Abstract
The yeast Zap1 transcription factor controls the expression of genes involved in zinc accumulation and storage. Zap1 is active in zinc-limited cells and repressed in replete cells. Zap1 has two activation domains, AD1 and AD2, which are both regulated by zinc. AD2 function was mapped to a region containing two Cys2His2 zinc fingers, ZF1 and ZF2, that are not involved in DNA binding. More detailed mapping placed AD2 almost precisely within the endpoints of ZF2, suggesting a role for these fingers in regulating activation domain function. Consistent with this hypothesis, ZF1 and ZF2 bound zinc in vitro but less stably than did zinc fingers involved in DNA binding. Furthermore, mutations predicted to disrupt zinc binding to ZF1 and/or ZF2 rendered AD2 constitutively active. Our results also indicate that the repressed form of AD2 requires an intramolecular interaction between ZF1 and ZF2. These studies suggest that these zinc fingers play an unprecedented role as zinc sensors to control activation domain function.
Keywords: gene expression/homeostasis/regulation/transcriptional activation/zinc fingers
Introduction
The Cys2His2 (C2H2) zinc finger motif is ubiquitous in biology. This domain was first characterized in transcription factor IIIA (TFIIIA) by Klug and colleagues (Miller et al., 1985). Since then, hundreds of proteins containing these motifs have been identified. In the human genome alone, 3% of the ∼32 000 predicted open reading frames encode proteins with zinc fingers (Landers, 2001). Detailed characterization of a relatively small subset of these proteins has implicated zinc fingers in several functions. The most commonly recognized role of zinc fingers is in protein–DNA binding (Rhodes and Klug, 1993). Zinc fingers and related zinc-binding motifs have also been shown to act in the binding of proteins to RNA (Finerty and Bass, 1999), lipids (Gaullier et al., 1998) and other proteins (Mackay and Crossley, 1998). In this report, we present evidence for a novel role of these motifs as zinc sensors involved in regulating the activation domain of a transcription factor.
Zinc is an essential nutrient but can be toxic to cells if accumulated in excess amounts. To survive, cells have mechanisms to maintain intracellular zinc homeostasis. The precision of zinc homeostasis was recently highlighted by studies of Escherichia coli, where intracellular zinc levels are controlled by the transcriptional regulation of both uptake and efflux transporters (Patzer and Hantke, 1998; Brocklehurst et al., 1999). Recent studies of the transcription factors responsible for this regulation suggest that these cells strive to maintain little or no free cytoplasmic zinc (Outten and O’Halloran, 2001). Several reports suggest that eukaryotic cells also maintain very low levels of cytoplasmic labile zinc (Sensi et al., 1997; Cheng and Reynolds, 1998).
We know much about zinc homeostasis in eukaryotes through studies of the yeast Saccharomyces cerevisiae. In this yeast, zinc homeostasis is largely mediated by the regulation of uptake transporters and transporters involved in the intracellular storage of zinc in the vacuole. The Zrt1, Zrt2 and Fet4 proteins are metal ion transporters responsible for zinc uptake across the plasma membrane (Zhao and Eide, 1996a,b; Waters and Eide, 2002). Vacuolar zinc storage is controlled by the Zrc1 and Zrt3 transporters (MacDiarmid et al., 2000; Miyabe et al., 2001). All of the genes encoding these transporters are regulated at the transcriptional level and are induced in zinc-limited cells. This zinc-responsive gene regulation is mediated by the Zap1 transcriptional activator (Zhao and Eide, 1997). Zap1 plays a central role in zinc homeostasis by controlling the expression of these genes and ∼40 others in the yeast genome (Lyons et al., 2000).
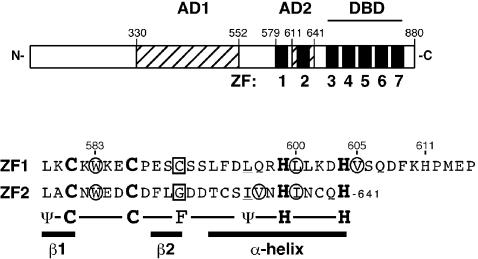
Zap1 is an 880 amino acid protein with seven C2H2 motifs. At its C-terminus is a DNA binding domain consisting of five C2H2 zinc fingers (designated ZF3–ZF7) (Figure 1) (Bird et al., 2000a; Evans-Galea et al., 2003). This domain binds specifically to a DNA element, the 11 bp zinc-responsive element or ZRE, found in one or more copies in the promoters of Zap1’s target genes (Zhao et al., 1998; Lyons et al., 2000). All five of the zinc fingers in the DNA binding domain are required for ZRE interaction. Zap1 also contains two activation domains that are rich in acidic residues (Bird et al., 2000b). One activation domain, called AD1, was mapped between amino acids 330 and 552. The second activation domain, AD2, was mapped between 552 and 705. Two additional zinc fingers, ZF1 and ZF2, are found within this latter region. ZF1 and ZF2 are not required for DNA binding, highlighting a possible role as zinc sensors.
Fig. 1. A depiction of the Zap1 protein. The positions of the seven Zap1 zinc fingers are shown with filled boxes and are numbered. The DNA binding domain (DBD) requires fingers 3–7. Zap1’s two activation domains (AD1 and AD2) are shown with hatched boxes; the location of AD2 reflects the detailed mapping data from Figure 2. The lower panel shows the sequence of Zap1 fingers 1 and 2 (residues 579–641). The positions of the β-strands and α-helices are indicated. Residues conserved in other zinc fingers are shown below the Zap1 sequence; ψ, hydrophobic. The C and G ‘finger core’ residues (C590 and G627) are boxed, the α-helical residues that also contribute to the hydrophobic core are underlined, and the residues proposed to make interfinger contacts are circled.
Recent results have indicated that Zap1 is regulated by zinc via four different mechanisms (Zhao and Eide, 1997; A.Bird, E.Blankman, D.R.Winge and D.J.Eide, in preparation). First, Zap1 controls its own expression through transcriptional autoregulation. Secondly, zinc controls Zap1 DNA binding activity. Overexpressing Zap1 overrides DNA binding control and results in constitutive ZRE occupancy. Under these conditions, we also found that zinc independently controls the activities of AD1 and AD2. In this report, we provide a molecular model for the zinc regulation of AD2. Our results demonstrate that ZF1 and ZF2 are critical for zinc regulation of AD2 and suggest a role for these fingers in zinc sensing and consequent regulation of Zap1 activity.
Results
A previous study mapped AD2 to the region of Zap1 between residues 552 and 705 (Bird et al., 2000b). Located within the 552–705 region are two C2H2-type zinc finger domains designated ZF1 and ZF2 (Figure 1) that are not involved in DNA binding (Bird et al., 2000a). These two domains have most of the conserved amino acids found in other zinc fingers. The consensus sequence for these domains is Ψ/Y-X-C-X2,4-C-X3-F-X5-Ψ-X2-H-X3-5-H, where Ψ denotes a hydrophobic amino acid (Berg and Godwin, 1997). ZF1 and ZF2 match this consensus, with the exception of C (C590) and G (G627) residues located in the position most commonly occupied by F at the end of the β2 strand of fingers 1 and 2, respectively. In most zinc fingers, this residue contributes to a hydrophobic core formed by the fold between the β2 strand and the α-helix. The other residue contributing to this hydrophobic core is the conserved hydrophobic residue in the α-helix; this position is conserved in ZF1 and ZF2 (L596 and I633, respectively).
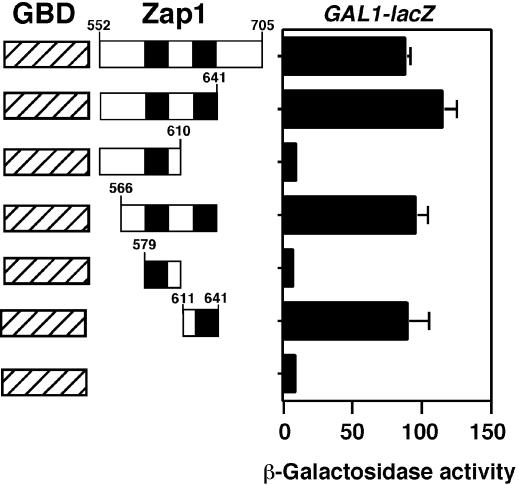
The association of ZF1 and ZF2 with the zinc-responsive AD2 activation domain suggested a role for these fingers in zinc sensing. Also consistent with this hypothesis, we found that AD2 function mapped to ZF2. This detailed mapping was performed using fusions of various portions of the Zap1 552–705 region to the Gal4 DNA binding domain (GBD). Expression of a GAL1-lacZ reporter in a gal4Δ mutant strain was then used to assess activation domain function of these fusions (Figure 2). The high level of activation domain function seen with the 552–705 fragment mapped completely to the subregion of amino acids 611–641, i.e. almost the precise endpoints of ZF2 (Figure 1). No activation domain function was detected in ZF1 or elsewhere in the 552–705 region.
