Abstract
Haloalcohol dehalogenases are bacterial enzymes that catalyze the cofactor-independent dehalogenation of vicinal haloalcohols such as the genotoxic environmental pollutant 1,3-dichloro-2-propanol, thereby producing an epoxide, a chloride ion and a proton. Here we present X-ray structures of the haloalcohol dehalogenase HheC from Agrobacterium radiobacter AD1, and complexes of the enzyme with an epoxide product and chloride ion, and with a bound haloalcohol substrate mimic. These structures support a catalytic mechanism in which Tyr145 of a Ser-Tyr-Arg catalytic triad deprotonates the haloalcohol hydroxyl function to generate an intramolecular nucleophile that substitutes the vicinal halogen. Haloalcohol dehalogenases are related to the widespread family of NAD(P)H-dependent short-chain dehydrogenases/reductases (SDR family), which use a similar Ser-Tyr-Lys/Arg catalytic triad to catalyze reductive or oxidative conversions of various secondary alcohols and ketones. Our results reveal the first structural details of an SDR-related enzyme that catalyzes a substitutive dehalogenation reaction rather than a redox reaction, in which a halide-binding site is found at the location of the NAD(P)H binding site. Structure-based sequence analysis reveals that the various haloalcohol dehalogenases have likely originated from at least two different NAD-binding SDR precursors.
Keywords: haloalcohol dehalogenase/SDR family/short-chain dehydrogenase/X-ray structure
Introduction
Dehalogenases are enzymes that are able to cleave carbon-halogen bonds (Janssen et al., 2001). Structural characterization of haloalkane dehalogenases and haloacid dehalogenases demonstrated that these hydrolytic enzymes are evolutionarily related to widespread esterase and phosphatase families (Ollis et al., 1992; Hisano et al., 1996; Ridder and Dijkstra, 1999). Haloalcohol dehalogenases, also known as halohydrin dehalogenases or halohydrin hydrogen-halide lyases, cannot be classified in these existing dehalogenase families. Instead, they show low sequence similarity to members of the short-chain dehydrogenase/reductase (SDR) family (Smilda et al., 2001; van Hylckama Vlieg et al., 2001). This family contains redox enzymes that depend on NAD(P)H, which is bound in a characteristic dinucleotide binding fold (Rossmann fold) (Rossmann et al., 1974). They have a conserved catalytic triad of Ser, Tyr and Lys/Arg residues (Jörnvall et al., 1995; Oppermann et al., 2003), which is also present in the haloalcohol dehalogenases (van Hylckama Vlieg et al., 2001). Many structures of short-chain dehydrogenases/reductases in complex with dinucleotides and substrates have revealed the structural details of the reactions catalyzed by them (Jörnvall et al., 1995; Filling et al., 2002; Oppermann et al., 2003). In addition, the structure of a dinucleotide-binding transcription factor that lacked the catalytic tyrosine and thus oxidoreductase activity showed that the SDR fold also functions in non-enzymatic activities (Stammers et al., 2001).
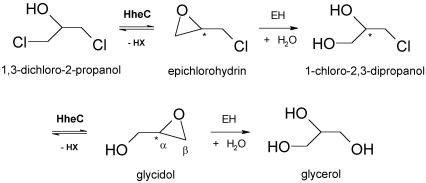
Haloalcohol dehalogenases catalyze the intramolecular displacement of a halogen by the vicinal hydroxyl group in 1,3-dichloro-2-propanol, yielding its corresponding epoxide, a halide ion and a proton (van Hylckama Vlieg et al., 2001). 1,3-dichloro-2-propanol is a well-known environmental pollutant that has extensively been applied in industry. It is mutagenic and genotoxic to bacterial and mammalian cells due to the spontaneous formation of the reactive epoxide epichlorohydrin (Hahn et al., 1991). Together with a well characterized epoxide hydrolase (Rink and Janssen, 1998; Nardini et al., 1999), the haloalcohol dehalogenase constitutes an efficient degradation pathway of 1,3-dichloro-2-propanol and epichlorohydrin into non-toxic metabolites, which can be used by the bacterium as a source of carbon and energy (Figure 1).
Fig. 1. Putative degradation route of 1,3-dichloro-2-propanol in A.radiobacter AD1. The epoxide hydrolysis steps are catalyzed by epoxide hydrolase (EH), whereas the dehalogenation steps are catalyzed by the haloalcohol dehalogenase HheC.
Haloalcohol dehalogenases have received increased attention due to their broad substrate specificity and enantioselectivity towards aliphatic as well as aromatic vicinal haloalcohols (van Hylckama Vlieg et al., 2001; Lutje Spelberg et al., 2002). They also efficiently catalyze the back reaction, the enantio- and β-regioselective epoxide ring opening by nucleophiles like azide and cyanide (Nakamura et al., 1994; Lutje Spelberg et al., 2001). These properties make them promising candidates to become tools in the synthesis of enantiopure aliphatic and aromatic epoxides and haloalcohols, as well as azido-, cyano- and other β-substituted alcohols (Lutje Spelberg et al., 1999, 2002).
X-ray crystallography and site-directed mutagenesis were used to provide insight into the catalytic machinery of the haloalcohol dehalogenase HheC from the soil bacterium Agrobacterium radiobacter AD1 (van Hylckama Vlieg et al., 2002) and its evolutionary relationships with SDRs. Our results extend our insight into the diversity of the superfamily of SDR-related proteins with a catalytically active member that has lost its NAD(P)H binding capability. Instead, the dinucleotide-binding site has become a spacious halide-binding site that facilitates haloalcohol dehalogenation by an SN2-type substitutive mechanism, rather than a redox reaction.
Results and discussion
Structure determination
Since previously studied hydrolytic dehalogenases contain a high affinity halide-binding site (Verschueren et al., 1993a; Ridder et al., 1997), we crystallized A.radiobacter haloalcohol dehalogenase HheC in the presence of bromide ions to use their anomalous signal for phasing (Dauter et al., 2001) (see Materials and methods). Its structure was subsequently elucidated via a three-wavelength MAD experiment on the Br-edge. Sixteen bromide ions with unusually high occupancies ranging from 0.7 to 0.95 could easily be located in the data, corresponding to the single high affinity halide-binding site in each of the sixteen molecules in the asymmetric unit. A 4-fold averaged 2.8 Å resolution electron density map obtained from the MAD data was used to build an initial model, which was subsequently refined against the high resolution data of a perfectly merohedrally twinned crystal. The resulting 1.8 Å resolution model contains one biologically functional tetramer of HheC per asymmetric unit, with each monomer consisting of the first 252 out of 254 amino acids, one bromide ion bound in the active site, and, in total, 291 water molecules. This model was the starting model for the structure elucidation of enzyme–substrate and enzyme–product complexes. A summary of the refinement and model statistics is shown in Table I.
