Abstract
Poly(A) polymerase (PAP) is a key enzyme responsible for the addition of the poly(A) at the 3′ end of pre-mRNA. The C-terminal region of mammalian PAP carries target sites for protein–protein interaction with the 25 kDa subunit of cleavage factor I and with splicing factors U1A and U2AF65. We used a yeast two-hybrid screen to identify 14-3-3ε as an additional protein binding to the C-terminal region of PAP. Interaction between PAP and 14-3-3ε was confirmed by both in vitro and in vivo binding assays. This interaction is dependent on PAP phosphorylation. Deletion analysis of PAP suggests that PAP contains multiple binding sites for 14-3-3ε. The binding of 14-3-3ε to PAP inhibits the polyadenylation activity of PAP in vitro, and overexpression of 14-3-3ε leads to a shorter poly(A) mRNA tail in vivo. In addition, the interaction between PAP and 14-3-3ε redistributes PAP within the cell by increasing its cytoplasmic localization. These data suggest that 14-3-3ε is involved in regulating both the activity and the nuclear/ cytoplasmic partitioning of PAP through the phosphorylation-dependent interaction.
Keywords: phosphorylation/poly(A) polymerase/polyadenylation activity/protein–protein interaction/14-3-3ε
Introduction
Most eukaryotic mRNA contains a poly(A) tail at the 3′ end. This tail is added following transcription by polyadenylation machinery composed of multiple trans-acting factors, including cleavage- and polyadenylation-specificity factor (CPSF), cleavage-stimulation factor (CstF), two cleavage factors (CFI and CFII), poly(A) polymerase (PAP) and poly(A)-binding protein II (PAB II) (Keller et al., 1991; Barabino and Keller, 1999; Zhao et al., 1999; Shatkin and Manley, 2000; Takagaki and Manley, 2000). The poly(A) tails of 200–250 residues in length are synthesized by PAP at specific pre-mRNA sites, which are generated by cleavage with an as yet unidentified enzyme. The role of the poly(A) tail is not firmly established; however, growing evidence suggests that it enhances the translation efficiency and stability of mRNA (Beelman and Parker, 1995; Tarun and Sachs, 1996). Therefore, regulation of polyadenylation could be one way by which cells can vary the expression level of a particular gene (Barabino and Keller, 1999), and this regulation can be achieved by directly influencing the activity of PAP. PAP activity can be regulated by phosphorylation during the cell cycle (Colgan et al., 1996, 1998). p34cdc2/cyclin B hyperphosphorylates PAP during mitosis and meiosis (M phase), thereby causing repression of PAP activity.
Mammalian PAP II (which is referred to as PAP throughout this paper) carries the catalytic domain in the N-terminal region (Martin and Keller, 1996; Martin et al., 2000), whereas the C-terminal region carries an RNA-binding domain, two nuclear localization signals (NLSs) and a serine/threonine-rich regulatory domain (Rabbe et al., 1994). The serine/threonine-rich domain contains multiple cyclin-dependent kinase (cdk) sites, which are phosphorylated in vitro and in vivo by p34cdc2/cyclin B (Colgan et al., 1998). An isoform of PAP, human PAPγ, has been also known as a bona fide PAP carrying both non-specific and CPSF/AAUAAA-dependent polyadenylation activity (Kyriakopoulou et al., 2001). PAP activity can be regulated through interactions between PAP and various regulatory proteins. For instance, it is repressed by interaction between the U1 snRNP A protein (U1A) and the C-terminal region of PAP (Gunderson et al., 1997). U1A also inhibits poly(A) tail addition to the IgM secretory mRNA in B cells (Phillips et al., 2001). The C-terminal region of PAP also serves as a platform for another splicing factor, U2AF65 (Vagner et al., 2000), as well as the 25 kDa subunit of CFI, a component of the mammalian polyadenylation machinery (Kim and Lee, 2001). Therefore, it is possible that other regulatory proteins are involved in the regulation of PAP functions via interaction with the C-terminal region.
We set out to determine whether or not these regulatory proteins exist and, if so, how they might regulate the activity of PAP. To do this, proteins that could potentially interact with the C-terminal region of PAP were identified using a yeast two-hybrid assay. We identified 14-3-3ε, one of the members of the 14-3-3 protein family, as a candidate binding partner. The 14-3-3 proteins were originally isolated as highly abundant acidic proteins in brain extracts (Moore and Perez, 1967). Seven isoforms (β, γ, ε, η, σ, τ and ζ) have been identified in mammals. The 14-3-3 proteins interact with various cellular proteins (Tzivion and Avruch, 2002), which requires target protein phosphorylation in many, but not all, cases (Liao and Omary, 1996; Muslin et al., 1996; Craparo et al., 1997; Liu et al., 1997; Michael et al., 1997; Piwncia, 1997). 14-3-3 proteins are involved in various cellular processes, including signal transduction, cell cycle progression and apoptosis (Freed et al., 1994; Chan et al., 1999; Kimura et al., 2001; van Hemert et al., 2001). They are thought to alter the stability, activity and/or subcellular localization of the target protein via protein–protein interaction within the cell (Rittinger et al., 1999; Tzivion and Avruch, 2002). Here, we show that 14-3-3ε interacts with the C-terminal regulatory region of PAP and that this interaction is dependent on PAP phosphorylation. Multiple binding motifs for this interaction reside in the serine/threonine-rich domain of PAP. We also examine the effects of binding to 14-3-3ε on the polymerization activity and subcellular localization of PAP. Our findings suggest that 14-3-3ε plays a role in modulating the PAP activity and the nuclear/cytoplasmic localization through a phosphorylation-dependent interaction.
Results
Identification of 14-3-3ε as a PAP-binding protein
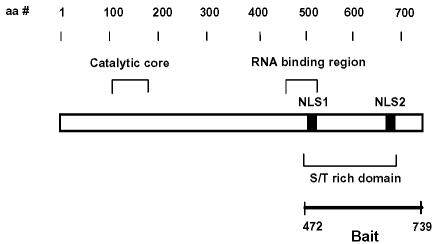
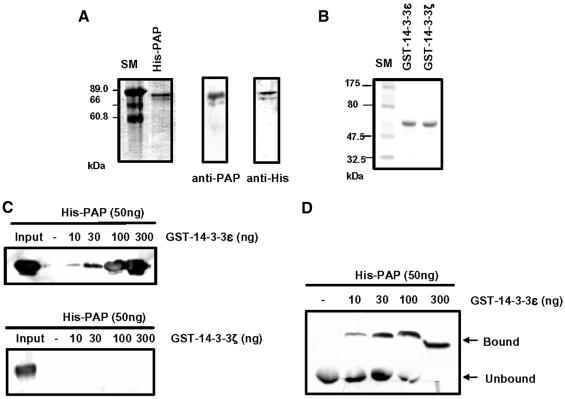
To identify proteins that regulate PAP activity via protein–protein interactions, we performed a LexA-based yeast two-hybrid screen using the C-terminal 268 residues of mouse PAP as bait (Figure 1). Approximately two million colonies from a human HeLa cell cDNA library were screened. One of the clones identified as an interacting sequence encodes the human 14-3-3ε protein. We obtained the coding sequence of mouse 14-3-3ε using PCR amplification from a mouse kidney cDNA library. This mouse 14-3-3ε sequence was used to analyze the interaction with mouse PAP throughout the study. His-PAP and GST–14-3-3ε, which were expressed in Sf9 insect cells and Escherichia coli cells, respectively, were purified. The purified His-PAP showed two bands (one major and one minor) on an SDS–polyacrylamide gel (Figure 2A). Since both bands were reacted with anti-PAP antibody and anti-His antibody, we concluded that His-PAP was purified as two forms of PAP. On the other hand, GST–14-3-3ε was purified as a single band on an SDS–polyacrylamide gel (Figure 2B). We analyzed in vitro interaction between His-PAP and GST–14-3-3ε by both coimmunoprecipitation and gel mobility shift assays. Prior to the gel mobility shift assay, we checked whether His-PAP would enter native gels. The purified His-PAP migrated into a native gel, whereas the RNase-treated His-PAP did not (data not shown). These data suggest that the purified His-PAP might have some residual RNA essential for its entrance into the native gel. Both the coimmunoprecipitation analysis (Figure 2C) and the gel mobility shift assay (Figure 2D) indicated that PAP and 14-3-3ε interact in vitro.