Fig. 2. Mapping AD2 within the 552–705 region. The indicated regions of Zap1 were fused to the GBD and expressed in gal4Δ cells (ABY29) co-transformed with the GAL1-lacZ reporter. Cells were grown to exponential phase in low zinc conditions (LZM + 3 µM ZnCl2) prior to β-galactosidase activity assays. A representative experiment is shown and each value is the mean of three replicates. Error bars represent 1 SD.
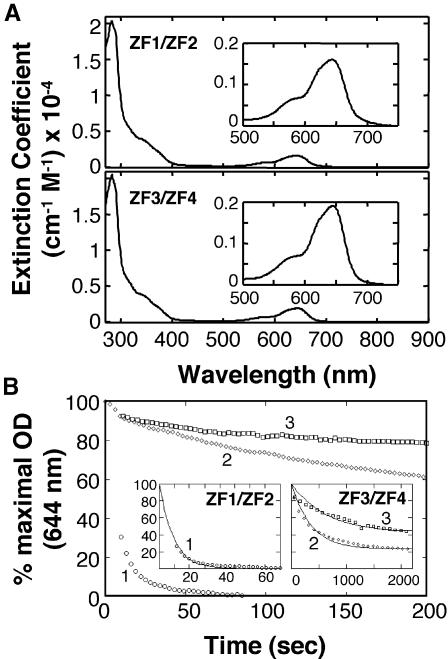
To test whether ZF1 and ZF2 bind zinc, we determined the zinc stoichiometry, affinity and stability of zinc binding in a Zap1 fragment (amino acids 575–643) containing these fingers. For comparison, we also examined Zn2+ binding to a fragment (residues 700–766) containing ZF3 and ZF4 from the Zap1 DNA binding domain. ZF3 and ZF4, both required for binding of Zap1 to DNA in zinc-limited cells, are likely to be representative of high affinity zinc sites in other zinc-dependent proteins (e.g. TFIIIA). Following their purification from E.coli, both ZF1/ZF2 and ZF3/ZF4 fragments were found to have zinc bound with a stoichiometry of ∼2 mol eq of Zn2+ [2.3 ± 0.6 and 2.1 ± 0.6 (n = 4) for ZF1/ZF2 and ZF3/ZF4, respectively]. These results suggested that the metal was bound by both fingers in each polypeptide fragment.
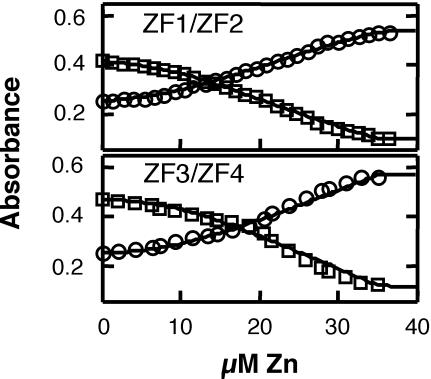
To determine the relative affinity of ZF1/ZF2 verses ZF3/ZF4 peptides for Zn2+, we used a competition assay with the fluorescent indicator Fura-2 (VanZile et al., 2000) Fura-2 binds Zn2+ in a 1:1 complex with a dissociation constant of 3 nM (Atar et al., 1995). Upon Zn binding to Fura-2, an absorbance shift occurs in the maxima from ∼369 to ∼339 nm, with the difference spectrum showing maximal loss of absorbance at ∼381 nm and maximal increase of absorbance at ∼332 nm (data not shown). Figure 3 shows the results of representative titration of ZnCl2 into a solution of 15 µM Fura-2, 10 µM apo-protein, 100 mM Tris–Cl, pH 7.5, with the fits calculated by the program DYNAFIT (Kuzmic, 1996). The best fits for both ZF1/ZF2 and ZF3/ZF4 are consistent with each peptide containing two Zn-binding sites of differing affinity. The apparent KD values for ZF1/ZF2 sites are 5.3 ± 2.2 and 0.3 ± 0.1 nM. The apparent KD values for ZF3/ZF4 sites are 3.0 ± 1.7 and 0.2 ± 0.0 nM.
Fig. 3. Determining the affinity of Zn2+ binding by Zap1 zinc fingers. The ability of ZF1/ZF2 (top panel) or ZF3/ZF4 (bottom panel) peptides to compete with the indicator Fura-2 for Zn2+ was tracked by following the loss of absorbance at 381 nm (squares indicate decreasing apo-Fura-2 concentration) and the increase of absorbance at 332 nm (circles indicate increasing Zn-Fura-2 concentration). The solution contained 15 µM Fura-2, 10 µM apo-protein, 100 µM Tris–Cl, pH 7.5. The Zn2+ titrations were performed by adding 4 µl aliquots of ZnCl2 by Hamilton syringe through an oxygen-free sealed cuvette septum. After mixing, the absorbance spectrum was scanned from 240 to 560 nm before the next titration. The final absorbance values and Zn2+ concentrations were corrected for dilution. The data was fit by the program DYNAFIT with all parameters assigned except the dissociation constants of the zinc finger pairs. A representative of three independent experiments is shown in each panel.
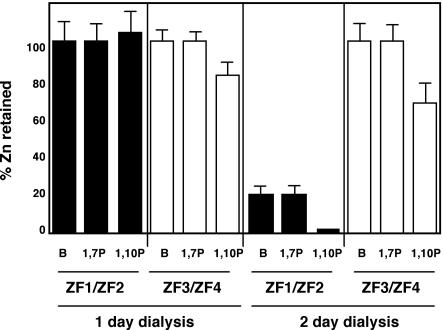
Because the finger pairs exhibited similar affinities for Zn2+, we then assessed whether the finger pairs differed in relative stabilities of zinc binding. First, these Zn2+–zinc finger complexes were extensively dialyzed against buffer or buffer plus a zinc chelator, 1,10-phenanthroline [stability constants of 1012.2/M and 1017.1/M for the Zn(phen)2 and Zn(phen)3 complexes, respectively] (NIST Database 46: Critical Stability Constants; http://www.nist.gov/srd/nist46.htm) or a related compound, 1,7-phenanthroline, which does not bind zinc. The amount of zinc retained by these peptides after dialysis was then determined (Figure 4). After 1 day of dialysis, zinc was largely retained by both ZF1/ZF2 and ZF3/ZF4 peptides. After dialysis for 2 days, little if any zinc was removed from the ZF3/ZF4 peptide during dialysis in buffer or buffer plus 1,7-phenanthroline. The chelator removed only ∼40% of the zinc from the ZF3/ZF4 fragment under the same conditions. In contrast, dialysis of ZF1/ZF2 in buffer alone or 1,7-phenanthroline removed 80% of the bound zinc. Dialysis of ZF1/ZF2 in 1,10-phenanthroline removed almost all of the zinc. The resulting Zn-depleted ZF1/ZF2 peptide was poorly soluble. Similar results were obtained by dialysis of the ZF1/ZF2 and ZF3/ZF4 fragments against another zinc chelator, 4-(2)-(pyridylazo)resorcinol (PAR, stability constant = 1017.1/M). Dissociation of Zn2+ from ZF1/ZF2 in the presence of PAR had a t1/2 of 1.4 days while zinc loss from ZF3/ZF4 was much slower (t1/2 >14 days) (data not shown). These data indicate that ZF1 and ZF2 do bind zinc but less stably than ZF3 and ZF4.
Fig. 4. Lability of zinc binding by ZF1/ZF2 and ZF3/ZF4 peptides. Peptides containing the indicated fingers and with 2 mol eq Zn2+ bound initially were extensively dialyzed against buffer alone (B) or buffer containing the indicated compound (1,7P, 1, 7-phenanthroline; 1,10P, 1,10-phenanthroline) (10 µM). After dialysis for 1 or 2 days, the zinc and protein content was determined. One hundred percent is defined as zinc content of the sample prior to dialysis. The averages of two sample experiments are shown and the error bars represent ±1 SD.