Table I. Data collection and refinement statistics.
| Data collection and statistics of MAD on Br– | Peak λ = 0.9190 Å | Inflection λ = 0.9196 Å | Remote λ = 0.8551 Å |
|---|---|---|---|
| Space group | P21212 | ||
| Unit cell [Å (°)] | a = 118.0, b = 292.9, c = 146.4 (α = β = γ = 90) | ||
| No. of molecules per ASU | 16 | ||
| No. of Br– sites (halide-binding site/other sites) | 16 (14) | ||
| Resolution (outer shell) (Å) | 25–2.7 (2.8–2.7) | 25–2.7 (2.8–2.7) | 25–2.8 (2.9–2.8) |
| Rsym (%) overall (outer shell) | 5.5 (14.5) | 5.8 (12.7) | 6.8 (19.0) |
| Completeness (%) overall (outer shell) | 95.9 (84.5) | 95.9 (85.1) | 89.4 (62.0) |
| I/σ overall (outer shell) | 15.7 (4.9) | 14.9 (4.9) | 10.3 (2.5) |
| Reflections total (unique) | 1 718 858 (139 737) | 1 726 074 (139 618) | 1 058 025 (138 855) |
| Overall phasing power (centrics:ISO/ANO) | –/– | 2.36/– | 2.25/– |
| Overall phasing power (acentrics: ISO/ANO) | –/0.76 | 3.46/1.28 | 3.41/0.98 |
| <FOM> (centric/acentric) | 0.26/0.35 | ||
| Data collection of complexes | HheC·Br– (twinned, α = 0.48) | HheC·Cl–·(R)-SO | HheC·(R)-pNPAE |
| Space group | P43 | P43212 | C2 |
| No. of molecules/ASU | 4 | 2 | 4 |
| Unit cell [Å (°)] | a = b = 103.2 c = 117.4 (α = β = γ = 90) | a = b = 103.9 c = 119.4 (α = β = γ = 90) | a = 146.7 b = 71.8 c = 97.3 (α = 90 β = 92.8 γ = 90) |
| Resolution (outer shell) (Å) | 35–1.8 (1.9–1.8) | 30–2.6 (2.7–2.6) | 25–1.9 (2.0–1.9) |
| Rsym (%) overall (outer shell) | 6.6 (17.0) | 9.6 (41.0) | 5.6 (18.5) |
| Completeness (%) overall (outer shell) | 95.2 (46.4) | 96.3 (91.7) | 98.7 (93.9) |
| I/σ overall (outer shell) | 23.4 (3.2) | 10.8 (2.9) | 23.5 (5.8) |
| Reflections total (unique) | 1 561 167 (63 725) | 297 117 (20 596) | 1 029 109 (78 669) |
| |
|
|
|
| Refinement | |||
| Total atoms/water | 7889/291 | 3916/64 | 8630/898 |
| R | 23.3 (twinned) | 24.6 | 16.9 |
| Rfree | 30.9 (twinned) | 29.6 | 19.6 |
| R.m.s.d. bonds (Å)/angles (°) | 0.008/1.51 | 0.007/1.42 | 0.005/1.37 |
| R.m.s.d. B (Å2) (main chain/side chain) | 0.85/1.19 | 0.91/1.61 | 0.68/1.51 |
| Ramachandran plot (%) | |||
| Favoured | 84.1 | 85.6 | 85.2 |
| Allowed | 14.0 | 13.0 | 13.4 |
| Generously allowed | 1.4 | 0.5 | 0.5 |
| Disallowed | 0.5 | 0.9 | 0.9 |
aRsym = Σ|I – <I>|/ΣI, where I is the observed intensity and <I> the average intensity.
bPhasing power = r.m.s. (|FX|/E), where |FX| can be the calculated isomorphous (ISO) or anomalous (ANO) structure factor and E is the residual lack of closure.
c<FOM> = mean figure of merit over all resolution bins.
dR = R based on 95% of the data used in refinement. Rfree = R based on 5% of the data withheld for the cross-validation test.
Overall structure of haloalcohol dehalogenase HheC
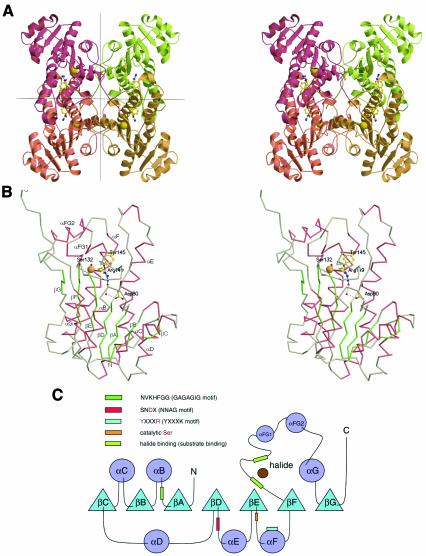
HheC is a tetramer with 222 point group symmetry that can be regarded as a dimer of dimers (Figure 2A). The different subunits of the tetramer superimpose, with root mean square deviations (r.m.s.d.s) between 0.45 and 0.55 Å for 252 Cα atoms. Each subunit consists of a seven- stranded parallel β-sheet flanked on both sides by α-helices, which resembles the characteristic Rossmann fold (Figure 2B and C). Two monomers form a dimer, in which the longest α-helices αE and αF and part of their connecting loops associate into an intermolecular, anti-parallel, four-helix bundle. Two of these dimers make up the full tetramer through contacts of β-strand βG and α-helix αG, and their connecting loops. The β-strands come together in an anti-parallel fashion and mainly interact through hydrophobic and electrostatic interactions of their side chains.
Fig. 2. (A) Stereo view of the tetrameric structure of the HheC·Br– complex. Tight dimer interactions are formed by two pairs of helices that form a four-helix bundle in the central horizontal plane. Two dimers associate by packing along the vertical plane to form the final tetramer. The location of the active site in each monomer is indicated by the catalytic serine, tyrosine and arginine in ball-and-stick representation, and the bound bromide ion as a yellow sphere. (B) Stereo view of the Cα trace of a monomer of the HheC·Br– complex. The catalytic residues Ser132, Tyr145, Arg149, Asp80 and two internal water molecules are depicted in ball-and-stick representation. The bromide ion is shown as a yellow sphere. The hydrogen-bonding pattern that extends from the catalytic base tyrosine to Asp80 at the surface of the molecule is shown as black dashed lines. (C) Secondary structure topology diagram showing the location of the sequence motifs that are responsible for halide binding, substrate binding and proton relay. The corresponding sequence motifs of the SDR enzyme family are shown in parentheses.
The fold and dimerization interfaces of HheC are typical for members of the SDR family, which mainly occur as tetramers or dimers (Jörnvall et al., 1995). The C-terminal end of each HheC monomer binds the 2-fold symmetry-related subunit that lies opposite in the other dimer, and contributes a tryptophan residue (Trp249) to its active site (Figures 2A and 3). Similar interactions of C-terminal residues with the active site of another subunit have been observed in structures of other members of the SDR family (Benach et al., 1998). The relevance of this interaction for HheC is not known. Sequence alignment of the available haloalcohol dehalogenases shows that some of them lack the last nine C-terminal residues, suggesting that the interaction of the C-terminus with the opposite active site is not essential.
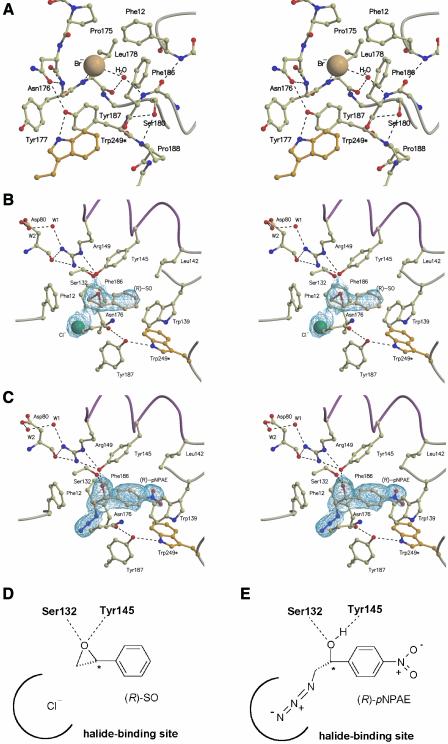
Fig. 3. (A) A detailed overview (in stereo) of the halide binding site in the HheC·Br– complex. Residues are shown in ball-and-stick representation and hydrogen bonds are indicated by black dashed lines. Trp249* is the only residue that is contributed by a opposite monomer in the tetramer. (B and C) Detailed schematic representations of the active site of the HheC·Cl– (R)-SO complex (B) and HheC·(R)-pNPAE complex (C). Portions of a 2Fo – Fc simulated annealing electron density omit map are shown (Brünger et al., 1997), contoured at 1σ and covering the bound molecules. (D and E) Simplified schematic drawing of the bound epoxide and azidoalcohol molecule in the HheC·Cl–·(R)-SO complex (D) and the HheC·(R)-pNPAE complex (E).