Fig. 1. Schematic diagram presenting the significant features of poly(A) polymerase (PAP). The catalytic domain, the serine/threonine-rich region, two nuclear localization sequences (NLS) and an RNA-binding region are indicated.
Fig. 2. Interaction of poly(A) polymerase (PAP) with 14-3-3ε in vitro. (A) Purification of the recombinant His-PAP from insect Sf9 cells. Left: the purified His-PAP was electrophoresed on a 10% SDS–polyacrylamide gel and visualized with Coomassie Blue. Right: the protein gel was blotted with anti-PAP or anti-His antibody. (B) Purification of the recombinant 14-3-3 proteins from E.coli. The purified proteins were electrophoresed on a 10% SDS–polyacrylamide gel and visualized with Coomassie Blue. (C and D) The recombinant His-PAP protein of 50 ng was incubated with the indicated amounts of GST–14-3-3ε in 30 µl of EBC solution. The protein mixture was pull downed with glutathione–Sepharose beads. The pull-downed proteins were resolved on a 10% SDS–polyacrylamide gel and blotted with anti-His antibody (C). GST–14-3-3ζ was used as a control protein (see Figure 4). Alternatively, complex formation between two proteins was analyzed by a gel mobility shift assay on an 8% native polyacrylamide gel. The protein gel was blotted by anti-His antibody (D).
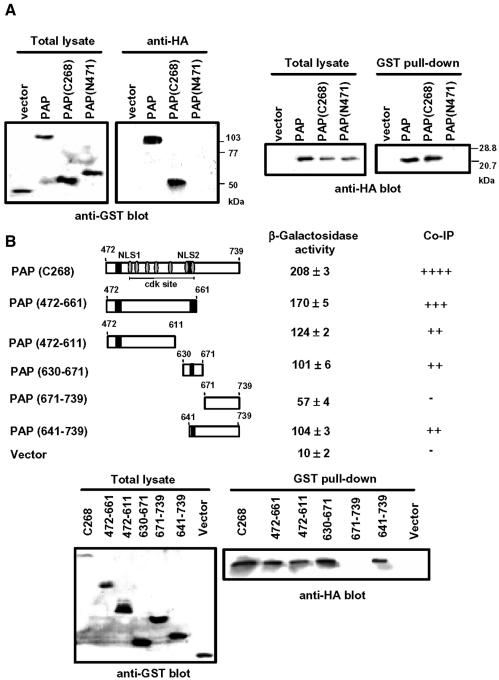
To determine whether the interaction between PAP and 14-3-3ε occurs in vivo, we performed a coimmunoprecipitation experiment using NIH 3T3 cells cotransfected with both the GST–PAP and the HA-14-3-3ε constructs (Figure 3A). The coimmunoprecipitation data indicated that the PAP–14-3-3ε interaction occurs in mammalian cells. Coimmunoprecipitation experiments were also performed with the C-terminal or N-terminal catalytic domain of PAP instead of the intact PAP (Figure 3A). 14-3-3ε binds to the C-terminal 268 residues of PAP, but not to the remaining N-terminal region, confirming the C-terminal region as containing the motif(s) for interaction with 14-3-3ε. To further delineate the binding domain in the C-terminal region of PAP for 14-3-3ε, we generated several truncated mutants of PAP. These mutant proteins were tested for their ability to bind 14-3-3ε by both yeast two-hybrid and coimmunoprecipitation analyses (Figure 3B). Strong interaction was observed with various fragments over the 472–671 region. Interestingly, the 472–611 fragment showed the interaction comparable with that of the non-overlapping 641–671 fragment. This finding suggests that PAP contains more than one binding site for 14-3-3ε. We measured slight β-galactosidase activity generated by the C-terminal 69 residues (671–739) in the yeast two-hybrid assay. However, no coimmunoprecipitation was detected between 14-3-3ε and the 671–739 fragment of PAP. Therefore, it is unlikely that this region includes a binding site for 14-3-3ε in mammalian cells. Although we have not examined this discrepancy further, it might be due to the difference in protein phosphorylation between yeast and mammalian systems.
Fig. 3. In vivo interaction of poly(A) polymerase (PAP) with 14-3-3ε. (A) Coimmunoprecipitation of GST–PAP with HA-14-3-3ε. Cell lysates from NIH 3T3 cells transfected with the indicated construct DNAs were immunoprecipitated with anti-HA or incubated with glutathione–Sepharose beads. The immunoprecipitates were resolved on a 10% SDS–polyacrylamide gel and analyzed by reciprocal immunoblot. Left: total lysates (2% input) or their immunoprecipitates with anti-HA antibody were blotted with anti-GST antibody. Right: total lysates (2% input) or bound proteins on glutathione–Sepharose beads (GST pull-down) were blotted with anti-HA antibody. The cell number equivalents were 1.5 × 106 per lane. (B) Regions of PAP required for interaction with 14-3-3ε. We analyzed the interaction between the PAP derivatives and full length 14-3-3ε using the yeast two-hybrid system and by coimmunoprecipitation. For the coimmunoprecipitation experiments, lysates of NIH 3T3 cells cotransfected with GST–PAP and HA-14-3-3ε were incubated with glutathione–Sepharose beads, and the GST pull-downed proteins were blotted with anti-HA antibody. The total lysates (2% input) were also blotted with anti-GST antibody for ensuring the expression of GST–PAP derivatives. The western blot data were semi-quantitated by calculating the relative amount of GST–PAP pull downed from the total lyasate (Co-IP).
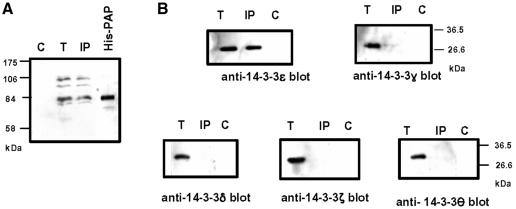
We carried out coimmunoprecipitation experiments to see whether endogenous PAP and 14-3-3ε interact in mammalian cells. The untranfected NIH 3T3 cells lysates were immunoprecipitated with anti-14-3-3ε antibody, and the immunoprecipitaes were blotted with anti-PAP (Figure 4A). The converse experiment was performed using anti-PAP and anti-14-3-3ε as the precipitating antibody and the blotting antibody, respectively (Figure 4B). The immunoblot indicated that the interaction between the two proteins occurs naturally in vivo. Since 14-3-3 proteins are a family of highly conserved proteins and there are seven isoforms in mammals, we determined whether PAP displayed an isoform-specific interaction with 14-3-3ε. We performed the same coimmunoprecipitation experiment with other isoform-specific antibodies. Figure 4B shows that PAP interacts with isoform ε, but not with isoforms δ, γ, θ and ζ. This result indicates that the interaction between PAP and 14-3-3ε is isoform specific.
Fig. 4. Association of endogenous poly(A) polymerase (PAP) with 14-3-3 proteins in vivo. (A) Immunoprecipitations were performed on lysates of untransfected NIH 3T3 cells using anti-14-3-3ε. Proteins were detected by western blot analysis using anti-PAP antibody. (B) The same lysates were immunoprecipitated with anti-PAP and immunoblotted with the indicated anti-14-3-3-isoform-specific antibodies. T, total lysate (2% input); C, preimmune used as a control antibody; His-PAP, the purified His-PAP used as a size marker; IP, immunoprecipitates with anti-14-3-3 proteins or anti-PAP.
