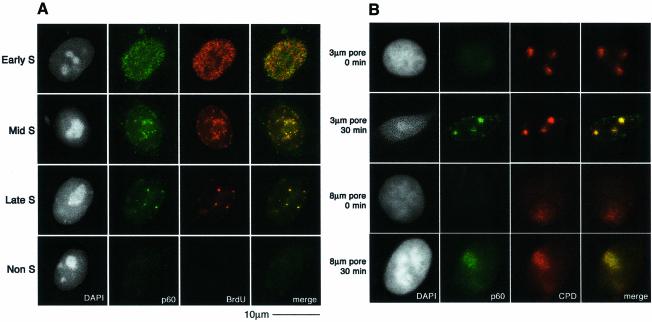
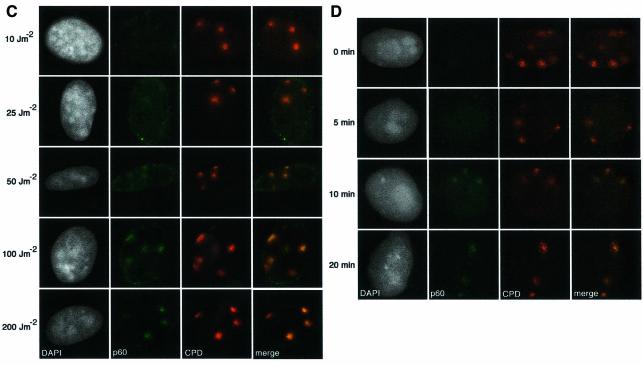
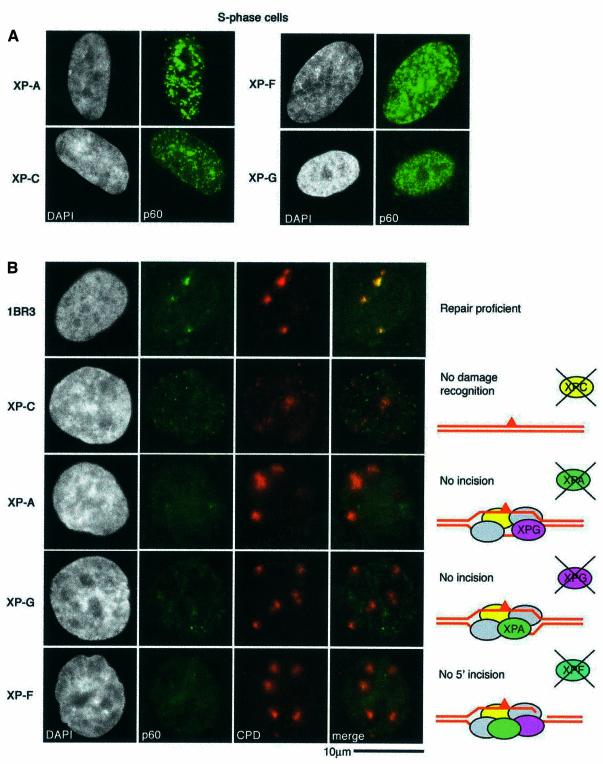
CAF-1 is a heterotrimeric protein comprising subunits of 48, 60 and 150 kDa. The p48 subunit (also called RbAp48) is a component of several different protein complexes (Ridgway and Almouzni, 2000), whereas the other subunits are specific for CAF-1. A proportion of the total cellular pool of CAF-1 in vivo is tightly associated with sites of replication, and this can be revealed by removal of the soluble fraction with a detergent extraction procedure prior to fixation (Krude, 1995; Martini et al., 1998; Taddei et al., 1999). In unirradiated, asynchronous HeLa cells, the nuclear localization of Triton-insoluble CAF-1 p60 closely coincides with the sites of DNA replication, marked by a 10 min pulse of BrdU incorporation. In this way it is possible to classify the cells as not in S-phase (Non S) or in early-, mid- or late-S phase of the cell cycle (Figure 2A). The low levels of CAF-1 in Triton-extracted non-S-phase cells allowed us to unambigously analyse the recruitment of CAF-1 p60 to sites of DNA damage in these cells. We irradiated (100 J/m2) HeLa cells growing on coverslips though a filter with 3 µm pores (Figure 2B). When the cells were Triton extracted and fixed immediately after the irradiation, we did not detect specific localization of p60 at damage sites. However, after a 30 min post-irradiation incubation period we observed a specific recruitment of the CAF-1 p60 subunit to sites of UV damage, with adjacent undamaged regions remaining free of p60 staining (Figure 2B). This pattern of CAF-1 p60 observed after irradiation through a filter is qualitatively distinct from any of the replication patterns observed during S phase (compare Figure 2A and B). Moreover, in order to confirm that the local concentration of CAF-1 p60 at the damage sites could not be confused with replication patterns, we repeated the irradiation using filters with 8 µm pores. In this case, CAF-1 p60 was localized in larger regions, colocalizing with the damage, confirming that this pattern of CAF-1 p60 was damage specific (Figure 2B). This p60 recruitment was also dose dependent; the signal was stronger at higher doses, which resulted in more damage events per irradiated region (Figure 2C). Similar staining patterns were observed using two independent polyclonal antibodies, and a monoclonal antibody that specifically recognizes a phosphorylated form of p60 (data not shown). Importantly, we also observed recruitment of CAF-1 p60 to localized sites of UV damage in MCF7 cells (data not shown), and in 1BR3 primary human fibroblasts (see Figure 5B), demonstrating that this phenomenon is not restricted to HeLa cells. The detection threshold is a limitation of any immunofluorescence technique; a certain concentration of specific epitopes is necessary to generate a visible signal over background. This probably explains why the recruitment of CAF-1 p60 to damage sites was more difficult to detect at doses <50 J/m2. Analysis of p60 recruitment after a fixed dose of 150 J/m2 showed that it was detectable 10 min after irradiation (Figure 2D).