To further explore the stability of metal binding by Zap1 zinc finger domains, Co2+ was titrated into apo-peptides of ZF1/ZF2 and ZF3/ZF4. The energies of the d–d transitions in the visible range were consistent with the predicted C2H2 coordination (Figure 5A). Displacement of this bound Co2+ by Zn2+ can then be used to assess the kinetic stability of metal binding (Buchsbaum and Berg, 2000). Therefore, these Co2+–peptide complexes were incubated with 2 mol eq Zn2+ and the kinetics of the Zn2+-displacement of the Co2+ d–d transitions were monitored at 644 nm. As can be seen in Figure 5B (curve 1), the Co2+ ions in ZF1/ZF2 were rapidly displaced by added Zn2+ with significant displacement occurring during the mixing time prior to the first measurement (0–10 s). The rate of Co2+ displacement from ZF1/ZF2 fit well to a single exponential with a t1/2 of 5.9 s (Figure 5B, inset). In contrast, the Zn2+-induced displacement of Co2+ in the ZF3/ZF4 peptide was much slower (Figure 5B, curve 3). The t1/2 determined from the fit to a single exponential was ∼441 s under the conditions where [Co2+]tot = [Zn2+]tot. Even when the displacement of Co2+ by Zn2+ was driven by a 10-fold higher concentration of Zn2+ than Co2+ (curve 2), the t1/2 for ZF3/ZF4 sample was ∼253 s, i.e. significantly longer than the equilibration t1/2 of ZF1/ZF2 (Figure 5B, inset). Attempts to exchange Co2+ into Zn2+–ZF1/ZF2 or Zn2+–ZF3/ZF4 complexes were not successful, confirming that Zn2+ binds more avidly to the Zap1 fingers than Co2+.
Fig. 5. Co2+ and Zn2+ titration of Zap1 ZF1/ZF2 and ZF3/ZF4 peptides. (A) Spectra of Co2+-ZF1/ZF2 and Co2+-ZF3/ZF4 were measured with samples containing 100 µM Co2+ and 50 µM of the indicated peptide. Addition of Co2+ to either metal-free ZF1/ZF2 or ZF3/ZF4 peptides resulted in the formation of a peak with a maximum at 644 nm and a shoulder at 580 nm. Addition of higher concentrations of Co2+ did not change these spectra (data not shown). (B) After addition of Zn2+ to a solution containing the indicated Co2+–protein complex, the loss of absorbance at 644 nm was monitored over time. The final concentrations were 100 µM Co2+, 100 µM Zn2+, 50 µM ZF1/ZF2 peptide (curve 1), 100 µM Co2+, 100 µM Zn2+, 50 µM ZF3/ZF4 peptide (curve 3), or 100 µM Co2+, 1000 µM Zn2+, 50 µM ZF3/ZF4 peptide (curve 2). The data were fit to single exponential curves (insets), giving exchange t1/2 values of ∼5.9 s for ZF1/ZF2 (curve 1), ∼441 s for ZF3/ZF4 (curve 3) or ∼253 s for ZF3/ZF4, where the exchange is driven by extremely high [Zn2+] (curve 2).
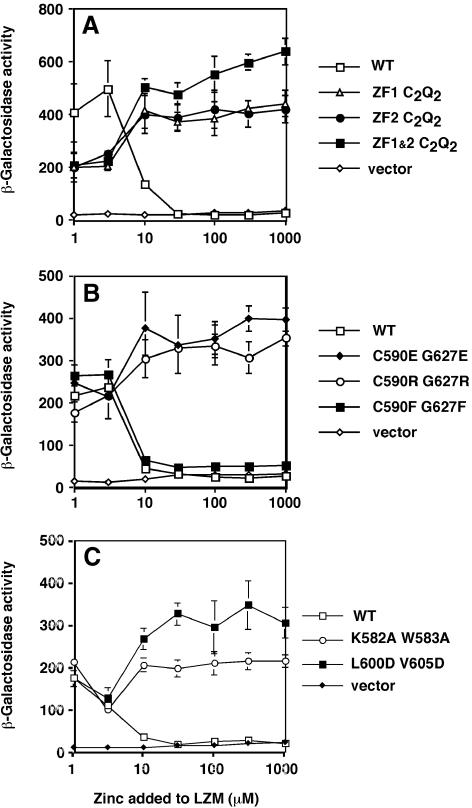
Their greater lability of Zn2+ binding is consistent with ZF1 and ZF2 acting as zinc sensors in regulating AD2 function. If this hypothesis is correct, mutations predicted to block zinc binding by ZF1 and/or ZF2 would also impair zinc-responsive gene regulation. GBD–Zap1552–705 fusions provided a useful assay to determine the effects of ZF1 and ZF2 mutations on zinc regulation of AD2 function. Previous studies had suggested that repression of AD2 by zinc required the presence of the Zap1 DNA binding domain (Bird et al., 2000b). Upon re-examination, we found that AD2 is regulated by zinc independently of other domains of Zap1 (see Discussion). As shown in Figure 6A, the wild-type GBD–Zap1552–705 fusion was active in zinc-limited cells and completely repressed in low zinc medium (LZM) medium supplemented with 30 µM ZnCl2. To test the role of ZF1 and ZF2 in this regulation, we first introduced mutations in which the two histidyl ligands were substituted with glutamines (i.e. C2H2→C2Q2). Such mutations, which disrupt zinc binding in other zinc fingers (Bird et al., 2000a), were generated in either ZF1, ZF2 or both. Cells expressing these C2Q2 mutant GBD–Zap1552–705 fusion proteins displayed strong activation domain function in low zinc (Figure 6A). However, in contrast to wild type, the activity of the mutant proteins was not repressed by zinc. These data suggest that zinc binding by both ZF1 and ZF2 is required for repression of AD2 function. The 2-fold increase in activity observed with zinc repletion of strains expressing the mutant fusions is similar to that seen with promoters not regulated by zinc (e.g. HIS4, CYC1) (Zhao and Eide, 1996a) and probably reflects a general decrease in expression in zinc-deficient cells that is alleviated as zinc levels rise to repletion.
Fig. 6. Strain ABY29 (gal4Δ) was transformed with the GAL1-lacZ reporter and plasmids expressing the indicated ZAP1 mutations in the GBD–Zap1552–705 fusion protein or the vector-only control. These cells were grown to exponential phase in LZM medium plus the indicated concentration of ZnCl2. Representative experiments are shown and each value is the mean of three replicates. Error bars represent ±1 SD.
We also sought to disrupt zinc binding in ZF1 and ZF2 by generating mutants in which the C590 and G627 finger core residues (Figure 1) were substituted with other amino acids. Studies of other zinc finger peptides have shown that mutations altering this residue greatly increase the flexibility of the domain and lower the stability of Zn2+ binding (Berg and Godwin, 1997). ZF1 and ZF2 mutations were constructed in which large, charged residues (i.e. C,G→E,E; C,G→R,R) were substituted into the finger core position. These changes are likely to disrupt zinc binding by destabilizing the protein fold. When assayed for zinc responsiveness in vivo, these mutations also caused constitutive AD2 function (Figure 6B).
Because the lability of zinc binding by wild type ZF1 and ZF2 could be due to the non-canonical C590 and G627 finger core residues, we also substituted these residues with phenylalanines, the residue most commonly found in this position in other zinc finger proteins. If the function of the finger core C,G residues was to reduce the zinc binding stability of ZF1 and ZF2, we predicted that the C,G→F,F substitutions would be repressed by even lower concentrations of Zn2+ than the wild-type fragment. In vivo, the ZF1 and ZF2 C,G→F,F mutant was regulated similar to wild type (Figure 6B). These results indicate that the non-canonical C and G finger core residues of ZF1 and ZF2 do not solely determine a regulatory set-point of these fingers. Immunoblotting indicated that the wild-type fusion and the C,G→F,F mutant proteins accumulated to similarly high levels (data not shown). Surprisingly, the mutants predicted to be defective for Zn2+ binding were destabilized in vivo such that steady-state protein levels were lower. Therefore, the failure of these mutant proteins to be regulated by zinc is not due to their overexpression relative to the wild-type fusion.