The halide binding site of haloalcohol dehalogenase HheC
The HheC·Br– binary complex highlights the active site, which is located in a loop-rich cavity that contains the Ser132-Tyr145-Arg149 catalytic triad and a bound bromide ion (Figure 2B and C). Tyr145 is hydrogen bonded to Arg149 in the next turn of helix αF. The third residue of the catalytic triad, Ser132, is located at the tip of strand βE, with its hydroxyl group making hydrogen bonds with the side-chain oxygen atom and the backbone NH group of Thr134. A loop starting from β-strand βF and a small loop between β-strand βA and helix αB form a spacious halide binding site. The interactions of the halide ion are more extensive and of a more hydrophobic nature than those previously found for haloalkane dehalogenases and haloacid dehalogenases (Verschueren et al., 1993a; Ridder et al., 1997). The bromide ion interacts with the peptide nitrogen atoms of Tyr177 and Leu178, with the side chains of Pro175 and Leu178, the Cα atom of Asn176, and with the aromatic rings of Phe12, Phe186 and Tyr187 (Figure 3A).
The halide ion also interacts with a water molecule that is bound in the back of the halide-binding site (Figure 3A), where it bridges the backbone carbonyl group of Leu178 and the amide group of the cis-peptide bond between Tyr185 and Phe186. This cis-peptide is stabilized further by a hydrogen bond between the backbone carbonyl oxygen atom of Tyr185 and the peptide nitrogen atom of Phe12. Other contacts that stabilize the overall conformation of the halide-binding site are hydrogen bonds between the side chains of Asn176 and Tyr187, and between Ser180 and the NH group of Tyr187. The tyrosine hydroxyl group further provides the docking site for the indole NH function of Trp249 from the opposite 2-fold symmetry-related subunit.
Product binding in haloalcohol dehalogenase HheC
The equilibrium of the dehalogenation reaction lies almost completely to the side of the epoxide (van Hylckama Vlieg et al., 2001). Therefore, to gain insight into epoxide product binding, HheC was crystallized in the presence of NaCl and a racemic mixture of styrene oxide. This resulted in a 2.6 Å resolution ternary complex of the enzyme with a chloride ion and (R)-styrene oxide [(R)-SO] (Figure 3B and D).
In this complex, the chloride ion occupies the halide-binding site, which shows no large conformational changes compared with its bromide-bound state in the HheC·Br– complex. The aromatic ring of styrene oxide interacts with the side chains of several aromatic and aliphatic residues that line the active site cleft. A water molecule bound between Ser132 and Tyr145 in the HheC·Br– complex has been replaced by the epoxide ring of (R)-SO. The Ser132 and Tyr145 side chains make hydrogen bonds of 2.7 Å with the epoxide oxygen atom, which are arranged in an approximately tetrahedral configuration with the two CO bonds of the epoxide ring. They position the epoxide Cβ atom close to the chloride ion bound in the halide-binding site, which could explain the high β-regioselectivity of nucleophilic epoxide ring opening reactions catalyzed by HheC (Lutje Spelberg et al., 2002).
Binding of a substrate mimic in haloalcohol dehalogenase HheC
Aromatic azidoalcohols or cyanoalcohols, the products of the epoxide ring opening reaction catalyzed by HheC in vitro, can be used to study the binding of haloalcohol substrates. They can be regarded as mimics of haloalcohols, as their azide and cyanide function have similar binding properties to halides (Norne et al., 1975; Verschueren et al., 1993b). The equilibrium of the nucleophilic ring opening of an epoxide by azide lies completely at the side of the azidoalcohol product (Lutje Spelberg et al., 2002). Therefore, co-crystallization of HheC in the presence of (R)-1-para-nitro-phenyl-2-azido-ethanol [(R)-pNPAE] resulted in a stable HheC· azidoalcohol complex at 1.9 Å resolution (Figure 3C and E). The azide group of the compound is bound in the halide-binding site of the enzyme, making similar contacts with the backbone amide groups, aromatic amino acid side chains and the structural water molecule as observed for bound halides. The secondary hydroxyl moiety of the chiral Cα atom of the azidoalcohol is firmly bound to Ser132 and Tyr145 by hydrogen bonds of ∼2.6 Å.
The epoxide and azidoalcohol complexes superimpose with a very low r.m.s.d. of only 0.20 Å for 1902 atoms of a HheC monomer. The conformations of the active site residues are virtually the same, showing that the substrate mimic and product are equally well accommodated by the active site. This is in agreement with HheC being able to catalyze both the back and forward reaction. The non-specific binding mode of the epoxide and azidoalcohol side chain in the complexes could explain the broad acceptance of the enzyme of substituted aliphatic and aromatic haloalcohol and epoxide side chains (van Hylckama Vlieg et al., 2001).
Structural relationships between haloalcohol dehalogenases and short-chain dehydrogenases/reductases
A structural similarity search for HheC against the Protein Data Bank (PDB) using the SCOP server (Murzin et al., 1995) yielded 20 SDR structures. The four highest scoring structures were 7α-hydroxysteroid dehydrogenase from Escherichia coli (PDB accession code 1FMC; Tanaka et al., 1996), trihydroxynaphtalene reductase from Magnaporthe grisea (accession code 1YBV; Andersson et al., 1996), and the plant tropinone reductases I and II from Datura stramonium [accession codes 1AE1 (Nakajima et al., 1998) and 2AE1 (Yamashita et al., 1999), respectively]. These four enzymes are all NAD(P)H-dependent enzymes of ∼250 residues that catalyze the dehydrogenation of a hydroxyl group or the reduction of a carbonyl group. All four structures superimpose on the structure of the HheC·Cl–·(R)-SO complex, with r.m.s. differences between 1.15 and 1.5 Å for ∼190 Cα atoms, whereas their amino acid sequences share only 20–25% sequence identity. High structural similarity, despite low sequence identity, has previously been recognized as a general property of the SDR family (Jörnvall et al., 1995).