Fig. 2. Recruitment of CAF-1 p60 to replication and damage sites. (A) Characteristic patterns of CAF-1 p60 staining throughout the cell cycle. Cells were pulsed with BrdU followed by double labelling with a polyclonal against p60 (green) and a rat monoclonal against BrdU after denaturation with 4 M HCl (red). (B) CAF-1 p60 is locally recruited to sites of UV damage. Cells were irradiated with 100 J/m2 through filters with either 3 µm or 8 µm pores, with or without post-irradiation incubation, as indicated. Indirect immunofluorescence was performed with a polyclonal against p60 (green) and the anti-thymine dimer mouse monoclonal antibody (red). (C) Damage sites and CAF-1 p60 were visualized as in (B), 30 min after irradiation at the doses indicated. (D) Damage sites and CAF-1 p60 were visualized as in (B), at different times after a dose of 150 J/m2.
Fig. 5. Steps in repair processing required for CAF-1 recruitment. (A) XP fibroblasts show normal S-phase staining for CAF-1. Asynchronous cells were fixed and CAF-1 p60 was revealed by indirect immunofluorescence (green). (B) Repair-proficient primary human fibroblasts and those derived from XP patients defective at different stages of NER, as depicted in the cartoons on the right, were locally irradiated at 100 J/m2. Recovery was at 37°C for 1 h before processing for indirect immunofluorescence using the rabbit polyclonal antibody against p60 (green) and the mouse anti-CPD monoclonal (red).
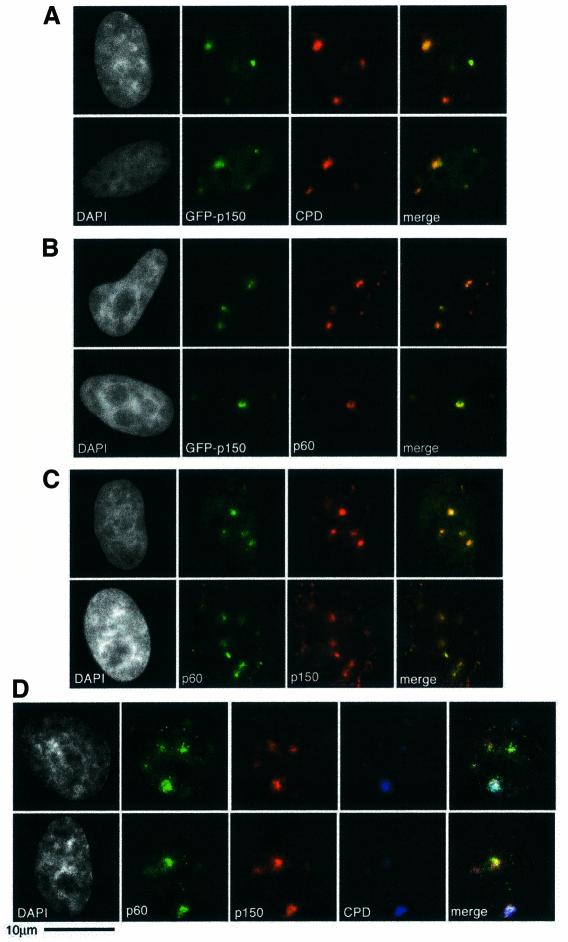
The p150 subunit of CAF-1 behaved in a similar manner to the p60 subunit (Figure 3). A transiently expressed green fluorescent protein (GFP)–p150 fusion protein was enriched at sites of DNA damage 30 min after localized irradiation at 100 J/m2 (Figure 3A). In non-S-phase cells with characteristic spots of p60 corresponding to damage sites, we were able to detect GFP signal colocalizing with p60 (Figure 3B). These observations were confirmed by colocalization of p60 and the endogenous p150 protein using two different antibodies against p150 (Figure 3C and data not shown). Finally, a triple labelling strategy demonstrated that both endogenous subunits colocalize within the same region of damage (Figure 3D). The pool of CAF-1 present in a soluble form within the nucleus (Martini et al., 1998), which is removed by our detergent extraction procedure, provides a readily available source for this rapid recruitment to repair sites. In all cases tested we have noted that the recruitment of the p60 and p150 subunits are coupled; we have never detected recruitment of one without the other. Our interpretation is either that CAF-1 p60 and p150 exist as a preformed complex, ready to be recruited to sites of action, or that two separate recruitment events occur that are extremely tightly coordinated. The localized nature of the recruitment of the p60 and p150 proteins is consistent with CAF-1 acting locally to the damage sites, and not in response to global chromatin alterations, such as chromatin relaxation for damage detection. However, we do not exclude the possibility that CAF-1 could have indirect effects at a global level resulting from cross talk between local and global events.
Fig. 3. Recruitment of CAF-1 p150 to damage sites. (A) Transient transfection of HeLa cells with a plasmid expressing the large subunit of CAF-1 tagged with GFP (GFP-p150) was used to visualize recruitment of GFP-p150 (green) to damage sites detected by indirect immunofluorescence (red) 30 min after irradiation at 100 J/m2. (B) Under the same conditions, GFP-p150 also colocalizes with endogenous p60 (red), detected by indirect immunofluorescence using the rabbit polyclonal antibody. (C) Endogenous p150 and p60 subunits were detected by indirect immunofluoresence 1 h after irradiation at 150 J/m2, using the rabbit polyclonal against p60 (green) and the mouse monoclonal against p150 (red). (D) A triple marking strategy was used to demonstrate the colocalization of endogenous p60 and p150 at damage sites 30 min after 150 J/m2. The proteins were detected by indirect immunofluorescence (p60 in green and p150 in red), with direct detection of the damage using the anti-damage antibody covalently coupled to AlexaFluor647 (blue); in this case a colocalization of all three antibodies results in a white signal in the merged view.