Interaction between 14-3-3 and PAP depends on phosphorylation of PAP
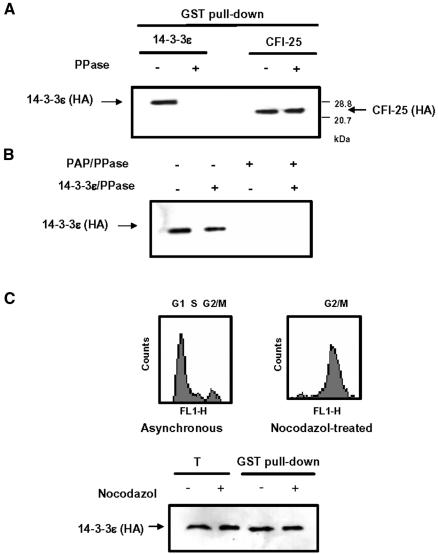
Since binding of 14-3-3 proteins requires phosphorylation of target proteins in many cases (Liao and Omary, 1996; Muslin et al., 1996; Craparo et al., 1997; Liu et al., 1997; Michael et al., 1997; Piwncia, 1997), we next examined whether the interaction between PAP and 14-3-3ε was phosphorylation dependent. Prior to immunoprecipitation, the GST–PAP and HA-14-3-3ε cotransfected cell lysates were treated with a non-specific phosphatase, λ phosphatase. This treatment resulted in loss of the interaction between PAP and 14-3-3ε (Figure 5A). To exclude the possibility that the phosphatase would indirectly affect the interaction, we performed a control experiment with cells cotranstected with GST–PAP and HA-CFI--25 fusion constructs because the C-terminal region of PAP also interacts with CFI-25 (Kim and Lee, 2001). The phosphatase treatment did not affect the PAP–CFI-25 interaction, confirming that the interaction between PAP and 14-3-3ε was phosphorylation dependent. We also transfected cells with GST–PAP or HA-14-3-3ε alone and separately treated the resulting cell lysates with the phosphatase. The two cell lysates were then mixed and analyzed by coimmunoprecipitation. The phosphatase treatment of the GST–PAP, but not the HA-14-3-3ε lysates, affected the PAP–14-3-3ε interaction (Figure 5B). This finding indicates that the PAP–14-3-3ε interaction depends on the phosphorylation state of PAP.
Fig. 5. Effects of dephosphorylation of poly(A) polymerase (PAP) on interaction with 14-3-3ε. (A) We performed coimmunoprecipitation experiments as in Figure 3A. Prior to the incubation with glutathione–Sepharose beads (GST pull-down), cell lysates of NIH 3T3 cells cotransfected with GST–PAP and either HA-14-3-3ε or HA-CFI-25 DNAs (Kim and Lee, 2001) were treated with λ phosphatase. A plus sign (+) indicates the phosphatase (PPase) treatment. (B) Lysates from NIH 3T3 cells transfected with GST–PAP or HA-14-3-3ε construct alone were also treated with λ phosphatase. A plus sign (+) indicates the phosphatase (PPase) treatment. The two lysates were mixed for coimmunoprecipitation. (C) HeLa cells cotransfected with GST–PAP and HA-14-3-3ε were treated with nocodazol (40 ng/ml) to arrest at the M phase. Top: FACScan profiles of the cells were collected for DNA content analysis with propidium iodide staining. Bottom: the x-axis (FL1-H) and the y-axis (Counts) represent linear scale of fluorescence and cell number, respectively; the lysate of the nocodazol treated cells were subjected to coimmunoprecipitation. T, total lysate (2% input).
It is known that PAP is hyperphosphorylated and its activity is repressed in M-phase cells (Colgan et al., 1998). Thus, we investigated whether the interaction between GST–PAP and HA-14-3-3ε is related to this hyperphosphorylation using M-phase arrested HeLa cells by nocodazol. Although we did not observed any significant difference in gel mobility between GSP–PAP proteins expressed in the M-phase arrested and unsynchronized cells (data not shown), we confirmed, by flow cytrometric analysis, that the nocodazol-treated cells were arrested at the M phase (Figure 5C). The interaction of GST–PAP with 14-3-3ε in the M-phase arrested cells differed little from the interaction in unsynchronized cells (Figure 5C), suggesting that the hyperphosphorylation status of PAP is not relevant to the interaction.
14-3-3ε inhibits the activity of PAP
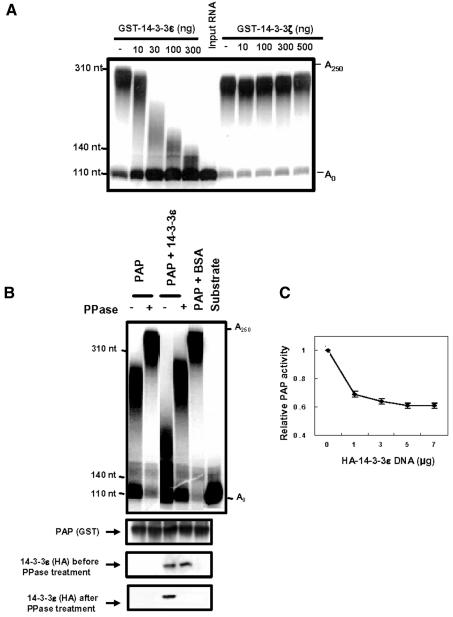
To examine how the binding of 14-3-3ε to PAP affects the function(s) of PAP, we measured the non-specific polyadenylation activity of PAP in a Mn2+-containing reaction buffer (Zhao and Manley, 1996). The purified His-PAP activity was assayed with c-mos mRNA as a substrate in the presence of GST–14-3-3ε. The trichloroacetic acid precipitation assay showed that the polyadenylation activity of His-PAP was inhibited by GST–14-3-3ε (Table I). The maximum decrease of the His-PAP activity by GST–14-3-3ε was ∼40%. When the same reaction products were electrophoresed on a gel, we found that poly(A) tails became shorter as more GST–14-3-3ε proteins were included in the reaction mixture (Figure 6A). These data implicate 14-3-3ε in the formation of shorter poly(A) tails by inhibiting PAP activity.
Table I. Inhibition of His-tagged poly(A) polymerase (His-PAP) activity by GST–14-3-3ε in vitro.
| His-PAP (ng)a | GST–14-3-3ε (ng)a | Relative PAP activity |
|---|---|---|
| 50 | 0 | 1.00 |
| 50 | 10 | 0.89 ± 0.08 |
| 50 | 30 | 0.75 ± 0.04 |
| 50 | 100 | 0.70 ± 0.08 |
| 50 | 300 | 0.62 ± 0.02 |
aHis-PAP and GST–14-3-3ε were purified as recombinant proteins from Sf9 cells and E.coli cells, respectively. The polyadenylation reaction was carried out as in Figure 6A, and the reaction products were analyzed by the trichloroacetic acid precipitation assay.
Fig. 6. Effects of interaction of poly(A) polymerase (PAP) with 14-3-3ε on non-specific polyadenylation. We performed non-specific polyadenylation assays with His-PAP of GST–PAP. The c-mos mRNA substrate was incubated with His-PAP or GST–PAP in a reaction buffer containing 0.5 mM MnCl2 and [α-32P]ATP. The PAP reaction products were analyzed by electrophoresis on a 5% polyacrylamide gel containing 8 M urea. The c-mos mRNA substrate (S) was labeled at the 3′ end with [32P]pCp and used as a size control. (A) The purified His-PAP was used for non-specific polyadenylation assays in the presence of the indicated amounts of GST–14-3-3ε. GST–14-3-3ζ, which showed no binding to His-PAP (Figure 2), was used as a negative control. (B) GST–PAP was purified from the lysates of NIH 3T3 cells transfected with both GST–PAP and HA-14-3-3ε DNAs, or GST–PAP DNA singly, by immobilization on glutathione–Sepharose beads. The GST–PAP bound glutathione beads were used for non-specific polyadenylation assays. The amount of GST–PAP or HA-14-3-3ε present in the purified fraction was semi-quantitatively determined by immunoblotting with anti-GST or anti-HA antibody. (C) The non-specific polyadenylation activity of GST–PAP from cells transfected with increasing amounts of HA-14-3-3ε was measured by the trichloroacetic acid precipitation assay, as in Table II.