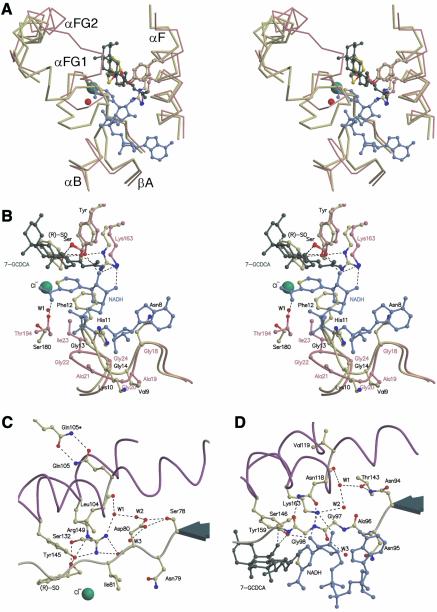
A structure-based sequence alignment shows that the Ser-Tyr-Arg/Lys catalytic triads are strictly conserved, whereas two regions show high divergence between HheC and related dehalogenases on the one hand and the four most similar SDR enzymes on the other (Figure 4). The first divergent region lies between residues 175 and 189 of HheC, which corresponds to the loop between strand βF and helix αFG2 that participates in cofactor and substrate binding in the SDR enzymes. In HheC, this loop forms most of the halide-binding site (Figure 5A). In the SDR enzymes, the amino group of the carboxylamine-pyridine function of the nicotinamide adenine dinucleotide (NADH) cofactor is stabilized by a mostly conserved threonine (Thr194 of 7α-HSD) (Filling et al., 2002), whereas the structurally equivalent Ser180 in HheC stabilizes the backbone conformation of the halide-binding loop. This residue is not conserved in the other haloalcohol dehydrogenases (Figure 4), which suggests that Ser180 plays no essential role in the dehalogenases. However, the corresponding loop contains the cis-peptide between Phe185 and Tyr186 that binds the halide-bound water molecule. These residues are conserved in other haloalcohol dehalogenase sequences, which may indicate a general stabilizing role for the cis-peptide in the architecture of the halide-binding site.
Fig. 4. Sequence alignment of the haloalcohol dehalogenases HalB (Agrobacterium tumefaciens, unpublished, DDBJ/EMBL/GenBank accession No. AAD34609), HheAAD2 (Arthrobacter sp. AD2; van Hylckama-Vlieg et al., 2002), HheA (Corynebacterium sp. N-1074; Yu et al., 1994), HheBGP1 (Mycobacterium sp. GP1; Poelarends et al., 1999) and HheB (Corynebacterium sp. strain N-1074; Yu et al., 1994), combined with a 3D-structure-based alignment of HheC (A.radiobacter AD1; van Hylckama-Vlieg et al., 2001), and the four homologous SDR structures with the highest structural similarity to HheC [7α-hydroxysteroid dehydrogenase from E.coli, PDB accession code 1FMC (Tanaka et al., 1996); trihydroxynaphtalene reductase from M.grisea, PDB accession code 1YBV (Andersson et al., 1996); and tropinone reductases I and II from D.stramonium, PDB accession codes 1AE1 and 2AE1, respectively (Nakajima et al., 1998; Yamashita et al., 1999)]. The colored amino acids correspond to the sequence motifs in Figure 2C and share the same color coding. The underlined bold amino acids correspond to the conserved asparagine in SDR enzymes (Asn118 in 7α-HSD) (Filling et al., 2002). The red- and blue-colored boxes, respectively, indicate the possible positions of acidic and basic residues that define the NADH or NADPH cofactor specificity in the SDR family (Persson et al., 2003).
Fig. 5. (A) Stereo view of a superposition of a Cα trace of the two divergent loops of the HheC·Cl–·(R)-SO complex (yellow) and the ternary complex of 7α-hydroxysteroid dehydrogenase (pink) with NADH (blue) and 7-oxoglycochenodeoxycholic acid (7-GCDCA) (dark green). The chloride ion and a water molecule are shown as green and red spheres, respectively. (B) Stereo view of a superposition of the active site regions of the HheC·Cl–·(R)-SO complex (yellow) and the ternary complex of 7α-hydroxysteroid dehydrogenase (pink) with NADH (blue) and 7-GCDCA (dark green). Residues, substrates and cofactor are shown in ball-and-stick representation, and hydrogen bonds are indicated by black dashed lines. Labels of haloalcohol dehalogenase HheC are colored black, labels of 7α-hydroxysteroid dehydrogenase are colored pink. (C) Representation of the proposed proton relay pathway in the HheC·Cl–·(R)-SO complex from the catalytic base Tyr145 to a water molecule (W3) in the first solvation shell of the HheC tetramer. Cα traces of the central parts of the helices αE and αF are shown in purple. Amino acids and epoxide substrate are shown in ball-and-stick representation. A chloride ion and water molecules are shown as green and red spheres, respectively. (D) A detailed schematic representation of the proton relay pathway in the ternary complex of 7α-hydroxysteroid dehydrogenase (pink) with NADH (blue) and 7-GCDCA (dark green).
The second and most striking difference is in the loop that connects βA and αB and that contains the highly conserved nucleotide-binding Gly-X-Gly-X3-Gly motif of the SDR enzymes (Jörnvall et al., 1995). In HheC, this motif is replaced by a NVKHFGG pattern (residues 8–14), and its larger side chains occupy the space where in the SDR enzymes part of the NADH cofactor is bound (Figures 4 and 5B). The glycine-replacing Phe12 makes important backbone–backbone interactions with the cis-peptide between Phe185 and Tyr186. Phe12 also has stacking interactions with the aromatic side chain of Phe185. These interactions cause the two described loops that each interact with the cofactor in SDR enzymes to pack closely together and to form the spacious halide-binding site in HheC.
Catalytic mechanism of haloalcohol dehalogenases
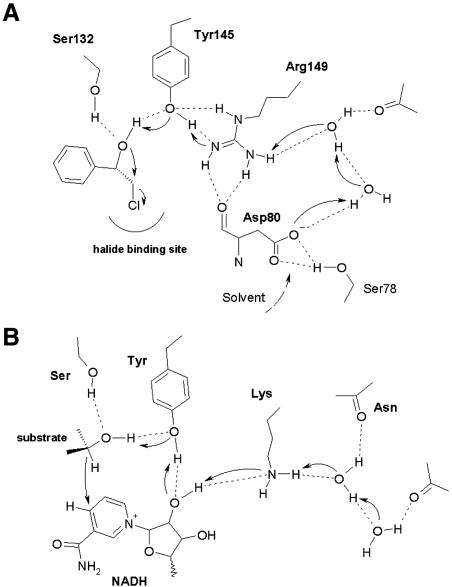
The structural correspondence of HheC to the SDR suggests a key role for the conserved catalytic residues in the dehalogenase mechanism. The epoxide and azidoalcohol complexes of HheC show that the Ser132 and Tyr145 side chains make hydrogen bonds with the oxygen atom of the epoxide product and with the hydroxyl group of a haloalcohol substrate (Figure 6A). The direct interaction of Arg149 with Tyr145 in the otherwise hydrophobic active-site cavity likely polarizes the tyrosine hydroxyl group and lowers its pKa. Deprotonation of the tyrosine would allow this residue to act as the catalytic base that accepts a proton from the hydroxyl group of a vicinal haloalcohol substrate to generate an intramolecular nucleophile, which concertedly substitutes the halogen. Ser132, Tyr145 and Arg149 were previously shown to be important for the catalytic mechanism, as a Ser132Ala, a Tyr145Phe and a Arg149Asn mutant are >10 000-fold less active than the wild-type enzyme (Table II) (van Hylckama Vlieg et al., 2001).
Fig. 6. (A) A schematic representation of the proposed catalytic mechanism of dehalogenation in HheC. The catalytic base Tyr145 activates the hydroxyl group adjacent to the halogen atom for nucleophilic attack on the halogen-bearing carbon atom. The proton is released to the solvent via Arg149, the water molecules and Asp80. (B) A schematic representation of the proposed catalytic mechanism of SDR enzymes (Filling et al., 2002; Oppermann et al., 2003). The secondary hydroxyl group is deprotonated by the catalytic tyrosine residue, with concurrent transfer of the hydride ion to NAD+. Proton release from the catalytic tyrosine is proposed to take place via the hydroxyl group of the cofactor ribose moiety, the conserved lysine amine function, and water molecules bound in the core of the enzyme.