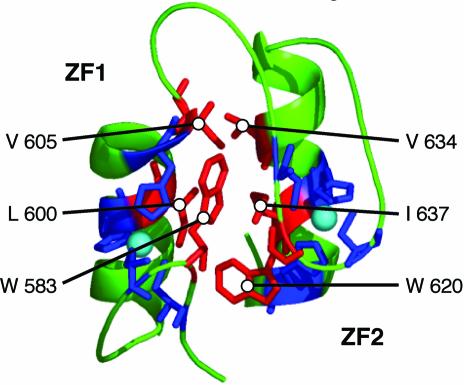
The results shown in Figure 6A indicate that while AD2 function mapped to ZF2, both ZF1 and ZF2 are required to repress AD2. One explanation for this requirement is that an intramolecular interaction occurs between these two fingers to form a conformation that represses AD2 function. That such a finger–finger interaction occurs was suggested by the amino acid sequence of these domains. Zap1 fingers 1 and 2 resemble the first two of the five zinc fingers in the Gli protein (Pavletich and Pabo, 1993). These two Gli fingers make intramolecular protein–protein contacts with each other. The packing interface between these fingers consists of two W residues in the β-hairpin loops between the cysteinyl ligands and hydrophobic packing between the two α-helices. The β-hairpin loops of Zap1 ZF1 and ZF2 contain similarly positioned W residues (W583 and W620) and the nonpolar residues contributing to interhelical hydrophobic packing in Gli are also conserved in Zap1 ZF1 and ZF2 (L600, V605, V634, I637) (Figure 1). To test the hypothesis that ZF1 and ZF2 interact to mask AD2, we constructed mutant alleles of ZF1 predicted to disrupt this proposed interaction. First, W583 was mutated to alanine. The adjacent K582 was also mutated to A in this allele (K582A, W583A) because it could potentially form an interaction-stabilizing salt bridge with E621 of ZF2. In a second mutant, the two hydrophobic residues in the α-helix of ZF1, L600 and V605, were mutated to aspartates (L600D, V605D). The effects of these mutations on AD2 regulation were determined using GBD–Zap1552–705 fusions. As predicted if zinc-responsive repression of AD2 required an interaction between ZF1 and ZF2, these mutations totally disrupted zinc regulation of AD2 activity (Figure 6C). Similar zinc non-responsiveness was observed when the W and adjacent E residues in ZF2 predicted to participate in the interaction were mutated (W620A, E621K) (data not shown).
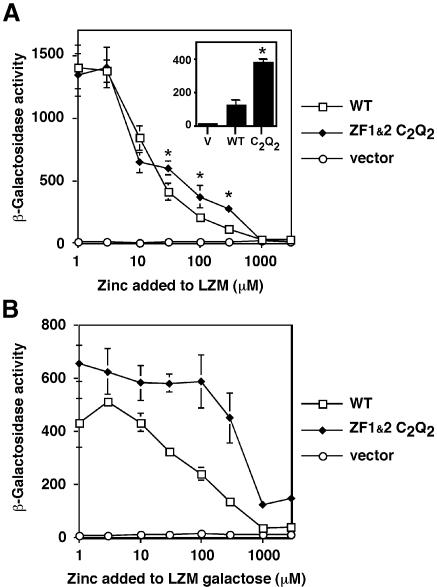
The analysis of AD2 regulation presented thus far has considered the behavior of AD2 in the non-native context of GBD–Zap1 fusions. If ZF1 and ZF2 are indeed involved in zinc regulation of Zap1, mutations affecting these domains should have some effect on the zinc-responsiveness of the full-length Zap1 protein. We introduced the ZF1/ZF2 C2Q2 mutations into full-length Zap1 expressed from the GAL1 promoter. The GAL1 promoter allows assessment of Zap1 function at low levels of expression (i.e. in glucose-grown cells) where the independent control of DNA binding by zinc occurs, and at high Zap1 levels (i.e. in galactose-grown cells) where ZRE occupancy is constitutive (A.Bird, E.Blankman, M.Evans-Galea, D.R.Winge and D.J.Eide, in preparation). These experiments were performed in a zap1Δ strain to allow analysis of the activity of the plasmid-encoded allele on a ZRE-lacZ reporter. Figure 7A shows Zap1 activity when DNA binding control is zinc-responsive. In the context of full-length Zap1, the ZF1/ZF2 C2Q2 mutant showed a reproducible defect in zinc repression at media zinc concentrations from 30 to 300 µM. The scale in Figure 7A tends to obscure the magnitude of this effect. As shown in Figure 7A (inset), expression at these concentrations (e.g. LZM + 300 µM ZnCl2) can be almost 3-fold higher in the mutant than the wild type. In the absence of DNA binding control, i.e. when ZRE occupancy is rendered constitutive by Zap1 overexpression, an even more striking result was obtained (Figure 7B). Little or no repression of the mutant Zap1 activity by zinc was observed up to 300 µM zinc, while the wild type showed repression with as little as 30 µM. These results demonstrate that ZF1 and ZF2 are important for the zinc responsiveness of Zap1 when the protein is bound to DNA. The residual regulation observed for the constitutively bound mutant protein may be largely due to regulation of AD1, independent of AD2 (Bird et al., 2000b).
Fig. 7. Effects of ZF1/ZF2 mutations on the zinc responsiveness of full-length Zap1. ZHY6 (zap1Δ) cells transformed with the ZRE-lacZ (pDg2) reporter and pYef2 (vector), full-length wild-type Zap1 (pMyc-Zap1), or full-length Zap1 with the ZF1/ZF2 C2Q2 mutations (pZF1/21–880) were grown in LZM medium plus the indicated concentration of ZnCl2. (A) Low-level expression of Zap1 from the GAL1 promoter in glucose-grown cells. DNA binding control occurs normally under these conditions. The inset shows the data for these strains grown in LZM + 300 µM ZnCl2. The asterisks indicate a significant difference from wild type (P < 0.05) as estimated by ANOVA. (B) High-level expression of Zap1 from the GAL1 promoter in galactose-grown cells. ZRE occupancy is constitutive under these conditions. A representative experiment of three separate experiments is shown and each value is the mean of three replicates. Error bars represent ±1 SD.
Finally, we examined the effects of ZF1/ZF2 C2Q2 mutations on zinc-responsiveness when Zap1 was expressed from its own promoter. A strain was engineered in which the ZF1/ZF2 C2Q2 mutations were introduced into the chromosomal ZAP1 gene. In this context, Zap1 expression is subject to transcriptional autoregulation, DNA binding control, and control of AD1 and AD2. Zinc-responsive gene expression was assayed by measuring mRNA levels generated by the chromosomal ZRT1 gene by S1 nuclease protection assay. In wild-type cells, ZRT1 expression was greatly repressed by zinc at concentrations of 30 µM and higher (Figure 8A). ZRT1 mRNA levels were quantified along with CMD1 (calmodulin) as a loading control. The ratio of ZRT1:CMD1 mRNA is plotted in Figure 8B. In contrast, zinc regulation of the chromosomal ZAP1ZF1&2 C2Q2 mutant was greatly impaired. These results demonstrate the importance of ZF1 and ZF2 in regulating Zap1 activity on a native target promoter.