CAF-1 localization at damage is concomitant with PCNA recruitment
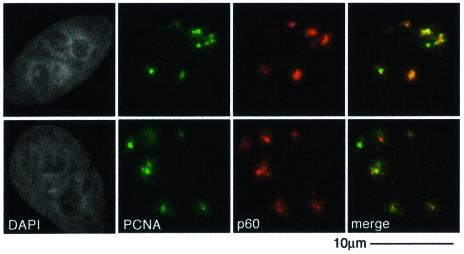
PCNA is required for the repair synthesis stage of NER. In vitro the p150 subunit of CAF-1 associates with PCNA and these two proteins can be immunoprecipitated from cell extracts (Shibahara and Stillman, 1999; Moggs et al., 2000). Given this, and the local recruitment of PCNA to damage sites (Figure 1E), it is possible that PCNA recruits CAF-1 to such sites. Importantly, we detected a detergent-resistant colocalization between PCNA and CAF-1 (both p60 and p150) in defined areas of locally irradiated, non S-phase cells (Figure 4 and data not shown). In all repair-competent cell lines tested, HeLa, MCF7 and 1BR3, we have not found conditions where we can separate the recruitment of CAF-1 and PCNA, arguing for a tight coupling between them. However, PCNA can be recruited to replication foci in cells treated with siRNA directed against CAF-1 p150 (J.-P.Quivy, personal communication).
Fig. 4. PCNA recruitment concomitant with CAF-1. Locally recruited, detergent-insoluble PCNA, detected with a mouse monoclonal against PCNA (green), colocalized with p60, detected with a rabbit polyclonal antibody (red) 30 min after localized irradiation at 150 J/m2.
Recruitment of CAF-1 and of PCNA is dependent on NER activity
The recruitment of CAF-1 locally to damage sites raised questions as to its role at these sites, and also the signal that initiates this recruitment. Because CAF-1 is a histone chaperone, one attractive theory was that it could act not only to restore chromatin structure but it could also act as a histone sink, to locally trap histones displaced during any repair-associated rearrangements. This would provide a means by which the epigenetic information contained within histone modifications could be maintained, as the original histones could be used to restore the structures. In this case we might expect CAF-1 recruitment to occur as an early event, prior to, or concomitant with, repair. With regard to PCNA, which may act to bring CAF-1 to the damage sites, there has previously been no extensive in vivo analysis of the proteins that are required for its recruitment to NER sites. Although the activity of PCNA is required at a late stage of NER, some recruitment of PCNA to an insoluble nuclear fraction was detected in UV irradiated XP-A cells, although this was qualitatively different from that found in repair-competent cells (Aboussekhra and Wood, 1995; Li et al., 1996). Furthermore, in the base excision repair pathway, PCNA is required not just at the synthesis stage, but for efficient excision (Gary et al., 1999). To determine whether NER was required for the recruitment of CAF-1 and PCNA, and if so to assess when such recruitment occurs, we filter-irradiated primary fibroblasts derived from patients with xeroderma pigmentosum (XP), defective at defined stages of the NER pathway, as depicted in Figure 5B. Importantly, the S-phase-dependent localization of CAF-1 and PCNA to replication foci was normal in these lines (Figure 5A and data not shown), demonstrating that CAF-1 and PCNA are present in these cells and can be recruited normally to replication forks. This was further confirmed by pulse-labelling replicating cells with BrdU and co-staining replication sites with CAF-1 (Supplementary figure 1, available at The EMBO Journal Online). The recruitment of CAF-1 can readily be detected in 1BR3 repair-proficient primary fibroblasts 1 h after irradiation at 100 J/m2 (Figure 5B). In none of the XP cells tested (XP-A, XP-C, XP-F or XP-G) could we detect recruitment of CAF-1 to sites of UV damage (Figure 5B). Crucially, neither did we detect recruitment of PCNA to the damage in any of these lines (data not shown), again highlighting that CAF-1 and PCNA recruitment are linked. The lack of recruitment of CAF-1 and PCNA to damage in all these cell lines suggests that initial damage recognition and open complex formation are not sufficient to initiate recruitment; the excision of the damage-containing oligonucleotide by the XPG and XPF/ERCC1 enzymes is required. This step will leave a double-stranded to single-stranded transition that in vitro would be an appropriate substrate for the loading of PCNA by the replication factor C complex. The interaction between CAF-1 and PCNA would then ensure that chromatin assembly activity is directed to repair sites at which DNA synthesis is ongoing. This late recruitment of CAF-1 does not necessarily exclude the possibility that CAF-1 utilizes its histone chaperone activity to act as a local histone sink. As we suggested previously, nucleosome rearrangement may be a multistep process, operating at different stages throughout the repair process (Green and Almouzni, 2002). It remains possible that CAF-1 gathers histones displaced during the DNA synthesis stage of NER and then reassembles nucleosomes on the newly synthesized DNA behind the polymerase, using the original histones.
Discussion
Access requirements for DNA repair in vivo: local and/or global responses?