To examine effects of in vivo interaction of PAP with 14-3-3ε on non-specific polyadenylation, we performed the same non-specific polyadenylation assay with GST–PAP purified by immobilization on glutathione–Sepharose beads from NIH 3T3 cells transfected with the GST–PAP fusion construct alone or from cells cotransfected with GST–PAP and HA-14-3-3ε. We confirmed the presence of GST–PAP or a complex of GST–PAP/HA-14-3-3ε in the purified fraction by immunoblot analysis. GST–PAP purified from the cotransfected cells added shorter poly(A) tails to the 3′ end of c-mos mRNA than did GST–PAP from the cells transfected with the GST–PAP construct alone (Figure 6B). The trichloroacetic acid precipitation assay of the same reaction products showed that the activity of GST–PAP purified from the cotransfected cells was reduced to 70% of the activity of GST–PAP from the GST–PAP singly transfected cells (Table II). The inhibition of GST–PAP activity by HA-14-3-3ε was not due to a non-specific protein–protein interaction, because the presence of excess bovine serum albumin (BSA) in the reaction mixture had little effect on the polyadenylation activity (Figure 6B, Table II). In addition, GST–PAP from cells transfected with increasing amounts of HA-14-3-3ε construct showed a dose-dependent decrease in activity (Figure 6C). The maximum inhibition of 40% in the dose-dependent experiment is comparable with the inhibition level of the activity of the purified His-PAP in the presence of excess GST–14-3-3ε (Table I). We also found that the GST–PAP activity was increased by treatment with phosphatase in the presence of HA-14-3-3ε (Figure 6B, Table II). Although a small increase of PAP activity by the treatment of phosphatase in the absence of 14-3-3ε was observed, this increase was much less than that in the presence of 14-3-3ε. Therefore, we postulate that the effect of the phosphatase treatment in the presence of 14-3-3ε was due to disruption of the interaction with 14-3-3ε through dephosphorylation of PAP. Together, these results indicate that the interaction between PAP and 14-3-3ε inhibits the polyadenylation activity of PAP.
Table II. In vitro activity of GST fusion poly(A) polymerase (GST–PAP) isolated from transfected NIH 3T3 cells.
| Proteina | λ phosphataseb | Relative PAP activityc |
|---|---|---|
| GST–PAP | – | 1.00 |
| GST–PAP | + | 1.08 ± 0.13 |
| GST–PAP + 14-3-3ε | – | 0.70 ± 0.02 |
| GST–PAP + 14-3-3ε | + | 0.90 ± 0.10 |
| GST–PAP + BSAd | – | 0.95 ± 0.14 |
| yPAP (positive control)e | – | 1.28 ± 0.08 |
| GST (negative control)f | – | 0.03 ± 0.00 |
aGST–PAP was purified by immobilization on glutathione–Sepharose beads from NIH 3T3 cells transfected with either both GST–PAP and HA-14-3-3ε constructs or GST–PAP alone. The polyadenylation reaction was carried out as in Figure 6B, and the reaction products were analyzed by the trichloroacetic acid precipitation assay.
bTreatment of the purified GST–PAP with λ phosphatase is indicated with +.
cThe relative PAP activity represents the amount of [α-32P]ATP incorporated by each protein fraction relative to that by GST–PAP without the λ phosphatase treatment.
dBSA of 2 µg was added in the reaction mixture.
eWe used 5 U of yeast PAP (yPAP; Invitrogen) as a positive control.
fWe purified the GST control protein from NIH 3T3 cells transfected with the GST host vector DNA as a negative control.
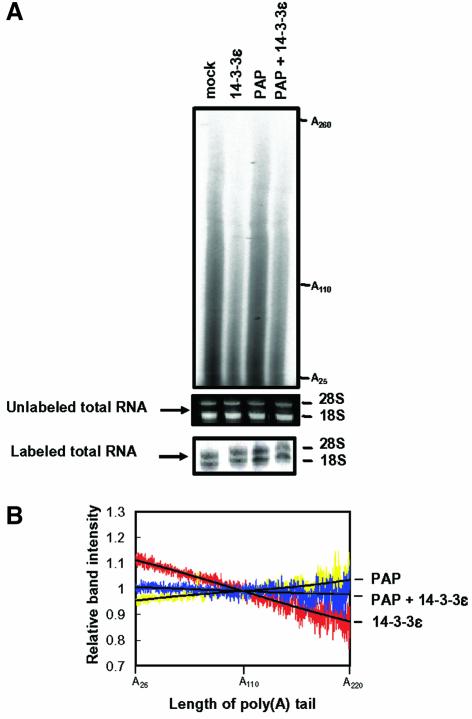
We performed additional experiments to assess how the inhibition of PAP activity by 14-3-3ε in vitro may impact on the poly(A) synthesis of pre-mRNA in vivo. We determined the length distribution of poly(A) tails in mRNA, which was isolated from NIH 3T3 cells transfected with the HA-14-3-3ε construct. The total RNA was labeled at the 3′ end and subjected to both RNase A and RNase T1 digestion to leave only poly(A) tails intact (Preker et al., 1995). The digested poly(A) tails were analyzed by gel electrophoresis (Figure 7A). We also represented these data as the relative length distribution by calculating the relative band intensity of poly(A) tails at a specific poly(A) length (Figure 7B). In the cells overexpressing HA-14-3-3ε, the length of the poly(A) tails was shorter than in the control cells. Since 14-3-3ε may be involved in various cellular functions (Tzivion and Avruch, 2002), the shortening of the poly(A) tails might have resulted from its indirect effect. For example, the population of mRNA carrying A25–A260 appeared to decrease by the overexpression of HA-14-3-3ε. We found that cotransfection with the GST–PAP construct rescued the poly(A) tail-shortening, although the population of mRNA was not fully restored. Therefore, this result supports a notion that the inhibition of in vivo activity of PAP through its interaction with overexpressed HA-14-3-3ε may be responsible for the shortening of the poly(A) tails.
Fig. 7. Effect of overexpressing 14-3-3ε on the length distribution of poly(A) tails in vivo. (A) Total RNA was purified from NIH 3T3 cells transfected with the indicated constructs and labeled at the 3′ end with [32P]pCp. Total RNA from untranfected cells (mock) was also used as a control. We confirmed the quality of RNA by observing intact 28S and 18S rRNAs on the gels before and after the labeling. The labeled RNA was extensively digested with RNase A and RNase T1. The products were analyzed on a 5% sequencing gel. (B) RNA band intensities were determined by an image analyzer BAS-1500 (Fuji) and the sum of peak integration was normalized to that of control cells (mock). The relative intensity is represented as a ratio of the normalized intensity of the transfected cells to the intensity of control cells at a specific poly(A) length. The relative length distribution of poly(A) tails of mRNA isolated from cells transformed with GST poly(A) polymerase (PAP) (yellow). HA-14-3-3ε (red) or both GST–PAP and HA-14-3-3ε (blue) are shown. The black line represents a third-degree polynomial fit of each relative length distribution. The data are representative of three separate experiments.
The subcellular localization of PAP is regulated by 14-3-3ε
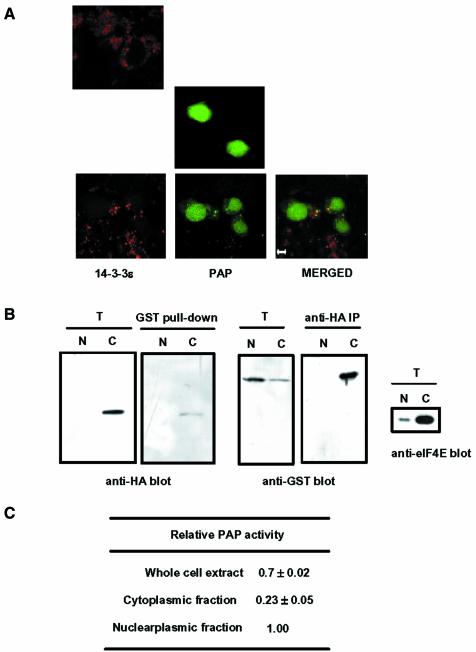
We used confocal fluorescence microscopy to investigate how the interaction of PAP with 14-3-3ε affects the intracellular localization of PAP (Figure 8A). Cells cotransfected with GFP–PAP and HA-14-3-3ε cDNAs showed a dispersed GFP–PAP fluorescence mainly in the nucleus, but with areas of staining also in the cytoplasm. HA-14-3-3ε appeared as fine punctate staining throughout the cytoplasm, and the merged images suggest that GFP–PAP and HA-14-3-3ε are colocalized in the cytoplasm. However, most of GFP–PAP proteins remained in the nucleus, and free HA-14-3-3ε proteins were shown in the cytoplasm, indicating that colocalization of two proteins in the cytoplasm is limited. We next compared the subcellular distribution of PAP in the absence and presence of 14-3-3ε. We randomly chose at least 50 cells for each case in three different experiments and quantified the subcellular distribution of PAP from the confocal-scanning microscope images. In the absence of 14-3-3ε, 8 ± 1.5% of the GFP–PAP signal was found in the cytoplasm, whereas the presence of 14-3-3ε increased the cytoplasmic localization of PAP to 18 ± 2.5%.