Table II. Steady-state kinetic parameters of wild-type and mutant HheC with the substrate para-nitro-2-bromo-1-phenylethanol.
| Enzyme | Km (mM) | kcat (s–1) | kcat/Km (s–1M–1) |
|---|---|---|---|
| Wild typea | 0.009 ± 0.003 | 22 ± 1 | 2.4 × 106 |
| Ser132Alab | –d | –d | –d |
| Tyr145Pheb | 0.13 | 0.072 | 5.5 × 102 |
| Arg149Lysb | 0.056 | 0.33 | 5.9 × 103 |
| Arg149Asnb | >0.40 | >0.15 | 3.8 × 102 |
| Asp80Asnc | 0.02 ± 0.002 | 0.1 ± 0.002 | 5.0 × 103 |
| Asp80Alac | –d | –d | –d |
aData taken from Tang et al. (2003).
bData taken from van Hylckama Vlieg et al. (2001).
cThis study.
dNo activity detectable.
In short-chain dehydrogenases, the catalytic tyrosine deprotonates the hydroxyl group of a substrate, and transfers the proton to the catalytic lysine via the oxygen atom of a ribose hydroxyl group of the cofactor (Figures 5D and 6B) (Liao et al., 2001; Filling et al., 2002). In HheC, the backbone carbonyl group of Asp80 is at the position of the ribose hydroxyl group, which precludes proton transfer from the substrate to the equivalent Arg149 via this route. Instead, Arg149 directly contacts the tyrosine hydroxyl group and thus can directly take up a proton from the tyrosine. Mutation of this arginine into a lysine yields an enzyme that is 400-fold less active than the wild-type enzyme (Table II) (van Hylckama Vlieg et al., 2001). Impaired proton transfer between the tyrosine and the lysine could provide a plausible explanation for this, although the possibility of an effect on the pKa of Tyr145 or on the integrity of the active site can not be excluded.
In the short-chain dehydrogenases, the catalytic lysine further interacts with one of two buried water molecules that is bound to the backbone carbonyl oxygen atom of a conserved asparagine, located at a kink in helix αF (Asn118 in 7α-HSD). The side chain of the asparagine is, in turn, stabilized by an interaction with the conserved NNAG motif (residues 94–97 in 7α-HSD). These water molecules are present in the majority of structurally characterized SDR enzymes and have been proposed to mediate proton relay to the solvent (Figures 5D and 6B) (Benach et al., 1998; Filling et al., 2002). In HheC, a similar water-filled enclosure is present near a kink in helix αF at Leu104, which aligns with Asn118 of 7α-HSD (Figure 5C). The bound water molecules form a continuous pattern of hydrogen bonds from Arg149 to the partly solvent-exposed side chain of Asp80. This aspartate is part of an SNDX pattern (residues 78–81) that superimposes on the NNAG motif of the SDR family. This suggests that the side chain of Asp80 could play a critical role in proton relay from Arg149 to the solvent. Strikingly, the pH optima of the dehalogenation and the epoxide ring opening activities are significantly different: pH 8–9 and 4–5, respectively. During a dehalogenation reaction a proton needs to be removed, which would be easiest at physiological pH or higher, where the aspartate is charged. In contrast, for epoxide ring opening reactions protons are needed, and their provision would benefit from an acidic pH, where Asp80 is mostly protonated. Therefore, the observed proton relay system offers a simple explanation for the different pH optima of the dehalogenation and epoxide ring opening reactions catalyzed by HheC. Strikingly, when Asp80 is mutated into an alanine, no activity can be detected (Table II). Moreover, an Asp80Asn mutant of HheC has a 220-fold reduced kcat for the dehalogenation reaction with respect to the wild-type enzyme, whereas the Km is approximately the same. The similar reduction in kcat of the Asp80Asn and Arg149Lys mutant further supports the importance of the arginine and aspartate for catalysis.
On the origin of haloalcohol dehalogenase activity
The structure of HheC reveals that its fold resembles that of members of the NAD(P)H-dependent SDR family. Together with the clear homology of HheC to the SDR enzymes (20–25% identity; van Hylckama Vlieg et al., 2001), this provides evidence for a divergent evolutionary relationship. The SDR family is characterized by conserved sequence motifs for the catalytic residues, the NAD(P)H-binding site, and the proton relay system (Filling et al., 2002; Oppermann et al., 2003). HheC has only retained the catalytic residues, whereas the sequence segments that correspond to the SDR cofactor-binding site and NNAG motifs define the halide-binding site and proton relay system in HheC. However, these sequences are not strictly conserved in other haloalcohol dehalogenases, which suggests diversity in halide binding and proton relay.
The SDR family is old in its origins and widespread in all kingdoms of life, with >60 different enzymes in humans and up to 46 copies in the sequenced genomes of various microbial species, for example (Jörnvall et al., 1999; Kallberg et al., 2002a). In contrast, only six haloalcohol dehalogenase genes have been isolated from selected strains of soil bacteria that degrade 1,3-dichloro-2-propanol and 1,2-dibromoethane (van Hylckama Vlieg et al., 2001). Whereas haloalkane dehalogenases are found in the genomes of several microbial species (Jesenská et al., 2000), a BLAST search (Altschul et al., 1997) with the haloalcohol dehalogenase sequences against microbial genomes did not find any haloalcohol dehalogenase sequence, but only retrieved sequences of 15–35% identity that contain the characteristic SDR NAD(P)H-binding Gly-rich motif. The rare and exclusive occurrence of haloalcohol dehalogenases in pollutant-degrading soil bacteria raises questions with respect to their evolutionary origin.
SDR enzymes have been grouped into seven different coenzyme-based subfamilies (Kallberg et al., 2002b; Persson et al., 2003) on the basis of the nature of the residues at positions 32, 33, 34 and 15 (HheC numbering). The C-type (HheC and HalB) and A-type (HheA and HheAAD2) haloalcohol dehalogenases contain an aspartic acid at position 32, suggesting that they have evolved from an NAD-binding precursor, rather than an NADP-dependent enzyme (Figure 4). Intriguingly, HheB and HheBGP1 have the characteristics of a different NAD-binding subfamily (with an aspartic acid at position 34), suggesting that these latter enzymes have originated from a different precursor than the A- and C-type dehalogenases. Furthermore, the B-type dehalogenases (HheB and HheBGP1) share only 24% sequence identity with the A- and C-type dehalogenases, which themselves are 33% identical, and, as a major difference, they lack the sequence segment that corresponds to β-strand βC and α-helix αD in HheC (Figure 4). This deletion in the Rossmann fold (Rossmann et al., 1974) has previously been overlooked as a result of misalignment of the sequences (Yu et al., 1994; Smilda et al., 2001; van Hylckama Vlieg et al., 2001).
Unfortunately, it is impossible to reconstruct the evolutionary history of the isolated haloalcohol dehalogenases without knowledge of their specific precursor enzymes. It is conceivable, however, that an NAD-dependent SDR precursor could have accommodated the halogen of a haloalcohol substrate in the apolar nicotinamide binding site of the cofactor and used the conserved Ser-Tyr-Lys/Arg catalytic triad to promote the substitution of the halogen. Some SDR enzymes could thus fortuitously have favored a rudimentary haloalcohol dehalogenating activity next to an NAD-dependent redox activity. Evidence is accumulating that alternative enzymatic reactions, distinct from the normally catalyzed ones, may play an important role in the diversification of enzymes (see for example O’Brien and Herschlag, 1999). The recruitment of NAD-binding SDR enzymes for such a haloalcohol dehalogenase activity may have occurred only very recently in response to elevated concentrations of halogenated pollutants in the environment. Although it can be argued that the low sequence identities of haloalcohol dehalogenases to the currently known bacterial SDR sequences (15–35%) may suggest that they diverged a long time ago, the enormous diversity of soil bacteria in the environment (Torsvik and Øvreås, 2002) leaves open the possibility that more closely related SDR enzymes exist that have not been isolated so far. It might be possible to test whether a limited number of mutations can convert an SDR enzyme into a dehalogenase in the laboratory by directed evolution experiments that select for haloalcohol dehalogenating activity.