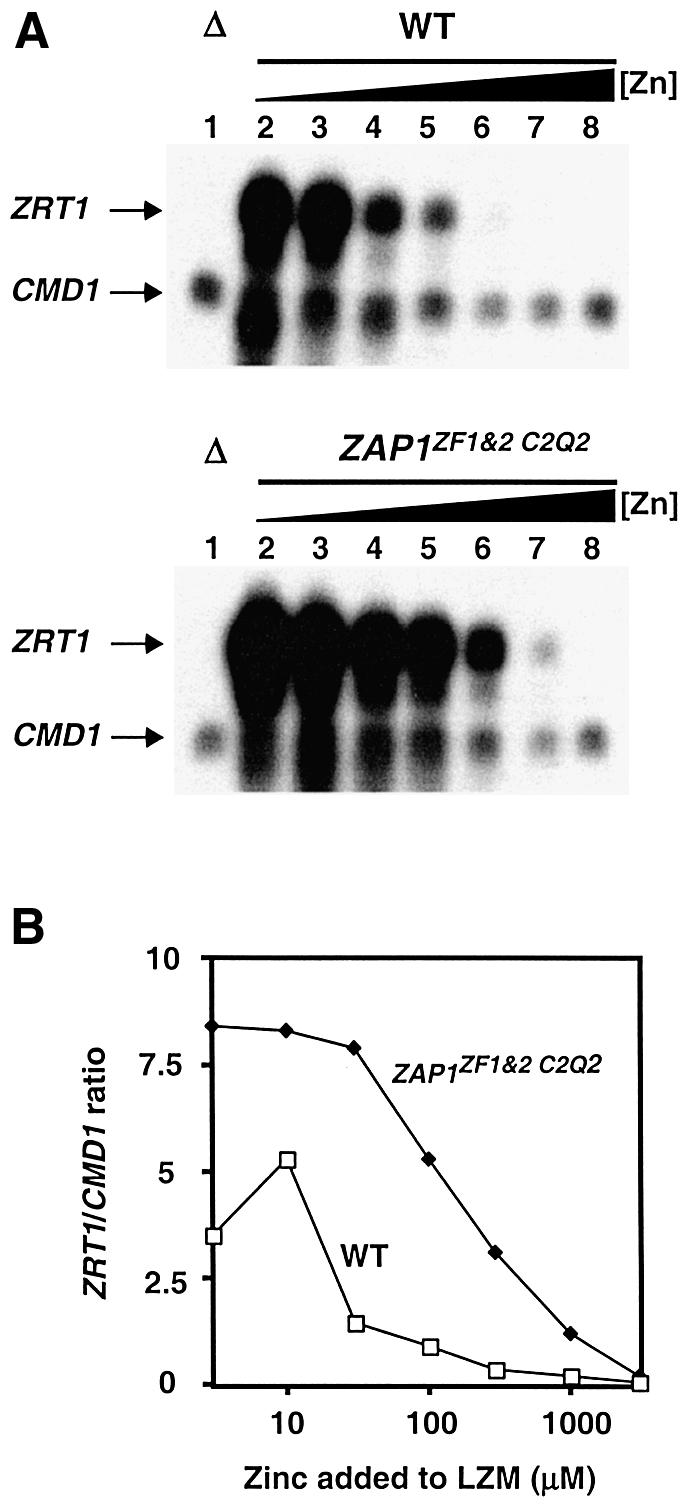
Fig. 8. Effects of ZF1/ZF2 mutations in the chromosomal ZAP1 gene on the regulation of gene expression. (A) Total RNA was extracted from exponential-phase cultures of the zap1 mutant strain ZHY6 (Δ) grown in LZM media supplemented with 3000 µM Zn2+ (lane 1) and from the wild-type strain, DY1457 and a chromosomal ZAP1ZF1&2 C2Q2 mutant, grown in LZM media supplemented with 3, 10, 30, 100, 300, 1000 and 3000 µM Zn2+ (lanes 2–8, respectively). The levels of ZRT1 mRNA were compared to the loading control CMD1 mRNA using S1 nuclease protection assays. Arrows indicate ZRT1 and CMD1 S1 nuclease protection products. (B) The band intensities in (A) were quantified and are plotted as the ratio of ZRT1:CMD1 mRNA levels at each zinc concentration. A representative of three independent experiments is shown.
Discussion
Our results indicate that Zap1 zinc fingers 1 and 2 serve an unprecedented regulatory role in controlling an activation domain. The hypothesis that ZF1 and ZF2 form canonical zinc fingers is supported by the close match between their sequences and the zinc finger consensus sequence. Studies by Berg and colleagues have established that the identity and position of the conserved residues in the consensus sequence (Figure 1) are all that is required to form the zinc finger structure (Michael et al., 1992). Moreover, the 2:1 zinc:peptide stoichiometry and characteristic d–d transition energies of the Co2+-bound ZF1/ZF2 fragment indicates that metal binding does occur in these fingers.
Support for the role of ZF1 and ZF2 in regulating Zap1 activity came first from our earlier characterization of the Zap1 DNA binding domain. ZF1 and ZF2 are not required for DNA binding (Bird et al., 2000a,b). In contrast, ZF3–ZF7 are each essential for high affinity ZRE interactions. We have shown that ZF1 and ZF2 are closely associated with AD2. In fact, the entire activation domain function of AD2 mapped within the approximate endpoints of ZF2. Thus, ZF2 and the nearby ZF1 are well positioned to exert an effect on AD2 function. Our results also show that several mutations in ZF1 and ZF2 predicted to disrupt Zn2+ binding (e.g. C2H2→C2Q2) render AD2 function constitutive.
Metal binding studies of Zap1 zinc fingers also support a regulatory role of ZF1 and ZF2. One would predict that regulatory zinc sites would have a lower stability and/or affinity for zinc binding than would structural sites. Lower stability of binding would allow Zap1 to respond quickly to changes in zinc status by binding or releasing zinc. A lower affinity for zinc would allow repression of activation domain function when intracellular zinc levels rise high enough to saturate structural zinc sites that have higher affinity for the metal. We found that the stability of ZF1 and ZF2 zinc binding was much lower than that of ZF3 and ZF4, i.e. likely representatives of structural zinc sites. Displacement of Co2+ by Zn2+ in a model zinc finger peptide was previously shown to likely occur by a dissociative mechanism where the rate-limiting release of Co2+ occurs prior to Zn2+ binding (Buchsbaum and Berg, 2000). If also true for the Zap1 zinc fingers, these data confirm that lability of metal binding is much greater in the ZF1/ZF2 peptide than for ZF3/ZF4.
The higher lability of Zn2+ binding by a regulatory domain of Zap1 is consistent with recent studies of zinc regulation in E.coli. O’Halloran and colleagues demonstrated that the Zn-responsive transcription factors Zur and ZntR of E.coli have very high affinities for Zn2+ in vitro, suggesting that zinc regulation in bacteria is under kinetic control (Outten and O’Halloran, 2001). The increased lability of ZF1 and ZF2 in Zn2+ binding likely makes them responsive to changes in the intracellular zinc level. The low nanomolar affinities of ZF1 and ZF2 suggest that AD2 responds when free zinc in the cell is in that concentration range. Zap1 is localized to the nucleus and a concentration of 5 nM, the apparent KD for the lower affinity zinc finger in AD2, corresponds to ∼10 atoms of free nuclear Zn2+ per cell. However, given the similar zinc binding affinities of the Zap1 ZF1/ZF2 and ZF3/ZF4 peptides compared with the large differences in stability of zinc binding, our data strongly suggest that the kinetics of zinc binding dominates regulation of AD2 activity.
A previous study suggested that zinc repression of AD2 required the presence of the Zap1 DNA binding domain (Bird et al., 2000b). We now know that AD2 is autonomously regulated by zinc. The explanation for this apparent paradox is in the genotype of the strains used to assess Zap1 activity. The previous study examined regulation of the GBD–Zap1552–705 fusion in a gal4Δ zap1Δ mutant strain that, because of the zap1Δ mutation, has a reduced ability to accumulate zinc. The zinc-replete growth condition (LZM + 1000 µM ZnCl2) chosen for these experiments was a concentration of zinc that restored wild-type growth rates. However, we now recognize that these mutant cells have decreased zinc accumulation even under these zinc-replete conditions. When assayed in ZAP1 (gal4Δ) cells where zinc homeostasis is normal, the GBD–Zap1552–705 fusion was strongly zinc regulated (Figure 6). This regulation of GBD–Zap1552–705 did not depend on the presence of chromosomal Zap1 per se, but rather on acquiring sufficient zinc to repress AD2 function. This was evident because the GBD–Zap1552–705 fusion was also repressed by zinc in a zap1Δ mutant where zinc accumulation was aided by expressing the Zrt1 zinc uptake transporter from the strong PGK1 promoter (data not shown).
How might zinc binding in ZF1 and ZF2 inhibit AD2 function? The simplest model is that zinc binding induces a conformation in AD2 that is no longer capable of transcriptional activation. This model is also supported by structural studies of activation domains in other proteins. Activation domains rich in acidic residues have little organized structure in solution but are more ordered when interacting with target factors (e.g. TFIID) (Uesugi et al., 1997; Lee et al., 2000). Similarly, in the absence of metal, zinc finger domains have little organized structure (Frankel et al., 1987). Upon zinc binding, the zinc finger conformation is formed and this disorder is lost. Therefore, it is easy to imagine that zinc binding to ZF1 and ZF2 constrains AD2 such that it is no longer capable of productively interacting with downstream transcription factors.