The Access Repair Restore model for DNA damage repair in vivo postulates that chromatin structures, which are inhibitory for repair processing, must be altered to allow repair and then restored after such repair (Green and Almouzni, 2002). This model is applicable not only to NER, but also to other repair pathways that are inhibited by chromatin structures. In a simplified model, rearrangements of chromatin would occur locally to the damaged sites. This could involve chromatin modifying activities such as histone acetyltransferases or chromatin remodelling factors that are recruited to damage sites; the fact the we have not yet detected such recruitments during NER may suggest that such interactions are transient. Alternatively, repair factors themselves could cause chromatin rearrangement; particularly good candidates for this type of function in the NER pathway are the transcription-coupled repair factor CSB, which has homology to SWI/SNF chromatin remodelling proteins, and the TFIIH complex that contains the helicase subunits XPD and XPB (Thoma, 1999). However, a non-mutually exclusive suggestion is that global chromatin relaxation increases accessibility over the whole genome in response to damage in order to expose the individual damage sites for recognition (Rubbi and Milner, 2003). Although large macromolecules can diffuse freely even within compact heterochromatic domains, it remains likely that the detection of any damage is hindered by nucleosomal structure. A system in which the detection of a lesion in a readily accessible area, such as a transcribed gene, acts as a signal to initiate chromatin relaxation and subsequent lesion detection at distant sites, might therefore be generally valid. Indeed, a similar dual system of both local and global chromatin changes may exist in the case of double strand break (DSB) repair, where the phosphorylation of histone H2AX occurs locally to DSB sites (Redon et al., 2002), and longer range chromatin changes have been postulated to act as a signal amplification mechanism (Bakkenist and Kastan, 2003).
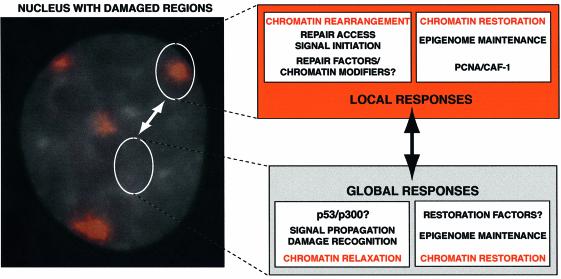
Localized UV irradiation enables analysis of NER in chromatin in vivo, from initial damage recognition to elimination of the damage, and allows us to distinguish between events that occur locally at damage sites and those that are global responses to damage detection. Our demonstration that CAF-1 is locally recruited to damage sites indicates that the activities of this protein are required in the vicinity of the damage, and not as a global response to DNA damage. This suggests that the function of CAF-1 is directly linked to local chromatin rearrangements, not global chromatin relaxation. As summarized in Figure 6, within a chromatin-based view of the cellular response to UV damage, a clear role for CAF-1 can be assigned in the local responses. This does not exclude, however, crosstalk between the two classes of responses, which may indirectly involve CAF-1, perhaps as a component of a signalling system. We do propose, however, that if global chromatin alterations such as relaxation do occur during NER, a factor other than CAF-1 should be involved in reversing them. The recent characterization of a chromatin assembly pathway that operates independently of DNA synthesis (Ray-Gallet et al., 2002) highlights the fact that chromatin assembly is an ongoing process, even in non-replicating cells, and provides possible means for a restoration of global chromatin alterations.
Fig. 6. A chromatin-based view of the cellular response to UV damage. The local irradiation technique enables the independent analysis of local and global responses to UV damage. The image shows a nucleus stained with DAPI, with damage sites, visualized by immunofluorescence, in red. Although each damaged region contains many individual lesions, this can be used as a model for events that occur at a single lesion surrounded by undamaged DNA (the more physiologically relevant case). Here, we highlight that the damage response involves both local and global events and that there will be crosstalk between these two systems; a local event can initiate signals that propagate to have global effects, and such global effects can in turn impinge on events occurring local to the damage. Alterations of the chromatin structure, whether global or local, will need to be reversed once damage is complete to ensure stability of the epigenome. In this work we have demonstrated that CAF-1 is likely to perform this role at a local level.
Because NER is a multi-step pathway, it was important to determine at which stage CAF-1 and PCNA are recruited. We used XP-A, XP-C, XP-F and XP-G cell lines to show two things: first, that NER is absolutely required for the local recruitment of CAF-1 and PCNA to damage sites; the presence of damage per se is not sufficient. Secondly, that only damage sites that have undergone the dual incision stage of repair processing are competent to recruit both PCNA and CAF-1. No recruitment of PCNA or CAF-1 is seen in XP-F cells, even though these are competent to recruit all the other NER factors and have the capacity to make the first (3′) incision. This argues for a late role of CAF-1 in NER, consistent with the Access Repair Restore model, in which CAF-1 is required to restore chromatin structures locally disturbed during repair, as a final stage of the repair process. It should be noted that, similar to the situation in XP-C cells, we have not detected the recruitment of CAF-1 to local damage produced in mouse fibroblasts (A.Gontijo, C.M.Green and G.Almouzni, unpublished data). Since in both cases GGR is specifically impaired (Hanawalt, 2001), this raises the intriguing possibility that this pathway is predominantly required for CAF-1 recruitment. Further investigation will be required to compare the role of CAF-1 in mouse and human cells.
Obviously, the connection with NER described here does not exclude additional involvement of CAF-1 during other repair processes. Recruitment of CAF-1 to SSB in vivo has recently been reported (Okano et al., 2003), reinforcing the previous in vitro studies (Moggs et al., 2000). Interestingly however, to date we have been unable to detect a specific enrichment of CAF-1 protein at ionizing radiation (IR)-induced foci of phosphorylated H2AX, where DSBs are postulated to be processed and repaired (unpublished data). It is therefore possible that any chromatin structural alterations required for DSB repair are not restored by CAF-1. Consistent with this, budding yeast cells lacking the CAF-1 orthologues are not sensitive to IR; other factors, such as ASF1, may be involved in the maintenance of chromatin to allow IR survival in this organism (Tyler et al., 1999). Extending these studies to include a more thorough analysis of chromatin structural alterations and restoration in response to DSBs and other damage in higher eukaryotic cells will be a future challenge.