Fig. 8. Subcellular localization of poly(A) polymerase (PAP) and 14-3-3ε. (A) NIH 3T3 cells were transfected with the GFP–PAP construct, the HA-14-3-3ε construct or both together. To visualize the HA-14-3-3ε localization, we incubated the transfected cells with an anti-HA antibody followed by a rhodamine-conjugated anti-rat antibody (Sigma). GFP–PAP and HA-14-3-3ε were visualized with a confocal microscope for green and red fluorescence, respectively. Cells transfected with HA-14-3-3ε alone (top), GFP–PAP alone (middle) or with both GFP–PAP and HA-14-3-3ε (bottom) demonstrate typical localization of the recombinant proteins. The bottom panel also shows a merged image of the two fluorescence emissions of the same image. Scale bar: 5 µm. (B) HeLa cells were transfected the GST–PAP and HA-14-3-3ε constructs. Nucleoplasmic and cytoplasmic fractions were isolated and each fraction was subjected to coimmunprecipitation analysis, as in Figure 3A. The same amount in the cell number was used for each fraction. The preparation of the nucleoplasmic and cytoplasmic fractions was ensured by immunoblotting with anti-eIF2E antibody. T, total lysates (2% input); N, nucleoplasmic fraction; C, cytoplasmic fraction. (C) GST–PAP of the nucleoplasmic and cytoplasmic fractions was purified by immobilization on glutathione–Sepharose beads. Non-specific polyadenylation activity of GST–PAP immobilized on the beads was determined by the trichloroacetic acid precipitation assay, as in Table II. The relative PAP activity represents the amount of [α-32P]ATP incorporated by GST–PAP from the whole cell extract or the cytoplasmic fraction relative to that by GST–PAP from the nucleoplasmic fraction. The equivalent of GST–PAP in the cell number was used for each fraction.
To confirm the colocalization of PAP and 14-3-3ε in the cytoplasm, we performed coimmunoprecipitation experiments using nucleoplasmic and cytoplamic fractions of HeLa cells cotransfected with the GST–PAP and HA-14-3-3ε constructs. The binding of GST–PAP with HA-14-3-3ε was observed in the cytoplasmic fraction, not in the nucleoplasmic faction (Figure 8B). GST–PAP was purified by immobilization on glutathione–Sepharose beads from the total cell lysate, nucleoplasmic fraction and cytoplasmic fraction, separately, and assayed for its non-specific polyadenylation activity (Figure 8C). In this assay, differences in recovery during the preparation of nuclear and cytoplasmic extracts could affect the quantitation of the PAP activity of each subcellular fraction. However, the data clearly showed that the activity of GST–PAP isolated from the whole cell extract was less than the sum of the activities of nucleoplasmic and cytoplasmic GST–PAP, despite the possibility of incomplete recovery during the preparation of the subcellular extracts. This finding suggests that free GST–PAP and free HA-14-3-3ε exist in the nucleus and cytoplasm, respectively, and that these free proteins can interact when the nuclear membrane barrier is disrupted during the preparation of the total cell lysate. Therefore, this result agrees well with the limited colocalization of GST–PAP and 14-3-3ε proteins shown in the confocal fluorescence images.
Discussion
PAP, a key enzyme of the polyadenylation machinery, is responsible for the addition of the poly(A) at the 3′ end of pre-mRNA. Since this poly(A) tail is implicated in the stability, nuclear export and translation efficiency of mRNA (Beelman and Parker, 1995; Huang and Carmichael, 1996; Tarun and Sachs, 1996), control of its addition by PAP could be an important mechanism to regulate the level of gene expression. We performed a LexA-based yeast two-hybrid screen to identify interaction partners for PAP that may be involved in the regulation of PAP functions. We identified 14-3-3ε, a member of the 14-3-3 protein family, as a potential partner. The interaction between 14-3-3ε and PAP was confirmed both biochemically and in cells. The binding of 14-3-3ε to PAP inhibits the polyadenylation activity of PAP. Thus, PAP activity is regulated by 14-3-3ε through protein–protein interaction. PAP is related to PAPγ in sequences of the region containing the 14-3-3ε binding sites (30.0% identity in the 472–611 region and 38.1% identity in the 641–661 region), and monoclonal antibodies raised against human PAP were reported to crossreact with human PAPγ (Kyriakopoulou et al., 2001). Therefore, we do not exclude the possibility of PAPγ contamination in our in vitro assays.
It was previously shown that PAP activity can be regulated within the pre-mRNA 3′ end processing machinery by CPSF-160 in mammals (Murthy and Manley, 1995) and by Fip1p in yeast (Preker et al., 1995; Zhelkovsky et al., 1998). 14-3-3ε has not been implicated as a component of the 3′ processing machinery. Thus, the basis of regulation of PAP activity by 14-3-3ε must differ from that involved in the 3′ processing machinery. 14-3-3 proteins are known to play a role in the regulation of diverse cellular processes, such as signal transduction, cell cycle regulation, apoptosis, stress responses, cytoskeletal organization and malignant transformation (reviewed by van Hemert et al., 2001). Therefore, 14-3-3ε could serve in some way to link polyadenylation with other cellular events. It has also been reported that 14-3-3 interacts with transcription factors and cofactors (Songqin et al., 1999; Kanai et al., 2000; Thorsten et al., 2000), which indicates a possible link between transcription and polyadenylation.
The deletion analysis performed in this study suggests that there are at least two 14-3-3ε-binding sites in the C-terminal region of PAP and that these binding sites are located within the serine/threonine-rich region. However, none of the known 14-3-3ε-binding motifs can be found in this region. Since most 14-3-3-binding sites require a specific phosphoserine residue, phosphorylation of particular serine residues in the region is most likely to be responsible for the binding of PAP to 14-3-3ε. Indeed, we observed that the interaction between PAP and 14-3-3ε was dependent on phosphorylation of PAP. Since this interaction inhibits the polyadenylation activity of PAP, phosphorylation–dephosphorylation of PAP at the 14-3-3ε-binding sites appears to be an important means of regulating PAP activity. It is known that phosphorylation of PAP affects its activity in vivo and in vitro (Ballantyne et al., 1995; Colgan et al., 1996). The serine/threonine-rich region contains multiple cdk sites. These sites are phosphorylated in vitro and in vivo by p34cdc2/cyclin B (Colgan et al., 1998). Hyperphosphorylation of PAP by p34cdc2/cyclin B is required for inhibition of PAP activity in the M phase of the cell cycle (Colgan et al., 1998). However, we showed that the hyperphosphorylation of PAP was not relevant to its interaction with 14-3-3ε. Therefore, regulation of PAP through its interaction with 14-3-3ε in a phosphorylation-dependent manner may function in phases other than the M phase of the cell cycle. The phosphorylation target sites on PAP for interaction with 14-3-3ε and the corresponding kinase remain to be identified.
The interaction between 14-3-3ε and PAP also redistributes PAP within the cell by increasing its cytoplasmic localization. It is not clear at this time whether interaction of PAP with 14-3-3ε is necessary for the modulation of nuclear import or nuclear export. The serine/threonine-rich region of PAP responsible for binding to 14-3-3ε includes two NLSs (NLS1 and NLS2). Therefore, it is possible that the binding of 14-3-3ε to this region interferes with the function of these NLSs and keeps some PAP in the cytoplasm. Alternatively, 14-3-3ε may participate actively in the nuclear export of PAP. 14-3-3 proteins contain a nuclear export sequence in their C-terminal region (Rittinger et al., 1999). Therefore, PAP could be relocalized into the cytoplasm by active export out of the nucleus on binding to 14-3-3ε.