Material and methods
Crystallization of HheC
Haloalcohol dehalogenase HheC was purified as described elsewhere (van Hylckama Vlieg et al., 2001). Crystals of HheC were prepared as described previously (de Jong et al., 2002). Bromide-derivatized crystals were prepared by co-crystallization. NaBr was added to a final concentration of 50 mM to the standard reservoir solution. Hanging drops were prepared of 2 µl of a protein solution of 7.5 mg/ml and 2 µl of the reservoir solution. Crystals grew in a few days at room temperature.
Crystals of a HheC·Cl–·(R)-SO complex were also obtained by co-crystallization. Sulfuric acid was used to prepare dialysis and crystallization buffers to ensure that no halides were present in the solutions. A racemic mixture of (R)- and (S)-SO solubilized in a small amount of dimethylsulfoxide (DMSO) (<1 µl) was added to 50 µl of the purified and dialyzed protein solution (7.5 mg/ml) to a final concentration of 20 mM. A small volume (1 µl) of a concentrated NaCl solution was added to a final concentration of 10 mM. After equilibrating the protein solution for 30 min, non-solubilized SO was spun down at 3000 g to prevent solid material being transferred to the crystallization medium. Hanging drops of a total volume of 3 µl were prepared by mixing 1.5 µl of the saturated protein–substrate mixture with 1.5 µl of the reservoir solution described above. Bipyramidal crystals in the tetragonal space group P43212 appeared in a few days. A HheC·azidoalcohol complex was obtained by following the same solubilization and crystallization procedure as described above for the HheC·Cl–·(R)-SO complex, using a final concentration of 25 mM of (R)-pNPAE (97% enantiomeric excess). Plate-like crystals in space group C2 grew in 3–5 days. In all cases a cryosolution was made by replacing the water in the well solution with a 20% glycerol solution.
Structure determination and refinement
A crystal of space group P21212 (cell dimensions: a = 117.5 Å, b = 293.1 Å, c = 146.5 Å), containing four HheC tetramers per asymmetric unit, was used in a three-wavelength anomalous dispersion experiment collected on a MAR CCD detector at the European Synchrotron Radiation Facility (ESRF) synchrotron beam line BM14. The wavelength of the X-rays was tuned around the absorption K-edge of bromide deduced from an edge-scan of the crystal. Wavelengths of the peak and inflection point dataset were set to 0.9190 and 0.9196 Å, respectively. Data were collected to 2.8 Å resolution over a 90° rotation of the crystal, with an interval of 0.2°. Thirty frames of the peak and inflection point datasets were recollected using an interval of 0.1° due to severe overlaps in this region of the data. A remote wavelength dataset (λ = 0.8551 Å) was collected over 90° with a 0.2° interval. All data were processed using DENZO and SCALEPACK (Table I) (Otwinowski and Minor, 1997). The resulting scaled anomalous intensities collected at the peak wavelength were used to search for bromide positions with the program Shake-and-Bake (Weeks and Miller, 1999). Coordinates and occupancies of the bromide sites were subsequently refined against the data of all three wavelengths with the program SHARP (Table I) (De la Fortelle and Bricogne, 1997). The positions of the 16 highest occupied sites per asymmetric unit formed approximately tetrahedral clusters of four atoms when expanded over the whole unit cell, suggesting that they corresponded to 16 bromide ions bound to the halide-binding sites of 16 HheC monomers. These 16 sites were subsequently used to find the NCS operators between the corresponding tetrameric HheC molecules using FINDNCS (CCP4, 1994). Three of the four NCS operators were refined in an averaging procedure using DM (Cowtan, 1994), using a combination of averaging, solvent flattening and histogram matching. A spherical mask with a radius of 35 Å around the center of gravity of the four-atom clusters made with the program NCSMASK (CCP4, 1994) was used in these calculations. The resulting 3-fold averaged electron density maps were of high quality and allowed the manual building of 90% of the residues of a HheC monomer.
This initial model was subsequently used as a search model for a molecular replacement experiment using data of a near-perfectly merohedrally twinned crystal of HheC in space group P43 [as indicated by TRUNCATE (CCP4, 1994) and Todd Yeates’ Twinning Server (Yeates, 1997)]. The data were collected at the Elettra synchrotron in Triëste (Italy) to 1.8 Å resolution at 100 K using an X-ray wavelength of 0.93 Å over a 180° rotation of the crystal with an interval of 0.5°. The twin-related reflections (twinning fraction α = 0.48) were averaged with the program SFTOOLS (CCP4, 1994). Four HheC monomers in each twin domain were found by molecular replacement in AMORE (Navaza and Saludijan, 1997). One twin domain was subsequently refined using twinning scripts available in the program suite CNX (Accelrys Inc., San Diego, CA).
Diffraction data of crystals of the HheC·Cl–·(R)-SO complex (2.6 Å resolution) and the HheC·(R)-pNPAE complex (1.9 Å resolution) were collected at the ID14-2 beam line at the ESRF in Grenoble (Table I). All data from crystals of the complexes were collected at 100 K using an X-ray wavelength of 0.93 Å. Initial phases for these data sets were obtained either from rigid body fitting in CNX or, in the case of the new crystal form, by molecular replacement calculations in AMORE (Navaza and Saludijan, 1997). Model coordinates for the substrate molecules were obtained by building and minimizing them in the program QUANTA (Accelrys Inc.). Parameter and topology files for these compounds were obtained from the Hic-Up Server (Kleywegt and Jones, 1998). All data processing was carried out using DENZO and SCALEPACK (Otwinowski and Minor, 1997). All refinements were carried out using the program suite CNX (Accelrys Inc.). All manual building of protein structures was carried out in QUANTA (Accelrys Inc.) and Xtalview (McRee, 1999). Figures were prepared using MOLSCRIPT (Kraulis, 1991), and RASTER3D (Merritt and Bacon, 1997).
Site-directed mutagenesis and kinetic characterization
Site-directed mutagenesis, purification of mutant enzymes and the measurement of the activity of the Asp80Asn mutant were performed routinely as described previously (van Hylckama Vlieg et al., 2001).
Structural alignment and sequence analysis
Iterative structural alignment of HheC and the four most similar crystal structures of SDR enzymes and the subsequent sequence alignment of other available haloalcohol dehalogenase sequences on the structural alignment were performed using the program SEQUOIA (Bruns et al., 1999).
Accession numbers
The coordinates and structure factor amplitudes of the HheC·Br–, HheC·Cl–·(R)-SO and HheC·(R)-pNPAE complexes have been deposited in the PDB under accession codes 1PWX, 1PWZ and 1PW0, respectively.
Acknowledgments
Acknowledgements
We thank Andy Thompson and the staff of the ESRF synchrotron in Grenoble, France, and the Elettra synchrotron in Triëste, Italy, for help with data collection. We also thank Jeffrey Lutje Spelberg for providing purified samples of the epoxide product and the azidoalcohol substrate mimic. This research was supported by The Netherlands Foundation of Chemical Research (CW), with financial aid from The Netherlands Foundation for Scientific Research (NWO).