CAF-1 and the maintenance of epigenetic information
Our data also have strong implications for genome function and the maintenance of cellular identity. Cellular systems for maintaining such information during DNA replication and cell division have been the subject of much study in recent years, but how this information is maintained during other events that transiently alter chromatin structure, including DNA repair, as discussed here, must also be considered. In this context, processes adapted to the scale of the chromatin changes in the nucleus will be critical. The early in vitro data (Gaillard et al., 1996) were a first hint that CAF-1 was a good candidate for this type of function during NER in cell extracts. Here, our study provides the first strong in vivo evidence for a direct connection between CAF-1 and NER. Furthermore, this work allows us to define when, within the sequential repair process, this recruitment of CAF-1 occurs, and the scale on which CAF-1 is acting. An immediate consequence of a defect in chromatin restoration after repair would be ‘epigenetic instability’, manifesting as changes in transcription profiles and silenced states. In fact, in budding yeast, the CAF-1 orthologs are required for maintenance of silenced states at telomeres and mating type loci (Enomoto et al., 1997; Kaufman et al., 1997; Enomoto and Berman, 1998). Furthermore, normal heterochromatin structures are required for proper chromosome segregation in fission yeast and in mammalian cells (Ekwall et al., 1997; Bernard et al., 2001; Taddei et al., 2001), hence ‘epigenetic instability’ can in turn lead to genetic instability. The use of dominant-negative strategies has demonstrated the grave consequences of perturbing CAF-1 function in higher eukaryotic cells (Quivy et al., 2001; Ye et al., 2003). In Arabidopsis, deletion of Fasciata, the homologue of p150, results in defects in proliferative tissues (Kaya et al., 2001). The cellular defects observed after interference with CAF-1 function, both in yeast and in higher eukaryotes, are currently attributed to the lack of proper chromatin assembly at the replication fork. However, the UV sensitivity of yeast strains defective in CAF-1 function suggests that a failure to restore proper chromatin structures after repair can also have serious consequences for cell survival after DNA damage (Kaufman et al., 1997). It is important to stress that the differences between the mutant phenotypes in S.cerevisiae and the effect of interfering with CAF-1 in higher eukaryotes are currently not completely understood. It may be that chromatin assembly during replication is not so crucial for the majority of the S.cerevisiae genome. However, it will be critical to evaluate to what extent the use of dominant-negative strategies compared with gene deletion can impinge on the final phenotype.
The development of new tools, both to abrogate and to assess CAF-1 function in vivo in various systems, will be essential for our understanding of its integrated role at the crossroads of DNA replication/repair and heterochromatin. Insight into the crosstalks between these processes and their spatio-temporal organization in living cells will be of significance for both fundamental research and medicine, considering that these processes are highly compromised in many cancer cases.
Materials and methods
Cell culture
HeLa and MCF7 cultures were grown in DMEM plus 10% fetal calf serum (FCS), in 5% CO2. Primary fibroblasts derived from XP patients were grown in F10 (Ham) media, plus 12% FCS, in 5% CO2. The 1BR3 line was grown in MEM plus 15% FCS in 5% CO2. All media were from Gibco-BRL and also contained 2 mM l-glutamine, 10 mg/ml penicillin and streptomycin. BrdU pulses were for 10 min in normal medium with 40 µM BrdU. Transient transfection of HeLa cells was performed using Effectene transfection reagent according to the manufacturer’s protocol (Qiagen).
Local irradiation
Glass coverslips were pre-treated with a solution of 20 µg/ml collagen and 1 µg/ml fibronectin in phosphate-buffered saline (PBS) for 1 h at 37°C. These were then rinsed in PBS and cells seeded at the appropriate density the evening before the experiment was due to be performed. Coverslips were rinsed in PBS then individually covered with a piece of Isopore 3.0 or 8.0 µm filter (Millipore), and irradiated at 254 nm using a 6 W VL-6.MC lamp at a fluence rate of 6 J/m2/s as measured with a VLX3W dosimeter (Vilber Lourmat). Doses of between 10 and 200 J/m2 were applied by varying the time of irradiation. Mock irradiated samples were processed identically, except the lamp was switched off. The filters were removed and the coverslips returned to the original medium at 37°C for post-irradiation incubation.
In situ detergent extraction and immunofluorescence analysis
After post-irradiation incubation, coverslips were rinsed with PBS then washed twice in CSK buffer (100 mM NaCl, 300 mM sucrose, 10 mM PIPES pH 7.0, 3 mM MgCl2) and soaked in CSK plus 0.2% Triton X-100 for 5 min at room temperature. After two further washes in CSK, cells were fixed by incubating in 2% paraformaldehyde (PAF) for 15 min at room temperature. Blocking was for 15 min in 5% bovine serum albumin (BSA) in PBT (PBS containing 0.1% Tween 20). Incubation with the appropriate primary antibodies, diluted in blocking buffer as described below, was for 1 to 2 h at room temperature, followed by three washes in PBT. Secondary antibodies against rabbit, mouse or rat immunoglobulin Gs (IgGs), and coupled to AlexaFluor488 (Molecular probes) or Texas Red (Interchim-Jackson Laboratories), were used at a dilution of 1/1000 for 45 min to 1 h. DAPI was added at a final concentration of 0.5 µg/ml for the final 5 min of incubation. Four washes of PBT and a final rinse in PBS were performed before the coverslips were mounted onto slides using vectashield mounting media (Vector Laboratories). For visualization of incorporated BrdU, Triton extraction, fixation and blocking were performed as described above, followed by incubation with the primary antibody against the protein to be co-stained. After three washes in PBT, a second fixation was performed in 2% PAF for 15 min. The DNA was then denatured by 10 min incubation in 4 M HCl at room temperature, followed by neutralization with four washes in PBS. A second blocking step of 10 min in 5% BSA in PBT was peformed, followed by the incubation in primary antibody against BrdU and then washing and incubation in secondary antibodies as above.