The biological relevance for the inhibition of PAP activity by 14-3-3ε is unclear. We found that overexpression of 14-3-3ε caused a shortening of the poly(A) tail length of pre-mRNA. Since 14-3-3ε may be involved in various cellular metabolisms, the shortening of the poly(A) tail length might be the indirect effect of the 14-3-3ε overexpression. However, the accompanying overexpression of PAP rescued the shortening of the poly(A) tail length, suggesting that the interaction between PAP and 14-3-3ε may play a role in regulating poly(A) tail length control. Since the interaction of PAP with 14-3-3ε appears to cause a redistribution of PAP into the cytoplasm, the shortening of poly(A) tails may be due to the decreased availability of PAP in the nucleus. The 14-3-3ε-bound PAP in the cytoplasm may be dephosphorylated when the PAP activity is needed in the cytoplasm. It is known that cytoplasmic control of poly(A) mRNA length plays a key role in activating or repressing gene expression during meiosis and early embryonic development when transcription is silent (Richter, 1999). Therefore, it is possible that the redistributed PAP has a chance to participate in cytoplasmic polyadenylation when cytoplasmic control of the length of poly(A) tails is needed. A recent finding that Cid13, a novel cytoplasmic PAP found in yeast, is associated with 14-3-3 proteins may support this possibility (Saitoh et al., 2002).
Since 14-3-3 isoforms are highly homologous, their PAP-binding properties may be conserved. However, the individual 14-3-3 isoforms are differentially regulated (Testerink et al., 1999; Rosenquist et al., 2000) and they differ in their ability to bind synthetic peptides and proteins (Vincenz and Dixit, 1996; Yaffe et al., 1997; Tang et al., 1998; Mils et al., 2000; van der Hoeven et al., 2000). These findings suggest that 14-3-3 proteins have isoform-specific roles in diverse cellular metabolisms. We found that PAP interacts with 14-3-3ε, but not isoforms δ, γ, θ and ζ, suggesting that the regulation of PAP by 14-3-3ε is isoform specific.
In summary, we identified 14-3-3ε as a protein that binds to the C-terminal region of PAP. This interaction depends on the phosphorylation status of PAP, and the binding of 14-3-3ε to PAP inhibits the polyadenylation activity of PAP in vitro. The PAP–14-3-3ε interaction may also play a role in controlling poly(A) mRNA tail length in vivo. The binding of 14-3-3ε to PAP causes the redistribution of PAP from the nucleus into the cytoplasm. These data provide in vitro and in vivo evidence that 14-3-3ε has a dual role in the regulation of PAP (i.e. inhibition of PAP activity and cytoplasmic localization of PAP), which could be an important mechanism for regulating the level of gene expression.
Materials and methods
Yeast two-hybrid analysis
We used the Matchmaker LexA two-hybrid system (Clontech), with the C-terminal 268 residues of mouse PAP as the bait in a pEQ202 LexA fusion-plasmid vector. A human HeLa cDNA library was prepared with the B42 transactivating domain fusion-plasmid vector and screened from 2 × 106 primary transformants of yeast strain EGY48. Yeast cells were grown in SD media lacking histidine, tryptophan and uracil. Clones expressing β-galactosidase activity were screened on the plates by using 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-gal) as a substrate. One of the selected clones was identified as a cDNA coding for 14-3-3ε. The coding sequence of mouse 14-3-3ε cDNA (DDBJ/EMBL/GenBank accession No. D87663) was obtained by PCR amplification from a mouse kidney cDNA library.
Protein–protein interactions in the liquid culture were determined from the β-galactosidase activity of yeast cells using the substrate o-nitrophenyl-β-d-galactopyranoside, as described previously (Kim and Lee, 2001).
Preparation of recombinant cDNA constructs
For the yeast two-hybrid analysis, different LexA–PAP fusion constructs encoding full-length or defined regions of mouse PAP were prepared in pEQ202 plasmid vectors. A plasmid expressing a full-length mouse 14-3-3ε fused to the B42 transactivating domain was constructed by cloning the corresponding DNA fragment into pJG4-5. To express PAP with an N-terminal histidine tag (His-PAP) in Sf9 insect cells, the coding sequence of full-length mouse PAP was cloned into pFASTBACHTa (Invirogen). Escherichia coli expression plasmids pGST–14-3-3ε and pGST–14-3-3ζ were constructed by cloning the coding sequence of full-length mouse 14-3-3ε and 14-3-3ζ into the pGEX4T-1 vector (Amersham Pharmacia Biotech), respectively. For expression of GST–PAP or its derivatives in mammalian cells, full-length or defined regions of PAP were cloned into pEBG (Kim and Lee, 2001). The HA-14-3-3ε construct was obtained by cloning the 14-3-3ε coding sequence into mammalian expression vector pSRα (Kim and Lee, 2001). A plasmid expressing GFP–PAP was constructed by cloning the PAP coding region into pEGFP-C1 (Clontech).
Expression and purification of the recombinant His-PAP and GST–14-3-3 proteins
The recombinant mouse His-PAP was expressed in Sf9 insect cells. Sf9 cells were infected with 1 p.f.u. of the recombinant baculovirus carrying the PAP coding sequence, as described in the Bac-to-Bac Baculovirus Expression System Manual (Gibco-BRL). The cell lysate was subjected to Ni-NTA affinity chromatography. The recombinant His-PAP-containing fraction was further purified by an FPLC system with HiPrep 16/60 Sephacryl S-200 column chromatography (Amersham Pharmacia). The recombinant GST–14-3-3 proteins were was expressed in E.coli and purified, as described previously (Kim and Lee, 2001).
Antibodies
Antiserum to bovine PAP was a gift from Dr G.Martin. Antisera specific for 14-3-3 proteins (Santa Cruz), GST (Santa Cruz), HA (Roche), His (Qiagen) and eIF4E (Sigma) were purchased.
In vitro binding assay
The purified recombinant His-PAP and GST–14-3-3 proteins were used for an in vitro binding assay. His-PAP of 50 ng was incubated with various amounts of GST–14-3-3ε or GST–14-3-3ζ in 30 µl of EBC solution (Kim and Lee, 2001) at 4°C for 1 h. The protein mixture of His-PAP and GST–14-3-3 proteins was electrophoresed on an 8% native polyacrylamide gel. The protein gel was blotted with anti-His antibody. Alternatively, the protein mixture was pull downed with glutathione beads, and the pull-downed proteins were immunoblotted with anti-His antibody, as described previously (Kim and Lee, 2001).
In vivo binding assay
NIH 3T3 cells were maintained in Dulbecco’s modified Eagle’s medium supplemented with 10% fetal calf serum. Cell lysates were prepared from NIH 3T3 cells (∼1 × 108) transfected with mammalian expression plasmid DNAs (1 µg of each), as described previously (Kim and Lee, 2001). The cell lysate (1 ml) was incubated at 4°C for 1 h with 25 µl of a 50% slurry solution of either anti-HA antibody (2 µg) bound protein G-agarose, or glutathione–Sepharose. Phosphatase treatments of the lysates were carried out prior to the pull-down experiment by incubating with λ phosphatase at a final concentration of 100 U/ml at 30°C for 1 h. EDTA was used to inactivate the phosphatase. The proteins bound to the beads were resolved on a 10% SDS–polyacrylamide gel and subjected to immunoblot analysis.
Similarly, cell lysates from untransfected NIH 3T3 cells were prepared and subjected to coimmunoprecipitation analysis using anti-PAP and anti-14-3-3 antibodies.
The in vivo binding assay was also performed using HeLa cells cotransfected with both the GST–PAP and HA-14-3-3ε. For isolation of M-phase HeLa cells, nocodazol (Sigma) was added to a final concentration of 40 ng/ml. To confirm that the nocodazol-treated cells were arrested at the M phase, the cells were subjected to fluorescence-activated cell sorting using a FACSCalibur flow cytometer (Beckton Dickinson), as described previously (Shin and Manley, 2002). Subcellular fractionation of HeLa cells was performed as described previously (Siomi et al., 1997).