It should be noted that while AD2 function mapped to ZF2, both ZF1 and ZF2 are required to repress AD2. This observation suggested that ZF1 and ZF2 interact with one another to mask AD2 function. A model for the zinc-repressed form of Zap1 AD2, based on the structural model of Gli zinc fingers, is presented in Figure 9. This model is supported by the finding that the amino acids that form a packing interface in fingers 1 and 2 of Gli are conserved in Zap1 ZF1 and ZF2. Moreover, mutation of these potential interface residues in Zap1 ZF1 or ZF2 resulted in constitutive AD2 function. These mutations are unlikely to interfere with zinc binding (Michael et al., 1992). Therefore, we propose that the repressed state of AD2 is a folded structure in which ZF1 and ZF2 interact to mask AD2. Mutation of the potential interface residues in the α-helix of ZF2 (V634 and I637) resulted in the loss of activation domain function (data not shown), suggesting that this surface is important for AD2’s ability to activate transcription.
Fig. 9. Model for the zinc-repressed form of Zap1 activation domain 2. Shown is a ribbon diagram of the structural model of ZF1 and ZF2 of Gli (Pavletich and Pabo, 1993). The zinc-binding cysteine and histidine residues are colored in blue and the Zn2+ atoms are colored in cyan. These two Gli fingers make intramolecular protein–protein contacts with each other. The amino acids that contribute to the intramolecular packing interface between these fingers are colored in red. These consist of two W residues in the β-hairpin loops between the cysteinyl ligands and two hydrophobic residues in each of the two α-helices. Zap1 ZF1 and ZF2 contain similarly positioned W residues and hydrophobic residues in the α-helices. These residues are labeled in the model with the position and the identity of the corresponding Zap1 amino acids.
While this model is attractive in its simplicity, other hypotheses are possible. For example, zinc binding in ZF1 and ZF2 could generate a conformation competent for protein–protein interactions with a co-repressor protein. Precedence for this model comes from the numerous examples of C2H2 zinc fingers acting in protein–protein interactions (Mackay and Crossley, 1998). Genetic analyses have thus far failed to identify a co-repressor (unpublished data).
Zap1 is regulated by zinc at multiple post-translational levels. Zap1 DNA binding is regulated by zinc and, independently, zinc controls AD1 and AD2 function (Bird et al., 2000b). The results shown in Figures 7 and 8 demonstrate the importance of AD2 in this control, but also highlight the contribution of these multiple regulatory mechanisms to overall Zap1 zinc responsiveness. These data raise an intriguing question regarding the different roles of these multiple modes of regulation. One possibility is that each mechanism ‘fine-tunes’ overall Zap1 activity in response to zinc. These different mechanisms may, in combination, give Zap1 an optimal profile of zinc responsiveness that best provides for zinc homeostasis in the cell. Alternatively, these regulatory mechanisms may contribute differently during transitions in zinc status. Our preliminary results suggest that during the transition from zinc-deficient to replete conditions, loss of DNA binding occurs very slowly suggesting a strong reliance on regulation of AD1 and AD2 function early in this transition (A.Bird, unpublished observation).
It has been proposed previously that zinc fingers could play a sensory role in controlling gene expression in response to zinc status. This model has often been presented in the context of zinc-regulated DNA binding via zinc finger DNA binding domains (O’Halloran, 1993). Indeed, this is a prevalent model for how zinc controls the activity of the MTF-1 transcription factor (Andrews, 2001). In response to high zinc, MTF-1 up-regulates genes involved in zinc homeostasis. MTF-1 has a DNA binding domain with six zinc fingers. Recent studies have suggested that fingers 5 and 6 of MTF-1 may play a regulatory role by stabilizing the interaction of MTF-1 with its MRE binding site (Chen et al., 1998, 1999). Consistent with this role, fingers 5 and 6 have a weaker apparent affinity for zinc (Chen et al., 1999). Analogously, we presented evidence here for a novel role of zinc fingers as zinc sensors to control activation domain function.
Materials and methods
Strains and growth conditions
The yeast strains used are DY1457 (MATα ade6 can1 his3 leu2 trp1 ura3), ZHY6 (DY1457 zap1Δ::TRP1), ABY29 (MATα ade6 can1 his3 leu2 trp1 ura3 gal4Δ::kan), ABY31 (ABY29 zap1Δ::TRP1) and ABY46 (DY1457 ZAP1ZF1&2 C2Q2). ABY46 was created by integrating the Candida albicans URA3MX4 cassette (Goldstein et al., 1999) into the wild-type strain DY1457 at the ZAP1 locus such that the 171 bp region encoding amino acids 583–639 was deleted. The region encoding Zap1552–705 was amplified by PCR using the template pGBD–Zap1mZnf1/2 (Bird et al., 2000a) and the resulting product used to transform ABY45 (DY1457 zap1::CaURA3). Transformants were selected by complementation of the zinc-limited growth phenotype of ABY45 and the presence of the Q substitutions was confirmed by sequencing. Yeast were grown in either YP medium supplemented with 2% glucose or synthetic defined (SD) medium supplemented with 2% glucose and the appropriate auxotrophic supplements. LZM was prepared as described previously (Gitan et al., 1998) with either 2% glucose (LZM) or 2% galactose (LZM galactose) for the carbon source. LZM is zinc-limiting because it contains 1 mM EDTA as a metal buffer.
Plasmid constructions
All clones were confirmed by sequencing. Fusions of Zap1 to the GBD were constructed in pMA424, which uses the ADH1 promoter for expression (Ma and Ptashne, 1987). Plasmids pGBD–Zap1552–880 and pGBD–Zap1552–705 were as described previously (Bird et al., 2000b). Related plasmids pGBD–Zap1552–641, pGBD–Zap1552–610, pGBD–Zap1566–641, pGBD–Zap1579–610 and pGBD–Zap1611–641 were created by amplifying the indicated ZAP1 region using PCR. PCR products were then cloned into EcoRI–BamHI-digested pMA424 by homo logous recombination (Ma et al., 1987). Site-directed mutagenesis was performed by two-step overlap PCR (Ho et al., 1989). GBD–Zap1552–705 plasmids containing H599Q H604Q (ZF1 C2Q2) and/or H636Q H641Q (ZF2 C2Q2) were made similarly using pGBD–Zap1mZnF1, pGBD–Zap1mZnF2 or pGBD–Zap1mZnF1/2 (Bird et al., 2000b) as templates. Plasmid pMyc-Zap1 expresses ZAP1 with six myc epitope tags from the GAL1 promoter (Bird et al., 2000b). To create pZF1/21–880, the region encoding Zap1 amino acids 1–551 was introduced into the EcoRI site of pGBD–Zap1mZnF1/2 by homologous recombination. A ZAP1 BstXI fragment containing the mutations was then cloned into BstXI-digested pMyc-Zap1. The reporter constructs used in this study are pDg2 ZRE-lacZ reporter (Zhao et al., 1998) and pGAL1-lacZ constructed in YEp353 (Myers et al., 1986).
β-galactosidase assays
Cells were grown for 15–20 h to exponential phase (A600 0.3–0.7) in LZM media supplemented with the indicated amount of ZnCl2. β-galactosidase activity was measured as described by Guarente (1983), and activity units were calculated as follows: (ΔA420 × 1000)/(min × ml of culture × absorbance of the culture at 595 nm).
S1 nuclease assays
RNA was extracted from cells grown to mid-log phase using the hot acid phenol method and S1 analysis was performed as described previously (Dohrmann et al., 1992). For each reaction, 12 µg of total RNA were hybridized to a 32P end-labeled DNA oligonucleotide probe before digestion with S1 nuclease and separation on an 8% polyacrylamide/8 M urea polyacrylamide gel. All data was quantified by PhosphorImager Analysis using Quantity One Software before exposure to X-ray film.