Slides were observed using a DMR epifluorescence microscope (Leica). Images shown are representative mid-plane sections captured using a CCD camera (Hamamatsu) and Metamorph software, and artificially coloured using Adobe Photoshop software.
Antibodies
Antibodies were used at the following dilutions: anti-thymine dimer mouse monoclonal (Kamiya Biomedical) 1/2000, after denaturation in 0.5 M NaOH for 5 min prior to blocking; anti-XPC rabbit polyclonal 1/1000; anti-HA epitope rat monoclonal (Roche) 1/100; anti-GCN5 (Santa Cruz) 1/200; anti-p300 (Santa Cruz) 1/200; anti-histone H3 acetylated on lysine 9 (Euromedex) 1/200; anti-histone H3 phosphorylated on serine 10 (Upstate Biotechnologies) 1/200; anti-histone H4 acetylated on lysine 5 and anti-histone H4 acetylated on lysine 12 (B.Turner) 1/1000; anti-CAF-1 p60 rabbit polyclonal (made by AgroBio using antigen supplied by D.Roche) or mouse monoclonal (Novus-Abcam) 1/1000; anti-CAF-1 p150 affinity purified rabbit polyclonal (made by AgroBio using antigen supplied by J.-P.Quivy) 1/100 or mouse monoclonal (Novus-Abcam) 1/1000; anti-PCNA mouse monoclonal (DAKO) 1/1000 or rabbit polyclonal (Santa Cruz) 1/20, both after 15 min incubation in 100% methanol at –20°C after PAF fixation; and anti-BrdU rat monoclonal antibody (Becton Dickinson) 1/200. For triple marking, the anti-thymine dimer monoclonal was coupled to AlexaFluor647 using a kit (Molecular Probes). After incubation with primary and secondary antibodies to detect the two CAF-1 subunits, a blocking step of 10 mg/ml mouse IgG in PBT was performed, followed by crosslinking in 4% PAF. Incubation with the AlexaFluor-647-coupled anti-damage antibody was for 30 min, followed by washing and mounting.
Supplementary data
Supplementary data are available at The EMBO Journal Online.
Acknowledgments
Acknowledgements
We thank Wim Vermeulen for X-P lines, Pat Nakatani and Regina Groisman for tagged-DDB2, Fumio Hanaoka for anti-XPC antibodies, Danièle Roche for technical advice, J.-P.Quivy for GFP-p150, members of UMR218 for discussion, and Ethel Moustacchi and Manuel Buchwald for critical reading of the manuscript. C.M.G. was supported by a European Community Marie Curie Fellowship (HPMF-CT-2000-00976) (disclaimer: the European commission is not responsible for any views or results expressed) and initially by a TMR network grant to G.A. (ERBF MRX CT-98-0191). G.A.’s team is supported by la Ligue Nationale contre le Cancer (Equipe labellisée la Ligue), Euratom (FIGH-CT1999-00010), the Commissariat à l’Energie Atomique (LRC No. 26) and a Programme Incitatif et Collaboratif between Institut Curie and the Commissariat à l’Energie Atomique.
References
- Aboussekhra A. and Wood,R.D. (1995) Detection of nucleotide excision repair incisions in human fibroblasts by immunostaining for PCNA. Exp. Cell Res., 221, 326–332. [DOI] [PubMed] [Google Scholar]
- Bakkenist C.J. and Kastan,M.B. (2003) DNA damage activates ATM through intermolecular autophosphorylation and dimer dissociation. Nature, 421, 499–506. [DOI] [PubMed] [Google Scholar]
- Batty D.P. and Wood,R.D. (2000) Damage recognition in nucleotide excision repair of DNA. Gene, 241, 193–204. [DOI] [PubMed] [Google Scholar]
- Bernard P., Maure,J.F., Partridge,J.F., Genier,S., Javerzat,J.P. and Allshire,R.C. (2001) Requirement of heterochromatin for cohesion at centromeres. Science, 294, 2539–2542. [DOI] [PubMed] [Google Scholar]
- Biggerstaff M. and Wood,R.D. (1999) Assay for nucleotide excision repair protein activity using fractionated cell extracts and UV-damaged plasmid DNA. Methods Mol. Biol., 113, 357–372. [DOI] [PubMed] [Google Scholar]
- Brand M., Moggs,J.G., Oulad-Abdelghani,M., Lejeune,F., Dilworth,F.J., Stevenin,J., Almouzni,G. and Tora,L. (2001) UV-damaged DNA-binding protein in the TFTC complex links DNA damage recognition to nucleosome acetylation. EMBO J., 20, 3187–3196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrier F. et al. (1999) Gadd45, a p53-responsive stress protein, modifies DNA accessibility on damaged chromatin. Mol. Cell Biol., 19, 1673–1685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Laat W.L., Jaspers,N.G and Hoeijmakers J.H. (1999) Molecular mechanism of nucleotide excision repair. Genes Dev., 13, 768–785. [DOI] [PubMed] [Google Scholar]
- Ekwall K., Olsson,T., Turner,B.M., Cranston,G. and Allshire,R.C. (1997) Transient inhibition of histone deacetylation alters the structural and functional imprint at fission yeast centromeres. Cell, 91, 1021–1032. [DOI] [PubMed] [Google Scholar]
- Enomoto S. and Berman,J. (1998) Chromatin assembly factor I contributes to the maintenance, but not the re-establishment, of silencing at the yeast silent mating loci. Genes Dev., 12, 219–232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enomoto S., McCune-Zierath,P.D., Gerami-Nejad,M., Sanders,M.A. and Berman,J. (1997) RLF2, a subunit of yeast chromatin assembly factor-I, is required for telomeric chromatin function in vivo. Genes Dev., 11, 358–370. [DOI] [PubMed] [Google Scholar]