References
- Altschul S.F., Madden,T.L., Schäffer,A.A., Zhang,J., Zhang,Z., Miller,W. and Lipman,D.J. (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res., 25, 3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersson A., Jordan,D., Schneider,G. and Lindqvist,Y. (1996) Crystal structure of the ternary complex of 1,3,8-trihydroxynaphthalene reductase from Magnaporthe grisea with NADPH and an active-site inhibitor. Structure, 4, 1161–1170. [DOI] [PubMed] [Google Scholar]
- Benach J., Atrian,S., Gonzàles-Duarte,R. and Ladenstein,R. (1998) The refined crystal structure of Drosophila lebanonensis alcohol dehydrogenase at 1.9 Å resolution. J. Mol. Biol., 282, 383–399. [DOI] [PubMed] [Google Scholar]
- Brünger A.T., Adams,P.D. and Rice,L.M. (1997) New applications of simulated annealing in X-ray crystallography and solution NMR. Structure, 5, 325–336. [DOI] [PubMed] [Google Scholar]
- Bruns C.M., Hubatsch,I., Ridderström,M., Mannervik,B. and Tainer,J.A. (1999) Human gluthathione transferase A4-4 crystal structures and mutagenesis reveal the basis of the high catalytic efficiency with toxic lipid peroxidation products. J. Mol. Biol., 288, 427–439. [DOI] [PubMed] [Google Scholar]
- CCP4 (1994) The CCP4 suite: programs for protein crystallography. Acta Crystallogr. D, 50, 760–763. [DOI] [PubMed] [Google Scholar]
- Cowtan K. (1994) An automated procedure for phase improvement by density modification. Joint CCP4 ESF-EACBM Newslett. Protein Crystallogr., 31, 34–38. [Google Scholar]
- Dauter Z., Li,M. and Wlodawer,A. (2001) Practical experience with the use of halides for phasing macromolecular structures: a powerful tool for structural genomics. Acta Crystallogr. D, 57, 239–249. [DOI] [PubMed] [Google Scholar]
- de Jong R.M., Rozeboom,H.J., Kalk,K.H., Tang,L., Janssen,D.B. and Dijkstra,B.W. (2002) Crystallization and preliminary X-ray analysis of an enantioselective halohydrin dehalogenase from Agrobacterium radiobacter AD1. Acta Crystallogr. D, 58, 176–178. [DOI] [PubMed] [Google Scholar]
- De la Fortelle E. and Bricogne,G. (1997) Maximum-likelyhood heavy atom refinement for multiple isomorphous replacement and multiwavelength anomalous dispersion methods. Methods Enzymol., 276, 472–494. [DOI] [PubMed] [Google Scholar]
- Filling C., Berndt,K.D., Benach,J., Knapp,S., Prozorovski,T., Nordling,E., Ladenstein,R., Jörnvall,H. and Oppermann,U. (2002) Critical residues for structure and catalysis in short-chain dehydrogenases/reductases. J. Biol. Chem., 277, 25677–25684. [DOI] [PubMed] [Google Scholar]
- Hahn H., Eder,E. and Deininger,C. (1991) Genotoxicity of 1,3-dichloro-2-propanol in the SOS chromotest and in the Ames test. Elucidation of the genotoxic mechanism. Chem. Biol. Interact., 80, 73–88. [DOI] [PubMed] [Google Scholar]
- Hisano T., Hata,Y., Fujii,T., Liu,J.Q., Kurihara,T., Esaki,N. and Soda,K. (1996) Crystal structure of L-2-haloacid dehalogenase from Pseudomonas sp. YL. An α/β hydrolase structure that is different from the alpha/beta hydrolase fold. J. Biol. Chem., 271, 20322–20330. [DOI] [PubMed] [Google Scholar]
- Janssen D.B., Oppentocht,J.E. and Poelarends,G.J. (2001) Microbial dehalogenation. Curr. Opin. Biotech., 12, 254–258. [DOI] [PubMed] [Google Scholar]
- Jesenská A., Sedláèek,I. and Damborský,J. (2000) Dehalogenation of haloalkanes by Mycobacterium tuberculosis H37Rv and other mycobacteria. Appl. Environ. Microbiol., 66, 219–222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jörnvall H., Persson,B., Krook,M., Atrian,S., Gonzalez Duarte,R., Jeffery,J. and Ghosh,D. (1995) Short-chain dehydrogenases/reductases (SDR). Biochemistry, 34, 6003–6013. [DOI] [PubMed] [Google Scholar]
- Jörnvall H., Höög,J.O. and Persson,B. (1999) SDR and MDR: completed genome sequences show these protein families to be large, of old origin, and of complex nature. FEBS Lett., 445, 261–264. [DOI] [PubMed] [Google Scholar]
- Kallberg Y., Oppermann,U., Jörnvall,H. and Persson,B. (2002a) Short-chain dehydrogenase/reductase (SDR) relationships: A large family with eight clusters common to human, animal and plant genomes. Protein Sci., 11, 636–641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kallberg Y., Oppermann,U., Jörnvall,H. and Persson,B. (2002b) Short-chain dehydrogenases/reductases (SDRs): coenzyme-based classification used in predictions on human and other completed genomes, Eur. J. Biochem., 269, 4409–4417. [DOI] [PubMed] [Google Scholar]
- Kleywegt G.J. and Jones,T.A. (1998) Databases in protein crystallography. Acta Crystallogr. D, 54, 1119–1131. [DOI] [PubMed] [Google Scholar]
- Kraulis P.J. (1991) MOLSCRIPT: a program to produce both detailed and schematic plots of protein structures. J. Appl. Crystallogr., 24, 946–950. [Google Scholar]
- Liao D.I., Thompson,J.E., Fahnestock,S., Valent,B. and Jordan,D.B. (2001) A structural account of substrate and inhibitor specificity differences between two naphthol reductases. Biochemistry, 40, 8696–8704. [DOI] [PubMed] [Google Scholar]
- Lutje Spelberg J.H., van Hylckama Vlieg,J.E.T., Bosma,T., Kellogg,R.M. and Janssen,D.B. (1999) A tandem enzyme reaction to produce optically active halohydrins, epoxides and diols. Tetrahedron: Asymmetry, 10, 2863–2870. [Google Scholar]
- Lutje Spelberg J.H., van Hylckama Vlieg,J.E.T., Tang,L., Janssen,D.B. and Kellogg,R.M. (2001) Highly enantioselective and regioselective biocatalytic azidolysis of aromatic epoxides. Org. Lett., 3, 41–43. [DOI] [PubMed] [Google Scholar]
- Lutje Spelberg J.H., Tang,L., van Gelder,M., Kellogg,R.M. and Janssen,D.B. (2002) Exploration of the biocatalytic potential of a halohydrin dehalogenase using chromogenic substrates. Tetrahedron: Asymmetry, 13, 1083–1089. [Google Scholar]
- McRee D.E. (1999) Xtalview/Xfit: a versatile program for manipulating atomic coordinates and electron density. J. Struct. Biol., 125, 156–165. [DOI] [PubMed] [Google Scholar]
- Merritt E.A. and Bacon,D.J. (1997) Raster3D photorealistic molecular graphics. Methods Enzymol., 277, 505–524. [DOI] [PubMed] [Google Scholar]
- Murzin A.G., Brenner,S.E., Hubbard,T. and Chothia,C. (1995) SCOP: a structural classification of proteins database for the investigation of sequences and structures. J. Mol. Biol., 247, 536–540. [DOI] [PubMed] [Google Scholar]