Analysis of metal binding by zinc finger peptides
The Zap1 ZF1/ZF2 and ZF3/ZF4 fragments were generated by amplifying the ZAP1 open reading by PCR with primers containing either EcoRI or NcoI sites at their 5′ ends. The products were inserted into EcoRI–NcoI-digested pET32a vector (Novagen). Zap1 truncates were expressed in BL21(DE3) pLysS cells grown at 37°C to an OD600 of 0.4. Expression was induced with 0.9 mM IPTG for 3 h at 30°C in the presence of 500 µM ZnCl2. Cells were harvested by centrifugation, washed in 0.25 M sucrose and frozen at –70°C. After thawing, pellets were resuspended in lysis buffer (20 mM NaH2PO4, 3.6 mM KH2PO4, pH 7.3, 280 mM NaCl, 5.4 mM KCl, 10 mM β-mercaptoethanol, 10 mM imidazole), sonicated and centrifuged at 100 000 g for 40 min at 4°C. Proteins were purified on Ni-NTA Superflow (Qiagen) columns. The columns were washed with 5 vol. of wash buffer (lysis buffer with 20 mM imidazole) and eluted with 450 mM imidazole in lysis buffer. Proteins were dialyzed into buffer [20 mM NaH2PO4, 3.6 mM KH2PO4, pH 7.3, 280 mM NaCl, 5.4 mM KCl, 10% glycerol, 5 mM dithiothreitol (DTT)] using Slide-A-Lyzer cassettes (MWCO 10 000) (Pierce). The His tag was removed by passing over a Ni-NTA column after incubating with thrombin (∼0.01 U/µg protein for 16 h at 4°C). The elutant was dialyzed as above and protein concentrations determined by amino acid analysis. Samples were hydrolyzed in 5.7 N HCl, 0.1% phenol in vacuo at 110°C before analysis on a Beckman 6300 analyzer. Zinc concentrations were measured by atomic absorption spectrophotometry.
Plasticware, dialysis membranes (Spectra/Por 6 Membranes, MWCO 3500), and clips were acid-washed, soaked in a 5 mM EDTA solution overnight and rinsed with deionized water (18 MΩ). Chelation buffer was prepared in cleaned plasticware using deionized water and 15 mM 3-(N-morpholino)propanesulfonic acid (MOPS; Sigma) pH 7.0, with 10 µM of 1,7-phenanthroline (Aldrich) or 1,10-phenanthroline (Aldrich). The protein samples (10 nmol in 2 ml buffer) were dialyzed at room temperature for 48–50 h. Samples were then transferred to microfuge tubes, centrifuged and transferred to new tubes. Protein content was quantified by amino acid analysis and zinc measured by atomic absorption spectrophotometry.
For the Co2+ binding studies, the Zap1 575–643 (ZF1/ZF2) and Zap1 700–766 (ZF3/ZF4) peptides were purified as described above. DTT was added to the ∼200–300 µM 7 ml protein solution, to a final concentration of 5 mM DTT and incubated at room temperature for 2 h. The pH was adjusted to 2.0 for the sample to dissociate the bound Zn2+ ions. After clarification, the samples were concentrated using a YM-3,000 Centricon (Millipore Corp.) to ∼2.5 ml. The samples were desalted on a PD-10 Column (prepacked Sephadex G-25 M column; Amersham Biosciences) equilibrated with 0.1 N HCl. The apo-protein concentration was calculated by determining the concentration of reactive thiols with the 2,2′-dithiodipyridine (DTDP; Sigma) method (Grassetti and Murray, 1967). The protein was diluted in ddH2O such that the final concentration of the protein after addition of all final buffer components was 50 µM in 1 ml. Next, 100 µl 1 M Tris–HCl, pH 7.5, was added and the solution was vortexed briefly. The solution was transferred to a 1 ml cuvette and 58.9 µl 100 p.p.m. Co2+, was added for a final Co2+ concentration of twice the protein concentration. Immediately following the Co2+ addition, the sample was mixed, placed in a Beckman DU 640 spectrophotometer, and the absorbance read from 270 to 900 nm or at 644 nm once per second. Once the absorbance had stabilized on a single value, 65.4 µl of either 100 p.p.m. Zn2+ or 1000 p.p.m. Zn2+, was added for a final Zn2+ concentration of either twice the protein concentration or 20 times the protein concentration. Immediately following the Zn2+ addition, the sample was, placed in a Beckman DU 640 spectrophotometer, and the absorbance read at 644 nm once per second.
Zn2+ competition binding studies were carried out using Fura-2 (Molecular Probes). All sample constituents were degassed and transferred to an anaerobic chamber. An aliquot of 1 mM metal-free Fura-2, quantified from ε369 = 28.300/cm/M, was mixed with an aliquot of ∼100–200 µM apo-protein in 100 mM Tris, pH 7.5, to give final concentrations of 15 µM Fura-2 and 10 µM apo-protein, in 1.3 ml. Due to the extremely low affinity of Tris for Zn2+ (dissociation constant of ∼5 mM) the presence of this buffer does not significantly affect the experimental results (calculations not shown). This sample was then placed in a sealed cuvette and the optical spectrum taken from 240 to 560 nm. Aliquots of degassed 0.2 mM ZnCl2 were added through the cuvette’s septum with a Hamilton syringe. Following each addition of ZnCl2 (typically 4 µl), the sample was mixed by shaking for 10 s and then allowed to sit in the instrument for 15 s before the optical spectrum was again taken from 240 to 560 nm. Examination of the 0 Zn and maximal Zn difference spectra showed maximal loss of absorbance at 381 nm and maximal increase of absorbance at 332 nm in the absence of protein, as well as in the presence of ZF1/ZF2 or ZF3/ZF4. The absorbance changes at 332 and 381 nm were followed, with the data corrected for dilution during the titration. The data were fit using DYNAFIT (Kuzmic, 1996). The parameters for the iterative fit by the DYNAFIT program were all set at the known concentrations and absorbance values. The dissociation constant of Fura-2 was also fixed and only the dissociation constants of the zinc fingers were allowed to vary during the fit process.
Acknowledgments
Acknowledgements
We thank Elizabeth Rogers and Mick Petris for helpful discussions. We also gratefully acknowledge the assistance of Dr D.Giedroc in using DYNAFIT. This work was supported by NIH grant RO1 GM56285.
References
- Andrews G.K. (2001) Cellular zinc sensors: MTF-1 regulation of gene expression. Biometals, 14, 223–237. [DOI] [PubMed] [Google Scholar]
- Atar D., Backx,P.H., Appel,M.M., Gao,W.D. and Marban,E. (1995) Excitation–transcription coupling mediated by zinc influx through voltage-dependent calcium channels. J. Biol. Chem., 270, 2473–2477. [DOI] [PubMed] [Google Scholar]
- Berg J.M. and Godwin,H.A. (1997) Lessons from zinc binding peptides. Ann. Rev. Biophys. Biomol. Struct., 26, 357–371. [DOI] [PubMed] [Google Scholar]
- Bird A.J., Evans-Galea,M., Blankman,E., Zhao,H., Luo,H., Winge,D.R. and Eide,D.J. (2000a) Mapping the DNA binding domain of the Zap1 zinc-responsive transcriptional activator. J. Biol. Chem., 275, 16160–16166. [DOI] [PubMed] [Google Scholar]
- Bird A.J., Zhui,H., Luo,H., Jensen,L.T., Srinivasan,C., Evans-Galea,M., Winge,D.R. and Eide,D.J. (2000b) A dual role for zinc fingers in both DNA binding and zinc sensing by the Zap1 transcriptional activator. EMBO J., 19, 3704–3713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brocklehurst K.R., Hobman,J.L., Lawley,B., Blank,L., Marshall,S.J., Brown,N.L. and Morby,A.P. (1999) ZntR is a Zn(II)-responsive MerR-like transcriptional regulator of zntA in Escherichia coli. Mol. Microbiol., 31, 893–902. [DOI] [PubMed] [Google Scholar]
- Buchsbaum J.C. and Berg,J.M. (2000) Kinetics of metal binding by a zinc finger peptide. Inorg. Chim., 297, 217–219. [Google Scholar]
- Chen X., Agarwal,A. and Giedroc,D.P. (1998) Structural and functional heterogeneity among the zinc fingers of human MRE-binding transcription factor-1. Biochemistry, 37, 11152–11161. [DOI] [PubMed] [Google Scholar]