- Ford J.M. and Hanawalt,P.C. (1997) Expression of wild-type p53 is required for efficient global genomic nucleotide excision repair in UV-irradiated human fibroblasts. J. Biol. Chem., 272, 28073–28080. [DOI] [PubMed] [Google Scholar]
- Fyodorov D.V. and Kadonaga,J.T. (2001) The many faces of chromatin remodeling: SWItching beyond transcription. Cell, 106, 523–525. [DOI] [PubMed] [Google Scholar]
- Gaillard P.H., Martini,E.M., Kaufman,P.D., Stillman,B., Moustacchi,E. and Almouzni,G. (1996) Chromatin assembly coupled to DNA repair: a new role for chromatin assembly factor I. Cell, 86, 887–896. [DOI] [PubMed] [Google Scholar]
- Game J.C. and Kaufman,P.D. (1999) Role of Saccharomyces cerevisiae chromatin assembly factor-I in repair of ultraviolet radiation damage in vivo. Genetics, 151, 485–497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gary R., Kim,K., Cornelius,H.L., Park,M.S. and Matsumoto,Y. (1999) Proliferating cell nuclear antigen facilitates excision in long-patch base excision repair. J. Biol. Chem., 274, 4354–4363. [DOI] [PubMed] [Google Scholar]
- Green C.M. and Almouzni,G. (2002) When repair meets chromatin: First in series on chromatin dynamics. EMBO Rep., 3, 28–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groisman R. et al. (2003) The ubiquitin ligase activity in the DDB2 and CSA complexes is differentially regulated by the COP9 signalosome in response to DNA damage. Cell, 113, 357–367. [DOI] [PubMed] [Google Scholar]
- Hanawalt P.C. (2001) Revisiting the rodent repairadox. Environ. Mol. Mutagen, 38, 89–96. [DOI] [PubMed] [Google Scholar]
- Hara R. and Sancar,A. (2002) The SWI/SNF chromatin-remodeling factor stimulates repair by human excision nuclease in the mononucleosome core particle. Mol. Cell. Biol., 22, 6779–6787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hara R., Mo,J. and Sancar,A. (2000) DNA damage in the nucleosome core is refractory to repair by human excision nuclease. Mol. Cell. Biol., 20, 9173–9181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hey T., Lipps,G., Sugasawa,K., Iwai,S., Hanaoka,F. and Krauss,G. (2002) The XPC-HR23B complex displays high affinity and specificity for damaged DNA in a true-equilibrium fluorescence assay. Biochemistry, 41, 6583–6587. [DOI] [PubMed] [Google Scholar]
- Hwang B.J., Ford,J.M., Hanawalt,P.C. and Chu,G. (1999) Expression of the p48 xeroderma pigmentosum gene is p53-dependent and is involved in global genomic repair. Proc. Natl Acad. Sci. USA, 96, 424–428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenuwein T. and Allis,C.D. (2001) Translating the histone code. Science, 293, 1074–1080. [DOI] [PubMed] [Google Scholar]
- Kaufman P.D., Kobayashi,R. and Stillman,B. (1997) Ultraviolet radiation sensitivity and reduction of telomeric silencing in Saccharomyces cerevisiae cells lacking chromatin assembly factor-I. Genes Dev., 11, 345–357. [DOI] [PubMed] [Google Scholar]
- Kaya H., Shibahara,K.I., Taoka,K.I., Iwabuchi,M., Stillman,B. and Araki,T. (2001) FASCIATA genes for chromatin assembly factor-1 in Arabidopsis maintain the cellular organization of apical meristems. Cell, 104, 131–142. [DOI] [PubMed] [Google Scholar]
- Krude T. (1995) Chromatin assembly factor 1 (CAF-1) colocalizes with replication foci in HeLa cell nuclei. Exp. Cell Res., 220, 304–311. [DOI] [PubMed] [Google Scholar]
- Li R., Hannon,G.J., Beach,D. and Stillman,B. (1996) Subcellular distribution of p21 and PCNA in normal and repair-deficient cells following DNA damage. Curr. Biol., 6, 189–199. [DOI] [PubMed] [Google Scholar]
- Martini E., Roche,D.M., Marheineke,K., Verreault,A. and Almouzni,G. (1998) Recruitment of phosphorylated chromatin assembly factor 1 to chromatin after UV irradiation of human cells. J. Cell Biol., 143, 563–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell D.L., Haipek,C.A. and Clarkson,J.M. (1985) (6-4)Photo products are removed from the DNA of UV-irradiated mammalian cells more efficiently than cyclobutane pyrimidine dimers. Mutat. Res., 143, 109–112. [DOI] [PubMed] [Google Scholar]