- Nakajima K., Yamashita,A., Akama,H., Nakatsu,T., Kato,H., Hashimoto,T., Oda,J. and Yamada,Y. (1998) Crystal structures of two tropinone reductases: different reaction stereospecificities in the same protein fold. Proc. Natl Acad. Sci. USA, 95, 4876–4881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura T., Nagasawa,T., Yu,F., Watanabe,I. and Yamada,H. (1994) A new enzymatic synthesis of (R)-γ-chloro-β-hydroxynitrile. Tetrahedron, 50, 11821–11826. [Google Scholar]
- Nardini M., Ridder,I.S., Rozeboom,H.J., Kalk,K.H., Rink,R., Janssen,D.B. and Dijkstra,B.W. (1999) The X-ray structure of epoxide hydrolase from Agrobacterium radiobacter AD1. An enzyme to detoxify harmful epoxides. J. Biol. Chem., 274, 14579–14586. [PubMed] [Google Scholar]
- Navaza J. and Saludjian,P. (1997) AMoRe: an automated molecular replacement program package. Methods Enzymol., 276, 581–594. [DOI] [PubMed] [Google Scholar]
- Norne J.E., Lilja,H., Lindman,B., Einarsson,R. and Zeppezauer,M. (1975) Pt(CN)2–4 and Au(CN)–2: potential general probes for anion-binding sites of proteins. 35Cl and 81Br nuclear-magnetic-resonance studies. Eur. J. Biochem., 59, 463–473. [DOI] [PubMed] [Google Scholar]
- O’Brien P.J. and Herschlag,D. (1999) Catalytic promiscuity and the evolution of new enzymatic activities. Chem. Biol., 6, R91–R105. [DOI] [PubMed] [Google Scholar]
- Ollis D.L. et al. (1992) The α/β-hydrolase fold. Protein Eng., 5, 197–211. [DOI] [PubMed] [Google Scholar]
- Oppermann U. et al. (2003) Short-chain dehydrogenases/reductases (SDR): the 2002 update. Chem. Biol. Interact., 143–144, 247–253. [DOI] [PubMed] [Google Scholar]
- Otwinowski Z. and Minor,W. (1997) Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol., 276, 307–326. [DOI] [PubMed] [Google Scholar]
- Persson B., Kallberg,Y., Oppermann,U., Jörnvall,H. (2003) Coenzyme-based functional assignments of short-chain dehydrogenases reductases (SDRs). Chem. Biol. Interact., 143–144, 271–278. [DOI] [PubMed] [Google Scholar]
- Poelarends G.J., van Hylckama Vlieg,J.E.T., Marchesi,J.R., Freitas Dos Santos,L.M. and Janssen,D.B. (1999) Degradation of 1,2-dibromoethane by Mycobacterium sp. strain GP1. J. Bacteriol., 181, 2050–2058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ridder I.S. and Dijkstra,B.W. (1999) Identification of the Mg2+-binding site in the P-type ATPase and phosphatase members of the HAD (haloacid dehalogenase) superfamily by structural similarity to the response regulator protein CheY. Biochem. J., 339, 223–226. [PMC free article] [PubMed] [Google Scholar]
- Ridder I.S., Rozeboom,H.J., Kalk,K.H., Janssen,D.B. and Dijkstra,B.W. (1997) Three-dimensional structure of L-2-haloacid dehalogenase from Xanthobacter autotrophicus GJ10 complexed with the substrate-analogue formate. J. Biol. Chem., 272, 33015–33022. [DOI] [PubMed] [Google Scholar]
- Rink R. and Janssen,D.B. (1998) Kinetic mechanism of the enantioselective conversion of styrene oxide by epoxide hydrolase from Agrobacterium radiobacter AD1. Biochemistry, 37, 18119–18127. [DOI] [PubMed] [Google Scholar]
- Rossmann M.G., Moras,D. and Olsen,K.W. (1974) Chemical and biological evolution of a nucleotide-binding protein. Nature, 250, 194–199. [DOI] [PubMed] [Google Scholar]
- Smilda T., Kamminga,A.H., Reinders,P., Baron,W., van Hylckama Vlieg,J.E.T. and Beintema,J.J. (2001) Enzymatic and structural studies on Drosophila alcohol dehydrogenase and other short-chain dehydrogenase/reductases. J. Mol. Evol., 52, 457–466. [DOI] [PubMed] [Google Scholar]
- Stammers D.K., Ren,J., Leslie,K., Nichols,C.E., Lamb,H.K., Cocklin,S., Dodds,A. and Hawkins,A.R. (2001) The structure of the negative transcriptional regulator NmrA reveals a structural superfamily which includes the short-chain dehydrogenase/reductases. EMBO J., 20, 6619–6626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka N., Takamasa,N., Tanabe,T., Tadashi,Y., Tsuru,D. and Mitsui,Y. (1996) Crystal structures of 7α-hydroxysteroid dehydrogenase from Escherichia coli. Biochemistry, 35, 7715–7730. [DOI] [PubMed] [Google Scholar]
- Tang L., Lutje Spelberg,J.H., Fraaije,M.W. and Janssen,D.B. (2003) Kinetic mechanism and enantioselectivity of halohydrin dehalogenase from Agrobacterium radiobacter. Biochemistry, 42, 5378–5386. [DOI] [PubMed] [Google Scholar]
- Torsvik V. and Øvreås,L. (2002) Microbial diversity and function in soil: from genes to ecosystems. Curr. Opin. Microbiol., 5, 240–245. [DOI] [PubMed] [Google Scholar]
- van Hylckama Vlieg J.E.T., Tang,L., Lutje Spelberg,J.H., Smilda,T., Poelarends,G.J., Bosma,T., van Merode,A.E.J., Fraaije,M.W. and Janssen,D.B. (2001) Halohydrin dehalogenases are structurally and mechanistically related to short-chain dehydrogenases/reductases. J. Bacteriol., 183, 5058–5066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verschueren K.H., Seljée,F., Rozeboom,H.J., Kalk,K.H. and Dijkstra,B.W. (1993a) Crystallographic analysis of the catalytic mechanism of haloalkane dehalogenase. Nature, 363, 693–698. [DOI] [PubMed] [Google Scholar]
- Verschueren K.H., Franken,S.M., Rozeboom,H.J., Kalk,K.H. and Dijkstra,B.W. (1993b) Non-covalent binding of the heavy atom compound (Au(CN)2)– at the halide binding site of haloalkane dehalogenase from Xanthobacter autotrophicus GJ10. FEBS Lett., 323, 267–270. [DOI] [PubMed] [Google Scholar]
- Weeks C.M. and Miller,R. (1999) The design and implementation of SnB v2.0. J. Appl. Crystallogr., 32, 120–124. [Google Scholar]
- Yamashita A., Kato,H., Wakatsuki,S., Tomizaki,T., Nakatsu,T., Nakajima,K., Hashimoto,T., Yamada,Y. and Oda,J. (1999) Structure of tropinone reductase-II complexed with NADP+ and pseudotropine at 1.9 Å resolution: implication for stereospecific substrate binding and catalysis. Biochemistry, 38, 7630–7637. [DOI] [PubMed] [Google Scholar]
- Yeates T.O. (1997) Detecting and overcoming crystal twinning. Methods Enzymol., 276, 344–358. [PubMed] [Google Scholar]
- Yu F., Nakamura,T., Mizunashi,W. and Watanabe,I. (1994) Cloning of two halohydrin hydrogen-halide lyase genes from Corynebacterium sp. strain N-1074 and structural comparison of the genes and gene products. Biosci. Biotechnol. Biochem., 58, 1451–1457. [DOI] [PubMed] [Google Scholar]