- Chen X., Chua,M. and Giedroc,D.P. (1999) MRE-binding transcription factor-1: Weak zinc-binding finger domains 5 and 6 modulate the structure, affinity and specificity of the metal response element complex. Biochemistry, 38, 12915–12925. [DOI] [PubMed] [Google Scholar]
- Cheng C. and Reynolds,I.J. (1998) Calcium-sensitive fluorescent dyes can report increases in intracellular free zinc concentration in cultured forebrain neurons. J. Neurochem., 71, 2401–2410. [DOI] [PubMed] [Google Scholar]
- Dohrmann P.R., Butler,G., Tamai,K., Dorland,S., Greene,J.R., Thiele,D.J. and Stillman,D.J. (1992) Parallel pathways of gene regulation: homologous regulators SWI5 and ACE2 differentially control transcription of HO and chitinase. Genes Dev., 6, 93–104. [DOI] [PubMed] [Google Scholar]
- Evans-Galea M.V., Blankman,E., Myszka,D.G., Bird,A.J., Eide,D.J. and Winge,D.R. (2003) Two of the five zinc fingers in the Zap1 transcription factor DNA binding domain dominate site-specific DNA binding. Biochemistry, 42, 1053–1061. [DOI] [PubMed] [Google Scholar]
- Finerty P.J. Jr and Bass,B.L. (1999) Subsets of the zinc finger motifs in dsRBP-ZFa can bind double-stranded RNA. Biochemistry, 38, 4001–4007. [DOI] [PubMed] [Google Scholar]
- Frankel A.D., Berg,J.M. and Pabo,C.O. (1987) Metal-dependent folding of a single zinc finger from transcription factor IIIA. Proc. Natl Acad. Sci. USA, 84, 4841–4845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaullier J.M., Simonsen,A., D’Arrigo,A., Bremnes,B., Stenmark,H. and Aasland,R. (1998) FYVE fingers bind PtdIns(3)P. Nature, 394, 432–433. [DOI] [PubMed] [Google Scholar]
- Gitan R.S., Luo,H., Rodgers,J., Broderius,M. and Eide,D. (1998) Zinc-induced inactivation of the yeast ZRT1 zinc transporter occurs through endocytosis and vacuolar degradation. J. Biol. Chem., 273, 28617–28624. [DOI] [PubMed] [Google Scholar]
- Goldstein A.L., Pan,X. and McCusker,J.H. (1999) Heterologous URA3MX cassettes for gene replacement in Saccharomyces cerevisiae. Yeast, 15, 507–511. [DOI] [PubMed] [Google Scholar]
- Grassetti D.R. and Murray,J.F.,Jr (1967) Determination of sulfhydryl groups with 2,2′- or 4,4′-dithiodipyridine. Arch. Biochem. Biophys., 119, 41–49. [DOI] [PubMed] [Google Scholar]
- Guarente L. (1983) Yeast promoters and lacZ fusions designed to study expression of cloned genes in yeast. Methods Enzymol., 101, 181–191. [DOI] [PubMed] [Google Scholar]
- Ho S.N., Hunt,H.D., Horton,R.M., Pullen,J.K. and Pease,L.R. (1989) Site-directed mutagenesis by overlap extension using the polymerase chain reaction. Gene, 77, 51–59. [DOI] [PubMed] [Google Scholar]
- Kuzmic P. (1996) Program DYNAFIT for the analysis of enzyme kinetic data: application to HIV proteinase. Anal. Biochem., 237, 260–273. [DOI] [PubMed] [Google Scholar]
- Landers E. et al. (2001) Initial sequencing and analysis of the human genome. Nature, 409, 860–921. [DOI] [PubMed] [Google Scholar]
- Lee H. et al. (2000) Local structural elements in the mostly unstructured transcriptional activation domain of human p53. J. Biol. Chem., 275, 29426–29432. [DOI] [PubMed] [Google Scholar]
- Lyons T.J., Gasch,A.P., Gaither,L.A., Botstein,D., Brown,P.O. and Eide,D.J. (2000) Genome-wide characterization of the Zap1p zinc-responsive regulon in yeast. Proc. Natl Acad. Sci. USA, 97, 7957–7962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma H., Kunes,S., Schatz,P.J. and Botstein,D. (1987) Plasmid construction by homologous recombination in yeast. Gene, 58, 201–216. [DOI] [PubMed] [Google Scholar]
- Ma J. and Ptashne,M. (1987) Deletion analysis of GAL4 defines two transcriptional activating segments. Cell, 48, 847–853. [DOI] [PubMed] [Google Scholar]
- MacDiarmid C.W., Gaither,L.A. and Eide,D. (2000) Zinc transporters that regulate vacuolar zinc storage in Saccharomyces cerevisiae. EMBO J., 19, 2845–2855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackay J.P. and Crossley,M. (1998) Zinc fingers are sticking together. Trends Biochem. Sci., 23, 1–4. [DOI] [PubMed] [Google Scholar]
- Michael S.F., Kilfoil,V.J., Schmidt,M.H., Amann,B.T. and Berg,J.M. (1992) Metal binding and folding properties of a minimalist Cys2His2 zinc finger peptide. Proc. Natl Acad. Sci. USA, 89, 4796–4800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller J., McLachlan,A.D. and Klug,A. (1985) Repetitive zinc-binding domains in the protein transcription factor IIIA from Xenopus oocytes. EMBO J., 4, 1609–1614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyabe S., Izawa,S. and Inoue,Y. (2001) Zrc1 is involved in zinc transport system between vacuole and cytosol in Saccharomyces cerevisiae. Biochem. Biophys. Res. Commun., 282, 79–83. [DOI] [PubMed] [Google Scholar]
- Myers A.M., Tzagoloff,A., Kinney,D.M. and Lusty,C.J. (1986) Yeast shuttle and integrative vectors with multiple cloning sites suitable for construction of lacZ fusions. Gene, 45, 299–310. [DOI] [PubMed] [Google Scholar]
- O’Halloran T.V. (1993) Transition metals in control of gene expression. Science, 261, 715–725. [DOI] [PubMed] [Google Scholar]
- Outten C.E. and O’Halloran,T.V. (2001) Femtomolar sensitivity of metalloregulatory proteins controlling zinc homeostasis. Science, 292, 2488–2492. [DOI] [PubMed] [Google Scholar]
- Patzer S.I. and Hantke,K. (1998) The ZnuABC high affinity zinc uptake system and its regulator Zur in Escherichia coli. Mol. Microbiol., 28, 1199–1210. [DOI] [PubMed] [Google Scholar]
- Pavletich N.P. and Pabo,C.O. (1993) Crystal structure of a five-finger GLI–DNA complex: new perspectives on zinc fingers. Science, 261, 1701–1707. [DOI] [PubMed] [Google Scholar]
- Rhodes D. and Klug,A. (1993) Zinc fingers. Sci. Am., 268, 56–65. [DOI] [PubMed] [Google Scholar]
- Sensi S.L., Canzoniero,L.M., Yu,S.P., Ying,H.S., Koh,J.Y., Kerchner, G.A. and Choi,D.W. (1997) Measurement of intracellular free zinc in living cortical neurons: routes of entry. J. Neurosci., 17, 9554–9564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uesugi M., Nyanguile,O., Lu,H., Levine,A.J. and Verdine,G.L. (1997) Induced alpha helix in the VP16 activation domain upon binding to a human TAF. Science, 277, 1310–1313. [DOI] [PubMed] [Google Scholar]
- VanZile M.L., Cosper,N.J., Scott,R.A. and Giedroc,D.P. (2000) The zinc metalloregulatory protein Synechococcus PCC7942 SmtB binds a single zinc ion per monomer with high affinity in a tetrahedral coordination geometry. Biochemistry, 39, 11818–11829. [DOI] [PubMed] [Google Scholar]
- Waters B.M. and Eide,D.J. (2002) Combinatorial control of yeast FET4 gene expression in response to iron, zinc and oxygen. J. Biol. Chem., 277, 33749–33757. [DOI] [PubMed] [Google Scholar]
- Zhao H. and Eide,D. (1996a) The yeast ZRT1 gene encodes the zinc transporter of a high affinity uptake system induced by zinc limitation. Proc. Natl Acad. Sci. USA, 93, 2454–2458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao H. and Eide,D. (1996b) The ZRT2 gene encodes the low affinity zinc transporter in Saccharomyces cerevisiae. J. Biol. Chem., 271, 23203–23210. [DOI] [PubMed] [Google Scholar]
- Zhao H. and Eide,D.J. (1997) Zap1p, a metalloregulatory protein involved in zinc-responsive transcriptional regulation in Saccharomyces cerevisiae. Mol. Cell. Biol., 17, 5044–5052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao H., Butler,E., Rodgers,J., Spizzo,T. and Duesterhoeft,S. (1998) Regulation of zinc homeostasis in yeast by binding of the ZAP1 transcriptional activator to zinc-responsive promoter elements. J. Biol. Chem., 273, 28713–28720. [DOI] [PubMed] [Google Scholar]