- Moggs J.G., Grandi,P., Quivy,J.P., Jonsson,Z.O., Hubscher,U., Becker,P.B. and Almouzni,G. (2000) A CAF-1-PCNA-mediated chromatin assembly pathway triggered by sensing DNA damage. Mol. Cell. Biol., 20, 1206–1218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moné M.J., Volker,M., Nikaido,O., Mullenders,L.H., van Zeeland,A.A., Verschure,P.J., Manders,E.M. and van Driel,R. (2001) Local UV-induced DNA damage in cell nuclei results in local transcription inhibition. EMBO Rep., 2, 1013–1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okano S., Lan,L., Caldecott,K.W., Mori,T. and Yasui,A. (2003) Spatial and temporal cellular responses to single-strand breaks in human cells. Mol. Cell. Biol., 23, 3974–3981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quivy J.P., Grandi,P. and Almouzni,G. (2001) Dimerization of the largest subunit of chromatin assembly factor 1: importance in vitro and during Xenopus early development. EMBO J., 20, 2015–2027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramanathan B. and Smerdon,M.J. (1986) Changes in nuclear protein acetylation in u.v.-damaged human cells. Carcinogenesis, 7, 1087–1094. [DOI] [PubMed] [Google Scholar]
- Ray-Gallet D., Quivy,J.P., Scamps,C., Martini,E.M., Lipinski,M. and Almouzni,G. (2002) HIRA is critical for a nucleosome assembly pathway independent of DNA synthesis. Mol. Cell, 9, 1091–1100. [DOI] [PubMed] [Google Scholar]
- Redon C., Pilch,D., Rogakou,E., Sedelnikova,O., Newrock,K. and Bonner,W. (2002) Histone H2A variants H2AX and H2AZ. Curr. Opin. Genet. Dev., 12, 162–169. [DOI] [PubMed] [Google Scholar]
- Ridgway P. and Almouzni,G. (2000) CAF-1 and the inheritance of chromatin states: at the crossroads of DNA replication and repair. J. Cell Sci., 113, 2647–2658. [DOI] [PubMed] [Google Scholar]
- Rubbi C.P. and Milner,J. (2003) p53 is a chromatin accessibility factor for nucleotide excision repair of DNA damage. EMBO J., 22, 975–986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shibahara K. and Stillman,B. (1999) Replication-dependent marking of DNA by PCNA facilitates CAF-1-coupled inheritance of chromatin. Cell, 96, 575–585. [DOI] [PubMed] [Google Scholar]
- Shivji M.K., Moggs,J.G., Kuraoka,I. and Wood,R.D. (1999) Dual-incision assays for nucleotide excision repair using DNA with a lesion at a specific site. Methods Mol. Biol., 113, 373–392. [DOI] [PubMed] [Google Scholar]
- Smerdon M.J. and Lieberman,M.W. (1978) Nucleosome rearrangement in human chromatin during UV-induced DNA-repair synthesis. Proc. Natl Acad. Sci. USA, 75, 4238–4241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith S. and Stillman,B. (1989) Purification and characterization of CAF-I, a human cell factor required for chromatin assembly during DNA replication in vitro. Cell, 58, 15–25. [DOI] [PubMed] [Google Scholar]
- Svejstrup J.Q. (2002) Mechanisms of transcription-coupled DNA repair. Nat. Rev. Mol. Cell. Biol., 3, 21–29. [DOI] [PubMed] [Google Scholar]
- Taddei A., Roche,D.,. Sibarita,J.-B., Turner,B.M. and Almouzni,G. (1999) Duplication and maintenance of heterochromatin domains. J. Cell Biol., 147, 1153–1166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taddei A., Maison,C., Roche,D. and Almouzni,G. (2001) Reversible disruption of pericentric heterochromatin and centromere function by inhibiting deacetylases. Nat. Cell. Biol., 3, 114–120. [DOI] [PubMed] [Google Scholar]
- Thoma F. (1999) Light and dark in chromatin repair: repair of UV-induced DNA lesions by photolyase and nucleotide excision repair. EMBO J., 18, 6585–6598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner B.M. (2002) Cellular memory and the histone code. Cell, 111, 285–291. [DOI] [PubMed] [Google Scholar]
- Tyler J.K., Adams,C.R., Chen,S.R., Kobayashi,R., Kamakaka,R.T. and Kadonaga,J.T. (1999) The RCAF complex mediates chromatin assembly during DNA replication and repair. Nature, 402, 555–560. [DOI] [PubMed] [Google Scholar]
- Ura K. et al. (2001) ATP-dependent chromatin remodeling facilitates nucleotide excision repair of UV-induced DNA lesions in synthetic dinucleosomes. EMBO J., 20, 2004–2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volker M. et al. (2001) Sequential assembly of the nucleotide excision repair factors in vivo. Mol. Cell, 8, 213–224. [DOI] [PubMed] [Google Scholar]
- Wakasugi M., Kawashima,A., Morioka,H., Linn,S., Sancar,A., Mori,T., Nikaido,O. and Matsunaga,T. (2002) DDB accumulates at DNA damage sites immediately after UV irradiation and directly stimulates nucleotide excision repair. J. Biol. Chem., 277, 1637–1640. [DOI] [PubMed] [Google Scholar]
- Wang Q., Zhu,Q., Wani,M.A., Wani,G., Chen,J. and Wani,A.A. (2003) Tumor suppressor p53 dependent recruitment of nucleotide excision repair factors XPC and TFIIH to DNA damage. DNA Repair (Amst), 2, 483–499. [DOI] [PubMed] [Google Scholar]
- Wolffe A.P. (1998) Chromatin: Structure and Function. Academic Press, New York, NY. [Google Scholar]
- Wood R.D. and Shivji,M.K. (1997) Which DNA polymerases are used for DNA repair in eukaryotes? Carcinogenesis, 18, 605–610. [DOI] [PubMed] [Google Scholar]
- Ye X., Franco,A.A., Santos,H., Nelson,D.M., Kaufman,P.D. and Adams,P.D. (2003) Defective S phase chromatin assembly causes DNA damage, activation of the S phase checkpoint and S phase arrest. Mol. Cell, 11, 341–351. [DOI] [PubMed] [Google Scholar]