In vitro polyadenylation assay
The recombinant His-PAP and GST–14-3-3ε were used for in vitro polyadenylation assay. We determined the activity of His-PAP in the presence of various amounts of GST–14-3-3ε by measuring incorporation of AMP from [α-32P]ATP into c-mos RNA lacking a polyadenylation signal. This was performed in the presence of MnCl2, as described previously (Zhao and Manley, 1996). The reaction mixture (30 µl) containing 50 ng of His-PAP was incubated at 37°C for 30 min. The reaction products were precipitated with 5% trichloroacetic acid, and the precipitates were analyzed by a scintillation counter. Alternatively, the products were analyzed on a 5% polyacrylamide gel containing 8 M urea. The in vitro polyadenylation assay was also performed with GST–PAP isolated from transfected NIH 3T3 cells. The whole-cell lysate (1 ml) was incubated at 4°C for 1 h with 25 µl of 50% slurry solution of glutathione–Sepharose. GST–PAP-bound glutathione beads were extensively washed with EBC solution and directly used for the in vitro polyadenylation assay. To assess dose dependency of 14-3-3ε, the expression plasmid DNA of HA-14-3-3ε was increased from 1 to 7 µg. If necessary, the lysates were treated with λ phosphatase, before the purification of GST–PAP. The activity of GST–PAP was measured, as described for His-PAP. The total protein used in each reaction was about 100 ng, and their identity was confirmed by SDS–polyacrylamide gel electrophoresis followed by immunoblot analysis.
GST–PAP from the nucleoplasmic and cytoplasmic fractions of HeLa cells was purified by immobilization on glutathione–Sepharose beads and its in vitro polyadenylation activity was determined by the trichloroacetic acid precipitation assay, as described above.
Determination of the length of poly(A) tails in vivo
NIH 3T3 cells were transfected with mammalian expression plasmid DNAs (1 µg of each). Total RNA was purified from the transfected cells using the TRISOL protocol (Roche). The purified RNA (2 µg) was labeled at the 3′ end with [32P]pCp. The labeled RNA was digested with 3.5 µg of RNase A and 2.5 U of RNase T1 in 350 µl of the reaction buffer (10 mM Tris–HCl pH 7.5, 5 mM EDTA, 300 mM sodium acetate) at 30°C for 30 min. The reaction was stopped by adding 10 µl of 20% SDS and 2.5 µl of proteinase K (20 mg/ml) for 15 min at 37°C. The products were analyzed on a 5% polyacrylamide gel containing 8 M urea.
Confocal laser scanning microscopy
NIH 3T3 cells were transfected with cDNAs (1 µg of each) expressing GFP–PAP and/or HA-14-3-3ε. The transfected cells were fixed with 2% formaldehyde diluted in phosphate-buffered saline (PBS) for 20 min at 25°C. The GFP–PAP protein localization was directly visualized on a Zeiss LSM510 confocal microscope (Carl Zeiss). To visualize the HA-14-3-3ε protein localization, however, anti-HA antibody and a TRITC (rhodamine)-conjugated secondary antibody (Sigma) were used. The preparations were mounted using DABCO (Sigma). We blocked the non-specific binding sites with 1% (w/v) BSA and 1% (v/v) normal goat serum in PBS. Imaging for HA-14-3-3ε was conducted on the same Zeiss LSM510 confocal microscope. Negative control staining was performed by incubating without the primary antibody. The subcellular distribution of GFP–PAP was scored according to the manufacturer’s manual.
Acknowledgments
Acknowledgements
We thank Dr G.Martin for providing the PAP antibody, and Dr J.Lee for critical review of the manuscript. This work was supported by a grant from the Molecular Medicine Research Program (M1-0106-04-0001), the Center for Molecular Design and Synthesis at KAIST, and the Brain Korea 21 Project.
References
- Ballantyne S., Bilger,A., Astrom,J., Virtanen,A. and Wickens,M. (1995) Poly(A) polymerases in the nucleus and cytoplasm of frog oocytes: dynamic changes during oocyte maturation and early development. RNA, 1, 64–78. [PMC free article] [PubMed] [Google Scholar]
- Barabino S.M. and Keller,W. (1999) Last but not least: regulated poly(A) tail formation. Cell, 99, 9–11. [DOI] [PubMed] [Google Scholar]
- Beelman C.A. and Parker,R. (1995) Degradation of mRNA in eukaryotes. Cell, 81, 179–183. [DOI] [PubMed] [Google Scholar]
- Chan T.A., Hermeking,M.H., Lengaucer,C., Kinzler,K.W. and Volgelstain,B. (1999) 14-3-3σ is required to prevent mitotic catastrophe after DNA damage. Nature, 401, 616–620. [DOI] [PubMed] [Google Scholar]
- Colgan D.F., Murthy,K.G., Prives,C. and Manley,J.L. (1996) Cell-cycle related regulation of poly(A) polymerase by phosphorylation. Nature, 384, 282–285. [DOI] [PubMed] [Google Scholar]
- Colgan D.F., Murthy,K.G., Zhao,W., Prives,C. and Manley,J.L. (1998) Inhibition of poly(A) polymerase requires p34cdc2/cyclin B phosphorylation of mutiple consensus and nonconsnensus sites. EMBO J., 17, 1053–1062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craparo A., Freund,R. and Gustafson,T.A. (1997) 14-3-3 (epsilon) interacts with the insulin-like growth factor I receptor and insulin receptor substrate I in a phosphoserine-dependent manner. J. Biol. Chem., 272, 11663–11669, [DOI] [PubMed] [Google Scholar]
- Freed E., Symons,M., Macdonald,S.G., Mccormick,F. and Ruggieri,R. (1994) Binding of 14-3-3 proteins to the proteins to protein kinase Raf and effects in its activation. Science, 265, 1713–1716. [DOI] [PubMed] [Google Scholar]
- Gunderson S.I., Vagner,S. Polycarpou-Scwarz,M. and Mattaj,I.W. (1997) Involvement of the carboxyl terminus of vertebrate poly(A) polymerase in U1A autoregulation and in the coupling of splicing and polyadenylation. Genes Dev., 11, 761–773. [DOI] [PubMed] [Google Scholar]
- Huang Y. and Carmichael,G.C. (1996) Role of polyadenylation in nucleocytoplasmic transport of mRNA. Mol. Cell. Biol., 16, 1534–1542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanai F. et al. (2000) TAZ: a novel transcriptional co-activator regulated by interactions with 14-3-3 and PDZ domain proteins. EMBO J., 19, 6778–6791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller W., Bienroth,S., Lang,K.M. and Christofori,G. (1991) Cleavage and polyadenlyation factor CPF specifically interacts with the pre-mRNA 3′ processing signal AAUAAA. EMBO J., 10, 4241–4249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H. and Lee,Y. (2001) Interaction of poly(A) polymerase with the 25-kDa subunit of cleavage factor I. Biochem. Biophys. Res. Commun., 289, 513–518. [DOI] [PubMed] [Google Scholar]
- Kimura M.T., Irie,S., Shoji-Hoshino,S., Mukai,J., Nadano,D., Oshimura,M. and Sato,T.A. (2001) 14-3-3 is involved in p75 neurotrophin receptor-mediated signal transduction. J. Biol. Chem., 276, 17291–17300. [DOI] [PubMed] [Google Scholar]
- Kyriakopoulou C.B., Nordvarg,H. and Virtanen,A. (2001) A novel nuclear poly(A) polymerase (PAP), PAP γ. J. Biol. Chem., 276, 33504–33511. [DOI] [PubMed] [Google Scholar]
- Liao J. and Omary,M.B. (1996) 14-3-3 proteins associate with phosphorylated simple epithelial keratins during cell cycle progression and act as a solubility cofactor. J. Cell Biol., 133, 345–357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y.C., Liu,Y., Elly,C., Yoshida,H., Lipkowitz,S. and Altman,A. (1997) Serine phosphorylation of Cbl induced by phorbol ester enhances its association with 14-3-3 proteins in T cells via a novel serine-rich 14-3-3 binding motif. J. Biol. Chem., 272, 9979–9985. [DOI] [PubMed] [Google Scholar]
- Martin G. and Keller,W. (1996) Mutational analysis of mammalian poly(A) polymerase identifies a region for primer binding and a catalytic domain, homologous to the family X polymerase, and to other nucleotidyltransferase. EMBO J., 15, 2593–2603. [PMC free article] [PubMed] [Google Scholar]
- Martin G., Keller,W. and Doublie,S. (2000) Crystal structure of mammalian poly(A) polymerase in complex with an analog of ATP. EMBO J., 19, 4193–4203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michael B.Y., Katrin,R., Stefano,V., Paul,R.C., Alastair,A., Henrik,L., Steven,J.G., Stephen,J.S. and Lewis,C.C. (1997) The structure basis for 14-3-3: phosphopeptide binding specificity. Cell, 91, 961–971. [DOI] [PubMed] [Google Scholar]
- Mils V., Baldin,V., Goubin,F., Pinta,I., Papin,C., Waye,M., Eychene,A. and Ducommun,B. (2000) Specific interaction between 14-3-3 isoforms and the human CDC25B phosphatase. Oncogene, 19, 1257–1265. [DOI] [PubMed] [Google Scholar]
- Moore B. and Perez,V.J. (1967) Specific proteins of the nervous system. In Carlson,F.D. (ed.), Physiological and Biochemical Aspects of Nervous Integration. Prentice-Hall, Englewood Cliffs, NJ, pp. 343–359. [Google Scholar]
- Murthy K.G. and Manley,J.L. (1995) The 160 kD subunit of human cleavage-polyadenylation specificity factor coordinates pre-mRNA 3′ end formation. Genes Dev., 9, 2672–2683. [DOI] [PubMed] [Google Scholar]
- Muslin A.J., Tanner,J.W., Allen,P.M. and Shaw,A.S. (1996) Interaction of 14-3-3 with signaling proteins is mediated by the recognition of phosphoserine. Cell, 84, 889–897. [DOI] [PubMed] [Google Scholar]
- Phillips C., Jung,S. and Gunderson,S.I. (2001) Regulation of nuclear poly(A) addition controls the expression of immunoglobulin M secretory mRNA. EMBO J., 20, 6443–6452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piwncia W.H. (1997) Mitotic and G2 checkpoint control regulation of 14-3-3 protein binding by phosphorylation of Cdc25c on serine 216. Science, 277, 1501–1505. [DOI] [PubMed] [Google Scholar]
- Preker P.J., Lingner,J., Minvielle-Sebastia,L. and Keller,W. (1995) The FIP1 gene encodes a component of yeast pre-mRNA polyadenylation factor that directly interacts with poly(A) polymerase. Cell, 81, 379–389. [DOI] [PubMed] [Google Scholar]
- Rabbe T., Murthy,K.G. and Manley.J.L. (1994) Poly(A) polymerase contains multiple functional domain. Mol. Cell. Biol., 14, 2946–2957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richter J.D. (1999) Cytoplasmic polyadenylation in development and beyond. Microbiol. Mol. Biol. Rev., 63, 446–456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rittinger K., Budman,J., Xu,J., Volinia,S., Cantley,L.C., Smerdon,S.J., Gamblin,S.J. and Yaffe,M.B. (1999) Structural analysis of 14-3-3 phosphopeptide complexes identifies a dual role for the nuclear export signal of 14-3-3 in ligand binding. Mol. Cell, 4, 153–166. [DOI] [PubMed] [Google Scholar]
- Rosenquist M., Sehnke,P., Ferl,R.J., Sommarin,M. and Larsson,C. (2000) Evolution of the 14-3-3 protein family: does the large number of isoforms in multicellular organisms reflect functional specificity? J. Mol. Evol., 51, 446–458. [DOI] [PubMed] [Google Scholar]
- Saitoh S., Chabes,A., McDonald,W.H., Thelander,L., Yates,J.R. and Russell,P. (2002) Cid13 is a cytoplasmic poly(A) polymerase that regulates ribonucleotide reductase mRNA. Cell, 109, 563–573. [DOI] [PubMed] [Google Scholar]
- Shatkin A.J. and Manley,J.L. (2000) The ends of the affair: capping and polyadenylation. Nat. Struct. Biol., 7, 838–842. [DOI] [PubMed] [Google Scholar]
- Shin C. and Manley,J.L. (2002) The SR protein SRp38 represses splicing in M phase cells. Cell, 111, 407–417. [DOI] [PubMed] [Google Scholar]
- Siomi M., Eder,P.S., Kataoka,N., Wan,L., Liu,Q. and Dreyfuss,G. (1997) Transportin-mediated nuclear import of heterogeneous nuclear RNP proteins. J. Cell Biol., 138, 1181–1192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Songqin P., Paul,C.S., Robert,J.F. and William,B.G. (1999) Specific interactions with TBP and TFIIB in vitro suggest that 14-3-3 proteins may participate in the regulation of transcription when part of a DNA binding complex. Plant Cell, 11, 1591–1602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takagaki Y. and Manley,J.L. (2000) Complex protein interactions within the human polyadenylation machinery identify a novel component. Mol. Cell. Biol., 20, 1515–1525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang S.J., Suen,T.C., McInnes,R.R. and Buchwald,M. (1998) Association of the TLX-2 homeodomain and 14-3-3η signaling proteins. J. Biol. Chem., 273, 25356–25363. [DOI] [PubMed] [Google Scholar]
- Tarun S.Z. and Sachs,A.B. (1996) Association of the yeast poly(A) tail binding protein with translation initiation factor eIF-4G. EMBO J., 15, 7168–7177. [PMC free article] [PubMed] [Google Scholar]
- Testerink C., van der Meulen,R.M., Oppedijk,B.J., de Boer,A.H., Heimovaara-Dijkstra,S., Kijne,J.W. and Wang,M. (1999) Differences in spatial expression between 14-3-3 isoforms in germinating barley embryos. Plant Physiol., 121, 81–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorsten H., Nico,S. and Wolfgang,W. (2000) Transcriptional activation by the PHD finger is inhibited through an adjacent leucine zipper that binds 14-3-3 proteins. Nucleic Acids Res., 28, 3542–3550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tzivion G. and Avruch,J. (2002) 14-3-3 proteins: active cofactors in cellular regulation by serine/threonine phosphorylation. J. Biol. Chem., 277, 3061–3064. [DOI] [PubMed] [Google Scholar]
- Vagner S., Vagner,C. and Mattaj,I.W. (2000) The carboxyl terminus of vertebrate poly(A) polymerase interacts with U2AF65 to couple 3′ end processing and splicing. Genes Dev., 14, 403–413. [PMC free article] [PubMed] [Google Scholar]
- van der Hoeven P.C., van der Wal,J.C., Ruurs,P., van Dijk,M.C. and van Blitterswijk,J. (2000) 14-3-3 isotypes facilitate coupling of protein kinase C-ζ to Raf-1: negative regulation by 14-3-3 phosphorylation. Biochem. J., 345, 297–306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Hemert M.J., Steensma,H.Y. and van Heusden,G.P.H. (2001) 14-3-3 proteins: key regulators of cell division, signalling and apoptosis. Bioessays, 23, 936–946. [DOI] [PubMed] [Google Scholar]
- Vincenz C. and Dixit,V.M. (1996) 14-3-3 proteins associate with A20 in an isoform-specific manner and function both as chaperone and adapter molecules. J. Biol. Chem., 271, 20029–20034. [DOI] [PubMed] [Google Scholar]
- Yaffe M.B., Rittinger,K., Volinia,S., Caron,P.R., Aitken,A., Leffers,H., Gamblin,S.J., Smerdon,S.J. and Cantley,L.C. (1997) The structure basis for 14-3-3: phosphopeptide binding specificity. Cell, 91, 961–971. [DOI] [PubMed] [Google Scholar]
- Zhao W. and Manley,J.L. (1996) Complex alternative RNA processing generates an unexpected diversity of poly(A) polymerase isoforms. Mol. Cell. Biol., 16, 2378–2386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J., Hyman,L. and Moore,C. (1999) Formation of mRNA 3′ ends in eukaryotes: mechanism, regulation, and interrelationships with other steps in mRNA synthesis. Microbiol. Mol. Biol. Rev., 63, 405–455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhelkovsky A., Helmling,S. and Moore,C. (1998) Processivity of the Saccharomyces cerevisiae poly(A) polymerase requires interactions at the carboxyl-terminal RNA binding domain. Mol. Cell. Biol., 18, 5942–5951. [DOI] [PMC free article] [PubMed] [Google Scholar]